Abstract
Diet analysis is an important aspect when investigating the ecology of fish‐eating animals and essential for assessing their functional role in food webs across aquatic and terrestrial ecosystems. The identification of fish remains in dietary samples, however, can be time‐consuming and unsatisfying using conventional morphological analysis of prey remains. Here, we present a two‐step multiplex PCR system, comprised of six assays, allowing for rapid, sensitive and specific detection of fish DNA in dietary samples. This approach encompasses 78 fish and lamprey species native to Central European freshwaters and enables the identification of 31 species, six genera, two families, two orders and two fish family clusters. All targeted taxa were successfully amplified from 25 template molecules, and each assay was specific when tested against a wide range of invertebrates and vertebrates inhabiting aquatic environments. The applicability of the multiplex PCR system was evaluated in a feeding trial, wherein it outperformed morphological prey analysis regarding species‐specific prey identification in faeces of Eurasian otters. Additionally, a wide spectrum of fish species was detected in field‐collected faecal samples and regurgitated pellets of Common Kingfishers and Great Cormorants, demonstrating the broad applicability of the approach. In conclusion, this multiplex PCR system provides an efficient, easy to use and cost‐effective tool for assessing the trophic ecology of piscivores in Central Europe. Furthermore, the multiplex PCRs and the primers described therein will be applicable wherever DNA of the targeted fish species needs to be detected at high sensitivity and specificity.
Keywords: Alcedo atthis, diagnostic fish primers, Lutra lutra, molecular scatology, Phalacrocorax carbo sinensis, piscivory
Introduction
The borders between aquatic and terrestrial ecosystems exhibit extraordinary diversity, productivity and manifold interactions between organisms (Polis et al. 1997). Movements of nutrients, prey and predators influence the structure and productivity of both the donor and the recipient system (Burdon & Harding 2007). In this regard, piscivores play a pivotal role as consumers and vectors (Polis et al. 2004; Ellis et al. 2006). They induce trophic cascades or general top‐down effects (Schmitz et al. 2010) and thus alter aquatic and terrestrial ecosystems at individual, population, community and ecosystem level (Polis et al. 2004; Baxter et al. 2005; Knight et al. 2005).
Piscivores such as the Common Kingfisher (Alcedo atthis, further on ‘kingfisher’), Bald Eagle (Haliaeetus leucocephalus) and Eurasian otter (Lutra lutra, further on ‘otter’) serve as ecosystem indicators and even flagship species for nature conservation (Entwistle & Dunstone 2000; Clucas et al. 2008). They provide key supporting and regulating ecosystem services (Green & Elmberg 2014) and indicate protection worthy, high levels of biodiversity (Sergio et al. 2006). In Europe, such piscivores and the freshwater ecosystems they inhabit are protected under the Council Directive 92/43/EEC and the Directive 2009/147/EC of the European Union (Council of the European Union 1992; European Parliament & Council of the European Union 2009). In contrast, piscivores with high population densities such as cormorants, herons or pinnipeds have long been perceived as food competitors by humans (Duffy 1995; Gosch et al. 2014) resulting in shootings, culling and repellent measures (Boudewijn & Dirksen 1999; Bowen & Lidgard 2013). Any management of piscivores aiming at either protecting or regulating these animals depends on a sound understanding of their feeding ecology. Consequently, a low‐cost method, enabling the identification of prey species at high taxonomic resolution, is a sorely needed tool to analyse non‐invasively gained dietary samples and shape conservation and management efforts along European freshwaters.
Dietary studies on piscivores in the traditional sense involve stomach content analysis of killed specim‐ens (Labansen et al. 2007; Bostrom et al. 2012), stomach flushing (Hull 1999; Alonso et al. 2014), direct observation (Gonzalez‐Solis et al. 1997), as well as morphological hard part analysis of indigestible prey remains in regurgitated pellets (Gonzalez‐Solis et al. 1997; Dias et al. 2012) and faeces (Jedrzejewska et al. 2001; Sinclair & Zeppelin 2002). The morphological analysis of prey remains found in pellets and faeces permits conclusions about the number and size of the consumed fish prey (Mariano‐Jelicich & Favero 2006; Gagliardi et al. 2007). There are, however, numerous disadvantages to these approaches such as the limited applicability of stomach flushing, digestion bias due to prey size and species, and often species‐specific identification is not possible (reviewed and discussed in Barrett et al. 2007; Bowen & Iverson 2012). DNA‐based prey identification can overcome many of these limitations, and a variety of molecular methods have been developed over the past 15 years enabling the investigation of feeding interactions across trophic levels and ecosystems at unprecedented resolution (King et al. 2008; Pompanon et al. 2012; Traugott et al. 2013; Symondson & Harwood 2014). Hence, even morphologically unidentifiable prey remains can be exploited to track trophic links (Casper et al. 2007; Bowen & Iverson 2012). Molecular techniques have also been used to assess the diet of piscivores with a focus on marine predators such as pinnipeds (Deagle et al. 2009; Marshall et al. 2010), squids (Deagle et al. 2005) and seabirds (Bowser et al. 2013; Jarman et al. 2013), but to this point, they have only been scarcely applied in freshwater ecosystems (e.g. Bradford et al. 2014; Brandl et al. 2014).
Whilst sequence‐based methods such as next‐generation sequencing (further on ‘NGS’) provide information on the prey range at high taxonomic resolution, they are time‐consuming and expensive, especially when dealing with high sample numbers (Pompanon et al. 2012). Diagnostic multiplex PCR provides a valuable alternative to sequence‐based approaches when a defined set of prey organisms is to be detected: multiplexing of taxon‐specific primers allows the identification of several prey taxa within one reaction, based on differences in amplicon size (Harper et al. 2005; Sint et al. 2012). Depending on the information needed, the taxonomic level of prey identification can be selected and through balancing primer concentrations, equal sensitivity can be reached across the targeted taxa (Sint et al. 2012). Diagnostic multiplex PCR works best for investigating trophic interactions in an environment with a limited and predictable number of prey species (Symondson & Harwood 2014), as for example, fish in Central European freshwaters. Nevertheless, the number of fish species in this environment exceeds the number of targets possible in one multiplex PCR. For such situations, we propose a novel two‐step approach where targets are initially identified at a high taxonomic level and species‐specific identification is carried out in the respective follow‐up PCRs (Caballero et al. 2012).
The aims of the present study were threefold: (I) to develop a two‐step multiplex PCR system for efficient, easy and sensitivity‐balanced identification of DNA from Central European freshwater fish in dietary samples and thus providing an alternative to the more time‐intensive and expensive sequence‐based methods, (II) to compare the performance of the new system with morphological scat analysis in a feeding trial with otters and (III) to apply the molecular detection system to field‐collected faecal samples of kingfishers and cormorants as well as to cormorant pellets.
Materials and methods
Origin of reference samples for DNA extraction and sequence generation
Between 2011 and 2013, tissue samples of 78 fish and lamprey species occurring in Central Europe (Rhine and Danube catchment) were collected. Species inhabiting coastal and brackish waters, except the Atlantic salmon, were not included. For very rare or endangered species, tissue samples or DNA extracts were provided by museums and scientific collections (Table S1, not included red‐listed species see Appendix S1, Supporting information). Additionally, nontarget organisms occurring in lotic and lentic freshwaters including gastropods, amphibians, reptiles and arthropods were collected (Table S2, Supporting information). Tissue samples of piscivorous mammals and birds were provided by the zoological collection of the Tiroler Landesmuseen (Table S2, Supporting information). All samples were DNA extracted using the DNeasy blood and tissue kit (QIAGEN, Hilden, Germany) following the manufacturer's instructions.
Fragments of the mitochondrial 16S rRNA (16S) gene and cytochrome c oxidase subunit I (COI) gene were amplified for all target species (Table S1, Supporting information) using the forward primer 16Sar plus the reverse primer 16Sbr for 16S (Gleason & Burton 2012) and the forward primer FishF1 plus the reverse primer FishR1 for COI (Ward et al. 2005), respectively. Purified PCR products were sequenced by Eurofins MWG Operon (Munich, Germany); the sequences were edited using BioEdit (Hall 1999) and their identity checked by blast (NCBI website http://blast.ncbi.nlm.nih.gov/Blast.cgi). Thereafter, 16S and COI consensus sequences were created for each species, using the sequences derived from the DNA extracts in combination with sequences already available on GenBank (NCBI) or BOLD (http://www.boldsystems.org/) and aligned across species in BioEdit.
Primer design and multiplex PCR development
Using Primer Premier 5 (PREMIER Biosoft International, Palo Alto, CA, USA), 82 primers with melting temperatures as close as possible to 60 °C were designed and arranged in six multiplex PCR assays. Within each assay, primer pairs were employed whose amplicons exhibited at least a 20‐bp difference in length to ensure proper separation in electrophoresis (see Appendix S2 for exception, Supporting information).
Initially, the functioning of all primer pairs was tested in singleplex PCRs. Thereafter, they were combined to multiplex PCR assays whose optimal annealing temperatures were determined by gradient PCR and the primer concentrations were adjusted to balance the sensitivity across primer pairs within each assay using standardized DNA templates (Sint et al. 2012). These target fish species DNA templates were generated from PCR products also used for sequencing. All reactions were based on the Multiplex PCR Kit (QIAGEN) using additional bovine serum albumin (BSA) to prevent PCR inhibition (Juen & Traugott 2006) and tetramethylammonium chloride (TMAC) to enhance specificity (Chevet et al. 1995). Standardized DNA templates of target species (Table S1, Supporting information) were used to determine the sensitivity of each primer pair within the six multiplex PCRs in the presence of ~300 ng cormorant DNA to simulate field‐collected dietary samples. An extensive specificity testing was carried out by applying the multiplex PCR assays to 632 DNA extracts of the targeted fish species (Table S1, Supporting information) and 113 nontarget samples representing 61 taxa (Table S2, Supporting information) to rule out false‐positive amplifications. Any additional PCR amplicons produced by DNA extracts during specificity testing were sequenced and confirmed to be contaminations in the extracts. These were subsequently replaced by uncontaminated DNA extracts generated from fish muscle tissue.
qiaxcel, an automatic capillary electrophoresis system with the corresponding software qiaxcel biocalculator version 3.2 (Method AL320; QIAGEN), was used for PCR product separation and analysis. All DNA fragments of the expected fragment size producing a signal strength above ≥0.07 relative fluorescent units (RFU) were deemed positive. Finally, in silico PCRs were carried out for all multiplex PCR assays with CLC Main Workbench 7 (CLC bio, Aarhus, Denmark) using the ‘Find Binding Sites and Create Fragments’ tool. The 16S and COI sequences of European freshwater Mollusca, Ephemeroptera, Plecoptera, Trichoptera, Zygoptera and Chironomidae available online at GenBank were used as a basis for these calculations (see Table S4 for detailed settings, Supporting information).
Multiplex PCR evaluation via feeding trial and field‐collected samples
In November 2013, a feeding trial with three Eurasian otters (Lutra lutra) was conducted at the Alpenzoo (Innsbruck, Austria). Otters were housed in a 142 m2 enclosure including a large pond (⅔ of enclosure surface), rocks and tree trunks to rest on, and a holt accessible from underwater. The otters were a family comprised of mother, father and son and at that time eight, six and one year old, respectively. Three days prior to the trial, they were fed a fish‐free diet consisting of day‐old chicks, chinchillas and guinea pigs (~2000 g per day). Thereafter, four different fish species were offered, one per evening, starting with 2500 g rainbow trout (Oncorhynchus mykiss) followed by 2025 g roach (Rutilus rutilus), 917 g perch (Perca fluviatilis) and 552 g whitefish (Coregonus spp.). All fish had been gilled and thoroughly rinsed under flowing water prior to the trial. The following 3 days the otters’ diet was kept fish‐free again and consisted of day‐old chicks and cattle heart (~2000 g per day). Five faecal samples (spraints) were collected each evening starting 1 day before rainbow trout was provided and ending 3 days after whitefish was offered. All spraints and field‐collected samples were individually collected in plastic bags or reaction tubes using gloves, frozen in cooling boxes in the zoo or field and stored at −80 °C until DNA extraction.
Concerning field‐collected dietary samples, on 11 and 12 June 2011 seven kingfisher faeces were collected on the riverbanks of Danube, March and Thaya in Germany and Austria after observing the birds defecate (see Table S3 for locations, Supporting information). Forty‐five faecal samples of cormorants were collected on 20 December 2012 under roosting trees along the Chiemsee shoreline (N47.85964, E12.51174, Germany), and 45 regurgitated cormorant pellets were collected on 1 February 2013 on a small island in the Chiemsee (N47.869092, E12.416847, Germany).
Processing of scat samples and pellets
All zoo‐ and field‐collected samples were lysed with a mixture of TES‐buffer (0.1 m TRIS, 10 mm EDTA, 2% sodium dodecyl sulphate; pH 8) and proteinase K (20 mg/ml) in a ratio of 190:1. The amount of lysis buffer added to the sample depended on its size: 6 ml for small (5 to 10 cm³), 8 ml for large (10 to 20 cm³) otter spraints and 300 μl for kingfisher faeces. Cormorant pellets were separated into three size classes with 3 ml for small, 5 ml for medium and 8 ml for large samples. Likewise, 300 μl, 500 μl and 3 ml of lysis buffer were used for small, medium and large cormorant faeces, respectively. After adding the lysis buffer, all samples were vortexed and incubated overnight at 56 °C in a rocking platform.
DNA extraction was carried out with the BioSprint 96 instrument (QIAGEN) using the BioSprint 96 DNA blood Kit (QIAGEN) in accordance with the manufacturer's instructions. Each otter spraint lysate was extracted three times to maximize the chances of DNA detection (Oehm et al. 2011). Per BioSprint run 92 lysates and four blank extraction controls were processed. Controls contained TES‐buffer instead of lysate and were checked with the family‐specific (FishTax) multiplex PCR assay (see Results section) for cross‐contamination potentially occurring during the DNA extraction process. All of them resulted negative. The obtained DNA extracts were subsequently analysed with the two‐step multiplex PCR system (see Results section), and samples testing negative in the FishTax multiplex PCR (see Results section) were spiked with ~50 ng perch DNA to test for PCR inhibition. For some spraints, prey identification was only possible at order level. These PCR amplicons produced by order‐specific primer pairs were sequenced to resolve fish species identity via sequencing.
Prior to the morphological analysis, the dissolved otter spraints were strained and rinsed with distilled water. Identifiable fish hard parts such as lenses, scales, vertebras, chewing pads and jaws were sorted out and identified using the keys of Knolleisen (1996) and Čech (2006) as well as using fish bone reference collections provided by Dr. Werner Suter (Swiss Federal Research Institute, Birmensdorf, Switzerland), Dr. Josef Trauttmansdorff (Otto‐König Institute, Stockerau, Austria) and the Bavarian State Collection of Zoology (Munich, Germany). A chi‐squared test was calculated to check for significant differences between the molecular and the morphological approach regarding species‐specific prey detection in the otter spraints using MS Excel 2010.
Results
Multiplex PCR system
For the design of the multiplex PCR system, 311 fish DNA sequences encompassing 78 species were generated and the highest quality sequences uploaded to GenBank (Accession nos in Table S1, Supporting information). This includes 20 and four species with not so far public 16S and COI sequences (GenBank, as of 10 February 2015), respectively (Table S1, Supporting information). Based on these and online available sequence information, six multiplex PCRs were set up for identi‐fication of 31 fish species, six genera, two families, two orders and two fish family clusters using primer pairs amplifying DNA fragments between 77 bp and 405 bp (Figs 1 and 2, Table 1). In the first, family‐specific multiplex PCR assay (‘FishTax’) each of the 78 fish and lamprey species is assigned to one of nine target groups. These include one fish family, two orders and two family clusters resulting from DNA sequence dissimilarities between these target groups. The target groups are Petromyzontidae, Siluriformes, Salmoniformes, Cobitidae/Nemacheilidae/Cyprinidae (further on ‘Cypriniformes’; lowest shared taxonomic level) and Gobiidae/Gasterosteidae/Cottidae/Centrarchidae/Percidae (further on ‘Percomorphaceae’; lowest shared taxonomic level). Four fish species, which are genetically separated from all other taxa examined here, are targeted species specifically in the FishTax assay, namely sturgeon (Acipenser ruthenus), European eel (Anguilla anguilla), burbot (Lota lota) and pike (Esox lucius). Samples testing positive with the group‐specific primers in the FishTax assay are to be subjected to the respective second‐step multiplex PCR(s). These five follow‐up assays for species‐specific identification are ‘SalForm’, ’PercMorph’, ‘CypForm 1’, ‘CypForm 2’ and ‘CypForm 3’ (Fig. 1). The SalForm assay identifies species within the Salmoniformes (Fig. 1), except Coregonus and Salvelinus for which identification is limited to genus level, as well as the species combination of Salmo trutta and Salmo labrax. The ‘PercMorph’ multiplex PCR identifies four species, one genus and one family within the Percomorphaceae (Fig. 1); sticklebacks (Gasterosteidae) are in practice displayed as one 135‐ to 136‐bp diagnostic band, as amplicon sizes of Pungitius pungitius and Gasterosteus spp. differ by only 1 bp. For the species‐rich Cypriniformes, three assays (CypForm 1–3) were set up, identifying 19 species and two genera (Fig. 1).
Figure 1.
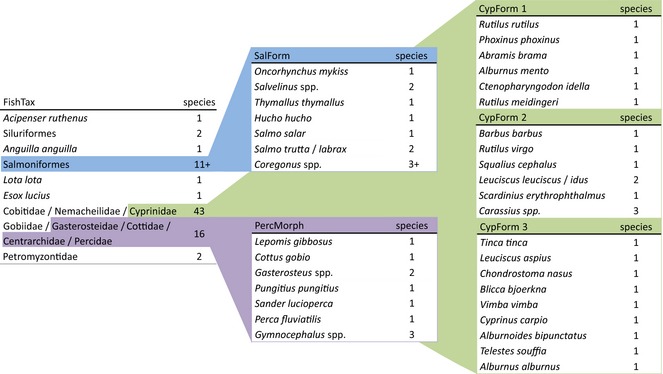
The two‐step multiplex PCR system comprising six assays (FishTax, SalForm, PercMorph, CypForm 1–3) to identify fish DNA in dietary samples, depicting the assays and the identity and number of the target taxa. Coloured areas indicate which target groups from the FishTax assay are subjected to further identification.
Figure 2.
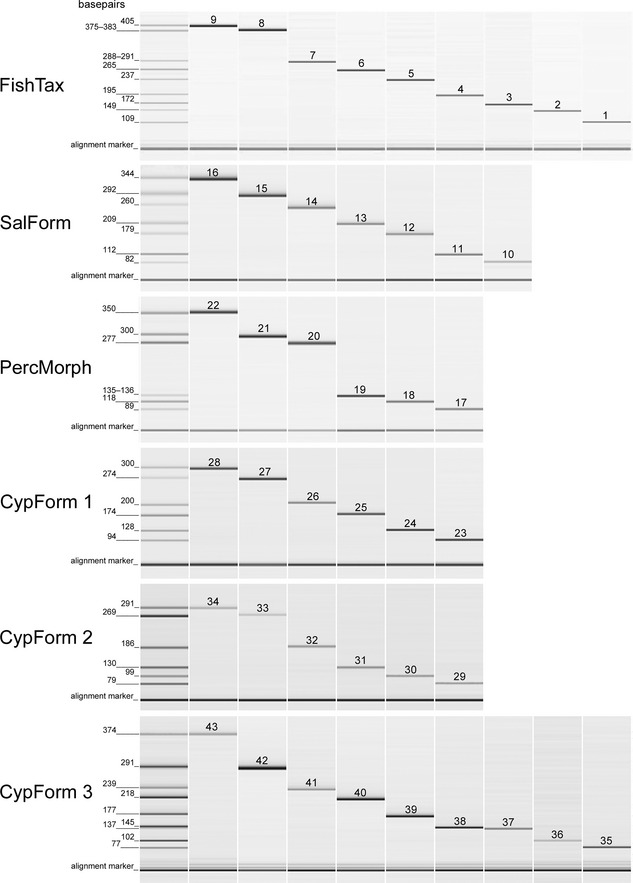
qiaxcel gel view of amplicons generated by the diagnostic multiplex PCR assays. The leftmost lane shows a mixture of all targeted taxa per reaction with equal target DNA concentrations and the amplicon lengths in base pairs. The single bands displayed in the other lanes were generated with ~150 double strands of target template DNA in the presence of ~300 ng nontarget DNA (Phalacrocorax carbo sinensis). FishTax: 1: Acipenser ruthenus, 2: Siluriformes, 3: Anguilla anguilla, 4: Salmoniformes, 5: Lota lota, 6: Esox lucius, 7: Cobitidae/Nemacheilidae/Cyprinidae, 8: Gobiidae/Gasterosteidae/Cottidae/Centrarchidae/Percidae, 9: Petromyzontidae. SalForm: 10: Oncorhynchus mykiss, 11: Salvelinus spp., 12: Thymallus thymallus, 13: Hucho hucho, 14: Salmo salar, 15: Salmo trutta/labrax, 16: Coregonus spp. PercMorph: 17: Lepomis gibbosus, 18: Cottus gobio, 19: Gasterosteus spp./Pungitius pungitius, 20: Sander lucioperca, 21: Perca fluviatilis, 22: Gymnocephalus spp. CypForm 1: 23: Rutilus rutilus, 24: Phoxinus phoxinus, 25: Abramis brama, 26: Alburnus mento, 27: Ctenopharyngodon idella, 28: Rutilus meidingeri. CypForm 2: 29: Barbus barbus, 30: Rutilus virgo, 31: Squalius cephalus, 32: Leuciscus leuciscus/idus, 33: Scardinius eryhthrophthalmus, 34: Carassius spp. CypForm 3: 35: Tinca tinca, 36: Leuciscus aspius, 37: Chondrostoma nasus, 38: Blicca bjoerkna, 39: Vimba vimba, 40: Cyprinus carpio, 41: Alburnoides bipunctatus, 42: Telestes souffia, 43: Alburnus alburnus.
Table 1.
A summary of the multiplex PCR assays: the sensitivity of each multiplex PCR in DNA double strands (ds) necessary to reliably detect a target taxon in a sample with mixed target and nontarget DNA is provided. Target taxa, primer names and sequences, genes targeted by the primers and their corresponding amplicon sizes, primer concentrations in PCR and the number of target species per primer pair are specified too
| Assay name | Sensitivity (DNA ds) | Target taxa | Primer name | Primer sequence (5′–3′) | Target gene | Fragment size (bp) | Primer concentration in PCR (μ m) | Number of target species |
|---|---|---|---|---|---|---|---|---|
| FishTax | 17 | Acipenser ruthenus | Gen‐mix‐S628 | AATGAAGACCTGTATGAATGGCAT | 16S | 109 | 1.2 | 1 |
| Aci‐rut‐A633 | CTTCTCGTCTTATGGGGTTATGCT | 16S | 0.2 | |||||
| Siluriformes | Sil‐for‐S629 | CGCCTCCTGCAAAAATCAAY | 16S | 149 | 0.2 | 2 | ||
| Sil‐for‐A637 | AGACAGTTAAGCCCTCGTTCCA | 16S | 0.2 | |||||
| Anguilla anguilla | Gen‐mix‐S628 | AATGAAGACCTGTATGAATGGCAT | 16S | 172 | a | 1 | ||
| Ang‐ang‐A638 | TGTTCCTTTTGGTTGGTTTGGT | 16S | 0.15 | |||||
| Salmoniformes | Gen‐mix‐S628 | AATGAAGACCTGTATGAATGGCAT | 16S | 195 | a | 11+ | ||
| Sal‐for‐A631 | CATAKGGGCTAGGGGTCACTG | 16S | 0.4 | |||||
| Lota lota | Gen‐mix‐S628 | AATGAAGACCTGTATGAATGGCAT | 16S | 237 | a | 1 | ||
| Lot‐lot‐A635 | CCACATGGGGGTTGTGTTTTA | 16S | 0.3 | |||||
| Esox lucius | Gen‐mix‐S628 | AATGAAGACCTGTATGAATGGCAT | 16S | 265 | a | 1 | ||
| Eso‐luc‐A636 | CGTGGTTATAAGGAGGTTTTCCTT | 16S | 0.2 | |||||
| Cobitidae, Nemacheilidae, Cyprinidae | Cyp‐cgo‐S627 | GAGGTCCAGCCTGCCCA | 16S | 288–291 | 0.7 | 43 | ||
| Cyp‐cgo‐A630 | CGCCCCAACCGAAGGTAA | 16S | 0.7 | |||||
| Gobiidae, Gasterosteidae, Cottidae, Centrarchidae, Percidae | Gen‐mix‐S628 | AATGAAGACCTGTATGAATGGCAT | 16S | 375–383 | a | 16 | ||
| Per‐ngc‐A634 | CCTTGTCGATRTGRGCTCTAAA | 16S | 0.3 | |||||
| Petromyzontidae | Pet‐myz‐S630 | ATCGCCTATTGGAGGCAAGA | 16S | 405 | 0.5 | 2 | ||
| Pet‐myz‐A639 | GGGGTAACTTGTTTCGTTAGGCA | 16S | 0.5 | |||||
| SalForm | 22 | Oncorhynchus mykiss | Onc‐myk‐S655 | TCTCCCTTCATTTAGCTGGAATC | COI | 82 | 0.15 | 1 |
| Onc‐myk‐A655 | GCTGGAGGTTTTATGTTAATAATGGTC | COI | 0.15 | |||||
| Salvelinus spp. | Sal‐vel‐S651 | ATAGTCGGCACCGCCCTT | COI | 112 | 0.15 | 2 | ||
| Sal‐vel‐A651 | TAACGAAGGCATGGGCTGTT | COI | 0.15 | |||||
| Thymallus thymallus | Thy‐thy‐S653 | ATCAAATTTATAATGTGATCGTCACG | COI | 179 | 0.15 | 1 | ||
| Thy‐thy‐A653 | AAGAAAGGACGGGGGAAGC | COI | 0.15 | |||||
| Hucho hucho | Huc‐huc‐S650 | TGATTTAACTATCTTCTCTCTCCACTTG | COI | 209 | 0.15 | 1 | ||
| Huc‐huc‐A650 | GGTCTGTAAGTAGTATAGTGATGCCC | COI | 0.15 | |||||
| Salmo salar | Sal‐sal‐S656 | TGGCGCCCTTCTGGGA | COI | 260 | 0.40 | 1 | ||
| Sal‐sal‐A656 | GGGGGTAGACTGTTCATCCG | COI | 0.40 | |||||
| Salmo trutta/labrax | Sal‐tru‐S654 | CTTTGGAAACTGATTAATCCCTCTC | COI | 292 | 0.70 | 2 | ||
| Sal‐tru‐A654 | TGGGGGTTTTATGTTAATAATGGTC | COI | 0.70 | |||||
| Coregonus spp. | Cor‐spp‐S652 | GATCAGATTTATAATGTAATCGTCACG | COI | 344 | 0.50 | 3+ | ||
| Cor‐spp‐A652 | ATAAAATTAACGGCCCCCAAG | COI | 0.50 | |||||
| PercMorph | 25 | Lepomis gibbosus | Lep‐gib‐S633 | GGAGCTTTAGACGCCTGAGTAAG | 16S | 89 | 0.48 | 1 |
| Lep‐gib‐A642 | CCCCAACCAAAGACATGGA | 16S | 0.48 | |||||
| Cottus gobio Cottus gobio | Cot‐gob‐S632 | GAATAAAGGACTAAACCAAGTGGG | 16S | 118 | 0.48 | 1 | ||
| Cot‐gob‐A641 | GCTGTAGCTCTCAGTTGTAGGAAAA | 16S | 0.48 | |||||
| Gasterosteus spp. | Gas‐ter‐S631 | ACAAGATGGAACCCACCCTG | 16S | 136 | 0.24 | 2 | ||
| Gas‐ter‐A649 | GATCTTTTTGGTCAGAAATTCTGTTTA | 16S | 0.4 | |||||
| Pungitius pungitius | Pun‐pun‐S640 | CCAAATGGAACCCACCCTG | 16S | 135 | 0.5 | 1 | ||
| Gas‐ter‐A649 | GATCTTTTTGGTCAGAAATTCTGTTTA | 16S | b | |||||
| Sander lucioperca | San‐luc‐S658 | CTCCTTGCTTCCTCAGGGGTA | COI | 277 | 0.16 | 1 | ||
| San‐luc‐A658 | CGGCAAGTACGGGGAGC | COI | 0.16 | |||||
| Perca fluviatilis | Per‐flu‐S671 | GTACCGGGTGAACTGTATATCCG | COI | 300 | 0.16 | 1 | ||
| Per‐flu‐A671 | CAGGGTCAAAGAAAGTTGTGTTC | COI | 0.16 | |||||
| Gymnocephalus spp. | Gym‐spp‐S677 | CTCCTTGCTTCCTCAGGAGTA | COI | 350 | 0.4 | 3 | ||
| Gym‐spp‐A657 | AATGTTGGTAGAGGATGGGATCR | COI | 0.4 | |||||
| CypForm 1 | 13 | Rutilus rutilus | Rut‐rut‐S665 | TTCYGGTGTTGAGGCCGGT | COI | 94 | 0.25 | 1 |
| Rut‐rut‐A665 | TGTTAAATCTACTGATGCCCCG | COI | 0.25 | |||||
| Phoxinus phoxinus | Pho‐Pho‐S639 | CGTGCAGAAGCGGATATAAATAC | 16S | 128 | 0.175 | 1 | ||
| Pho‐Pho‐A648 | CCAACCGAAGGTAAAGTCTTATTG | 16S | 0.175 | |||||
| Abramis brama | Abr‐bra‐S638 | GGAGCTTAAGGTACAAAATTTAACCAT | 16S | 174 | 0.15 | 1 | ||
| Abr‐bra‐A647 | CAGATGTTCTGCGGCTTATAGG | 16S | 0.15 | |||||
| Alburnus mento | Alb‐men‐S662 | TTTCTGACTCCTTCCGCCG | COI | 200 | 0.6 | 1 | ||
| Alb‐men‐A662 | TGGTGGTAATGAAGTTGACTGCA | COI | 0.6 | |||||
| Ctenopharyngodon idella | Cte‐ide‐S635 | CGCCTCCTGCAATCAAACTC | 16S | 274 | 0.4 | 1 | ||
| Cte‐ide‐A644 | CTTTTTATTGAGTTGCTTAACGTGA | 16S | 0.4 | |||||
| Rutilus meidingeri | Rut‐mei‐S666 | CTACCCCCATCATTCCTATTATTGT | COI | 300 | 0.2 | 1 | ||
| Rut‐mei‐A666 | GGCAGCTAGCACTGGTAGTGAC | COI | 0.2 | |||||
| CypForm 2 | 25 | Barbus barbus | Bar‐bar‐S637 | CGTGCAGAAGCGGGTATAATAT | 16S | 79 | 0.4 | 1 |
| Bar‐bar‐A646 | TTGCTTGACGTGGTTGATCTTTA | 16S | 0.4 | |||||
| Rutilus virgo | Rut‐vir‐S667 | CCTACCCCCATCATTCCTATTATTAC | COI | 99 | 0.4 | 1 | ||
| Rut‐vir‐A667 | GCGAGGTTGCCTGCAAGC | COI | 0.4 | |||||
| Squalius cephalus | Squ‐cep‐S669 | CAGTATACCCACCGCTTGCG | COI | 130 | 0.2 | 1 | ||
| Squ‐cep‐A669 | TTAATAATTGTGGTAATGAAGTTGACC | COI | 0.2 | |||||
| Leuciscus leuciscus/idus | Leu‐lid‐S663 | CATCTCCCAGTATCAAACACCG | COI | 186 | 0.2 | 2 | ||
| Leu‐lid‐A663 | AATCAGAATAAGTGTTGGTACAGGATC | COI | 0.2 | |||||
| Scardinius eryhtrophthalmus | Sca‐ery‐S668 | GAGTTTCTGACTTCTCCCTCCG | COI | 269 | 0.1 | 1 | ||
| Sca‐ery‐A668 | CCAGTACGGCTCATACAAACAGC | COI | 0.1 | |||||
| Carassius spp. | Car‐ass‐S659 | GAGCTGGCACCGGATGG | COI | 291 | 0.25 | 3 | ||
| Car‐ass‐A659 | TGGTGTTAAGATTTCGATCTGTTAAA | COI | 0.25 | |||||
| CypForm 3 Chondrostoma nasus | 17 | Tinca tinca | Tin‐tin‐S636 | GTACAAAATTCAACCACGTCAAGA | 16S | 77 | 0.4 | 1 |
| Tin‐tin‐A691 | CCAACCGAAGGTAAAAGTTCATAA | 16S | 0.4 | |||||
| Leuciscus aspius | Leu‐asp‐S641 | CACGTTAAACGACTCCGCAC | 16S | 102 | 0.2 | 1 | ||
| Leu‐asp‐A692 | CCAATCCACTCGGAGGCTC | 16S | 0.2 | |||||
| Chondrostoma nasus | Cho‐nas‐S678 | CTTCTACCCCCCTCATTCCTCT | COI | 137 | 0.4 | 1 | ||
| Cho‐nas‐A678 | GAGAAGATTGTTAAATCTACTGATGCA | COI | 0.4 | |||||
| Blicca bjoerkna | Bli‐bjo‐S675 | GGTCACTTTTAGGCGATGACCAG | COI | 145 | 0.15 | 1 | ||
| Bli‐bjo‐A676 | GCCATATCAGGCGCACCG | COI | 0.15 | |||||
| Vimba vimba | Vim‐vim‐S676 | AATCTCGCCCATGCTGGC | COI | 177 | 0.4 | 1 | ||
| Vim‐vim‐A677 | GACGGCTGTTACTAGTACGGCC | COI | 0.7 | |||||
| Cyprinus carpio | Cyp‐car‐S660 | CCCACCTCTTGCAGGGAACT | COI | 218 | 0.5 | 1 | ||
| Cyp‐car‐A660 | AAACAGGTAATGATAGAAGGAGCAAT | COI | 0.5 | |||||
| Alburnoides bipunctatus | Alb‐bip‐S673 | TGACTACTACCTCCATCATTTTTGC | COI | 239 | 0.2 | 1 | ||
| Alb‐bip‐A674 | GGTGTTTGATACTGGGAGATAGCC | COI | 0.2 | |||||
| Telestes souffia | Tel‐sou‐S642 | CATCGCCTCCTGCAACTAATC | 16S | 291 | 0.2 | 1 | ||
| Tel‐sou‐A693 | CACCACTAAGTTCGTGCTTTCTATC | 16S | 0.2 | |||||
| Alburnus alburnus | Alb‐alb‐S672 | CACGAATAAATAACATGAGTTTCTGG | COI | 374 | 0.15 | 1 | ||
| Alb‐alb‐A673 | AAGAATGTGGTATTAAGATTACGATCC | COI | 0.1 |
Total concentration: 1.2 μ m; equals 6 × 0.2 μ m.
Total concentration: 0.4 μ m; equals 2 × 0.2 μ m.
The 10 μl PCRs were performed using the Multiplex PCR Kit (QIAGEN) including 3.2 μl of DNA extract except for the FishTax assay, which contained 1.5 μl. One‐time reaction mix, 5 μg BSA, 30 mm TMAC, primers in respective concentrations (Table 1) and PCR‐grade water (FishTax assay only) were used in each reaction. The optimized thermocycling conditions were 15 min at 95 °C, 35 cycles of 30 s at 94 °C, 90 s at 64 °C (FishTax, SalForm, PercMorph, CypForm 2) or 66 °C (CypForm 1, CypForm 3), 1 min at 72 °C and 10 min at 72 °C once.
Within the multiplex PCRs described above, each primer was specific for its target taxon as no cross‐amplification with the wide set of nontarget taxa occurred. Occasionally, additional PCR products, which were clearly distinguishable from the target bands, were observed (see Appendix S2, Supporting information). The assays also proved to be highly sensitive: in the presence of cormorant DNA, 25 or less double‐stranded DNA template molecules (Table 1) were sufficient to generate amplicons with a signal strength above 0.07 RFU in the qiaxcel system. The in silico PCRs showed that of 7585 16S sequences, none produced an amplicon with any of the multiplex PCRs. Of the 59 202 COI sequences, 102 in theory produced an amplicon (Table S4, Supporting information). All of these sequences originate from samples collected in Canada, the United States or Australia, and in case the sequence is assigned to a species, its distribution does not include Europe.
Detecting fish prey in otter spraints
The multiplex PCR system allowed detecting DNA from the consumed fish species 1 day after the respective feeding event: of the five spraint samples collected per evening, DNA of rainbow trout, roach, perch and whitefish was detected in four, three, four and one spraint, respectively (Fig. 3). Rainbow trout was also detected in one spraint each, collected prior and 2 days after the rainbow trout meal (Fig. 3). Moreover, DNA of Salmoniformes was detected in altogether seven spraints collected on evenings one, three, five, six and seven. Sequencing of these PCR products showed that of the seven spraints testing positive for Salmoniformes (Fig. 3), the two samples collected on evening five contained DNA of whitefish, whilst all others contained DNA of rainbow trout. The morphological analysis of the otter spraints enabled species‐specific perch identification, whereas roach and whitefish remains could be assigned to family/order level only; nonidentifiable fish remains were found in another eight spraint samples (Fig. 3). Overall, species‐specific prey detection was significantly higher for the molecular (65%) compared to morphological analysis (20%; χ2 = 4.14; P = 0.042). All samples testing negative with the FishTax assay produced an amplicon in the spike PCR.
Figure 3.
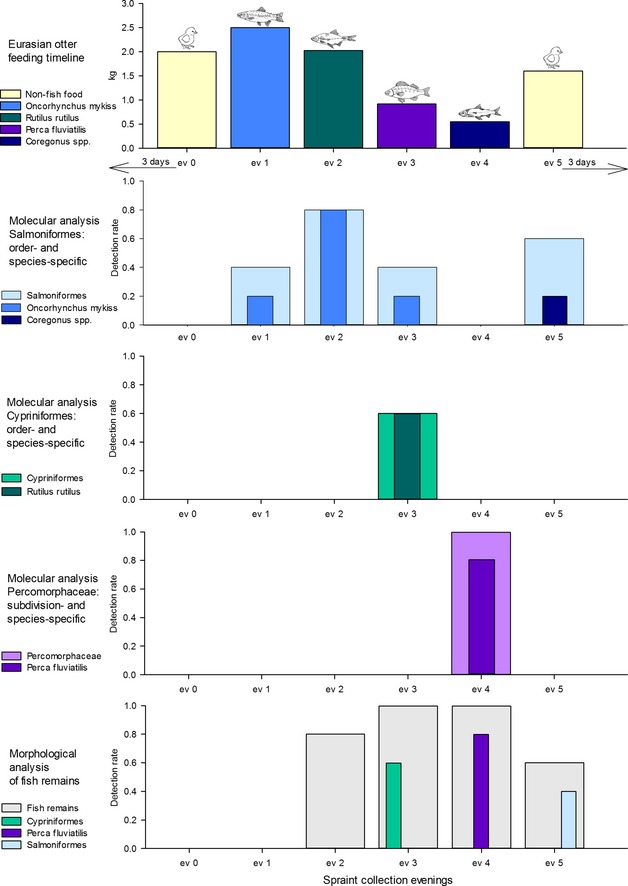
Molecular and morphological identification of fish prey in spraints of the Eurasian otter within a feeding trial. Top panel: X‐axis shows the order of the different prey species fed; Y‐axis provides the total mass of the prey items. Lower panels: X‐axis indicates spraint collection during evenings; Y‐axis displays the detection rate of fish prey (molecular or morphological). Note that molecular detections of Salmoniformes in samples collected on evenings six (0.2) and seven (0.4) are not shown.
Detecting fish DNA in field‐collected dietary samples
The six multiplex PCR assays enabled the detection of semidigested fish DNA in field‐collected dietary samples of kingfishers and cormorants. Of seven kingfisher faeces, four yielded amplicons in the FishTax assay including one and three samples positive for pike and Cypriniformes, respectively. When applying the three CypForm assays to the latter samples, six species were identified with asp (Leuciscus aspius) being present in all of them (Fig. 4).
Figure 4.
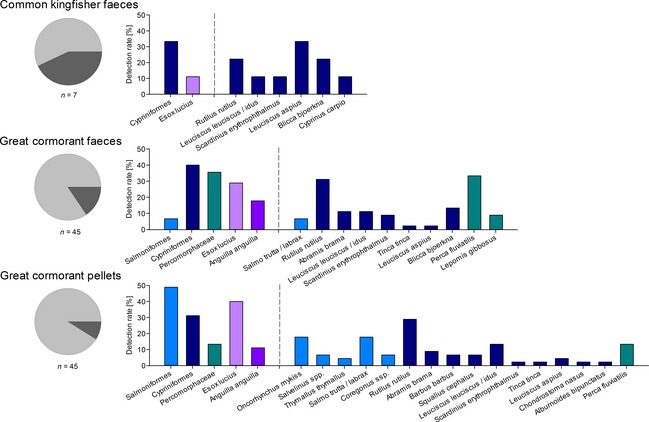
Fish DNA detected in field‐collected faeces (Common Kingfisher, Great Cormorant) and pellets (Great Cormorant) via the multiplex PCR system. Pie charts display the percentage of positive (light grey) and negative (dark grey) samples with amplifiable fish DNA, whilst bar charts show detection rates (%) per target taxon within the FishTax and the follow‐up assays left and right of the dotted line, respectively. Note that species‐specific bars do not add up to the detection rate at family level as one sample can test positive for more than one species.
Testing 45 cormorant faecal samples with the FishTax assay resulted in 84% of samples positive for at least one target taxon; Cypriniformes were the most frequently detected group. Additionally, DNA of pike, European eel, Salmoniformes and Percomorphaceae was amplified. Subjecting the respective samples to the CypForm 1–3, PercMorph and SalForm assays resulted in the identification of seven cyprinid taxa, perch and common sunfish (Lepomis gibbosus), and S. trutta/S. labrax, respectively (Fig. 4). Of 45 cormorant pellet samples, 91% tested positive for fish DNA in the FishTax assay, and detections were allocated to the same five target groups as found in cormorant faeces. Regarding pellet extracts, Salmoniformes were most frequently detected including five different genera/species (Fig. 4). Whilst only perch was amplified within the PercMorph assay, ten different cyprinid species were detected through assays CypForm 1–3 (Fig. 4).
To verify the identification of fish prey by diagnostic PCR, up to five PCR products per detected fish genus or species were sequenced. In all cases, the obtained sequences matched the targeted taxon. With the exception of one kingfisher faeces, all samples testing negative with the FishTax assay produced an amplicon in the spike PCR.
Discussion
Multiplex PCR system
The two‐step multiplex PCR approach presented here provides an alternative to work‐intensive and expensive sequence‐based methods of prey identification. It is ideal for situations in which a defined set of prey taxa needs to be examined within a large number of individual dietary samples. The two‐step system, where prey is first identified in PCR at a high taxonomic level followed by PCRs for species‐specific identification within the respective taxon, permits cost‐efficient screening for a large number of taxa. This extends the application of diagnostic multiplex PCR to research in environments where higher numbers of prey species need to be examined such as studies on the diet of piscivores in Central European freshwaters.
Our multiplex PCR system encompasses 78 Central European fish and lamprey species and enables the detection and identification of 31 species, six genera, two families, two orders and two fish family clusters. The detection system focuses on characteristic lotic and lentic species of the Alpine foreland, their companion species, and fish being relevant for commercial and recreational fishing according to the Water Framework Directive of the European Union (European Parliament & Council of the European Union 2000). Fish species which were targeted only by group‐specific primers in the FishTax assay and red‐listed fish species which were not considered in this study (Appendix S1, Supporting information) are either endangered, invasive or are small bottom‐dwelling fish occurring in large rivers such as Danube or Rhine. According to their limited distribution, these species are unlikely to constitute a frequently used prey. NGS techniques, already implemented for dietary analyses of marine piscivores such as penguins (Jarman et al. 2013) or seals (Deagle et al. 2009), have the potential to detect DNA of these species (Glenn 2011; Pompanon et al. 2012; Shokralla et al. 2012). In case consumption of rare species needs to be assessed, such NGS techniques can complement the presented diagnostic system. The multiplex PCR approach, even when applied in a stepwise manner, will be stretched to its limits when an even greater variety of fish species need to be identified in dietary samples. Under such circumstances, NGS technologies, which are rapidly evolving with decreasing costs per sample (Glenn 2014), will provide a valuable alternative. These sequence‐based approaches of prey DNA identification will also benefit from the new sequences generated in this study.
Our diagnostic system, nevertheless, is broadly applicable to assess the feeding ecology of fish‐eating invertebrates and vertebrates in Central Europe. It has the potential to be used in other regions such as northern and western Europe too, as several of the presented primers should also work with species outside the Central European range. For example, the genus‐specific primer pair targeting whitefish is based on three whitefish species occurring in the Alpine foreland of Austria and Germany. Yet all online available COI sequences of 24 whitefish species match well with the designed primers (forward and reverse primers have one and two mismatches maximum, respectively). Correspondingly, other primers presented here might be suitable for use outside the study area. Testing their specificity against herein not considered nontarget taxa, which are native to the respective study area, is strongly recommended to avoid false‐positive amplifications. This includes, as outlined in this study, sequencing of PCR products generated form field‐collected samples to confirm amplicon identity. Furthermore, primers which produced an amplicon during the in silico PCRs should not be applied in North America/Australia without further specificity tests. The application of the six assays will also not be restricted to dietary samples of the three species examined here; they will be technically working with at least seven other Central European piscivores whose DNA was included in our nontarget testing. Amongst these are five bird species, the northern raccoon (Procyon lotor), currently invading Central Europe (Michler et al. 2009) and the grass snake (Natrix natrix).
The sensitivity of the multiplex PCR assays was high across the board, enabling successful amplification based on as little as 25 template molecules. The high sensitivity combined with the balanced primer concentrations, ensuring similar amplification efficacies for each target taxon, safeguards against detection dropouts caused by differing amounts of prey species’ DNA in dietary samples (Sint et al. 2012). Moreover, the high assay sensitivity should counteract the lower detection probability of longer prey DNA fragments which are usually present in minute quantities (Deagle et al. 2006).
The multiplex PCR system presented here provides also a straightforward approach in terms of its practical implementation: once the DNA has been extracted from the samples, these can be analysed quickly and at comparably low cost. For example, it took one person 2 days to subject the 90 cormorant samples to the FishTax assay and the second‐step PCRs, to run the electrophoresis and to tabulate the screening results. The average screening costs per sample for consumables was about € 3.5, and all work can be performed with basic molecular laboratory equipment.
Otter feeding trial and spraint analysis
In the otter feeding trial, the molecular detection system outperformed the morphological analysis of prey remains with regard to species‐specific prey identification. However, only on evenings two and four the molecular assays reached a prey detection rate of 80% (rainbow trout (Oncorhynchus mykiss) and perch (Perca fluviatilis), respectively) in the spraints. The question arises, as why the highly sensitive assays could not detect the respective fish DNA in all of the samples. Amplicon size seems to be negligible in this regard, because the primer pair producing the longest fragment (Percomorphaceae; 375–383 bp) scored the highest detection rate. PCR inhibitors, which could get copurified during DNA extraction of the spraint samples, were also ruled out as amplification was not blocked in the spike PCR. Other factors which could explain our findings include differences in protein and lipid content of the fed fish species, different meal sizes and the occurrence of empty spraints. A high lipid content in prey fish seems to reduce mitochondrial DNA degradation through digestion as shown by Thomas et al. (2014) in a feeding trial on harbour seals. This observation fits to the presented findings as Schreckenbach et al. (2001) found higher crude fat proportions in rainbow trout (11.57%) compared to roach (1.94%) and whitefish (6.39%). The small whitefish meal (552 g, 4 fish) could explain the low detection rates of this prey, because it is likely that not all otters had a share of this meal. Finally, in a feeding trial on captive otters, Carss & Parkinson (1994) found approximately one‐third of the collected otter spraints (n = 1544) to be anal jelly secretions not containing any morphologically identifiable fish remains. In the presented trial, three spraints, one from evening two and two from evening five, neither contained morphologically identifiable fish remains nor was fish DNA detected. Presumably, these spraints were anal jelly secretions and such samples should be excluded in future dietary studies based on otter spraints.
DNA of Salmoniformes, precisely rainbow trout, was detected in some spraints collected before and more than 1 day after the otters were fed this species. Whilst the detection event 2 days postfeeding could be attributed to differences in gut passage times between otters, environmental DNA contamination is a likely explanation for the other detections as rainbow trout constitutes a major part of the otters’ usual diet at the Alpenzoo. Furthermore, otters sometimes hide their prey (Ruiz‐Olmo 1995), thus remains of previous meals could have contributed to the contamination of spraints with environmental DNA.
Field‐collected dietary samples
Fish species such as pike, roach and bleak, detected in the kingfisher faeces by the two‐step multiplex PCR system, have been previously identified as part of this bird's diet in Central Europe (Čech & Čech 2011) and strengthen the species’ image as a generalist fish eater (Vilches et al. 2013). The fish were most likely juveniles, caught in still waters in the alluvial forests along the main riverbeds, as the maximum prey size of the kingfisher is 100 mm fork length (Cramp 1985). The seven faecal samples were contaminated with soil when they were collected; as soil material is known to cause problems with molecular prey detection in bird droppings (Oehm et al. 2011), this is a likely explanation for the negative result in the spike PCR. For future studies, an optimization of the DNA extraction protocol could prevent negative results caused by inhibition (Zarzoso‐Lacoste et al. 2013). Additionally, performing the multiplex PCRs more than once on a subset of samples could help to determine the robustness of the approach for field‐collected samples.
The application of the six multiplex PCR assays to field‐collected cormorant faeces and pellets lead to the detection of 12 and 18 fish species/genera, respectively. This reflects the broad prey spectrum cormorants utilize in Alpine foreland freshwaters which are characterised by a diverse fish fauna (Marzano et al. 2013). Previous studies applying morphological prey identification to pellets have struggled to identify cyprinid species as their hard parts are usually not species‐specifically distinguishable (Keller 1998). This problem is remedied by our DNA‐based approach that identified nine cyprinid species and one genus in the 45 tested pellets. In 9% of the pellets, no fish DNA could be detected. This does not come as a surprise as cormorants are known to produce empty pellets as juveniles, at food shortage, or under stress (Zijlstra & Vaneerden 1995). Likewise, the cormorant faeces wherein no fish DNA was detected (16%) most likely contained urea as main component and hardly any prey DNA.
Conclusion and outlook
The two‐step multiplex PCR approach presented here provides an efficient, easy to use and cost‐effective tool to examine the diet of piscivores in great detail. Although the system has been developed for Central Europe, it will be applicable to other regions where the targeted fish species occur; however, we strongly recommend evaluating specificity a priori. Furthermore, the application of the multiplex PCR system is not restricted to prey identification, but the assays or single primer pairs will be useful to any approach where fish DNA needs to be identified with high specificity and sensitivity such as environmental monitoring, studies on environmental DNA (Rees et al. 2014) or species‐specific identification of fish eggs, larvae and carcasses (Hubert et al. 2015). Finally, the use of blocking primers (Vestheim et al. 2011) in combination with the here presented multiplex PCR assays could promote dietary studies on fish themselves.
B.T., J.O. and M.T. conceived and designed the study. J.O. generated the reference sample collection with the help of B.T. B.T. designed the primers, established the molecular assays and performed laboratory work together with H.M., A.O. and C.Z. B.T. and A.O. carried out the feeding trial; J.O. and A.O. carried out the morphological analysis. B.T. analysed the data, compiled tables and figures and wrote the manuscript which was revised and improved by J.O. and M.T.
Data accessibility
DNA sequences: GenBank Accession nos KR476824–KR476986 for 16S and KR476987–KR477299 for COI sequences. The screening results of the otter feeding trial and of the field‐collected samples as well as 16S and COI alignments of all sequences submitted to GenBank have been archived in the Dryad Digital Repository (doi:10.5061/dryad.1vf74).
Supporting information
Table S1. The taxonomy of all Central European fish species targeted by the multiplex PCR assays including the number of DNA extracts generated from tissue samples and their origin.
Appendix S1. Red list freshwater fish species of Central Europe not included in this study.
Table S2. Non‐target animals used to evaluate the specificity of the multiplex PCR assays including the number of DNA extracts tested per taxon.
Appendix S2. Notes on the multiplex PCR assays.
Table S3. Collection locations of the Common Kingfisher (Alcedo atthis) a faeces.
Table S4. In silico PCR conditions and results.
Acknowledgements
This research was funded by the Austrian Science Fund (FWF): P24059 ‘The feeding ecology of the Great Cormorant’. We also acknowledged financial support through grants from Hypo Tirol Bank, Swarovski and PhD scholarships (University of Innsbruck) for BT and JO. We are very grateful to F. Lassacher for collecting and preserving kingfisher faecal samples and thank the Alpenzoo (Innsbruck, Austria), for allowing and helping us with the otter feeding trial; the Zoological Research Museum A. Koenig (Bonn, Germany), the Bavarian State Collection of Zoology (Munich, Germany), the Tiroler Landesmuseen (Innsbruck, Austria), F. Bonell, M. Matzinger, C. Ratschan, C. Gumpinger, W. Mark, R. Schwarzenberger, H. Daxner, J. Wanzenböck and D. Sint, for providing tissue samples and DNA extracts. Finally, we thank the members of the Applied and Trophic Ecology research group for constructive feedback and valuable discussions on early versions of the manuscript.
References
- Alonso H, Granadeiro JP, Waap S et al (2014) An holistic ecological analysis of the diet of Cory's shearwaters using prey morphological characters and DNA barcoding. Molecular Ecology, 23, 3719–3733. [DOI] [PubMed] [Google Scholar]
- Council of the European Union (1992) Council Directive 92/43/EEC of 21 May 1992 on the conservation of natural habitats and of wild fauna and flora, OJ L 206, 22/07/1992, 7–50.
- European Parliament, Council of the European Union (2000) Directive 2000/60/EC of the European Parliament and of the Council of 23 October 2000 establishing a framework for Community action in the field of water policy, OJ L 327, 22/12/2000, 1–73.
- European Parliament, Council of the European Union (2009) Directive 2009/147/EC of the European Parliament and of the Council of 30 November 2009 on the conservation of wild birds, OJ L 20, 26/01/2010, 1–17.
- Barrett RT, Camphuysen K, Anker‐Nilssen T et al (2007) Diet studies of seabirds: a review and recommendations. Ices Journal of Marine Science, 64, 1675–1691. [Google Scholar]
- Baxter CV, Fausch KD, Carl Saunders W (2005) Tangled webs: reciprocal flows of invertebrate prey link streams and riparian zones. Freshwater Biology, 50, 201–220. [Google Scholar]
- Bostrom MK, Ostman O, Bergenius MAJ, Lunneryd SG (2012) Cormorant diet in relation to temporal changes in fish communities. Ices Journal of Marine Science, 69, 175–183. [Google Scholar]
- Boudewijn TJ, Dirksen S (1999) Review of deterrent, scaring and other methods to prevent cormorant foraging In: The Assessment of the Effectiveness of Management Measures to Control Damage by Fish‐Eating Birds to Inland Fisheries in England and Wales. (eds McKay H, Furness R, Russell I, Parrott D, Rehfisch M, Watola G, Packer J, Armitage M, Gill E, Robertson P.), pp. a1–a27. MAFF, London. [Google Scholar]
- Bowen WD, Iverson SJ (2012) Methods of estimating marine mammal diets: a review of validation experiments and sources of bias and uncertainty. Marine Mammal Science, 29, 719–754. [Google Scholar]
- Bowen WD, Lidgard D (2013) Marine mammal culling programs: review of effects on predator and prey populations. Mammal Review, 43, 207–220. [Google Scholar]
- Bowser AK, Diamond AW, Addison JA (2013) From puffins to plankton: a DNA‐based analysis of a seabird food chain in the northern Gulf of Maine. PLoS ONE, 8, e83152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford TM, Humphreys WF, Austin AD, Cooper SJB (2014) Identification of trophic niches of subterranean diving beetles in a calcrete aquifer by DNA and stable isotope analyses. Marine and Freshwater Research, 65, 95–104. [Google Scholar]
- Brandl S, Schumer G, Schreier BM et al (2014) Ten real‐time PCR assays for detection of fish predation at the community level in the San Francisco Estuary‐Delta. Molecular Ecology Resources, 15, 278–284. [DOI] [PubMed] [Google Scholar]
- Burdon FJ, Harding JS (2007) The linkage between riparian predators and aquatic insects across a stream‐resource spectrum. Freshwater Biology, 53, 330–346. [Google Scholar]
- Caballero S, Cardenosa D, Soler G, Hyde J (2012) Application of multiplex PCR approaches for shark molecular identification: feasibility and applications for fisheries management and conservation in the Eastern Tropical Pacific. Molecular Ecology Resources, 12, 233–237. [DOI] [PubMed] [Google Scholar]
- Carss DN, Parkinson SG (1994) Errors associated with otter Lutra lutra faecal analysis. 1. Assessing general diet from spraints. Journal of Zoology, 238, 301–317. [Google Scholar]
- Casper RM, Jarman SN, Deagle BE, Gales NJ, Hindell MA (2007) Detecting prey from DNA in predator scats: a comparison with morphological analysis, using Arctocephalus seals fed a known diet. Journal of Experimental Marine Biology and Ecology, 347, 144–154. [Google Scholar]
- Čech M (2006) Keys of Fish Head Identification Bones. Biology Centre AS CR, Institute of Hydrobiology, České Budějovice, Czech Republic, http://www.hbu.cas.cz/fishecu/staffmembers/cech/Key%20of%20diagnostic%20bones.pdf. [Google Scholar]
- Čech M, Čech P (2011) Diet of the Common Kingfisher (Alcedo atthis) in relation to habitat type: a summary of results from the Czech Republic. Sylvia, 47, 33–47. [Google Scholar]
- Chevet E, Lemaitre G, Katinka MD (1995) Low concentrations of tetramethylammonium chloride increase yield and specificity of PCR. Nucleic Acids Research, 23, 3343–3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clucas B, McHugh K, Caro T (2008) Flagship species on covers of US conservation and nature magazines. Biodiversity and Conservation, 17, 1517–1528. [Google Scholar]
- Cramp S (1985) Handbook of the Birds of Europe, the Middle East and North Africa: The Birds of the Western Palearctic. Oxford University Press, Oxford. [Google Scholar]
- Deagle BE, Jarman SN, Pemberton D, Gales NJ (2005) Genetic screening for prey in the gut contents from a giant squid (Architeuthis sp.). Journal of Heredity, 96, 417–423. [DOI] [PubMed] [Google Scholar]
- Deagle BE, Eveson JP, Jarman SN (2006) Quantification of damage in DNA recovered from highly degraded samples – a case study on DNA in faeces. Frontiers in Zoology, 3, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deagle BE, Kirkwood R, Jarman SN (2009) Analysis of Australian fur seal diet by pyrosequencing prey DNA in faeces. Molecular Ecology, 18, 2022–2038. [DOI] [PubMed] [Google Scholar]
- Dias E, Morais P, Leopold M, Campos J, Antunes C (2012) Natural born indicators: great cormorant Phalacrocorax carbo (Aves: Phalacrocoracidae) as monitors of river discharge influence on estuarine ichthyofauna. Journal of Sea Research, 73, 101–108. [Google Scholar]
- Duffy DC (1995) Why is the Double‐crested Cormorant a problem? Insights from Cormorant ecology and human sociology. Colonial Waterbirds, 18, 25–32. [Google Scholar]
- Ellis JC, Farina JM, Witman JD (2006) Nutrient transfer from sea to land: the case of gulls and cormorants in the Gulf of Maine. Journal of Animal Ecology, 75, 565–574. [DOI] [PubMed] [Google Scholar]
- Entwistle A, Dunstone N (2000) Priorities for the conservation of mammalian diversity. Has the panda had its day? Conservation Biology Series (Cambridge), 3, i–xvi, 1‐455. [Google Scholar]
- Gagliardi A, Martinoli A, Preatoni D, Wauters LA, Tosi G (2007) From mass of body elements to fish biomass: a direct method to quantify food intake of fish eating birds. Hydrobiologia, 583, 213–222. [Google Scholar]
- Gleason LU, Burton RS (2012) High‐throughput molecular identification of fish eggs using multiplex suspension bead arrays. Molecular Ecology Resources, 12, 57–66. [DOI] [PubMed] [Google Scholar]
- Glenn TC (2011) Field guide to next‐generation DNA sequencers. Molecular Ecology Resources, 11, 759–769. [DOI] [PubMed] [Google Scholar]
- Glenn T (2014) NGS Field Guide: Overview. An update to Glenn 2011 (Mol. Ecol. Resou. 11, 759‐769), http://www.molecularecologist.com/next-gen-fieldguide-2014/. [DOI] [PubMed]
- Gonzalez‐Solis J, Oro D, Pedrocchi V, Jover L, Ruiz X (1997) Bias associated with diet samples in Audouin's gulls. Condor, 99, 773–779. [Google Scholar]
- Gosch M, Hernandez‐Milian G, Rogan E, Jessopp M, Cronin M (2014) Grey seal diet analysis in Ireland highlights the importance of using multiple diagnostic features. Aquatic Biology, 20, 155–167. [Google Scholar]
- Green AJ, Elmberg J (2014) Ecosystem services provided by waterbirds. Biological Reviews of the Cambridge Philosophical Society, 89, 105–122. [DOI] [PubMed] [Google Scholar]
- Hall TA (1999) BioEdit: a user‐friendly biological sequence alignment editor and analysis program for Windows 95 ⁄ 98 ⁄NT. Nucleic Acids Symposium Series, 41, 95–98. [Google Scholar]
- Harper GL, King RA, Dodd CS et al (2005) Rapid screening of invertebrate predators for multiple prey DNA targets. Molecular Ecology, 14, 819–827. [DOI] [PubMed] [Google Scholar]
- Hubert N, Espiau B, Meyer C, Planes S (2015) Identifying the ichthyoplankton of a coral reef using DNA barcodes. Molecular Ecology Resources, 15, 57–67. [DOI] [PubMed] [Google Scholar]
- Hull CL (1999) Comparison of the diets of breeding royal (Eudyptes schlegeli) and rockhopper (Eudyptes chrysocome) penguins on Macquarie Island over three years. Journal of Zoology, 247, 507–529. [Google Scholar]
- Jarman SN, McInnes JC, Faux C et al (2013) Adelie penguin population diet monitoring by analysis of food DNA in scats. PLoS ONE, 8, e82227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jedrzejewska B, Sidorovich VE, Pikulik MM, Jedrzejewski W (2001) Feeding habits of the otter and the American mink in Bialowieza Primeval Forest (Poland) compared to other Eurasian populations. Ecography, 24, 165–180. [Google Scholar]
- Juen A, Traugott M (2006) Amplification facilitators and multiplex PCR: tools to overcome PCR‐inhibition in DNA‐gut‐content analysis of soil‐living invertebrates. Soil Biology & Biochemistry, 38, 1872–1879. [Google Scholar]
- Keller T (1998) The food of Cormorants (Phalacrocorax carbo sinensis) in Bavaria. Journal Fur Ornithologie, 139, 389–400. [Google Scholar]
- King RA, Read DS, Traugott M, Symondson WO (2008) Molecular analysis of predation: a review of best practice for DNA‐based approaches. Molecular Ecology, 17, 947–963. [DOI] [PubMed] [Google Scholar]
- Knight TM, McCoy MW, Chase JM, McCoy KA, Holt RD (2005) Trophic cascades across ecosystems. Nature, 437, 880–883. [DOI] [PubMed] [Google Scholar]
- Knolleisen M (1996) Fischbestimmungsatlas als Grundlage für nahrungsökologische Untersuchungen. Institute of Wildlife Biology and Game Management, University of Natural Resources and Life Sciences, Vienna, Austria. [Google Scholar]
- Labansen AL, Lydersen C, Haug T, Kovacs KM (2007) Spring diet of ringed seals (Phoca hispida) from northwestern Spitsbergen, Norway. Ices Journal of Marine Science, 64, 1246–1256. [Google Scholar]
- Mariano‐Jelicich R, Favero M (2006) Assessing the diet of the black skimmer through different methodologies: is the analysis of pellets reliable? Waterbirds, 29, 81–87. [Google Scholar]
- Marshall HD, Hart KA, Yaskowiak ES et al (2010) Molecular identification of prey in the stomach contents of Harp Seals (Pagophilus groenlandicus) using species‐specific oligonucleotides. Molecular Ecology Resources, 10, 181–189. [DOI] [PubMed] [Google Scholar]
- Marzano M, Carss DN, Cheyne I (2013) Managing European cormorant‐fisheries conflicts: problems, practicalities and policy. Fisheries Management and Ecology, 20, 401–413. [Google Scholar]
- Michler F‐U, Koehnemann BA, Roth M et al (2009) Mortality of radio tracked raccoons in the Mueritz‐Nationalpark (Germany). Beitraege zur Jagd‐ und Wildforschung, 34, 339–355. [Google Scholar]
- Oehm J, Juen A, Nagiller K, Neuhauser S, Traugott M (2011) Molecular scatology: how to improve prey DNA detection success in avian faeces? Molecular Ecology Resources, 11, 620–628. [DOI] [PubMed] [Google Scholar]
- Polis GA, Anderson WB, Holt RD (1997) Toward an integration of landscape and food web ecology: the dynamics of spatially subsidized food webs. Annual Review of Ecology and Systematics, 28, 289–316. [Google Scholar]
- Polis GA, Power ME, Huxel GR (2004) Food Webs at the Landscape Level. University of Chicago Press, Chicago & London. [Google Scholar]
- Pompanon F, Deagle BE, Symondson WOC et al (2012) Who is eating what: diet assessment using next generation sequencing. Molecular Ecology, 21, 1931–1950. [DOI] [PubMed] [Google Scholar]
- Rees HC, Maddison BC, Middleditch DJ et al (2014) The detection of aquatic animal species using environmental DNA – a review of eDNA as a survey tool in ecology. Journal of Applied Ecology, 51, 1450–1459. [Google Scholar]
- Ruiz‐Olmo J (1995) Observations on the predation behaviour of the otter Lutra lutra in NE Spain. Acta Theriologica, 40, 175–180. [Google Scholar]
- Schmitz OJ, Hawlena D, Trussell GC (2010) Predator control of ecosystem nutrient dynamics. Ecology Letters, 13, 1199–1209. [DOI] [PubMed] [Google Scholar]
- Schreckenbach K, Knosche R, Ebert K (2001) Nutrient and energy content of freshwater fishes. Journal of Applied Ichthyology, 17, 142–144. [Google Scholar]
- Sergio F, Newton IAN, Marchesi L, Pedrini P (2006) Ecologically justified charisma: preservation of top predators delivers biodiversity conservation. Journal of Applied Ecology, 43, 1049–1055. [Google Scholar]
- Shokralla S, Spall JL, Gibson JF, Hajibabaei M (2012) Next‐generation sequencing technologies for environmental DNA research. Molecular Ecology, 21, 1794–1805. [DOI] [PubMed] [Google Scholar]
- Sinclair EH, Zeppelin TK (2002) Seasonal and spatial differences in diet in the western stock of Steller sea lions (Eumetopias jubatus). Journal of Mammalogy, 83, 973–990. [Google Scholar]
- Sint D, Raso L, Traugott M (2012) Advances in multiplex PCR: balancing primer efficiencies and improving detection success. Methods in Ecology and Evolution, 3, 898–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symondson WOC, Harwood JD (2014) Special issue on molecular detection of trophic interactions: unpicking the tangled bank. Molecular Ecology, 23, 3601–3604. [DOI] [PubMed] [Google Scholar]
- Thomas AC, Jarman SN, Haman KH, Trites AW, Deagle BE (2014) Improving accuracy of DNA diet estimates using food tissue control materials and an evaluation of proxies for digestion bias. Molecular Ecology, 23, 3706–3718. [DOI] [PubMed] [Google Scholar]
- Traugott M, Kamenova S, Ruess L, Seeber J, Plantegenest M (2013) Empirically characterising trophic networks: what emerging dna‐based methods, stable isotope and fatty acid analyses can offer In: Ecological Networks in an Agricultural World (eds Woodward G, Bohan DA.), pp. 177–224. Academic Press, Amsterdam, the Netherlands. [Google Scholar]
- Vestheim H, Deagle BE, Jarman SN (2011) Application of blocki ng oligonucleotides to improve signal‐to‐noise ratio in a PCR. Methods in Molecular Biology, 687, 265–274. [DOI] [PubMed] [Google Scholar]
- Vilches A, Arizaga J, Miranda R, Ibbotson A (2013) Impact of Common Kingfisher on a salmon population during the nestling period in southern England. Knowledge and Management of Aquatic Ecosystems, 410, 28–35. [Google Scholar]
- Ward RD, Zemlak TS, Innes BH, Last PR, Hebert PD (2005) DNA barcoding Australia's fish species. Philosophical Transactions of the Royal Society of London. Series B, Biological sciences, 360, 1847–1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarzoso‐Lacoste D, Corse E, Vidal E (2013) Improving PCR detection of prey in molecular diet studies: importance of group‐specific primer set selection and extraction protocol performances. Molecular Ecology Resources, 13, 117–127. [DOI] [PubMed] [Google Scholar]
- Zijlstra M, Vaneerden MR (1995) Pellet production and the use of otoliths in determining the diet of cormorants Phalacrocorax carbo sinensis: trials with captive birds. Ardea, 83, 123–131. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. The taxonomy of all Central European fish species targeted by the multiplex PCR assays including the number of DNA extracts generated from tissue samples and their origin.
Appendix S1. Red list freshwater fish species of Central Europe not included in this study.
Table S2. Non‐target animals used to evaluate the specificity of the multiplex PCR assays including the number of DNA extracts tested per taxon.
Appendix S2. Notes on the multiplex PCR assays.
Table S3. Collection locations of the Common Kingfisher (Alcedo atthis) a faeces.
Table S4. In silico PCR conditions and results.
Data Availability Statement
DNA sequences: GenBank Accession nos KR476824–KR476986 for 16S and KR476987–KR477299 for COI sequences. The screening results of the otter feeding trial and of the field‐collected samples as well as 16S and COI alignments of all sequences submitted to GenBank have been archived in the Dryad Digital Repository (doi:10.5061/dryad.1vf74).


