Abstract
T helper 17 (Th17)-associated cytokines are integral to the immune responses to tuberculosis, initiating both protective and harmful inflammatory responses. The aim of the present study was to evaluate applied aspects of interleukin-17 (IL-17) biology in the context of Mycobacterium bovis infection of cattle. Using transcriptome sequencing (RNA-Seq), numerous Th17-associated cytokine genes (including IL-17A, IL-17F, IL-22, IL-19, and IL-27) were upregulated >9-fold in response to purified protein derivative stimulation of peripheral blood mononuclear cells from experimentally M. bovis-infected cattle. Protective vaccines elicited IL-17A, IL-17F, IL-22, and IL-27 responses. Reduced IL-17A responses by vaccine recipients, compared to nonvaccinated animals, at 2.5 weeks after M. bovis challenge correlated with reduced disease burdens. Additionally, IL-17A and interferon gamma (IFN-γ) responses were highly correlated and exhibited similar diagnostic capacities. The present findings support the use of Th17-associated cytokines as biomarkers of infection and protection in the immune responses to bovine tuberculosis.
INTRODUCTION
Tuberculosis (TB) in humans and animals may result from exposure to bacilli within the Mycobacterium tuberculosis complex (i.e., Mycobacterium tuberculosis, Mycobacterium bovis, Mycobacterium africanum, Mycobacterium pinnipedii, Mycobacterium microti, Mycobacterium caprae, Mycobacterium orygis, or Mycobacterium canetti) (1). Mycobacterium bovis is the species most often isolated from tuberculous cattle and may infect humans. The global median proportion of human TB cases due to M. bovis is relatively small (∼1.4%); however, higher rates are reported for continental Africa (∼2.8%), Mexico (∼7.6%), Turkey (∼5.3%), and the West Bank (∼6.5%) (2). Therefore, bovine TB is one of the seven most neglected endemic zoonotic diseases and is particularly devastating in resource-poor settings, due to limited regulatory control, consumption of nonpasteurized milk and noninspected meat, and comorbidities affecting host susceptibility and disease severity. Compared to M. tuberculosis, M. bovis has a relatively wide host range, including several wildlife reservoir hosts, which significantly hinders control of the disease in cattle (3). Even with intensive control efforts, bovine TB remains a costly disease for producers and regulatory agencies, with estimates of 50 million cattle being infected worldwide, at a cost of 3 billion dollars per year (4). Other major consequences of this disease include adverse effects on the trade of animals and animal products, animal health, producer livelihoods, natural resources (e.g., preservation of endangered wildlife species), and public health.
Strategies for the control of bovine TB rely largely on antemortem testing and slaughter inspection to identify herds at risk. With cattle, the principal antemortem tests for presumptive diagnosis of bovine TB are immunoassays to detect cell-mediated responses, including tuberculin skin test (TST) procedures and interferon gamma (IFN-γ) release assays (IGRAs). TSTs are used almost exclusively as the primary tests and IGRAs may be used as confirmatory tests in cattle (4). In the United States, the caudal fold test (CFT) (intradermal injection of M. bovis purified protein derivative [PPD] in the caudal skin fold adjacent to the tail) is used as a primary test and the comparative cervical test (CCT) (intradermal injection of Mycobacterium avium and M. bovis PPDs at separate sites in the neck) and the Bovigam assay (Prionics Ag, Schlieren, Switzerland) (an IGRA) are used as secondary confirmatory tests. While TSTs are the mainstays for bovine TB antemortem testing, blood-based tests are attractive, as they require only a single visit to the farm, provide the opportunity for immediate repeat testing, and yield results that are less subjective than those of TSTs. Additional biomarkers have recently emerged as potential candidates for use in blood-based TB tests for humans (reviewed by Walzl et al. [5] and Salgame et al. [6]) and cattle, including interleukin-1β (IL-1β), IL-2, tumor necrosis factor alpha (TNF-α), nitric oxide, IL-17, and IFN-γ-induced protein 10 (IP-10) (7–13). Of these, IL-17 is particularly appealing as a biomarker for bovine TB, as IL-17A mRNA responses determined using real-time quantitative PCR (RT-qPCR) may be predictive of both vaccine efficacy (11, 14, 15) and lesion severity (7) when measured after vaccination and infection, respectively.
In the present study, we performed an extensive analysis of IL-17A responses (protein and mRNA), both as a measure of vaccine efficacy and as a diagnostic biomarker, using samples from experimental infection and vaccine efficacy studies. Using transcriptome sequencing (RNA-Seq) and RT-qPCR, we also evaluated the expression of other T helper 17 (Th17)-associated genes, particularly IL-17F, IL-22, IL-23p19, and IL-27. Also, IL-17A responses were directly compared to IFN-γ responses, given the widespread use of IGRAs for TB diagnosis, vaccine, and pathogenicity studies.
MATERIALS AND METHODS
Study overview and aerosol infection with Mycobacterium bovis.
Samples from four independent studies were included, i.e., one study that compared the virulence of two field strains of M. bovis in calves, two vaccine efficacy studies, and a M. bovis infection-only study (for RNA-Seq samples). An overview of the studies, including treatment groups, challenge strains and doses, and timing of treatments, is provided in Table 1. Two strains of M. bovis were used for challenge inocula in the various studies, i.e., 95-1315 (Michigan white-tailed deer isolate) (16) and 10-7428 (Colorado Holstein isolate) (17). Challenge inocula from frozen stocks were prepared in Middlebrook 7H9 liquid medium (Becton Dickinson, Franklin Lakes, NJ) supplemented with 10% oleic acid-albumin-dextrose complex (OADC) plus 0.05% Tween 80 (Sigma, St. Louis, MO), following standard techniques (18). Holstein steers were obtained from TB-free herds in Iowa and housed in a biosafety level 3 (BSL3) facility at the National Animal Disease Center (Ames, IA), according to institutional biosafety and animal care and use committee guidelines and oversight (i.e., formal review and approval of studies). For experimental infection, Holstein steers received virulent M. bovis by aerosol, as described previously (19). Strict biosafety protocols were followed to protect personnel from exposure to M. bovis throughout the study, including BSL3 containment upon initiation of M. bovis challenge in animal rooms and standard BSL3 laboratory practices for handling of M. bovis cultures and samples from M. bovis-infected animals.
TABLE 1.
Experimental design and efficacy parameters
| Study and group | Vaccinationa |
M. bovis challengeb |
Pathology score (mean ± SEM)c | Culture (log CFU/g) (mean ± SEM)d | |||
|---|---|---|---|---|---|---|---|
| Age (wk) | Interval (mo) | Strain | Dose (CFU) | Interval (mo) | |||
| Comparative virulence studye | |||||||
| No infection (n = 7) | NAf | NA | NA | NA | 0 ± 0 | 0 ± 0 | |
| M. bovis strain 95-1315 infection (n = 8) | NA | 95-1315 | 104 | 4 | 16.4 ± 2.5g | 3.25 ± 0.33g | |
| M. bovis strain 10-7428 infection (n = 8) | NA | 10-7428 | 104 | 4 | 20.5 ± 1.3g | 3.53 ± 0.20g | |
| Vaccine efficacy study from 2014 | |||||||
| No vaccination (n = 10) | NA | 10-7428 | 600 | 4.5 | 12.0 ± 1.6 | 3.86 ± 0.28 | |
| BCG vaccination (n = 9) | 3 | 3.5 | 10-7428 | 600 | 4.5 | 0.6 ± 0.4g | 0.34 ± 0.34g |
| BCG mutant vaccination (n = 10) | 3 | 3.5 | 10-7428 | 600 | 4.5 | 0.9 ± 0.4g | 1.74 ± 0.72g |
| Vaccine efficacy study from 2007h | |||||||
| No vaccination (n = 11) | NA | 95-1315 | 103 | 4.5 | 14.1 ± 1.4 | 3.95 ± 0.13 | |
| BCG vaccination (n = 11) | 2 | 3 | 95-1315 | 103 | 4.5 | 3.4 ± 0.9g | 2.59 ± 0.42g |
| M. bovis ΔRD1 vaccination (n = 10) | 2 | 3 | 95-1315 | 103 | 4.5 | 6.0 ± 1.8g | 2.62 ± 0.59g |
| RNA-Seq studyi | |||||||
| M. bovis strain 95-1315 infection (n = 6) | NA | 95-1315 | 8 × 103 | 11 | All had lesions | All were culture positive | |
Age of vaccination and interval between vaccination and aerosol challenge with virulent M. bovis.
Strain and dose of virulent strain administered by aerosol and interval between challenge and necropsy.
Total gross pathology scores, which include scores for tracheobronchial and mediastinal (i.e., pulmonary) lymph nodes and lung lobes.
M. bovis CFU per gram of tracheobronchial lymph node.
See reference 37 for additional details of this study.
NA, not applicable (i.e., animals were not vaccinated or challenged).
Differs from the noninfected or nonvaccinated group within each study (P < 0.05, ANOVA followed by Tukey's multiple-comparison test).
See reference 20 for additional details of this study.
Pathology scoring and quantitative culture were not applied; however, all animals had tuberculous lesions and M. bovis was isolated from each animal.
Vaccine efficacy studies.
Two independent vaccine efficacy studies were performed. The age of calves at vaccination and the vaccine and challenge intervals are provided in Table 1. Briefly, calves were vaccinated subcutaneously at 2 to 3 weeks of age, challenged with virulent M. bovis at ∼4 months of age, and euthanized at ∼8 months of age. In the 2007 study (20), the vaccine treatment groups were as follows: no vaccination (n = 11), 106 CFU M. bovis bacillus Calmette-Guerin (BCG) Danish (n = 11), and 106 CFU M. bovis Ravenel ΔRD1 (n = 10). In the 2014 study, the vaccine treatment groups were as follows: no vaccination (n = 10), 106 CFU M. bovis BCG Danish (n = 9), and 106 CFU (total dose) of a cocktail of four BCG Danish deletion strains, i.e., BCG Δfdr8, BCG ΔleuCD Δpks16, BCG ΔmmaA4 (21), and BCG ΔmetA (22) (n = 10). All four BCG Danish deletion derivatives (Δfdr8, ΔleuCD Δpks16, ΔmetA, and ΔmmaA4) are more attenuated and safer than the parental BCG strain in immunocompromised mice (21; L. Berney-Meyer, M. Larsen, and W. R. Jacobs, unpublished data). In immunocompetent mice, the BCG deletions Δfdr8, ΔmmaA4, and Δpks16 each result in enhanced mycobacterial immunogenicity through enhanced cross-presentation of mycobacterial antigens (Δfdr8), cytokine modulation (ΔmmaA4), and biofilm formation (Δpks16), compared to the parental BCG (21; Berney-Meyer et al., unpublished). BCG mutants, such as these, may also be used as vaccine vectors to promote epitope-specific responses (e.g., BCG Δpks12 for enhanced CD8 responses) (23).
Assessment of mycobacterial lesions and colonization.
All calves were euthanized ∼4 to 4.5 months (Table 1) after challenge, by intravenous administration of sodium pentobarbital. Tissues were examined for gross lesions and processed for microscopic analysis and isolation of M. bovis. Tissues collected included lung, liver, and mandibular, parotid, medial retropharyngeal, mediastinal, tracheobronchial, hepatic, and mesenteric lymph nodes. Lymph nodes were sectioned at 0.5-cm intervals and examined. Each lung lobe was sectioned at 0.5- to 1.0-cm intervals and examined separately. Lungs and lymph nodes (mediastinal and tracheobronchial) were evaluated using a semiquantitative gross pathology scoring system adapted from the report by Vordermeier et al. (24). Head and abdominal lymph nodes were not included in the pathology scoring analysis because the route (aerosol) and duration (4 to 4.5 months) of experimental infection resulted in lesions focused primarily in the lungs and lung-associated lymph nodes. Lung lobes (left cranial, left caudal, right cranial, right caudal, middle, and accessory) were individually assessed with the following scoring system: 0, no visible lesions; 1, no external gross lesions but lesions seen after slicing; 2, <5 gross lesions <10 mm in diameter; 3, >5 gross lesions <10 mm in diameter; 4, >1 distinct gross lesion >10 mm in diameter; 5, gross coalescing lesions. Scoring of lymph node pathology was based on the following system: 0, no necrosis or visible lesions; 1, small focus (1 to 2 mm in diameter); 2, several small foci; 3, extensive necrosis. Gross pathology data are presented as total gross pathology scores (mean ± standard error of the mean [SEM]), including scores for each lung lobe as well as the tracheobronchial and mediastinal lymph nodes.
Tissues collected for microscopic analysis were fixed by immersion in 10% neutral buffered formalin. For microscopic examination, formalin-fixed tissues were processed with standard paraffin-embedment techniques, cut in 5-μm sections, and stained with hematoxylin and eosin. Adjacent sections from samples containing caseonecrotic granulomata, suggesting tuberculosis, were stained with the Ziehl-Neelsen technique for identification of acid-fast bacteria. Microscopic tuberculous lesions were staged (stage I to IV) based on a scoring system developed by Wangoo et al. (25). Data are presented as mean ± SEM for the sum of the number of granulomas representing each stage (stage I to IV) individually and the total for all stages observed in mediastinal lymph nodes, tracheobronchial lymph nodes, and lung histological sections.
Quantitative assessments of mycobacterial burdens were evaluated as described previously (26). Briefly, tracheobronchial lymph nodes were removed, examined for gross lesions, and weighed, and entire lymph nodes (other than an ∼1-g section for histological assessment) were homogenized in phenol red nutrient broth using a blender (Oster, Shelton, CT). Logarithmic dilutions (100 to 10−9) of homogenates in phosphate-buffered saline (PBS) were plated in 100-μl aliquots on Middlebrook 7H11 selective agar plates (Becton Dickinson) and incubated for 8 weeks at 37°C for determination of log10 CFU per gram of tissue. IS6110 real-time PCR, as described by Thacker et al. (27), was used to confirm that colonies were M. bovis.
Whole-blood stimulation.
Duplicate 250-μl heparinized whole-blood aliquots were distributed in 96-well plates with RPMI 1640 medium (Sigma) alone, 1 μg/ml recombinant early secretory antigenic target 6 (rESAT-6):culture filtrate protein 10 (CFP10) (a gift from Chris Minion, Iowa State University), 1 μg/ml each of recombinant Ag85A (rAg85A) and recombinant TB10.4 (rTB10.4) (Lionex Diagnostics and Therapeutics GmbH, Braunschweig, Germany), 10 μg/ml M. bovis PPD (CSL; Prionics Ag), or 1 μg/ml pokeweed mitogen (PWM) (Sigma) and were incubated at 39°C in 5% CO2 for 18 h for cytokine analysis by enzyme-linked immunosorbent assay (ELISA). The normal body temperature of cattle (Bos taurus) is 39°C and incubation of human blood at 39°C, rather than 37°C, augments cytokine responses (28).
Cell culture.
Peripheral blood mononuclear cells (PBMCs) were isolated by density gradient centrifugation of peripheral blood buffy coat fractions collected into 2× acid-citrate-dextrose, as described previously (12). PBMCs were seeded into 96-well round-bottom microtiter plates (Falcon; Becton-Dickinson, Lincoln Park, NJ) at 1 × 106 cells in a total volume of 200 μl of complete RPMI 1640 (RPMI 1640 with 10% [vol/vol] fetal bovine serum [FBS] [Atlanta Biologics, Lawrenceville, GA], 2 mM l-glutamine, 25 mM HEPES buffer, 100 units/ml penicillin, 0.1 mg/ml streptomycin, 1% nonessential amino acids, 2% essential amino acids, 1% sodium pyruvate, and 50 μM 2-mercaptoethanol [all reagents in complete RPMI 1640 medium from Sigma except for FBS]). Wells contained medium alone (nonstimulated), 1 μg/ml each of rAg85A and rTB10.4 (Lionex Diagnostics and Therapeutics GmbH), 1 μg/ml rESAT-6:CFP10, or 10 μg/ml M. bovis PPD (Prionics Ag). Cultures were incubated at 39°C in 5% CO2 for 16 h for cytokine mRNA analysis in cell lysates or protein analysis in supernatants.
RNA isolation and analysis of cytokine gene expression by real-time PCR.
Isolation and reverse transcription of RNA in PBMCs were performed as described previously (29). Briefly, PBMCs were harvested by centrifugation and lysed with 150 μl/well buffer RLT (Qiagen, Valencia, CA), according to the manufacturer's directions. Replicate wells were combined, and samples were stored at −80°C. RNA was isolated using an RNAeasy minikit (Qiagen), according to the manufacturer's directions, and was eluted from the column with 50 μl RNase-free water (Ambion, Austin, TX). Contaminating DNA was enzymatically removed by treating RNA with DNA-free (Ambion). One microgram of RNA was reverse transcribed in a 50-μl reaction mixture using SuperScript II (Invitrogen, Carlsbad, CA) with 0.5 μg of oligo(dT)12–18 and 40 units of RNaseOut (Invitrogen), according to the manufacturer's directions. Samples were heated to 70°C for 5 min and then reverse transcribed at 42°C for 60 min. The resulting cDNA was stored at −80°C until used in real-time PCR assays. Real-time PCR assays were performed using a TaqMan gene expression assay kit (Applied Biosystems, Foster City, CA), according to the manufacturer's directions. Applied Biosystems primers and probes for cytokine genes are presented in Table 2, and amplification conditions were established according to the manufacturer's directions. Reactions were performed on an Applied Biosystems 7300 real-time PCR system (Life Technologies, Grand Island, NY). Relative gene expression was expressed as 2−ΔΔCT (30), with eukaryotic 18S rRNA (catalogue no. 4333760; Applied Biosystems) as the endogenous control, and the medium-only (i.e., no stimulation) sample from each animal was used as the calibrator for evaluation of PBMC responses.
TABLE 2.
Cytokine primers/probes for RT-qPCR identification
| Cytokine | Entrez Gene IDa | Assay IDb |
|---|---|---|
| IL-17A | 282863 | Bt03210252_m1 |
| IL-17F | 506030 | Bt04309062_m1 |
| IL-22 | 507778 | Bt03261459_m1 |
| IL-23p19 subunit | 511022 | Bt04284624_m1 |
| IL-27 | 614927 | Bt04298832_m1 |
| IFN-γ | 281237 | Bt03212723_m1 |
Entrez Gene (http://www.ncbi.nlm.nih.gov/gene) identification (ID) number.
Applied Biosystems product number.
RNA-Seq analysis.
Whole blood was collected from six M. bovis-infected calves prior to and 9 weeks after challenge (Table 1). PBMCs were isolated and stimulated with PPD as described previously (29). According to the manufacturer's directions, RNA was isolated from stimulated PBMCs using an RNAeasy Maxi kit (Qiagen) and treated with DNase (DNA-free; Ambion). RNA was concentrated using 30K Microcon centrifugal filter devices (Millipore). Samples were quantitated, and the RNA integrity was checked using an Agilent 2100 Bioanalyzer (Agilent Technologies), according to the manufacturer's directions. All samples had a RNA integrity number greater than 7.0. Each sample (3.3 μg RNA) was randomly added to one of two pooled samples for each time point. Three time points were chosen for sequencing, i.e., prior to infection, 1 month postinfection, and 2 months postinfection. Pooled samples were sequenced at the Iowa State University DNA Sequencing Facility. Each pool was sequenced on an Illumina Genome Analyzer II, using a 75-base run. Sequences were analyzed using FastQC (version 0.10.0) (http://www.bioinformatics.babraham.ac.uk/projects/fastqc), and low-quality reads were trimmed with PRINSEQ-lite (31). Reads were aligned to the Ensmbl Btau 4.0 version of the cattle genome. Reads mapping to identified genes were counted using HTseq (version 0.5.3p3) (32). The counts per gene for each sample were collated using a database developed in-house using MySQL. Genes that had combined expression of <20 reads were removed from further analysis. Differentially expressed genes were identified using EdgeR (version 2.8) (33–35). A total of 348 genes with adjusted P values of <0.01 were considered significant using tag-wise dispersion. Data are presented as genes upregulated (i.e., >9-fold) after M. bovis infection versus before M. bovis infection. Gene expression did not differ (<0.01) for 1 month versus 2 months postinfection; therefore, postinfection data were analyzed as a single data point.
IFN-γ and IL-17A ELISAs.
IFN-γ and IL-17A concentrations in stimulated plasma from whole blood or supernatants from PBMC cultures were determined using commercial ELISA-based kits (Bovigam [Prionics Ag] and bovine IL-17A ELISA VetSet [Kingfisher Biotech Inc., Saint Paul, MN]), according to the manufacturers' instructions. Absorbance values for standards (recombinant bovine IFN-γ [Endogen, Rockford, IL] and recombinant bovine IL-17A [Kingfisher Biotech Inc.]) and test samples were determined at 450 nm using an ELISA plate reader (Molecular Devices, Menlo Park, CA). Duplicate samples for individual treatments were analyzed, and both IL-17A and IFN-γ data are presented as nanograms of protein per milliliter of plasma.
IL-17 ELISPOT assay.
The protocol for the IL-17A enzyme-linked immunosorbent spot (ELISPOT) assay was as described previously (36). Briefly, 2 × 105 PBMCs were added to polyvinylidene difluoride 96-well assay plates (Millipore, Watford, United Kingdom) that had been coated with anti-bovine IL-17A polyclonal antibodies (5 μg/ml; Kingfisher Biotech) and were incubated in the presence or absence of 10 μg/ml M. bovis PPD, 3 μg/ml rAg85A-rTB10.4, or 3 μg/ml rESAT-6:CFP10. Plates were incubated for 18 h, washed, and incubated for 2 h with biotinylated anti-bovine IL-17A detection antibodies (5 μg/ml; Kingfisher Biotech). Plates were washed and developed using a Vectastain ABC-AP standard kit and a Vector blue alkaline phosphatase substrate kit (both from Vector Laboratories, Burlingame, CA), according to the manufacturer's instructions. Plates were read and analyzed using a standard ELISPOT reader (Cellular Technology).
Statistical analysis.
Data were analyzed by analysis of variance (ANOVA) followed by Tukey's multiple-comparison test, Student's t test, or Spearman's correlation, using commercially available software (Prism 6.0c; GraphPAD Software, La Jolla, CA). Comparisons with P values of <0.05 were considered significant.
RESULTS
Characterization of tuberculous lesions and M. bovis colonization in vaccine efficacy trials.
In the comparative virulence study, the levels of M. bovis colonization, distributions of lesions, and severity of gross and microscopic lesions were similar (P > 0.05) for the M. bovis strain 95-1315- and 10-7428-challenged groups (Table 1) (37). Tuberculous lesions were not detected and M. bovis was not isolated from any of the animals in the noninfected control group. Similarly, infection of 6-month-old Holstein steers for samples used in the RNA-Seq study (Table 1) resulted in tuberculous lesions and M. bovis colonization typical of aerosol M. bovis infection, as described by Palmer et al. (19). Samples from these studies were used to evaluate IL-17A responses as a biomarker of M. bovis infection in cattle.
Vaccination of neonatal calves with BCG, M. bovis ΔRD1, or BCG mutants (i.e., the Δfdr8, ΔleuCD Δpks16, ΔmetA, and ΔmmaA4 mutants) resulted in significant (P < 0.05) protection against challenge with virulent M. bovis, as determined by assessment of gross pathology and M. bovis colonization (Table 1) (20), as well as microscopic staging of lesions (Table 3). For TB, organ weight is often associated with the degree of lesion severity. Lung-associated lymph nodes (i.e., tracheobronchial and mediastinal lymph nodes) from vaccine recipients weighed less (P < 0.05) than the respective lymph nodes from nonvaccinated animals (data not shown). Lesion severity and M. bovis colonization did not differ (P > 0.05) between vaccinated groups (i.e., BCG versus M. bovis ΔRD1 or BCG versus BCG mutants), compared within the two vaccine efficacy trials (Tables 1 and 3) (20). Samples from these studies were used to evaluate IL-17A responses as a correlate of protection.
TABLE 3.
Histological evaluation of lesion severity in 2014 vaccine efficacy study
| Treatment group | No. of granulomas (mean ± SEM)a |
||||
|---|---|---|---|---|---|
| Stage I | Stage II | Stage III | Stage IV | Total (stages I–IV) | |
| Nonvaccinated (n = 10) | 12.2 ± 2.5 | 2.9 ± 1.0 | 4.3 ± 1.2 | 4.0 ± 1.3 | 23.4 ± 4.4 |
| BCG-vaccinated (n = 9) | 1.9 ± 1.2b | 1.3 ± 0.9b | 0.1 ± 0.1b | 0 ± 0b | 3.3 ± 1.9b |
| BCG mutant-vaccinated (n = 10) | 1.1 ± 0.6b | 0.8 ± 0.5b | 0.8 ± 0.7b | 0.1 ± 0.1b | 2.8 ± 1.7b |
Microscopic tuberculous lesions were staged (stage I to IV) as described by Wangoo et al. (25). Data are presented as the mean ± SEM of the sum of the number of granulomas representing each stage observed in mediastinal and tracheobronchial lymph nodes and lung histological sections.
Value differs from that for nonvaccinated animals (same stage or total) (P < 0.05, ANOVA followed by Tukey's multiple-comparison test).
RNA-Seq analysis and gene expression of Th17-associated cytokines.
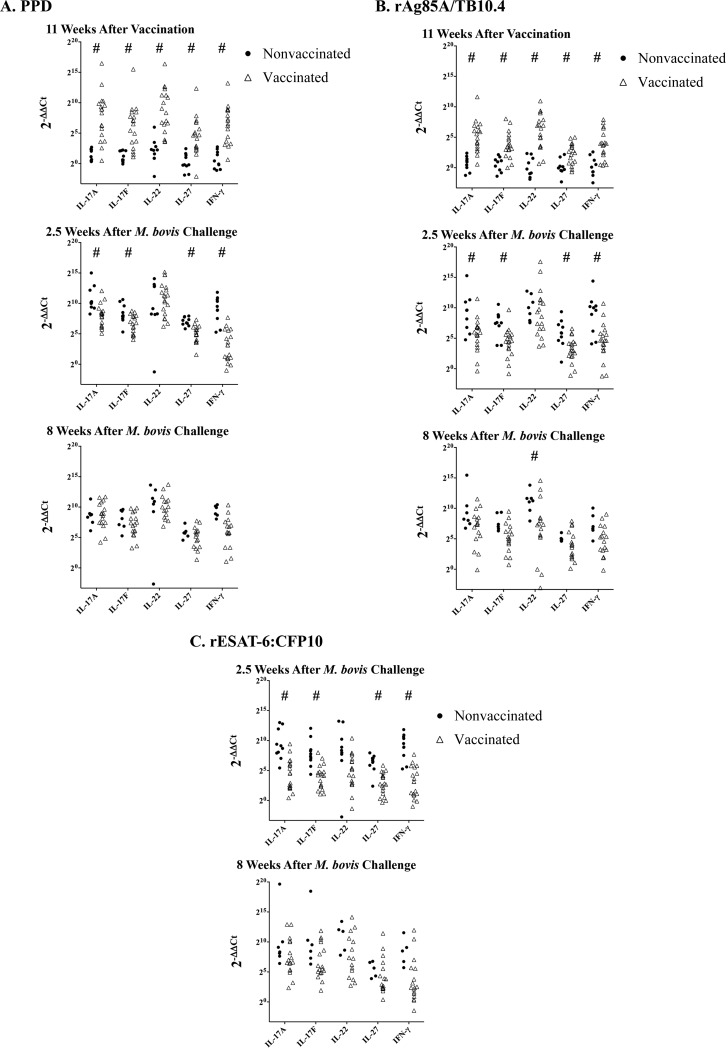
Before and after M. bovis challenge, PBMCs were stimulated with 1 μg/ml M. bovis PPD for 16 h and mRNA was isolated for whole-transcriptome sequencing. Compared to preinfection, 200 genes were >9-fold upregulated and 142 genes were >9-fold downregulated in response to PPD after M. bovis infection. Upregulated genes included numerous genes related to T cell function, especially Th1-related (i.e., IFN-γ, lymphotoxin α, TNF-α, IL-12Rβ2, and IL-12Rα) and Th17-related (i.e., IL-17A, IL-17F, IL-22, and IL-19) cytokine genes and IL-27, a cytokine that limits IL-17 responses through inhibition of the transcription factor retinoic acid receptor (RAR)-related orphan receptor γ (RORγ) (38). Upregulated cytokine and transcriptional regulatory network genes associated with in vitro differentiation of Th17 cells, as defined by Ciofani et al. (39), are presented in Table 4. As measured by RT-qPCR, BCG vaccination elicited Th17-associated cytokine responses to PPD (Fig. 1A) and rAg85A-rTB10.4 (Fig. 1B). At 2.5 weeks after M. bovis challenge, IL-17A, IL-17F, and IL-27 responses by nonvaccinated animals to PPD (Fig. 1A), rAg85A-rTB10 (Fig. 1B), and rESAT-6:CFP10 (Fig. 1C) exceeded (P < 0.05) the respective responses by vaccinated animals; however, IL-22 responses by nonvaccinated animals and vaccine recipients did not differ at that time point. At 8 weeks after challenge, IL-22 responses to M. bovis antigens by vaccinated and nonvaccinated animals did not differ, with one exception (i.e., responses to rAg85A-rTB10) (Fig. 1B). IL-17A gene expression and protein (ELISA) responses were correlated (Spearman's ρ = 0.60, with analysis including rESAT-6:CFP10, rAg85A-rTB10.4, and PPD stimulations; data not shown). IL-23p19 was also evaluated (data not shown); however, antigen-specific changes in gene expression were not detected for this cytokine, which is associated with expansion of Th17 responses.
TABLE 4.
Th17-associated genes upregulated >9-fold in response to M. bovis PPD after M. bovis infection
| Genea | Fold changeb | Rank |
|---|---|---|
| IL-22 | 91,019 | 3 |
| LIF | 6,162 | 10 |
| LTA | 2,652 | 14 |
| IL-19 | 2,149 | 17 |
| IL-17A | 1,229 | 20 |
| IL-17F | 328 | 29 |
| TNF | 89 | 47 |
| TBX21 | 56 | 66 |
| IL-27 | 51 | 71 |
| IRF4 | 20 | 120 |
| IFN-γc | 13,147 | 7 |
The Th17 global transcriptional regulatory network was defined by Ciofani et al. (39). LIF, leukemia inhibitory factor; LTA, lymphotoxin α; IRF4, interferon regulatory factor 4.
Genes were differentially expressed (upregulated >9-fold) in response to M. bovis PPD stimulation after versus before M. bovis infection.
IFN-γ was used for comparison.
FIG 1.
Antigen-specific gene expression of Th17-associated cytokines in response to vaccination and subsequent challenge with virulent M. bovis. Relative gene expression levels were calculated using the 2−ΔΔCT method, using nonstimulated cells as the calibrator and eukaryotic 18S rRNA as the endogenous control. Data are presented as individual animal responses to PPD (A), rAg85A-rTB10.4 (B), or rESAT-6:CFP10 (C) in nonvaccinated animals and vaccinated animals, at the indicated time points. Responses did not differ (P > 0.05) between animals vaccinated with BCG mutants versus BCG; thus, these two groups were pooled as vaccinated. #, responses differ between nonvaccinated animals and vaccinated animals for the respective cytokine (P < 0.05, Student's t test, using ΔΔCT values for comparisons).
IL-17A responses to M. bovis infection and comparisons with IFN-γ responses.
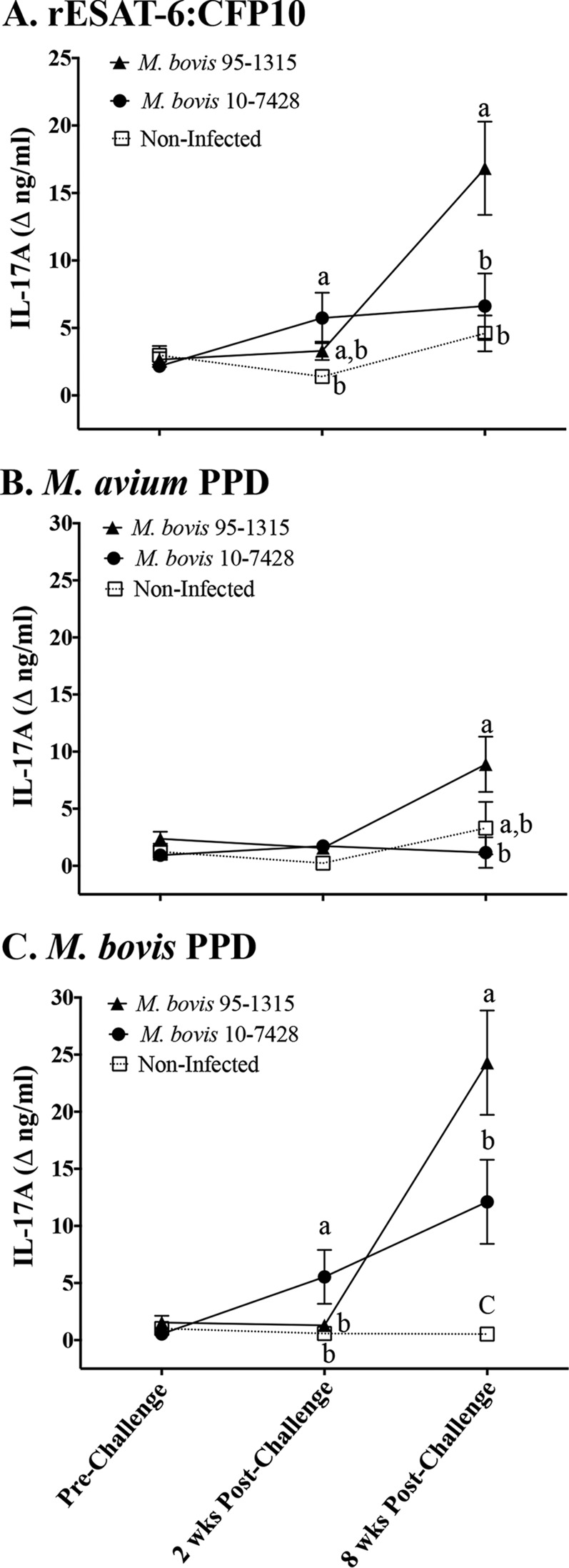
Experimental infection of cattle with M. bovis strain 95-1315 or 10-7428 elicited IL-17A responses to M. bovis antigens (Fig. 2). The response to M. bovis PPD by strain 10-7428-infected calves exceeded (P < 0.05) the response by 95-1315-infected calves at 2 weeks after challenge, whereas responses to rESAT-6:CFP10 and M. bovis PPD by 95-1315-infected calves exceeded (P < 0.05) the responses by 10-7428-infected calves at 8 weeks after challenge. As with IFN-γ responses (40), IL-17A responses to M. bovis PPD generally exceeded the responses to M. avium PPD (Fig. 2), and the responses to PPDs from two commercial sources were similar (M. avium PPD, CSL versus Lelystad, Spearman's ρ = 0.69; M. bovis PPD, CSL versus Lelystad, ρ = 0.82). Using a standard diagnostic algorithm of the response to M. bovis PPD minus the response to M. avium PPD, IL-17A responses were comparable to IFN-γ responses (Table 5). Considering all in vitro treatments (i.e., medium alone, PWM, rESAT-6:CFP10, M. bovis PPDs, and M. avium PPDs), IL-17A and IFN-γ responses were highly correlated (ρ = 0.74).
FIG 2.
IL-17A responses (protein) to M. bovis infection of cattle. Treatment groups included noninfected (n = 7), strain 95-1315-infected (white-tailed deer M. bovis isolate; n = 8), and strain 10-7428-infected (Holstein M. bovis isolate; n = 8) calves, with the experimental design described in Table 1. Whole blood was collected into heparinized tubes and stimulated with 1 μg/ml rESAT-6:CFP10 (A), 20 μg/ml M. avium PPD (Lelystad; Prionics Ag) (B), 20 μg/ml M. bovis PPD (Lelystad; Prionics Ag) (C), or medium alone (no stimulation) for 16 h at 39°C. Plasma was harvested for IL-17A analysis by ELISA (bovine IL-17A ELISA VetSet; Kingfisher Biotech). Data (mean ± SEM) are presented as the change in nanograms per milliliter (i.e., antigen stimulation minus medium alone) for each treatment group at the indicated time points relative to challenge. a to c, different letters indicate that responses differ for the given time point (P < 0.05, ANOVA followed by Tukey's multiple-comparison test).
TABLE 5.
Diagnostic capacity 8 weeks after M. bovis challenge (M. bovis PPD minus M. avium PPD)
| Treatment group | IL-17Aa |
IFN-γ |
||||||
|---|---|---|---|---|---|---|---|---|
| CSLb |
Lelystadb |
CSL |
Lelystad |
|||||
| ΔOD (mean ± SEM) |
No. positive | ΔOD (mean ± SEM) | No. positive | ΔOD (mean ± SEM) | No. positive | ΔOD (mean ± SEM) | No. positive | |
| No infection (n = 7) | 0.1 ± 0.04 | 2 | −0.1 ± 0.11 | 0 | 0.07 ± 0.01 | 1 | −0.1 ± 0.07 | 0 |
| M. bovis 95-1315 infection (n = 8) | 0.6 ± 0.07c | 8 | 0.7 ± 0.12c | 8 | 1.0 ± 0.12c | 7 | 1.7 ± 0.20c | 8 |
| M. bovis 10-7428 infection (n = 8) | 0.6 ± 0.15c | 8 | 0.7 ± 0.17c | 8 | 0.7 ± 0.30c | 8 | 1.4 ± 0.30c | 8 |
Data are presented as mean ± SEM of changes in optical densities (ODs) (i.e., M. bovis PPD minus M. avium PPD) and number positive (i.e., change in optical density of >0.1).
Sources of PPD.
Differs from the response by the noninfected group (P < 0.05, ANOVA followed by Tukey's multiple-comparison test).
IL-17A responses to vaccination and subsequent challenge with virulent M. bovis.
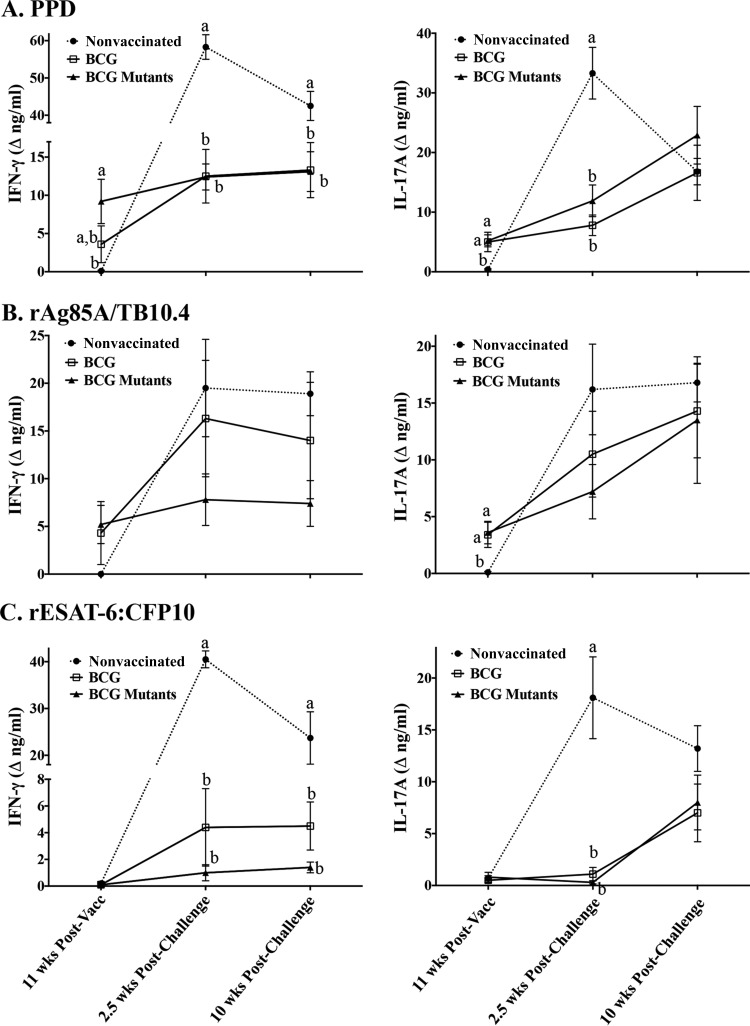
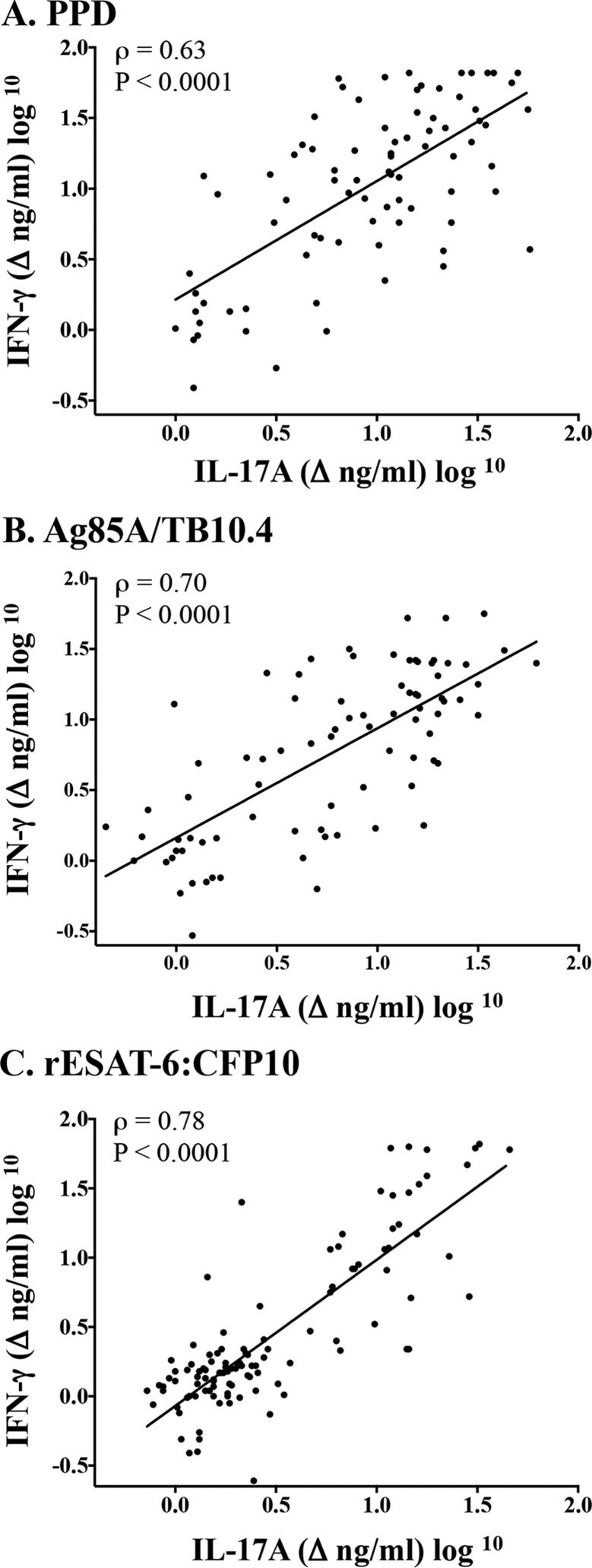
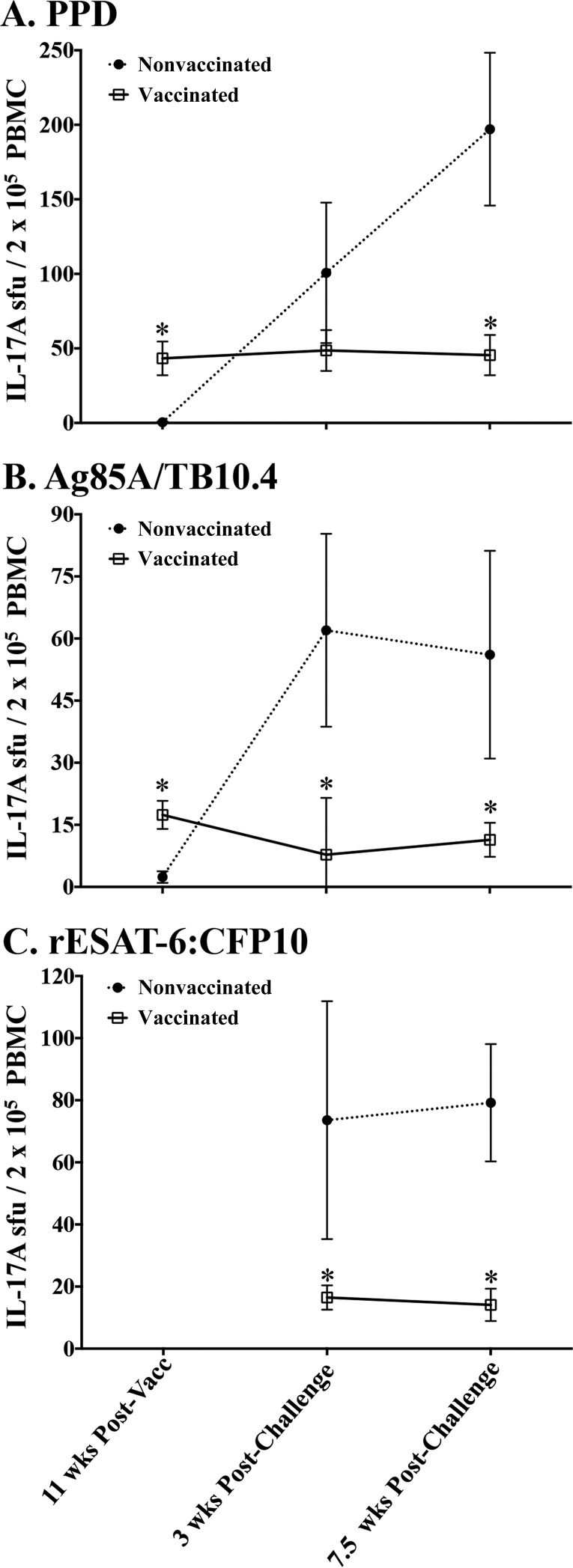
Vaccination with BCG or BCG mutants elicited IL-17A responses to PPD and rAg85A-rTB10.4 (Fig. 3). As expected, responses were not detected after stimulation with rESAT-6:CFP10 (i.e., antigens encoded within the RD1 region of virulent M. bovis and absent in BCG) prior to challenge with virulent M. bovis (Fig. 3C). At 2.5 weeks after M. bovis challenge, IL-17A and IFN-γ responses to rESAT-6:CFP10 and PPD increased dramatically in nonvaccinated calves, greatly exceeding (P < 0.05) the respective responses in vaccinated animals (Fig. 3). As in the comparative virulence study, IL-17A and IFN-γ responses were highly correlated (Spearman's ρ = 0.63 to 0.78, depending on the antigen) (Fig. 4). Analysis by ELISPOT assays corroborated the ELISA data, demonstrating both IL-17A responses to vaccination (i.e., to PPD and rAg85A-rTB10.4) and greater (P < 0.05) IL-17A responses to all 3 antigens by nonvaccinated animals at 3 and 7.5 weeks after challenge, compared to the respective responses by vaccinated animals (Fig. 5; see also Fig. S1 in the supplemental material). As in the BCG mutant study, IL-17A responses to PPD were elicited by vaccination with either M. bovis Ravenel ΔRD1 or BCG in the 2007 vaccine efficacy study (see Fig. S2 in the supplemental material). Also, at 3.5 weeks after challenge, responses to rESAT-6:CFP10 and PPD by nonvaccinated animals exceeded (P < 0.05) the respective responses by both BCG- and M. bovis Ravenel ΔRD1-vaccinated animals. It should be noted that levels of IL-17A were ∼5-fold lower in PBMC culture supernatants from the 2007 study, compared to stimulated plasma from whole blood from the 2014 study; however, direct comparison of samples collected at the same time point (i.e., in the comparative virulence study) demonstrated that the responses determined using the two culture methods were correlated (ρ = 0.6) (see Fig. S3 in the supplemental material). Together, these findings demonstrate that protective TB vaccines elicit IL-17A responses and these responses are dampened at 2.5 weeks after infection, compared to responses by nonvaccinated animals, likely coincident with reduced antigen loads (41) associated with protective vaccination.
FIG 3.
IFN-γ and IL-17A responses to vaccination and subsequent challenge with virulent M. bovis. Treatment groups included nonvaccinated animals (n = 10), BCG-vaccinated animals (n = 9), and animals vaccinated with BCG mutants (i.e., BCG Δfdr8, BCG ΔleuCD Δpks16, BCG ΔmetA, and BCG ΔmmaA4) (n = 10). The virulent M. bovis strain 10-7428 was administered by aerosol to all calves 3.5 months after vaccination, and calves were euthanized 4.5 months after challenge (Table 1). Whole blood was collected into heparinized tubes and stimulated with 20 μg/ml M. bovis PPD (Lelystad; Prionics Ag) (A), 1 μg/ml rAg85A-rTB10.4 (B), 1 μg/ml rESAT-6:CFP10 (C), or medium alone (no stimulation) for 16 h at 39°C. Plasma was harvested for IFN-γ (left) and IL-17A (right) analyses using commercial ELISA kits (Bovigam [Prionics Ag] and bovine IL-17A ELISA VetSet [Kingfisher Biotech]). Data (mean ± SEM) are presented as the change in nanograms per milliliter (i.e., antigen stimulation minus medium alone) for each treatment group at the indicated time points relative to vaccination (Vacc) or challenge. a to c, different letters indicate that responses differ for the given time point (P < 0.05, ANOVA followed by Tukey's multiple-comparison test).
FIG 4.
Correlation of IL-17A and IFN-γ responses. Treatment groups included nonvaccinated animals (n = 10), BCG-vaccinated animals (n = 9), and animals vaccinated with BCG mutants (i.e., BCG Δfdr8, BCG ΔleuCD Δpks16, BCG ΔmetA, and BCG ΔmmaA4) (n = 10). Virulent M. bovis strain 10-7428 was administered by aerosol to all calves 3.5 months after vaccination, and calves were euthanized 4.5 months after challenge (Table 1). Whole blood was collected into heparinized tubes from all calves 11 weeks after vaccination, 2.5 weeks after M. bovis challenge, and 10 weeks after M. bovis challenge and were stimulated with 20 μg/ml M. bovis PPD (Lelystad; Prionics Ag) (A), 1 μg/ml rAg85A-rTB10.4 (B), 1 μg/ml rESAT-6:CFP10 (C), or medium alone (no stimulation) for 16 h at 39°C, and plasma was harvested for IFN-γ and IL-17A analyses using ELISA kits (Bovigam [Prionics Ag] and bovine IL-17A ELISA VetSet [Kingfisher Biotech]). Data represent the changes in nanograms per milliliter (log10) (i.e., antigen stimulation minus medium alone) for IFN-γ versus IL-17A responses for each individual animal at each time point (n = 87). Prior to analysis and graphing, data were transformed for positive skewness with zero values using the following formula: new x = log10(x + 1).
FIG 5.
Dampening of IL-17A responses (ELISPOT assay) to M. bovis infection with prior BCG vaccination. Treatment groups included nonvaccinated animals (n = 10) and vaccinated animals (n = 19). The vaccinated group consisted of animals vaccinated with BCG (n = 9) or BCG mutants (i.e., BCG Δfdr8, BCG ΔleuCD Δpks16, BCG ΔmetA, and BCG ΔmmaA4; n = 10). Virulent M. bovis strain 10-7428 was administered by aerosol to all calves 3.5 months after vaccination, and calves were euthanized 4.5 months after M. bovis challenge (Table 1). For IL-17A ELISPOT analysis, PBMCs (2 × 105 PBMCs/well) were stimulated with 10 μg/ml M. bovis PPD (A), 3 μg/ml rAg85A-rTB10.4 (B), or 3 μg/ml rESAT-6:CFP10 (C) for 18 h prior to spot development and counting, as described in Materials and Methods. Results (mean ± SEM) are expressed as spot-forming units (sfu) per 2 × 105 cells for each treatment group at the indicated time points relative to vaccination (Vacc) or challenge. *, response differs from that for nonvaccinated animals at that time point (P < 0.05, as determined by ANOVA followed by Tukey's multiple-comparison test).
Correlations with protection.
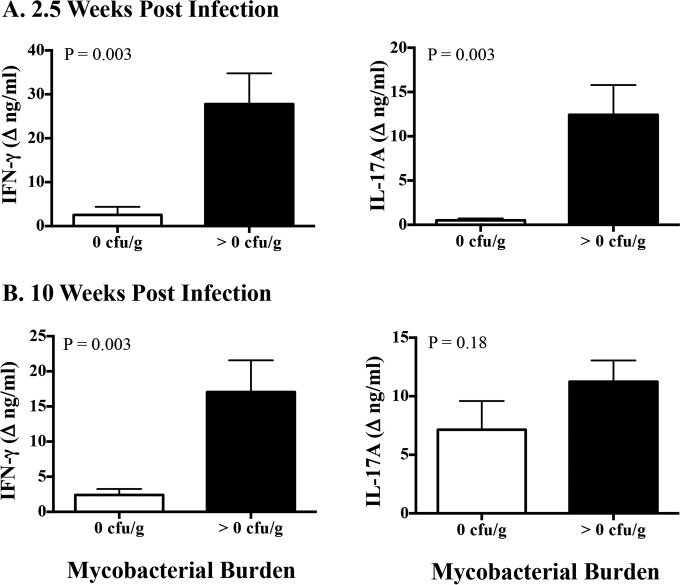
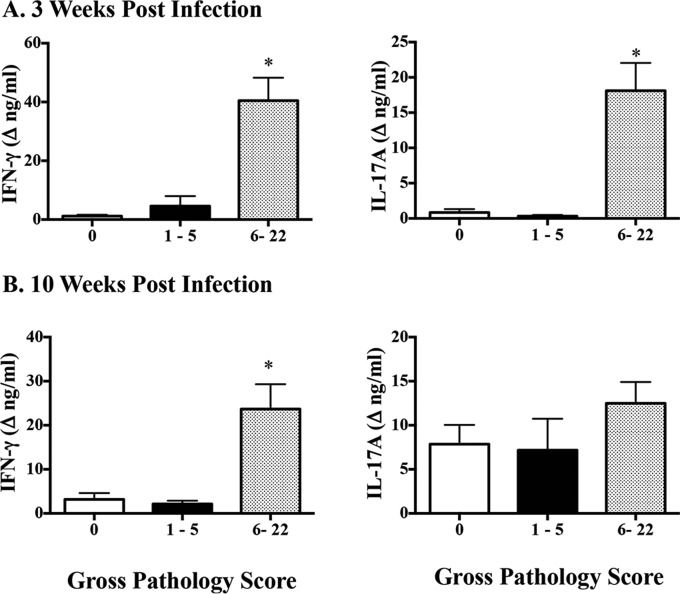
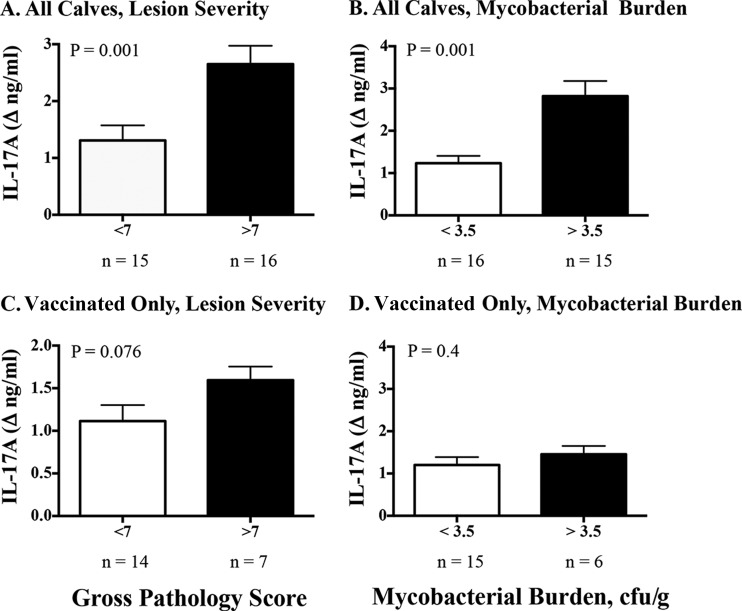
BCG, BCG mutants, and M. bovis ΔRD1 vaccines elicited IL-17A responses to PPD and/or rAG85A-rTB10.4, as detected 11 to 12 weeks after vaccination and prior to challenge (Fig. 3 and 5; see also Fig. S1 and S2 in the supplemental material). In the 2014 efficacy study, vaccination with BCG mutants and BCG afforded exquisite protection, with 2- to 3.5-log10 reductions in mycobacterial colonization and greatly reduced lesion severity (Table 1) at necropsy 4.5 months after challenge. Lower (P < 0.05) IL-17A and IFN-γ responses to PPD and rESAT-6:CFP10 at 2.5 weeks after infection were associated with no detectable M. bovis by quantitative culture (Fig. 6A) and low pathology scores (Fig. 7A) at necropsy 4.5 months after challenge. Significantly lower (P < 0.05) IFN-γ responses, but not IL-17A responses, at 10 weeks after infection were also associated with no detectable M. bovis by quantitative culture (Fig. 6B) and low pathology scores (Fig. 7B) at necropsy 4.5 months after challenge. In general, lesion severity and mycobacterial burdens in both the vaccinated and nonvaccinated groups were greater in the 2007 efficacy study than in the 2014 efficacy study, possibly due to a slightly higher M. bovis challenge dose (Table 1). In the 2007 vaccine efficacy study, greater (P < 0.05) IL-17A responses at 8 weeks after M. bovis challenge were positively associated with lesion severity (Fig. 8A) and mycobacterial burdens (Fig. 8B) determined at necropsy 4.5 months after challenge. Furthermore, greater (P < 0.05) IL-17A responses were associated with increased (P = 0.076) lesion severity among vaccinated animals (Fig. 8C) but not mycobacterial burdens (Fig. 8D) determined at necropsy 4.5 months after challenge.
FIG 6.
Association of ESAT-6:CFP10-specific IFN-γ and IL-17A responses with mycobacterial burdens in the 2014 vaccine efficacy study. Treatment groups included nonvaccinated animals, BCG-vaccinated animals, and animals vaccinated with BCG mutants (i.e., BCG Δfdr8, BCG ΔleuCD Δpks16, BCG ΔmetA, and BCG ΔmmaA4). Virulent M. bovis strain 10-7428 was administered by aerosol to all calves 3.5 months after vaccination, and calves were euthanized 4.5 months after M. bovis challenge (Table 1). Whole blood from all calves was collected into heparinized tubes at 2.5 and 10 weeks after challenge and stimulated with 1 μg/ml rESAT-6:CFP10 or medium alone (no stimulation) for 16 h at 39°C, and plasma was harvested for IFN-γ and IL-17A analyses using commercial ELISA kits (Bovigam [Prionics Ag] and bovine IL-17A ELISA VetSet [Kingfisher Biotech]). Mycobacterial burdens were determined by serial dilution culture of entire tracheobronchial lymph node homogenates and are presented as culture-forming units per gram of tissue. Groups were split based on mycobacterial burdens of 0 CFU/g (n = 14) or >0 CFU/g (n = 15). All 14 animals in the 0-CFU/g group were vaccinated animals. In the >0-CFU/g group, 5/15 animals were vaccinated animals and 10/15 were nonvaccinated animals. Data (mean ± SEM) are presented as IFN-γ (left) and IL-17A (right) responses (changes in nanograms per milliliter, i.e., antigen stimulation minus medium alone) to rESAT-6:CFP10 at 3 weeks after M. bovis challenge (A) and 10 weeks after M. bovis challenge (B), as related to mycobacterial burdens determined at necropsy 4.5 months after challenge. Similar results were obtained for responses to M. bovis PPD (data not shown). Student's t test P values are provided in the upper left corner of each graph.
FIG 7.
Association of ESAT-6:CFP10-specific IFN-γ and IL-17A responses with lesion severity (i.e., gross pathology scores) in the 2014 vaccine efficacy study. Treatment groups included nonvaccinated animals, BCG-vaccinated animals, and animals vaccinated with BCG mutants (i.e., BCG Δfdr8, BCG ΔleuCD Δpks16, BCG ΔmetA, and BCG ΔmmaA4). Virulent M. bovis strain 10-7428 was administered by aerosol to all calves 3.5 months after vaccination, and calves were euthanized 4.5 months after M. bovis challenge (Table 1). Whole blood from all calves was collected into heparinized tubes at 2.5 and 10 weeks after challenge and stimulated with 1 μg/ml rESAT-6:CFP10 or medium alone (no stimulation) for 16 h at 39°C, and plasma was harvested for IFN-γ and IL-17A analyses using ELISA kits (Bovigam [Prionics Ag] and bovine IL-17A ELISA VetSet [Kingfisher Biotech]). The lesion severity for lungs and lymph nodes (i.e., mediastinal and tracheobronchial lymph nodes) was determined using a semiquantitative scoring system adapted from that described by Vordermeier et al. (24). Groups were split based on gross pathology scores of 0 (n = 12; all vaccinated animals), 1 to 5 (n = 7; all vaccinated animals), or 6 to 22 (n = 10; all nonvaccinated animals). Data (mean ± SEM) are presented as IFN-γ (left) and IL-17A (right) responses (changes in nanograms per milliliter, i.e., antigen stimulation minus medium alone) to rESAT-6:CFP10 at 3 weeks after M. bovis challenge (A) and 10 weeks after M. bovis challenge (B), as related to gross pathology scores determined at necropsy 4.5 months after challenge. Similar results were obtained for responses to M. bovis PPD (data not shown). *, response differs from the other responses in the graph (P < 0.05, ANOVA followed by Tukey's multiple-comparison test).
FIG 8.
Correlation of ESAT-6:CFP10-specific IL-17A responses at 8 weeks after M. bovis challenge with lesion severity and mycobacterial burden in the 2007 vaccine efficacy study. Treatment groups included nonvaccinated, BCG-vaccinated, and M. bovis ΔRD1-vaccinated animals. Virulent M. bovis strain 95-1315 was administered by aerosol to all calves 3 months after vaccination, and calves were euthanized 4.5 months after M. bovis challenge (Table 1). Eight weeks after M. bovis challenge, PBMCs (2 × 105 PBMCs/well) were stimulated with 1 μg/ml rESAT-6:CFP10 or medium alone at 39°C for 16 h, and supernatants were harvested for IL-17A analysis by ELISA (bovine IL-17A ELISA VetSet; Kingfisher Biotech). For evaluation of gross pathology, lungs and lymph nodes (mediastinal and tracheobronchial) were evaluated using a semiquantitative scoring system (24). Mycobacterial burdens were determined by culture of a series of dilutions of entire tracheobronchial lymph node homogenates and are presented as CFU per gram of tissue (26). Data (mean ± SEM) are presented as changes in nanograms per milliliter (i.e., antigen stimulation minus medium alone) for each treatment group based on gross pathology scores (A and C) or mycobacterial burdens (B and D). Student's t test P values are provided in the upper left corner of each graph.
DISCUSSION
Significant IL-17 responses are elicited by M. tuberculosis infection of mice (42) and humans (43, 44), as well as M. bovis infection of cattle (7, 11). With M. tuberculosis infection of mice, early expression of IL-17 in response to vaccination is required for the rapid accumulation of protective memory cells in the lungs (45). Responding quickly upon aerosol challenge with M. tuberculosis, IL-17-producing cells recruit other effector cells that limit pathogen growth (42). PPD-specific IL-17 responses to BCG vaccination are also associated with reduced disease burdens upon subsequent M. tuberculosis infection of cynomolgus macaques (46). With BCG plus virus-vectored Ag85A vaccination of cattle, vaccine-elicited IL-17 mRNA responses to Ag85A stimulation at 10 weeks after vaccination correlated with reduced TB-associated pathology (11). Rizzi et al. (15) also demonstrated that IL-17 mRNA responses in cattle vaccinated with a BCG strain overexpressing Ag85B correlated with reduced lesion severity after experimental M. bovis infection. Using RNA-Seq analysis followed by RT-qPCR analysis, Bhuju et al. (47) demonstrated that IL-22 responses to PPD after vaccination correlated with protection in cattle. The present findings support and extend those prior studies, demonstrating that protective bovine TB vaccines elicited IL-17F and IL-27 mRNA responses, IL-17A, IL-17F, and IL-27 responses at 2.5 weeks after M. bovis infection were reduced in vaccinated animals versus nonvaccinated animals, IL-17A protein levels measured by ELISAs or ELISPOT assays correlated with IL-17A mRNA responses in both the kinetics and levels of responses, and higher IL-17A and IFN-γ levels at 2.5 weeks after M. bovis infection correlated with increased lesion severity and mycobacterial burdens upon postmortem inspection 4 months after challenge. Evaluating a M. bovis Δmce2 deletion mutant, Blanco et al. (14) also demonstrated reduced IL-17 mRNA responses in vaccinated animals versus nonvaccinated animals after M. bovis challenge; however, the responses differed 160 days after challenge, and time points earlier than 80 days postchallenge were not evaluated. In the present study, diminished IL-17-related cytokine and IFN-γ responses early after infection likely result from reduced antigen loads at the site of aerosol-delivered M. bovis infection, due to protective responses initiated by vaccination; however, further studies are required to confirm this notion. Also, as with IFN-γ and delayed-type hypersensitivity (DTH) responses (48, 49), it is likely that many TB vaccines will induce IL-17 responses but not all IL-17-inducing vaccines will be protective.
In prior studies, we demonstrated, with samples from a limited number of animals, that CD4+ and γδ+ T cells from M. bovis-infected cattle produced IL-17A in response to M. bovis PPD or rESAT-6:CFP10 (36). PPD-specific IL-17 mRNA responses at 60 and 90 days after experimental M. bovis infection correlated with the presence of gross tuberculous lesions at necropsy 4 months after challenge (7). Using laser capture microdissection followed by qPCR, Aranday-Cortes et al. demonstrated increased IL-17A and IL-22 expression within tuberculous granulomas versus nonaffected tissues from experimentally infected cattle, particularly in more advanced lesions (50). In the present study, M. bovis infection also elicited IL-17A protein responses that correlated with infection, similar to IFN-γ responses (Table 5). Infection also elicited IL-17F, IL-22, and IL-27 responses. IL-27 is a known inhibitor of Th17 responses in mice and humans (51, 52); however, both IL-27 (44) and IL-17 (41, 43) are associated with active disease in M. tuberculosis infections in humans. Thus, perhaps it is not too surprising that IL-27 and Th17 cytokine responses followed similar kinetics. Antigen-specific IL-23p19 mRNA expression was not detectable in response to either infection or vaccination, possibly due to the duration of culture, poorly represented dendritic cell and macrophage populations within PBMCs, or the lack of “danger signals” (e.g., Toll-like receptors or nucleotide-binding oligomerization domain-like signals) within the antigen preparations. Similarly, Blanco et al. (7) did not detect IL-12p35 expression with similar protocols. In contrast to IL-17A, IL-17F, and IL-27, IL-22 responses were not different between vaccinated animals and nonvaccinated animals at 2.5 weeks after M. bovis infection, suggesting that this cytokine is less affected by antigen loads or that vaccine-elicited IL-22 responses are more durable than other Th17-associated cytokine responses. Together, these findings indicate that IL-17A and potentially other Th17-associated cytokines, such as IL-17F, IL-22, and IL-27, may prove useful as biomarkers for M. bovis infections in cattle.
The Th17 lineage is known for its plasticity and instability, that is, IL-17 expression may cease over time (53). Th17 cells can start expressing cytokines typical of other T helper subsets as a result of a nonresolving immune response (switch to a Th1 phenotype), chronic inflammation or autoimmunity (switch to a T regulatory 1 [Tr1] phenotype), Nippostrongylus brasiliensis infection (switch to a Th2 or Tr1 phenotype), or Staphylococcus aureus infection (switch to a Tr1 phenotype), thereby providing a mechanism to contribute to resolution of inflammation (53). With aerosol BCG infection of mice, IL-17A produced by Vγ4+ and Vγ6+ γδ+ T cells is necessary for appropriate maturation of granulomas (54), and early IL-17 produced by γδ+ T cells occurs prior to αβ+ T cell priming, thus biasing the ensuing adaptive response (55). However, excessive IL-17 responses may be detrimental; repeated BCG vaccination of M. tuberculosis-infected mice exacerbated inflammation due to infection, and this exaggerated response was not detected in mice treated with anti-IL-17 blocking antibody or in IL-23p19-deficient mice, demonstrating the IL-17 dependence of the damaging response (56). Also, treatment regimens that block IL-17 responses (e.g., RORγ inhibitors to promote a more favorable IL-17/IFN-γ balance via inhibition of IL-17 production) are being considered for inclusion in treatment regimens for M. tuberculosis infections in humans (57). Together, these findings suggest that the timing and amounts of IL-17 production in response to TB infections are critical for the balance between responses that support control of the bacilli and detrimental inflammatory responses. Given the plasticity of the responses, Th17 cells may transdifferentiate into phenotypes not expressing IL-17 or Th17-associated cytokines. In future studies, it will be critical to evaluate the responses at sites of M. bovis infection and over the course of infection, to account for local environmental factors associated with chronic inflammation that affect the plasticity of Th17 responses. Studies utilizing mycobacterial antigens or live bacteria in subcutaneous biopolymers as in vivo models of granuloma formation and maturation (58) may provide additional insights into the kinetics of Th17 responses. In conclusion, the present findings support the use of IL-17-associated cytokines as biomarkers of infection and protection in the immune responses to bovine tuberculosis.
Supplementary Material
ACKNOWLEDGMENTS
Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture.
Funding Statement
This project was supported by Agriculture and Food Research Initiative Competitive Grant no. 2011-67015-30736 from the USDA National Institute of Food and Agriculture to W. Ray Waters and Mitchell V. Palmer. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/CVI.00637-15.
REFERENCES
- 1.Smith NH, Hewinson RG, Kremer K, Brosch R, Gordon SV. 2009. Myths and misconceptions: the origin and evolution of Mycobacterium tuberculosis. Nat Rev Microbiol 7:537–544. doi: 10.1038/nrmicro2165. [DOI] [PubMed] [Google Scholar]
- 2.Müller B, Dürr S, Alonso S, Hattendorf J, Laisse CJ, Parsons SD, van Helden PD, Zinsstag J. 2013. Zoonotic Mycobacterium bovis-induced tuberculosis in humans. Emerg Infect Dis 19:899–908. doi: 10.3201/eid1906.120543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Palmer MV. 2013. Mycobacterium bovis: characteristics of wildlife reservoir hosts. Transbound Emerg Dis 60(Suppl 1):S1–S13. [DOI] [PubMed] [Google Scholar]
- 4.Schiller I, Oesch B, Vordermeier HM, Palmer MV, Harris BN, Orloski KA, Buddle BM, Thacker TC, Lyashchenko KP, Waters WR. 2010. Bovine tuberculosis: a review of current and emerging diagnostic techniques in view of their relevance for disease control and eradication. Transbound Emerg Dis 57:205–220. [DOI] [PubMed] [Google Scholar]
- 5.Walzl G, Ronacher K, Hanekom W, Scriba TJ, Zumla A. 2011. Immunological biomarkers of tuberculosis. Nat Rev Immunol 11:343–354. doi: 10.1038/nri2960. [DOI] [PubMed] [Google Scholar]
- 6.Salgame P, Geadas C, Collins L, Jones-López E, Ellner JJ. 2015. Latent tuberculosis infection: revisiting and revising concepts. Tuberculosis (Edinb) 95:373–384. doi: 10.1016/j.tube.2015.04.003. [DOI] [PubMed] [Google Scholar]
- 7.Blanco FC, Bianco MV, Meikle V, Garbaccio S, Vagnoni L, Forrellad M, Klepp LI, Cataldi AA, Bigi F. 2011. Increased IL-17 expression is associated with pathology in a bovine model of tuberculosis. Tuberculosis (Edinb) 91:57–63. doi: 10.1016/j.tube.2010.11.007. [DOI] [PubMed] [Google Scholar]
- 8.Goosen WJ, Cooper D, Miller MA, van Helden PD, Parsons SD. 2015. IP-10 is a sensitive biomarker of antigen recognition in whole blood stimulation assays used for the diagnosis of Mycobacterium bovis infection in African buffaloes (Syncerus caffer). Clin Vaccine Immunol 22:974–978. doi: 10.1128/CVI.00324-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jones GJ, Pirson C, Hewinson RG, Vordermeier HM. 2010. Simultaneous measurement of antigen-stimulated interleukin-1β and gamma interferon production enhances test sensitivity for the detection of Mycobacterium bovis infection in cattle. Clin Vaccine Immunol 17:1946–1951. doi: 10.1128/CVI.00377-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rhodes SG, McKinna LC, Steinbach S, Dean GS, Villarreal-Ramos B, Whelan AO, Pirson C, Jones GJ, Clifford D, Vordermeier HM. 2014. Use of antigen-specific interleukin-2 to differentiate between cattle vaccinated with Mycobacterium bovis BCG and cattle infected with M. bovis. Clin Vaccine Immunol 21:39–45. doi: 10.1128/CVI.00522-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vordermeier HM, Villarreal-Ramos B, Cockle PJ, McAulay M, Rhodes SG, Thacker T, Gilbert SC, McShane H, Hill AV, Xing Z, Hewinson RG. 2009. Viral booster vaccines improve Mycobacterium bovis BCG-induced protection against bovine tuberculosis. Infect Immun 77:3364–3373. doi: 10.1128/IAI.00287-09 (Erratum, 79:2134, 2011.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Waters WR, Palmer MV, Whipple DL, Carlson MP, Nonnecke BJ. 2003. Diagnostic implications of antigen-induced IFN-γ, nitric oxide, and TNF-α production by blood mononuclear cells from Mycobacterium bovis-infected cattle. Clin Diagn Lab Immunol 10:960–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Waters WR, Thacker TC, Nonnecke BJ, Palmer MV, Schiller I, Oesch B, Vordermeier HM, Silva E, Estes DM. 2012. Evaluation of gamma interferon (IFN-γ)-induced protein 10 responses for detection of cattle infected with Mycobacterium bovis: comparisons to IFN-γ responses. Clin Vaccine Immunol 19:346–351. doi: 10.1128/CVI.05657-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blanco FC, Bianco MV, Garbaccio S, Meikle V, Gravisaco MJ, Montenegro V, Alfonseca E, Singh M, Barandiaran S, Canal A, Vagnoni L, Buddle BM, Bigi F, Cataldi A. 2013. Mycobacterium bovis Δmce2 double deletion mutant protects cattle against challenge with virulent M. bovis. Tuberculosis (Edinb) 93:363–372. doi: 10.1016/j.tube.2013.02.004. [DOI] [PubMed] [Google Scholar]
- 15.Rizzi C, Bianco MV, Blanco FC, Soria M, Gravisaco MJ, Montenegro V, Vagnoni L, Buddle B, Garbaccio S, Delgado F, Leal KS, Cataldi AA, Dellagostin OA, Bigi F. 2012. Vaccination with a BCG strain overexpressing Ag85B protects cattle against Mycobacterium bovis challenge. PLoS One 7:e51396. doi: 10.1371/journal.pone.0051396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schmitt SM, Fitzgerald SD, Cooley TM, Bruning-Fann CS, Sullivan L, Berry D, Carlson T, Minnis RB, Payeur JB, Sikarskie J. 1997. Bovine tuberculosis in free-ranging white-tailed deer from Michigan. J Wildl Dis 33:749–758. doi: 10.7589/0090-3558-33.4.749. [DOI] [PubMed] [Google Scholar]
- 17.Francisco TI, Orloski KA, Roberts NJ. 2014. Investigation of a Mycobacterium bovis outbreak in cattle at a Colorado dairy in 2010. J Am Vet Med Assoc 244:805–812. doi: 10.2460/javma.244.7.805. [DOI] [PubMed] [Google Scholar]
- 18.Larsen MH, Biermann K, Jacobs WR. 2007. Laboratory maintenance of Mycobacterium tuberculosis. Curr Protoc Microbiol 10:Unit 10A.1. [DOI] [PubMed] [Google Scholar]
- 19.Palmer MV, Waters WR, Whipple DL. 2002. Aerosol delivery of virulent Mycobacterium bovis to cattle. Tuberculosis (Edinb) 82:275–282. doi: 10.1054/tube.2002.0341. [DOI] [PubMed] [Google Scholar]
- 20.Waters WR, Palmer MV, Nonnecke BJ, Thacker TC, Scherer CF, Estes DM, Hewinson RG, Vordermeier HM, Barnes SW, Federe GC, Walker JR, Glynne RJ, Hsu T, Weinrick B, Biermann K, Larsen MH, Jacobs WR Jr. 2009. Efficacy and immunogenicity of Mycobacterium bovis ΔRD1 against aerosol M. bovis infection in neonatal calves. Vaccine 27:1201–1209. doi: 10.1016/j.vaccine.2008.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dao DN, Sweeney K, Hsu T, Gurcha SS, Nascimento IP, Roshevsky D, Besra GS, Chan J, Porcelli SA, Jacobs WR. 2008. Mycolic acid modification by the mmaA4 gene of M. tuberculosis modulates IL-12 production. PLoS Pathog 4:e1000081. doi: 10.1371/journal.ppat.1000081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Berney M, Berney-Meyer L, Wong KW, Chen B, Chen M, Kim J, Wang J, Harris D, Parkhill J, Chan J, Wang F, Jacobs WR Jr. 2015. Essential roles of methionine and S-adenosylmethionine in the autarkic lifestyle of Mycobacterium tuberculosis. Proc Natl Acad Sci U S A 112:10008–100013. doi: 10.1073/pnas.1513033112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Panas MW, Sixsmith JD, White K, Korioth-Schmitz B, Shields ST, Moy BT, Lee S, Schmitz JE, Jacobs WR Jr, Porcelli SA, Haynes BF, Letvin NL, Gillard GO. 2014. Gene deletions in Mycobacterium bovis BCG stimulate increased CD8+ T cell responses. Infect Immun 82:5317–5326. doi: 10.1128/IAI.02100-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vordermeier HM, Chambers MA, Cockle PJ, Whelan AO, Simmons J, Hewinson RG. 2002. Correlation of ESAT-6-specific gamma interferon production with pathology in cattle following Mycobacterium bovis BCG vaccination against experimental bovine tuberculosis. Infect Immun 70:3026–3032. doi: 10.1128/IAI.70.6.3026-3032.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wangoo A, Johnson L, Gough J, Ackbar R, Inglut S, Hicks D, Spencer Y, Hewinson G, Vordermeier M. 2005. Advanced granulomatous lesions in Mycobacterium bovis-infected cattle are associated with increased expression of type I procollagen, γδ (WC1+) T cells and CD 68+ cells. J Comp Pathol 133:223–234. doi: 10.1016/j.jcpa.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 26.Waters WR, Palmer MV, Nonnecke BJ, Thacker TC, Scherer CF, Estes DM, Jacobs WR Jr, Glatman-Freedman A, Larsen MH. 2007. Failure of a Mycobacterium tuberculosis ΔRD1 ΔpanCD double deletion mutant in a neonatal aerosol M. bovis challenge model: comparisons to responses elicited by M. bovis bacilli Calmette Guerin. Vaccine 25:7832–7840. doi: 10.1016/j.vaccine.2007.08.029. [DOI] [PubMed] [Google Scholar]
- 27.Thacker TC, Harris B, Palmer MV, Waters WR. 2011. Improved specificity for detection of Mycobacterium bovis in fresh tissues using IS6110 real-time PCR. BMC Vet Res 7:50. doi: 10.1186/1746-6148-7-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aabye MG, Ravn P, Johansen IS, Eugen-Olsen J, Ruhwald M. 2011. Incubation of whole blood at 39°C augments gamma interferon (IFN-γ)-induced protein 10 and IFN-γ responses to Mycobacterium tuberculosis antigens. Clin Vaccine Immunol 18:1150–1156. doi: 10.1128/CVI.00051-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thacker TC, Palmer MV, Waters WR. 2007. Associations between cytokine gene expression and pathology in Mycobacterium bovis infected cattle. Vet Immunol Immunopathol 119:204–213. doi: 10.1016/j.vetimm.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 30.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 31.Schmieder R, Edwards R. 2011. Quality control and preprocessing of metagenomic datasets. Bioinformatics 27:863–864. doi: 10.1093/bioinformatics/btr026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Anders S, Pyl PT, Huber W. 2015. HTSeq: a Python framework to work with high-throughput sequencing data. Bioinformatics 31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Robinson MD, Smyth GK. 2007. Moderated statistical tests for assessing differences in tag abundance. Bioinformatics 23:2881–2887. doi: 10.1093/bioinformatics/btm453. [DOI] [PubMed] [Google Scholar]
- 34.Robinson MD, Smyth GK. 2008. Small-sample estimation of negative binomial dispersion, with applications to SAGE data. Biostatistics 9:321–332. [DOI] [PubMed] [Google Scholar]
- 35.Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McGill JL, Sacco RE, Baldwin CL, Telfer JC, Palmer MV, Waters WR. 2014. Specific recognition of mycobacterial protein and peptide antigens by γδ T cell subsets following infection with virulent Mycobacterium bovis. J Immunol 192:2756–2769. doi: 10.4049/jimmunol.1302567. [DOI] [PubMed] [Google Scholar]
- 37.Waters WR, Thacker TC, Nelson JT, DiCarlo DM, Maggioli MF, Greenwald R, Esfandiari J, Lyashchenko KP, Palmer MV. 2014. Virulence of two strains of Mycobacterium bovis in cattle following aerosol infection. J Comp Pathol 151:410–419. doi: 10.1016/j.jcpa.2014.08.007. [DOI] [PubMed] [Google Scholar]
- 38.Diveu C, McGeachy MJ, Boniface K, Stumhofer JS, Sathe M, Joyce-Shaikh B, Chen Y, Tato CM, McClanahan TK, de Waal Malefyt R, Hunter CA, Cua DJ, Kastelein RA. 2009. IL-27 blocks RORc expression to inhibit lineage commitment of Th17 cells. J Immunol 182:5748–5756. doi: 10.4049/jimmunol.0801162. [DOI] [PubMed] [Google Scholar]
- 39.Ciofani M, Madar A, Galan C, Sellars M, Mace K, Pauli F, Agarwal A, Huang W, Parkurst CN, Muratet M, Newberry KM, Meadows S, Greenfield A, Yang Y, Jain P, Kirigin FK, Birchmeier C, Wagner EF, Murphy KM, Myers RM, Bonneau R, Littman DR. 2012. A validated regulatory network for Th17 cell specification. Cell 151:289–303. doi: 10.1016/j.cell.2012.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bass KE, Nonnecke BJ, Palmer MV, Thacker TC, Hardegger R, Schroeder B, Raeber AJ, Waters WR. 2013. Clinical and diagnostic developments of a gamma interferon release assay for use in bovine tuberculosis control programs. Clin Vaccine Immunol 20:1827–1835. doi: 10.1128/CVI.00519-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Basile JI, Geffner LJ, Romero MM, Balboa L, Sabio Y, García C, Ritacco V, García A, Cuffré M, Abbate E, López B, Barrera L, Ambroggi M, Alemán M, Sasiain MC, de la Barrera SS. 2011. Outbreaks of Mycobacterium tuberculosis MDR strains induce high IL-17 T-cell response in patients with MDR tuberculosis that is closely associated with high antigen load. J Infect Dis 204:1054–1064. doi: 10.1093/infdis/jir460. [DOI] [PubMed] [Google Scholar]
- 42.Khader SA, Cooper AM. 2008. IL-23 and IL-17 in tuberculosis. Cytokine 41:79–83. doi: 10.1016/j.cyto.2007.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jurado JO, Pasquinelli V, Alvarez IB, Peña D, Rovetta AI, Tateosian NL, Romeo HE, Musella RM, Palmero D, Chuluyán HE, García VE. 2012. IL-17 and IFN-γ expression in lymphocytes from patients with active tuberculosis correlates with the severity of the disease. J Leukoc Biol 91:991–1002. doi: 10.1189/jlb.1211619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Torrado E, Fountain JJ, Liao M, Tighe M, Reiley WW, Lai RP, Meintjes G, Pearl JE, Chen X, Zak DE, Thompson EG, Aderem A, Ghilardi N, Solache A, McKinstry KK, Strutt TM, Wilkinson RJ, Swain SL, Cooper AM. 2015. Interleukin 27R regulates CD4+ T cell phenotype and impacts protective immunity during Mycobacterium tuberculosis infection. J Exp Med 212:1449–1463. doi: 10.1084/jem.20141520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Khader SA, Bell GK, Pearl JE, Fountain JJ, Rangel-Moreno J, Cilley GE, Shen F, Eaton SM, Gaffen SL, Swain SL, Locksley RM, Haynes L, Randall TD, Cooper AM. 2007. IL-23 and IL-17 in the establishment of protective pulmonary CD4+ T cell responses after vaccination and during Mycobacterium tuberculosis challenge. Nat Immunol 8:369–377. doi: 10.1038/ni1449. [DOI] [PubMed] [Google Scholar]
- 46.Wareham AS, Tree JA, Marsh PD, Butcher PD, Dennis M, Sharpe SA. 2014. Evidence for a role for interleukin-17, Th17 cells and iron homeostasis in protective immunity against tuberculosis in cynomolgus macaques. PLoS One 9:e88149. doi: 10.1371/journal.pone.0088149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bhuju S, Aranday-Cortes E, Villarreal-Ramos B, Xing Z, Singh M, Vordermeier HM. 2012. Global gene transcriptome analysis in vaccinated cattle revealed a dominant role of IL-22 for protection against bovine tuberculosis. PLoS Pathog 8:e1003077. doi: 10.1371/journal.ppat.1003077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Black GF, Weir RE, Floyd S, Bliss L, Warndorff DK, Crampin AC, Ngwira B, Sichali L, Nazareth B, Blackwell JM, Branson K, Chaguluka SD, Donovan L, Jarman E, King E, Fine PE, Dockrell HM. 2002. CG-induced increase in interferon-gamma response to mycobacterial antigens and efficacy of BCG vaccination in Malawi and the UK: two randomised controlled studies. Lancet 359:1393–1401. doi: 10.1016/S0140-6736(02)08353-8. [DOI] [PubMed] [Google Scholar]
- 49.Waters WR, Palmer MV, Buddle BM, Vordermeier HM. 2012. Bovine tuberculosis vaccine research: historical perspectives and recent advances. Vaccine 30:2611–2622. doi: 10.1016/j.vaccine.2012.02.018. [DOI] [PubMed] [Google Scholar]
- 50.Aranday-Cortes E, Hogarth PJ, Kaveh DA, Whelan AO, Villarreal-Ramos B, Lalvani A, Vordermeier HM. 2012. Transcriptional profiling of disease-induced host responses in bovine tuberculosis and the identification of potential diagnostic biomarkers. PLoS One 7:e30626. doi: 10.1371/journal.pone.0030626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Batten M, Li J, Yi S, Kljavin NM, Danilenko DM, Lucas S, Lee J, de Sauvage FJ, Ghilardi N. 2006. Interleukin 27 limits autoimmune encephalomyelitis by suppressing the development of interleukin 17-producing T cells. Nat Immunol 7:929–936. doi: 10.1038/ni1375. [DOI] [PubMed] [Google Scholar]
- 52.Liu H, Rohowsky-Kochan C. 2011. Interleukin-27-mediated suppression of human Th17 cells is associated with activation of STAT1 and suppressor of cytokine signaling protein 1. J Interferon Cytokine Res 31:459–469. doi: 10.1089/jir.2010.0115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gagliani N, Vesely MC, Iseppon A, Brockmann L, Xu H, Palm NW, de Zoete MR, Licona-Limón P, Paiva RS, Ching T, Weaver C, Zi X, Pan X, Fan R, Garmire LX, Cotton MJ, Drier Y, Bernstein B, Geginat J, Stockinger B, Esplugues E, Huber S, Flavell RA. 2015. Th17 cells transdifferentiate into regulatory T cells during resolution of inflammation. Nature 523:221–225. doi: 10.1038/nature14452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Okamoto Yoshida Y, Umemura M, Yahagi A, O'Brien RL, Ikuta K, Kishihara K, Hara H, Nakae S, Iwakura Y, Matsuzaki G. 2010. Essential role of IL-17A in the formation of a mycobacterial infection-induced granuloma in the lung. J Immunol 184:4414–4422. doi: 10.4049/jimmunol.0903332. [DOI] [PubMed] [Google Scholar]
- 55.Lockhart E, Green AM, Flynn JL. 2006. IL-17 production is dominated by γδ T cells rather than CD4 T cells during Mycobacterium tuberculosis infection. J Immunol 177:4662–4669. doi: 10.4049/jimmunol.177.7.4662. [DOI] [PubMed] [Google Scholar]
- 56.Cruz A, Fraga AG, Fountain JJ, Rangel-Moreno J, Torrado E, Saraiva M, Pereira DR, Randall TD, Pedrosa J, Cooper AM, Castro AG. 2010. Pathological role of interleukin 17 in mice subjected to repeated BCG vaccination after infection with Mycobacterium tuberculosis. J Exp Med 207:1609–1616. doi: 10.1084/jem.20100265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Baures PW. 2012. Is RORγ a therapeutic target for treating Mycobacterium tuberculosis infections? Tuberculosis (Edinb) 92:95–99. doi: 10.1016/j.tube.2011.11.009. [DOI] [PubMed] [Google Scholar]
- 58.Plattner BL, Huffman EL, Hostetter JM. 2013. Gamma-delta T-cell responses during subcutaneous Mycobacterium avium subspecies paratuberculosis challenge in sensitized or naive calves using matrix biopolymers. Vet Pathol 50:630–637. doi: 10.1177/0300985812463404. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.