Abstract
Current clinically applicable tissue and organ replacement therapies are limited in the field of cardiovascular regenerative medicine. The available options do not regenerate damaged tissues and organs, and, in the majority of the cases, show insufficient restoration of tissue function. To date, anticoagulant drug‐free heart valve replacements or growing valves for pediatric patients, hemocompatible and thrombus‐free vascular substitutes that are smaller than 6 mm, and stem cell‐recruiting delivery systems that induce myocardial regeneration are still only visions of researchers and medical professionals worldwide and far from being the standard of clinical treatment. The design of functional off‐the‐shelf biomaterials as well as automatable and up‐scalable biomaterial processing methods are the focus of current research endeavors and of great interest for fields of tissue engineering and regenerative medicine. Here, various approaches that aim to overcome the current limitations are reviewed, focusing on biomaterials design and generation methods for myocardium, heart valves, and blood vessels. Furthermore, novel contact‐ and marker‐free biomaterial and extracellular matrix assessment methods are highlighted.
Keywords: cardiovascular tissue engineering, biomaterials, heart valves, blood vessels, myocardium, Raman spectroscopy, biomimetic tissues
1. Introduction
Cardiovascular diseases (CVDs) such as coronary heart disease, rheumatic heart disease, congenital heart disease, myocardial infarction (MI) and strokes remain the number one cause of death in western countries. Despite the development of new therapies and the approach of risk‐reducing strategies to prevent CVDs, the World Health Organization estimates an increase from 17.3 million deaths in 2008 to 23.3 million by 2030.1 To date, there are no effective therapies to fully restore cardiovascular organ function after damage, and due to an insufficient number of organ donors,2, 3 the design of regenerative strategies such as stem‐cell‐based therapies and in vitro‐engineered tissues are in great demand. One focus in the field of regenerative medicine is the generation and improvement of functional biomaterials. These materials can be bioactive; releasing drugs, proteins, growth factors and extracellular matrix (ECM) components, or they can have an improved mechanical functionality. Today, there are several different types of biomaterials used to induce tissue regeneration or remodeling, and to support tissue function or even replace damaged structures. These biomaterials include metals, metal alloys and also synthetic and natural occurring polymers.4 Enormous progress has been made in generating and designing biomaterials for tissue repair. Highly promising are the so‐called biomimetic materials, which mimic ECM architecture and provide potentially controllable in vivo‐like microenvironments for cells.5 In vivo, cell behavior includes survival or controlled‐death, migration, proliferation and differentiation, which can be all controlled or impacted by the three‐dimensional (3D) ECM.5, 6, 7 Accordingly, physical, chemical and biological cues from the ECM must be investigated and subsequently recapitulated.
In this article, we provide an overview of functional biomaterials that are currently investigated for the use in the fields of cardiovascular tissue engineering and regenerative medicine, mainly focusing on myocardial repair, heart valves and blood vessels. Furthermore, promising contact‐ and marker‐free assessment methods are discussed for quality and pre‐implantation control or biomaterial assessment.
2. Biomaterials Used for Myocardial Repair
Multiple clinical and preclinical trials have investigated the effect of stem cell injections to induce cardiac regeneration after MI.8, 9, 10, 11, 12 Although the injection of a cell suspension showed promising results regarding improvement of cardiac function, there exist major obstacles such as poor cell retention and hence consistent efficacy cannot be achieved.13 To increase cell survival and to enable a localized delivery of cells without mechanical washout, biomaterials are used (Table 1 ).
Table 1.
Overview of biomaterials and fabrication methods used for myocardial repair
| Biomaterial | Fabrication Method | Cells/Molecules included | Application and Results | Ref. |
|---|---|---|---|---|
| PEGylated fibrinogen | Hydrogel formation | Cardiomyocytes, Embryonic stem cells | in vivo: improved fractional shortening | 15 |
| Chitosan | Hydrogel formation | Brown adipose derived stem cells | in vivo: enhanced cardiomyocyte differentiation and increased angiogenesis | 16 |
| Chitosan | Hydrogel formation | Adipose‐derived mesenchymal stem cells | in vivo: increased cell survival | 17 |
| Fibrin glue | Hydrogel formation | Adipose‐derived stem cells | in vivo: improved heart function | 18 |
| Alginate‐RGD | Hydrogel formation | Human mesenchymal stem cells | in vivo: reduced infarct size and improved cell survival | 20 |
| PEG‐vinylsulfone | Hydrogel formation | ‐ | in vitro: directs differentiation of pluripotent cardioprogenitors | 23 |
| Alginate | Hydrogel formation | B16‐F10 cells | shape memory gels in vivo: enhanced cell survival | 24 |
| Collagen type I | Hydrogel formation | miR‐29B | in vivo and in vitro: reduced wound contraction, improved ECM remodeling | 27 |
| HEMA – hyaluronic acid | Hydrogel formation | Neuregulin‐1β | in vivo: enhanced left ventricular ejection fraction | 28 |
| RADA16 | Self‐assembling hydrogel | VEGF and heparin | in vivo: enhanced cardiac function | 29 |
| Poly(ethylene argininylaspartate diglyceride) | Self‐assembling hydrogel | sonic hedgehoc | in vitro: upregulation of growth factors in a cardiac fibroblast cell culture | 30 |
| PEG | Hydrogel formation | SDF1‐GPVI | in vitro: release studies in vivo: cell immobilization via SDF‐1‐GPVI | 3, 32 |
| ECM | Cell sheets | ‐ | in vivo: improved cardiac function and neovascularization | 35 |
| Myocardial ECM | Hydrogel formation | ‐ | in vivo (large animal): increased cardiac function and reduced infarct fibrosis | 36 |
| Xylan/PVA | Electrospinning | ‐ | in vivo: increased cardiac cell proliferation | 38 |
| Pullan‐Dextran | Salt‐leaching | ‐ | in vitro: hECFC culture | 39 |
| Hyaluronic acid | Hydrogel formation | ‐ | in vivo: increased wall thickness | 40 |
| Alginate | Hydrogel formation | ‐ | Clinical trial: preserved LV function | 45 |
| PU‐aniline pentamer with PCL | Molding | oligoanilines | in vitro: cytocompatibility and conductivity tests | 49 |
| PLGA‐Gelatin | Electrospinning | ‐ | in vitro: integration of cardiomyocytes | 50 |
| PGS | Molding | ‐ | matches physical (mechanical) properties of the heart | 51 |
| PCL | Electrospinning | ‐ | in vitro: cardiomyocyte attachment and beating on day 3 | 52 |
| PCU or PLGA | Electrospinning on a textile‐template | ‐ | in vitro: adhesion and proliferation of H9C2 cardiacmyoblasts cell line and beating on day 10 on PCU | 58 |
| PCL‐Gelatin | Electrospinning | Mesenchymal stem cells | in vivo: reduced scar size and microvessel formation | 59 |
| PGS‐Fibrinogen | Electrospinning | VEGF and mesenchymal stem cells | in vivo (large animal): improved ejection fraction | 60 |
| PEUU | Phase separation | ‐ | in vivo: improved cardiac remodeling and contractile function | 63 |
| PLLA (on a PCLA sponge) | Knitting | Vascular smooth muscle cells | in vivo: improved LV function | 64 |
| Alginate | Hydrogel formation | gold nanoparticles | in vitro: improved electrical communication between neonatal ventricular myocytes | 65 |
| PLA | Electrospinning | carbon nanotubes | in vitro: mesenchymal stem cell differentiation to cardiomyocytes | 66 |
| PANI‐PLGA | Electrospinning | ‐ | in vitro: synchronized beating of cardiomyocytes | 67 |
| Algisyl‐LVR | Hydrogel formation | ‐ | Clinical trial: improved LV function | 70 |
2.1. Injectable Hydrogels
Immobilizing cells in a 3D hydrogel is a common approach.14 Depending on the required gelation time, cell type, site of injury and desired time of degradation, various material types and different gelation mechanisms can be considered. The hydrogels used in preclinical studies include for example synthetic Polyethylene glycol (PEG)15 or natural occurring chitosan,16, 17 fibrin,18 collagens19 and alginate.20 Wang et al. demonstrated cardiomyocyte differentiation and enhanced cell survival by injecting brown adipose tissue‐derived stem cells that were immobilized in a thermo‐responsive chitosan gel.16 Further gelation mechanisms described for cardiac applications are photopolymerization,15 Ca2+,20 pH21 or phosphate16, 17 dependent gelation, or assembly due to physical interactions like in the case of fibrin.18 Kraehenbuehl et al. developed a PEG‐vinylsulfone hydrogel system that undergoes Michael‐type addition reaction in situ for crosslinking.22 Combined with cells and bioactive factors, these gels were able to preserve the contractile function of cardiomyocytes and resulted in decreased infarct sizes.23 An advantage of in situ‐forming hydrogels is the possibility of a minimal invasive delivery, utilizing suitable catheters. For this approach, the hydrogel should form immediately after injection to the site of injury, whereas catheter blockage due to premature gelation has to be avoided.13 In order to overcome this problem, Bencherif et al. described a method to generate a preformed but still injectable hydrogel with shape memory effect.24 Here, alginate served as the base material, which was metharcylated to allow later radical polymerization. The at –20 °C polymerized, nanoporous, preformed alginate hydrogel can be pushed through a needle or catheter. As soon as the applied shear force is removed, the hydrogel returns to its original preformed shape.24
For applications in the heart, hydrogels can either be seeded with cells, or they can be loaded with bioactive agents in order to induce (stem) cell migration or differentiation, and thus tissue regeneration (Figure 1 ). However, despite promising results designing injectable cell‐loaded hydrogels, the amount of cells that effectively differentiated into cardiomyocytes in these studies was insufficient.13 Nevertheless, a cell‐free strategy is highly attractive in regards of regulatory aspects, and since this approach is not limited to a specific patient, an off‐the‐shelf up‐scalable production is possible. Recent in vitro and in vivo studies focused on the application of bioactive drug‐releasing hydrogels. Currently used and entrapped bioactive molecules include small molecules like prostaglandins,25 RNA,26, 27 growth factors28, 29 or various other factors such as sonic hedgehog30 and bone morphogenetic protein‐2.31
Figure 1.

Biomaterials for cardiac applications. Injectable hydrogels but also cardiac patches are used as treatment options for cardiac damage. Both materials can either be cell‐seeded or loaded with bioactive molecules such as RNA, small molecules, growth factors or proteins.
Improvement in wound healing and increased ECM remodeling has been described when encapsulating micro RNA (miR)‐29B in a collagen hydrogels.27 In this system, a collagen solution was incubated with a four‐arm PEG terminated succinimidyl glutarate in order to induce gelation.27 Another approach aimed for myocardial repair and regeneration after MI damage by the sustained release of stromal derived factor‐1 (SDF‐1)‐GPVI from a photopolymerizable PEG‐based hydrogel in order to attract circulating endothelial progenitor cells (EPCs).32 In this study, the released protein was able to bind extracellular occurring collagen with the GPVI domain, and EPCs that expressed CXCR4 were recruited by SDF‐1.33 Due to the non‐fouling properties of PEG, the encapsulated proteins do not bind to the PEG‐molecules and can therefore be easily released from the gel.34 Furthermore, drug delivery systems can be designed by using self‐assembling peptides. Guo et al. introduced a heparin‐binding domain to the self‐assembling peptide RADA16. The heparin‐binding vascular endothelial growth factor (VEGF) was entrapped in the biomaterial by first binding heparin to the newly designed RADA16 peptide before adding VEGF.29 In vivo MI studies confirmed effectiveness of the generated biomaterial, since microvessel formation, cardiomyocyte survival as well as an increased cardiac function was observed.29 An enhanced left ventricular ejection fraction (EF) in a model of ischemic cardiomyopathy was described in a study, where neuregulin‐1b growth factor was encapsulated in a hydrogel consisting of hyaluronic acid macromers with HEMA group modifications.28 Very interesting results have been also achieved when injecting natural occurring biomaterials like ECM or selected ECM components.35, 36 It has been speculated that these materials are advantageous due to the presence of native structures and architecture; however, chemically or enzymatically treated tissues lack of biochemically important matricellular proteins and proteoglycans, as well as microfibrillar structures.37 Furthermore, natural occurring polysaccharides such as xylan,38 dextran,39 pullan,39 chitosan,16 hyaluronan,40 and alginates41, 42, 43 have been investigated for myocardial applications. Existing patents related to polysaccharide‐based strategies are nicely summarized by Silva et al.44 A successful first‐in‐man clinical trial has been performed with alginate (IK‐5001). Interestingly, the alginate hydrogel alone preserved left ventricular function and left ventricular ejection fraction.45
When designing hydrogels for cardiac applications, not only hydrogel composition and ingredients impact in vivo performance, but also the mechanical strength and stiffness of the hydrogel are important parameters. MI for example leads to wall thinning46 and therefore a hydrogel is needed to mechanically support the damaged zone. However, it should be considered that hydrogels with too much stiffness can cause diastolic dysfunctions.47 Therefore, it is important to specify adequate mechanical properties. In addition, it is known that cells, including stem cells, respond to their microenvironment, and hydrogel stiffness has been shown to play a major role in stem cell differentiation.5, 48 Mesenchymal stem cells (MSCs) were seeded onto hydrogels with different stiffness. Neural differentiation occurred when the cells were cultured on a 0.1 kPa hydrogel, whereas 11 kPa resulted in myogenic tissue, and 34 kPa were necessary to induce osteogenic differentiation.5 To date, no best‐practice guidelines for optimal parameters with regards to therapeutic efficacy such as concentrations of bioactive agents, degradation rate and stiffness have been defined for biomaterials design.13 Future studies must focus much more on optimizing the biomaterials design by considering mechanical aspects.
Although injectable hydrogels are currently under intense investigation, there are also other biomaterials‐based strategies available to support cardiac repair. For example, solid preformed scaffolds, so‐called patches, can be applied epicardially onto the damaged heart. Just like hydrogels, these scaffolds can either be seeded with cells or they can be loaded with bioactive molecules (Figure 1). Patches do not only have a therapeutic effect by releasing bioactive molecules or delivering cells, they also provide mechanical support and are able to reduce dilation.49 However, certain requirements need to be considered when engineering a cardiac patch: The patch should be stable but flexible and provide mechanical strength; however, too stiff materials can induce diastolic dysfunctions and can lead to non‐contractile constructs.50 In most cases, it is desired that the patch degrades after sufficient remodeling.49, 50 Although the selection of an adequate material is a crucial issue, many studies do not consider the mechanical properties of the chosen material. The stiffness or E‐modulus of human myocardium has been described with 0.02–0.5 MPa, and the tensile strength was specified with 3–15 kPa.51 Prabhakaran et al. generated a nanofibrous composite scaffold of poly(dl‐lactide‐co‐glycolide) (PLGA) and gelatin via electrospinning. The authors impressively showed that the stiffness of the material could be manipulated by introducing the natural occurring polymer gelatin. Gelatin decreased the E‐modulus and furthermore enabled in vitro integration of cardiomyocytes.50 In addition, nanofibrous poly‐ε‐caprolactone (PCL) was identified as a beneficial scaffold for cardiomyocyte attachment.52 The range of material stiffness can also be influenced by synthesizing the polymer at different temperatures. This phenomenon has been proven with molded poly (glycerol sebacate) (PGS) foils, which were polymerized at 110 °C, 120 °C and 130 °C.51
It is important to notice that scaffold or patch characteristics highly vary between multiple manufacturing methods that are used for scaffold fabrication.
2.2. Electrospinning to Generate a Cardiac Patch
Electrospinning for example is a well described method for creating fiber‐containing and highly porous scaffolds.53 The principle of electrospinning is schematically displayed in Figure 2 . Briefly, a polymer is dissolved in a solvent and pumped through a syringe. The droplet on the tip of the syringe first forms a cone when a high voltage is applied. As soon as the electrical field is increased, a fiber ejects from the droplet, which travels in spinning motions to the counter electrode where the solvent evaporates and a fibrous scaffold is formed.54, 55, 56 By adjusting parameters such as polymer concentration and molecular weight, solvent volatility and conductivity, electrode distance, needle size, flow rate and voltage, the fiber shape, fiber diameter and pore size can be influenced.57 Nanofibrous scaffolds of electrospun PCL have been shown to be beneficial for cardiac applications.52 Furthermore, electrospun polycarbonate urethane scaffolds exhibited comparable mechanical properties such as the native myocardium.58 In vivo data indicated that a cell‐seeded electrospun PCL‐gelatin patch can provide sufficient mechanical strength to the myocardium, promotes angiogenesis and decreases scar size after MI.59 In addition, large animal in vivo studies confirmed that an electrospun PGS‐fibrinogen patch modified with VEGF can lead to an improved ejection fraction.60 With electrospinning it is not only possible to mimic the structural and mechanical properties of the native heart muscle, our group has shown that it is also possible to introduce crucial biochemical cues like proteoglycans to the electrospun fiber.61
Figure 2.
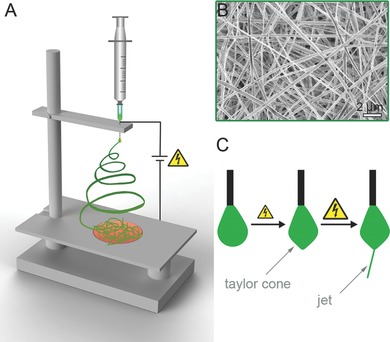
Electrospinning of patches for regenerative medicine and tissue engineering applications. A) General electrospinning set up: A polymer solution is pumped through a nozzle and forms a drop on the tip. The ejected fiber travels to the collector in spinning motions. Since the solvent evaporates, a randomly oriented solid fiber mat is deposed on the collector. B) Scanning electron microscopic image of an electrospun scaffold. C) The droplet on the needle tip forms a cone in an electrical field. As soon as the electrical field strength exceeds the surface energy of the droplet, a thin fiber is ejected.
2.3. Further Patch‐forming Approaches
Another method used to generate cardiac patches is thermally induced phase separation. With this technique it is possible to create well‐defined interconnected micropores utilizing simple equipment.62 To improve cardiac remodeling, polyester urethane urea (PEUU) has been processed into patches using phase separation techniques.63 Furthermore, Matsubyashi et al. engineered a muscle graft by knitting. Here, the authors reinforced a caprolactone‐co‐l‐lactide (PCLA) sponge with knitted poly‐l‐lactide (PLLA) in order to increase cardiac function.64
Another interesting approach is the generation of conductive biomaterials that support the cardiomyocytes' ability to contract. Several groups intensively studied these highly functional biomaterials in order to engineer an optimal cardiac patch.49, 65, 66, 67 Accordingly, a contracting patch could be obtained when using conductive polymers like polyaniline (PANI),67 or by manipulating non‐conductive polymers with conducting elements such as carbon nanotubes66 or biocompatible gold nanoparticles.65 In terms of cytocompatibility, conducting polymers including PANI,49 polypyrrole,68 and polythiophene69 have been successfully tested with various cell types, resulting in enhanced proliferation, adhesion and differentiation. Although these materials seem to be adequate for cardiac patch generation, a key limiting factor is that they are non‐degradable. In order to address this issue, low molecular weight oligoanilines have been examined.49 Molded films were prepared by synthesizing a biodegradable polyurethane‐containing aniline pentamer, which was then blended with PCL.49 Despite promising in vitro studies, these materials need to be further investigated in appropriate in vivo models. In regards to clinical translation, the innocuousness of the degradation products has to be carefully monitored.
2.4. Future Directions for Usage of Biomaterials for Myocardial Repair
To date, various methods to treat the injured myocardium have been studied. Cardiac patches showed encouraging results; however, the surgical procedure, which is required for implantation represents a drawback of this approach. In addition, conflicting results regarding the success of cardiac patches were generated, so there is a need for further long‐term in vivo studies. Moreover, since there are difficulties of finding the right cell source for myocardial tissue engineering, injectable cell‐free biomaterials may be more suitable for clinical use. It has been shown that injectable biomaterials provide mechanical strength70 and therefore prevent negative remodeling.47 Since too stiff materials lead to diastolic dysfunctions, it is necessary to carefully examine in future studies the mechanical properties needed for sufficient myocardial treatment. Furthermore, the question of, how the materials can be safely applied to patients needs to be answered. Alginate and fibrin are promising injectable biomaterials, which have already underwent clinical trials. However, the long‐term effects are not examined and in our opinion further investigations need to be performed.
3. Biomaterials Used for Heart Valve Tissue Engineering
Four valves regulate the blood flow in the normal heart: the semilunar pulmonary and aortic valves, and the atrioventricular tricuspid and mitral valves.71 These valves enable a unidirectional blood flow through the heart by opening and closing approximately 100 000 times a day.72, 73 This translates in more than 3 billion opening and closing movements over an average life cycle, where the heart valve leaflets are exposed to mechanical forces such as flow, tension and flexure.71 Accordingly, engineering a material that withstands these loads but maintains the hemodynamics of the heart is quite challenging. Native heart valve leaflets have a highly sophisticated histoarchitecture in order to enable a lifelong performance. In detail, three layers form a heart valve leaflet: the fibrosa, which is mainly composed of collagens, the glycosaminoglycan‐rich spongiosa and the elastic‐fiber containing ventricularis.74, 75, 76 It is hypothesized that the complex architecture of a heart valve leaflet is formed and maintained mainly by two different cell types, the ECM‐producing valvular interstitial cells (VICs), which are located predominantly in the spongiosa layer, and the valvular endothelial cells (VECs) that form a confluent endothelium as a barrier to the blood stream.72, 74 The current gold standard to treat diseased heart valves is the replacement using mechanical or biological valves. Mechanical valves are advantageous since they have an excellent durability compared to bioprosthetic valves, which are less durable and in addition prone to degradation; however, the risk of thromboembolism is higher with mechanical valves.77 A bioprosthetic substitute needs a glutaraldehyde fixation prior to implantation, a process that stiffens the matrix and inhibits repopulation.77 None of these replacement options offers the potential of growth or remodeling after implantation.72, 74, 75 A tissue‐engineered valve would be a promising possibility to address these limitations; however, most of the currently designed tissue‐engineered valves were pre‐clinically or clinically unsuccessful due to problems with calcification or fibrosis.75 A non‐thrombogenic and non‐calcific prosthesis with adequate mechanical properties and sufficient strength that maintains the native valve hemodynamics has so far not been designed.72 There are two different approaches to create a tissue‐engineered heart valve substitute. One is the traditional idea of seeding autologous cells onto a 3D scaffold in vitro prior to implantation. The other approach involves tissue regeneration and relies on implanting a cell‐free construct, which enables material‐guided reseeding in vivo.72
It has been proposed that the most adequate substrate in terms of mechanical and functional behavior is a decellularized xeno‐ or homograft.78, 79, 80 These substrates already exhibit the complex native valvular structure and architecture, but their restricted availability (homograft) as well as their risk of transferring zoonoses are limiting factors and major drawbacks of these constructs.80 Synthetic and natural polymers have been demonstrated to be an alternative for homografts and xenografts (Table 2 ). The techniques used to generate a valve‐shaped 3D scaffold range from simple molding methods like freeze molding collagen type I and elastin to form a two‐layered leaflet,81 to complex multi‐step approaches. Salt leaching is an interesting method to generate porous pre‐formed heart valve substitutes by adding sodium chloride crystals to a polymeric solution.82 After the polymer hardens, the salt crystals can be washed out and a porous scaffold is formed. Polyhydroxyalkanoate (PHA) for example has been described to be favorable to fabricate a tri‐leaflet heart valve with salt leaching, which can be successfully seeded with vascular cells.82 Another in vitro study described poly (glycerol sebacate) (PGS) and PCL as a suitable polymer combination to support the growth of MSCs and VICs, and in addition mimics the anisotropic mechanical properties of native heart valve leaflets.83 In order to create such a material, the combination of different fabrication methods was necessary. First, PGS was molded, and in a second step PCL was electrospun onto both sides of the PGS sheet resulting in a three‐layered scaffold with good mechanical properties.83 Combining different biomaterials but also using variable fabrication techniques in order to generate an optimal and complex cytocompatible scaffold is an increasing trend in current studies. Weber et al. created a tubular fibrin gel loaded with umbilical artery cells. They further strengthened the construct with a textile co‐scaffold.84 In contrast to the molded fibrin, the textile was a thermostabilized wrap‐knitted PET scaffold. With this technique it was possible to reproduce the valvular geometry and the mechanical strength.84
Table 2.
Overview of biomaterials and fabrication methods suitable for heart valve tissue engineering
| Biomaterial | Fabrication Method | Cells/Molecules included | Application and Results | Ref. |
|---|---|---|---|---|
| ECM | Decellularization | Endothelial cells and myofibroblasts | in vitro: matrix characterization and reseeding in a bioreactor | 76 |
| Homograft | Decellularization | ‐ | Clinical trial: excellent function, no thrombus formation | 79 |
| Elastin and collagen | Molding | ‐ | in vitro: bi‐layered material characterization and cell‐matrix interaction studies | 81 |
| PHA | Salt leaching | ‐ | in vitro: viable ECM formation in a bioreactor | 82 |
| PGS‐PCL | Micromolding – Electrospinning | ‐ | in vitro: 3‐layered construct supported growth of VICs and MSCs, ECM deposition | 83 |
| Fibrin gel and PET mesh | Hydrogel formation and knitting | Umbilical artery smooth muscle cells/myofibroblasts | in vitro: enhanced mechanical properties and tissue formation in a bioreactor | 84 |
| PEG‐PLA | Elektrospinning | ‐ | in vitro: biomimicking scaffold, cytocompatible with VICs and VECs | 85 |
| ECM | Cell sheets | Human fibroblasts | in vitro: matrix characterization | 86 |
| ECM | Decellularization | Endothelial progenitor cell‐derived endothelial cells OR CD 133 antibody | in vivo (large animal): CD 133‐conjugated leaflets exhibited a progressive recellularization across the entire leaflet, no calcification | 87 |
| PEG | Hydrogel formation and micropatterning | RGDS peptide | in vitro: controllable morphology and activation of VICs via micropatterns | 88 |
| PGA mesh‐P4HB ‐ECM | Decellularization after ECM production with vascular derived cells | Mesenchymal stem cells | in vitro: mechanical and biochemical characterization in vivo (primate): moderate valvular insufficiency, rapid cellular repopulation | 89, 90 |
| ECM | Decellularization | Umbilical cord‐derived endothelial cells | in vitro: complete recellularization in a bioreactor (Mitral valve) | 92 |
| Fibrin gel and PET mesh | Hydrogel formation and knitting | Umbilical vein smooth muscle cells/fibroblasts | in vitro: tissue development in a bioreactor, recapitualtes the native structure (Mitral valve) | 95 |
The idea of mimicking nature is a popular approach when aiming for an optimized heart valve substitute. The first step towards a bio‐inspired tissue is to obtain comprehensive data that describes the tissue's ultrastructure including cells and ECM as well as biomechanical properties. As soon as these critical parameters are defined, an adequate material and material generation technique can be selected. In a recently published study we identified the mechanical, structural and biochemical properties of a native heart valve leaflet. Based on this blueprint, we generated a scaffold by electrospinning a blend of PLA and photocrosslinkable PEG.85 In this study, the E‐modulus was determined in every single leaflet layer—the fibrosa, spongiosa and ventricularis—by applying atomic force microscopy (AFM) on unfixed cryosections.85 Another material fabrication strategy is to utilize ECM‐producing cells as scaffold producers. Fibroblasts for example produce a significant amount of ECM in the form of sheets, when cultured in vitro.86 It has been shown that these ECM‐sheets can be produced, and subsequently layered in order to increase strength and to shape a valve.86 Advantageous in this case is that the ECM‐sheets can be produced patient‐tailored using autologous cells.
In order to induce a variety of cellular responses, scaffolds can be functionalized with bioactive molecules. It was reported that substrates, which were functionalized with a conjugated anti‐CD133 antibody, attracted autologous cells and showed complete cellular ingrowth after implantation in a sheep model.87 In vitro studies using RGD‐modified micro‐patterned PEG hydrogels demonstrated VIC activation.88 The results of this study indicate that both, biochemical as well as topographical modifications are important modulators when designing a heart valve replacement. Dijkman et al. described the fabrication of a non‐woven PGA‐coated scaffold utilizing poly‐4‐hydroxybutyrate (P4HB). The scaffold was seeded with ovine vascular cells using fibrin as a cell carrier.89 Interestingly, exposing the cell‐material construct to dynamic strains by applying increasing transvalvular pressure differences enabled tissue maturation. The authors further described the possibility to minimally invasive deliver such an off‐the‐shelf valve.89 Later studies with the PGA‐P4HB valve showed remarkable cellular repopulation after implantation in primates.90 Such off‐the‐shelf cell‐free medical products are quite advantageous in terms of reaching clinical approval, since they are potentially more cost‐efficient and available at any time. The current trend of producing off‐the‐shelf materials and to combine it with minimally invasive implantation strategies is of high interest for the field of regenerative medicine.
To date, the majority of studies that reported on tissue engineering of heart valves focused on the semilunar (pulmonary and aortic) valves. Since it has been described that it is essential to mimic the shape of the mitral valve,91 methods such as decellularizing atrioventricular heart valves have been implemented in order to achieve or maintain a proper function.92 In addition to the mechanical characteristics, the ECM as well as the residing cells of the mitral valve were intensively investigated.93, 94 Moreira et al. demonstrated an elegant way to fabricate a textile‐reinforced mitral valve including annulus, asymmetric leaflets (anterior and posterior), and chordae tendineae. With this artificial mitral valve it is possible to maintain the native valve function and hemodynamics.95
3.1. Bioreactors for Valve Tissue Maturation
Bioreactors have been used in the field of tissue engineering to mimic the biophysical signals that are present in the native organo‐physiological environment. Depending on the organ or the cell type, various forces such as strain, pressure, torsion or flow occur in a tissue. Cellular behavior like migration, proliferation and differentiation are highly dependent on external forces and can be regulated by inducing physical stimuli with bioreactor systems.96 This has been shown to be also advantageous when generating a tissue‐engineered heart valve in vitro.75 For example, an increased ECM production by vascular cells was observed on a porous scaffold within a pulsatile flow bioreactor.82 An increased collagen and elastin production but also a significantly improved recellularization of the heart valve has been described under pulsatile conditions.78 These results indicate that not only the 3D scaffold but also the forces that are applied on the scaffold are important in order to obtain a highly functional tissue‐engineered construct.
3.2. Future Directions for Utilization of Biomaterials for Heart Valve Tissue Engineering Applications
Decellularized human valves have already been successfully used in clinical trials.79 Despite promising in vitro and in vivo experimental results, no tissue‐engineered heart valve construct has found its way into clinical reality. Similar to what we have described for myocardial tissue repair attempts, finding an adequate cell source is the major challenge. Interesting results were obtained by culturing MSCs on synthetic materials;83 however, it seems like there is a trend to generate cell‐free off‐the‐shelf substrates. In our opinion, it is important to mimic the mechanical, structural and biochemical properties of a native heart valve in order to overcome issues like limited tissue growth in pediatric patients. To prevent calcification and to enable cell attachment after implantation, it will be necessary to further investigate the biomaterial‐tissue‐interface and to screen for suitable surface modifications. As reviewed here, many in vitro studies were successfully performed showing sufficient cell attachment, cytocompatibility and mechanical graft stability; however, long‐term in vivo studies are needed in order to determine the true performance of the tissue‐engineered valves.
4. Biomaterials Used for Blood Vessel Engineering
Blood vessels are responsible for the nutrient and oxygen supply of all organs. As a reason of lifestyle or genetic abnormalities, vascular diseases such as artherosclerosis or aneurysms occur. Current treatments to address these problems are the transplantation of autologous grafts from other regions of the body,97 or the implantation of stents and synthetic grafts.97, 98 Clinically applied synthetic polymer substitutes are made of expanded‐polytetrafluoroethylene (ePTFE), Dacron or polyurethane.99, 100 Vessels with a diameter larger than 6 mm can be successfully replaced with these materials. However, due to thrombus formation in vessels of smaller sizes, there is an ongoing need for vascular alternatives.97 To date, there are different strategies pursued in order to engineer a functional blood vessel. These strategies are either tissue‐engineered cell‐seeded scaffolds or bioactive, cell‐free approaches (Table 3 ).
Table 3.
Overview of biomaterials and fabrication methods used to engineer blood vessel substitutes
| Biomaterial | Fabrication Method | Cells/Molecules included | Application and Results | Refs |
|---|---|---|---|---|
| ECM | Cell sheets | Fibroblasts | in vivo (small animal and primate): anti‐thrombogenic, mechanically stable, tissue integration Clinical trials | 101, 106 |
| PGA mesh with ECM | woven structure with decellularized ECM | Endothelial progenitor cells | in vivo (large animal): resistance to clotting and intimal hyperplasia | 102 |
| ECM | Cell sheets from human induced pluripotent stem cells | ‐ | in vitro: smooth muscle cell differentiation and collagenous matrix generation in a bioreactor | 103 |
| PGA‐PCLLA | Solvent casting | Heparin with VEGF OR CD34 antibody | in vivo: increased endothelial cell attachment | 107 |
| PLLA‐PLCL | Phase separation | Heparin | in vitro: improved anticoagulation properties in vivo: neovascularization (subcutaneous) | 108 |
| Elastin‐like protein‐ collagen I | Hydrogel formation | ‐ | In vitro: mechanical characterization in vivo: limited early inflammatory response | 109 |
| PET‐PGA | woven | ‐ | in vivo: mechanical integration with the aorta | 110 |
| PCL reinforced with PET | freez‐dried tube reinforced with knitted PET | ‐ | Increased mechanical properties | 111 |
| PGA | woven | Mononuclear cells | Clinical trial: no clacification or infection but stenosis in 4 cases | 112 |
| PLA | ||||
| PCL‐collagen I – ECM | Elektrospinning Cell sheet | Smooth muscle cells | in vitro: improved cell viability | 114 |
| Agarose | Bioprinting | Mouse embryonic fibroblasts | Development of a self‐supporting structure | 115 |
Tissue‐engineered cell‐seeded constructs have been intensively studied in order to replace a vein or artery. The inner layer of a blood vessel, the so‐called intima, is lined with endothelial cells.80 A major drawback in engineering small diameter blood vessels is thrombus formation and inflammation due to non‐sufficient endothelialization.80, 101 Therefore, either the seeding technique needs to be improved or more functional materials have to be developed. One approach to generate more functional implants is the culture of cell‐seeded scaffolds under defined and highly physiological conditions in a bioreactor system prior to implantation.102, 103 Regarding the biomaterial tube itself; many different materials have been described.80, 104 An interesting idea is the generation of native ECM sheets produced by smooth muscle cells (SMCs) from another species105 or from human induced‐pluripotent stem cell‐derived SMCs.103 The engineered vessels were then decellularized, leaving behind a robust ECM graft. In a final step, the decellularized grafts were seeded with endothelial cells of the graft recipient.102, 103 L′Heureux et al. published a similar ECM‐sheet technique but utilized human fibroblasts. This in vitro‐engineered blood vessel showed complete integration after implantation into nude rats.101 The approach has already been transferred into clinics, where a successful implantation of the tissue‐engineered blood vessel was performed.106 However, when aiming to generate a graft for bypass surgery, the time it takes to produce autologous tissue‐engineered implants is rather long. Therefore, off‐the‐shelf small diameter blood vessels are more favorable. To achieve in vivo endothelialization of these acellular grafts, scaffold functionalization with chemokines and growth factors like SDF‐1 and VEGF can be performed.98, 107 Figure 3 summarizes various possibilities to introduce bioactive agents to a scaffold. Wang et al. covalently linked heparin as an anticoagulant to a thermally induced phase separated scaffold made of PLLA and poly (l‐lactide‐co‐ε‐caprolactone) (PLCL). Interestingly, the scaffold enabled endothelial cell attachment, but showed low protein attachment, which is important to avoid stenosis, thrombus or neointima formation.108 Another strategy for designing an acellular but functional scaffold for blood vessel replacement is the application of structural ECM components, such as collagens and elastin.109 In addition to biochemical aspects, vascular grafts need to fulfill mechanical requirements in order to perform in vivo without complications. Due to the high blood pressure especially in arteries, both, biomaterial and scaffold design techniques need to be carefully selected. Woven core sheath fibers made of polyethylene terephthalate (PET) and polyglycolic acid (PGA) were used to enhance the aortic wall strength for aneurism repair.110 In this study, the PGA component was replaced over time with native tissue in vivo, hereby enabling integration of the remaining scaffold, which helped stabilizing the vessel.110 In another study, mechanical properties such as tensile strength, E‐modulus, compression recovery and radial compliance were significantly improved by reinforcing a PCL scaffold with a knitted PET fabric.111 Woven PLA and PGA grafts seeded with mononuclear cells were implanted in patients without signs of calcification or infection; however, in four cases stenosis occurred.112 Besides molding, weaving, phase separation and knitting, electrospinning has been used to generate vascular grafts. By utilizing a rotating mandrel as the collector, tubular scaffolds can be electrospun.113 Also the combination of electrospinning and cell sheet engineering has been described as a promising vascular graft generation technique.114 Another scaffold generation method that has been studied intensively for generating macro‐ and microvascular structures is the computer‐controlled layer‐by‐layer deposition technique named printing.115 It is distinguished between three fabrication techniques: laser‐based, printer‐based and nozzle‐based.116 Laser‐based systems such as stereolithography and multiphoton polymerization are used to generate scaffolds from a bath of photosensitive polymers, enabling vessel sizes down to the sub‐micrometer range.117 Printer‐based or drop‐on‐demand systems include thermal and piezoelectric inkjet printers, which are also able to directly print cells.118 The third technique is direct writing of polymers using pressure‐assisted nozzle‐based systems.119 Depending on the system, various natural and synthetic biomaterials can be processed into a 3D tubular construct.116
Figure 3.
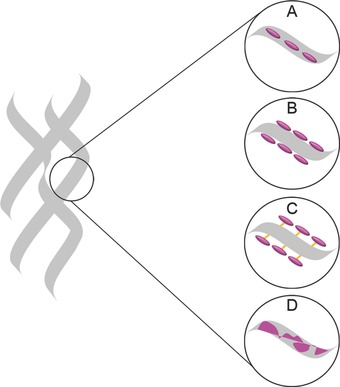
Schematic depiction of scaffold design strategies in combination with bioactive cues. The polymeric scaffold is shown in grey. Bioactive molecules (pink) can be delivered by A) encapsulation, B) physical adsorption on the surface, C) chemical binding (orange) to the surface or (C) by blending two materials.
4.1. Elastic Fiber Generation in Tissue‐Engineered Vascular Grafts
Although considerable progress has been made in the field of blood vessel tissue engineering, there is still a major limitation for all of these constructs – they lack of the presence of functional elastic fibers. Elastic fibers are responsible for tissue elasticity and resilience, especially in load bearing tissues such as heart valves, skin and also blood vessels.120 The elastin content in blood vessels is around 30–50%.121 Successful elastic fiber generation in tissue‐engineered constructs has been described in animal models;101 however, mature human cell‐based elastic fibers have so far not been realized. There have been a few reports using antibody staining, western blots or PCRs to detect elastin/tropoelastin protein or gene expression.122 However, the elastin protein is not the only protein forming a functional elastic fiber. Mecham et al. thoroughly investigated the processes that lead to normal elastic fiber assembly and found that cells secrete the soluble precursor tropoelastin that subsequently agglomerates in the extracellular space.123, 124 Simultaneously, fibrillin containing micro‐fibrils are assembled, to which in a next step the tropoelastin agglomerates are placed. Finally, lysyl oxidase (LOX) crosslinks the deposited elastin molecules to form the elastic fiber.123, 124, 125 To enable the process of elastogenesis, further proteins such as fibulins, elastin microfibril interface located protein 1 (EMILIN‐1), fibronectin, latent TGF‐beta binding protein 4 (LTBP‐4) are necessary.123, 126 The process of elastogenesis is highly complex and still not completely understood; however, it has been shown that these events already play a major role in early human cardiovascular development.127 Therefore, when aiming for a nature‐mimicking highly functional biomaterial scaffold, elastic fibers and the elastic fiber‐associated proteins must be considered.
4.2. Future Directions for the Use of Biomaterials in Vascular Graft Design
The most challenging part in blood vessel tissue engineering is the generation of small diameter grafts (<6 mm). Today, decellularized human vascular grafts are commercially available; however, the limiting factor is the lack of donors. The cell sheet technique seems to be quite promising,106 but long production times as well as high costs are unfavorable. Vascular graft design based on the use of solely synthetic biomaterials can be limited due to low hemocompatibility or unfavorable biomechanical profiles, either too stiff or too weak, which can potentially lead to graft leakage or dilation. In order to address these problems, we believe that mimicking the biological and mechanical properties of the native vessel will be important next steps towards a tissue‐engineered blood vessel. The use of hybrid substrates manufactured from synthetic materials and natural ECM proteins is highly promising. Further studies will be required to improve our knowledge of normal and pathological vascular ECM development and remodeling.
5. Monitoring of Biomaterials
Histological, histochemical and immunohistological methods are currently the gold standard for analyzing detailed 3D architectures of cells and ECM components.128, 129 Although, this information is of high value to evaluate failure or success of an experimental approach, histological processing is not suitable for online process monitoring, as it requires the sacrificing of samples at each time point of the experiment. Since in vitro‐generated advanced therapy medicinal products (ATMPs) or biomaterials are designated to be implanted into a patient, non‐destructive methods are required to control their quality, structure and composition. Implementation of techniques that allow the monitoring of the progress of tissue formation and maturation time‐dependently in vitro is a goal not only for the field of tissue engineering.128 Clinically approved imaging technologies that visualize tissues and organs within the human body to diagnose, treat and monitor disease stages represent a promising approach that could be adapted to be employed for the screening of tissue‐engineered materials.128, 130 Computer tomography (CT), magnetic resonance imaging (MRI) and ultrasound (US) are imaging technologies that can provide depth‐resolved information of structural tissue features and are therefore of high interest for the label‐free assessment of in vitro‐engineered ATMPs and biomaterials.131 In addition, novel optical techniques employing advanced laser systems to excite specifically light scattering, multiphoton absorption or intrinsic fluorescence were already adapted to provide valuable information of 3D tissue compositions and architectures.132 In the following, we will discuss principles, advantages and limitations of optical techniques suitable to assess biomaterials and ATMPs in a label‐free fashion (Table 4 ).
Table 4.
In vivo and in vitro applications of established and novel optical techniques
| Technique | Energy | Source of contrast | Penetration depth | In vivo application | In vitro application | Ref. |
|---|---|---|---|---|---|---|
| CT | X‐rays | Absorption | Throughout the human body | Bone fractures Tumors | Mineralization in cell cultures, Scaffold structure | 128, 130, 133, 134 |
| US imaging | Ultrasonic waves | Reflection, attenuation | 1–3 cm | Fibrosis Tumors | ECM remodeling | 128, 137, 138, 139 |
| MRI | Radio‐frequency pulses | Relaxation times of dipolar molecules | Throughout the human body | Presurgical imaging | ‐ | 128, 144, 145 |
| OCT | Non‐coherent light | Reflection | 1–3 mm | Cardiology Tumors Ophthalmic | Scaffold structure ECM remodeling | 128, 129, 132, 151, 152 |
| Raman microspectroscopy | NIR laser beam | Molecular vibrations | 100–300 μm | Gastric neoplasia Skin cancer | Cell and ECM identification and monitoring | 162, 163, 164, 165, 166 |
| Mutiphoton absorption SHG FLIM | Pulsed NIR laser beam | Auto‐fluorescence and decay times of intrinsic fluorophores | 200 μm | Skin cancer | Fibers and fibrils in tissues, metabolic profiling, Scaffold structures | 63, 132, 180, 187, 189, 190, 194, 195 |
5.1. Adaption of Clinically Approved Optical Techniques for In Vitro Applications
Electromagnetic radiation is employed for imaging of the human organs or even the whole body; measurements of spatially resolved absorption, refraction, attenuation or scattering of the incident energy are then transferred into image contrast depicting physical tissue properties.128, 130, 131, 132 In clinics, X‐rays, light, US or electromagnetic fields are employed as radiation source.128, 130, 131 X‐rays, the oldest imaging modality, utilize an electronic beam to irradiate tissues.130 From its historical development till today, X‐ray beams were predominantly employed for displaying bone tissues and fractures since mineralized tissues highly absorb the electronic beam resulting in excellent image contrast.130 The computational reconstitution of 2D and 3D X‐ray absorber maps, denoted as CT, vastly increases image contrast and can for instance display small tumors and pulmonary alveoli.130 By miniaturizing the electronic beam, μ‐CT can achieve spatial resolutions up to nanoscale levels.128, 130 For in vitro applications, μ‐CT has been successfully implemented to monitor and quantify cell‐mediated mineralization processes, spatially resolved and even under dynamic conditions in a bioreactor system.133 Additionally, μ‐CT was employed on manufactured scaffold materials to provide quantitative measures of fibers, pore sizes and porosity.128, 134 However, strongly different attenuation times of X‐rays are difficult to resolve; hence, multiple biomaterial composites such as natural ECM deposition within porous scaffolds could not be distinguished using μ‐CT.135 In summary, μ‐CT is neither optimal to resolve soft tissue architectures nor is it capable to distinguish biochemical ECM components.128, 131, 136
In contrast, US imaging employs acoustic waves and provides good soft tissue contrast. It is clinically established to detect fibrotic tissues and tumors.128, 130 Dependent on the acoustic wave frequency, spatial resolutions can exceed 1 μm, although this showed a negative impact on the penetration depth.128, 130 Nevertheless, ultrasound is considered a promising technology to monitor ECM formation and remodeling in tissue‐engineered constructs.128, 137, 138, 139 Cell death events, biomaterial degradation and tissue development have been shown to affect the output US image.128, 138, 139, 140 However, treatment of cell cultures with low‐intensity ultrasonic waves has been indicated to promote cell proliferation and differentiation in cultured stem cells.141, 142 Therefore, continuous US imaging during cell and tissue culture could impact the quality of tissue‐engineered constructs. Moreover, ultrasonic imaging is highly sensitive to air bubbles, speckled‐noise and therefore error‐prone for dynamic culture systems.128, 143
MRI excites nuclear magnetic resonance effects and transfers these into an image contrast. Resonant radiofrequency pulses are generated by an electromagnetic field, displacing the orientation of protons in dipolar molecules and measuring their relaxation times.130 These parameters are highly tissue‐specific, resulting in excellent soft tissue contrast in optical sections that can be acquired throughout the human body.128, 130 MRI is therefore regarded as the most powerful modality for pre‐surgical and diagnostic imaging.144, 145 In vitro high‐resolution MRI has been applied to visualize tissue ultrastructure and to assess biochemical measures such as the glycosaminoglycan content in engineered cartilage.128 Gründer et al. depicted a correlation between histological polarized light microscopy and MRI images and demonstrated that collagen fiber orientation is visible in MRI.146 However, resolution of MRI is still limited, requiring the application of contrast enhancing magnetic iron particles to optimize tissue contrast and to resolve cellular structures.128, 145 In addition, MRI acquisition is time consuming and expensive,144 making it therefore currently unsuitable for real‐time monitoring experiments.
Optical coherence tomography (OCT) employs near‐infrared low‐coherent light sources and measures its optical reflectance to spatially display cross‐sections of tissue microstructures.130, 147, 148 Resolution of OCT can achieve up to 1 μm and exceeds US imaging.149 Altered tissue morphologies were visualized and could be assigned to disease stages in correlation to histological and histopathological diagnosis.149 In clinics, OCT is utilized in cardiology and oncology, as well as to diagnose dermal inflammations and ophthalmic diseases.129, 130, 150 Due to different refractive indices of polymers and water, pores in polymeric scaffolds can appear darker in OCT images, which then allow the quantification of pore sizes.151 Penetration depth and resolution of OCT enables the discrimination of different tissue layers such as the epidermal and dermal layer of skin.129, 147 Monitoring of ECM deposition, remodeling, cell migration, and proliferation was accomplished on tissue‐engineered constructs in vitro by utilizing OCT.128, 129, 132 Bagnaninchi et al. implemented OCT into a perfusion bioreactor system and detected an increase in ECM formation under shear stress conditions.152 Currently, penetration depth presents the limiting factor for OCT.128, 149 In addition, the information gained by OCT does not provide much detail and it is therefore not sufficient to resolve cellular morphologies and characteristics.129
5.2. Laser‐Based Strategies to Assess Biomaterials and Tissue Architectures
Raman spectroscopy is a non‐destructive method, established in pharmaceutical and polymer industries to identify substances based on their specific molecular vibrational patterns.153 Laser light is employed for exciting these intrinsic molecular vibrations. Each molecule within the sample triggers specific frequency shifts relative to the incident light that is then detected within the Raman spectrum.154, 155 Water has a minor Raman scattering signal, which does not interfere with the peaks of organic substances, making the technology advantageous for biological systems.156, 157 For tissues and viable specimens, near‐infrared (NIR) lasers are utilized for Raman excitation, thereby problems of photo degradation, sample heating or fluorescence emission are nearly excludable and collection of signals from deeper tissue regions is facilitated due to a low level of light absorbance.156, 157 Raman microspectroscopy, an approach where the laser system is coupled to a microscope, can be conducted on very small sample volumes.157, 158 Spatially resolved molecular information can be provided by Raman microspectroscopy facilitating the analysis of different tissue structures and the analysis of single cells under physiological conditions158, 159, 160, 161, 162, 163 (Figure 4 ). Components of the ECM such as collagens, elastin and proteoglycans exhibit according to their molecular diversity specific Raman fingerprint spectra allowing for their distinction.162, 164, 165, 166 The enzymatic degradation of collagen fibrils in aortic heart valves has been shown to result in a down‐regulation of collagen‐specific Raman signals.164 Due to the non‐invasive and contact‐free measurement mode, Raman microspectroscopy is suitable to be implemented into a bioreactor system and was accomplished for time course monitoring of ECM formation under dynamic conditions.167, 168 As tissue resident cells exhibit specific signals for nucleic acids, which are absent in ECM structures, single cells and even subcellular structures are resolvable by Raman microspectroscopy.163, 167 In addition, changes of the cell phenotype such as altered protein expression result in a shift of the cell‐specific Raman fingerprint spectrum, empowering the technique as a method to characterize cells after isolation and to monitor the cellular phenotype in 2D and 3D culture systems.159, 162, 169, 170 The molecular sensitivity of Raman microspectroscopy further facilitates to monitor cell death modalities and to distinguish apoptotic and necrotic cell death in vitro.160, 161, 171 Acquiring Raman spectra simultaneously with standard fluorescence images allows the identification of defined spectral patterns and assists with the evaluation of the technology with established marker‐based methods.160, 172 Moreover, comparing molecular vibrational spectra of native and in vitro‐engineered tissues allows the definition of qualitative and quantitative quality criteria for tissue assessment.173 Scanning whole tissue sections by Raman microspectroscopy and transferring the resultant spatially resolved biochemical information into image contrast has been shown to enable highly sensitive marker‐free depiction of different tissue components and structures, analogously to histological staining174, 175, 176 (Figure 5 ). Here, resolutions of up to 1 μm can be obtained;175 however, for scanning large tissue areas, long acquisition times are required due to the low cross‐section of Raman scattering.177 Currently, many research groups focus on the combination of Raman spectroscopy with other optical tools, or to identify strategies how to improve the Raman efficiency by utilizing specific enhancement effects.177, 178, 179 Compared to spontaneous Raman spectroscopy, fluorescence signals have multiple folds higher signal efficiencies.132, 177 Employing confocal microscopy allows the acquisition of optical sections that can display the spatial distribution and networks of multiple structures in 3D tissue‐engineered constructs.129, 132 While various fluorescence dyes have been designed for live cell imaging, and genetic modifications can achieve the expression of fluorescence reporter proteins,129 none of these strategies fulfills the requirement of being completely non‐invasive and label‐free. Imaging endogenous fluorophores employing multiphoton effects represents a completely label‐free approach.56, 180 Two multiphoton mechanisms can be discriminated – second harmonic generation (SHG) and two‐photon excited fluorescence (TPEF).56, 180 In both, NIR laser pulses excite the tissue sample, having the advantage of increased axial resolution, penetration depth and low levels of photodamage on the sample.132, 180 Multiphoton signals are shifted towards shorter wavelengths compared to the incident light.132, 180 SHG visualizes non‐centrosymmetric molecular structures and results in precise signals at exactly half of the incident laser wavelength.181 Three‐dimensional organization of fibrillar collagens, microtubuli structures, and myosin in muscle tissues have been successfully monitored using SHG.181, 182 TPEF, which results in broader emission spectra, was employed to visualize and characterize metabolic states in cell cultures, based on the detection of intrinsic enzymatic co‐factors such as NADPH, porphyrins, and flavins.183, 184, 185, 186 In addition, elastic fiber networks in native tissues are excitable by TPEF.56 It has been shown that coupling SHG and TPEF microscopy to a multimodal imaging system can provide qualitatively and quantitatively information on 3D collagen and elastic fiber networks.56, 76, 187, 188 Multiphoton microscopy can therefore detect damaged ECM structures and represents a possibility to monitor fiber assembly, maturation and remodeling in time lapse.76, 187, 189, 190 Based on multiphoton signals of cardiac myosin and collagen fibrils, the level of fibrosis in rat hearts after myocardial infarction was quantitatively assessed.182 Moreover, it was demonstrated that stem cell differentiation and stem cell fate commitment can result in changes of multiphoton signals.56, 185, 191, 192, 193 Stem cell‐derived cardiomyocytes have been identified by visualizing their mature myosin filaments using SHG.191, 192 Buschke et al. detected an increased TPEF signal of NADH due to cardiac commitment in embryonic bodies.193
Figure 4.
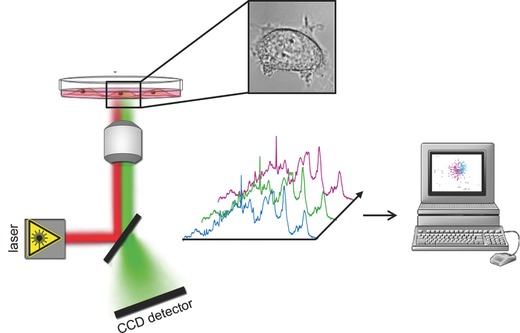
Principle of Raman microspectroscopy for characterizing cells under physiological conditions. A NIR laser is focused through a microscopic objective and directed onto a single cell or different subcellular regions. Resultant Raman spectra are analyzed using computer‐based multivariate algorithms.
Figure 5.
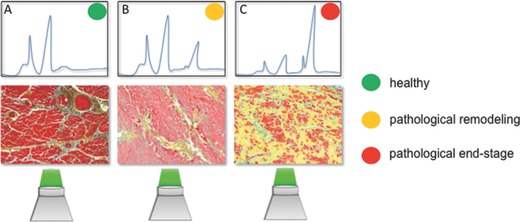
Schema of the principle of label‐free identification of different tissue stages based on molecular vibrational signals obtained by Raman spectroscopy: A) healthy tissues, B) early onset of pathological remodeling, and C) end‐stage pathological tissues show different signal patterns, which correlate to the histological findings (Movat's pentachrome staining).
Polymeric scaffolds made of synthetic materials, which are widely used for tissue engineering approaches, exhibit broad fluorescence emission in both, SHG and TPEF channels.129, 180 Although, the integrity and structure of polymeric scaffolds can be assessed by multiphoton imaging,132, 180 it is challenging to separate strong polymeric autofluorescence signals from those of natural ECM deposits and cellular metabolites.132 Fluorescence lifetime imaging (FLIM) offers a possibility to improve the resolution for multiple intrinsic fluorophores, exhibiting similar emission spectra.184, 186 Time‐ and spatially resolved analysis of fluorescence signals generates a new dimension to characterize the architectures of in vitro‐engineered tissues.186 FLIM represents a time‐resolved analysis of fluorescence signals that are transferred into image contrast.56, 186 Measured fluorescence time decays enable the distinction of different fluorescence molecules, which can further be impacted when the (bio)chemical environment surrounding a fluorophore is changed due to induced manipulation or disease.56, 186 FLIM has been demonstrated to be a potential technique to non‐invasively monitor cell fate commitment and stem cell differentiation.194, 195 Furthermore, FLIM depicts metabolic parameters supporting the assessment of cell viability, which is of great interest when studying cell‐biomaterial interactions.32
In summary, non‐invasive molecular‐sensitive monitoring techniques are suggested to be more robust and reliable than biochemical assays.32 These new technologies may complement, or in the near future even potentially replace, histological, histochemical or immunohistological screening, which make it necessary to sacrifice tissues for analyses at a certain time‐point of the manufacturing process.129, 132 However, to date, validation with classical invasive assays is still required for the interpretation and evaluation of multidimensional datasets obtained by these new techniques. In the future, a device that combines several non‐destructive optical methods could gather multidimensional quality parameters and support continuous monitoring of biological manufacturing processes.
Acknowledgements
The authors thank Shannon Lee Layland (Fraunhofer IGB Stuttgart, Germany) for his appreciated comments on the manuscript, and the Fraunhofer‐Gesellschaft Internal programs (Attract, 692263), the Ministry of Science, Research and the Arts of Baden‐Württemberg (33–729.55–3/214–1,2,3 and SI‐BW 01222–91), the BMBF (0316059), the Deutsche Forschungsgemeinschaft (SCHE 701/7–1 and/10–1), IZST‐Industry on Campus (IOC) funds (IOC‐102), the EU 7th Framework Programme (AMCARE, NMP3‐SME‐2013–604531), the Fraunhofer IGB Stuttgart and the Medical Faculty of the Eberhard Karls University Tübingen for their financial support (all to K.S.L.).
The copyright line for this article was changed on 23 June 2015 after original online publication.
Biographies
Svenja Hinderer received her Ph.D. in chemistry in 2014 from the University of Stuttgart, Germany. During her Ph.D. thesis, she worked in the field of tissue engineering and biomaterials design with focus on cardiovascular applications. Since 2014, she is a Post‐Doc at the Fraunhofer IGB in Stuttgart, Germany in the Department of Cell and Tissue Engineering. Her research focuses on biomaterials design, electrospinning, and establishment of biocompatibility assays.

Eva Brauchle received her Ph.D. in biology in 2015 from the Eberhard Karls University Tübingen, and works as a Post‐Doc at the Fraunhofer IGB, the IGVP of the University of Stuttgart and the Research Institute for Women's Health at the Eberhard Karls University Tübingen, Germany. Her research focus is to apply Raman spectroscopy as a non‐invasive and label‐free tool in biomedical research. Employing this technology, she studies cell death modalities, stem cell differentiation, and tissue architectures of native healthy and diseased, as well as in vitro‐generated tissues.

Katja Schenke‐Layland is the Professor of Biomaterials in Regenerative Medicine at the Research Institute for Women's Health, Eberhard Karls University Tübingen, the Head of the Department of Cell and Tissue Engineering at Fraunhofer IGB in Stuttgart, Germany, and Adjunct Associate Professor at the Department of Medicine/Cardiology at UCLA, USA. Her expertise is in biomaterials and medical product design, advanced therapy medicinal product (ATMP) certification processes, the development of human‐based three‐dimensional (3D) in vitro test systems, stem cell and extracellular matrix biology, and (non‐invasive) biomedical imaging.

The copyright line for this article was changed on 23 June 2015 after original online publication.
References
- 1. W. H. O. (WHO), 2014, http://www.who.int/mediacenter/factsheets/fs317/en/.
- 2. Patra C., Boccaccini A. R., Engel F. B., Thromb Haemost. 2014, 113, DOI: 10.1160/TH14–05–0480. [DOI] [PubMed] [Google Scholar]
- 3. Langone A. J., Helderman J. H., N. Engl. J. Med. 2003, 349, 704. [DOI] [PubMed] [Google Scholar]
- 4. Jaganathan S. K., Supriyanto E., Murugesan S., Balaji A., Asokan M. K., BioMed. Res. Int. 2014, 2014, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kim T. G., Shin H., Lim D. W., Adv. Funct. Mater. 2012, 22, 2446. [Google Scholar]
- 6. Nakayama K. H., Hou L., Huang N. F., Adv. Healthcare Mater. 2014, 3, 628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Karoly J., Cyrille N., Francoise M., Keith M., Gordana V.‐N., Gabor F., Biofabrication 2010, 2, 022001.20811127 [Google Scholar]
- 8. Laflamme M. A., Chen K. Y., Naumova A. V., Muskheli V., Fugate J. A., Dupras S. K., Reinecke H., Xu C., Hassanipour M., Police S., O'Sullivan C., Collins L., Chen Y., Minami E., Gill E. A., Ueno S., Yuan C., Gold J., Murry C. E., Nat. Biotechnol. 2007, 25, 1015. [DOI] [PubMed] [Google Scholar]
- 9. Behfar A., Yamada S., Crespo‐Diaz R., Nesbitt J. J., Rowe L. A., Perez‐Terzic C., Gaussin V., Homsy C., Bartunek J., Terzic A., J. Am. Col. Cardiol. 2010, 56, 721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Haider H. K., Lei Y., Ashraf M., Curr. Opin. Mol. Ther. 2008, 10, 611. [PMC free article] [PubMed] [Google Scholar]
- 11. Smits P. C., van Geuns R.‐J. M., Poldermans D., Bountioukos M., Onderwater E. E. M., Lee C. H., Maat A. P. W. M., Serruys P. W., J. Am. Col. Cardiol. 2003, 42, 2063. [DOI] [PubMed] [Google Scholar]
- 12. van Berlo J. H., Kanisicak O., Maillet M., Vagnozzi R. J., Karch J., Lin S.‐C. J., Middleton R. C., Marban E., Molkentin J. D., Nature 2014, 509, 337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hastings C. L., Roche E. T., Ruiz‐Hernandez E., Schenke‐Layland K., Walsh C. J., Duffy G. P., Adv. Drug Delivery Rev. 2014, doi: 10.1016/j.addr.2014.08.006. [DOI] [PubMed] [Google Scholar]
- 14. Russo V., Young S., Hamilton A., Amsden B. G., Flynn L. E., Biomaterials 2014, 35, 3956. [DOI] [PubMed] [Google Scholar]
- 15. Habib M., Shapira‐Schweitzer K., Caspi O., Gepstein A., Arbel G., Aronson D., Seliktar D., Gepstein L., Biomaterials 2011, 32, 7514. [DOI] [PubMed] [Google Scholar]
- 16. Wang H., Shi J., Wang Y., Yin Y., Wang L., Liu J., Liu Z., Duan C., Zhu P., Wang C., Biomaterials 2014, 35, 3986. [DOI] [PubMed] [Google Scholar]
- 17. Liu Z., Wang H., Wang Y., Lin Q., Yao A., Cao F., Li D., Zhou J., Duan C., Du Z., Wang Y., Wang C., Biomaterials 2012, 33, 3093. [DOI] [PubMed] [Google Scholar]
- 18. Zhang X., Wang H., Ma X., Adila A., Wang B., Liu F., Chen B., Wang C., Ma Y., Exp. Biol. Med. 2010, 235, 1505. [DOI] [PubMed] [Google Scholar]
- 19. Dai W., Wold L. E., Dow J. S., Kloner R. A., J. Am. Col. Cardiol. 2005, 46, 714. [DOI] [PubMed] [Google Scholar]
- 20. Yu J., Du K. T., Fang Q., Gu Y., Mihardja S. S., Sievers R. E., Wu J. C., Lee R. J., Biomaterials 2010, 31, 7012. [DOI] [PubMed] [Google Scholar]
- 21. Bastings M. M. C., Koudstaal S., Kieltyka R. E., Nakano Y., Pape A. C. H., Feyen D. A. M., van Slochteren F. J., Doevendans P. A., Sluijter J. P. G., Meijer E. W., Chamuleau S. A. J., Dankers P. Y. W., Adv. Healthcare Mater. 2014, 3, 70. [DOI] [PubMed] [Google Scholar]
- 22. Kraehenbuehl T. P., Zammaretti P., Van der Vlies A. J., Schoenmakers R. G., Lutolf M. P., Jaconi M. E., Hubbell J. A., Biomaterials 2008, 29, 2757. [DOI] [PubMed] [Google Scholar]
- 23. Kraehenbuehl T. P., Ferreira L. S., Hayward A. M., Nahrendorf M., van der Vlies A. J., Vasile E., Weissleder R., Langer R., Hubbell J. A., Biomaterials 2011, 32, 1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bencherif S. A., Sands R. W., Bhatta D., Arany P., Verbeke C. S., Edwards D. A., Mooney D. J., Proc. Natl. Acad. Sci. U.S.A. 2012, 109, 19590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hsueh Y. C., Wu J. M. F., Yu C. K., Wu K. K., Hsieh P. C. H., EMBO Mol. Med. 2014, 6, 496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zangi L., Lui K. O., von Gise A., Ma Q., Ebina W., Ptaszek L. M., Spater D., Xu H., Tabebordbar M., Gorbatov R., Sena B., Nahrendorf M., Briscoe D. M., Li R. A., Wagers A. J., Rossi D. J., Pu W. T., Chien K. R., Nat. Biotechnol. 2013, 31, 898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Monaghan M., Browne S., Schenke‐Layland K., Pandit A., Mol. Ther. 2014, 22, 786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cohen J. E., Purcell B. P., MacArthur J. W., Mu A., Shudo Y., Patel J. B., Brusalis C. M., Trubelja A., Fairman A. S., Edwards B. B., Davis M. S., Hung G., Hiesinger W., Atluri P., Margulies K. B., Burdick J. A., Woo Y. J., Circ. Heart Fail. 2014, 7, 619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Guo H.‐d., Cui G.‐h., Yang J.‐j., Wang C., Zhu J., Zhang L.‐s., Jiang J., Shao S.‐j., Biochem. Biophys. Res. Commun. 2012, 424, 105. [DOI] [PubMed] [Google Scholar]
- 30. Johnson N. R., Wang Y., PLoS One 2013, 8, e63075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ebelt H., Hillebrand I., Arlt S., Zhang Y., Kostin S., Neuhaus H., Müller‐Werdan U., Schwarz E., Werdan K., Braun T., Shock 2013, 39, 353. [DOI] [PubMed] [Google Scholar]
- 32. Schesny M. K., Monaghan M., Bindermann A. H., Freund D., Seifert M., Eble J. A., Vogel S., Gawaz M. P., Hinderer S., Schenke‐Layland K., Biomaterials 2014, 35, 7180. [DOI] [PubMed] [Google Scholar]
- 33. Ziegler M., Elvers M., Baumer Y., Leder C., Ochmann C., Schönberger T., Jürgens T., Geisler T., Schlosshauer B., Lunov O., Engelhardt S., Simmet T., Gawaz M., Circulation 2012, 125, 685. [DOI] [PubMed] [Google Scholar]
- 34. Bridges A. W., Garcia A. J., J. Diabetes Sci. Technol. 2008, 2, 984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Masumoto H., Ikuno T., Takeda M., Fukushima H., Marui A., Katayama S., Shimizu T., Ikeda T., Okano T., Sakata R., Yamashita J. K., Sci. Rep. 2014, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Seif‐Naraghi S. B., Singelyn J. M., Salvatore M. A., Osborn K. G., Wang J. J., Sampat U., Kwan O. L., Strachan G. M., Wong J., Schup‐Magoffin P. J., Braden R. L., Bartels K., DeQuach J. A., Preul M., Kinsey A. M., DeMaria A. N., Dib N., Christman K. L., Sci. Transl. Med. 2013, 5, 173ra25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hinderer S., Schenke‐Layland K., Expert Rev. Med. Devices 2013, 10, 33. [DOI] [PubMed] [Google Scholar]
- 38. Venugopal J., Rajeswari R., Shayanti M., Sridhar R., Sundarrajan S., Balamurugan R., Ramakrishna S., Mater. Sci. Eng. C Mater. Biol. Appl. 2013, 33, 1325. [DOI] [PubMed] [Google Scholar]
- 39. Lavergne M., Derkaoui M., Delmau C., Letourneur D., Uzan G., Le Visage C., Macromol. Biosci. 2012, 12, 901. [DOI] [PubMed] [Google Scholar]
- 40. Ifkovits J. L., Tous E., Minakawa M., Morita M., Robb J. D., Koomalsingh K. J., Gorman J. H., Gorman R. C., Burdick J. A., Proc. Natl. Acad. Sci. U.S.A. 2010, 107, 11507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Yu J., Gu Y., Du K. T., Mihardja S., Sievers R. E., Lee R. J., Biomaterials 2009, 30, 751. [DOI] [PubMed] [Google Scholar]
- 42. Tsur‐Gang O., Ruvinov E., Landa N., Holbova R., Feinberg M. S., Leor J., Cohen S., Biomaterials 2009, 30, 189. [DOI] [PubMed] [Google Scholar]
- 43. Hao X., Silva E. A., Månsson‐Broberg A., Grinnemo K. H., Siddiqui A. J., Dellgren G., Wärdell E., Brodin L.Å., Mooney D. J., Sylvén C., Cardiovasc. Res. 2007, 75, 178. [DOI] [PubMed] [Google Scholar]
- 44. Silva A. K. A., Juenet M., Meddahi‐Pellé A., Letourneur D., Carbohydr. Polym. 2015, 116, 267. [DOI] [PubMed] [Google Scholar]
- 45. Frey N., Linke A., Süselbeck T., Müller‐Ehmsen J., Vermeersch P., Schoors D., Rosenberg M., Bea F., Tuvia S., Leor J., Circ. Cardiovasc. Interv. 2014, 10.1161/circinterventions.114.001478. [DOI] [PubMed] [Google Scholar]
- 46. Mann D. L., Circulation 1999, 100, 999. [DOI] [PubMed] [Google Scholar]
- 47. Christman K. L., Lee R. J., J. Am. Col. Cardiol. 2006, 48, 907. [DOI] [PubMed] [Google Scholar]
- 48. Hopkins A. M., De Laporte L., Tortelli F., Spedden E., Staii C., Atherton T. J., Hubbell J. A., Kaplan D. L., Adv. Funct. Mater. 2013, 23, 5140. [Google Scholar]
- 49. Baheiraei N., Yeganeh H., Ai J., Gharibi R., Azami M., Faghihi F., Mater. Sci. Eng. C Mater. Biol. Appl. 2014, 44, 24. [DOI] [PubMed] [Google Scholar]
- 50. Molamma P. P., Dan K., Laleh G.‐M., Seeram R., Biomed. Mater. 2011, 6, 055001.21813957 [Google Scholar]
- 51. Chen Q.‐Z., Bismarck A., Hansen U., Junaid S., Tran M. Q., Harding S. E., Ali N. N., Boccaccini A. R., Biomaterials 2008, 29, 47. [DOI] [PubMed] [Google Scholar]
- 52. Shin M., Ishii O., Sueda T., Vacanti J. P., Biomaterials 2004, 25, 3717. [DOI] [PubMed] [Google Scholar]
- 53. Kumbar S. G., James R., Nukavarapu S. P., Laurencin C. T., Biomed. Mater. 2008, 3, 034002. [DOI] [PubMed] [Google Scholar]
- 54. Greiner A., Wendorff J. H., Angew. Chem. 2007, 119, 5770. [Google Scholar]
- 55. Sill T. J., von Recum H. A., Biomaterials 2008, 29, 1989. [DOI] [PubMed] [Google Scholar]
- 56. Schenke‐Layland K., J. Biophotonics 2008, 1, 451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Agarwal S., Wendorff J. H., Greiner A., Adv. Mater. 2009, 21, 3343. [DOI] [PubMed] [Google Scholar]
- 58. Şenel Ayaz H. G., Perets A., Ayaz H., Gilroy K. D., Govindaraj M., Brookstein D., Lelkes P. I., Biomaterials 2014, 35, 8540. [DOI] [PubMed] [Google Scholar]
- 59. Kai D., Wang Q.‐L., Wang H.‐J., Prabhakaran M. P., Zhang Y., Tan Y.‐Z., Ramakrishna S., Acta Biomater. 2014, 10, 2727. [DOI] [PubMed] [Google Scholar]
- 60. Ravichandran R., Venugopal J. R., Subramanian S., Mukherjee S., Ramakrishna S., Tissue Eng. Part A 2015, 10.1089/ten.TEA.2014.0265. [Google Scholar]
- 61. Hinderer S., Schesny M., Bayrak A., Ibold B., Hampel M., Walles T., Stock U. A., Seifert M., Schenke‐Layland K., Biomaterials 2012, 33, 5259. [DOI] [PubMed] [Google Scholar]
- 62. Liu S., He Z., Xu G., Xiao X., Mater. Sci. Eng. C Mater. Biol. Appl. 2014, 44, 201. [DOI] [PubMed] [Google Scholar]
- 63. Fujimoto K. L., Tobita K., Merryman W. D., Guan J., Momoi N., Stolz D. B., Sacks M. S., Keller B. B., Wagner W. R., J. Am. Col. Cardiol. 2007, 49, 2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Matsubayashi K., Fedak P. W. M., Mickle D. A. G., Weisel R. D., Ozawa T., Li R.‐K., Circulation 2003, 108, II. [DOI] [PubMed] [Google Scholar]
- 65. Dvir T., Timko B. P., Brigham M. D., Naik S. R., Karajanagi S. S., Levy O., Jin H., Parker K. K., Langer R., Kohane D. S., Nat. Nanotechnol. 2011, 6, 720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Mooney E., Mackle J. N., Blond D. J. P., O'Cearbhaill E., Shaw G., Blau W. J., Barry F. P., Barron V., Murphy J. M., Biomaterials 2012, 33, 6132. [DOI] [PubMed] [Google Scholar]
- 67. Hsiao C.‐W., Bai M.‐Y., Chang Y., Chung M.‐F., Lee T.‐Y., Wu C.‐T., Maiti B., Liao Z.‐X., Li R.‐K., Sung H.‐W., Biomaterials 2013, 34, 1063. [DOI] [PubMed] [Google Scholar]
- 68. Lee J.‐W., Serna F., Schmidt C. E., Langmuir 2006, 22, 9816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Guimard N. K. E., Sessler J. L., Schmidt C. E., Macromolecules 2008, 42, 502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Lee R. J., Hinson A., Helgerson S., Bauernschmitt R., Sabbah H. N., Cell Transplantation 2013, 22, 529. [DOI] [PubMed] [Google Scholar]
- 71. Schoen F. J., Annu. Rev. Patho.‐Mech. Dis. 2012, 7, 161. [DOI] [PubMed] [Google Scholar]
- 72. Brody S., Pandit A., J. Biomed. Mater. Res. B Appl. Biomater. 2007, 83B, 16. [DOI] [PubMed] [Google Scholar]
- 73. Howard I., Patterson E., Yoxall A., J. Med. Eng. Technol. 2003, 27, 259. [DOI] [PubMed] [Google Scholar]
- 74. Schoen F. J., Curr. Opin. Biotechnol. 2011. [Google Scholar]
- 75. Rippel R., Ghanbari H., Seifalian A., World J. Surg. 2012, 36, 1581. [DOI] [PubMed] [Google Scholar]
- 76. Schenke‐Layland K., Stock U. A., Nsair A., Xie J., Angelis E., Fonseca C. G., Larbig R., Mahajan A., Shivkumar K., Fishbein M. C., MacLellan W. R., Eur. Heart J. 2009, 30, 2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Jana S., Tefft B. J., Spoon D. B., Simari R. D., Acta Biomater. 2014, 10, 2877. [DOI] [PubMed] [Google Scholar]
- 78. Schenke‐Layland K., Opitz F., Gross M., Doring C., Halbhuber K. J., Schirrmeister F., Wahlers T., Stock U. A., Cardiovasc Res. 2003, 60, 497. [DOI] [PubMed] [Google Scholar]
- 79. Bobylev D., Breymann T., Boethig D., Haverich A., Ono M., Ann. Thorac. Surg. 2014, 97, 1792. [DOI] [PubMed] [Google Scholar]
- 80. Bouten C. V., Dankers P. Y., Driessen‐Mol A., Pedron S., Brizard A. M., Baaijens F. P., Adv. Drug Delivery Rev. 2011, 63, 221. [DOI] [PubMed] [Google Scholar]
- 81. Chen Q., Bruyneel A., Carr C., Czernuszka J., Proc. Eng. 2013, 59, 247. [Google Scholar]
- 82. Sodian R., Hoerstrup S. P., Sperling J. S., Daebritz S. H., Martin D. P., Schoen F. J., Vacanti J. P., Mayer J. E. Jr., Ann. Thorac. Surg. 2000, 70, 140. [DOI] [PubMed] [Google Scholar]
- 83. Masoumi N., Annabi N., Assmann A., Larson B. L., Hjortnaes J., Alemdar N., Kharaziha M., Manning K. B., Mayer J. E. Jr., Khademhosseini A., Biomaterials 2014, 35, 7774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Weber M., Heta E., Moreira R., Gesché V., Schermer T., Frese J., Jockenhoevel S., Mela P., Tissue Eng. Part C 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Hinderer S., Seifert J., Votteler M., Shen N., Rheinlaender J., Schäffer T. E., Schenke‐Layland K., Biomaterials 2014, 35, 2130. [DOI] [PubMed] [Google Scholar]
- 86. Dubé J., Bourget J.‐M., Gauvin R., Lafrance H., Roberge C. J., Auger F. A., Germain L., Acta Biomater. 2014, 10, 3563. [DOI] [PubMed] [Google Scholar]
- 87. Jordan J. E., Williams J. K., Lee S.‐J., Raghavan D., Atala A., Yoo J. J., J. Thorac. Cardiovasc. Surg. 2012, 143, 201. [DOI] [PubMed] [Google Scholar]
- 88. Zhang X., Xu B., Puperi D. S., Yonezawa A. L., Wu Y., Tseng H., Cuchiara M. L., West J. L., Grande‐Allen K. J., Acta Biomater. 2015, 14, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Dijkman P. E., Driessen‐Mol A., Frese L., Hoerstrup S. P., Baaijens F. P., Biomaterials 2012, 33, 4545. [DOI] [PubMed] [Google Scholar]
- 90. Weber B., Dijkman P. E., Scherman J., Sanders B., Emmert M. Y., Grünenfelder J., Verbeek R., Bracher M., Black M., Franz T., Kortsmit J., Modregger P., Peter S., Stampanoni M., Robert J., Kehl D., van Doeselaar M., Schweiger M., Brokopp C. E., Wälchli T., Falk V., Zilla P., Driessen‐Mol A., Baaijens F. P. T., Hoerstrup S. P., Biomaterials 2013, 34, 7269. [DOI] [PubMed] [Google Scholar]
- 91. Stevanella M., Krishnamurthy G., Votta E., Swanson J. C., Redaelli A., Ingels N. B. Jr., J. Biomech. 2011, 44, 2229. [DOI] [PubMed] [Google Scholar]
- 92. Weymann A., Radovits T., Schmack B., Li S., Korkmaz S., Soós P., Istók R., Veres G., Chaimow N., Karck M., Szabó G., Artif Organs 2014, 38, E118. [DOI] [PubMed] [Google Scholar]
- 93. Flanagan T. C., Black A., O'Brien M., Smith T. J., Pandit A. S., Cells Tissues Organs 2006, 183, 12. [DOI] [PubMed] [Google Scholar]
- 94. Grande‐Allen K. J., Liao J., Curr. Cardiol. Rep. 2011, 13, 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Moreira R., Gesche V. N., Hurtado‐Aguilar L. G., Schmitz‐Rode T., Frese J., Jockenhoevel S., Mela P., Tissue Eng. Part C Methods 2014, 20, 741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Pusch J., Votteler M., Göhler S., Engl J., Hampel M., Walles H., Schenke‐Layland K., Biomaterials 2011, 32, 7469. [DOI] [PubMed] [Google Scholar]
- 97. Li S., Sengupta D., Chien S., Wiley Interdiscip. Rev. Syst. Biol. Med. 2014, 6, 61. [DOI] [PubMed] [Google Scholar]
- 98. Muylaert D. E., Fledderus J. O., Bouten C. V., Dankers P. Y., Verhaar M. C., Heart 2014, 100, 1825. [DOI] [PubMed] [Google Scholar]
- 99. Zdrahala R. J., J. Biomater. Appl. 1996, 10, 309. [DOI] [PubMed] [Google Scholar]
- 100. Zdrahala R. J., J. Biomater. Appl. 1996, 11, 37. [DOI] [PubMed] [Google Scholar]
- 101. L'Heureux N., Dusserre N., Konig G., Victor B., Keire P., Wight T. N., Chronos N. A. F., Kyles A. E., Gregory C. R., Hoyt G., Robbins R. C., McAllister T. N., Nat. Med. 2006, 12, 361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Quint C., Kondo Y., Manson R. J., Lawson J. H., Dardik A., Niklason L. E., Proc. Natl. Acad. Sci. U.S.A. 2011, 108, 9214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Sundaram S., One J., Siewert J., Teodosescu S., Zhao L., Dimitrievska S., Qian H., Huang A. H., Niklason L., Stem Cells Transl. Med. 2014, DOI: 10.5966/sctm.2014–0065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Tranquillo R. T., Ann. NY Acad. Sci. 2002, 961, 251. [DOI] [PubMed] [Google Scholar]
- 105. Quint C., Arief M., Muto A., Dardik A., Niklason L. E., J. Vasc. Surg. 2012, 55, 790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. L'Heureux N., McAllister T. N., de la Fuente L. M., N. Engl. J. Med. 2007, 357, 1451. [DOI] [PubMed] [Google Scholar]
- 107. Melchiorri A. J., Hibino N., Yi T., Lee Y. U., Sugiura T., Tara S., Shinoka T., Breuer C., Fisher J. P., Biomacromolecules 2014, 10.1021/bm501853s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Wang W., Hu J., He C., Nie W., Feng W., Qiu K., Zhou X., Gao Y., Wang G., J. Biomed. Mater. Res. A 2014, DOI: 10.1002/jbm.a.35315. [DOI] [PubMed] [Google Scholar]
- 109. Kumar V. A., Caves J. M., Haller C. A., Dai E., Liu L., Grainger S., Chaikof E. L., Acta Biomater. 2013, 9, 8067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Takeuchi M., Kuratani T., Miyagawa S., Shirakawa Y., Shimamura K., Kin K., Yoshida T., Arai Y., Hoashi T., Teramoto N., Hirakawa K., Kawaguchi N., Sawa Y., J. Thorac. Cardiovasc. Surg., 2014, 148, 1719. [DOI] [PubMed] [Google Scholar]
- 111. Wang F., Mohammed A., Li C., Ge P., Wang L., King M. W., Biomed. Mater. Eng. 2014, 24, 2127. [DOI] [PubMed] [Google Scholar]
- 112. Hibino N., McGillicuddy E., Matsumura G., Ichihara Y., Naito Y., Breuer C., Shinoka T., J. Thorac. Cardiovasc. Surg. 2010, 139, 431. [DOI] [PubMed] [Google Scholar]
- 113. Rocco K. A., Maxfield M. W., Best C. A., Dean E. W., Breuer C. K., Tissue Eng. Part B Rev 2014, DOI: 10.1089/ten.TEB.2014.0123. [DOI] [PubMed] [Google Scholar]
- 114. Ahn H., Ju Y. M., Takahashi H., Williams D. F., Yoo J. J., Lee S. J., Okano T., Atala A., Acta Biomater. DOI: 10.1016/j.actbio.2015.01.030. [DOI] [PubMed] [Google Scholar]
- 115. Kucukgul C., Ozler S. B., Inci I., Karakas E., Irmak S., Gozuacik D., Taralp A., Koc B., Biotechnol. Bioeng. 2014, doi: 10.1002/bit.25493. [DOI] [PubMed] [Google Scholar]
- 116. Hoch E., Tovar G. E. M., Borchers K., Eur. J. Cardiothorac. Surg. 2014, 46, 767. [DOI] [PubMed] [Google Scholar]
- 117. Stefan B., Franziska N., Ligon S. C., Anneliese N., Helga B., David B., Jürgen S., Robert L., Biomed. Mater. 2011, 6, 055003.21849722 [Google Scholar]
- 118. Hoch E., Hirth T., Tovar G. E. M., Borchers K., J. Mater. Chem. B 2013, 1, 5675. [DOI] [PubMed] [Google Scholar]
- 119. Chang C. C., Boland E. D., Williams S. K., Hoying J. B., J. Biomed. Mater. Res. B Appl. Biomater. 2011, 98B, 160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Waterhouse A., Wise S. G., Ng M. K., Weiss A. S., Tissue Eng. Part B Rev. 2011, 17, 93. [DOI] [PubMed] [Google Scholar]
- 121. Bashur C. A., Venkataraman L., Ramamurthi A., Tissue Eng. Part B 2012, 18, 203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Venkataraman L., Bashur C. A., Ramamurthi A., Tissue Eng. Part A 2014, DOI: 10.1089/ten.tea.2013.0294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Mecham R. P., Curr. Protoc. Cell Biol. 2001, 57. [DOI] [PubMed] [Google Scholar]
- 124. Wagenseil J. E., Mecham R. P., Birth Defects Res. C Embryo Today 2007, 81, 229. [DOI] [PubMed] [Google Scholar]
- 125. Kielty C. M., Stephan S., Sherratt M. J., Williamson M., Shuttleworth C. A., Philos. Trans. R. Soc. Lond. B. Biol. Sci. 2007, 362, 1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Baldwin A. K., Simpson A., Steer R., Cain S. A., Kielty C. M., Expert Rev. Mol. Med. 2013, 15, 1. [DOI] [PubMed] [Google Scholar]
- 127. Votteler M., Berrio D. A. C., Horke A., Sabatier L., Reinhardt D. P., Nsair A., Aikawa E., Schenke‐Layland K., Development 2013, 140, 2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Appel A. A., Anastasio M. A., Larson J. C., Brey E. M., Biomaterials 2013, 34, 6615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Smith L. E., Smallwood R., Macneil S., Microsc. Res. Tech. 2010, 73, 1123. [DOI] [PubMed] [Google Scholar]
- 130. Haidekker M., Medical Imaging Technology, Springer, New York: 2013. [Google Scholar]
- 131. Pancrazio J. J., Wang F., Kelley C. A., Biosens. Bioelectron. 2007, 22, 2803. [DOI] [PubMed] [Google Scholar]
- 132. Georgakoudi I., Rice W. L., Hronik‐Tupaj M., Kaplan D. L., Tissue Eng. Part B Rev. 2008, 14, 321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Porter B. D., Lin A. S., Peister A., Hutmacher D., Guldberg R. E., Biomaterials 2007, 28, 2525. [DOI] [PubMed] [Google Scholar]
- 134. Ho S. T., Hutmacher D. W., Biomaterials 2006, 27, 1362. [DOI] [PubMed] [Google Scholar]
- 135. Jones J. R., Atwood R. C., Poologasundarampillai G., Yue S., Lee P. D., J. Mater. Sci. Mater. Med. 2009, 20, 463. [DOI] [PubMed] [Google Scholar]
- 136. Kerckhofs G., Sainz J., Wevers M., Van de Putte T., Schrooten J., Eur. Cell Mater. 2013, 25, 179. [DOI] [PubMed] [Google Scholar]
- 137. Kreitz S., DOhmen G., Hasken S., Schmitz‐Rode T., Mela P., Jockenhoevel S., Tissue Eng. Part C Methods 2011, 17, 1021. [DOI] [PubMed] [Google Scholar]
- 138. Rice M. A., Waters K. R., Anseth K. S., Acta Biomater. 2009, 5, 152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Fite B. Z., Decaris M., Sun Y., Sun Y., Lam A., Ho C. K., Leach J. K., Marcu L., Tissue Eng. Part C Methods 2011, 17, 495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Czarnota G. J., Kolios M. C., Abraham J., Portnoy M., Ottensmeyer F. P., Hunt J. W., Sherar M. D., Br. J. Cancer 1999, 81, 520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Lv Y., Zhao P., Chen G., Sha Y., Yang L., Biotechnol. Lett. 2013, 35, 2201. [DOI] [PubMed] [Google Scholar]
- 142. Xu P., Gul‐Uludag H., Ang W. T., Yang X., Huang M., Marquez‐Curtis L., McGann L., Janowska‐Wieczorek A., Xing J., Swanson E., Chen J., Biotechnol. Lett. 2012, 34, 1965. [DOI] [PubMed] [Google Scholar]
- 143. Sun Y., Responte D., Xie H., Liu J., Fatakdawala H., Hu J., Athanasiou K. A., Marcu L., Tissue Eng. Part C Methods 2012, 18, 215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Hu X., Norris D. G., Annu. Rev. Biomed. Eng. 2004, 6, 157. [DOI] [PubMed] [Google Scholar]
- 145. Struys T., Ketkar‐Atre A., Gervois P., Leten C., Hilkens P., Martens W., Bronckaers A., Dresselaers T., Politis C., Lambrichts I., Himmelreich U., Cell Transplant 2013, 22, 1813. [DOI] [PubMed] [Google Scholar]
- 146. Grunder W., NMR Biomed. 2006, 19, 855. [DOI] [PubMed] [Google Scholar]
- 147. Huang D., Swanson E. A., Lin C. P., Schuman J. S., Stinson W. G., Chang W., Hee M. R., Flotte T., Gregory K., Puliafito C. A., Fujimoto J. G., Science 1991, 254(5035), 1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Matcher S. J., Methods Mol. Biol. 2011, 695, 261. [DOI] [PubMed] [Google Scholar]
- 149. Fujimoto J. G., Nat. Biotechnol. 2003, 21, 1361. [DOI] [PubMed] [Google Scholar]
- 150. Boustany N. N., Boppart S. A., Backman V., Annu. Rev. Biomed. Eng. 2010, 12, 285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Yang Y., Dubois A., Qin X. P., Li J., El Haj A., Wang R. K., Phys. Med. Biol. 2006, 51, 1649. [DOI] [PubMed] [Google Scholar]
- 152. Bagnaninchi P. O., Yang Y., Zghoul N., Maffulli N., Wang R. K., Haj A. J., Tissue Eng. 2007, 13, 323. [DOI] [PubMed] [Google Scholar]
- 153. Das R. S., Agrawal Y. K., Vibrational Spectrosc. 2011, 57, 163. [Google Scholar]
- 154. Brauchle E., Schenke‐Layland K., Biotechnol. J. 2013, 8, 288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Raman C. V., Krishnan K. S., Nature 1928, 501–5, 121. [Google Scholar]
- 156. LaPlant F., in Emerging Raman Applications and Techniques in Biomedical and Pharmaceutical Fields, (Eds: Matousek P., Morris M. D.), Springer, Berlin, Heidelberg: 2010, 1. [Google Scholar]
- 157. Manoharan R., Wang Y., Feld M. S., Spectrochim. Acta Part A: Mol. Biomol. Spectrosc. 1996, 52, 215. [Google Scholar]
- 158. Puppels G. J., de Mul F. F., Otto C., Greve J., Robert‐Nicoud M., Arndt‐Jovin D. J., Jovin T. M., Nature 1990, 347, 301. [DOI] [PubMed] [Google Scholar]
- 159. Pudlas M., Koch S., Bolwien C., Thude S., Jenne N., Hirth T., Walles H., Schenke‐Layland K., Tissue Eng. Part C Methods 2011, 17, 1027. [DOI] [PubMed] [Google Scholar]
- 160. Brauchle E., Thude S., Brucker S. Y., Schenke‐Layland K., Sci. Rep. 2014, 4, 4698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Brauchle E., Noor S., Holtorf E., Garbe C., Schenke‐Layland K., Busch C., Clin. Exp. Dermatol. 2014, 39, 636. [DOI] [PubMed] [Google Scholar]
- 162. Pudlas M., Brauchle E., Klein T. J., Hutmacher D. W., Schenke‐Layland K., J. Biophotonics 2013, 6(2): 205–11, doi:10.1002/jbio.201200064. [DOI] [PubMed] [Google Scholar]
- 163. Bonifacio A., Beleites C., Vittur F., Marsich E., Semeraro S., Paoletti S., Sergo V., Analyst 2010, 135, 3193. [DOI] [PubMed] [Google Scholar]
- 164. Votteler M., Carvajal Berrio D. A., Pudlas M., Walles H., Stock U. A., Schenke‐Layland K., J. Biophotonics 2012, 5, 47. [DOI] [PubMed] [Google Scholar]
- 165. Frushour B. G., Koenig J. L., Biopolymers 1975, 14, 379. [DOI] [PubMed] [Google Scholar]
- 166. Ellis R., Green E., Winlove C. P., Connect Tissue Res. 2009, 50, 29. [DOI] [PubMed] [Google Scholar]
- 167. Pully V. V., Lenferink A., van Manen H. J., Subramaniam V., van Blitterswijk C. A., Otto C., Anal. Chem. 2010, 82, 1844. [DOI] [PubMed] [Google Scholar]
- 168. Ghita A., Pascut F. C., Sottile V., Notingher I., Analyst 2014, 139, 55. [DOI] [PubMed] [Google Scholar]
- 169. Pudlas M., Berrio D. A. C., Votteler M., Koch S., Thude S., Walles H., Schenke‐Layland K., Med. Laser Appl. 2011, 26, 119. [Google Scholar]
- 170. Brauchle E., Johannsen H., Nolan S., Thude S., Schenke‐Layland K., Biomaterials 2013, 34, 7401. [DOI] [PubMed] [Google Scholar]
- 171. Notingher I., Green C., Dyer C., Perkins E., Hopkins N., Lindsay C., Hench L. L., J. R. Soc. Interface 2004, 1, 79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Puppels G. J., Garritsen H. S., Segers‐Nolten G. M., de Mul F. F., Greve J., Biophys. J. 1991, 60, 1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Leroy M., Labbe J. F., Ouellet M., Jean J., Lefevre T., Laroche G., Auger M., Pouliot R., Acta Biomater. 2014, 10, 2703. [DOI] [PubMed] [Google Scholar]
- 174. Beljebbar A., Bouche O., Diebold M. D., Guillou P. J., Palot J. P., Eudes D., Manfait M., Crit Rev Oncol. Hematol. 2009, 72, 255. [DOI] [PubMed] [Google Scholar]
- 175. Koljenovic S., Bakker Schut T. C., van Meerbeeck J. P., Maat A. P., Burgers S. A., Zondervan P. E., Kros J. M., Puppels G. J., J. Biomed. Opt. 2004, 9, 1187. [DOI] [PubMed] [Google Scholar]
- 176. Kunstar A., Leijten J., van Leuveren S., Hilderink J., Otto C., van Blitterswijk C. A., Karperien M., van Apeldoorn A. A., J Biomed Opt 2012, 17, 116012. [DOI] [PubMed] [Google Scholar]
- 177. Kong K., Rowlands C. J., Varma S., Perkins W., Leach I. H., Koloydenko A. A., Williams H. C., Notingher I., Proc. Natl. Acad. Sci. U.S.A. 2013, 110, 15189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Chan J. W., J. Biophotonics 2013, 6, 36.23175434 [Google Scholar]
- 179. Stetciura I. Y., Markin A. V., Ponomarev A. N., Yakimansky A. V., Demina T. S., Grandfils C., Volodkin D. V., Gorin D. A., Langmuir 2013, 29, 4140. [DOI] [PubMed] [Google Scholar]
- 180. Sun Y., Tan H. Y., Lin S. J., Lee H. S., Lin T. Y., Jee S. H., Young T. H., Lo W., Chen W. L., Dong C. Y., Microsc. Res. Tech. 2008, 71, 140. [DOI] [PubMed] [Google Scholar]
- 181. Vanzi F., Sacconi L., Cicchi R., Pavone F. S., J. Biomed. Opt. 2012, 17, 060901. [DOI] [PubMed] [Google Scholar]
- 182. Caorsi V., Toepfer C., Sikkel M. B., Lyon A. R., MacLeod K., Ferenczi M. A., PLoS One 2013, 8, e56136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183. Zoumi A., Yeh A., Tromberg B. J., Proc. Natl. Acad. Sci. U.S.A. 2002, 99, 11014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184. Georgakoudi I., Quinn K. P., Annu. Rev. Biomed. Eng. 2012, 14, 351. [DOI] [PubMed] [Google Scholar]
- 185. Quinn K. P., Sridharan G. V., Hayden R. S., Kaplan D. L., Lee K., Georgakoudi I., Sci. Rep. 2013, 3, 3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186. Seidenari S., Arginelli F., Bassoli S., Cautela J., French P. M., Guanti M., Guardoli D., Konig K., Talbot C., Dunsby C., Dermatol. Res. Pract. 2012, 2012, 810749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187. Schenke‐Layland K., Madershahian N., Riemann I., Starcher B., Halbhuber K. J., Konig K., Stock U. A., Ann. Thorac. Surg. 2006, 81, 918. [DOI] [PubMed] [Google Scholar]
- 188. Opitz F., Schenke‐Layland K., Cohnert T. U., Starcher B., Halbhuber K. J., Martin D. P., Stock U. A., Cardiovasc. Res. 2004, 63, 719. [DOI] [PubMed] [Google Scholar]
- 189. Bancelin S., Decenciere E., Machairas V., Albert C., Coradin T., Schanne‐Klein M. C., Aime C., Soft Matter 2014, 10, 6651. [DOI] [PubMed] [Google Scholar]
- 190. Toki F., Honkura N., Shirakata Y., Imamura T., Higashiyama S., Nanba D., Cell Struct. Funct. 2013, 38, 227. [DOI] [PubMed] [Google Scholar]
- 191. Wallace S. J., Morrison J. L., Botting K. J., Kee T. W., J. Biomed. Opt. 2008, 13, 064018. [DOI] [PubMed] [Google Scholar]
- 192. Awasthi S., Matthews D. L., Li R. A., Chiamvimonvat N., Lieu D. K., Chan J. W., J. Biophotonics 2012, 5, 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193. Buschke D. G., Squirrell J. M., Vivekanandan A., Rueden C. T., Eliceiri K. W., Ogle B. M., Cytometry Part A 2014, 85, 353. [DOI] [PubMed] [Google Scholar]
- 194. Stringari C., Sierra R., Donovan P. J., Gratton E., J. Biomed. Opt. 2012, 17, 046012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195. Stringari C., Cinquin A., Cinquin O., Digman M. A., Donovan P. J., Gratton E., Proc. Natl. Acad. Sci. U.S.A. 2011, 108, 13582. [DOI] [PMC free article] [PubMed] [Google Scholar]


