Abstract
Detection of a broad range of chemosensory signals is necessary for the survival of multicellular organisms. Chemical signals are the main facilitators of foraging, escape, and social behaviors. To increase detection coverage, animal sensory systems have evolved to create a large number of neurons with highly specific functions. The olfactory system, much like the nervous system as a whole, is astonishingly diverse.1‐3 The mouse olfactory system has millions of neurons with over a thousand classes, whereas the more compact Drosophila genome has approximately 80 odorant receptor genes that give rise to 50 neuronal classes and 1300 neurons in the adult.4 Understanding how neuronal diversity is generated remains one of the central questions in developmental neurobiology. Here, we review the current knowledge on the development of the adult Drosophila olfactory system and the progress that has been made toward answering this central question. WIREs Dev Biol 2015, 4:609–621. doi: 10.1002/wdev.197
For further resources related to this article, please visit the WIREs website.
STRUCTURE OF THE DROSOPHILA OLFACTORY SYSTEM
Adult flies have two main olfactory appendages, the antennae and the maxillary palps. The surfaces of both structures are covered with hair‐like structures called sensilla.1, 5, 6, 7 Each sensillum houses 1–4 olfactory receptor neurons (ORNs, Figure 1(a)).1, 5, 6, 7 Sensilla can be divided into three morphological classes, trichoids, ceoloconics, and basiconics. They can be further divided into subtypes based upon the invariable combinations of ORNs they house6 (Figure 1(b)). Each sensillum is named by two letters and a number, representing the olfactory appendage, the morphological class, and the subtype number, respectively. For example, the at4 sensilla are the 4th subtype of the antennal trichoid sensilla. ORNs in the basiconic and trichoid sensilla express olfactory receptor (Or) genes, whereas coeloconic ORNs express the ionotropic receptors (Irs).4, 8 There are also ORNs, such as the CO2 sensory neurons on the antennae, which express genes that resemble gustatory receptor (Gr) more than Or genes.9, 10, 11 ORN cell bodies reside at the base of sensilla where they are surrounded by supporting cells6 (Figure 1(c)). Each ORN extends its dendrites into the sensillar protrusion where it is exposed to the environment through the cuticular pores that enable diffusion of odorants into the sensilla7 (Figure 1(c)). Axonal projections of ORNs from the antennae enter the antennal lobe in the brain via the antenna nerve, whereas projections from the maxillary palp ORNs enter via the labial nerve.6, 12, 13 In the antennal lobe, axonal terminals of each ORN class synapse with the dendrites of projection neurons (PNs) within class‐specific glomeruli12, 13, 14 (Figure 1(d)). The antennal lobe is the first relay station of olfactory processing and PNs then send this information into higher brain structures, by projecting their axon terminals into primarily the mushroom body calyx and lateral horn.7 The Drosophila olfactory system serves as an excellent model for general nervous system development with a few distinct advantages. First, because each neuron expresses a single receptor and connects to a single glomerulus, it is possible to more carefully understand the relationship between neuronal identity and axonal connectivity. Second, the terminal fate of ORNs generated from each individual precursor can be clearly defined as they cluster within the same sensillum. Using the unique genetic toolkit available in Drosophila, it is possible to trace the fate of each precursor and interrogate the function of different proteins in assembling this diverse circuitry.
Figure 1.
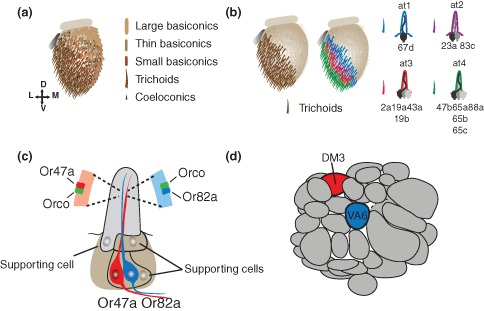
Structure of the Drosophila olfactory system. (a) The antenna is covered by sensory hairs called sensilla that can be divided into three classes based upon their morphology, basiconics, trichoids, and coeloconics. (b) Within morphological classes, sensilla can be divided into subtypes defined by the combination of olfactory receptor neurons (ORNs) they house. There are four classes of trichoid sensilla, at1‐4. Each houses a unique set of ORNs. (c) An example of a basiconic sensillum. ORNs project their dendrites into sensillum and express their receptors on the surface of their dendrites where the receptors dimerize with the olfactory coreceptor, Orco, and are exposed to the environment. (d) The antennal lobe of the fly brain is divided into class‐specific glomeruli, where ORN axons synapse with projection neuron (PN) dendrites. Each glomerulus is distinguished by its size position and shape. The glomeruli corresponding to ORNs from panel c are highlighted.
REGULATION OF OLFACTORY RECEPTOR EXPRESSION
One of the major questions in understanding the development of the Drosophila olfactory system is how Or genes are regulated in different ORN classes, where each ORN class expresses a single receptor from the large number of possibilities in the genome. What are the intrinsic and extrinsic factors that instruct each ORN to express a particular receptor? In Drosophila, Or expression at the periphery appears to be highly zonal and deterministic.7, 15 The mammalian olfactory epithelium is also divided into several zones, each containing ORNs that express distinct sets of olfactory receptors.16 However, in contrast to Drosophila, mammalian neurons within each zone appear to stochastically express a single allele of a single receptor.15 This system of selection certainly has its advantages for mammals, which can have over 1000 Or genes and over a million ORNs. The amount of regulatory factors needed to deterministically specify each fate in the mammalian system would be staggering, so some stochasticity likely eases the regulatory burden on the organism. In addition, the dynamics of neuron turnover in mammalian olfactory system also can benefit from a stochastic selection process as new neurons are integrated into existing circuits. Specifying the ∼60 ORN classes from the ∼80 Or genes that exist in Drosophila, however, is much more manageable by comparison, while maintaining a highly diverse system. The determinism seen in the Drosophila olfactory system may make it a better model for general nervous system development, which is less likely to rely on stochastic mechanisms where precise fate decisions are required.
Larval and Pupal Patterning Factors
The antenna and maxillary palps, like many other adult structures in the fly, both arise from an imaginal disc, specifically the eye‐antennal disc.17, 18 Imaginal discs are small epithelial sacs that are put aside during embryonic development that give rise to adult structures during pupal metamorphosis. Each disc is specified into its respective structure by the expression of homeotic genes.17 Homothorax is the primary homeotic factor that controls antennal fate, although it is also known to have other functions in the leg disc.19, 20 The legs and antennae have long been thought of as analogous organs. Both appendages are segmented, ventral organs, unlike unsegmented, dorsal organs such as the wing. Further, gain of function mutations of the homeotic gene antennapedia converts the antennae into legs.21 This suggests that the antennae and the legs may share a common developmental strategy for diversifying cell types, which is particularly interesting because the legs are gustatory organs that are covered by sensilla that house diverse sets of gustatory receptor neurons expressing Gr and Ir genes.18, 22, 23, 24 Most imaginal discs, including the leg and antennal discs are patterned by the expression of several genes and signaling pathways. For example, engrailed (en) is expressed early in disc development in the posterior compartment of the discs and activates hedgehog, which then signals to the anterior compartment of the disc18 (Figure 2(a)). Hedgehog (Hh) then activates both Wingless (Wg) and Decapentaplegic (Dpp), which are expressed in wedge‐like patterns on opposite sides of the disc25 (Figure 2(a)). The reason that these wedge‐like patterns form is that in the posterior compartment of the disc that the downstream signaling pathway of Hh is repressed by En, thereby restricting the activation of Wg and Dpp. Wg and Dpp then diffuse outward to form signaling gradients, which specify the ventral and dorsal fate, respectively, thereby, layering a new axis on top of the anterior–posterior compartments. At the center of the disc, where the expression of Wg and Dpp meet, the combination of these signaling pathways establish the proximal–distal axis of the antenna through activation of vein, an EGF receptor ligand, and distal‐less (dll) (Figure 2(a)). Both are expressed in the center of the disc25 and give rise to the more distal regions of the antenna, namely the entire third segment. Epidermal growth factor receptor (EGFR) signaling is critical for delineating the proximal–distal axis of the antenna, with the highest amount of activity in the center of the disc (distal) and gradually decreasing toward the outermost region (proximal).20, 25
Figure 2.
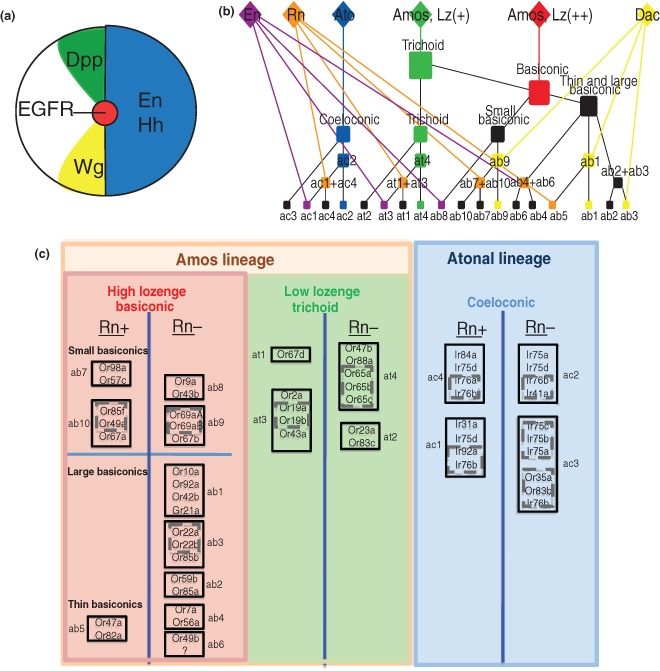
Control of olfactory receptor neuron (ORN) identity by larval prepatterning factors. (a) Schematic of morphogen signaling in the antennal disc. Engrailed (En) and Hedgehog (Hh) are expressed in the posterior compartment of the disc and Hh diffuses and signals to the anterior compartment. Wingless (Wg) and Decapentaplegic (Dpp) signaling are activated by Hh and establish the dorsoventral axis. In the center of the disc, where Wg and Dpp signaling events meet, epidermal growth factor receptor (EGFR) is activated to establish the proximal–distal axis. (b) Decision tree of sensory organ precursor (SOP) identity based upon combinatorial expression of prepatterning factors. Each SOP expresses a combination of prepatterning factors that control its fate in a nested and hierarchical fashion. The expression any given factor modifies the fate of a given SOP based upon the previous or concurrent expression of other prepatterning factors. (c) Expression of lozenge (lz), amos, and atonal define sensillar morphological classes. Atonal expressing precursors develop into coeloconic sensilla. Amos expression is required for both basiconic and trichoid sensilla. The level of lz expression then divides out the two classes, with basiconics and trichoids expressing high and low lz, respectively. Rn expression creates two populations of precursors within each morphological class, thereby increasing the number of possible sensilla fates by a factor of 2.
Control of Morphological Identity
Several genes are expressed in larval‐ and pupal‐antennal discs that lay down an initial patterning that is necessary for proper antennal development. Three of the factors expressed at these stages, lozenge (lz), amos, and atonal, control the morphological identity of sensilla26, 27, 28, 29 (Figure 2(b) and (c)). Lz, a member of the AML‐1/Runt transcription factor family, is the first of these factors to be expressed, beginning at the third instar larval stage and controls both basiconic and trichoid fates, most of which generate ORNs that express Ors.26, 29 Interestingly, it has been proposed that the trichoid and basiconic identities are controlled by the level of expression of lz.29 Hypomorphic mutations in lz lead to antennae that lack basiconic sensilla, whereas strong lz mutants lack both basiconic and trichoid sensilla, suggesting different thresholds for Lz function for determining basiconic and trichoid fates.29 These data have led to the idea that a subset of lz+ precursors expresses a high level of lz and become basiconics and another express a low level of lz and become trichoids (Figure 2(b) and (c)). Lz is also a positive regulator of amos, a bHLH transcription factor that controls morphological class identity.26 Like lz, amos is required for both basiconic and trichoid sensilla.26 Lz mutants lack amos expression and broad activation of lz correspondingly leads to broad expression of amos.26 As such, amos expression begins after lz, around the initiation of puparium formation (0 h APF, after puparium formation) and peaks around 6–8 h APF.26 It is worth noting that basiconic sensilla have their own morphological subclasses named large, thin, and small basiconics. While both lz and amos are required for all three types of basiconics, the factors that divide each subtype are not known. The development of coeloconic sensilla, which express primarily Irs as opposed to Ors, requires another bHLH transcription factor, atonal, which is highly related to amos 28 (Figure 2(b)). Atonal expression starts before amos, but generally overlaps with it in early pupal development as sensory organ precursors are being selected.28 Amos and Atonal both function as proneural genes and control precursor selection and identity, which underlie their role in specifying sensillar morphological identity.26, 28
Control of Sensillar Subtype
Diversification decisions regarding sensillar subtype identity are regulated by additional factors. For example, unlike the regulators of morphological divisions of sensilla (lz, amos, and atonal), Rotund (Rn), Dachshund (Dac) and Engrailed (En) specify sensillar subtype identity.30 Dac specifies several sensilla subtypes within basiconic sensilla, whereas En regulates specification within a subset within each morphological class.31, 32 Rn, similar to En, is also required to specify half of the subtypes within each morphological class30 (Figure 2(b) and (c)). However, unlike en mutants, rn mutants convert certain sensilla subtypes to others within each morphological sensilla type. For example, in trichoid sensilla both at1 and at3 precursors express rn, whereas precursors for at4 and at2 do not.30 In rn mutants, at1 and at3 sensilla are converted to at4 sensilla.30 In this model, Rn diversifies both at1 and at3 fates from the default at4. In other words, each trichoid sensilla type can be defined as rn+ or rn−. Lineage tracing experiments show that rn+ precursors do in fact give rise to not only half of the subtypes within trichoids but also basiconics and coeloconics (Figure 2(b) and (c)), suggesting that it functions to specify several fates within each morphological class.30 Indeed, in rn mutants, rn+ sensilla and the ORNs housed in them are converted to one of the default rn− sensilla and ORN identities within all sensilla morphological types30 (Figure 2(b) and (c)). These results suggest that Rn regulates the combinations of ORN identities that can be generated from a given multipotent precursor cell. It is intriguing that rn is expressed in the very early stages of olfactory system development and is turned off prior to the onset of receptor expression. In addition, Rn does not bind to Or/Ir promoters, thus, how Rn regulates the ORN differentiation potential of precursors and sensilla identity is still not clear.30 Thus, it is likely that Rn regulates the Or expression patterns indirectly via induction of transcription factors, which can either directly interact with OR promoters or modify chromatin around Or promoters affecting Or expression.
Notch Signaling
Once the precursor potentials are determined by the early patterning factors described in the previous section, each multipotent precursor undergoes several rounds of asymmetric divisions to generate 1–4 terminally differentiated ORNs in the same sensillum.33 Each consecutive asymmetric division from a single multipotent precursor is associated with binary segregation of possible cell fates. Notch signaling is a common pathway that is utilized for such binary segregation, and also contributes to proper segregation of ORN fates within each sensillum through lateral inhibition33 (Figure 3). The current model suggests the initial precursor, pI, undergoes an asymmetric division and generates two daughters, pIIa and pIIb33 (Figure 3). pIIa, a Notch‐on precursor, generates outer, supporting cells, whereas pIIb, a Notch‐off precursor generates the ORNs of each sensillum.33 pI, pIIa, and pIIb all express senseless (sens), a marker of precursor identity.33 pIIa and pIIb then each undergo a second round of asymmetric division to create the transit‐amplifying (or intermediate precursor) cells pOa and pOb, pNa and pNb, respectively.33 pO cells express cut, whereas pN cells continue to express sens.33 As the number of ORNs in different sensilla vary from 1 to 4 cells, this division pattern is thought to be complemented by mechanisms, such as cell death or adoption of glial fates, which determine the total number of ORNs per sensillum.34 And finally, ORN classes within a given sensillum can be classified as Notch‐on or Notch‐off based upon their requirement for Notch for their identity.33 For example, mutations in the positive effector of Notch signaling, mastermind (mam), leads to duplication of one ORN identity (Notch‐off) at the expense of another (Notch‐on) within a sensillum.33 In contrast, numb mutants, a protein that antagonizes Notch signaling, the Notch‐on ORN is duplicated at the expense of the Notch‐off33 (Figure 3). Thus, Notch signaling governs binary fate segregation of specific combinations of ORN identities from each precursor divisions. Mutations in early patterning factors, like rn mutants, cause regions of the antenna to lose pools of ORNs housed in sensilla from rn‐positive precursors and are rather covered by sensilla from rn‐negative precursors, keeping the ORN pairing and fate segregation appropriate. These results suggest that precursor identity must be multipotent yet be restricted in its potential to give rise to a restricted set of ORNs. Once this potential is set by early patterning factors, Notch signaling acts on each precursor division in a context‐dependent manner to segregate binary fate decisions toward terminally differentiated ORNs. It is unclear, however, what molecularly defines the restricted ORN potentials of different precursors. It is likely that the transcriptional and chromatin profiles of fields of cells in the antennal disc patterned during development are what define the precursor cell potentials.
Figure 3.
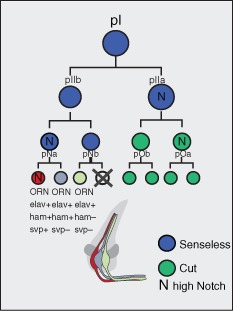
Generation of a sensillum from a single SOP through the use of Notch signaling. The initial SOP, initial precursor (pI), divides to create two daughter cell pIIa and pIIb, which will generate supporting cells and olfactory receptor neurons (ORNs), respectively. pIIb is Notch‐off and laterally inhibits pIIa from adopting a neural precursor fate through Notch signaling. pIIa and pIIb undergo two more rounds of division each before generating terminally differentiated cells. Blue cells express sens, green cells express cut, and N represents a Notch‐On state. Each ORN within a sensillum can also be defined by the combinatorial expression of elav, ham, and seven‐up (svp).
Epigenetic Regulation of Or Genes
Because Notch signaling is used broadly in all sensilla, each precursor must then have a restricted differentiation potential or allowable combination of Or genes that can be expressed by its daughters. Thus, the intermediate precursor cells must retain a cellular memory for Notch signaling to act on during each asymmetric division. It is plausible to imagine that the mechanisms governing these processes will likely include chromatin regulation.
Indeed, recent work has connected the mechanism of Notch signaling in ORN specification to the chromatin modifiers Hamlet (Ham, a homolog of Prdm16) and C‐terminal‐binding protein (CtBP).35 Hamlet is expressed in a subset of ORNs within each sensillum and is required for regulating Or expression.35 Ham functions as a repressor of Notch signaling and complexes with CtBP to reduce the amount of activating H3K4 methylation and increase the amount of repressive H3K27 methylation around Notch target genes.35 Further, the transcriptional corepressor Atrophin (Atro) has been shown to regulate Or expression in Notch‐on ORNs.36 Loss of Atro leads to the derepression of Notch‐on ORs in other Notch‐on ORNs, and likewise, overexpression of atro represses Notch‐on Ors.36 Interestingly, atro overexpression leads to a reduction in the amount of H3 acetylation in ORNs and the loss of Or expression in atro overexpression can be rescued by the loss of the histone deacetylase hdac3.36 These data suggest that modulation of chromatin and epigenetic states are critical for proper segregation of alternate ORN fates. As Notch signaling is used broadly across all sensilla types, it is not clear how Notch signaling during asymmetric divisions regulate Or expression among the ORNs to be generated from each precursor in a context independent manner. One attractive possibility is that the prepatterning factors discussed earlier modify each precursor to create a unique set of Or expression competency on which Notch signaling can then act to segregate out individual ORNs.
Recent work has also connected regulation of the CO2 receptors Gr21a and Gr63a expression to chromatin modifying proteins, specifically the MMB/dREAM complex.37 The MMB/dREAM complex is composed of several transcription factors and chromatin remodeling factors. Two members of the dREAM complex, Myb and Mip130, positively regulate Gr63a expression, whereas other members, Mip120 and E2F2, repress Gr63a expression in inappropriate ORNs.37 dREAM complex antagonizes H3K9 methylation to regulate Or expression in the appropriate olfactory appendages and ORNs.37 These data present a new layer of complexity to what is known about Or regulation that may lead to many new insights. However, a true interrogation of chromatin states at Or promoters at multiple developmental stages will be necessary to truly understand how epigenetic modifications contribute to Or regulation.
Late Transcription Factors
There are several transcription factors that are expressed in later stages of ORN development and regulate Or expression in postmitotic ORNs. This set of transcription factors are similar to the terminal selector genes proposed by Oliver Hobert, that function by directly binding to the promoters of Or genes, as the terminal differentiation genes.38 The more extensively studied of these factors is acj6, which is required for Or expression in a subset of ORN classes.39 Acj6 is a POU domain transcription factor that is expressed in adult maxillary palps and antennae.39 In acj6 mutants, the response profiles of a large number of ORNs are changed suggesting a change in ORN identity.40 Later research showed that expression of both antennal and palp Or genes are affected in acj6 mutants.40, 41 In addition to presence of Acj6‐binding sites upstream of these Or genes, most of the work on the molecular function of Acj6 in Or regulation was carried out for palp Or genes.42 These studies identified 13 different acj6 splice isoforms, each of which differentially regulates subsets of Ors in the maxillary palp.42 Expression of a single isoform of acj6 leads to the rescue expression of a subset Ors regulated by Acj6 in an acj6 mutant and individual isoforms can have both activation and repression functions.42 The molecular details of how each isoform of acj6 functions are still unclear but it is possible that each isoform binds to unique binding motifs or complexes with a unique set of other proteins. Pdm3 is also POU domain transcription factor, whose expression and function are similar to that of acj6.43 Pdm3 has also primarily been investigated in the palps, where it is required for activation of Or42a.43 Although pdm3 is expressed broadly in the antenna and maxillary palp, its function in each ORN class is not clear. In the context of Or42a, Pdm3 is known to work cooperatively with Acj6, suggesting they work in combination or possibly dimerize to regulate expression of their target Or genes.43
Given the large number of Or genes, and the singular expression of an Or gene in each ORN, the most parsimonious model suggests that a combinatorial code of transcription factors that control expression of different receptors in Drosophila. Most recently, Jafari et al. identified combinatorial function for Acj6 and six novel transcription factors (zf30c, sim, xbp1, fer1, E93, and onecut), that are required for proper Or expression in the antenna.41 Of these six proteins, Xbp1 and E93, function to both activate and restrict Or expression and all others serve solely as activators.41 These factors, as well as Acj6 and Pdm3, support the combinatorial regulation model of Or expression. However, there is evidence to suggest that these TFs alone cannot entirely explain how ORNs select a particular Or to express. For example, analysis of TF‐binding motifs shows that binding motifs for specific TFs are often present upstream of many Or genes, yet the expression of only a subset of OR genes are affected in the TF mutants.41 This might be due to combinatorial or competitive interactions at specific promoter elements, or the neuron‐specific patterns of TF expression. This set of TFs also act only in late stages of pupal development around the onset of Or expression. The only exception being zf30c, whose expression is required in both early and late stages of development.41 We anticipate that a thorough lineage map of TF expression patterns in the context of ORN development with mutant analysis as well as chromatin landscape of different ORNs and precursors will help reconcile these contradictions in the future.
Cis‐Regulatory Elements
A critical step in ORN specification is the selection of the olfactory receptor. In Drosophila, artificial Or promoter reporter constructs are able to faithfully recapitulate Or expression, even when not inserted in the endogenous locus. This observation has led to the conclusion that all of the information required for regulation of Or expression is contained within Or promoters. These sites are thought to encode information regarding which olfactory appendage, sensilla type, and subtype a given Or gene will be expressed in. The decision determining which olfactory appendage a given Or gene will be expressed requires two motifs, Dyad1 and Oligo1.2 Dyad 1 is required for expression of Or71a, Or46a, and Or85e in the maxillary palps.2 Conversely, Oligo1 is required to repress antennal expression of the same maxillary palp Ors.2 Other TFs that regulate OR expression in late stages, such as acj6 and pdm3, also have known binding motifs upstream of OR genes that are required for proper expression of specific set of Or genes.44 Examination of the trichoid Or promoter structure of Or47b, Or88a, Or65a, and Or67d has identified both activator and repressor elements.45 Or47b, Or88a, and Or65a are expressed in at4 sensilla, whereas at1 sensilla have a single ORN expressing Or67d. Miller and Carlson identified a GCAATTA motif common to the Or47b, Or88a, Or65a, and Or67d promoters they examined.45 Even though this motif served both as an activator and a repressor depending on the position of the motif within the promoter, the TF interacting with this specific motif is still unknown.45 In addition, both Or47b and Or67d promoters contain repressor elements that restrict their expression to trichoid sensilla zone on the antennae.45 Deletion of these elements leads to Or47b and Or67d expression in basiconic sensilla, both in antennae and maxillary palps.45 Interestingly, not only do Or47b and Or67d promoter deletion transgenes show ectopic expression in different subsets of basiconic sensilla, but Or67d expression was also detected in non‐neuronal cells, suggesting that there are other factors repressing the expression of specific trichoid Ors within subsets of basiconic sensilla.45
These data point to two prevailing theories of Or regulation: larval patterning and restriction of precursor cell potentials that determine the allowable combinations of Or genes to be expressed in a given sensilla, and a combinatorial code of terminal selector TFs that control Or expression during or after the asymmetric precursor divisions. Both mechanisms work in conjunction with each other to specify ORN fates. This model would also allow for more evolutionary flexibility as evolutionary processes can add or eliminate regulatory factors at different precursor decision points to alter Or expression or ORN diversity. What is currently missing is a clear link between these two theories. Unification of these two theories could also explain some of the inconsistencies between sets of results. For example, why are there Acj6‐binding motifs present upstream of Ors that Acj6 does not regulate? As discussed earlier, this can be due to either differences in the expression pattern of TFs or the differences in chromatin landscape around Or promoters in specific precursors set up by early prepatterning genes. It is plausible to think that the expression of the appropriate Or gene requires the presence of both the appropriate factor and the accessibility of the chromatin. So even though all Ors might have the binding site for a given transcription factor, their expression might not be affected by that transcription factor because of chromatin modifications that are a result of programs that pattern and restrict the differentiation potential of precursors in specific lineages. Understanding the mechanisms that underlie the patterning of precursors will likely be critical to establishing this link and pushing the field forward, as well as leading to key insights into the developmental strategies for generating cellular and neuronal diversity.
MicroRNA Control of ORN Circuit Assembly
In addition to the layers of transcriptional regulation of ORN specification described above, the microRNA miR‐279 has also been implicated in the control of ORN specification, specifically as a regulator of the position of CO2 sensory neurons.46 In wild‐type flies, the CO2 receptors, Gr21a and Gr63a, are coexpressed in the ab1 sensilla on the antenna and connect to the most ventral glomerulus in the antennal lobe. In miR‐279 mutants, however, Gr21a and Gr63a expressing ORNs are found in the maxillary palp and connect to a medial glomerulus.46 The expression pattern and connectivity of these ectopic neurons resembles that of a hybrid between mosquitoes which are attracted to CO2 and Drosophila, which normally avoid CO2. 46 Further work has demonstrated that miR‐279 functions in the development of maxillary palp olfactory sensilla precursors. miR‐279 is expressed in sensory organ precursors in the maxillary palps and miR‐279 mutants have an extra neuron that is generated within particular palp sensilla.46, 47 This is likely due to derepression of miR‐279 targets that function in the developmental program of the sensillum, which eventually determines the number of neurons. In the presence of miR‐279, these factors are downregulated in Drosophila leading to the elimination of CO2 ORNs from the maxillary palps.46, 47 The transcription factor Prospero may be a key regulator of this pathway, as it has been shown to activate miR‐279.47 This work has revealed a new level of post‐transcriptional regulation of the development of the Drosophila olfactory system, highlighting the complicated nature of generating such a diverse system.
AXONAL TARGETING
Not only must ORNs choose a receptor to express in a stereotyped manner, they must also connect to their class‐specific glomeruli in the antennal lobe. ORN axons in the antenna form three major bundles that fasciculate to form the antennal nerve.48 Axons from the antennal nerve enter the antennal lobe beginning around 18–20 h APF, and once in the brain the antennal nerve defasciculates into a ventromedial and a dorsolateral pathway48 (Figure 4). ORN axons then cross the midline via the dorsal commissure and connect to the appropriate glomerulus in both contralateral and ipsilateral antennal lobes.13, 14 Maxillary palp ORN axons enter the antennal lobe via labial nerve around 30 h APF and are guided to their glomerular target regions through interactions with antennal ORN axons that are already within the lobe13, 49, 50 (Figure 4). The rest of pupal development of the antennal lobe is devoted to refining and segregating the glomeruli, which are easily identifiable by size, position, and structure. In order to achieve the class‐specific connectivity of 50 different ORN classes to nonoverlapping glomeruli, ORN axons of the same type must converge onto the same glomerulus and synapse with the proper PNs as well as repel axons of all other classes (Figure 4). One simple solution to this problem is the use of olfactory receptors as instructive cues to govern glomerular targeting in order to compartmentalize connectivity in such a diverse system. Indeed, this strategy has been adopted in mammalian olfactory system.51, 52, 53 In mammals, Or genes encode G protein‐coupled receptors, and differences in agonist independent signaling from each OR leads to differences in the levels and combinations of cell surface molecules, which signal to sort out ORN axons as they innervate the olfactory bulb.52, 53 Despite the organizational similarities of olfactory system in mammals and Drosophila, Drosophila ORNs do not require receptor function for glomerular targeting. This suggests that in Drosophila programs for regulation of Or expression and glomerular connectivity are distinct, but must be coupled during ORN development. Indeed some of the early patterning factors, such as Hh, also pattern the axonal projections and glomerular connectivity of ORNs.54 In addition, molecular mechanisms that regulate Or expression, such as Notch signaling, and the transcription factors Acj6 and Pdm3, also play a significant roles in controlling glomerular targeting.33, 43, 55 For example, disrupting Notch signaling not only leads to conversion of sensory fates but also glomerular targeting.33 Both Acj6 and Pdm3 function to control targeting for different subsets of ORNs, with their loss producing ectopic glomeruli as well as glomeruli with diffuse boundaries.43, 55 Because both Acj6 and Pdm3 are transcription factors, it is likely that they regulate expression of cell surface proteins and/or guidance molecules that control wiring identity. It is thought that combinations of cell surface and guidance molecules expressed by ORN classes ensure proper connectivity in stepwise fashion that includes ORN axon trajectory selection, interglomerular and intraglomerular interactions that establish glomerular boundaries, and ORN–PN matching that ensures ORN wiring specificity.
Figure 4.
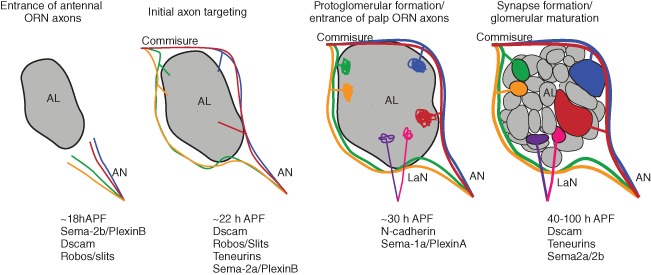
Development of the Drosophila antennal lobe. Olfactory receptor neuron (ORN) axons enter the antennal lobe around 18 h APF (after puparium formation) via the antennal nerve, guided by Dscam and Robos, and defasciculate into two major bundles based upon Semaphorin‐2b (Sema‐2b)/Plexin‐B signaling. Around 22 h APF ORN axons begin to target their respective glomeruli controlled by Dscam, Robos teneurins, and Sema‐2b, and cross the midline at the commissure to reach the contralateral antennal lobe. Around 30 h APF maxillary palp ORN axons enter the antennal lobe and protoglomerular formation begins, under the control of N‐cadherin (Ncad) and Semaphorin‐1a (Sema‐1a). From 40 h APF, glomeruli segregate and form distinct boundaries and ORN axons synapse with their projection neuron (PN) dendrites, under the guidance of Dscam, teneurins, and Sema2a/2b.
Coordination of Wiring among Neurons within the Same Sensillum
As discussed earlier, cell bodies of ORNs are clustered within individual sensilla, which develop through asymmetric divisions of a single precursor cell. As ORNs from the same sensilla enter the antennal lobe they take separate trajectories either dorsolateral or ventromedial bundles. Despite the well‐known role of Notch signaling on binary segregation of ORN fate decisions regarding olfactory receptor expression and targeting programs, until recently the molecular mechanisms working downstream of Notch to specifically regulate axonal trajectory selection were unclear. Recent work has connected Semaphorin‐2b (Sema‐2b) and Plexin‐B (PlexB) signaling to proper bundle segregation during this process.56 Sema‐2b is member of a family of secreted molecule, which signals through PlexB receptors. Sema‐2b is expressed in ORNs whose axons project along the ventromedial bundle and sema‐2b mutants lose the binary bundle separation and exclusively project through the dorsolateral bundle.56 This evidence suggests that Sema‐2b attracts and consolidates ORN axons into the ventromedial bundle. Notch‐δ signaling, in addition to controlling olfactory receptor expression among the ORNs, also segregates Sema‐2b expression within ORNs in the same sensillum.56 Notch signaling negatively regulates Sema‐2b expression, where Notch‐off neurons are positive for sema‐2b expression, suggesting Notch signaling negatively regulates sema‐2b.56 Perturbations to Notch signaling lead to inappropriate expression of sema‐2b accounting for the glomerular targeting defects observed in mam mutants. This is consistent with the observation that glomeruli targeted by Notch‐on and Notch‐off ORNs are segregated within subsections of the antennal lobe.
ORN Axon Guidance
Once in the antennal lobes, ORNs must navigate to specific regions to ultimately connect to their appropriate PN partners. General axon guidance molecules such as Dscam and Robo receptors were both shown to be required for this process.57, 58 Both Dscam and Robo receptors signal through the SH2/SH3 adaptor protein Dreadlocks (Dock) and the serine/threonine kinase Pak, which suggests that the action of these two molecules is coordinated.57, 58 Drosophila has three Robo receptors (Robo, Robo2, and Robo3), all of which are expressed in the olfactory system.58 Robo is broadly expressed across the majority of ORN axons, whereas expressions of robo2 and robo3 are more restricted.58 Robo2 is enriched in and near the commissure, and robo2 mutants are unable to properly cross the midline and form ectopic glomeruli near the commissure.58 Robo3 is expressed primarily in axons that follow the ventromedial trajectory, and loss of robo3 in these axons causes mistargeting and the formation of ectopic glomeruli, although some axons are still able to find the correct glomerulus.58 Suppression of Robo function leads to broad defects in glomerular structure, likely because many axons follow incorrect trajectories and choose incorrect targets leading to indistinct glomeruli.58 Similar to Robos, Dscam is also required for proper glomerular targeting. Dscam is a repulsive cell surface molecule and a member of the IgG superfamily.59 Dscam shows extraordinary molecular diversity through generation of approximately 38,000 alternative splice isoforms.59 Work on dscam in other contexts has suggested that stochastic alternative splicing within neurons leads to neuron‐specific expression of a subset of dscam isoforms, which show isoform‐specific homophilic binding.59 In the olfactory system, loss of dscam is associated with ectopic glomerular targeting and defects in both ipsilateral and contralateral ORN projections.57 These data suggest that Dscam is critical for proper targeting and guidance of ORN axons, however, this function of Dscam is diversity independent.57 Dscam diversity, on the other hand, is required for the proper class‐specific convergence of ORN axons within glomeruli.59 Even though dscam null mutants show ectopic glomerular targeting, they can still converge to form proper glomerular structures.57 In contrast, in flies that express only a single dscam isoform, ORN axons also are unable for converge into distinct glomeruli in addition to mistargeting defects.59 It is possible that the diversity independent function of Dscam regulates the initial axon guidance to specific regions within the antennal lobe, whereas Dscam diversity is required for class‐specific sorting and convergence into glomerular structures.57, 58
Intraclass Attraction
One of the critical steps in ORN connectivity is the class‐specific convergence and arborization of ORN axon terminals within a single glomerulus. This requires recognition among the ORN axons of the same class to confine them to the same glomerulus. As discussed above, Dscam diversity is required for proper glomerular convergence, as diversity compromised mutant ORN axons arrive at the antennal lobe yet are unable to converge and establish glomerular boundaries. This likely occurs due to repulsion of ORN axons of the same class in single dscam isoform mutants. In addition to Dscam, N‐cadherin (Ncad), which belongs to the calcium‐dependent cell‐adhesion molecule family, also contributes to the glomerular convergence.60, 61 In the olfactory system, ncad mutant ORN axons correctly target distinct regions within the antennal lobe, but they are not able to properly condense into protoglomeruli during mid pupal development leading to defects in the adult glomerular formation.60 These data suggest that Ncad is required for intraclass attraction during class‐specific convergence of ORNs into a single glomerulus. It is not entirely clear whether Ncad also mediates interactions between ORNs or between ORNs and PNs. Ncad is also broadly expressed across most ORN classes and as such ORN axons must have a way to distinguish and repel different classes.
Interclass Repulsion
Establishing 50 nonoverlapping glomeruli for class‐specific connections of 50 ORN classes requires interclass repulsion, which ensures that axon terminals from different ORN classes do not intermingle. Semaphorin‐1a (Sema‐1a) has been shown to be largely responsible for repulsion between classes of ORN axons. Unlike Sema‐2b, Sema‐1a is a transmembrane protein that acts through the Plexin‐A receptor. Sema‐1a is required for segregating different ORNs into distinct glomeruli.62 Mutants for sema‐1a show an intermingling of different ORN classes.62 Mutant axons are able to follow the correct trajectories into and through the antennal lobe, but then fail to properly sort into the correct glomeruli, and instead stay intermingled.62 Unlike ncad, clonal analysis of sema‐1a mutants demonstrates that it acts noncell autonomously, which suggests that its primary role is to repel axons of different classes. This repulsive function is critical for the formation of distinct glomerular boundaries.62 Sema‐1a is broadly expressed in most ORNs and different classes express sema‐1a at different levels, but it is not clear how differential expression levels of sema‐1a lead to glomerular segregation.62 However, Sema‐1a signaling acts in short ranges in the olfactory system, which could explain how it can be broadly expressed but lead to fine tuning of glomerular boundaries.
ORN–PN Matching
The final step in establishment of ORN circuits is proper class‐specific matching of ORNs with appropriate PNs for synaptic connectivity. It is plausible to imagine that cell surface molecules regulating ORN guidance will ultimately regulate ORN–PN matching. For example, in addition to its function in ORN connectivity, Dscam and Ncad have also been shown to affect PN targeting.63 Loss of dscam in PNs inhibits proper arborization of PN dendrites within glomeruli leading to dendritic clumping, consistent with its self‐avoidance function identified in other dendritic structures.63 Overexpression of dscam in PNs, on the other hand, leads to more diffuse dendritic structures as well as positional shifts.63 Interestingly, the positional shift of PN dendrites also causes ORN axons to shift while maintaining ORN–PN matching.63 This suggests that ORN axons recognize the appropriate PN dendrites, likely through other transmembrane interactions. Indeed, gradients of Sema‐2a and Sema‐2b expressed by PNs are required to position both PN dendrites and incoming ORN axons to specific zones within the antennal lobe.50, 55 Ncad also contributes to ORN–PN matching. Ncad mutant axons of both ORNs and PNs ultimately retract, unable to arborize within the glomerulus, and instead remain on the surface of the neuropil.60 The Luo group has also shown that the synaptic partner matching between ORN axons and PN dendrites requires the cell surface molecules, tenuerins.64 Teneurin‐m (Ten‐m) and Teneurin‐a (Ten‐a) are homophilic attraction proteins, expressed in matching pairs of ORNs and PNs.64 However, the partially overlapping expression of both ten‐m and ten‐a suggests that additional cell surface molecules working in combination should be specifying synaptic matching for 50 different ORN–PN to establish the proper connectivity of the olfactory circuits in the antennal lobes. Indeed, more recently Toll receptors, especially Toll‐6 and Toll‐7 were also shown to contribute to ORN–PN matching.65 Toll receptors are involved in embryonic patterning and innate immunity, however, their role in connectivity in the olfactory circuit appears to be independent of Toll pathways involved in these processes. Toll receptors encode transmembrane proteins with leucine‐rich repeats (LRR) involved in heterophilic protein–protein interactions. These results suggest both hemophilic and heterophilic interactions govern synaptic partner selection between ORNs and PNs.
CONCLUDING REMARKS
Despite the considerable work that has been done to elucidate mechanisms controlling ORN specification and diversity, much of the molecular details coordinating regulation of OR expression and wiring decisions to establish ORN circuits remains unknown. First, what molecular mechanisms link expression of specific olfactory receptor and wiring identity? While it is clear from the overall structure of the olfactory system that both decisions must be coordinated, receptor function is not required for proper targeting. As of yet only notch, acj6, and pdm3 have been shown to control both processes, but the molecular mechanisms utilized by these proteins at distinct developmental decision points remain largely unknown. Second, how do ORNs and their precursors determine which OR to express? While evidence exists for models of prepatterning of precursors as well as a combinatorial code of terminal selector TF expression, no clear link has yet been established between these two theories. It is likely that stepwise restrictions on distinct fate programs through combinations of transcription factors and cell surface molecules diversify sensory and wiring identities as ORNs are generated from precursors. Future work focusing on identification of these pathways and understanding their molecular and cellular details will allow us to understand mechanisms of neuronal diversity in the nervous system and will likely be broadly applicable in other systems.
Conflict of interest: The authors have declared no conflicts of interest for this article.
REFERENCES
- 1. Brochtrup A, Hummel T. Olfactory map formation in the Drosophila brain: genetic specificity and neuronal variability. Curr Opin Neurobiol 2011, 21:85–92. [DOI] [PubMed] [Google Scholar]
- 2. Fuss SH, Ray A. Mechanisms of odorant receptor gene choice in Drosophila and vertebrates. Mol Cell Neurosci 2009, 41:101–112. [DOI] [PubMed] [Google Scholar]
- 3. Takeuchi H, Sakano H. Neural map formation in the mouse olfactory system. Cell Mol Life Sci 2014, 71:3049–3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Clyne PJ, Warr CG, Freeman MR, Lessing D, Kim J, Carlson JR. A novel family of divergent seven‐transmembrane proteins: candidate odorant receptors in Drosophila . Neuron 1999, 22:327–338. [DOI] [PubMed] [Google Scholar]
- 5. Vosshall LB, Amrein H, Morozov PZ, Rzhetsky A, Axel R. A spatial map of olfactory receptor expression in the Drosophila antenna. Cell 1999, 96:725–736. [DOI] [PubMed] [Google Scholar]
- 6. Couto A, Alenius M, Dickson BJ. Molecular, anatomical, and functional organization of the Drosophila olfactory system. Curr Biol 2005, 15:1535–1547. [DOI] [PubMed] [Google Scholar]
- 7. Rodrigues V, Hummel T. Development of the Drosophila olfactory system. Adv Exp Med Biol 2008, 628:82–101. [DOI] [PubMed] [Google Scholar]
- 8. Benton R, Vannice KS, Gomez‐Diaz C, and Vosshall LB. Variant ionotropic glutamate receptors as chemosensory receptors in Drosophila . Cell 2009, 136:149–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kwon JY, Dahanukar A, Weiss LA, Carlson JR. The molecular basis of CO2 reception in Drosophila . Proc Natl Acad Sci U S A 2007, 104:3574–3578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yao CA, Carlson JR. Role of G‐proteins in odor‐sensing and CO2‐sensing neurons in Drosophila . J Neurosci 2010, 30:4562–4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jones WD, Cayirlioglu P, Kadow IG, Vosshall LB. Two chemosensory receptors together mediate carbon dioxide detection in Drosophila . Nature 2007, 445:86–90. [DOI] [PubMed] [Google Scholar]
- 12. Fishilevich E, Vosshall LB. Genetic and functional subdivision of the Drosophila antennal lobe. Curr Biol 2005, 15:1548–1553. [DOI] [PubMed] [Google Scholar]
- 13. Vosshall LB, Wong AM, Axel R. An olfactory sensory map in the fly brain. Cell 2000, 102:147–159. [DOI] [PubMed] [Google Scholar]
- 14. Bhalerao S, Sen A, Stocker R, Rodrigues V. Olfactory neurons expressing identified receptor genes project to subsets of glomeruli within the antennal lobe of Drosophila melanogaster . J Neurobiol 2003, 54:577–592. [DOI] [PubMed] [Google Scholar]
- 15. Clowney EJ, LeGros MA, Mosley CP, Clowney FG, Markenskoff‐Papadimitriou EC, Myllys M, Barnea G, Larabell CA, Lomvardos S. Nuclear aggregation of olfactory receptor genes governs their monogenic expression. Cell 2012, 151:724–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ressler KJ, Sullivan SL, Buck LB. A zonal organization of odorant receptor gene expression in the olfactory epithelium. Cell 1993, 73:597–609. [DOI] [PubMed] [Google Scholar]
- 17. Aplin AC, Kaufman TC. Homeotic transformation of legs to mouthparts by proboscipedia expression in Drosophila imaginal discs. Mech Dev 1997, 62:51–60. [DOI] [PubMed] [Google Scholar]
- 18. Morata G. How Drosophila appendages develop. Nat Rev Mol Cell Biol 2001, 2:89–97. [DOI] [PubMed] [Google Scholar]
- 19. Yao L‐C, Liaw G‐J, Pai C‐Y, Sun SH. A common mechanism for antenna‐to‐leg transformation in Drosophila: suppression of homothorax transcription by four HOM‐C genes. Dev Biol 1999, 211:268–276. [DOI] [PubMed] [Google Scholar]
- 20. Dong PDS, Chu J, Panganiban G. Coexpression of the homeobox genes distal‐less and homothorax determines Drosophila antennal identity. Development 2000, 127:209–216. [DOI] [PubMed] [Google Scholar]
- 21. Schneuwly S, Klemenz R, Gehring WJ. Redesigning the body plan of Drosophila by ectopic expression of the homeotic gene Antennapedia. Nature 1987, 325:816–817. [DOI] [PubMed] [Google Scholar]
- 22. Stocker RF. The organization of the chemosensory system in Drosophila melanogaster: a review. Cell Tissue Res 1994, 275:3–26. [DOI] [PubMed] [Google Scholar]
- 23. Dahanukar A, Hallem EA, Carlson JR. Insect chemoreception. Curr Opin Neurobiol 2005, 15:423–430. [DOI] [PubMed] [Google Scholar]
- 24. Ling F, Dahanukar A, Weiss LA, Kwon JY, Carlson JR. The molecular and cellular basis of taste coding in the legs of Drosophila . J Neurosci 2014, 34:7148–7164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Royet J, Finkelstein R. Establishing primordia in the Drosophila eye‐antennal disc: the roles of Decapentaplegic, Wingless and hedgehog. Development 1997, 124:4793–4800. [DOI] [PubMed] [Google Scholar]
- 26. Goulding SE, zur Lage P, Jarman AP. amos, a proneural gene for Drosophila olfactory sense organs that is regulated by lozenge . Neuron 2000, 25:69–78. [DOI] [PubMed] [Google Scholar]
- 27. zur Lage PI, Prentice DR, Holohan EE, Jarman AP. The Drosophila proneural gene amos promotes olfactory sensillum formation and suppresses bristle formation. Development 2003, 130:4683–4693. [DOI] [PubMed] [Google Scholar]
- 28. Gupta BP, Rodrigues V. Atonal is a proneural gene for a subset of olfactory sense organs in Drosophila . Genes Cells 1997, 2:225–233. [DOI] [PubMed] [Google Scholar]
- 29. Gupta BP, Rodrigues V. Patterning an epidermal field: Drosophila lozenge, a member of the AML‐1/Runt family of transcription factors, specifies olfactory sense organ type in a dose‐dependent manner. Dev Biol 1998, 203:400–411. [DOI] [PubMed] [Google Scholar]
- 30. Li Q, Ha TS, Okuwa S, Wang Y, Wang Q, Millard SS, Smith DP, Volkan PC. Combinatorial rules of precursor specification underlying olfactory neuron diversity. Curr Biol 2013, 23:2481–2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Song E, de Bivor B, Dan C, Kunes S. Determinants of the Drosophila odorant receptor pattern. Dev Cell 2012, 22:363–376. [DOI] [PubMed] [Google Scholar]
- 32. Blagburn JM. Engrailed expression in subsets of adult Drosophila sensory neurons: an enhancer‐trap study. Invert Neurosci 2008, 8:133–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Endo K, Aoki T, Yoda Y, Kimura K, Hama C. Notch signal organizes the Drosophila olfactory circuitry by diversifying the sensory neuronal lineages. Nat Neurosci 2007, 10:153–160. [DOI] [PubMed] [Google Scholar]
- 34. Sen A, Kuruvilla D, Pinto L, Sarin A, Rodrigues V. Programmed cell death and context dependent activation of the EGF pathway regulate gliogenesis in the Drosophila olfactory system. Mech Dev 2004, 121:65–78. [DOI] [PubMed] [Google Scholar]
- 35. Endo K, Karim MR, Taniguchi H, Krejci A, Kinameri E, Siebert M, Ito K, Bray SJ, Moore AW. Chromatin modification of Notch targets in olfactory receptor neuron diversification. Nat Neurosci 2012, 15:224–233. [DOI] [PubMed] [Google Scholar]
- 36. Alkhori L, Ost A, Alenius M. The corepressor Atrophin specifies odorant receptor expression in Drosophila . FASEB J 2014, 28:1355–1364. [DOI] [PubMed] [Google Scholar]
- 37. Sim CK, Perry S, Tharadra SK, Lipsick JS, Ray A. Epigenetic regulation of olfactory receptor gene expression by the Myb‐MuvB/dREAM complex. Genes Dev 2012, 26:2483–2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hobert O. Regulatory logic of neuronal diversity: terminal selector genes and selector motifs. Proc Natl Acad Sci U S A 2008, 105:20067–20071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ayer RK Jr, Carlson J. Olfactory physiology in the Drosophila antenna and maxillary palp: acj6 distinguishes two classes of odorant pathways. J Neurobiol 1992, 23:965–982. [DOI] [PubMed] [Google Scholar]
- 40. Clyne PJ, Certel SJ, de Bruyne M, Zaslavsky L, Johnson WA, Carlson JR. The odor specificities of a subset of olfactory receptor neurons are governed by Acj6, a POU‐domain transcription factor. Neuron 1999, 22:339–347. [DOI] [PubMed] [Google Scholar]
- 41. Jafari S, Alkhori L, Schleiffer A, Brochtrup A, Hummel T, Alenius M. Combinatorial activation and repression by seven transcription factors specify Drosophila odorant receptor expression. PLoS Biol 2012, 10:e1001280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bai L, Carlson JR. Distinct functions of acj6 splice forms in odor receptor gene choice. J Neurosci 2010, 30:5028–5036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Tichy AL, Ray A, Carlson JR. A new Drosophila POU gene, pdm3, acts in odor receptor expression and axon targeting of olfactory neurons. J Neurosci 2008, 28:7121–7129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lee M‐H, Salvaterra PM. Abnormal chemosensory jump 6 is a positive transcriptional regulator of the cholinergic gene locus in Drosophila olfactory neurons. J Neurosci 2002, 22:5291–5299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Miller CJ, Carlson JR. Regulation of odor receptor genes in trichoid sensilla of the Drosophila antenna. Genetics 2010, 186:79–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Cayirioglu P, Kadow IG, Zhan X, Okamura K, Suh GSB, Gunning D, Lai EC, Zipursky SL. Hybrid neurons in a microRNA mutant are putative evolutionary intermediates in insect CO2 sensory systems. Science 2008, 319:1256–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hartl M, Loschek LF, Stephan D, Siju KP, Knappmeyer C, Kadow IC. A new Prospero and microRNA‐279 pathway restricts CO2 receptor neuron formation. J Neurosci 2011, 31:15660–15673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Jhaveri D, Sen A, Rodrigues V. Mechanisms underlying olfactory neuronal connectivity in Drosophila‐the atonal lineage organizes the periphery while sensory neurons and glia pattern the olfactory lobe. Dev Biol 2000, 226:73–87. [DOI] [PubMed] [Google Scholar]
- 49. Jefferis GS, Hummel T. Wiring specificity in the olfactory system. Semin Cell Dev Biol 2006, 17:50–65. [DOI] [PubMed] [Google Scholar]
- 50. Sweeney LB, Couto A, Chou YH, Berdnik D, Dickson BJ, Luo L, Komiyama T. Temporal target restriction of olfactory receptor neurons by Semaphorin‐1a/PlexinA‐mediated axon‐axon interactions. Neuron 2007, 53:185–200. [DOI] [PubMed] [Google Scholar]
- 51. Feinstein P, Bozza T, Rodriguez I, Vassalli A, Mombaerts P. Axon guidance of mouse olfactory sensory neurons by odorant receptors and the β2 adrenergic receptor. Cell 2004, 117:833–846. [DOI] [PubMed] [Google Scholar]
- 52. Imai T, Suzuki M, Sakano H. Odorant receptor‐derived cAMP signals direct axonal targeting. Science 2006, 314:657–661. [DOI] [PubMed] [Google Scholar]
- 53. Nakashima A, Takeuchi H, Imai T, Saito H, Abe T, Chen M, Weinstein LS, Yu CR, Storm DR, et al. Agonist‐independent GPCR activity regulates anterior‐posterior targeting of olfactory sensory neurons. Cell 2013, 154:1314–1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Chou YH, Zheng X, Beachy PA, Luo L. Patterning axon targeting of olfactory receptor neurons by coupled hedgehog signaling at two distinct steps. Cell 2010, 142:954–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Komiyama T, Carlson JR, Luo L. Olfactory receptor neuron axon targeting: intrinsic transcriptional control and hierarchical interactions. Nat Neurosci 2004, 7:819–825. [DOI] [PubMed] [Google Scholar]
- 56. Joo WJ, Sweeney LB, Liang L, Luo L. Linking cell fate, trajectory choice, and target selection: genetic analysis of Sema‐2b in olfactory axon targeting. Neuron 2013, 78:673–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Hummel T, Vasconcelos ML, Clemens JC, Fishilevich Y, Vosshall LB, Zipursky SL. Axonal targeting of olfactory receptor neurons in Drosophila is controlled by Dscam. Neuron 2003, 37:221–231. [DOI] [PubMed] [Google Scholar]
- 58. Jhaveri D, Saharan S, Sen A, Rodrigues V. Positioning sensory terminals in the olfactory lobe of Drosophila by Robo signaling. Development 2004, 131:1903–1912. [DOI] [PubMed] [Google Scholar]
- 59. Hattori D, Demir E, Kim HW, Viragh E, Zipursky SL, Dickson BJ. Dscam diversity is essential for neuronal wiring and self‐recognition. Nature 2007, 449:223–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Hummel T, Zipursky SL. Afferent induction of olfactory glomeruli requires N‐cadherin. Neuron 2004, 42:77–88. [DOI] [PubMed] [Google Scholar]
- 61. Zhu H, Luo L. Diverse functions of N‐cadherin in dendritic and axonal terminal arborization of olfactory projection neurons. Neuron 2004, 42:63–75. [DOI] [PubMed] [Google Scholar]
- 62. Lattemann M, Zierau A, Schulte C, Seidl S, Kuhlmann B, Hummel T. Semaphorin‐1a controls receptor neuron‐specific axonal convergence in the primary olfactory center of Drosophila . Neuron 2007, 53:169–184. [DOI] [PubMed] [Google Scholar]
- 63. Zhu H, Hummel T, Clemens JC, Berdnik D, Zipursky SL, Luo L. Dendritic patterning by Dscam and synaptic partner matching in the Drosophila antennal lobe. Nat Neurosci 2006, 9:349–355. [DOI] [PubMed] [Google Scholar]
- 64. Hong W, Mosca TJ, Luo L. Teneurins instruct synaptic partner matching in an olfactory map. Nature 2012, 484:201–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Ward A, Hong W, Favaloro V, Luo L. Toll receptors instruct axon and dendrite targeting and participate in synaptic partner matching in a Drosophila olfactory circuit. Neuron 2015, 85:1013–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]


