Abstract
Innate lymphoid cell (ILCs) subsets differentially populate various barrier and non-barrier tissues, where they play important roles in tissue homeostasis and tissue-specific responses to pathogen attack. Recent findings have provided insight into the molecular mechanisms that guide ILC migration into peripheral tissues, revealing common features among different ILC subsets as well as important distinctions. Recent studies have also highlighted the impact of tissue-specific cues on ILC migration, and the importance of the local immunological milieu. We review these findings here and discuss how the migratory patterns and tissue tropism of different ILC subsets relate to the development and differentiation of these cells, and to ILC-mediated tissue-specific regulation of innate and adaptive immune responses. In this context we outline open questions and important areas of future research.
Introduction
ILCs emerge from the lymphoid lineage and are characterized by the lack of expression of lymphocyte antigen receptors. ILCs have been divided into three different subsets – ILC1, ILC2, and ILC3 – according to their dependence on distinct lineage-determining transcription factors (reviewed in [1–3]). This classification scheme includes phenotypically and functionally distinct cells in common subsets; for example, the ILC3 subset includes both CCR6− (chemokine CC motif receptor) NKp46+/− (natural killer cell P46-related protein/NCR1) ILC3, and CCR6+ CD4+/− lymphoid tissue inducer (LTi) subsets [4,5]. It is clear that much remains to be understood with regard to how development of ILCs relates to their function in the periphery.
In terms of effector function, ILCs exhibit striking similarities to T cells. Similarly to CD8+ T cells, natural killer (NK) cells are cytotoxic to tumor cells and virus-infected cells. Signature cytokines of type 1 T helper (Th1), Th2, and Th17 cells are produced by ILC1, ILC2, and ILC3 cells, respectively. ILC1 produce interferon (IFN)-γ and tumor necrosis factor (TNF)-α; ILC2 produce interleukins (IL)-4, IL-5, IL-9, IL-13 and amphiregulin; and ILC3 produce LTα1β2, IL-17A, IL-22, granulocyte macrophage colony-stimulating factor (GM-CSF), and TNFα [1–3]. ILCs use these cytokines to fight infection by intracellular pathogens (ILC1), helminths (ILC2), and extracellular pathogens (ILC3). ILC1 and ILC3 have been associated with inflammatory disease, and ILC2 play central roles in Th2 type allergic inflammation and in the regulation of metabolic homeostasis [6–12]. T cell effector function is associated with migration to target tissues, which is preceded by migration of naïve T cells from the thymus to secondary lymphoid tissues (SLTs) [13–15]. Similarly, effector function of mature myeloid cells requires migration from the bone marrow, as either precursors or mature cells, to peripheral tissues [16]. ILC subsets have differential tissue distribution, as discussed further below, and the factors that determine this tissue-specific migration and residence, as well as the trafficking mechanisms involved, are an area of active investigation. In particular, given that ILCs have characteristics of both innate and adaptive immune cells, how do ILC migration programs relate to those of other immune cell subsets?
ILCs are widely distributed throughout barrier and non-barrier tissues including the skin, intestines, lungs, uterus, liver, spleen, and SLTs, and tissue localization is strongly associated to subset type [17–21]. Recent studies have revealed that some ILCs, specifically ILC1 and ILC3, express lymphoid tissue-homing receptors (HRs) to migrate into SLTs, and can switch expression of HRs to migrate to non-lymphoid tissues in a manner similar to T cells [22,23]. ILC2, on the other hand, appear to migrate directly from the hematopoietic site to target tissues, in a manner similar to myeloid cells and some innate T cells [22]. Trafficking receptors play important roles in ILC tissue tropism and interaction with other cell types [22,24,25], and recent evidence suggests that they may be important for the migration of bone marrow ILC progenitors to peripheral tissues [26]. Furthermore, specific tissue tropism of ILCs is important for their functions in immune regulation [22–25,27].
We review here current understanding of the migration programs that mediate the distribution of ILC subsets in different tissues. We begin by integrating evidence for differential tissue distribution of ILC subsets in both mice and human, and compare it, when relevant, to our understanding of T cells and innate immune cell migration programs. In this context, we outline common and distinct features of the migration programs of ILC subsets, discuss how they relate to ILC development and function, and outline areas requiring further investigation in this rapidly moving field.
Tissue Distribution of ILC Subsets
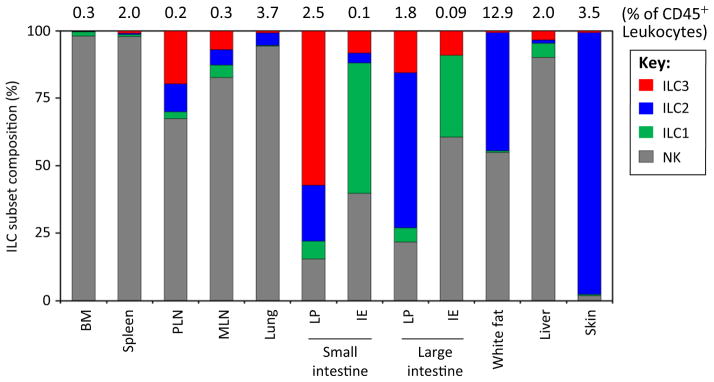
ILCs are widely distributed in the body, and a growing body of evidence suggests that ILC subsets are strategically localized in specific tissues in a manner that relates to their roles in immune and inflammatory responses [10,11,28–36]. Most studies to date focus on one or two ILC subsets and tissue types; to present a more comprehensive view we have integrated available data on ILC tissue distributions (Figure 1). NK cells are the dominant population in the bone marrow, spleen, lymph nodes, lungs and liver (>80% of all ILCs), whereas ILC2 are the dominant population in the lung. Lung ILC2 are found in collagen-rich interstitial tissues and can in some situations induce eosinophilia through the production of IL-5 and IL-13 [28,30–32]. Although smaller in number than ILC2, lung ILC3 can mediate airway hyper-reactivity through IL-17 production [29]. ILC2 are also found in significant numbers in lamina propria of the small intestine (SI LP), although the majority of ILCs in this locale are ILC3. Intestinal ILC2 mediate antihelminth responses through production of IL-5 and IL-13 [20,28]. Within the intestine, ILC3 are also found in intestinal cryptic patches, isolated lymphoid follicles, and the perifollicular areas of Peyer’s patches (PPs) [33,34].
Figure 1.
Tissue Distribution of ILC Subsets. Average percent frequencies of NK, ILC1, ILC2, and ILC3 in total ILCs in indicated tissues of C57BL/6 mice are shown (n ≥ 4; 7–10 weeks of age). Total ILC frequencies among CD45+ leukocytes are also shown. NK cells were identified based on CD3−CD19−CD127−NKp46+NK1.1+ (most tissues), CD3−CD19−RORγt−NKp46+NK1.1+ (intestinal LP), or CD3−CD19−CD127−RORγt−NKp46+NK1.1+ Eomes+ (IE) phenotype as described before [17,36]. ILC1 were identified by Lin−RORγt−CD127+NKp46+NK1.1+ (most tissues) or Lin−RORγt−CD127−NKp46+NK1.1+ (IE) phenotype as described before [17,18,36]. ILC2 were identified by Lin−CD127+CD90+KLRG1+GATA3+ (most tissues), Lin−CD90+CD25+GATA3+ (spleen and skin), or Lin−CD127+CD90+T1/ST2+GATA3+ (lungs and WAT) phenotype [10,20,37,47,93]. ILC3 were identified by Lin−GATA3−CD127+CD90+RORγt+ phenotype [20]. ILC3 subsets within the total ILC3 group were not examined separately. The ILC1 population in the bone marrow includes ILC1 progenitors. Abbreviations: BM, bone marrow; IE, intraepithelial compartment; ILC, innate lymphoid cell; LP, lamina propria; MLN, mesenteric lymph node; NK, natural killer cell; PLN, peripheral lymph node (inguinal LN).
ILC1 are the dominant ILC population in intestinal intraepithelial (IE) compartment (Figure 1). By contrast, ILC2 are the dominant ILC subset in the skin, where ILC2 production of IL-5 and IL-13 are important mediators of Th2 type immune responses [37]. ILC2 are also found in white adipose tissue (WAT) where they promote thermogenesis through a process termed ‘beiging’ [10,11]. ILC2 are present in significant numbers also in fat-associated lymphoid clusters in the intestinal mesentery, and mediate anti-helminth inflammatory responses [38].
Taken together, these findings suggest that tissue localization is intimately related to ILC phenotype and function. What processes underlie this differential distribution, and at what stage in ILC development are these mechanisms at work?
Origins of Peripheral ILCs
ILCs emerge from fetal progenitors and adult bone-marrow progenitors [39,40]. ILCs populate various tissues from mid to late stages of fetal development. In mice, fetal ILC progenitors with an LTi phenotype populate the intestine as early as embryonic day (E)12.5–13.5, and develop lymphoid structures such as PPs via the expression of LTα1β2 [41,42]. These progenitors also have the potential to become ILC1 and ILC2 in the gut. T-bet+ (T box 21/TBX21), RORγt+ [retinoic acid receptor (RAR)-related orphan receptor] and GATA3+ (GATA binding protein 3) cells are detected in the fetal gut at E15.5, indicating the presence of most ILC subsets at late-stage fetal development [41]. In humans, a subset of ILCs defined by the expression of CRTH2 (chemoattractant receptor expressed on Th2 cells) populate the fetal gut [43]. Together with other innate lymphocytes, such as γδ T and mucosal-associated invariant T (MAIT) cells [44], these fetal ILCs are thought to provide protection from pathogens during early life before the development of adult ILCs and antigen-specific lymphocytes.
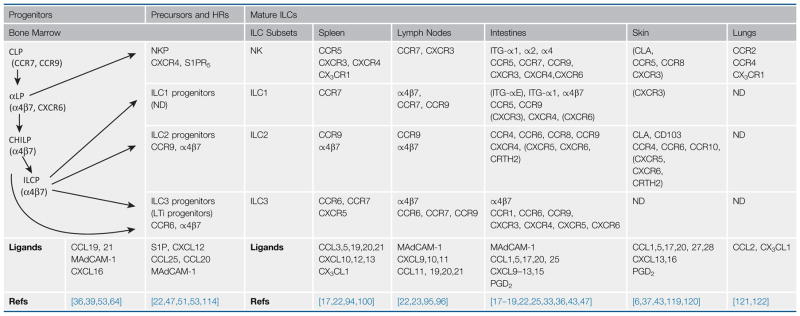
ILCs in the adult bone marrow undergo multiple stages of differentiation to become functionally mature ILCs (reviewed in [1]). Individual ILC subsets emerge from the common lymphoid progenitors (CLP) after a series differentiation stages that include the bipotent CXCR6+ (chemokine CXC motif receptor 6) ILC/NK progenitors (αLP) and the common ‘helper-like’ ILC precursors (CHILP), which become common ILC progenitors (ILCP) (Table 1, Key Table). The immediate precursors for NK, ILC1, ILC2, and ILC3 have been described [36,45–49].
Table 1.
Homing Receptors (HRs) of ILC Subsets and Their Progenitorsa
ILC subsets such as NK, ILC1, ILC2, and ILC3 emerge from a group of progenitor cells such as CLP, αLP, CHILP, and ILCP in the bone marrow or from fetal liver progenitors. These ILC progenitors or precursors express CCR7, α4β7, CCR9, CXCR6, and/or S1PR5, which enables their migration to the periphery. HRs expressed by human ILCs are shown in parentheses.
ND, not determined.
The generation of ILCs is regulated by distinct sets of transcription factors. Transcription factors such as nuclear factor interleukin 3 regulated (NFIL3), inhibitor of DNA binding 2 (ID2), thymocyte selection-associated high mobility group box protein (TOX), GATA3, and zinc finger and BTB domain-containing protein 16 (PLZF) regulate ILCP development [1,36]. Eomesodermin (Eomes), TOX, v-Ets avian erythroblastosis virus E26 oncogene homolog 1 (ETS1), T-bet, and Runt-related transcription factor 3 (RUNX3) regulate NK cell development. T-bet is required for ILC1, whereas GATA3, Notch, RORα, transcription factor 1 (TCF1), and growth factor-independent 1 (GFI1) regulate ILC2 development. RORγt, Notch, TCF1, RUNX1, and aryl hydrocarbon receptor (AHR) are required for ILC3 development. The transcriptional regulation of ILC development has been reviewed recently, and we refer the reader to these articles for further in-depth discussion [1,4,50].
ILC distribution in the body is determined, in part, by the migratory capacity of ILC progenitors from the fetal liver or bone marrow to peripheral tissues. Mature ILCs are generally distinguished from ILC progenitors by high levels of expression of subset-specific surface markers (NK1.1, NKp46, KLRG1, IL-2/IL-15Rβ, IL-23R, IL-33R, IL-25R, ICOS, CD4, and CCR6) and transcription factors (Eomes, T-bet, GATA3, and RORγt) [22,51,52]. Moreover, when these cells are activated by appropriate cytokines, they produce lineage-specific effector cytokines. While NK cells are present in the bone marrow, few non-NK mature ILCs are found in the bone marrow (Figure 1). For example, T-bet+ ILC1 and RORγt+ ILC3 are found in peripheral tissues, but few of these cells are present in the bone marrow [51,53]. GATA3+ ILC2 progenitors are found in the bone marrow, but these cells do not produce ILC2 effector cytokines such as IL-4, IL-5, and IL-13 [47,54]. Similarly, functionally immature ILC1 and ILC3 progenitors, but few mature ILCs, are found in the bone marrow or fetal liver [36,39,53]. Taken together, these findings argue against the notion that mature ILCs emerge from the bone marrow to populate peripheral tissues; instead, the majority of peripheral ILCs are likely to be the descendants of ILC progenitors or precursors that emigrated the bone marrow. In turn, this argues that tissue-selective migration of lineage-committed ILC precursors, such as progenitors for NK, ILC1, ILC2, and ILC3 cells, and selective differentiation and expansion of common ILC progenitors, such as αLP, CHILP and ILCP, in the periphery are key factors in determining ILC distribution in the body.
Homing Receptors for ILCPs
HRs, including integrins and chemoattractant receptors, regulate the migration of hematopoietic progenitors and mature cells [55,56]. The NK/ILC progenitors, αLP, express the integrin α4β7 and the chemokine receptor CXCR6 [52,53]. ILCPs also express α4β7 [39]. These receptors are implicated in immune cell migration and cell–cell interaction in peripheral tissues. MAdCAM-1 (mucosal vascular addressin cell adhesion molecule 1), the major binding partner for α4β7, is highly expressed by the endothelial cells of peripheral tissues such as gut-associated lymphoid tissues, intestinal lamina propria, and sinus-lining cells of the spleen marginal zone [57]. α4β7 also binds to vascular cell adhesion protein 1 (VCAM-1), which is expressed by activated endothelial cells [58,59]. An extracellular matrix protein, fibronectin, provides an additional substrate for α4β7 [60]. Thus, α4β7 has the potential to guide ILCP cells to both mucosal and non-mucosal tissues. Leukocytes undergo multistep processes (rolling, chemoattractant receptor activation, and firm adhesion) on blood vessels to enter tissues [55,61]. α4β7 and potentially other adhesion molecules would mediate the rolling and firm adhesion of ILC progenitors on endothelial cells. To trigger firm adhesion of lymphocytes on blood vessels, the activation of chemoattractant receptors and integrins is required [61]. Several chemokine receptors appear to regulate the migration of ILCPs from the bone marrow to peripheral tissues. CXCR6 regulates lymphocyte migration to various peripheral tissues (the liver, spleen red pulp, intestine, lungs, and skin) and promotes cell–cell interaction with dendritic cells and fibroblastic reticular cells [62,63]. CXCR6 appears to play regulatory roles in the migration of ILCPs. In CXCR6−/− mice, the number of Lin− IL-7Rα+ c-Kitmed RORγt+ ILC3 precursors was decreased in the fetal liver, and CXCR6−/− ILC3 precursors failed to populate peripheral tissues such as SI LP and liver [26]. By contrast, the numbers of bone marrow ILCP cells were increased in CXCR6−/− mice. Thus, it appears that CXCR6 regulates the bone marrow emigration and peripheral seeding of ILCPs.
Another potentially important HR for ILC progenitors or precursors is CCR9. Bone marrow CLPs and ILC2 progenitors express CCR9 [47,64]. CCR9 is a key HR for small intestine- and thymus-homing mature lymphocytes and progenitors, such as multipotent progenitors and CLPs [64–66]. It is a major HR to guide ILCPs into the gut [22]. It is expected that additional HRs are likely to play important roles in migration of ILCPs to other tissues. Indeed, CCR7 plays a role in the migration of ILCPs to SLTs. CCR7 is also utilized by naïve CD4+ T cells and dendritic cells to enter lymph nodes [67]. CCL21 (chemokine CC motif ligand 21) and CCL19, expressed by the high endothelial venules and T zone stromal cells of lymph nodes, respectively, activate CCR7 to recruit CCR7+ cells into lymphoid tissues [68,69]. CLPs and potentially other progenitors in the bone marrow express CCR7 [64]. Thus, multiple ILCPs have the capacity to migrate to various peripheral tissues.
Tissue factors for Differentiation and Population of ILCs in Peripheral Tissues
In general, the factors that induce the differentiation, expansion and contraction of ILCs have the potential to control ILC tissue distribution. Cytokines induce the proliferation and differentiation of ILCPs. IL-7 is widely expressed in the body and supports the generation of all ILCs, including non-NK ILCs and NK cells [36,70–72]. NK cell development from αLP is induced by IL-15 [73–75]. IL-7 is also required for the generation of T and B cells, and IL-15 is also required for the development of CD8+ T cells and γδ T cells [76]. In this regard, the cytokines that specifically generate individual ILC subsets from ILC progenitors or precursors remain unclear.
Cytokines also induce the proliferation and activation of mature ILCs. IL-15 and IL-18 activate NK cells for IFNγ production [75,77,78]. IL-12 and IL-15 activate ILC1 for IFN-γ production [18,19]. These cytokines are produced constitutively or in response to pro-inflammatory signals, such as Toll-like receptor (TLR) ligands and type I IFN, by antigen-presenting cells (APCs), stromal cells, fibroblasts, endothelial cells, and/or epithelial cells [73]. IL-2, IL-4, IL-9, IL-25, IL-33, thymic stromal lymphopoietin (TSLP), and TNF-like ligand 1A (TL1A) activate ILC2 [4]. These cytokines are produced constitutively or upon cellular stress by various immune cells and tissue cells. IL-1β and IL-23 activate ILC3 and are produced by myeloid cells and tissue cells after activation with inflammatory signals and TLR ligands [79–81]. Infection and inflammation can alter ILC composition in affected tissues. For example, the infection by an extracellular bacterial pathogen, Citrobacter rodentium, increases ILC3 but decreases ILC2 frequency, whereas helminthic infection increases ILC2 but decreases ILC3 frequency in the gut [20,82]. Differential TLR activation and cytokine production during infection or inflammatory conditions are likely to influence the size of ILC populations in peripheral tissues. More studies will be necessary to establish the roles of ILC-activating cytokines in the expansion and contraction of ILC subsets in diverse immunological contexts.
Nutritional status is another factor that affects ILC composition in peripheral tissues. The vitamin A metabolite, all-trans retinoic acid (RA), increases ILC3 but decreases ILC2 populations [20]. RA is highly produced in the intestine, particularly SI, and supports innate and adaptive immune responses [83,84]. RA also increases IL-22 production by ILC3 [81]. AHR drives the generation of ILC3 subsets such as LTi and NKp46+ ILCs in the intestine [85–87]. AHR is a transcription factor that induces the expression xenobiotic-metabolizing enzymes such as cytochrome P450. AHR activation is triggered by diverse synthetic and naturally occurring compounds such as dietary carotenoids and tryptophan derivatives [88,89]. Thus, AHR ligands act as another gut-specific tissue factor that induces the generation of ILC3. Gut microbiota selectively induces ILC3. In germ-free (GF) or Myd88−/− (myeloid differentiation primary response 88) mice, the numbers of IL-22-producing ILC3 were decreased [90,91]. By contrast, others have observed normal or increased numbers of ILC3 in GF mice [87,92], which suggests that the ILC3 response to microbiota can vary, perhaps because of the variations in microbiota composition. ILC1 and ILC2 are not significantly affected by microbiota [18,93]. This distinction suggests potentially variable but selective influence of microbiota on ILC differentiation or expansion. In addition, circadian rhythm and metabolic cues, such as caloric intake, control the production of effector cytokines such as IL-5 and IL-13 by ILC2 [28]. Thus, ILC numbers and activities in peripheral tissues are under the control of hematopoietic cytokines, inflammatory cytokines, nutrients, dietary materials, and other environmental cues.
Migration of Mature ILCs to Lymphoid Tissues
While the majority of ILCs in SLTs are NK cells, non-NK ILCs such as ILC1, ILC2, and ILC3 are also present in SLTs at detectable levels (Figure 1). The ILCs in SLTs have the potential to influence the activation and differentiation of T and B cells [80,94]. In addition, SLTs would provide a conducive environment for functional maturation of ILCs themselves through ILC-activating signals from dendritic cells. Mature ILCs in SLTs can be generated in situ from ILC progenitors or mature ILCs that migrated from other tissues. The presence of mature circulating ILCs in the blood circulation [35,43] suggests that ILCs actively migrate into SLTs such as the spleen and lymph nodes. T and B cells acquire new sets of trafficking receptors after activation in SLTs, and migrate to other tissues through blood circulation [13–15]. A recent study indicates that ILCs have the potential to undergo a similar process [22]. Lymphocytes migrate into SLTs through two different pathways. The first route is through specialized blood vessels such as high endothelial cells (HEVs) in lymph nodes. The second route is through afferent lymphatic vessels which drain immune cells along with tissue fluid. NK cells migrate to LNs through HEVs [95,96]. This migration is mediated by CCR7 or CXCR3. By producing IFN-γ, NK cells that migrated to LNs can promote T cell differentiation into Th1 cells [95]. ILCs have the potential to perform a similar function. ILC1 and ILC3 migrate to SLTs, and this migration also requires CCR7 [22]. Utilizing photoactivatable fluorescent proteins, it has been determined that LTi ILC3 migrate from the small intestine to the draining mesenteric lymph nodes (MLNs) in a CCR7-dependent manner [23]. This CCR7-dependent ILC3 exit from the small intestinal tissue is similar to that reported for effector T cells such as Th17 cells [97].
The lymphocyte localization in SLTs is largely determined by coordinated expression of CCR7 (the T zone HR) and CXCR5 (the B zone HR) [98]. In this regard, CXCR5+ LTi ILC3 are present in SLTs [99,100]. The CXCL13–CXCR5 axis recruits LTi ILC3 to lymphoid follicles and the spleen marginal zone [27,100,101]. A major function of these LTi ILC3 in LNs is to support stromal cells and maintain lymph node integrity through lymphotoxin production [102]. Thus, ILCs have the potential to migrate and localize to specialized areas of SLTs to regulate innate and adaptive immune responses, and maintain tissue integrity. More studies will be necessary to fully establish the migration mechanism and localization sites of ILCs within SLTs.
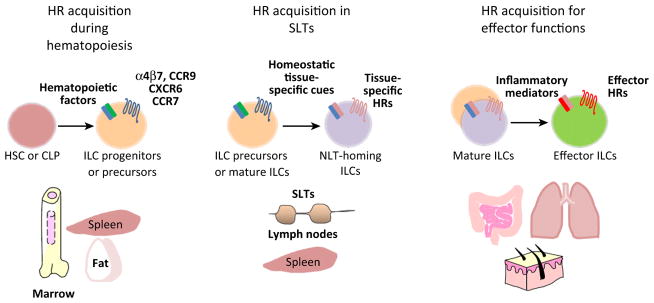
Trafficking Receptor Switches and the Establishment of ILC Tissue Residency in Non-Lymphoid Tissues
Naïve T cells acquire non-lymphoid tissue HRs while they undergo differentiation in SLTs. For example, naïve T cells acquire CCR9 and α4β7 expression in MLNs or PPs to migrate to the gut [103]. However, they acquire skin HRs such as cutaneous lymphocyte antigen (CLA), CCR4, CCR8, and CCR10 in skin-draining lymph nodes [104]. CCR4, CCR8, and CCR10 are implicated in lymphocyte trafficking to inflamed skin tissues [105–108]. This process is termed the ‘HR or trafficking receptor switch’ (Figures 2,3). It has been reported that ILCs undergo a similar HR switch. Spleen ILC3 highly express CCR7, but activation of these cells with IL-7 and RA downregulated CCR7 but upregulated CCR9 and α4β7 expression [22]. Once activated with RA ex vivo, spleen ILC3 efficiently migrated to the intestine. A similar switch occurs in T cells during their activation by antigens and RA presented by mucosal dendritic cells and macrophages [109]. In a manner similar to ILC3, ILC1 also undergo the HR switch to express gut HRs [22].
Figure 2.
Potential HR Switches for the Development and Effector Functions of ILCs. Three potential types of HR switches are shown. The first HR switch occurs during ILC development in hematopoietic tissues, such as bone marrow and spleen. ILC precursors and mature ILCs express peripheral tissue-HRs such as α4β7, CCR9, and CXCR6. The second type of HR switch occurs in lymphoid tissues where ILCs upregulate HRs for their migration to NLT. Tissue-specific cues in peripheral tissues induce distinct HRs on ILCs. The third type of HR switch occurs at the effector stage during infection or inflammatory responses. This type of HR switch upregulates HRs important for ILC effector functions. ILC subsets are apparently different from each other in these HR switches. Abbreviations: CCR, chemokine CC motif receptor; CLP, common lymphoid progenitors; CXCR, chemokine CXC motif receptor; HSC, hematopoietic stem cells; HR, homing receptor; ILC, innate lymphoid cell; SLT, secondary lymphoid tissue; NLT, non-lymphoid tissue.
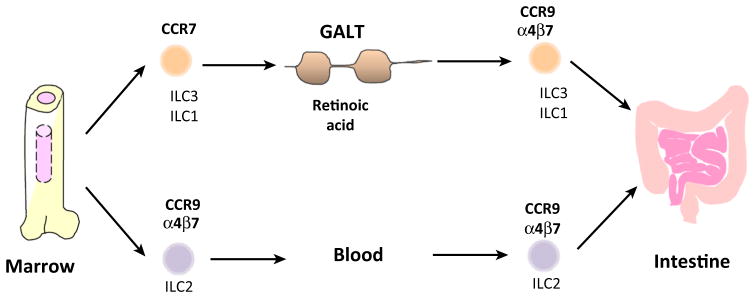
Figure 3.
Acquisition of Gut HRs by ILC Subsets. As an example for the second type of HR switch described in Figure 2, ILC1 and ILC3 in GALT, such as MLNs and PPs, upregulate CCR9 and α4β7 (two major gut HRs) in response to RA. At the same time, they lose CCR7 expression. This process is facilitated by mucosal dendritic cells, which produce both cytokines and RA. However, ILC2 acquire gut HRs during their development in hematopoietic tissues in a RA-independent manner. Abbreviations: CCR, chemokine CC motif receptor; GALT, gut-associated lymphoid tissue; HRs, homing receptors; ILC, innate lymphoid cell; MLNs, mesenteric lymph nodes; PPs, Peyer’s patches; RA, retinoic acid.
An important difference between ILC2 and other ILC subsets (i.e., ILC1 and ILC3) is that ILC2 do not need the RA-dependent HR switch to express gut HRs (Figure 3). All ILC2 in the bone marrow, spleen, and MLNs highly express CCR9 and α4β7 [22]. This indicates that gut HRs are upregulated at an ILC2 progenitor stage in the bone marrow rather than on mature ILC2 in SLTs. In addition, ILC2 do not significantly express CCR7, unlike ILC3 and ILC1 [22]. This indicates that ILC2 lineage cells may enter SLTs utilizing other HRs, or that they are made in situ in SLTs from progenitors.
ILC2 are the dominant ILC subset in the skin (Figure 1). Human skin ILC2 express CLA, CCR4, and CCR10 [6], which are commonly expressed by cutaneous or skin-homing circulating T cells. It has been reported that mouse skin ILC2 express some of the skin HRs and CD103 (the α chain of integrin αEβ7) [37]. Integrin αEβ7 promotes lymphocyte interaction with keratinocytes through E-cadherin, as demonstrated for epidermal γδ T cells [110]. CLA binds to the vascular lectin endothelial cell-leukocyte adhesion molecule 1 (ELAM-1), a major vascular addressin for the skin [111]. Thus, ILC2 have the necessary trafficking receptors to actively migrate into the skin. Vitamin D (1,25-dihydroxyvitamin D3) induces CCR10 on skin-homing T cells [112]. While it is not clear if vitamin D induces CCR10 on ILCs, it has been reported that it inhibited the expression of α4β7 on human ILCs [113], which can potentially suppress ILC migration to the gut. The site of skin HR upregulation by ILC2 should be determined along with the identity of the factors that induce HR expression.
Beyond the lymphoid tissue and tissue-specific HRs, ILC subsets express additional HRs. These HRs include CCR5, CCR6, CXCR3, CXCR4, CX3CR1, S1PR5 (sphingosine-1-phosphate receptor 5), and integrins (ITG-α2, αm, and α4) as shown in Table 1. These receptors support lymphocyte migration, localization, and cell–cell interaction in peripheral tissues. For example, intestinal CX3CR1+ myeloid cells express CXCL16 (the ligand for CXCR6) for functional interaction with CXCR6+ NKp46+ ILC3 [25]. This promotes the accumulation of ILC3 in the gut and their activation by DC-produced IL-23. CXCR6 also promotes NK cell migration to the liver [24]. CCR6 guides LTi cells to CCL20-producing stromal cells and epithelial cells in the gut [33,34]. S1PR5, a receptor for the bioactive lipid mediator sphingosine-1-phosphate, is required for tissue exit of NK cells from lymph nodes and the bone marrow [114]. The roles of S1P receptors in tissue exit by ILCs have not been established. A question of interest is if ILCs express group or subset-specific HRs. CD4+ T helper subsets characteristically express CCR5 and CXCR3 (Th1), CCR4 and CCR8 (Th2), and CCR6 (Th17) for polarized immune responses [115–117]. CCR6 fits this category of HRs in that it is characteristically expressed by LTi ILC3 subsets [118]. It remains to be determined if ILC subsets are similarly polarized in HR expression.
Concluding Remarks
The migration programs of ILCs have been unclear, but recent progress has revealed new insights into migration potential and tissue tropism. First, ILC progenitors or precursors in the bone marrow and fetal liver migrate to the periphery for development and maturation. ILCPs express α4β7, CCR7, CCR9, and/or CXCR6 for migration to barrier and non-barrier tissues, such as spleen, liver, lymph nodes, skin, and mucosal tissues. The migration of ILC progenitors or precursors to peripheral tissues would be important for tissue-specific generation of ILC subsets with diverse functions (Figure 2). Second, some ILC precursors and mature ILCs use HRs, such as CCR7, to actively migrate to lymphoid tissues in a manner similar to naïve and central memory T cells. These cells have the potential to circulate through the blood circulation, migrate into secondary lymphoid tissues, and colocalize with T cells and dendritic cells. Third, ILCPs or mature ILCs undergo HR switches in peripheral tissues to develop diverse tissue homing capacities. They lose original HRs but upregulate additional non-lymphoid tissue homing receptors (Figure 2). This HR switch appears to be heterogeneous and dependent on both tissue sites and ILC subsets. For example, ILC2 upregulate gut HRs in the bone marrow, while ILC3 and ILC1 acquire gut HRs in the periphery. Similarly, ILCs express HRs specific for the skin and potentially other tissues as well. Through heterogeneous HR switches, ILCs develop diverse trafficking potentials. This process ensures efficient ILC migration and interaction with other cell types, such as APCs and target cells, to effectively mount innate immunity and to regulate metabolism and adaptive immune responses.
Much remains to be understood regarding how the migratory behavior of ILC subsets relates to their development and function (see Outstanding Questions). A major obstacle in the study of ILC migration is the relative paucity of ILCs compared to other lymphocytes in the body. Another problem is lack of ILC-specific markers or fluorescent-tagging systems to track their migration. This makes it extremely difficult to perform intravital microscopy to study their migration in vivo. Development of animal models with increased ILC numbers or ILC subsets specifically tagged with fluorescent proteins will be highly useful. Utilization of parabiosis models are expected to be an effective method to study ILC migration between organs.
Outstanding Questions.
Do ILCs enter peripheral tissues as progenitors, precursors, or as mature ILCs?
Do ILCs recirculate between secondary lymphoid organs and non-lymphoid tissues, in a manner similar to some T cells?
Do ILCs enter lymph nodes through high endothelial venules or lymphatic vessels?
Do ILC subsets differ from each other in their migration behavior to peripheral tissues? What is the molecular basis for these differences?
What are the HRs that are specifically expressed by individual ILC subsets?
When and where do ILC subsets acquire tissue-specific HRs?
What are the factors and tissue-specific cues that induce the expression of key HRs?
What are the HRs that are functionally important for ILC effector functions?
What HRs are important for the tissue tropism of ILC1 in the intraepithelial compartment of the intestine?
Do ILC2 undergo tissue-specific HR switches for migration to skin, fat, and lungs?
We conclude by highlighting important areas of future research in ILC migration, tissue-specificity, and function. It will be important to determine the factors that impact on both the differentiation of ILC progenitors and the expression of HRs in different peripheral tissues. The mechanisms that control the expansion and contraction of ILC populations in response to infector or inflammatory insults, and how these relate to the migratory programs of ILCs, are important areas of investigation, as are the mechanisms – specifically the HRs – that regulate ILC recirculation between lymph nodes and blood circulation. An important step will be to examine whether ILC lineage-determining transcription factors and cytokines play a role in HR expression and in the regulation of ILC trafficking machinery. Deciphering these issues will be key to understanding the tissue- and pathogen-specific functions of ILC subsets in the body.
Trends.
ILC progenitors express homing receptors and actively migrate to peripheral tissues.
ILC subsets have distinct tissue tropisms, which are regulated in part by their selective expression of homing receptors.
ILC subsets have heterogeneous migration programs with similarities to those of both adaptive (T cells) and innate immune cells.
ILCs undergo homing receptor switches to develop specific tissue tropisms.
Acknowledgments
This work was supported, in part, from grants from the National Institutes of Health (R01AI0 74745, R01DK0 76616, 1S10RR 02829, and R01AI0 80769) and the National Multiple Sclerosis Society to C.H.K. The authors thank L. Friesen at Purdue University for critical reading of the manuscript.
References
- 1.Serafini N, et al. Transcriptional regulation of innate lymphoid cell fate. Nat Rev Immunol. 2015;15:415–428. doi: 10.1038/nri3855. [DOI] [PubMed] [Google Scholar]
- 2.Sonnenberg GF, Artis D. Innate lymphoid cells in the initiation, regulation and resolution of inflammation. Nat Med. 2015;21:698–708. doi: 10.1038/nm.3892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McKenzie AN, et al. Innate lymphoid cells in inflammation and immunity. Immunity. 2014;41:366–374. doi: 10.1016/j.immuni.2014.09.006. [DOI] [PubMed] [Google Scholar]
- 4.Artis D, Spits H. The biology of innate lymphoid cells. Nature. 2015;517:293–301. doi: 10.1038/nature14189. [DOI] [PubMed] [Google Scholar]
- 5.Montaldo E, et al. Group 3 innate lymphoid cells (ILC3s): origin, differentiation, and plasticity in humans and mice. Eur J Immunol. 2015;45:2171–2182. doi: 10.1002/eji.201545598. [DOI] [PubMed] [Google Scholar]
- 6.Salimi M, et al. A role for IL-25 and IL-33-driven type-2 innate lymphoid cells in atopic dermatitis. J Exp Med. 2013;210:2939–2950. doi: 10.1084/jem.20130351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Molofsky AB, et al. Interleukin-33 and interferon-gamma counter-regulate group 2 innate lymphoid cell activation during immune perturbation. Immunity. 2015;43:161–174. doi: 10.1016/j.immuni.2015.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Molofsky AB, et al. Innate lymphoid type 2 cells sustain visceral adipose tissue eosinophils and alternatively activated macrophages. J Exp Med. 2013;210:535–549. doi: 10.1084/jem.20121964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hams E, et al. Cutting edge: IL-25 elicits innate lymphoid type 2 and type II NKT cells that regulate obesity in mice. J Immunol. 2013;191:5349–5353. doi: 10.4049/jimmunol.1301176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brestoff JR, et al. Group 2 innate lymphoid cells promote beiging of white adipose tissue and limit obesity. Nature. 2015;519:242–246. doi: 10.1038/nature14115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee MW, et al. Activated type 2 innate lymphoid cells regulate beige fat biogenesis. Cell. 2015;160:74–87. doi: 10.1016/j.cell.2014.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goldberg R, et al. The unusual suspects – innate lymphoid cells as novel therapeutic targets in IBD. Nat Rev Gastroenterol Hepatol. 2015;12:271–283. doi: 10.1038/nrgastro.2015.52. [DOI] [PubMed] [Google Scholar]
- 13.Agace WW. T-cell recruitment to the intestinal mucosa. Trends Immunol. 2008;29:514–522. doi: 10.1016/j.it.2008.08.003. [DOI] [PubMed] [Google Scholar]
- 14.Kim CH. Migration and function of FoxP3+ regulatory T cells in the hematolymphoid system. Exp Hematol. 2006;34:1033–1040. doi: 10.1016/j.exphem.2006.03.014. [DOI] [PubMed] [Google Scholar]
- 15.Campbell DJ, et al. Chemokines in the systemic organization of immunity. Immunol Rev. 2003;195:58–71. doi: 10.1034/j.1600-065x.2003.00067.x. [DOI] [PubMed] [Google Scholar]
- 16.Geissmann F, et al. Development of monocytes, macrophages, and dendritic cells. Science. 2010;327:656–661. doi: 10.1126/science.1178331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Robinette ML, et al. Transcriptional programs define molecular characteristics of innate lymphoid cell classes and subsets. Nat Immunol. 2015;16:306–317. doi: 10.1038/ni.3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fuchs A, et al. Intraepithelial type 1 innate lymphoid cells are a unique subset of IL-12- and IL-15-responsive IFN-gamma-producing cells. Immunity. 2013;38:769–781. doi: 10.1016/j.immuni.2013.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bernink JH, et al. Human type 1 innate lymphoid cells accumulate in inflamed mucosal tissues. Nat Immunol. 2013;14:221–229. doi: 10.1038/ni.2534. [DOI] [PubMed] [Google Scholar]
- 20.Spencer SP, et al. Adaptation of innate lymphoid cells to a micronutrient deficiency promotes type 2 barrier immunity. Science. 2014;343:432–437. doi: 10.1126/science.1247606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Male V, et al. Immature NK cells, capable of producing IL-22, are present in human uterine mucosa. J Immunol. 2010;185:3913–3918. doi: 10.4049/jimmunol.1001637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim MH, et al. Retinoic acid differentially regulates the migration of innate lymphoid cell subsets to the gut. Immunity. 2015;43:107–119. doi: 10.1016/j.immuni.2015.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mackley EC, et al. CCR7-dependent trafficking of RORgamma+ ILCs creates a unique microenvironment within mucosal draining lymph nodes. Nat Commun. 2015;6:5862. doi: 10.1038/ncomms6862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paust S, et al. Critical role for the chemokine receptor CXCR6 in NK cell-mediated antigen-specific memory of haptens and viruses. Nat Immunol. 2010;11:1127–1135. doi: 10.1038/ni.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Satoh-Takayama N, et al. The chemokine receptor CXCR6 controls the functional topography of interleukin-22 producing intestinal innate lymphoid cells. Immunity. 2014;41:776–788. doi: 10.1016/j.immuni.2014.10.007. [DOI] [PubMed] [Google Scholar]
- 26.Chea S, et al. CXCR6 expression is important for retention and circulation of ILC precursors. Mediators Inflamm. 2015;2015:368427. doi: 10.1155/2015/368427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marchesi F, et al. CXCL13 expression in the gut promotes accumulation of IL-22-producing lymphoid tissue-inducer cells, and formation of isolated lymphoid follicles. Mucosal Immunol. 2009;2:486–494. doi: 10.1038/mi.2009.113. [DOI] [PubMed] [Google Scholar]
- 28.Nussbaum JC, et al. Type 2 innate lymphoid cells control eosinophil homeostasis. Nature. 2013;502:245–248. doi: 10.1038/nature12526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim HY, et al. Interleukin-17-producing innate lymphoid cells and the NLRP3 inflammasome facilitate obesity-associated airway hyperreactivity. Nat Med. 2014;20:54–61. doi: 10.1038/nm.3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klein Wolterink RG, et al. Essential, dose-dependent role for the transcription factor Gata3 in the development of IL-5+ and IL-13+ type 2 innate lymphoid cells. Proc Natl Acad Sci USA. 2013;110:10240–10245. doi: 10.1073/pnas.1217158110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hurst SD, et al. New IL-17 family members promote Th1 or Th2 responses in the lung: in vivo function of the novel cytokine IL-25. J Immunol. 2002;169:443–453. doi: 10.4049/jimmunol.169.1.443. [DOI] [PubMed] [Google Scholar]
- 32.Voehringer D, et al. Type 2 immunity reflects orchestrated recruitment of cells committed to IL-4 production. Immunity. 2004;20:267–277. doi: 10.1016/s1074-7613(04)00026-3. [DOI] [PubMed] [Google Scholar]
- 33.Luci C, et al. Influence of the transcription factor RORgammat on the development of NKp46+ cell populations in gut and skin. Nat Immunol. 2009;10:75–82. doi: 10.1038/ni.1681. [DOI] [PubMed] [Google Scholar]
- 34.Klose CS, et al. A T-bet gradient controls the fate and function of CCR6−RORgammat+ innate lymphoid cells. Nature. 2013;494:261–265. doi: 10.1038/nature11813. [DOI] [PubMed] [Google Scholar]
- 35.Teunissen MB, et al. Composition of innate lymphoid cell subsets in the human skin: enrichment of NCR+ ILC3 in lesional skin and blood of psoriasis patients. J Invest Dermatol. 2014;134:2351–2360. doi: 10.1038/jid.2014.146. [DOI] [PubMed] [Google Scholar]
- 36.Klose CS, et al. Differentiation of type 1 ILCs from a common progenitor to all helper-like innate lymphoid cell lineages. Cell. 2014;157:340–356. doi: 10.1016/j.cell.2014.03.030. [DOI] [PubMed] [Google Scholar]
- 37.Roediger B, et al. Cutaneous immunosurveillance and regulation of inflammation by group 2 innate lymphoid cells. Nat Immunol. 2013;14:564–573. doi: 10.1038/ni.2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moro K, et al. Innate production of T(H)2 cytokines by adipose tissue-associated c-Kit+Sca-1+ lymphoid cells. Nature. 2010;463:540–544. doi: 10.1038/nature08636. [DOI] [PubMed] [Google Scholar]
- 39.Constantinides MG, et al. A committed precursor to innate lymphoid cells. Nature. 2014;508:397–401. doi: 10.1038/nature13047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Serafini N, et al. Gata3 drives development of RORgammat+ group 3 innate lymphoid cells. J Exp Med. 2014;211:199–208. doi: 10.1084/jem.20131038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bando JK, et al. Identification and distribution of developing innate lymphoid cells in the fetal mouse intestine. Nat Immunol. 2015;16:153–160. doi: 10.1038/ni.3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van de Pavert SA, et al. Maternal retinoids control type 3 innate lymphoid cells and set the offspring immunity. Nature. 2014;508:123–127. doi: 10.1038/nature13158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mjosberg JM, et al. Human IL-25- and IL-33-responsive type 2 innate lymphoid cells are defined by expression of CRTH2 and CD161. Nat Immunol. 2011;12:1055–1062. doi: 10.1038/ni.2104. [DOI] [PubMed] [Google Scholar]
- 44.Vermijlen D, Prinz I. Ontogeny of innate T lymphocytes – some innate lymphocytes are more innate than others. Front Immunol. 2014;5:486. doi: 10.3389/fimmu.2014.00486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Constantinides MG, et al. PLZF expression maps the early stages of ILC1 lineage development. Proc Natl Acad Sci USA. 2015;112:5123–5128. doi: 10.1073/pnas.1423244112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tang Y, et al. Emergence of NK-cell progenitors and functionally competent NK-cell lineage subsets in the early mouse embryo. Blood. 2012;120:63–75. doi: 10.1182/blood-2011-02-337980. [DOI] [PubMed] [Google Scholar]
- 47.Hoyler T, et al. The transcription factor GATA-3 controls cell fate and maintenance of type 2 innate lymphoid cells. Immunity. 2012;37:634–648. doi: 10.1016/j.immuni.2012.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cherrier M, et al. Notch Id2, and RORgammat sequentially orchestrate the fetal development of lymphoid tissue inducer cells. J Exp Med. 2012;209:729–740. doi: 10.1084/jem.20111594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Montaldo E, et al. Human RORgammat+CD34+ cells are lineage-specified progenitors of group 3 RORgammat+ innate lymphoid cells. Immunity. 2014;41:988–1000. doi: 10.1016/j.immuni.2014.11.010. [DOI] [PubMed] [Google Scholar]
- 50.Klose CS, Diefenbach A. Transcription factors controlling innate lymphoid cell fate decisions. Curr Top Microbiol Immunol. 2014;381:215–255. doi: 10.1007/82_2014_381. [DOI] [PubMed] [Google Scholar]
- 51.Sawa S, et al. Lineage relationship analysis of RORgammat+ innate lymphoid cells. Science. 2010;330:665–669. doi: 10.1126/science.1194597. [DOI] [PubMed] [Google Scholar]
- 52.Yu X, et al. The basic leucine zipper transcription factor NFIL3 directs the development of a common innate lymphoid cell precursor. Elife. 2014;3:e04406. doi: 10.7554/eLife.04406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Possot C, et al. Notch signaling is necessary for adult, but not fetal, development of RORgammat+ innate lymphoid cells. Nat Immunol. 2011;12:949–958. doi: 10.1038/ni.2105. [DOI] [PubMed] [Google Scholar]
- 54.Hoyler T, et al. T-bet and Gata3 in controlling type 1 and type 2 immunity mediated by innate lymphoid cells. Curr Opin Immunol. 2013;25:139–147. doi: 10.1016/j.coi.2013.02.007. [DOI] [PubMed] [Google Scholar]
- 55.Kim CH. The greater chemotactic network for lymphocyte trafficking: chemokines and beyond. Curr Opin Hematol. 2005;12:298–304. doi: 10.1097/01.moh.0000166496.18773.e3. [DOI] [PubMed] [Google Scholar]
- 56.Lapidot T, et al. How do stem cells find their way home? Blood. 2005;106:1901–1910. doi: 10.1182/blood-2005-04-1417. [DOI] [PubMed] [Google Scholar]
- 57.Kraal G, et al. Expression of the mucosal vascular addressin MAdCAM-1, on sinus-lining cells in the spleen. Am J Pathol. 1995;147:763–771. [PMC free article] [PubMed] [Google Scholar]
- 58.Osborn L, et al. Direct expression cloning of vascular cell adhesion molecule 1, a cytokine-induced endothelial protein that binds to lymphocytes. Cell. 1989;59:1203–1211. doi: 10.1016/0092-8674(89)90775-7. [DOI] [PubMed] [Google Scholar]
- 59.Ruegg C, et al. Role of integrin alpha 4 beta 7/alpha 4 beta P in lymphocyte adherence to fibronectin and VCAM-1 and in homotypic cell clustering. J Cell Biol. 1992;117:179–189. doi: 10.1083/jcb.117.1.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Strauch UG, et al. Distinct binding specificities of integrins alpha 4 beta 7 (LPAM-1), alpha 4 beta 1 (VLA-4), and alpha IEL beta 7. Int Immunol. 1994;6:263–275. doi: 10.1093/intimm/6.2.263. [DOI] [PubMed] [Google Scholar]
- 61.Campbell JJ, et al. Chemokines and the arrest of lymphocytes rolling under flow conditions. Science. 1998;279:381–384. doi: 10.1126/science.279.5349.381. [DOI] [PubMed] [Google Scholar]
- 62.Matloubian M, et al. A transmembrane CXC chemokine is a ligand for HIV-coreceptor Bonzo. Nat Immunol. 2000;1:298–304. doi: 10.1038/79738. [DOI] [PubMed] [Google Scholar]
- 63.Hara T, et al. A transmembrane chemokine CXC chemokine ligand 16, expressed by lymph node fibroblastic reticular cells has the potential to regulate T cell migration and adhesion. Int Immunol. 2006;18:301–311. doi: 10.1093/intimm/dxh369. [DOI] [PubMed] [Google Scholar]
- 64.Zlotoff DA, et al. CCR7 and CCR9 together recruit hematopoietic progenitors to the adult thymus. Blood. 2010;115:1897–1905. doi: 10.1182/blood-2009-08-237784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kunkel EJ, et al. Lymphocyte CC chemokine receptor 9 and epithelial thymus-expressed chemokine (TECK) expression distinguish the small intestinal immune compartment: epithelial expression of tissue-specific chemokines as an organizing principle in regional immunity. J Exp Med. 2000;192:761–768. doi: 10.1084/jem.192.5.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schwarz BA, et al. Selective thymus settling regulated by cytokine and chemokine receptors. J Immunol. 2007;178:2008–2017. doi: 10.4049/jimmunol.178.4.2008. [DOI] [PubMed] [Google Scholar]
- 67.Worbs T, Forster R. A key role for CCR7 in establishing central and peripheral tolerance. Trends Immunol. 2007;28:274–280. doi: 10.1016/j.it.2007.04.002. [DOI] [PubMed] [Google Scholar]
- 68.Luther SA, et al. Coexpression of the chemokines ELC and SLC by T zone stromal cells and deletion of the ELC gene in the plt/plt mouse. Proc Natl Acad Sci USA. 2000;97:12694–12699. doi: 10.1073/pnas.97.23.12694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gunn MD, et al. A chemokine expressed in lymphoid high endothelial venules promotes the adhesion and chemotaxis of naive T lymphocytes. Proc Natl Acad Sci USA. 1998;95:258–263. doi: 10.1073/pnas.95.1.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Huang HY, Luther SA. Expression and function of interleukin-7 in secondary and tertiary lymphoid organs. Semin Immunol. 2012;24:175–189. doi: 10.1016/j.smim.2012.02.008. [DOI] [PubMed] [Google Scholar]
- 71.Vonarbourg C, Diefenbach A. Multifaceted roles of interleukin-7 signaling for the development and function of innate lymphoid cells. Semin Immunol. 2012;24:165–174. doi: 10.1016/j.smim.2012.03.002. [DOI] [PubMed] [Google Scholar]
- 72.Vosshenrich CA, et al. A thymic pathway of mouse natural killer cell development characterized by expression of GATA-3 and CD127. Nat Immunol. 2006;7:1217–1224. doi: 10.1038/ni1395. [DOI] [PubMed] [Google Scholar]
- 73.Marcais A, et al. Regulation of mouse NK cell development and function by cytokines. Front Immunol. 2013;4:450. doi: 10.3389/fimmu.2013.00450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lodolce JP, et al. IL-15 receptor maintains lymphoid homeostasis by supporting lymphocyte homing and proliferation. Immunity. 1998;9:669–676. doi: 10.1016/s1074-7613(00)80664-0. [DOI] [PubMed] [Google Scholar]
- 75.Kennedy MK, et al. Reversible defects in natural killer and memory CD8 T cell lineages in interleukin 15-deficient mice. J Exp Med. 2000;191:771–780. doi: 10.1084/jem.191.5.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ma A, et al. Diverse functions of IL-2 IL-15, and IL-7 in lymphoid homeostasis. Annu Rev Immunol. 2006;24:657–679. doi: 10.1146/annurev.immunol.24.021605.090727. [DOI] [PubMed] [Google Scholar]
- 77.Carson WE, et al. Interleukin (IL) 15 is a novel cytokine that activates human natural killer cells via components of the IL-2 receptor. J Exp Med. 1994;180:1395–1403. doi: 10.1084/jem.180.4.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chaix J, et al. Cutting edge: priming of NK cells by IL-18. J Immunol. 2008;181:1627–1631. doi: 10.4049/jimmunol.181.3.1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Buonocore S, et al. Innate lymphoid cells drive interleukin-23-dependent innate intestinal pathology. Nature. 2010;464:1371–1375. doi: 10.1038/nature08949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.von Burg N, et al. Activated group 3 innate lymphoid cells promote T-cell-mediated immune responses. Proc Natl Acad Sci USA. 2014;111:12835–12840. doi: 10.1073/pnas.1406908111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mielke LA, et al. Retinoic acid expression associates with enhanced IL-22 production by gammadelta T cells and innate lymphoid cells and attenuation of intestinal inflammation. J Exp Med. 2013;210:1117–1124. doi: 10.1084/jem.20121588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tait Wojno ED, Artis D. Innate lymphoid cells: balancing immunity, inflammation, and tissue repair in the intestine. Cell Host Microbe. 2012;12:445–457. doi: 10.1016/j.chom.2012.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Raverdeau M, Mills KH. Modulation of T cell and innate immune responses by retinoic acid. J Immunol. 2014;192:2953–2958. doi: 10.4049/jimmunol.1303245. [DOI] [PubMed] [Google Scholar]
- 84.Kim CH. Retinoic acid, immunity, and inflammation. Vitam Horm. 2011;86:83–101. doi: 10.1016/B978-0-12-386960-9.00004-6. [DOI] [PubMed] [Google Scholar]
- 85.Qiu J, et al. The aryl hydrocarbon receptor regulates gut immunity through modulation of innate lymphoid cells. Immunity. 2012;36:92–104. doi: 10.1016/j.immuni.2011.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kiss EA, et al. Natural aryl hydrocarbon receptor ligands control organogenesis of intestinal lymphoid follicles. Science. 2011;334:1561–1565. doi: 10.1126/science.1214914. [DOI] [PubMed] [Google Scholar]
- 87.Lee JS, et al. AHR drives the development of gut ILC22 cells and postnatal lymphoid tissues via pathways dependent on and independent of Notch. Nat Immunol. 2012;13:144–151. doi: 10.1038/ni.2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Stockinger B, et al. The aryl hydrocarbon receptor: multitasking in the immune system. Annu Rev Immunol. 2014;32:403–432. doi: 10.1146/annurev-immunol-032713-120245. [DOI] [PubMed] [Google Scholar]
- 89.Denison MS, Nagy SR. Activation of the aryl hydrocarbon receptor by structurally diverse exogenous and endogenous chemicals. Annu Rev Pharmacol Toxicol. 2003;43:309–334. doi: 10.1146/annurev.pharmtox.43.100901.135828. [DOI] [PubMed] [Google Scholar]
- 90.Satoh-Takayama N, et al. Microbial flora drives interleukin 22 production in intestinal NKp46+ cells that provide innate mucosal immune defense. Immunity. 2008;29:958–970. doi: 10.1016/j.immuni.2008.11.001. [DOI] [PubMed] [Google Scholar]
- 91.Reynders A, et al. Identity, regulation and in vivo function of gut NKp46+RORgammat+ and NKp46+RORgammat− lymphoid cells. EMBO J. 2011;30:2934–2947. doi: 10.1038/emboj.2011.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sawa S, et al. RORgammat+ innate lymphoid cells regulate intestinal homeostasis by integrating negative signals from the symbiotic microbiota. Nat Immunol. 2011;12:320–326. doi: 10.1038/ni.2002. [DOI] [PubMed] [Google Scholar]
- 93.Monticelli LA, et al. Innate lymphoid cells promote lung-tissue homeostasis after infection with influenza virus. Nat Immunol. 2011;12:1045–1054. doi: 10.1031/ni.2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Magri G, et al. Innate lymphoid cells integrate stromal and immunological signals to enhance antibody production by splenic marginal zone B cells. Nat Immunol. 2014;15:354–364. doi: 10.1038/ni.2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Martin-Fontecha A, et al. Induced recruitment of NK cells to lymph nodes provides IFN-gamma for T(H)1 priming. Nat Immunol. 2004;5:1260–1265. doi: 10.1038/ni1138. [DOI] [PubMed] [Google Scholar]
- 96.Mailliard RB, et al. IL-18-induced CD83+CCR7+ NK helper cells. J Exp Med. 2005;202:941–953. doi: 10.1084/jem.20050128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Brown MN, et al. Chemoattractant receptors and lymphocyte egress from extralymphoid tissue: changing requirements during the course of inflammation. J Immunol. 2010;185:4873–4882. doi: 10.4049/jimmunol.1000676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Haynes NM, et al. Role of CXCR5 and CCR7 in follicular Th cell positioning and appearance of a programmed cell death gene-1high germinal center-associated subpopulation. J Immunol. 2007;179:5099–5108. doi: 10.4049/jimmunol.179.8.5099. [DOI] [PubMed] [Google Scholar]
- 99.Ohl L, et al. Cooperating mechanisms of CXCR5 and CCR7 in development and organization of secondary lymphoid organs. J Exp Med. 2003;197:1199–1204. doi: 10.1084/jem.20030169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kim MY, et al. Heterogeneity of lymphoid tissue inducer cell populations present in embryonic and adult mouse lymphoid tissues. Immunology. 2008;124:166–174. doi: 10.1111/j.1365-2567.2007.02750.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ansel KM, et al. A chemokine-driven positive feedback loop organizes lymphoid follicles. Nature. 2000;406:309–314. doi: 10.1038/35018581. [DOI] [PubMed] [Google Scholar]
- 102.Scandella E, et al. Restoration of lymphoid organ integrity through the interaction of lymphoid tissue-inducer cells with stroma of the T cell zone. Nat Immunol. 2008;9:667–675. doi: 10.1038/ni.1605. [DOI] [PubMed] [Google Scholar]
- 103.Mora JR, et al. Selective imprinting of gut-homing T cells by Peyer’s patch dendritic cells. Nature. 2003;424:88–93. doi: 10.1038/nature01726. [DOI] [PubMed] [Google Scholar]
- 104.Sigmundsdottir H, Butcher EC. Environmental cues, dendritic cells and the programming of tissue-selective lymphocyte trafficking. Nat Immunol. 2008;9:981–987. doi: 10.1038/ni.f.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Campbell JJ, et al. The chemokine receptor CCR4 in vascular recognition by cutaneous but not intestinal memory T cells. Nature. 1999;400:776–780. doi: 10.1038/23495. [DOI] [PubMed] [Google Scholar]
- 106.Reiss Y, et al. CC chemokine receptor (CCR)4 and the CCR10 ligand cutaneous T cell-attracting chemokine (CTACK) in lymphocyte trafficking to inflamed skin. J Exp Med. 2001;194:1541–1547. doi: 10.1084/jem.194.10.1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Schaerli P, et al. A skin-selective homing mechanism for human immune surveillance T cells. J Exp Med. 2004;199:1265–1275. doi: 10.1084/jem.20032177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Homey B, et al. CCL27–CCR10 interactions regulate T cell-mediated skin inflammation. Nat Med. 2002;8:157–165. doi: 10.1038/nm0202-157. [DOI] [PubMed] [Google Scholar]
- 109.Iwata M, et al. Retinoic acid imprints gut-homing specificity on T cells. Immunity. 2004;21:527–538. doi: 10.1016/j.immuni.2004.08.011. [DOI] [PubMed] [Google Scholar]
- 110.Schon MP, et al. Dendritic epidermal T cells (DETC) are diminished in integrin alphaE(CD103)-deficient mice. J Invest Dermatol. 2002;119:190–193. doi: 10.1046/j.1523-1747.2002.17973.x. [DOI] [PubMed] [Google Scholar]
- 111.Picker LJ, et al. ELAM-1 is an adhesion molecule for skin-homing T cells. Nature. 1991;349:796–799. doi: 10.1038/349796a0. [DOI] [PubMed] [Google Scholar]
- 112.Sigmundsdottir H, et al. DCs metabolize sunlight-induced vitamin D3 to ‘program’ T cell attraction to the epidermal chemokine CCL27. Nat Immunol. 2007;8:285–293. doi: 10.1038/ni1433. [DOI] [PubMed] [Google Scholar]
- 113.Ruiter B, et al. Vitamins A and D have antagonistic effects on expression of effector cytokines and gut-homing integrin in human innate lymphoid cells. Clin Exp Allergy. 2015;45:1214–1225. doi: 10.1111/cea.12568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Jenne CN, et al. T-bet-dependent S1P5 expression in NK cells promotes egress from lymph nodes and bone marrow. J Exp Med. 2009;206:2469–2481. doi: 10.1084/jem.20090525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kim CH, et al. Rules of chemokine receptor association with T cell polarization in vivo. J Clin Invest. 2001;108:1331–1339. doi: 10.1172/JCI13543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.D’Ambrosio D, et al. Selective up-regulation of chemokine receptors CCR4 and CCR8 upon activation of polarized human type 2 Th cells. J Immunol. 1998;161:5111–5115. [PubMed] [Google Scholar]
- 117.Wang C, et al. The roles of CCR6 in migration of Th17 cells and regulation of effector T-cell balance in the gut. Mucosal Immunol. 2009;2:173–183. doi: 10.1038/mi.2008.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Takatori H, et al. Lymphoid tissue inducer-like cells are an innate source of IL-17 and IL-22. J Exp Med. 2009;206:35–41. doi: 10.1084/jem.20072713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Ottaviani C, et al. CD56brightCD16− NK cells accumulate in psoriatic skin in response to CXCL10 and CCL5 and exacerbate skin inflammation. Eur J Immunol. 2006;36:118–128. doi: 10.1002/eji.200535243. [DOI] [PubMed] [Google Scholar]
- 120.Ebert LM, et al. Homing and function of human skin gammadelta T cells and NK cells: relevance for tumor surveillance. J Immunol. 2006;176:4331–4336. doi: 10.4049/jimmunol.176.7.4331. [DOI] [PubMed] [Google Scholar]
- 121.Yu YR, et al. Defective antitumor responses in CX3CR1-deficient mice. Int J Cancer. 2007;121:316–322. doi: 10.1002/ijc.22660. [DOI] [PubMed] [Google Scholar]
- 122.van Helden MJ, et al. CCR2 defines a distinct population of NK cells and mediates their migration during influenza virus infection in mice. PLoS ONE. 2012;7:e52027. doi: 10.1371/journal.pone.0052027. [DOI] [PMC free article] [PubMed] [Google Scholar]