Abstract
Leaf primordia are born around meristem‐containing stem cells at shoot apices, grow along three axes (proximal–distal, adaxial–abaxial, medial–lateral), and develop into flat symmetric leaves with adaxial–abaxial polarity. Axis development and polarity specification of Arabidopsis leaves require a network of genes for transcription factor‐like proteins and small RNAs. Here, we summarize present understandings of adaxial‐specific genes, ASYMMETRIC LEAVES1 (AS1) and AS2. Their complex (AS1–AS2) functions in the regulation of the proximal–distal leaf length by directly repressing class 1 KNOX homeobox genes (BP, KNAT2) that are expressed in the meristem periphery below leaf primordia. Adaxial–abaxial polarity specification involves antagonistic interaction of adaxial and abaxial genes including AS1 and AS2 for the development of two respective domains. AS1–AS2 directly represses the abaxial gene ETTIN/AUXIN RESPONSE FACTOR3 (ETT/ARF3) and indirectly represses ETT/ARF3 and ARF4 through tasiR‐ARF. Modifier mutations have been identified that abolish adaxialization and enhance the defect in the proximal–distal patterning in as1 and as2. AS1–AS2 and its modifiers synergistically repress both ARFs and class 1 KNOXs. Repression of ARFs is critical for establishing adaxial–abaxial polarity. On the other hand, abaxial factors KANADI1 (KAN1) and KAN2 directly repress AS2 expression. These data delineate a molecular framework for antagonistic gene interactions among adaxial factors, AS1, AS2, and their modifiers, and the abaxial factors ARFs as key regulators in the establishment of adaxial–abaxial polarity. Possible AS1–AS2 epigenetic repression and activities downstream of ARFs are discussed. WIREs Dev Biol 2015, 4:655–671. doi: 10.1002/wdev.196
For further resources related to this article, please visit the WIREs website.
INTRODUCTION
Leaves develop as lateral organs from the peripheral zone of a shoot apical meristem (SAM) along three structural axes. A group of cells is initially patterned along the proximal–distal axis and then along the adaxial–abaxial axis. Subsequent cell proliferation along the medial–lateral axis results in flat and mediolateral symmetric leaves1, 2, 3, 4, 5, 6, 7, 8, 9 (Figure 1). The process of leaf differentiation is a good model to study organ development from stem cells. The SAM consists of stem cells in a central zone (CZ), which divide slowly and replenish a peripheral zone (PZ) of more rapidly dividing cells, in which leaf initiation occurs10 (Figure 1). Leaf primordia are detected as transcriptionally distinct groups of leaf founder cells before they become morphologically distinct from the SAM. This process was first clearly demonstrated as the disappearance of class‐1 KNOTTED‐like homeobox (class 1 KNOX) gene transcripts from the leaf primordia.11 In dicotyledonous plants, a leaf primordium 0 (p0) is initially contained entirely within the SAM (see Figure 1), and then begins to grow outward.12 It has been speculated that the primordium acquires adaxial–abaxial polarity in the radial dimension,13 soon after it becomes visible, which is between the p1 and p2 stages of development. Based on the observation that if adaxial–abaxial polarity is perturbed, filamentous shaped leaves are formed, Waites and Hudson14 proposed that cell proliferation might be induced at the boundary between the adaxial and abaxial domains, and result in the expansion of leaf lamina in the medial–lateral direction. Genetic and molecular studies of leaf development in dicotyledonous plants support their concept. The molecular mechanisms underlying the cell proliferation induced by the juxtaposition of these two domains, however, are not well understood.
Figure 1.
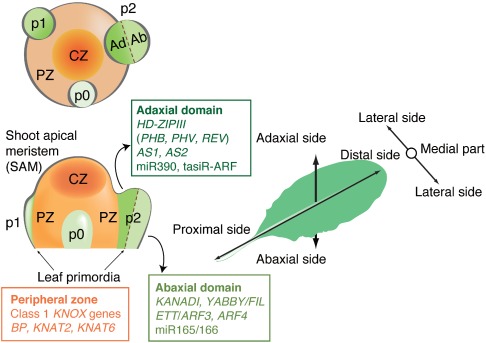
The leaf structure develops along three axes. Developmental compartments in the shoot apex around the apical meristem and the three structural leaf axes are schematically shown on the left‐hand side and in the middle, respectively (see details in text). CZ, central zone; PZ, peripheral zone; p0, primordium 0; p1, primordium 1; p2, primordium 2.
As summarized in Figure 1, genes involved in the adaxial–abaxial partitioning of leaves have been isolated and characterized in Arabidopsis thaliana. Analyses of these genes have shown that networks of several families of transcription factor‐like proteins and small RNAs must play critical roles in the specification of leaf polarity. The PHABULOSA (PHB), PHAVOLUTA (PHV), and REVOLUTA (REV) genes, which encode class III homeodomain‐leucine zipper (HD‐ZIPIII) proteins, determine adaxial cell fate,15 The accumulation of their transcripts in the adaxial domain is a consequence of their degradation in the abaxial domain by microRNA165 (miR165) and miR166 (miR165/166).16 The ASYMMETRIC LEAVES1 (AS1) and AS2 genes, which encode nuclear proteins with the MYB (SANT) domain17 and the plant‐specific AS2/LOB domain (https://www.arabidopsis.org/browse/genefamily/AS2.jsp), respectively18, 19, 20 (Figure 2(a)) were initially identified as factors involved in the formation of symmetric flat lamina of leaves.21 It has recently been shown, however, that they are related to the formation of proper morphology along three leaf axes including the adaxial–abaxial axis, which will be mainly discussed in this article.
Figure 2.
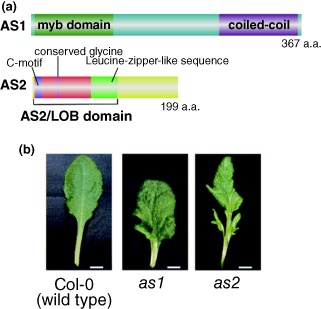
(a) Domain organization of AS1 and AS2 proteins. Both are nuclear proteins. AS2 belongs to the AS2/LOB plant‐specific protein family (42 members are designated as AS2 and ASL1–ASL41).18, 20 (b) Phenotypes of as1 and as2 leaves. Both as1 and as2 show pleiotropic phenotypes including asymmetrically curled leaf blades, asymmetric lobes, asymmetric secondary vein patterns, less prominent midveins, plump and swelled leaf laminas, and shorter petioles than seen in wild type leaves. Asymmetric leaflet‐like structures are formed in as2. Photograph (as2) is reproduced from Ref 5 (Development 2001, 128:1771–1783).
Members of the KANADI (KAN) gene family, which encode proteins with the GARP domain, and members of the YABBY (YAB)/FIL family (https://www.arabidopsis.org/browse/genefamily/C2C2YABBY.jsp), which encode proteins with a zinc finger and a HMG box‐like domain are involved in the specification of abaxial cell fate in the leaf lamina.6, 7, 8, 9 In addition to the abaxial development, it is also suggested that YAB genes are involved in developing the expanding flat leaves with diverse systems characterized by more lamina‐specific genetic programs that are related to marginal auxin flow and activation of a maturation schedule directing determinate growth.22 ETTIN/AUXIN RESPONSE FACTOR3 (ETT/ARF3) and ARF4 (https://www.arabidopsis.org/browse/genefamily/ARF.jsp) also specify both abaxial cell fate and lateral growth of leaf lamina.23 This result suggests that the lateral growth of the lamina could be related to the determination of adaxial–abaxial identity as proposed previously.14 Transcripts of both ETT/ARF3 and ARF4 are specifically degraded by the small RNA tasiR‐ARF in the presumptive adaxial domain, contributing to the determination of the adaxial cell fate.24, 25
The idea that leaf polarity is specified by antagonistic interactions between adaxial and abaxial genes was proposed on the basis of genetic and expression analyses of these genes.26 Such expression patterns change during leaf development. During this process, both adaxial and abaxial promoting genes are initially expressed throughout the primordium (see p0–p1 in Figure 1), and subsequently their expression patterns are restricted to their respective complementary domains (p2). The patterning of expression of the polarity genes is generated by the mutually exclusive actions of their protein products. For example, expression of the abaxial gene FIL/YAB is abolished by the ectopic expression of PHB (HD‐ZIPIII adaxial gene).27 Although leaf regions expressing PHB and KAN messenger RNAs (mRNAs) are mutually exclusive,15, 28 and these gene families act genetically in an antagonistic manner during embryo patterning,29 it has not been clearly demonstrated that KAN regulates HD‐ZIPIII expression directly.
Recently, Nakata et al.30 showed that PRESSED FLOWER/WUSCHEL‐RELATED HOMEOBOX3 (WOX3) and WOX1 (https://www.arabidopsis.org/browse/genefamily/wox.jsp), which are expressed in the middle domain between the adaxial and abaxial domains, function redundantly in lateral‐specific lamina outgrowth and leaf margin‐specific cell fate and, furthermore, that expression patterns of the two WOX genes are negatively and positively regulated by the KAN and YAB genes, respectively. They also propose a three‐domain model, in which these WOX genes would coordinate adaxial/abaxial patterning in cooperation with adaxial‐ and abaxial‐specific regulators, including the ASYMMETRIC LEAVES2 (AS2) and YAB3. YUCCA genes, responsible for auxin biosynthesis, are expressed in response to the juxtaposition of adaxial and abaxial domains, which is responsible at least in part for leaf margin expansion.31
AS1 and AS2, both of which are referred as to as adaxial genes and exhibit similar laminar abnormalities, repress the expression of abaxial genes, such as KAN2, YAB5, ETT/ARF3, and ARF4, but do not affect the HD‐ZIPIII family genes,32 suggesting that they are involved in the antagonistic interactions between genes that specify adaxial–abaxial polarity. The direct repression of AS2 by KAN was first reported by Wu et al.33 Our group has recently reported the direct repression of ETT/ARF3 by transcriptional gene silencing (TGS) through AS1–AS2 and indirect repression of both ARF3 and ARF4, a redundant member of the ARF gene family, by post TGS (PTGS) through AS1–AS2 functions.34 These results provide a molecular framework for the antagonistic interaction of genes involved in adaxial–abaxial specification. In this article, we will overview the recent results on molecular mechanisms for the opposing interplay of polarity‐related genes by AS1 and AS2 and discuss prospects for a novel epigenetic system of gene repression to guarantee the polarity specification necessary for leaf development.
CHARACTERIZATION OF ADAXIAL‐SPECIFIC AS1 AND AS2 GENES
The PHANTASTICA (PHAN) gene of Antirrhinum majus is involved in the growth and adaxial–abaxial determination of lateral organs and its expression is required early in the establishment of the proximal–distal axis.2, 14 Because plants with a mutation in PHAN are known to generate abaxialized filamentous leaves only when grown at 15–17 °C2, 14 or in the handlebars (hb) mutation background, it is proposed that a cold‐sensitive pathway and some other gene might be redundantly involved in the adaxial–abaxial determination of leaves together with PHAN (see the later section of ‘Modifier mutations’ of Arabidopsis).35 In the as1 mutant in A. thaliana, the PHAN MYB ortholog is disrupted.17
Both as1 and as2 mutants exhibit pleiotropic phenotypes5, 36, 37 (Figure 2(b)). These mutants produce petioles and leaf blades that are much shorter than those of the wild type, in addition to asymmetrically lobed and downwardly curled leaf blades, which are bilaterally asymmetric. Furthermore, as1 and as2 also often generate leaflet‐like structures from petioles in asymmetric positions and fail to produce a thick and distinct midvein; and as2 often generates leaflet‐like structures from petioles in asymmetric positions. Higher‐ordered veins are asymmetric and simplified. The observation that the leaf laminas of as1 and as2 are often plump and swelled at their base implies that adaxial development in the leaves of these mutants is slightly diminished.5 Thus, the AS1 and AS2 genes are involved not only in the symmetric development along the mediolateral axis, but also in development along the adaxial–abaxial axis and the proximal–distal axis. In addition, expression levels of many genes, including both class‐1 KNOX genes (BP, KNAT2, KNAT6) and abaxial‐determinant genes, such as ETT/ARF3, KAN2 and YAB5, are elevated.5, 18, 38
Both AS1 and AS2 transcripts accumulate in the early stage of above‐ground organ primordia, and AS1 and AS2 expression sites become restricted to middle/inner regions and the adaxial epidermis of cotyledonary and leaf primordia. AS2 transcripts are also accumulated in the columella root cap.32 After leaf maturation, the expression levels of these genes are reduced.32 AS2‐fused YFP (AS2‐YFP) proteins are localized to subnuclear bodies39, 40, 41 adjacent to the nucleoli in leaf cells, called AS2 bodies, and some are also dispersed in the nucleoplasm.39, 41 GFP‐fused AS1 proteins are located as speckles in the nucleoplasm39, 40, 41 and are also concentrated in the AS2 bodies by an AS2‐dependent process.39, 41 AS1 and AS2 form the AS1–AS2 complex,42, 43 which represses the expression of two class 1 KNOX genes, BP and KNAT2, by binding to their respective promoter regions, showing that these KNOX genes are direct targets of AS1–AS2.42 In addition, AS1–AS2 directly represses ETT/ARF3 by binding to its promoter region.34
Although genes that are predicted to encode AS1 orthologs and members of the AS2/LOB protein family are detected in genome databases of many plant species, genes that might encode amino acid sequences entirely homologous to the AS2 sequence are not detected in rice genome databases, and even complementary DNA (cDNAs) encoding AS2 orthologs have yet to be reported from monocotyledonous plants. Although AS2 homologues have been predicted to be present in various dicotyledonous plants, their roles in plants other than Arabidopsis are not yet intensively studied.
PROXIMAL–DISTAL POLARITY DEVELOPMENT OF LEAVES BY AS1–AS2
Genes involved in the formation of proximal–distal polarity were first identified in maize. While leaves of dicotyledonous plants are composed of stipule, petiole, and leaf blade along with proximal–distal axis, monocotyledonous plants such as maize and rice develop other distinct leaf features: the sheath in the proximal region of the leaf and the blade in its distal region. The sheath and blade are separated at their boundary by the auricle and ligule, which are not present in leaves of dicotyledonous plants (http://www.fsl.orst.edu/forages/projects/regrowth/print‐section.cfm?title=GrassStructures). Recessive mutants of the rough sheath2 (rs2) gene of maize, an ortholog of PHAN and AS1,44, 45 exhibit a disruption of the blade‐sheath boundary owing to disorganized cell growth and acropetal ligule displacement, and the semi‐bladeless phenotype of leaves.46 In rs2 mutants, class 1 KNOX genes are ectopically expressed, a condition that is also observed in some dominant mutants exhibiting phenotypes similar to those of rs2. Thus, rs2 is involved in the proximal–distal patterning of maize leaves through repression of class 1 KNOX genes.47
AS1 and AS2 of A. thaliana are also involved in regulation of the proximal–distal development of leaves through repression of class 1 KNOX genes (Figure 3). Although petiole and leaf lengths are markedly reduced both in as1 and as2, the extent of the reduction is more severe in as1.
Figure 3.
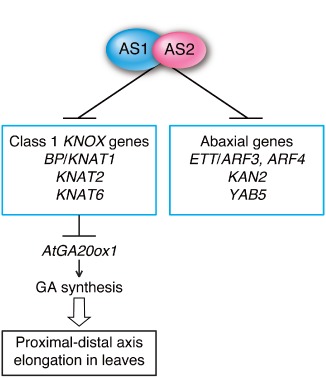
Roles of the AS1–AS2 complex in the regulation of class 1 KNOX, ETT/ARF3 and ARF4 genes in early stages of leaf primordia in Arabidopsis thaliana. The introduction of bp knat2 knat6 triple mutations into as1‐1 or as2‐1 efficiently suppressed the phenotypes of short petiole and leaf blade seen in Figure 2(b).48
To investigate effects of the elevated expression of class 1 KNOX genes on the phenotypes of as1 and as2 mutants, a number of studies have been performed using over‐ and ectopic‐expression systems of class 1 KNOXs under the control of the 35S promoter of Cauliflower mosaic virus. Overexpression of KNOX genes in tobacco and A. thaliana plants repressed transcription of the gibberellin‐synthetic (GA‐synthetic) genes that encode GA‐20 oxidase,49, 50 and application of GA partially suppressed the abnormal phenotypic features of PHANTASTICA‐antisense transgenic tobacco plants.51 Analyses of multiple loss‐of‐function mutants of KNOX genes (bp knat2 knat6) in as1 and as2 backgrounds show that the formation of shorter petioles and leaf blades is due to repression of GA‐synthetic genes by the upregulation of BP/KNAT1, KNAT2, and KNAT6 48 (Figure 3). Thus, elevated expression of KNOXs is responsible for limited numbers of as1 and as2 phenotypes including petiole and lamina sizes, the less prominent midvein, and the lower potential of root regeneration from leaf sections in in vitro culture. The formation of asymmetric leaf lobes, leaf curling, leaflet‐like structures from petioles, and the increased potential of shoot regeneration are, however, not due to upregulation of class 1 KNOXs.
PHAN in Antirrhinum is also involved in elaboration of the proximal–distal axis as well as the adaxial–abaxial polarity in leaves.2 NSPHAN of Nicotiana sylvestris is also proposed to be involved in proximal–distal development.47 Taken together, the KNOX‐repressive systems mediated by AS1 orthologs (PHAN and RS2), which appear to be involved in the proximal–distal polarity patterning, might be conserved at least in the plants mentioned in this section. Nevertheless, roles of AS2 orthologs in such patterning have not been determined in these plants other than Arabidopsis.
ADAXIAL–ABAXIAL POLARITY SPECIFICATION OF LEAVES BY AS1–AS2
Molecular Roles of AS1–AS2: Repression of Abaxial Genes
Gene expression analyses of as1 and as2 show that transcript levels of several abaxial side‐specific genes (ETT/ARF3, KAN2, YAB5) are significantly increased, whereas those of HD‐ZIPIII do not change.32 These results suggest that AS1 and AS2 directly or indirectly repress expression of the abaxial‐specific genes (Figure 3). In addition, systematic molecular and genetic analyses have identified a target gene, ETT/ARF3, which encodes an abaxial factor acting downstream of the AS1–AS2 complex.34 As schematically summarized in Figure 4, the AS1–AS2 complex represses ETT/ARF3 by the direct binding of AS1 to the ETT/ARF3 promoter and also indirectly induces accumulation of miR390 and tasiR‐ARF, which negatively regulate the expression of both ETT/ARF3 and ARF4. Thus, the complex dually represses the expression of ETT/ARF3. Several abnormalities of as2 plants are slightly suppressed by the introduction of an ett or arf4 single mutation into as2 plants. The introduction of ett arf4 double mutations into as2 efficiently suppresses these asymmetric leaf phenotypes34 (Figure 5(a)). These results are consistent with the observation that overexpression of a tasiR‐ARF‐insensitive ETT/ARF3 cDNA yields as2‐like phenotypes.52 Similarly, some lamina phenotypes of as1 were also rescued by the introduction of ett arf4 (Figure 5(a)). The phenotype of wrinkled lamina with patches of abaxialized cells on the adaxial surface, which indicates a slight increase in abaxialization,14, 53 was also rescued in both as1 and as2 by the introduction of ett arf4.34 These results suggest that several leaf abnormalities, including those related to adaxial–abaxial polarity defects in as1 and as2 plants, result from elevated expression of the abaxial genes ETT and ARF4 (Figure 4). Analyses of modifier mutations of as1 and as2 have further confirmed that repression of these ARFs by AS1–AS2 is important for the adaxialization of leaves. Although expression levels of KAN2 and YAB5 are increased in as1 and as2, they are indirectly regulated by AS1–AS2.34
Figure 4.
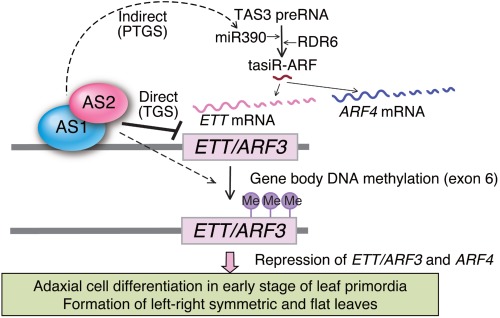
Dual regulation of ETT/ARF3 gene expression, including by the possibly epigenetic system of AS1–AS2. The AS1–AS2 complex represses ETT/ARF3 directly, and ETT/ARF3 and ARF4 indirectly, via stimulating the miR390 and tasiR‐ARF pathway. In addition, AS1 and AS2 maintain gene‐body DNA methylation of the ETT/ARF3 gene. Solid lines indicate direct regulation and dashed black lines indicate indirect regulation.
Figure 5.

(a) The ett and arf4 mutations suppressed major leaf phenotypes of as1‐1 and as2‐1. Representative gross morphology of 40‐day plants and magnified views of their leaves. Gross morphology of Col‐0 (wild type), as1‐1, as1‐1 ett‐13 arf4‐1, as2‐1, and as2‐1 ett‐13 arf4‐1 plants is shown. The genotype of each plant is indicated. Red arrowheads indicate leaf lobes and the arrow indicates a leaflet‐like structure. The introduction of ett arf4 double mutations into as1‐1 or as2‐1 efficiently suppressed the phenotypes of asymmetrically curled leaf blades, asymmetric lobes, and plump and swelled leaf laminas in both mutants34 in Figure 2(b). Scale bars: 5 mm (upper) and 2 mm (lower). (b) Gross morphology of typical double mutants (as2‐1 elo3‐27 and as2‐1 eal‐1/bob1). Introduction of ett and arf4 mutations into the double mutants efficiently suppressed the abaxialized leaf phenotypes to form flat symmetric leaves. See details of modifier mutations in Table 1. Scale bars: 5 mm. Plants were photographed at 28 days after sowing. White arrowheads indicate filamentous leaves. Scale bars: 5 mm. Higher magnification views of filamentous leaves are shown. Scale bars: 1 mm in higher magnification views. Photographs (a) and (b) are reproduced and modified from Ref 34 (Development 2013, 140:1958–1969) and Ref 69 (Plant Cell Physiol 2013, 54:418–431), respectively.
Although the systems whereby tasiR‐ARFs regulate ARF3 expression are conserved in both Arabidopsis and maize plants, the contribution of tasiR‐ARFs to adaxial–abaxial patterning in Arabidopsis seems to be different from that in maize; the extents of adaxial defects in mutations in tasiR‐ARF biogenesis components of Arabidopsis are subtle as compared with those of maize.13 This might be due to difference in the involvement of AS1 in repressing ARF3 expression in Arabidopsis from that of RS2 in repressing ARF3 in maize. Recently, loss‐of‐function mutants of small RNA biogenesis components (RDR6, SGS3, AGO7, and DCL4) in tomato (Solanum lycopersicum) have been isolated. In severe cases, they generate shoestring leaves that lack leaf blade expansion (wiry leaves).54 In the tomato mutants, levels of tasiR‐ARFs are reduced and ARF3 and ARF4 are upregulated, suggesting that the repressive system of ARF3 and ARF4 regulation by tasiR‐ARFs is also conserved in the pathway for adaxial–abaxial specification in leaves of tomato; increased activity of either of ARFs phenocopies results in wiry leaves. Interestingly, overexpression of these ARFs in Arabidopsis, tobacco (Nicotiana tabacum), and potato (Solanum tuberosum), however, fails to produce wiry leaves, suggesting that such a response in tomato is distinct from those in other dicotyledonous plants. The tomato system of adaxial–abaxial specification by tasiR‐ARFs appears to be somewhat similar to the developmental control of adaxial–abaxial patterning by the tasiR‐ARFs in maize.
Modifier Mutations Enhancing Leaf Polarity Defects of as Mutants
Many mutations that enhance leaf polarity defects of as1 and as2 have been isolated and characterized, which is reminiscent of the presence of the cold‐sensitive pathway in the original phan mutant of Antirrhinum and handlebars as the enhancer mutation of phan.2, 35 The causative genes are designated as modifiers of AS1 and AS2 34, 69 and, as listed in Table 1 24, 30, 34, 39, 58, 59, 62, 63, 64, 69, 70, 71, 72, 74, 75, 76, 77, 78, 79, 81, 82, 84, 88, 89, 90, 93, 95 they include those for biogenesis of tasiR‐ARF, biogenesis of ribosomes, chromatin modification and nucleosome assembly proteasome‐mediated protein degradation, genomic stability, and cell proliferation. Prominent phenotypes in almost all double mutants with as2 and a modifier mutation involve the generation of filamentous leaf‐like organs (Figure 5(b)), which are surrounded by an abaxialized epidermis and possess no or markedly premature vascular tissues. Double mutations in PRESSED FLOWER/WUSCHEL‐RELATED HOMEOBOX3 (WOX3) and WOX1 also cause the formation of severely abaxialized filamentous leaves in the as2 background.30, 89
Table 1.
Arabidopsis Gene Mutations, Which Act as Modifiers Enhancing Leaf Adaxial–Abaxial Abnormalities in as1 and as2
| 1. Gene (Mutation) | 2. AGI Code | 3. Protein | 4. Cellular Process and Status | 5. Subcellular Localization | 6. References |
|---|---|---|---|---|---|
| I. Biogenesis of small RNA | |||||
| RNA‐DEPENDENT RNA POLYMERASE6 (rdr6/sde1/sgs2) | AT3G49500 | RNA‐dependent RNA polymerase | Duplication of TAS3 mRNAs; biogenesis of ta‐siRNA | Cytoplasm,55, 56 nucleus57 | 24 58 |
| ARGONAUTE7 (ago7/zip) | AT1G69440 | ARGONAUTE family protein: RNA slicer | Biogenesis of miR390 for ta‐siRNA production | Cytoplasm55 | 24 59 |
| SUPPRESSOR OF GENE SILENCING3 (sgs3) | AT5G23570 | Unknown | Biogenesis of siRNA, stabilization of ta‐siRNA | Cytoplasm55, 56 | 24 59 |
| DICER‐LIKE4 (dcl4) | AT5G20320 | DICER‐LIKE protein: RNase III‐like enzyme | Processing of ta‐siRNA intermediates | Nucleus57, 60 | 24 59 |
| ARGONAUTE1 (ago1) | AT1G48410 | ARGONAUTE family protein: RNA slicer | Recruit of miRNA and siRNA to mRNAs to be degraded | Nucleus (D‐body) and cytoplasm61 | 59 62, 63 |
| II. Chromatin modification and remodeling | |||||
| PICKLE (pkl/gymnos) | AT2G25170 | Chromodomain helicase DNA‐binding (CHD3) family protein | Component of chromatin remodeling complex SWI/SNF | 64 | |
| SERRATE (se) | AT2G27100 | C2‐H2‐type zinc finger protein | miRNA‐mediated gene expression | Nucleus65 | 64 |
| HDT1 (hdt1/hd2a/hda3) | AT3G44750 | Histone deacetylase (plant‐specific class) | Deacetylation of nucleosomal histone H3, transcription of rDNAs | Nucleolus39, 66, 67 | 39 |
| HDT2 (hdt2/hd2b) | AT5G22650 | Histone deacetylase (plant‐specific class) | Deacetylation of nucleosomal histone H3, transcription of rDNAs | Nucleolus39, 66, 67 | 39 |
| ELONGATA3 (elo3/east1); ELO2 (elo2/elp1/abo1); ELONGATOR PROTEIN2 (elp2) | AT5G50320; AT5G13680; AT1G49540 | Histone acetyltransferase; scaffold proteins | Core subcomplex of holo‐elongator; stimulation of transcriptional elongation; DNA replication | Nucleus (predominant) and cytosol (lesser extent)68 | 69, 70, 71 |
| ELO1 (elo1/elp4); ELP5 (elp5); ELP6 (elp6) | AT3G11220; AT2G18410; AT4G10090 | Accessory subcomplex of holo‐elongator; stimulation of transcriptional elongation; DNA replication | 70 | ||
| ELONGATA4/DRL1 (elo4/drl1) | AT1G13870 | Associated protein of elongator complex | Stimulation of transcriptional elongation; DNA replication | 70 71 | |
| FASCIATA1 (fas1); FAS2 (fas2) | AT1G65470; AT5G64630 | H3 and H4 histone chaperone | p150 subunit of chromatin assembly factor‐1 (CAF‐1); p60 subunit of CAF1; chromatin replication | 71 72 | |
| III. Ribosomal protein (or biogenesis of ribosomes) | |||||
| RPL10a (rpl10a/pgy1); RPL9c (rpl9c/pgy2); RPL5a (pgy3/ae6/oli5); RPL28a (ae5); RPL24b (rpl24b/stv1); RPL5b (rpl5b/oli7) | AT2G27530; AT1G33140; AT3G25520; AT2G19730; AT3G53020; AT5G39740 | L10a; L9; L5; L28e; L24b; L5b | Subunits of ribosome; components of pre‐rRNA‐protein complex | Cytoplasm, nucleus, and nucelolus73 | 74, 75, 76, 77 |
| RPL4d (rpl4d); RPL7b (rpl7b); RPL18c (rpl18c); RPL38b (rpl38b); RPL39c (rpl39c); RPS6a (rps6a); RPS21b (rps21b); RPS24b (rps24b); RPS28b (rps28b); RPS15ab (rps15ab); RPL27ac (pgy6/rpl27ac); APICULATA2/RPL36AB (api2); RPL36aA (rpl36aa) | AT5G02870; AT2G01250; AT5G27850; AT3G59540; AT4G31985; AT4G31700; AT3G53890; AT5G28060; AT5G03850; AT2G19720; AT1G70600; AT4G14320; AT3G23390 | L4d (L1) family; L30/L7 family (translational regulation); L18e (L15) superfamily; L38e family; L39 family; S6; S21e; S24e; S28; S15AB; L18e/L15 superfamily; L44e | Subunits of ribosome; components of pre‐rRNA‐protein complex | Cytoplasm, nucleus, and nucelolus73 | 76, 77, 78, 79 |
| APUM23 (apum23) | AT1G72320 | Pumillio protein containing PUF domain | Pre‐rRNA processing and rRNA maturation | Nucleolus80 | 81 |
| IV. DNA replication and repair | |||||
| TEBICHI (tebichi) | AT4G32700 | A homologue of Drosophila MUS308 and mammalian DNA polymerase | Repair at damaged DNA | 82 | |
| ABA OVERLY SENSITIVE4 (abo4) | AT1G08260 | POL2A, DNA polymerase epsilon catalytic subunit | Interaction with PCNA; DNA‐directed DNA polymerase | 71 | |
| ASYMMETRIC LEAVES1/2 ENHANCER7 (ae7/duf59) | AT1G68310 | Nucleus and cytoplasm83 | 84 | ||
| V. Proteasome | |||||
| RPN8a (asymmetric leaves enhancer3/rpn8a); RPT2a (hir‐2/rpt2a); PBE1 (pbe1); RPT5a (rpt5a); RPT1a (rpt1a); RPN9a (rpn9a); RPT4a (rpt4a); PAD1 (pad1) | AT5G05780; AT4G29040; AT1G13060; AT3G05530; AT1G53750; AT5G45620; AT5G43010; AT3G51260 | 26S proteasome subunit; 20S β subunit; one of the six AAA‐ATPases of the proteasome; proteasome component domain; 20S proteasome α subunit | Component of 26S or 20S proteasome | Endoplasmic reticulum and golgi (RPT2a),85 cytoplasm and nucleus86, 87 | 88 |
| VI. Transcription factors | |||||
| PRESSED FLOWER (prs/wox3); WOX1(wox1) | AT2G28610; AT3G18010 | WUSCHEL‐related homeobox proteins | Transcription | Nucleus | 30 89 |
| VII. Others | |||||
| BOBBER1 (bobber1/enhancer of asymmetric leaves1) | AT5G53400 | A noncanonical small heat shock protein (HSP20‐like chaperone); NudC domain protein | Protein folding | Cytoplasm90, 91 | 69 90 |
| ANGUSTIFOLIA3 (an3) | AT5G28640 | Growth regulating factor1 (GRF1) INTERACTING FACTOR | Transcriptional coactivator | Nucleus92 | 93 |
| VIII. Plastid genes | |||||
| EMBRYO DEFECTIVE DEVELOPMENT1 (edd1/ddm1) | AT3G48110 | Glycine‐tRNA ligase | Glycil tRNA aminoacylation in mitochondria and chloroplasts | Chloroplast and mitochondrion94 | 95 |
| SCABRA3 (sca3) | AT2G24120 | DNA‐directed RNA polymerase | Transcription in plastids and mitochondria | Chloroplast and mitochondrion96 | 95 |
In the double mutants that have been examined, transcript levels of several abaxial‐specific genes (KAN2, YAB5, ETT/ARF3, ARF4) as well as class 1 KNOX genes are markedly increased; these genes are upregulated in the as2 single mutant and slightly upregulated in some of the modifier single mutants. When the double mutations of ETT/ARF3 and ARF4 (see Figure 4), are introduced into double mutants with as2 and one of the modifier mutations, such as elo3 and bob1/eal‐1, the phenotype of abaxialized filamentous leaves is restored to flat and expanded shapes34, 69 (Figure 5(b)). These results show that the upregulation of these ARF genes in the double mutants is responsible for the disappearance of adaxial specification of the mutants, which suggests that repression of these ARF genes by the synergistic action of AS1–AS2 and modifier proteins is critical for proper development of the adaxial domain.
How can the modifiers and AS1–AS2 synergistically repress expression of ARFs in a wild type plant? As mentioned in the previous section, ETT/ARF3 expression is regulated dually by AS1–AS2‐dependent TGS and tasiR‐ARF‐mediated PTGS through AS1–AS2, and ARF4 is regulated by the PTGS. Therefore, the synergistic repression of these ARFs is achieved by the two independent pathways, AS1–AS2 and factors involved in small RNA biogenesis such as RDR6, AGO7, and DCL4, as illustrated in Figure 4. Molecular mechanisms for the prominent repression by other modifiers and AS1–AS2 have yet to be elucidated, but they might be involved in such repression through independent pathways69, 70, 72, 90 (Figure 6). It might be worthwhile, however, to stress that modifier mutations so far identified are weak alleles of genes that are essential for cell viability or, conversely, strong alleles of one of the functionally redundant members of such essential gene families. In addition, it is also interesting to note that most of the proteins encoded by such modifier genes are localized in the nucleus or nucleus‐related structures or compartments, such as nucleoli and spindles for chromosome separation (Table 1). Only two genes, WOX1 and WOX3, for conventional transcription factor‐like proteins are identified as modifiers.
Figure 6.
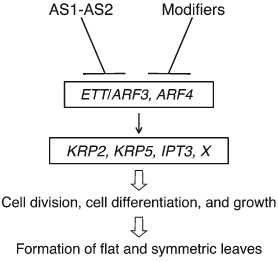
Model for repression of ETT/ARF3 and ARF4 by the AS1–AS2 complex and modifiers in the early stage of leaf primordia in Arabidopsis thaliana. Such repression events are crucial for the establishment of adaxial–abaxial polarity and then cell division and growth along the medial–lateral axis. Class 1 KNOX genes are similarly repressed by AS1–AS2 together with modifier genes, although that is not depicted in this figure.
It should be interesting to solve the question of how AS1–AS2, which is localized to nuclear bodies adjacent to the nucleolus, might repress coordinately ETT/ARF3, ARF4, and class 1 KNOX genes with modifier proteins, after they might complete roles in polarity development of leaves. As many modifiers are localized to the nucleus or nuclear compartments, they function in certain nuclear processes to repress directly or indirectly the expression of KNOXs and ARFs. In cases where modifiers might be involved in unidentified nuclear processes, including such known processes as chromatin assembly and ribosome biogenesis mediated by some modifiers, any gene‐repressive functions of AS1–AS2 must be associated with such unidentified processes.
Genes Downstream of the AS‐Abaxial Factor Pathway
Transcript levels of Kip‐related protein2 (KRP2), KRP5, and Isopentenyltransferase3 (IPT3) increase on the as2 and modifier (eal and elo3) backgrounds, and such upregulation events are canceled by the introduction of an ett arf4 double mutation into as2.69 These results suggest that expression of KRP2, KRP5, and IPT3 genes was negatively controlled by AS1–AS2 through repression of ARF3/ETT and ARF4 functions in the wild type plant. KRP2 and KRP5 of A. thaliana encode cyclin‐dependent kinase inhibitors (CKIs),69, 97 which interact with CDKs to inhibit their kinase activities and act as key regulators of cell cycle progression.98 It is possible that cell proliferation required for leaf formation might be achieved by the proper repressive control of KRP2 and KRP5 expression by AS1–AS2.
The IPT genes encode adenylate isopentenyltransferase, a cytokinin biosynthesis enzyme, in A. thaliana.99, 100, 101 Among members of the IPT family, IPT3 is expressed throughout a plant including the shoot apex and leaves.102 These data predict that the endogenous level of cytokinin might increase around the SAM in as1 and as2 mutants, which might affect developmental states of cells in the leaf primordia of these mutants.
These results suggest that the AS1‐AS2‐ETT pathway plays a critical role in controlling the cell cycle progression and the cytokinin level at the shoot apex for leaf growth and development. The relationship between the control of these downstream genes and adaxial development of leaves by AS1–AS2, however, has been unknown.
POTENTIAL EPIGENETIC REGULATION BY AS1–AS2
The AS1–AS2 complex targets the cis elements in the promoters of BP and KNAT2.42 AS1 and maize RS2, an AS1 ortholog, interact with the plant HIRA proteins which is predicted to be a histone chaperone that maintains KNOX gene silencing.103 Polycomb‐repressive complexes (PRCs) ensure the correct spatiotemporal expression of numerous key developmental regulators. Recently, it was shown that the AS1–AS2 complex physically interacts with PRC2 and recruits this complex to the homeobox genes BP and KNAT2 to stably silence in differentiating leaf cells.104 This recruitment mechanism resembles the Polycomb response element‐based recruitment of PRC2 originally defined in flies and provides the first such example in plants. These findings reveal a conserved paradigm to epigenetically regulate homeobox gene expression during development.
It has also been shown that levels of DNA methylation in exon 6 of ETT were depressed in both as1 and as2 mutants.34 It was reported that over one third of expressed genes in A. thaliana contain DNA methylation within their transcribed regions,105 and loss of such methylation results in enhanced levels of transcription.106 Recently, it has been verified that DNA demethylation increases ETT expression in a mutant for MET1.107 Levels of ETT transcripts increase in shoot apices of met1.34 There are clear parallel relationships between levels of the ETT gene‐body methylation and its transcript levels in as1 and as2,34 implying the involvement of AS1–AS2 in epigenetic control through DNA methylation.
CONCLUSION
In Arabidopsis and some other dicotyledonous plants, development of the adaxial domain requires two types of genes: genes for the HD‐ZIPIII protein family and those for the AS1–AS2 complex. In the present review, we have summarized two characteristic features of the AS1–AS2‐mediated adaxialization (Figures 6 and 7): (1) This complex dually represses expression of the abaxial‐specific ARF gene family, ETT/ARF3 and ARF4. These repression systems are experimentally demonstrated to be critical for development of the adaxial domain in Arabidopsis leaves. (2) The repression is further achieved by at least one other molecular system controlled by a modifier gene independently from the AS1–AS2 system. Although molecular mechanisms of the synergistic repression by AS1–AS2 and the modifiers have not been elucidated, their concerted actions should play a critical role in adaxial development. Events that repress the expression of these abaxial genes might occur in the presumptive adaxial domain of the leaf anlagen at its early developmental stages (p0–p1: Figure 7). Unlike the situation in maize, the contribution of tasiR‐ARFs to adaxialization is not apparent in Arabidopsis. In the presumptive abaxial domain at such early stages, AS2 is also directly repressed by KAN1 and KAN2 to induce abaxial specification.33 Considered collectively, AS1–AS2 is a central player in the antagonistic interactions of genes expressed in the process of adaxial–abaxial polarity specification.
Figure 7.
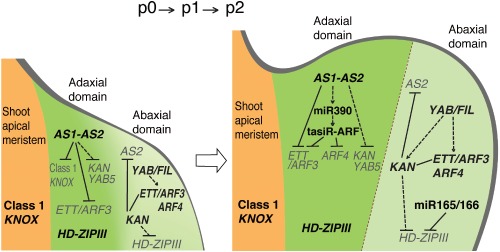
AS1–AS2 plays a central role in the antagonistic interaction of genes for adaxial–abaxial polarity specification. Solid lines indicate direct regulation and dashed lines indicate indirect regulation or unconfirmed interactions. Faded names of genes indicate those to be repressed.
Recently, Qi et al.108 have reported the existence of a transient low auxin zone in the adaxial domain of leaf primordia from p1 to p9, and suggested that auxin flow from leaf primordia to the SAM is responsible for the adaxial low auxin domain and, thus, acts as a signal influencing formation or maintenance of the leaf adaxial domain. The relationship between the auxin flow and the AS1‐AS2‐ETT pathway remains to be elucidated.
Antagonistic interaction has been proposed between KAN and YAB families and the HD‐ZIPIII family in leaf polarity development. Recently, many phytohormone‐related genes have been identified as candidates downstream of the respective KAN1 and REVOLUTA, a member of the HD‐ZIPIII family,109, 110, 111 suggesting the involvement of genes for phytohormone biosynthesis, response, and transport in the antagonistic interaction. Molecular networks of the interaction are, however, still not clear. Loss‐of‐function as2 mutations and double mutations of AS2 and its modifier genes do not significantly affect expression of HD‐ZIPIII genes,32, 70, 90 suggesting that the adaxialization mediated by HD‐ZIPIII is independent from that by AS1–AS2.
AS1–AS2 and various modifiers synergistically repress these developmentally important genes through certain nuclear processes. Taken together with these observations, it is likely that the repression system mediated by AS1–AS2 and modifier proteins might be, at least in part, involved in epigenetic processes. It will be intriguing to elucidate how coordinate actions of these molecules might determine epigenetic states of repression of KNOX and ARF family genes and how the MET1‐dependent ETT methylation might be involved in establishment of the epigenetic state during leaf polarity specification.
ACKNOWLEDGMENTS
The authors are grateful to Dr. Nanako Ishibashi (Nagoya University) for helpful discussion. This work was supported by four grants‐in aid: Scientific Research on Priority Areas (no. 19060003 to Y.M.); Scientific Research (C) (no. 24570061 to C.M.), Scientific Research on Innovative Areas (no. 26113519 to S.K.), and Scientific Research (B) (no. 26291056 to Y.M.) from the Ministry of Education, Science, Culture and Sports of Japan.
Conflict of interest: The authors have declared no conflicts of interest for this article.
References
REFERENCES
- 1. Steeves TA, Sussex IM. Patterns in Plant Development. Cambridge: Cambridge University Press; 1989. [Google Scholar]
- 2. Waites R, Selvadurai HR, Oliver IR, Hudson A. The PHANTASTICA gene encodes a MYB transcription factor involved in growth and dorsoventrality of lateral organs in Antirrhinum . Cell 1998, 93:779–789. [DOI] [PubMed] [Google Scholar]
- 3. Hudson A. Development of symmetry in plants. Annu Rev Plant Physiol Plant Mol Biol 2000, 51:349–370. [DOI] [PubMed] [Google Scholar]
- 4. Byrne ME, Timmermans M, Kidner C, Martienssen R. Development of leaf shape. Curr Opin Plant Biol 2001, 4:38–43. [DOI] [PubMed] [Google Scholar]
- 5. Semiarti E, Ueno Y, Tsukaya H, Iwakawa H, Machida C, Machida Y. The ASYMMETRIC LEAVES2 gene of Arabidopsis thaliana regulates formation of a symmetric lamina, establishment of venation and repression of meristem‐related homeobox genes in leaves. Development 2001, 128:1771–1783. [DOI] [PubMed] [Google Scholar]
- 6. Bowman JL, Floyd SK. Patterning and polarity in seed plant shoots. Annu Rev Plant Biol 2008, 59:67–88. [DOI] [PubMed] [Google Scholar]
- 7. Szakonyi D, Moschopoulos A, Byrne ME. Perspectives on leaf dorsoventral polarity. J Plant Res 2010, 123:281–290. [DOI] [PubMed] [Google Scholar]
- 8. Nakata M, Okada K. The leaf adaxial‐abaxial boundary and lamina growth. Plants 2013, 2:174–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tsukaya H. Leaf development. Arab Book 2013, 11:e0163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mauseth JD. Plant Anatomy. Menlo Park, CA: The Benjamin/Cummings; 1988. [Google Scholar]
- 11. Smith LG, Hake S. The initiation and determination of leaves. Plant Cell 1992, 4:1017–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lynn K, Fernandez A, Aida M, Sedbrook J, Tasaka M, Masson P, Barton MK. The PINHEAD/ZWILLE gene acts pleiotropically in Arabidopsis development and has overlapping functions with the ARGONAUTE1 gene. Development 1999, 126:469–481. [DOI] [PubMed] [Google Scholar]
- 13. Husbands AM, Chitwood DH, Plavskin Y, Timmermans MC. Signals and prepatterns: new insights into organ polarity in plants. Genes Dev 2009, 23:1986–1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Waites R, Hudson A. Phantastica: a gene required for dorsoventrality in leaves of Antirrhinum majus . Development 1995, 121:2143–2154. [Google Scholar]
- 15. McConnell JR, Emery J, Eshed Y, Bao N, Bowman JL, Barton MK. Role of PHABULOSA and PHAVOLUTA in determining radial patterning in shoots. Nature 2001, 411:709–713. [DOI] [PubMed] [Google Scholar]
- 16. Kidner CA, Martienssen RA. Spatially restricted microRNA directs leaf polarity through ARGONAUTE1. Nature 2004, 428:81–84. [DOI] [PubMed] [Google Scholar]
- 17. Byrne ME, Barley R, Curtis M, Arroyo JM, Dunham M, Hudson A, Martienssen RA. Asymmetric leaves1 mediates leaf patterning and stem cell function in Arabidopsis . Nature 2000, 408:967–971. [DOI] [PubMed] [Google Scholar]
- 18. Iwakawa H, Ueno Y, Semiarti E, Onouchi H, Kojima S, Tsukaya H, Hasebe M, Soma T, Ikezaki M, Machida C, et al. The ASYMMETRIC LEAVES2 gene of Arabidopsis thaliana, required for formation of a symmetric flat leaf lamina, encodes a member of a novel family of proteins characterized by cysteine repeats and a leucine zipper. Plant Cell Physiol 2002, 43:467–478. [DOI] [PubMed] [Google Scholar]
- 19. Shuai B, Reynaga‐Pena CG, Springer PS. The lateral organ boundaries gene defines a novel, plant‐specific gene family. Plant Physiol 2002, 129:747–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Matsumura Y, Iwakawa H, Machida Y, Machida C. Characterization of genes in the ASYMMETRIC LEAVES2/LATERAL ORGAN BOUNDARIES (AS2/LOB) family in Arabidopsis thaliana, and functional and molecular comparisons between AS2 and other family members. Plant J 2009, 58:525–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rédei GP, Hirono Y. Linkage studies. Arab Inf Serv 1964, 1:9. [Google Scholar]
- 22. Sarojam R, Sappl PG, Goldshmidt A, Efroni I, Floyd SK, Eshed Y, Bowman JL. Differentiating Arabidopsis shoots from leaves by combined YABBY activities. Plant Cell 2010, 22:2113–2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Pekker I, Alvarez JP, Eshed Y. Auxin response factors mediate Arabidopsis organ asymmetry via modulation of KANADI activity. Plant Cell 2005, 17:2899–2910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Garcia D, Collier SA, Byrne ME, Martienssen RA. Specification of leaf polarity in Arabidopsis via the trans‐acting siRNA pathway. Curr Biol 2006, 16:933–938. [DOI] [PubMed] [Google Scholar]
- 25. Chitwood DH, Nogueira FTS, Howell MD, Taiowa A, Montgomery TA, Carrington JC, Timmermans MCP. Pattern formation via small RNA mobility. Genes Dev 2009, 23:549–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Emery JF, Floyd SK, Alvarez J, Eshed Y, Hawker NP, Izhaki A, Baum SF, Bowman JL. Radial patterning of Arabidopsis shoots by class III HD‐ZIP and KANADI genes. Curr Biol 2003, 13:1768–1774. [DOI] [PubMed] [Google Scholar]
- 27. Siegfried KR, Eshed Y, Baum SF, Otsuga D, Drews GN, Bowman JL. Members of the YABBY gene family specify abaxial cell fate in Arabidopsis . Development 1999, 126:4117–4128. [DOI] [PubMed] [Google Scholar]
- 28. Kerstetter RA, Bollman K, Taylor RA, Bomblies K, Poethig RS. KANADI regulates organ polarity in Arabidopsis . Nature 2001, 11:706–709. [DOI] [PubMed] [Google Scholar]
- 29. Izhaki A, Bowman JL. KANADI and class III HD‐zip gene families regulate embryo patterning and modulate auxin flow during embryogenesis in Arabidopsis . Plant Cell 2007, 19:495–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nakata M, Matsumoto N, Tsugeki R, Rikirsch E, Laux T, Okada K. Roles of the middle domain‐specific WUSCHEL‐RELATED HOMEOBOX genes in early development of leaves in Arabidopsis . Plant Cell 2012, 24:519–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wang W, Xu B, Wang H, Li J, Huang H, Xu L. YUCCA genes are expressed in response to leaf adaxial‐abaxial juxtaposition and are required for leaf margin development. Plant Physiol 2011, 157:1805–1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Iwakawa H, Iwasaki M, Kojima S, Ueno Y, Soma T, Tanaka H, Semiarti E, Machida Y, Machida C. Expression of the ASYMMETRIC LEAVES2 gene in the adaxial domain of Arabidopsis leaves represses cell proliferation in this domain and is critical for the development of properly expanded leaves. Plant J 2007, 51:173–184. [DOI] [PubMed] [Google Scholar]
- 33. Wu G, Lin WC, Huang T, Poethig RS, Springer PS, Kerstetter RA. KANADI1 regulates adaxial–abaxial polarity in Arabidopsis by directly repressing the transcription of ASYMMETRIC LEAVES2 . Proc Natl Acad Sci USA 2008, 105:16392–16397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Iwasaki M, Takahashi H, Iwakawa H, Nakagawa A, Ishikawa T, Tanaka H, Matsumura Y, Pekker I, Eshed Y, Vial‐Pradel S, et al. Dual regulation of ETTIN (ARF3) gene expression by AS1‐AS2, which maintains the DNA methylation level, is involved in stabilization of leaf adaxial‐abaxial partitioning in Arabidopsis . Development 2013, 140:1958–1969. [DOI] [PubMed] [Google Scholar]
- 35. Waites R, Hudson A. The Handlebars gene is required with Phantastica for dorsoventral asymmetry of organs and for stem cell activity in Antirrhinum . Development 2001, 128:1923–1931. [DOI] [PubMed] [Google Scholar]
- 36. Serrano‐Cartagena J, Robles P, Ponce MR, Micol JL. Genetic analysis of leaf form mutants from the Arabidopsis information service collection. Mol Gen Genet 1999, 261:725–739. [DOI] [PubMed] [Google Scholar]
- 37. Byrne ME, Simorowski J, Martienssen RA. ASYMMETRIC LEAVES1 reveals KNOX gene redundancy in Arabidopsis . Development 2002, 129:1957–1965. [DOI] [PubMed] [Google Scholar]
- 38. Takahashi H, Iwakawa H, Nakao S, Ojio T, Morishita R, Morikawa S, Machida Y, Machida C, Kobayashi T. Knowledge‐based fuzzy adaptive resonance theory and the application to gene expression analysis of plants. J Biosci Bioeng 2008, 106:587–593. [DOI] [PubMed] [Google Scholar]
- 39. Ueno Y, Ishikawa T, Watanabe K, Terakura S, Iwakawa H, Okada K, Machida C, Machida Y. Histone deacetylases and asymmetric leaves2 are involved in the establishment of polarity in leaves of Arabidopsis . Plant Cell 2007, 19:445–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhu Y, Li Z, Xu B, Li H, Wang L, Dong A, Huang H. Subcellular localizations of AS1 and AS2 suggest their common and distinct roles in plant development. J Integr Plant Biol 2008, 50:897–905. [DOI] [PubMed] [Google Scholar]
- 41. Luo L, Ando S, Sasabe M, Machida C, Kurihara D, Higashiyama T, Machida Y. Arabidopsis asymmetric leaves2 protein required for leaf morphogenesis consistently forms speckles during mitosis of tobacco BY‐2 cells via signals in its specific sequence. J Plant Res 2012, 125:661–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Guo M, Thomas J, Collins G, Timmermans MC. Direct repression of KNOX loci by the ASYMMETRIC LEAVES1 complex of Arabidopsis . Plant Cell 2008, 20:48–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yang JY, Iwasaki M, Machida C, Machida Y, Zhou X, Chua NH. βC1, the pathogenicity factor of TYLCCNV, interacts with AS1 to alter leaf development and suppress selective jasmonic acid responses. Genes Dev 2008, 22:2564–2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Timmermans MC, Hudson A, Becraft PW, Nelson T. ROUGH SHEATH2: a Myb protein that represses knox homeobox genes in maize lateral organ primordia. Science 1999, 284:151–153. [DOI] [PubMed] [Google Scholar]
- 45. Tsiantis M, Schneeberger R, Golz JF, Freeling M, Langdale JA. The maize rough sheath2 gene and leaf development programs in monocot and dicot plants. Science 1999, 284:154–156. [DOI] [PubMed] [Google Scholar]
- 46. Schneeberger R, Tsiantis M, Freeling M, Langdale JA. The rough sheath2 gene negatively regulates homeobox gene expression during maize leaf development. Development 1998, 125:2857–2865. [DOI] [PubMed] [Google Scholar]
- 47. Byrne ME. Networks in leaf development. Curr Opin Plant Biol 2005, 8:59–66. [DOI] [PubMed] [Google Scholar]
- 48. Ikezaki M, Kojima M, Sakakibara H, Kojima S, Ueno Y, Machida C, Machida Y. Genetic networks regulated by ASYMMETRIC LEAVES1 (AS1) and AS2 in leaf development in Arabidopsis thaliana: KNOX genes control five morphological events. Plant J 2010, 61:70–82. [DOI] [PubMed] [Google Scholar]
- 49. Sakamoto T, Kamiya N, Ueguchi‐Tanaka M, Iwahori S, Matsuoka M. KNOX homeodomain protein directly suppresses the expression of a gibberellin biosynthetic gene in the tobacco shoot apical meristem. Genes Dev 2001, 15:581–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hay A, Kaur H, Phillips A, Hedden P, Hake S, Tsiantis M. The gibberellin pathway mediates KNOTTED1‐type homeobox function in plants with different body plans. Curr Biol 2002, 12:1557–1565. [DOI] [PubMed] [Google Scholar]
- 51. McHale NA, Koning RE. PHANTASTICA regulates development of the adaxial mesophyll in Nicotiana leaves. Plant Cell 2004, 16:1251–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Hunter C, Willmann MR, Wu G, Yoshikawa M, de la Luz G‐NM, Poethig SR. Trans‐acting siRNA‐mediated repression of ETTIN and ARF4 regulates heteroblasty in Arabidopsis . Development 2006, 133:2973–2981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Sawa S, Watanabe K, Goto K, Liu YG, Shibata D, Kanaya E, Morita EH, Okada K. FILAMENTOUS FLOWER, a meristem and organ identity gene of Arabidopsis, encodes a protein with a zinc finger and HMG‐related domains. Genes Dev 1999, 13:1079–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Yifhar T, Pekker I, Peled D, Friedlander G, Pistunov A, Sabban M, Wachsman G, Alvarez JP, Amsellem Z, Eshed Y. Failure of the tomato trans‐acting short interfering RNA program to regulate AUXIN RESPONSE FACTOR3 and ARF4 underlies the wiry leaf syndrome. Plant Cell 2012, 24:3575–3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Jouannet V, Moreno AB, Elmayan T, Vaucheret H, Crespi MD, Maizel A. Cytoplasmic Arabidopsis AGO7 accumulates in membrane‐associated siRNA bodies and is required for ta‐siRNA biogenesis. EMBO J 2012, 31:1704–1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kumakura N, Takeda A, Fujioka Y, Motose H, Takano R, Watanabe Y. SGS3 and RDR6 interact and colocalize in cytoplasmic SGS3/RDR6‐bodies. FEBS Lett 2009, 583:1261–1266. [DOI] [PubMed] [Google Scholar]
- 57. Hoffer P, Ivashuta S, Pontes O, Vitins A, Pikaard C, Mroczka A, Wagner N, Voelker T. Posttranscriptional gene silencing in nuclei. Proc Natl Acad Sci USA 2011, 108:409–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Li H, Xu L, Wang H, Yuan Z, Cao X, Yang Z, Zhang D, Xu Y, Huang H. The putative RNA‐dependent RNA polymerase RDR6 acts synergistically with ASYMMETRIC LEAVES1 and 2 to repress BREVIPEDICELLUS and microRNA165/166 in Arabidopsis leaf development. Plant Cell 2005, 17:2157–2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Xu L, Yang L, Pi L, Liu Q, Ling Q, Wang H, Poethig RS, Huang H. Genetic interaction between the AS1‐AS2 and RDR6‐SGS3‐AGO7 pathways for leaf morphogenesis. Plant Cell Physiol 2006, 47:853–863. [DOI] [PubMed] [Google Scholar]
- 60. Hiraguri A, Itoh R, Kondo N, Nomura Y, Aizawa D, Murai Y, Koiwa H, Seki M, Shinozaki K, Fukuhara T. Specific interactions between Dicer‐like proteins and HYL1/DRB‐family dsRNA‐binding proteins in Arabidopsis thaliana . Plant Mol Biol 2005, 57:173–188. [DOI] [PubMed] [Google Scholar]
- 61. Fang Y, Spector DL. Identification of Nuclear Dicing Bodies Containing Proteins for MicroRNA Biogenesis in Living Arabidopsis Plants. Curr Biol 2007, 17:818–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Kidner CA, Martienssen RA. The role of ARGONAUTE1 (AGO1) in meristem formation and identity. Dev Biol 2005, 280:504–517. [DOI] [PubMed] [Google Scholar]
- 63. Yang L, Huang W, Wang H, Cai R, Xu Y, Huang H. Characterizations of a hypomorphic argonaute1 mutant reveal novel AGO1 functions in Arabidopsis lateral organ development. Plant Mol Biol 2006, 61:63–78. [DOI] [PubMed] [Google Scholar]
- 64. Ori N, Eshed Y, Chuck G, Bowman JL, Hake S. Mechanisms that control knox gene expression in the Arabidopsis shoot. Development 2000, 127:5523–5532. [DOI] [PubMed] [Google Scholar]
- 65. Yang L, Liu Z, Lu F, Dong A, Huang H. SERRATE is a novel nuclear regulator in primary microRNA processing in Arabidopsis. Plant J 2006, 47:841–850. [DOI] [PubMed] [Google Scholar]
- 66. Lawrence RJ, Earley K, Pontes O, Silva M, Chen ZJ, Neves N, Viegas W, Pikaard CS. A concerted DNA methylation/histone methylation switch regulates rRNA gene dosage control and nucleolar dominance. Mol Cell 2004, 13:599–609. [DOI] [PubMed] [Google Scholar]
- 67. Zhou C, Labbe H, Sridha S, Wang L, Tian L, Latoszek‐Green M, Yang Z, Brown D, Miki B, Wu K. Expression and function of HD2‐type histone deacetylases in Arabidopsis development. Plant J 2004, 38:715–724. [DOI] [PubMed] [Google Scholar]
- 68. Nelissen H, De Groeve S, Fleury D, Neyt P, Bruno L, Bitonti MB, Vandenbussche F, Van der Straeten D, Yamaguchi T, Tsukaya H, et al. Plant elongator regulates auxin‐related genes during RNA polymerase II transcription elongation. Proc Natl Acad Sci USA 2010, 107:1678–1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Takahashi H, Iwakawa H, Ishibashi N, Kojima S, Matsumura Y, Prananingrum P, Iwasaki M, Takahashi A, Ikezaki M, Luo L, et al. Meta‐analyses of microarrays of Arabidopsis asymmetric leaves1 (as1), as2 and their modifying mutants reveal a critical role for the ETT pathway in stabilization of adaxial‐abaxial patterning and cell division during leaf development. Plant Cell Physiol 2013, 54:418–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Kojima S, Iwasaki M, Takahashi H, Imai T, Matsumura Y, Fleury D, Van Lijsebettens M, Machida Y, Machida C. Asymmetric leaves2 and Elongator, a histone acetyltransferase complex, mediate the establishment of polarity in leaves of Arabidopsis thaliana . Plant Cell Physiol 2011, 52:1259–1273. [DOI] [PubMed] [Google Scholar]
- 71. Xu D, Huang W, Li Y, Wang H, Huang H, Cui X. Elongator complex is critical for cell cycle progression and leaf patterning in Arabidopsis . Plant J 2012, 69:792–808. [DOI] [PubMed] [Google Scholar]
- 72. Ishibashi N, Machida C, Machida Y. ASYMMETRIC LEAVES2 and FASCIATA2 cooperatively regulate the formation of leaf adaxial‐abaxial polarity in Arabidopsis thaliana . Plant Biotech 2013, 30:411–415. [Google Scholar]
- 73. Carroll AJ, Heazlewood JL, Ito J, Millar AH. Analysis of the Arabidopsis cytosolic ribosome proteome provides detailed insights into its components and their post‐translational modification. Mol Cell Proteomics 2008, 7:347–369. [DOI] [PubMed] [Google Scholar]
- 74. Pinon V, Etchells JP, Rossignol P, Collier SA, Arroyo JM, Martienssen RA, Byrne ME. Three piggyback genes that specifically influence leaf patterning encode ribosomal proteins. Development 2008, 135:1315–1324. [DOI] [PubMed] [Google Scholar]
- 75. Yao Y, Ling Q, Wang H, Huang H. Ribosomal proteins promote leaf adaxial identity. Development 2008, 135:1325–1334. [DOI] [PubMed] [Google Scholar]
- 76. Horiguchi G, Mollá‐Morales A, Pérez‐Pérez JM, Kojima K, Robles P, Ponce MR, Micol JL, Tsukaya H. Differential contributions of ribosomal protein genes to Arabidopsis thaliana leaf development. Plant J 2011, 65:724–736. [DOI] [PubMed] [Google Scholar]
- 77. Byrne M. A role for the ribosome in development. Trends Plant Sci 2009, 14:513–519. [DOI] [PubMed] [Google Scholar]
- 78. Szakonyi D, Byrne ME. Ribosomal protein L27a is required for growth and patterning in Arabidopsis thaliana . Plant J 2011, 65:269–281. [DOI] [PubMed] [Google Scholar]
- 79. Casanova‐Sáez R, Candela H, Micol JL. Combined haploinsufficiency and purifying selection drive retention of RPL36a paralogs in Arabidopsis . Sci Rep 2014, 4:4122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Abbasi N, Kim HB, Park NI, Kim HS, Kim YK, Park YI, Choi SB. APUM23, a nucleolar Puf domain protein, is involved in pre‐ribosomal RNA processing and normal growth patterning in Arabidopsis. Plant J 2010, 64:960–976. [DOI] [PubMed] [Google Scholar]
- 81. Huang T, Kerstetter RA, Irish VF. APUM23, a PUF family protein, functions in leaf development and organ polarity in Arabidopsis . J Exp Bot 2014, 65:1181–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Inagaki S, Nakamura K, Morikami A. A link among DNA replication, recombination, and gene expression revealed by genetic and genomic analysis of TEBICHI gene of Arabidopsis thaliana . PLoS Genet 2009, 5:e1000613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Luo D, Bernard DG, Balk J, Hai H, Cui X. The DUF59 family gene AE7 acts in the cytosolic iron‐sulfur cluster assembly pathway to maintain nuclear genome integrity in Arabidopsis . Plant Cell 2012, 24:4135–4148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Yuan Z, Luo D, Li G, Yao X, Wang H, Zeng M, Huang H, Cui X. Characterization of the AE7 gene in Arabidopsis suggests that normal cell proliferation is essential for leaf polarity establishment. Plant J 2010, 64:331–342. [DOI] [PubMed] [Google Scholar]
- 85. Traverso JA, Micalella C, Martinez A, Brown SC, Satiat‐Jeunemaître B, Meinnel T, Giglione C. Roles of N‐terminal fatty acid acylations in membrane compartment partitioning: Arabidopsis h‐type thioredoxins as a case study. Plant Cell 2013, 25:1056–1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Fu H, Reis N, Lee Y, Glickman MH, Vierstra RD. Subunit interaction maps for the regulatory particle of the 26S proteasome and the COP9 signalosome. EMBO J 2001, 20:7096–7107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Yang P, Fu H, Walker J, Papa CM, Smalle J, Ju YM, Vierstra RD. Purification of the Arabidopsis 26 S proteasome: biochemical and molecular analyses revealed the presence of multiple isoforms. J Biol Chem 2004, 279:6401–6413. [DOI] [PubMed] [Google Scholar]
- 88. Huang W, Pi L, Liang W, Xu B, Wang H, Cai R, Huang H. The proteolytic function of the Arabidopsis 26S proteasome is required for specifying leaf adaxial identity. Plant Cell 2006, 18:2479–2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Nakata M, Okada K. The three‐domain model: a new model for the early development of leaves in Arabidopsis thaliana . Plant Signal Behav 2012, 7:1423–1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Ishibashi N, Kanamaru K, Ueno Y, Kojima S, Kobayashi T, Machida C, Machida Y. Asymmetric‐leaves2 and an ortholog of eukaryotic NudC domain proteins repress expression of Auxin‐response‐factor and class 1 KNOX homeobox genes for development of flat symmetric leaves in Arabidopsis . Biol Open 2012, 1:197–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Jurkuta RJ, Kaplinsky NJ, Spindel JE, Barton MK. Partitioning the apical domain of the Arabidopsis embryo requires the BOBBER1 NudC domain protein. Plant Cell 2009, 21:1957–1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Kawade K, Horiguchi G, Usami T, Hirai MY, Tsukaya H. ANGUSTIFOLIA3 signaling coordinates proliferation between clonally distinct cells in leaves. Curr Biol 2013, 23:788–792. [DOI] [PubMed] [Google Scholar]
- 93. Horiguchi G, Nakayama H, Ishikawa N, Kubo M, Demura T, Fukuda H, Tsukaya H. ANGUSTIFOLIA3 plays roles in adaxial/abaxial patterning and growth in leaf morphogenesis. Plant Cell Physiol 2011, 52:112–124. [DOI] [PubMed] [Google Scholar]
- 94. Duchêne AM, Peeters N, Dietrich A, Cosset A, Small ID, Wintz H. Overlapping destinations for two dual targeted glycyl‐tRNA synthetases in Arabidopsis thaliana and Phaseolus vulgaris . J Biol Chem 2001, 276:15275–15283. [DOI] [PubMed] [Google Scholar]
- 95. Moschopoulos A, Derbyshire P, Byrne ME. The Arabidopsis organelle‐localized glycyl‐tRNA synthetase encoded by embryo defective development1 is required for organ patterning. J Exp Bot 2012, 63:5233–5243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Hedtke B, Meixner M, Gillandt S, Richter E, Börner T, Weihe A. Green fluorescent protein as a marker to investigate targeting of organellar RNA polymerases of higher plants in vivo . Plant J 1999, 17:557–561. [DOI] [PubMed] [Google Scholar]
- 97. De Veylder L, Beeckman T, Beemster GT, Krols L, Terras F, Landrieu I, Van Der Schueren E, Maes S, Naudts M, Inzé D. Functional analysis of cyclin‐dependent kinase inhibitors of Arabidopsis . Plant Cell 2001, 13:1653–1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Sherr CJ, Roberts JM. CDK inhibitors: positive and negative regulators of G1‐phase progression. Genes Dev 1999, 13:1501–1512. [DOI] [PubMed] [Google Scholar]
- 99. Kakimoto T. Identification of plant cytokinin biosynthetic enzymes as dimethylallyl diphosphate: ATP/ADP isopentenyltransferases. Plant Cell Physiol 2001, 42:677–685. [DOI] [PubMed] [Google Scholar]
- 100. Takei K, Sakakibara H, Sugiyama T. Identification of genes encoding adenylate isopentenyl transferase, a cytokinin biosynthesis enzyme, in Arabidopsis thaliana . J Biol Chem 2001, 276:26405–26410. [DOI] [PubMed] [Google Scholar]
- 101. Sakakibara H, Kasahara H, Ueda N, Kojima M, Takei K, Hishiyama S, Asami T, Okada K, Kamiya Y, Yamaya T, et al. Agrobacterium tumefaciens increases cytokinin production in plastids by modifying the biosynthetic pathway in the host plant. Proc Natl Acad Sci USA 2005, 102:9972–9977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Miyawaki K, Matsumoto‐Kitanoand M, Kakimoto T. Expression of cytokinin biosynthetic isopentenyltransferase genes in Arabidopsis: tissue specificity and regulation by auxin, cytokinin, and nitrate. Plant J 2004, 37:128–138. [DOI] [PubMed] [Google Scholar]
- 103. Phelps‐Durr TL, Thomas J, Vahab P, Timmermans MC. Maize rough sheath2 and its Arabidopsis orthologue ASYMMETRIC LEAVES1 interact with HIRA, a predicted histone chaperone, to maintain knox gene silencing and determinacy during organogenesis. Plant Cell 2005, 17:2886–2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Lodha M, Marco CF, Timmermans MC. The ASYMMETRIC LEAVES complex maintains repression of KNOX homeobox genes via direct recruitment of polycomb‐repressive complex2. Genes Dev 2013, 27:596–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Zhang X, Yazaki J, Sundaresan A, Cokus S, Chan SW, Chen H, Henderson IR, Shinn P, Pellegrini M, Jacobsen SE, et al. Genome‐wide high‐resolution mapping and functional analysis of DNA methylation in Arabidopsis . Cell 2006, 126:1189–1201. [DOI] [PubMed] [Google Scholar]
- 106. Zilberman D, Gehring M, Tran RK, Ballinger T, Henikoff S. Genome‐wide analysis of Arabidopsis thaliana DNA methylation uncovers an interdependence between methylation and transcription. Nat Genet 2007, 39:61–69. [DOI] [PubMed] [Google Scholar]
- 107. Li W, Liu H, Cheng ZJ, Su YH, Han HN, Zhang Y, Zhang XS. DNA methylation and histone modifications regulate de novo shoot regeneration in Arabidopsis by modulating WUSCHEL expression and auxin signaling. PLoS Genet 2011, 7:e1002243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Qi J, Wang Y, Yu T, Cunha A, Wu B, Vernoux T, Meyerowitz E, Jiao Y. Auxin depletion from leaf primordia contributes to organ patterning. Proc Natl Acad Sci USA 2014, 111:18769–18774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Reinhart BJ, Liu T, Newell NR, Magnani E, Huang T, Kerstetter R, Michaels S, Barton MK. Establishing a framework for the ad/abaxial regulatory network of Arabidopsis: ascertaining targets of class III homeodomain leucine zipper and KANADI regulation. Plant Cell 2013, 25:3228–3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Merelo P, Xie Y, Brand L, Ott F, Weigel D, Bowman JL, Heisler MG, Wenkel S. Genome‐wide identification of KANADI1 target genes. PLoS One 2013, 8:e77341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Huang T, Harrar Y, Lin C, Reinhart B, Newell NR, Talavera‐Rauh F, Hokin SA, Barton MK, Kerstetter RA. Arabidopsis KANADI1 acts as a transcriptional repressor by interacting with a specific cis‐element and regulates auxin biosynthesis, transport, and signaling in opposition to HD‐ZIPIII factors. Plant Cell 2014, 26:246–262. [DOI] [PMC free article] [PubMed] [Google Scholar]


