Abstract
Accurate and rapid methods for the detection of DNA damage foci in eukaryotic cells are central to DNA repair studies, which identify differences in DNA repair capacity in cell lines. Such assays have been important in delineating mechanisms of DNA repair in human cells. Previously we were the first to demonstrate the use of imaging flow cytometry for the detection of γ‐H2AX foci in cells exposed to ionizing radiation causing the induction of DNA strand breaks. In this report we extend these studies and show an enhancement of foci quantitation and image resolution using next generation imaging flow cytometry with the Amnis ImagestreamX Mark II. We demonstrate using cell lines derived from normal individuals, and DNA double strand break repair defective cells that the number of foci observed is significantly increased when using 60× as compared to 40× magnification. Also, foci numbers and resolution is further increased with the application of the focus stacking (Extended Depth of Field–EDF) capacity activated. This report represents the first such demonstration of multimagnification and EDF for the enhanced quantitation of DNA damage in cells and provides a level of resolution, which near matches in situ microscopy methods for the detection of γ‐H2AX foci. © 2015 The Authors. Published by Wiley Periodicals Inc. on behalf of ISAC.
Keywords: γ‐H2AX foci, DNA repair, DNA damage, imaging flow cytometry, magnification, EDF
Nuclear localisation of γ‐H2AX foci has proven to be a useful method to indirectly quantify DNA double strand breaks (DSB) in eukaryotic cells. Induction of a DNA DSB leads to the phosphorylation of the minor histone protein H2AX on serine139 within the protein to form γ‐H2AX. Thousands of phosphorylations at the sites of DNA DSB lead formation of a focus and the number of foci correspond broadly to the level of DNA DSB 1. γ‐H2AX foci can be identified using immunological methods by the application of antibodies raised against the phosphorylated serine139 residue within the γ‐H2AX histone protein 2. Furthermore, using this approach individual cellular differences in DNA DSB repair capacity can be identified by measuring the kinetics of repair typically over a 24 h period. For example in cells derived from patients with the inherited disorder Ataxia Telangiectasia (A‐T), which is characterized by a defect in the repair of DNA DSB, there is a retention of γ‐H2AX foci as compared to cells derived from normal individuals 3. Recent years has seen the development of high throughput automated platforms for the quantitation of γ‐H2AX foci within cells. Traditionally and arguably the most accurate method is to use in situ methods using fluorescence microscopy. However, this approach requires significant user input and the quantitation of large cell numbers for statistical robustness is impossible. Alternatively, zero resolution flow cytometry permits the estimation of overall fluorescence levels in many thousands of cells but lacks the ability to quantify foci numbers in individual cells. Automated single cell microscopy analysis combined with laser image analysis provides accurate foci quantitation together with high cell number analysis 4, 5 and permits high throughput analysis and quantitation.
Alternatively and in more recent years the novel application of imaging flow cytometry provides high cell throughput capability whilst allowing the acquisition of cellular images on multiple channels simultaneously. In 2012 we reported in Cytometry A the use of imaging flow cytometry to identify γ‐H2AX foci in human cells derived from normal individuals and those from individuals with inherited defects in DNA DSB repair. Using an ImagestreamX Mark I (Amnis Corporation, Merck‐Millipore, Seattle, WA) with a 40× magnification objective, we demonstrated that in cells derived from an A‐T patient and from an individual with a defect in the DNA‐PKcs gene a defect in DSB repair was evidenced by a retention of γ‐H2AX foci in the nuclei of cells exposed to 2 Gy gamma radiation over a 24 h period 3. DNA‐PKcs is an essential component of the non‐homologous end joining (NHEJ) pathway central to DSB repair. This retention of foci corresponded to the defect in DNA DSB repair characteristic of these cell lines. This observation was gratifying given that we were able to detect subtle differences in DNA DSB repair in a variety of human cells types. Ultimately, we wish to use the ImagestreamX technology as a diagnostic platform for the identification of radiotherapy patients, which are likely to experience severe normal tissue toxicity due to occult defects in DNA DSB repair 6.
Within the same study we also compared the detection of foci using the ImagestreamX with foci numbers determined using in situ microscopy detection of γ‐H2AX. While we were able to detect quantitative differences in foci induction and persistence in various cell lines, we noticed that the number of observable foci in the nuclei of cells estimated using in situ microscopy was some 25–30% higher than that observed with imaging flow cytometry using a 40× magnification objective. This is not surprising given the higher level of resolution afforded by traditional microscopy approaches with a 100× magnification objective. Notwithstanding, imaging flow cytometry has allowed our group to identify novel DNA repair defects in radiation hypersensitive breast cancer patients 7 and revealed differences in DNA DSB repair in lymphoblastoid cells lines derived from individuals with heterozygous mutations in the BRCA1 and BRCA2 genes 8.
More recently we have acquired the ImagestreamX Mark II which provides a significant enhancement to imaging flow cytometry capability. For the quantitation of γ‐H2AX foci the ability to image foci at higher magnification (up to 60×) together with a focus stacking capability provided by the extended depth of field (EDF) function potentially allows a more accurate assessment of foci number throughout the complete nuclear region. To examine the efficiency of these novel functions within the ImagestreamX Mark II, we have conducted a follow‐up study to our 2012 (Cytometry A) investigation and have quantitated foci induction in normal and DNA DSB‐repair defective cell lines. Gamma‐H2AX foci were determined in two normal and two DNA DSB repair defective cell lines using 40× and 60× magnification with and without EDF image acquisition. We demonstrated that, (1) the use of higher magnification (60×) permitted the quantitation of increased foci numbers in individual cell nuclei and, (2) the use of EDF (focus stacking) further increased the resolution of foci within cell nuclei. Moreover, the EDF function improved image resolution, reduced background staining and perhaps most importantly allowed the quantitation of foci at similar levels to that achieved with in situ microscopy approaches 3.
In summary, the enhanced functions of imaging flow cytometry afforded by the Amnis ImagestreamX Mark II allows more accurate analysis of DNA damage foci and is a reliable and useful platform for the analysis of DNA damage and repair in eukaryotic cells.
Materials and Methods
Cell Lines
The XP14BRneo17 (SV40 immortalised) fibroblast cell line was derived from an individual with a defect in the NHEJ DNA DSB repair pathway and has been described in detail elsewhere 9. AT5BIVA SV40 immortalised fibroblasts are derived from a patient with the DNA DSB repair defective inherited disease ataxia telangiectasia (A‐T) and is well described by Murnane et al. 1985 10. The MRC5‐SV1 SV40 immortalized lung fibroblasts and 1BR.3‐G SV40 immortalised skin fibroblasts are derived from normal individuals 11.
Cell Culture
The cell lines were routinely cultured in T75 cell culture flasks or petri dishes (Sarstedt, Leicester, UK) in Dulbecco's modified Eagle's medium (Labtech, East Sussex, UK) supplemented with 10% foetal bovine serum, 2.0 mM l‐Glutamine (Labtech) and 100 U/ml Penicillin and Streptomycin (Scientific Laboratory Supplies, Leicester, UK). Cells were incubated at 37°C in a humidified atmosphere of 5% CO2 in air. At ∼80% confluence cells were routinely subcultured by trypsinization in trypsin/EDTA solution containing 170 U/ml trypsin, 0.2 mg/ml EDTA in phosphate buffered saline (Labtech).
Cell counts and viability were determined using a “Countess™” automated cell counter that utilises the trypan blue exclusion method (Life Technologies, Paisley, Scotland, UK). Cells were not used for experimentation if overall viability was <80%. To minimise genetic drift in culture all cells were used over a restricted range of 10 passages.
DNA Damage Induction by Gamma Radiation Exposure
Cells were plated into 10 cm dishes at concentrations of 2 × 106 cells/dish for the determination of DNA DSB by γ‐H2AX induction.
Two dishes of each cell line were retained as an unirradiated control; the remaining dishes (2 per time point) were irradiated with 2 Gy gamma radiation from a 60Cobalt source (Puridec irradiation Technologies, Oxon, UK) sited at a distance of 25 cm from the source with a dose rate of 0.8 Gy per minute. Two Gray gamma irradiation was chosen to correspond to fractionated doses of radiation used in clinical radiotherapy protocols. We have also demonstrated previously that the induction of DNA DSB by 2 Gy gamma irradiation represents a region of maximal γ‐H2AX induction 3. Following exposure the cells were returned to the incubator (incubation conditions described previously), and two dishes removed at 30 min, 3 h, 5 h, and 24 h after irradiation and fixed in methanol:acetone (50:50 v/v).
Cells were harvested by washing once with PBS (Severn Biotech, Worcestershire, UK) followed by trypsinization as described above. Cells were pelleted by centrifugation and washed in ice‐cold PBS and fixed in ice‐cold methanol:acetone (50:50 v/v). Two compensation samples for imaging flow cytometry were prepared in the same way at the 30 min time‐point. Samples were stored at −20°C until the immunocytochemistry stage. The immunocytochemistry was conducted within 5 days of irradiation.
Immunocytochemistry—Gamma H2AX Assay
The method for antibody staining of γ‐H2AX foci has been described elsewhere (e.g., 3, 8). In brief cells were resuspended in PBS then agitated gently for 5 min at room temperature in permeabilization buffer consisting of 0.5% Triton™ X‐100 (Sigma‐Aldrich, Dorset, UK) in PBS. Cells were incubated with gentle agitation at room temperature for 1 h in blocking buffer consisting of 5.0% rabbit serum (Labtech) with 0.1% Triton™ X‐100 in PBS. After the removal of the blocking buffer the cells were incubated for 24 h with gentle agitation at 4°C in primary antibody solution, consisting of an antiphospho‐histone H2AX (serine139) mouse monoclonal IgG1 antibody, clone JBW301 (Millipore, Watford, UK) diluted 1 in 10,000 in blocking buffer. Excess primary antibody was removed by washing twice in buffer consisting of 0.1% Triton™ X‐100 in PBS. A secondary antibody solution consisting of Alexa Fluor®488 rabbit antimouse IgG antibody (Invitrogen) diluted 1 in 1000 in blocking buffer was added to each sample, except the Draq 5 (Biostatus, Leicester, UK) compensation samples, and incubated with gentle agitation for 1 h at room temperature. Excess secondary antibody was removed by washing twice with wash buffer.
Cells were prepared for imaging flow cytometry by resuspending in PBS containing 2% foetal bovine serum (FBS) (Labtech) prior to the addition of 5 μM Draq 5. During imaging flow cytometry, images of between 5000 and 10,000 cells were acquired at 40× and 60× magnification with and without EDF using the 488 nm excitation laser set at 100 mW. For compensation samples images of 500 cells were acquired illuminated by the 488 nm laser with the brightfield and darkfield lasers inactivated.
Analysis of Cell Images—Calculation of Gamma H2AX Foci Number in Cells
Gamma‐H2AX foci were quantified in ∼5000–10,000 images of cells captured with the Inspire™ imaging flow cytometry software using a method described in detail elsewhere 3, 8.
In brief the Ideas™ software employs a series of predefined building blocks which first identifies cells that are in focus, followed by single cells based upon cell area and aspect ratio (Figs. 1 and 2, Supporting Information). The spot counting analysis wizard then prompts the user to define a population of cells with very few foci (<3) and very high foci numbers (>10), which are identified by the user to encompass spots of different sizes, shape, and fluorescence intensity. These user defined “Truth” populations are then used to create the most accurate foci mask to enumerate the number of foci in each cell for the whole population of cells (Fig. 3, Supporting Information). Examples of the masking workflow strategy used for each magnification level with and without EDF applied are included in Figure 1.
Figure 1.

This figure shows representative images of foci derived from the imaging flow cytometry data collection. Images of γ‐H2AX foci are shown at 40× magnification with and without EDF (panels A and B) and at 60× magnification with and without EDF (panels C and D). Under each image panel the Ideas workflow masking strategy is included to demonstrate how foci were quantified at each magnification level.
Statistical Analysis
At the point of maximal foci induction (30 min post gamma radiation exposure) we have compared the number of foci quantified at each magnification level with and without EDF. Comparisons were made by using a Student's unpaired t‐test and a P‐value equal to or less than 0.05 was regarded as being significantly different.
Results
In Figures 2, 3, 4 we display the number of foci in each cell line calculated using the Ideas™ software. We present the number of foci in untreated control cells and also in cells irradiated with 2 Gy gamma irradiation at 30 min, 3, 5, and 24 h post‐irradiation. These time points allow a period of repair to occur and provide a suitable time frame during which differences in repair dynamics in individual cell lines may be revealed 3. For each cell line we further present the number of foci calculated at 40× and 60× magnification with and without focus stacking provided by EDF.
Figure 2.
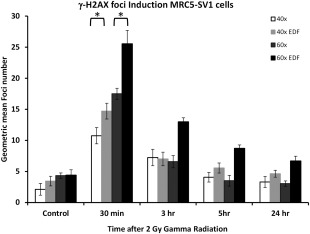
This figure shows the induction of γ‐H2AX in the nuclei of the DNA DSB repair competent MRC5‐SV1 cells over a 24 h time course following exposure to 2 Gy gamma radiation. Foci induction kinetics are shown for images captured at 40× and 60× magnification with and without the application of EDF. Data are derived from the capture of at least 5000–10,000 images of cells and error bars represent the standard deviation of foci numbers derived from the quantitated foci numbers. Statistical analysis (Student's unpaired t‐test) has been applied to the 30 min time point and shows a significant increase in foci detection between 40× and 40× with EDF and similarly with the 60× magnification comparison (P < 0.01).
Figure 3.
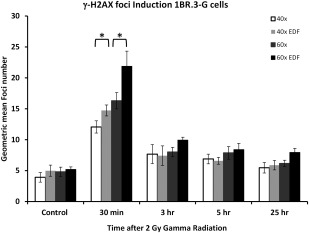
This figure shows the induction of γ‐H2AX in the nuclei of the DNA DSB repair competent 1BR.3‐G cells over a 24 h time course following exposure to 2 Gy gamma radiation. Foci induction kinetics are shown for images captured at 40× and 60× magnification with and without the application of EDF. Data are derived from the capture of at least 5000–10,000 images of cells and error bars represent the standard deviation of foci numbers derived from the quantitated foci numbers. Statistical analysis (Student's unpaired t‐test) has been applied to the 30 min time point and shows a significant increase in foci detection between 40× and 40× with EDF and similarly with the 60× magnification comparison (P < 0.01).
Figure 4.
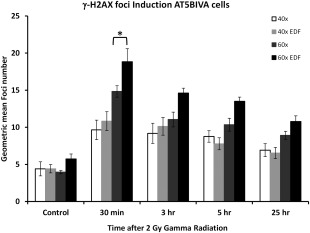
This figure shows the induction of γ‐H2AX in the nuclei of the DNA DSB repair defective AT5BIVA cells over a 24 h time course following exposure to 2 Gy gamma radiation. Foci induction kinetics are shown for images captured at 40× and 60× magnification with and without the application of EDF. Data are derived from the capture of at least 5000–10,000 images of cells and error bars represent the standard deviation of foci numbers derived from the quantitated foci numbers. Statistical analysis (Student's unpaired t‐test) has been applied to the 30 min time point and shows a significant increase in foci detection between 40× and 40× with EDF (P < 0.01).
γ‐H2AX Foci Induction in MRC5‐SV1 Cells
The MRC5‐SV1 SV40 immortalized lung fibroblast is derived from a clinically normal individual with normal repair kinetics. Figure 2 shows the induction of foci over a 24 h period. Furthermore, a normal repair kinetic is displayed in the cells over a 24 h period following the induction of DNA DSB by exposure to 2 Gy gamma radiation. In control (untreated) cells we observed a low number of foci, which range from 2.13 foci/cell at 40× magnification without EDF and increases to 4.41 foci/cells when images of the cells are captured at 60× magnification with EDF. At 30 min post‐irradiation, typically the time when foci induction is maximal, we observed an increase in foci number in each cell ranging from 10.75 foci/cell at 40× magnification to 25.54 foci per cells at 60× with EDF. From 30 min to 24 h there is an expected reduction in the number of foci to near normal levels (as would be expected in a DNA DSB repair competent cell line). Interestingly, the magnification at 60× with EDF displays a higher number of residual foci at each time point but a convincing repair kinetic is observed, which is similar to that observed in images of cells captured at other magnifications with and without EDF.
γ‐H2AX Foci Induction in 1BR.3‐G Cells
The 1BR.3‐G SV40 immortalized skin fibroblast cell line is derived from a phenotypically normal individual with a normal capacity for DNA DSB repair. Figure 3 shows the induction of γ‐H2AX foci over a 24 h period following exposure to 2 Gy gamma radiation. In untreated control cells, the residual level of foci ranges from an average 3.92–5.21 foci/cell in images captured at 40× magnification, which is similar at all levels of magnification with or without EDF capture. The maximum average number of foci was 4.21 foci/cells in those images captured at 60× magnification with EDF. Similar to the MRC5‐SV1 cells the average number of foci in the 1BR.3‐G cells increases dramatically at 30 min postradiation exposure. Here at 40× magnification 12.06 foci/cell are resolved and this steadily increases to a maximum of 20.88 foci per cells in those images captured at 60× with EDF. In a pattern similar to the repair competent MRC5‐SV1 cells the number of foci at 3, 5, and 24 h postexposure decline to near normal level but in all cases the number of foci observed is higher in those images captured with EDF.
γ‐H2AX Foci Induction in AT5BIVA Cells
The AT5BIVA SV40 immortalized fibroblast cells are derived from an individual with the genetic disease A‐T due to a mutation in the ataxia telangiectasia mutated gene (ATM), which functions as a cell cycle checkpoint control gene 12. As a result cells derived from these individuals display hypersensitivity to the lethal effects of ionising radiation because of a defect in DNA DSB repair. In comparison to the DSB repair competent MRC5‐SV1 and 1BR.3‐G cells this defect in DNA DSB repair is displayed by retention in the average number of γ‐H2AX foci per cell over the typical 24 h time course experiment conducted in this study (Figure 4). The residual numbers of foci are plotted for the untreated control cells and range between 3.996 and 4.733 foci/cell depending on the magnification objective used to capture images with or without EDF. This increases in a predictable manner at 30 min following exposure to ionizing radiation. However, over the 24 h time course the number of foci does not return to untreated levels as would be expected by the defect in DNA DSB repair. However, the number of foci detected in significantly higher in those cells captured with 60× magnification with EDF capability. Thus, here we have been able to demonstrate the DNA repair defect of the AT5BIVA cells and moreover shown how the enhanced magnification features together with EDF provides a more accurate estimation of foci number in the nuclei of cells.
γ‐H2AX Foci Induction in XP14BRneo17 Cells
The XP14BRneo17 cells are derived from a patient with two defects in distinct DNA repair pathways. The cells are derived from a patient with the cancer prone genetic disease xeroderma pigmentosum (XP), complementation group C, which is characterized by extreme clinical and cellular sensitivity to ultraviolet (UV) radiation. This is due to a defect in nucleotide excision repair. However, the patient was also characterized as having a defect in the DNA‐PKcs gene central to NHEJ 9. Correspondingly, we have previously demonstrated that these cells are hypersensitive to ionizing radiation due to a defect in DNA DSB repair 3, 9. Here this defect in DSB repair is confirmed by a persistence of γ‐H2AX foci in the nuclei of cells during a 24 h time course as shown in Figure 5. In untreated cells, residual foci levels range from 2.07 to 3.46 foci/cell depending on the magnification objective and the application of EDF. At 30 min post‐gamma radiation exposure, foci levels are at maximal levels. Similar to the repair defective AT5BIVA cells, a persistence of foci is observed over the 24 h period and at no time point does the level of foci return to untreated basal levels as expected by the DSB repair defect in this cell line. However, similar to the three other cell lines examined in this study, the use of higher magnification together with the introduction of EDF for imaging, we observe a substantially increased number of foci as compared to foci numbers observed with 40× magnification with or without EDF.
Figure 5.
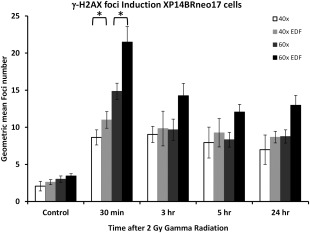
This figure shows the induction of γ‐H2AX in the nuclei of the DNA DSB repair defective XP14BRneo17 cells over a 24 h time course following exposure to 2 Gy gamma radiation. Foci induction kinetics are shown for images captured at 40× and 60× magnification with and without the application of EDF. Data are derived from the capture of at least 5000–10,000 images of cells and error bars represent the standard deviation of foci numbers derived from the quantitated foci numbers. Statistical analysis (Student's unpaired t‐test) has been applied to the 30 min time point and shows a significant increase in foci detection between 40× and 40× with EDF and similarly with the 60× magnification comparison (P < 0.01).
Discussion
In this report we present a fundamental and follow‐up study to our 2012 Cytometry A publication in which we demonstrated the use of imaging flow cytometry for the detection and quantitation of γ‐H2AX foci in eukaryotic cells. This was the first such report in which the use of imaging flow cytometry could be used for this application and further demonstrated that defects in DNA repair in individual cell lines could be distinguished from repair normal cells 3. Here we extend this study and demonstrate the use of two new capabilities of the ImagestreamX. These features include the multimagnification module affording 20×, 40×, and 60× magnification and the EDF focus stacking module. We have applied both modules to this study and have examined the effectiveness of increased magnification and focus stacking by EDF in the imaging and quantitation of γ‐H2AX foci in human cells with normal and defective DNA DSB repair capacity. In two repair normal cell lines (MRC5‐SV1 and 1BR.3‐G) we observed a typical induction of DNA DSB induced by exposure to 2 Gy gamma radiation measured by the number of foci in the nuclei of irradiated cells. Moreover, during a 24 h time course a typical and predictable profile of repair was observed where DNA DSB are repaired and foci levels return to near normal levels. In the DSB repair defective cell lines, AT5BIVA and XP14BRneo17 cells we also observed a typical induction of DSB caused by radiation exposure but a persistence of foci is observed over 24 h evidenced by the retention of foci in the nuclei of irradiated cells. These observations in themselves were fully expected and unsurprising and confirm our earlier observations 3. However, the use of higher magnification capability together with EDF revealed that in all cell lines a significantly increased number of foci can be observed at 60× magnification when compared to 40× capture and moreover, increased foci are observed with the application of EDF. While this might be regarded as a predictable result, this article provides the first such report where increased magnification with EDF demonstrates enhanced capacity for the resolution and quantitation of DNA damage in imaging flow cytometry. In addition, we observed higher resolution images of foci in the cells and this is especially evident with the use of EDF (Figure 1).
The increased foci numbers observed by greater magnification and EDF may be due to two fundamental reasons. First, the increased focal plane afforded by higher magnification will allow the resolution of foci that may, under lower magnification appear as a conjoined and therefore single focus. For example at 60× magnification with EDF applied, the average foci size is ∼1–1.5 μm (using the Ideas custom measuring tool) but at 40× magnification with EDF average foci size is ∼2 μm. However, in this latter case the possibility of conjoined spots cannot be eliminated. Second, the use of EDF allow a series of images at different focal length to be combined into a single stack, therefore foci at different locations within the three dimensional structure of the nucleus can be seen. Therefore, it is not surprising that the total focus count is further increased with EDF.
A major limitation that has been leveled at imaging flow cytometry for the quantitation and resolution of γ‐H2AX foci is that while the technique allows quantitation in high cell numbers, which affords favorable statistical value, the accuracy of the technique suffers due to low resolution and is unable to match the accuracy of in situ fluorescence microscopy. We conceded this in our 2012 Cytometry A article in which we compared γ‐H2AX foci quantitation using imaging flow and in situ microscopy. We demonstrated a 25–30% increased foci resolution with in situ microscopy methods as compared to imaging flow. However, our report here demonstrates that 60× magnification together with EDF begins to approach the same levels of resolution of foci when compared to in situ foci quantitation.
We show that with the use of higher magnification (60×) and EDF we are able to capture and quantify accurately up to 40–50% more foci in cell nuclei. Moreover, we also observe distinct differences in cell lines with varying capabilities in DNA DSB repair capacity.
Therefore, the application of both EDF and increased magnification will significantly increase the accuracy and resolution of γ‐H2AX foci measurements in cells and represent a welcome enhancement of imaging flow cytometric capability.
The application of imaging flow cytometry with EDF is a relatively new technical application and there are few reports in the literature documenting the use of this feature. However, there are reports for example, demonstrating the enhanced imaging of aneuploidy in cells using fluorescence in situ hybridization 13.
In summary, we welcome the use of these enhanced imaging flow cytometry features and will apply them to the development of diagnostic tests designed to predict the response of cancer patients to clinical radiotherapy. We have demonstrated using non‐imaging flow cytometry a persistence of γ‐H2AX‐associated nuclear fluorescence in blood lymphocytes of patients, which experienced severe normal tissue toxicity following radiotherapy 6. Therefore, higher magnification imaging flow cytometry together with EDF should enhance the accuracy of such diagnostic tests.
Supporting information
Supporting Information
Supporting Information
Supporting Information
Literature Cited
- 1. Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem 1998;273:5858–5868. [DOI] [PubMed] [Google Scholar]
- 2. Sedelnikova OA, Pilch DR, Redon C, Bonner WM. Histone H2AX in DNA damage and repair. Cancer Biol Ther 2003;2:233–235. [DOI] [PubMed] [Google Scholar]
- 3. Bourton EC, Plowman PN, Adam Zahir S, Ulus‐Senguloglu G, Serrai, S , Bottley G, Parris CN. Multispectral imaging flow cytometry reveals distinct frequencies of γ‐H2AX foci induction in double strand break repair defective human cell Lines. Cytom. Part A 2012;81A:130–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mistrik M, Oplustilova L, Lukas J, Bartek J. Low‐dose DNA damage and replication stress responses quantified by optimised automated single‐cell image analysis. Cell Cycle 2009;16:2592–2599. [DOI] [PubMed] [Google Scholar]
- 5. Furuta T, Hayward RI, Meng L‐H, Takemura H, Aune GJ, Bonner WM, Aladjem MI, Kohn KW, Pommier Y. p21CDKN1A allows the repair of replication‐mediated DNA double‐strand breaks induced by topoisomerase I and is activated by the checkpoint kinase inhibitor 7‐hydroxystaurosporine. Oncogene 2006;25:2839–2849. [DOI] [PubMed] [Google Scholar]
- 6. Bourton EC, Plowman PN, Smith D, Arlett CF, Parris CN. Prolonged expression of the γ‐H2AX DNA repair biomarker correlates with excess acute and chronic toxicity from radiotherapy treatment. Int J Cancer 2011;129:2928–2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ulus‐Senguloglu G, Arlett CF, Plowman PN, Parnell J, Patel N, Bourton EC, Parris CN. Elevated expression of artemis in human fibroblast cells is associated with cellular radiosensitivity and increased apoptosis. Br J Cancer 2012;107:1506–1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bourton EC, Plowman PN, Harvey AJ, Adam Zahir S, Parris CN. The PARP‐1 inhibitor olaparib causes retention of γ‐H2AX foci in brca1 heterozygote cells following exposure to gamma radiation. J Cancer Ther 2013;4:44–52. [Google Scholar]
- 9. Abbaszadeh F, Clingen PH, Arlett CF, Plowman PN, Bourton EC, Themis M, Makarov EM, Newbold RF, Green MHL, Parris CN. A novel splice variant of the DNA‐PKcs gene is associated with clinical and cellular radiosensitivity in a xeroderma pigmentosum patient. J Med Genet 2010;47:176–181. [DOI] [PubMed] [Google Scholar]
- 10. Murnane JP, Fuller LF, Painter RB. Establishment and characterization of a permanent pSV ori‐transformed ataxia‐telangiectasia cell line. Exp Cell Res 1985;158:119–126. [DOI] [PubMed] [Google Scholar]
- 11. Arlett CF, Green MHL, Priestley A, Harcourt SA, Mayne LV. Comparative human cellular radiosensitivity. The effect of SV40 immortalisation on the gamma‐radiation survival of skin derived fibroblasts from normal individuals and from ataxia telangiectasia patients and heterozygotes. Int J Radiat Biol 1988;549:911–928. [DOI] [PubMed] [Google Scholar]
- 12. Lavin MF, Shiloh Y. The genetic defect in ataxia‐telangiectasia. Annu Rev Immunol 1997;15:177–202. [DOI] [PubMed] [Google Scholar]
- 13. Minderman H, Humphrey K, Arcadi JK, Wierzbicki A, Maguire O, Wang ES, Block AW, Sait SNJ, George TC, Wallace PK. Image cytometry‐based detection of aneuploidy by fluorescence in situ hybridization in suspension. Cytometry Part A 2012;81A:776–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information
Supporting Information
Supporting Information


