Abstract
Background Information
Prostate cancer (PCa) is a common disease but only a small subset of patients are at risk of developing metastasis and lethal disease, and identifying which patients will progress is challenging because of the heterogeneity underlying tumour progression. Understanding this heterogeneity at the molecular level and the resulting clinical impact is a critical step necessary for risk stratification. Defining genomic fingerprint elucidates molecular variation and may improve PCa risk stratification, providing more accurate prognostic information of tumour aggressiveness (or lethality) for prognostic biomarker development. Therefore, we explored transcriptomic differences between patients with indolent disease outcome and patients who developed metastasis post‐radical prostatectomy using genome‐wide expression data in the post radical prostatectomy clinical space before metastatic spread.
Results
Based on differential expression analysis, patients with adverse pathological findings who are at higher risk of developing metastasis have a distinct transcriptomic fingerprint that can be detected on surgically removed prostate specimens several years before metastasis detection. Nearly half of the transcriptomic fingerprint features were non‐coding RNA highlighting their pivotal role in PCa progression. Protein‐coding RNA features in the fingerprint are involved in multiple pathways including cell cycle, chromosome structure maintenance and cytoskeleton organisation. The metastatic transcriptomic fingerprint was determined in independent cohorts verifying the association between the fingerprint and metastatic patients. Further, the fingerprint was confirmed in metastasis lesions demonstrating that the fingerprint represents early metastatic transcriptomic changes, suggesting its utility as a prognostic tool to predict metastasis and provide clinical value in the early radical prostatectomy setting.
Conclusions
Here, we show that transcriptomic patterns of metastatic PCa exist that can be detected early after radical prostatectomy. This metastatic fingerprint has potential prognostic ability that can impact PCa treatment management potentially circumventing the requirements for unnecessary therapies.
Keywords: Metastasis, Prostate, Radical prostatectomy, Transcriptomic fingerprint
Abbreviations
- BCR
biochemical recurrence
- MFD
median fold difference
- PCa
prostate cancer
- PSA
prostate‐specific antigen
- RP
radical prostatectomy
- RF
random forest
Introduction
Prostate cancer (PCa) is a molecularly and clinically heterogeneous cancer which renders its progression unpredictable (Nwosu et al., 2001; Boyd et al., 2012; Wyatt et al., 2013). PCas that appear clinically similar at surgery often exhibit a range of clinical outcomes that is not captured by existing risk categories (e.g. D'Amico criteria) or prognostic tools such as prostate‐specific antigen (PSA), Gleason grade or tumour stage (Erho et al., 2013). Although these tools continue to be important in risk assessment, they lack the precision required to guide decisions regarding treatment options. Nearly 50% of patients treated with surgery will have adverse pathology or subsequently develop a PSA rise and are therefore considered at increased risk for developing clinical recurrence (Nakagawa et al., 2008). However, according to Boorjian et al. (2011), 12% of patients who developed biochemical recurrence (BCR) after radical prostatectomy (RP), presented a systematic progression at certain point. Patients with PSA rise post‐surgery and no metastasis for at least 10 years were shown to be no different molecularly than patients with no evidence of developing PSA rise (Alshalalfa et al., 2015). Incorporating genomic data effectively stratified patients at risk of developing metastasis post PSA rise (Ross et al., 2014). Recently, the incorporation of genomic data with clinical risk assessment has shown potential to improve risk stratification at all stages of disease (Cooperberg et al., 2015).
The genomic alterations underlying the transition of PCa from an indolent state to a more aggressive nature have not been fully explored. With advances in high‐throughput technology, our understanding of the genomic changes responsible for the development and progression of PCa has expanded (Taylor et al., 2010; Berger et al., 2011; Barbieri et al., 2013). Large scale ‘omics’ approaches such as microarray and next‐gene sequencing have generated a large body of data that has enormously increased our knowledge of the functional role molecular alterations play in cancer progression and offers an opportunity to integrate molecular data to provide improved risk assessment (Erho et al., 2013). Genomics‐informed cancer medicine offers the potential for precision cancer treatment by delivering individualised information in the clinic that providers and patients can use to make personalised medical decisions. As an example, genomic fingerprinting may impact clinical decisions for localised PCa by providing individualised cancer progression risk estimates of disease recurrence after surgery that can inform at diagnostic or post‐operative treatment decisions (Davis, 2014). Likewise, several multi‐gene fingerprints have expanded the landscape of the breast cancer diagnostics (Ross et al., 2008).
Large‐scale omics approaches have enabled the discovery and development of diagnostic and prognostic biomarkers for personalised medicine (Yuan et al., 2014). An increasing number of biomarkers (Figure 1A) discovered at various stages of disease progression contribute to the early diagnosis of PCa (Leyten et al., 2014), predict BCR (Taylor et al., 2010) and identify high‐risk patients that may avoid unnecessary treatment (Karnes et al., 2013). Among the most common molecular events in early prostate tumour development is the rise of PSA (Tosoian and Loeb, 2010; Payne and Cornford, 2011; Prensner et al., 2012). Although highly sensitive, PSA measurement provides poor specificity for clinically significant disease detection and can result in unnecessary biopsies leading to over‐diagnosis and over‐treatment of indolent tumours that might be managed more conservatively. Currently, two sets of biomarkers have improved risk stratification on follow‐up biopsy: prostate cancer antigen 3 (PCA3) and methylation level of GSTP1, APC and RASSF1. PCA3 is commercially available as Progensa and GSTP1, APC and RASSF1 methylation as ConfirMDx. The PCA3 mRNA level in urine is positively associated with cancer detection and better able to predict the presence of cancer on repeat biopsy than PSA alone (Hessels and Schalken, 2009; Durand et al., 2011). ConfirMDx utilises the methylation pattern of these three genes to identify men at low risk on follow‐up biopsy (Trock et al., 2012). Other single gene diagnostic biomarkers like AMACR (from tissue) (Jiang et al., 2013) and TMPRSS2‐ERG gene fusions (in urine) were demonstrated to be specific PCa biomarkers (Tomlins et al., 2011). Multiple studies improved early detection of PCa by combined these biomarkers (Perdonà et al., 2013; Salami et al., 2013; Leyten et al., 2014).
Figure 1.
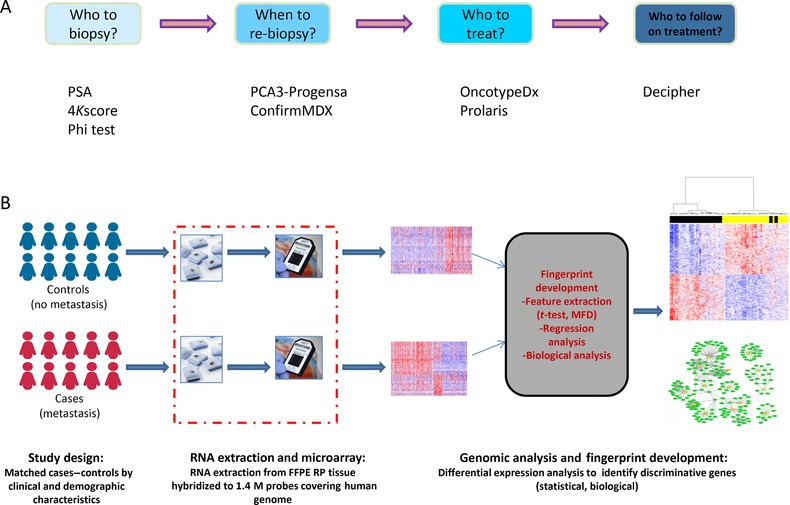
(A) Prostate cancer diagnostic and prognostic tools are commercially available at various stages of cancer. (B) A flow diagram of developing genome‐wide prognostic tools. It starts with the relevant clinical question, designing the study, using the most informative technology and applying the appropriate feature filtering methods.
At a positive cancer diagnosis, two commercially available gene panels (Cuzick et al., 2012; Klein et al., 2014) have been developed to discriminate between clinically indolent and potentially high‐risk patients to avoid overtreatment. Other gene panels such as the Penny gene signature (Penney et al., 2011) was developed using a gene array (∼6000 genes) capturing the difference between low and high Gleason scores and then evaluate it in intermediate Gleason score patients. Other gene panels (Markert et al., 2011; Irshad et al., 2013; Jin et al., 2014) were discovered and validated on single institute data sets and claim to be associated with lethal PCa, although only a few have made it into the clinic.
Gene expression profiling of tumour samples from RP has the capacity to characterise the tumour biology and help identify patients who may develop metastasis several years after RP (Erho et al., 2013). Identifying post‐RP patients with adverse pathology that can be safely monitored without further intervention and patients who should be put on a second line of treatment is a significant and unmet need. Characterising the biology underlying tumour aggressiveness is of paramount importance to characterise tumour outcome and develop biomarkers that offer prognostic information with the expectation of better post‐surgical treatment. This was the rationale for the development of the Decipher PCa genomic test to identify patients who will develop metastasis within 5 years after surgery (Erho et al., 2013).
In this work, we delineate the molecular biology underlying metastasis after RP to identify a transcriptomic fingerprint of metastatic PCa and then, using the Decipher pipeline as an example, go through a series of steps to demonstrate delivery of the fingerprint into the clinic as prognostic tool.
Results and discussion
Clinical characteristics of the study cohort
In order to develop and evaluate a metastatic transcriptomic fingerprint, 1449 RP patients were pooled from eight studies Table S1 that had samples profiled on Affymetrix Human Exon 1.0 ST arrays. The discovery cohort, a case‐control design, (Mayo Clinic I) is an intermediate‐high risk cohort of European Americans; 70% developed BCR and 40% developed clinical metastasis Table 1. BCR was defined as two successive increases in PSA measurement above 0.2 ng/ml with subsequent measure of 0.05 ng/ml above the first measurement. Clinical metastasis was confirmed by bone or CT scan. Seventeen percent developed metastasis within 5 years with a median follow up of 41 months, and 18% developed metastasis between 5 and 10 years with median follow up of 88 months. The eight cohorts used for this study represent the spectrum of PCa treated with radical prostatectomy from low to high‐risk localised disease. Overall, 61% of patients in the pooled cohort had one or more adverse pathology findings ranging from 5% to 89%, and 22% (on average) of patients developed metastasis ranging from 7% to 40% Table 1.
Table 1.
Clinicopathological characteristics of the patients used for discovering and evaluating the metastatic fingerprint
| Discovery | Evaluation | |
|---|---|---|
| Race | 545 | 904 |
| European American | 545(100%) | 734(81.2%) |
| African American | 0 | 57(6.31%) |
| Others | 0 | 10(1.11%) |
| NA | 0 | 103(11.4%) |
| Patient age (yr) | ||
| Median [range] | 66 [47, 79] | 61 [37.3, 83] |
| Pre‐operative PSA | ||
| Pre‐PSA<10 | 286(51.91%) | 371(41.04%) |
| 10<= pre‐PSA<20 | 118(21.42%) | 158(17.48%) |
| Pre‐PSA>20 | 132(23.96%) | 86(9.51%) |
| NA | 15(2.72%) | 289(31.97%) |
| Path GS | ||
| <=6 | 64(11.62%) | 170(18.81%) |
| 7 | 275(49.91%) | 521(57.63%) |
| 8 | 68(12.34%) | 100(11.06%) |
| >=9 | 144(26.13%) | 110(12.17%) |
| NA | 0(0%) | 3(0.33%) |
| EPE | ||
| Positive | 276(50.09%) | 378(41.81%) |
| NA | 0(0%) | 242(26.77%) |
| SVI | ||
| Positive | 177(32.12%) | 173(19.14%) |
| NA | 0(0%) | 239(26.44%) |
| SM | ||
| Positive | 270(49%) | 364(40.27%) |
| NA | 0(0%) | 99(10.95%) |
| LNI | ||
| Positive | 73(13.25%) | 60(6.64%) |
| NA | 0(0%) | 79(8.74%) |
| Met | ||
| Positive | 215(39.02%) | 148(16.37%) |
| NA | 0(0%) | 191(21.13%) |
| BCR | ||
| Positive | 393(71.32%) | 275(30.42%) |
| NA | 0(0%) | 191(21.13%) |
Overall, about 61% of patients manifest with adverse pathologic findings, and patients are mostly of European descent.
Development of transcriptomic fingerprint of metastasis
In this study, we sought to determine whether a genomic fingerprint of metastasis could be detected in the primary tumours of patients who develop metastatic disease through comparison with primary tumours of patients that do not develop evidence of metastasis even after long clinical follow up. First, we divided the Mayo Cohort I into metastatic (cases) and non‐metastatic (controls) and applied discriminative analysis to filter informative features (Figure 1B). Differential expression analysis identified 568 up‐regulated and 86 down‐regulated probesets in patients that developed metastasis (metastatic group), suggesting there is a transcriptomic fingerprint identifying patients at higher risk of developing metastasis after RP, long before the clinical detection of metastasis (Figure 2A). We investigated the representation of protein coding and non‐coding regions of the genome in the fingerprint detected in this analysis (metastatic fingerprint); approximately half of the 654 features in the fingerprint are non‐coding RNA including long non‐codingRNAs (lncRNAs) (11%), intergenic (16%) and anti‐sense probsets (13%) (Figure 2B; Table S2). lncRNAs including the SChLAP1 region, and NEAT1 were among the most up‐regulated features in the metastatic fingerprint (Figure 2C). On their own, SChLAP1 and NEAT1 have been demonstrated to be independent predictors of metastasis in two separate studies (Chakravarty et al., 2014; Prensner et al., 2014). Here, we confirmed those findings and add to the growing body of evidence supporting a key role for long non‐coding RNA in PCa disease progression. We found the lncRNA PCAT18 is down‐regulated in metastatic patients, although other studies have shown it to be subsequently up‐regulated in metastatic castration‐resistant disease (Crea et al., 2014), suggesting that PCAT18 may play a role in developing castration resistance. Several reports show that these lncRNAs are prostate specific, suggesting that they have specific role in PCa progression (Martens‐Uzunova et al., 2014). Delineating the functional role of these lncRNAs requires further investigation that is not within the scope of this investigation.
Figure 2.
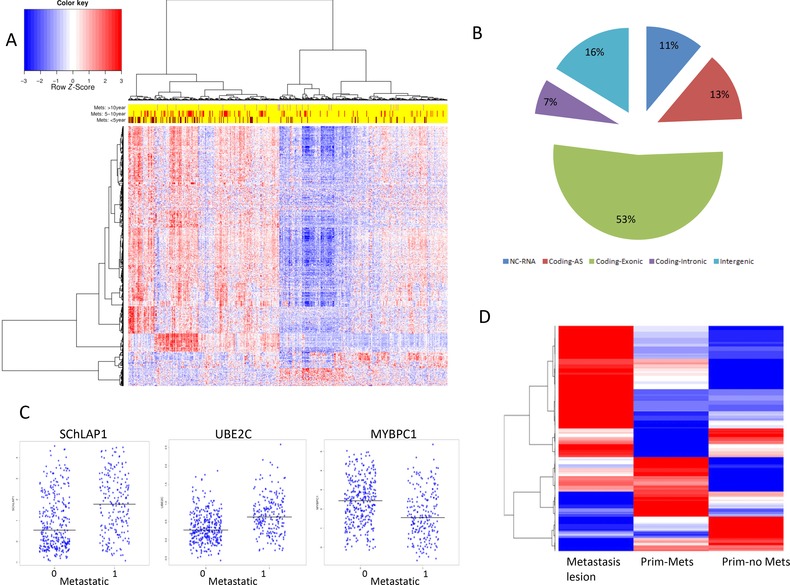
Metastatic fingerprint (n = 654) discovered from the Mayo Clinic I showing that most of the features are up‐regulated in the metastatic group
(A) Heatmap showing three main correlated groups of upregulated genes, among which is the SChLAP1 region. (B) Approximately half of the features are non‐coding (lncRNA, intronic, intergenic and anti‐sense). (C) SChLAP1 and UBE2C are the top upregulated lncRNA and coding RNA respectively in the metastatic group, and MYBPC1 is the most down‐regulated. (D) The metastatic fingerprint is representing the biology of metastasis tissues from MSKCC. Most of features up‐regulated in localised cancer with metastatic outcome (Prim‐Met) are also up‐regulated in metastasis lesions.
To gain more biological insights into the metastatic fingerprint, Gene Enrichment Analysis using DAVID web services (Dennis et al., 2003) showed that up‐regulated genes are highly enriched with cell cycle processes (P = 8E−9), cytoskeleton organisation (P = 2E−3) and ECM–receptor interaction (P = 1.3E−3), biological processes that are all involved in metastasis down‐regulated genes were associated with protein kinase cascade (P = 3.2E−3) and signal transduction (P = 0.02). The top down‐regulated genes were Myosin‐Binding Protein C (MYBPC1) (Figure 2C) and Anoctamin7 (ANO7); the first plays a structural role to preserve cell rigidity and the second maintains cell–cell interactions. On the other hand, the top up‐regulated genes were UBE2C, TOP2A, CAMK2N1, CENPF and ZWILCH. All of these genes are involved in cell cycle processes, mitotic spindle assembly and mitotic checkpoints, and have been previously reported to be associated with PCa recurrence (Cuzick et al. 2012) and castration‐resistant PCa (Chen et al., 2011). Likely, overexpression of these genes allows the cell to proliferate, re‐organise its skeleton and escape cell cycle control mechanisms thereby avoiding pro‐apoptotic signals.
The metastatic fingerprint in metastatic tissues
Further confirming that the metastatic fingerprint from the primary tumour represents transcriptomic changes that are also present in the metastatic lesions, we compared the expression data of the metastatic fingerprint from 19 metastatic lesions and 131 primary PCa as a control set from the MSKCC cohort (9 with metastatic outcome) (Figure 2D). We found that 30% of the metastatic fingerprint is overexpressed in the 19 metastatic lesions samples including TOP2A, TMSB10, SChLAP1, STMN1 and MKI67 confirming that the metastatic fingerprint represents the early stages of metastasis development. Functional analysis of these features showed that they are mainly involved in cell cycle, cytoskeleton protein binding and ECM–receptor interaction. We observed that not all the features contributed equally to distinguishing patients with metastasis cancer from patients with localised cancer (Figure 2D). Interestingly we found that NEAT1 expression did not change in the metastatic lesions patients, suggesting an early role of NEAT1 to drive metastasis. Approximately 10% of the genes were further down‐regulated in metastatic lesions, including MYBPC1, ANO7 and AZGP1, compared with primaries with metastasis outcome. These genes were mainly enriched for functions in focal adhesion. Approximately 10% of the features were down‐regulated in the metastasis lesions after being relatively overexpressed in the primary tumour with metastatic outcome including SFRP4, ASPN and GNPTAB. These genes enriched with ECM–receptor interaction, small cell lung cancer genes and glycosaminoglycan binding.
These results suggest that most of the features of the metastatic fingerprint play roles throughout the metastasis development process with only a small portion playing a role in early stages to drive metastasis.
This could indicate that the primary tumour contains cells with high metastatic potential that may have already spread to lymphatic system or bone; perhaps in a dormant state, until they are unmasked or revealed during the aging process. An alternative hypothesis could be that primary tumour has gained the early genomic changes needed for spreading and are in a transition state waiting upon endogenous or external signals to spread out, a model similar to that proposed in breast cancer metastasis (Weigelt et al., 2005)
Evaluating the metastatic fingerprint in RP cohorts
The most challenging and critical step for a fingerprint to be clinically adopted is validating the model in independent cohorts to show equivalent performance and demonstrate clinical impact on decision making (Figure 3A). To further verify the relevance of the metastatic fingerprint to metastatic patients, we evaluated the fingerprint in two independent datasets (Figure 3B) from Mayo Clinic and CCF. Most of the features of the fingerprint were prognostic in the validation cohorts (Table S2). Hierarchical clustering of the patients in each cohort using Pearson's correlation and Ward's method aggregated most metastatic patients together, verifying the associations between the fingerprint and metastasis. Patients in the two evaluation sets were subtyped into ERG+, ETS+, SPINK+ and TripleNeg groups to assess if the metastatic fingerprint is associated with certain subtype. We found the clustering of high risk patients to be independent of these subgroups (Figure 3B), demonstrating equivalent performance across different subtypes. Efficacy across common genetic subtypes is required for genomic tests to be part of standard clinical practice. For instance, the discrimination power of the Decipher test was found to be independent of the molecular subtypes (Figure 3C), suggesting that Decipher is robust in a heterogeneous population.
Figure 3.
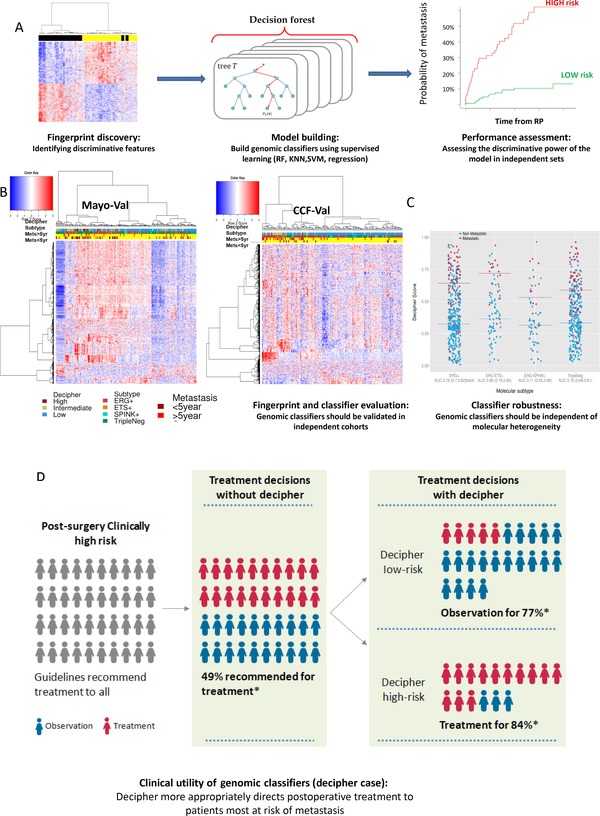
Delivering a fingerprint into the clinic requires appropriate modelling and effective performance in independent cohorts to validate the prognosis ability
(A) Modelling fingerprint to genomic classifiers is a critical step to show utility of the fingerprint. It requires external independent cohorts to assess its performance. (B) The metastatic fingerprint from this work is showing high relevance to metastatic groups in two independent cohorts, and to Decipher risk categories. Metastatic patients fall into ERG+, ETS+, SPINK+ and TripleNeg subgroups (annotation bar) suggesting a molecular heterogeneity underling metastasis. (C) Decipher was shown to be independent of molecular subtypes and effective prognostic tool in all subtypes. (D) Ultimate goal of each prognostic tool is to have an impact on treatment decision making. Decipher more appropriately directs post‐RP treatment to patients most at risk of metastasis.
Translating fingerprint into prognostic tools: The Decipher PCa classifier
Delivering a fingerprint into the clinic as a diagnostic or prognostic tool starts with asking the relevant question, appropriate study design, using the appropriate methodologies, validating it the right setting resulting in clinical benefit. We first designed the study to define a fingerprint from the discovery cohort and then evaluated it in independent cohorts using clustering analysis. As clustering is not an appropriate approach to be used to predict prognosis of each new patient, more rigid models such as weighted regression or random forest (RF) modelling are used to generate scores that are easier to apply cut‐points or categorise into risk groups based on an algorithm. In this case, we desired to show the relevance of the fingerprint to the metastatic group in independent cohorts. To provide a relevant example of translating a fingerprint into a prognostic tool, we use the Decipher classifier to demonstrate transitioning from fingerprint to clinical use, starting from transcriptomic fingerprint discovery to commercially available prognostic test.
Decipher is a transcriptomic fingerprint for metastasis that has been developed as a genomic prognostic test to predict the probability of metastasis after RP. Twenty‐two RNA biomarkers filtered from 1.4 M probesets covering most of the human genome constitute the Decipher test. Several filtering criteria including Lasso regression were adopted to reduce the discriminative features into the most informative, non‐redundant features, resulting in the 22 RNA biomarkers that best discriminated patients who developed metastasis within 5 years from patients that did not develop metastasis. These 22 RNA biomarker were modelled into a RF model that generates a score between 0 and 1; the higher the value the more likely the patients will develop metastasis. The model was validated in several cohorts and surpassed currently used clinical tools demonstrating clinical utility (Karnes et al., 2013; Den et al., 2014; Klein et al., 2014b). Combining the fingerprint genes into a single model with subsequent independent validation are the most critical in translating the fingerprint into a clinically meaningful score. Modelling the fingerprint requires a clean set of training data to maximise the discrimination power of the fingerprint features collectively. Likewise, the validation cohort should be clinically similar to the training data to properly assess the performance of the model. If a model demonstrates superior performance compared with existing tools, it goes through to the clinical impact evaluation in prospective studies. So far, no prognostic test for predicting metastasis after radical prostatectomy has passed this step.
The Decipher platform provides true representation of the transcriptomic landscape of PCa progression in the surgical setting and captures novel transcriptomic alterations, including poorly annotated regions such as lncRNAs that cannot be detected using other platforms. These lncRNAs have been linked to biological processes involved in tumorigenesis (Prensner and Chinnaiyan, 2011) and PCa (Cheng et al., 2013). It is essential to understand the molecular biology behind the Decipher biomarkers to properly understand the molecular mechanism of PCa progression. Characterising Decipher biomarker pathways is important to identify new key players that may act as therapeutic targets.
Twelve Decipher genes, including UBE2C, ZWILCH, NUSAP1, MYCBP1, ANO7, EPPK1, TNFRSF19, LASP1, THBS2, IQGAP3, CAMK2N1 and RABGAP1, were in the metastatic fingerprint defined in this study. To better understand the biological role of the Decipher biomarkers in the cellular context, the protein partners of the functionally annotated Decipher biomarkers were characterised through the functional protein association network tool STRING (Franceschini et al., 2013). Several Decipher biomarkers were related to PCa progression through the cell cycle and mitotic checkpoint, cell adhesion and apoptosis pathways. Ten modules capturing the function of the Decipher gene were identified (Figure 4), with the two major modules being involved in cell cycle process and mitosis and cell adhesion. Other modules are also involved key processes related to cancer, including apoptosis, cytoskeleton organisation, immune modulation, cell differentiation and transcription regulation.
Figure 4.
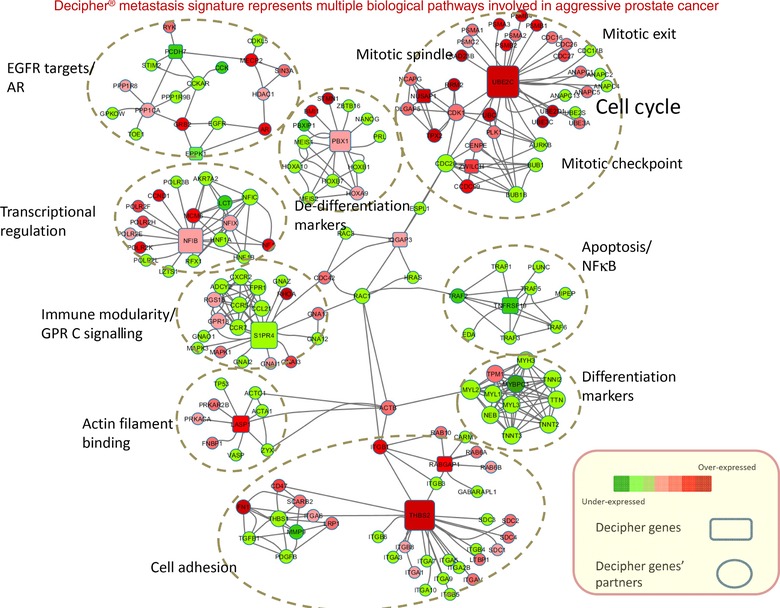
The systems biology landscape of the Decipher biomarkers shows modularity. The Decipher genes form 10 intertwined modules representing several pathways; mainly cell cycle, cell adhesion and cytoskeleton reorganisation. Other pathways are related to apoptosis, cell differentiation, immune modularity and inflammation.
Comparing Decipher signature to other PCa prognostic signature, several Decipher genes are among existing PCa prognostic signatures. For example, UBE2C, NUSAP1 and MYBPC1 are among the 86 gene Penny signature (Penney et al., 2011) that predicts high risk cases in Gleason 7. The three genes were highly differentially expressed between Gleason <= 6 and Gleason >= 8. NUSAP1 is among the 31 CCP genes that constitute the Prolaris signature (Cuzick et al., 2011), and BUB1B and CDC20, that are among the 31 CCP genes, are highly interacting with ZWILCH and UBE2C from Decipher suggesting the Decipher is capturing the cell cycle dysregulation aspect of tumour. Several Decipher biomarkers have been identified with links to breast cancer metastasis, bladder, and colon cancer. For example, UBE2C has been identified as a prognostic protein in high risk breast cancer (Psyrri et al., 2012), non‐muscle invasive bladder cancer (Fristrup et al., 2013) and node‐positive breast cancer (Loussouarn et al., 2009). LASP1 has been linked to breast cancer metastasis as a negative prognostic indicator of long‐term survival of breast cancer patients (Frietsch et al., 2010), ovarian cancer migration and proliferation by influencing zyxin localisation (Grunewald et al., 2007) and progression and metastasis dissemination of medulloblastoma (Traenka et al., 2010). Furthermore, PBX1 is a significant prognostic marker in non‐small lung carcinoma (Qiu et al., 2009). Several lines of evidence have shown that NFIB is associated with progression of several other cancers. It serves as an oncogene in small cell lung cancer (Dooley et al., 2011) and has elevated expression in metastatic Giant cell tumour of bone (Mosakhani et al., 2012) and is a potential target of oestrogen receptor‐negative breast cancer as it has a role in cell proliferation (Moon et al., 2011).
Characterising the association between the metastatic fingerprint and Decipher risk categories revealed its relevance to PCa risk. Additionally, patients that developed metastasis were found to fall in four subgroups (ERG+, ETS+, SPINK+ and TripleNeg) suggesting a molecular heterogeneity underlying metastasis (Figure 3B). A prognostic fingerprint should offer relatively equivalent performance across subtypes to ensure reliability and robustness. Here, we found the fingerprint is independent of the PCa molecular subtypes (Figure 3B) and the Decipher PCa classifier is an effective prognostic tool of metastasis in the four subgroups supporting that Decipher is robust transcriptomic fingerprint (Figure 3C). Decipher's use as a prognostic tool has made significant impact on decision making in several published reports (Figure 3D) (Badani et al., 2013), which is the ultimate goal of transforming lab developed fingerprints into clinically utilised tests.
Challenges of translating fingerprint into prognostic tools
Many attempts have been made to develop prostate genomic tests based on transcriptomic data for PCa risk stratification, but most of them failed to be clinically adopted (Irshad et al., 2013; Wu et al., 2013; Bismar et al., 2014). This failure can be attributed to multiple reasons. First, analytical validation in independent cohorts is essential and most developed signatures failed to be reproduced in independent cohorts. Second, the validation to predict the relevant clinical endpoint is critical; most developed gene panels are validated in different settings, different endpoints or different tissue. This could be because of the lack of the appropriate cohort for validation as only a handful of fully annotated PCa data sets are publically available. The last and most important requirement for clinical adoption is the equivalent and robust performance across a spectrum of disease subtypes that are related, but not limited, to race, age and molecular subtype.
Current emerging biomarkers aim to enable the determination of an appropriate treatment strategy for individual patients, detect advanced disease at an earlier stage, and predict metastatic cancer and recurring disease following prostatectomy. Because of the heterogeneity of the disease, no single biomarker will be diagnostic and prognostic for every patient. The next PCa diagnostic or prognostic test should employ multiple biomarkers that could be combined with current risk assessment tools. Whether the next tools are genomic, transcriptomic, proteomic, metabolomic or a combination of all, the test should be based on genome‐wide analysis to truly represent the biology underlying PCa. Robust prognostic tools better present the spectrum of tumour biology (Hanahan and Weinberg, 2011) to uncover the alterations at different levels of cancer progression. Decipher results are indicative of the dysregulation of multiple pathways and biological processes in the early stages of cancer progression, unlike other commercial tests that focus on single pathways, suggesting that Decipher provides a more complete perspective of PCa development, making it more sensitive to early molecular tumorgenic events. Currently, the Decipher test is the only prognostic test that was developed using a genome‐wide unbiased approach to define the fingerprint discriminating metastatic patients. The 22 genes comprising the Decipher showed to be representing the biology underling metastasis and impacting treatment decision.
Materials and methods
Patient population
A total of 1449 patient expression profiles were analysed from eight RP cohorts from Mayo Clinic (I and II) (Erho et al., 2013; Karnes et al., 2013), Thomas Jefferson University (TJU) (Den et al., 2014), Cleveland Clinic (CCF)(Klein et al., 2014b), Johns Hopkins (JHMI), Memorial Sloan Kettering (MSKCC) (Taylor et al., 2010), Erasmus MC (EMC) (Boormans et al., 2013) and the German National Cancer Registry (DKFZ) (Brase et al., 2011) (Table S1). Five cohorts utilised RNA extracted from FFPE RP specimens and three cohorts used RNA extracted from fresh‐frozen RP specimens. The Mayo Clinic I (n = 545) cohort with 215 patients developing metastasis was used as a discovery cohort to define the metastatic fingerprint, and the remaining sets used for evaluation.
Data generation and pre‐processing
Tumour specimens were obtained from archived paraffin blocks and RNA extraction was performed using the Decipher platform as described previously (Erho et al., 2013). RNA was amplified, labelled and hybridised to Human Exon 1.0 ST microarrays (Affymetrix) covering 1.4 million probesets. Annotation of features to genes was based on the alignment provided by xmapcore against the hg19 version of the human genome. Based on the proximity of a feature relative to the genes annotated Ensembl v62, features were categorised as either coding, UTR, intronic, anti‐sense and intergenic as previously described. Features overlapping with known non‐coding transcripts were annotated as non‐coding transcripts. More than 75% of the features on the Human Exon array cover regions annotated as non‐protein coding. The SCAN algorithm (Piccolo et al., 2012) was used for individual patient profile pre‐processing and normalisation.
Molecular subtyping of PCa patients
Patients in this study were classified into four molecularly distinct groups based on microarray‐based classifiers, where we developed a supervised RF model that was trained to predict ERG rearrangement assessed by FISH. Rearrangements in other genes such as ETV1, ETV4, ETV5 and SPINK1 were predicted using outlier analysis method. The four subgroups were defined based on the results of the microarray‐based classifiers (m‐ERG, m‐ETS and m‐SPINK1) models described in (unpublished data). Patient profiles with high m‐ERG score (m‐ERG+) and m‐ETV1─, m‐ETV4─, m‐ETV5─, m‐FLI1─ and m‐SPINK1─ were classified as ERG+ subtype, patient profiles that were m‐ETV1+, m‐ETV4+, m‐ETV5+ or m‐FLI1+ and m‐ERG− were classified as ERG−ETS+ subtype, patient profiles that were m‐SPINK1+ and m‐ERG─ were classified as ERG−SPINK1+ subtype, and patient profiles that are m‐ERG─, m‐ETV1─, m‐ETV4─, m‐ETV5─, m‐FLI1─ and m‐SPINK1─ were classified as the ‘Triple Negative’ subtype.
Expression analysis of the PCa metastatic transcriptome
Mayo Clinic I cohort (n = 545), a matched case control cohort, was used to develop a metastatic fingerprint. Differential expression analysis was applied to identify discriminative features between patients who developed metastasis after RP (40%) and patients who did not (60%). Two criteria were used to filter discriminative features; median fold difference (MFD) adjusted for the interquantile range across all patients (threshold 0.3), and Wilcoxon test (P = 0.001) after MFD filtering. Features that passed these criteria formed the metastatic fingerprint.
Author contribution
M.A. and E.D. conceptualised and designed the research; M.A., M.S., H.S., N.E. and E.D. performed the data analysis and interpretation; M.A., M.S., H.S. and E.D. wrote the article.
Conflict of interest statement
M.A., M.S., H.S., N.E. and E.D. are employees of GenomeDx Biosciences. E.D. is the president and CSO of GenomeDx Biosciences.
Supporting information
Supplementary Material
References
- Alshalalfa, M. , Crisan, A. , Vergara, I.A. , Ghadessi, M. , Buerki, C. , Erho, N. and Yousefi, K. (2015) Clinical and genomic analysis of metastatic prostate cancer progression in a background of post‐operative biochemical recurrence. BJU Int. doi:10.1111/bju13013. [DOI] [PubMed] [Google Scholar]
- Badani, K. , Thompson, D.J.S. , Buerki, C. , Davicioni, E. , Garrison, J. , Ghadessi, M. , Mitra, A.P. , Wood, P.J. and Hornberger, J. (2013) Impact of a genomic classifier of metastatic risk on postoperative treatment recommendations for prostate cancer patients: a report from the DECIDE study group. Oncotarget 4, 600–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbieri, C.E. , Bangma, C.H. , Bjartell, A. , Catto, J.W.F. , Culig, Z. , Grönberg, H. , Luo, J. , Visakorpi, T. and Rubin, M.A. (2013) The mutational landscape of prostate cancer. Eur. Urol. 64, 567–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger, M.F. , Lawrence, M.S. , Demichelis, F. , Drier, Y. , Cibulskis, K. , Sivachenko, A.Y. , Sboner, A. , Esgueva, R. , Pflueger, D. , Sougnez, C. , Onofrio, R. , Carter, S.L. , Park, K. , Habegger, L. , Ambrogio, L. , Fennell, T. , Parkin, M. , Saksena, G. , Voet, D. , Ramos, A.H. , Pugh, T.J. , Wilkinson, J. , Fisher, S. , Winckler, W. , Mahan, S. , Ardlie, K. , Baldwin, J. , Simons, J.W. , Kitabayashi, N. , MacDonald, T.Y. , Kantoff, P.W. , Chin, L. , Gabriel, S.B. , Gerstein, M.B. , Golub, T.R. , Meyerson, M. , Tewari, A. , Lander, E.S. , Getz, G. , Rubin, M.A. , Garraway, L.A. (2011) The genomic complexity of primary human prostate cancer. Nature 470, 214—220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bismar, T.A. , Alshalalfa, M. , Petersen, L.F. , Teng, L.H. , Gerke, T. , Bakkar, A. , Al‐Mami, A. , Liu, S. , Dolph, M. , Mucci, L.A. , Alhajj, R. (2014) Interrogation of ERG gene rearrangements in prostate cancer identifies a prognostic 10‐gene signature with relevant implication to patients’ clinical outcome. BJU Int. 113, 309–319 [DOI] [PubMed] [Google Scholar]
- Boorjian, S.A. , Thompson, R.H. , Tollefson, M.K. , Rangel, L.J. , Bergstralh, E.J. , Blute, M.L. and Karnes, R.J. (2011) Long‐term risk of clinical progression after biochemical recurrence following radical prostatectomy: the impact of time from surgery to recurrence. Eur. Urol. 59, 893–899 [DOI] [PubMed] [Google Scholar]
- Boormans, J.L. , Korsten, H. , Ziel‐Van Der Made, A.J.C. , Van Leenders, G.J.L.H. , De Vos, C.V. , Jenster, G. and Trapman, J. (2013) Identification of TDRD1 as a direct target gene of ERG in primary prostate cancer. Int. J. Cancer 133, 335–345 [DOI] [PubMed] [Google Scholar]
- Boyd, L.K. , Mao, X. and Lu, Y.‐J. (2012) The complexity of prostate cancer: genomic alterations and heterogeneity. Nat. Rev. Urol. 9, 652–664 [DOI] [PubMed] [Google Scholar]
- Brase, J.C. , Johannes, M. , Mannsperger, H. , Falth, M. , Metzger, J. , Kacprzyk, L.A. , Andrasiuk, T. , Gade, S. , Meister, M. , Sirma, H. , Sauter, G. , Simon, R. , Schlomm, T. , Beissbarth, T. , Korf, U. , Kuner, R. , Sultmann, H. (2011) TMPRSS2‐ERG ‐specific transcriptional modulation is associated with prostate cancer biomarkers and TGF‐beta signaling. BMC Cancer 11, 507. doi:10.1186/1471‐2407‐11‐507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravarty, D. , Sboner, A. , Nair, S.S. , Giannopoulou, E. , Li, R. , Hennig, S. , Mosquera, J.M. and Pauwels, J. (2014) The oestrogen receptor alpha‐regulated lncRNA NEAT1 is a critical modulator of prostate cancer. Nat. Commun. 5, 5383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Z. , Zhang, C. , Wu, D. , Chen, H. , Rorick, A. , Zhang, X. and Wang, Q. (2011) Phospho‐MED1‐enhanced UBE2C locus looping drives castration‐resistant prostate cancer growth. EMBO J. 30, 2405–2419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, W. , Zhang, Z. and Wang, J. (2013) Long noncoding RNAs: New players in prostate cancer. Cancer Lett. 339, 8–14 [DOI] [PubMed] [Google Scholar]
- Cooperberg, M.R. , Davicioni, E. , Crisan, A. , Jenkins, R.B. , Ghadessi, M. and Karnes, R.J. (2015) Combined value of validated clinical and genomic risk stratification tools for predicting prostate cancer mortality in a high‐risk prostatectomy cohort. Eur. Urol. 67, 326–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crea, F. , Watahiki, A. , Quagliata, L. , Xue, H. , Pikor, L. , Parolia, A. , Wang, Y. , Lin, D. , Lam, W.L. , Farrar, W.L. , Isogai, T. , Morant, R. , Castori‐Eppenberger, S. , Chi, K.N. , Wang, Y. , Helgason, C.D. , (2014) Identification of a long non‐coding RNA as a novel biomarker and potential therapeutic target for metastatic prostate cancer. Oncotarget 5, 764–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuzick, J. , Swanson, G.P. , Fisher, G. , Brothman, A.R. , Berney, D.M. , Reid, J.E. , Mesher, D. , Speights, V.O. , Stankiewicz, E. , Foster, C.S. , Møller, H. , Scardino, P. , Warren, J.D. , Park, J. , Younus, A. , Flake, D.D. , Wagner, S. , Gutin, A. , Lanchbury, J.S. , Stone, S. , (2011) Prognostic value of an RNA expression signature derived from cell cycle proliferation genes in patients with prostate cancer: a retrospective study. Lancet Oncol. 12, 245–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuzick, J. , Berney, D.M. , Fisher, G. , Mesher, D. , Møller, H. , Reid, J.E. , Perry, M. , Park, J. , Younus, A. , Gutin, A. , Foster, C.S. , Scardino, P. , Lanchbury, J.S. , Stone, S. , (2012) Prognostic value of a cell cycle progression signature for prostate cancer death in a conservatively managed needle biopsy cohort. Br. J. Cancer 106, 1095–1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis, J. (2014) Novel commercially available genomic tests for prostate cancer: a roadmap to understanding their clinical impact. BJU Int. 114, 320–322 [DOI] [PubMed] [Google Scholar]
- Den, R.B. , Feng, F.Y. , Showalter, T.N. , Mishra, M. V. , Trabulsi, E.J. , Lallas, C.D. , Gomella, L.G. , Kelly, W.K. , Birbe, R.C. , McCue, P.A. , Ghadessi, M. , Yousefi, K. , Davicioni, E. , Knudsen, K.E. , Dicker, A.P. (2014) Genomic prostate cancer classifier predicts biochemical failure and metastases in patients after postoperative radiation therapy. Int. J. Radiat. Oncol. 89, 1038–1046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis, G. , Sherman, B.T. , Hosack, D.A. , Yang, J. , Gao, W. , Lane, H.C. and Lempicki, R.A. (2003) DAVID: Database for annotation, visualization, and integrated discovery. Genome Biol. 4, P3 [PubMed] [Google Scholar]
- Dooley, A.L. , Winslow, M.M. , Chiang, D.Y. , Banerji, S. , Stransky, N. , Dayton, T.L. , Snyder, E.L. , Senna, S. , Whittaker, C.A. , Bronson, R.T. , Crowley, D. , Barretina, J. , Garraway, L. , Meyerson, M. , Jacks, T. , (2011) Nuclear factor I/B is an oncogene in small cell lung cancer. Genes Dev. 25, 1470–1475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand, X. , Moutereau, S. , Xylinas, E. and de la Taille, A. (2011) ProgensaTM PCA3 test for prostate cancer. Expert Rev. Mol. Diagn. 11, 137–144 [DOI] [PubMed] [Google Scholar]
- Erho, N. , Crisan, A. , Vergara, I.A. , Mitra, A.P. , Ghadessi, M. , Buerki, C. , Bergstralh, E.J. , Kollmeyer, T. , Fink, S. , Haddad, Z. , Zimmermann, B. , Sierocinski, T. , Ballman, K. V , Triche, T.J. , Black, P.C. , Karnes, R.J. , Klee, G. , Davicioni, E. , Jenkins, R.B. , (2013) Discovery and validation of a prostate cancer genomic classifier that predicts early metastasis following radical prostatectomy. PLoS One 8, e66855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franceschini, A. , Szklarczyk, D. , Frankild, S. , Kuhn, M. , Simonovic, M. , Roth, A. , Lin, J. , Minguez, P. , Bork, P. , von Mering, C. , Jensen, L.J. , (2013) STRING v9.1: protein‐protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 41, D808–D815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frietsch, J.J. , Grunewald, T.G.P. , Jasper, S. , Kammerer, U. , Herterich, S. , Kapp, M. , Honig, A. and Butt, E. (2010) Nuclear localisation of LASP‐1 correlates with poor long‐term survival in female breast cancer. Br. J. Cancer 102, 1645–1653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fristrup, N. , Birkenkamp‐Demtrder, K. , Reinert, T. , Sanchez‐Carbayo, M. , Segersten, U. , Malmstrm, P.U. , Palou, J. , Alvarez‐Múgica, M. , Pan, C.C. , Ulhi, B.P. , Borre, M. , Rntoft, T.F. , Dyrskjt, L. , (2013) Multicenter validation of cyclin D1, MCM7, TRIM29, and UBE2C as prognostic protein markers in non‐muscle‐invasive bladder cancer. Am. J. Pathol. 182, 339–349 [DOI] [PubMed] [Google Scholar]
- Grunewald, T.G.P. , Kammerer, U. , Winkler, C. , Schindler, D. , Sickmann, A. , Honig, A. and Butt, E. (2007) Overexpression of LASP‐1 mediates migration and proliferation of human ovarian cancer cells and influences zyxin localisation. Br. J. Cancer 96, 296–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan, D. and Weinberg, R.A. (2011) Hallmarks of cancer: The next generation. Cell 144, 646–674 [DOI] [PubMed] [Google Scholar]
- Hessels, D. and Schalken, J.A. (2009) The use of PCA3 in the diagnosis of prostate cancer. Nat. Rev. Urol. 6, 255–261 [DOI] [PubMed] [Google Scholar]
- Irshad, S. , Bansal, M. , Castillo‐Martin, M. , Zheng, T. , Aytes, A. , Wenske, S. , Le Magnen, C. , Guarnieri, P. , Sumazin, P. , Benson, M.C. , Shen, M.M. , Califano, A. , Abate‐Shen, C. (2013) A molecular signature predictive of indolent prostate cancer. Sci. Transl. Med. 5, 202ra122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, N. , Zhu, S. , Chen, J. , Niu, Y. and Zhou, L. (2013) A‐Methylacyl‐CoA Racemase (AMACR) and prostate‐cancer risk: A meta‐analysis of 4,385 participants. PLoS One 8, e74386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, R. , Yi, Y. , Yull, F.E. , Blackwell, T.S. , Clark, P.E. , Koyama, T. , Smith, J. A. and Matusik, R.J. (2014) NF‐κB gene signature predicts prostate cancer progression. Cancer Res. 74, 2763–2772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karnes, R.J. , Bergstralh, E. , Davicioni, E. , Ghadessi, M. , Buerki, C. , Mitra, A. , Crisan, A. , Erho, N. , Vergara, I. , Lam, L. , Carlson, R. , Thompson, D. , Haddad, Z. , Zimmermann, B. , Sierocinski, T. , Triche, T. , Kollmeyer, T. , Ballman, K. , Black, P. , Klee, G. , Jenkins, R. , (2013) Validation of a genomic classifier that predicts metastasis following radical prostatectomy in an at risk patient population. J. Urol. 190, 2047–2053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein, E.A. , Cooperberg, M.R. , Magi‐Galluzzi, C. , Simko, J.P. , Falzarano, S.M. , Maddala, T. , Chan, J.M. , Li, J. , Cowan, J.E. , Tsiatis, A.C. , Cherbavaz, D.B. , Pelham, R.J. , Tenggara‐Hunter, I. , Baehner, F.L. , Knezevic, D. , Febbo, P.G. , Shak, S. , Kattan, M.W. , Lee, M. , Carroll, P.R. , (2014) A 17‐gene assay to predict prostate cancer aggressiveness in the context of gleason grade heterogeneity, tumor multifocality, and biopsy undersampling. Eur. Urol. 66, 550–560 [DOI] [PubMed] [Google Scholar]
- Klein, E.A. , Yousefi, K. , Haddad, Z. , Choeurng, V. , Buerki, C. , Stephenson, A.J. , Li, J. , Kattan, M.W. , Magi‐Galluzzi, C. and Davicioni, E. (2014b) A genomic classifier improves prediction of metastatic disease within 5 years after surgery in node‐negative high‐risk prostate cancer patients managed by radical prostatectomy without adjuvant therapy. Eur. Urol. 67, 778–786 [DOI] [PubMed] [Google Scholar]
- Leyten, G.H.J.M. , Hessels, D. , Jannink, S.A. , Smit, F.P. , De Jong, H. , Cornel, E.B. , De Reijke, T.M. , Vergunst, H. , Kil, P. , Knipscheer, B.C. , Van Oort, I.M. , Mulders, P.F.A. , Hulsbergen‐Van De Kaa, C.A. , Schalken, J.A. , (2014) Prospective multicentre evaluation of PCA3 and TMPRSS2‐ERG gene fusions as diagnostic and prognostic urinary biomarkers for prostate cancer. Eur. Urol. 65, 534–542 [DOI] [PubMed] [Google Scholar]
- Loussouarn, D. , Campion, L. , Leclair, F. , Campone, M. , Charbonnel, C. , Ricolleau, G. , Gouraud, W. , Bataille, R. and Jézéquel, P. (2009) Validation of UBE2C protein as a prognostic marker in node‐positive breast cancer. Br. J. Cancer 101, 166–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markert, E.K. , Mizuno, H. , Vazquez, A. and Levine, A.J. (2011) Molecular classification of prostate cancer using curated expression signatures. Proc. Natl. Acad. Sci. U.S.A. 108, 21276–21281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens‐Uzunova, E.S. , Böttcher, R. , Croce, C.M. , Jenster, G. , Visakorpi, T. and Calin, G.A. (2014) Long noncoding RNA in prostate, bladder, and kidney cancer. Eur. Urol. 65, 1140–1151 [DOI] [PubMed] [Google Scholar]
- Moon, H.G. , Hwang, K.T. , Kim, J.A. , Kim, H.S. , Lee, M.J. , Jung, E.M. , Ko, E. , Han, W. and Noh, D.Y. (2011) NFIB is a potential target for estrogen receptor‐negative breast cancers. Mol. Oncol. 5, 538–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosakhani, N. , Pazzaglia, L. , Benassi, M.S. , Borze, I. , Quattrini, I. , Picci, P. and Knuutila, S. (2012) MicroRNA expression profiles in metastatic and non‐metastatic giant cell tumor of bone. Histol. Histopathol. 28, 671–678 [DOI] [PubMed] [Google Scholar]
- Nakagawa, T. , Kollmeyer, T.M. , Morlan, B.W. , Anderson, S.K. , Bergstralh, E.J. , Davis, B.J. , Asmann, Y.W. , Klee, G.G. , Ballman, K.V. and Jenkins, R.B. (2008) A tissue biomarker panel predicting systemic progression after PSA recurrence post‐definitive prostate cancer therapy. PLoS One 3, e2318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nwosu, V. , Carpten, J. , Trent, J.M. and Sheridan, R. (2001) Heterogeneity of genetic alterations in prostate cancer: evidence of the complex nature of the disease. Hum. Mol. Genet. 10, 2313–2318 [DOI] [PubMed] [Google Scholar]
- Payne, H. and Cornford, P. (2011) Prostate‐specific antigen: an evolving role in diagnosis, monitoring, and treatment evaluation in prostate cancer. Urol. Oncol. 29, 593–601 [DOI] [PubMed] [Google Scholar]
- Penney, K.L. , Sinnott, J.A. , Fall, K. , Pawitan, Y. , Hoshida, Y. , Kraft, P. , Stark, J.R. , Fiorentino, M. , Perner, S. , Finn, S. , Calza, S. , Flavin, R. , Freedman, M.L. , Setlur, S. , Sesso, H.D. , Andersson, S.‐O. , Martin, N. , Kantoff, P.W. , Johansson, J.‐E. , Adami, H.‐O. , Rubin, M.A. , Loda, M. , Golub, T.R. , Andrén, O. , Stampfer, M.J. , Mucci, L.A. , (2011) mRNA expression signature of Gleason grade predicts lethal prostate cancer. J. Clin. Oncol. 29, 2391–2396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perdonà, S. , Bruzzese, D. , Ferro, M. , Autorino, R. , Marino, A. , Mazzarella, C. , Perruolo, G. , Longo, M. , Spinelli, R. , Di Lorenzo, G. , Oliva, A. , De Sio, M. , Damiano, R. , Altieri, V. , Terracciano, D. , (2013) Prostate health index (phi) and prostate cancer antigen 3 (PCA3) significantly improve diagnostic accuracy in patients undergoing prostate biopsy. Prostate 73, 227–235 [DOI] [PubMed] [Google Scholar]
- Piccolo, S.R. , Sun, Y. , Campbell, J.D. , Lenburg, M.E. , Bild, A.H. and Johnson, W.E. (2012) A single‐sample microarray normalization method to facilitate personalized‐medicine workflows. Genomics 100, 337–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prensner, J.R. and Chinnaiyan, A.M. (2011) The emergence of lncRNAs in cancer biology. Cancer Discov. 1, 391–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prensner, J.R. , Rubin, M.A. , Wei, J.T. and Chinnaiyan, A.M. (2012) Beyond PSA: the next generation of prostate cancer biomarkers. Sci. Transl. Med. 4, 127rv3–rv127rv3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prensner, J.R. , Shuang, Z. , Erho, N. , Schipper, M. , Lyer, M.K. , Dhanasekaran, S.M. and Magi‐Galluzzi, C. (2014) RNA biomarkers associated with metastatic progression in prostate cancer: a multi‐institutional high‐throughput analysis of SChLAP1. Lancet Oncol. 15, 1469–1480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Psyrri, A. , Kalogeras, K.T. , Kronenwett, R. , Wirtz, R.M. , Batistatou, A. , Bournakis, E. , Timotheadou, E. , Gogas, H. , Aravantinos, G. , Christodoulou, C. , Makatsoris, T. , Linardou, H. , Pectasides, D. , Pavlidis, N. , Economopoulos, T. , Fountzilas, G. , (2012) Prognostic significance of UBE2C mRNA expression in high‐risk early breast cancer. A Hellenic Cooperative Oncology Group (HeCOG) study. Ann. Oncol. 23, 1422–1427 [DOI] [PubMed] [Google Scholar]
- Qiu, Y. , Morii, E. , Tomita, Y. , Zhang, B. , Matsumura, A. , Kitaichi, M. , Okumura, M. and Aozasa, K. (2009) Prognostic significance of pre B cell leukemia transcription factor 2 (PBX2) expression in non‐small cell lung carcinoma. Cancer Sci. 100, 1198–1209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross, AE. , Feng, F.Y. , Ghadessi, M. , Erho, N. , Crisan, A. , Buerki, C. , Sundi, D. , Mitra, A.P. , Vergara, I.A. , Thompson, D.J.S. , Triche, T.J. , Davicioni, E. , Bergstralh, E.J. , Jenkins, R.B. , Karnes, R.J. , Schaeffer, E.M. (2014) A genomic classifier predicting metastatic disease progression in men with biochemical recurrence after prostatectomy. Prostate Cancer Prostatic Dis. 17, 64–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross, J.S. , Hatzis, C. , Symmans, W.F. , Pusztai, L. and Hortob'agyi, G.N. (2008) Commercialized multigene predictors of clinical outcome for breast cancer. Oncologist 13, 477–493 [DOI] [PubMed] [Google Scholar]
- Salami, S.S. , Schmidt, F. , Laxman, B. , Regan, M.M. , Rickman, D.S. , Scherr, D. , Bueti, G. , Siddiqui, J. , Tomlins, S.A. , Wei, J.T. , Chinnaiyan, A.M. , Rubin, M.A. , Sanda, M.G. (2013) Combining urinary detection of TMPRSS2: ERG and PCA3 with serum PSA to predict diagnosis of prostate cancer. Urol. Oncol. Semin. Orig. Investig. 31, 566–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor, B.S. , Schultz, N. , Hieronymus, H. , Gopalan, A. , Xiao, Y. , Carver, B.S. , Arora, V.K. , Kaushik, P. , Cerami, E. , Reva, B. , Antipin, Y. , Mitsiades, N. , Landers, T. , Dolgalev, I. , Major, J.E. , Wilson, M. , Socci, N.D. , Lash, A.E. , Heguy, A. , Eastham, J.A. , Scher, H.I. , Reuter, V.E. , Scardino, P.T. , Sander, C. , Sawyers, C.L. , and Gerald, W.L. (2010) Integrative genomic profiling of human prostate cancer. Cancer Cell 18, 11–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlins, S.A. , Aubin, S.M.J. , Siddiqui, J. , Lonigro, R.J. , Sefton‐Miller, L. , Miick, S. , Williamsen, S. , Hodge, P. , Meinke, J. , Blase, A. , Penabella, Y. , Day, J.R. , Varambally, R. , Han, B. , Wood, D. , Wang, L. , Sanda, M.G. , Rubin, M.A. , Rhodes, D.R. , Hollenbeck, B. , Sakamoto, K. , Silberstein, J.L. , Fradet, Y. , Amberson, J.B. , Meyers, S. , Palanisamy, N. , Rittenhouse, H. , Wei, J.T. , Groskopf, J. , Chinnaiyan, A.M. (2011) Urine TMPRSS2:ERG fusion transcript stratifies prostate cancer risk in men with elevated serum PSA. Sci. Transl. Med. 3, 94ra72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tosoian, J. and Loeb, S. (2010) PSA and beyond: the past, present, and future of investigative biomarkers for prostate cancer. Scientific World J. 10, 1919–1931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traenka, C. , Remke, M. , Korshunov, A. , Bender, S. , Hielscher, T. , Northcott, P.A. , Witt, H. , Ryzhova, M. , Felsberg, J. , Benner, A. , Riester, S. , Scheurlen, W. , Grunewald, T.G.P. , von Deimling, A. , Kulozik, A.E. , Reifenberger, G. , Taylor, M.D. , Lichter, P. , Butt, E. , Pfister, S.M. (2010) Role of LIM and SH3 protein 1 (LASP1) in the metastatic dissemination of medulloblastoma. Cancer Res. 70, 8003–8014 [DOI] [PubMed] [Google Scholar]
- Trock, B.J. , Brotzman, M. , Mangold, L. , Bigley, J. , et al. (2012) Evaluation of GSTP1 and APC methylation as indicators for repeat biopsy in a high‐risk cohort of men with negative initial prostate biopsies. BJU Int. 110, 56–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigelt, B. , Peterse, J.L. and van 't Veer, L.J. (2005) Breast cancer metastasis: markers and models. Nat. Rev. Cancer 5, 591–602 [DOI] [PubMed] [Google Scholar]
- Wu, C.‐L. , Schroeder, B.E. , Ma, X.‐J. , Cutie, C.J. , Wu, S. , Salunga, R. , Zhang, Y. , Kattan, M.W. , Schnabel, C.A. , Erlander, M.G. , McDougal, W.S. (2013) Development and validation of a 32‐gene prognostic index for prostate cancer progression. Proc. Natl. Acad. Sci. U.S.A. 110, 6121–6126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyatt, A.W. , Mo, F. , Wang, Y. and Collins, C.C. (2013) The diverse heterogeneity of molecular alterations in prostate cancer identified through next‐generation sequencing. Asian J. Androl. 15, 301–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan, Y. , Van Allen, E.M. , Omberg, L. , Wagle, N. , Amin‐Mansour, A. , Sokolov, A. , Byers, L. a, Xu, Y. , Hess, K.R. , Diao, L. , Han, L. , Huang, X. , Lawrence, M.S. , Weinstein, J.N. , Stuart, J.M. , Mills, G.B. , Garraway, L.A. , Margolin, A.A. , Getz, G. , Liang, H. (2014) Assessing the clinical utility of cancer genomic and proteomic data across tumor types. Nat. Biotechnol. 32, 644–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material


