Abstract
MicroRNAs (miRNAs) are small, non-coding RNAs that play important roles in post-transcriptional regulation of their target genes, yet the transcriptional regulation of plant miRNAs by promoter is poorly understood. Here, we firstly clone pri-miR475b cDNA and its native promoter from P. suaveolens, and characterize Psu-MIR475b as class-II gene transcribed by RNA polymerase II. By 5′ deletion analysis of Psu-miR475b promoter in a series of promoter-GUS chimeric vectors, we functionally identify three positive regulatory regions and multiple cis-acting elements responsible for Psu-miR475b promoter activity in response to freezing stress and exogenous hormone treatment. Moreover, the Psu-miR475b promoter activity displays a tissue-specific manner, negatively regulated by freezing stress and positively by MeJA, SA or GA treatment. Importantly, we comparatively analyze the time-course transcriptional profiles of Psu-miR475b and its targets in Psu-miR475b over-expression transgenic plants controlled by Psu-miR475b-specific promoter or CaMV 35S constitutive promoter, and explore the regulatory mechanism of Psu-miR475b promoter controlling transcriptional expressions of Psu-MIR475b and its targets in response to freezing stress and exogenous hormone treatment. Our results reveal that Psu-miR475b promoter-mediated transcriptions of Psu-MIR475b and its targets in response to freezing stress may be involved in a cross-talk between freezing response and stress signaling process.
Low temperature, especially freezing (<0 °C), is one of the major environmental stresses that seriously influence in the growth, development, distribution and productivity of plants1,2. Freezing tolerance and cold acclimation are highly complex process involved in physiological and metabolic modifications for cold response and a multiple gene expression network controlling plant tolerance to cold stress1,2,3,4,5,6,7,8,9. However, the regulatory networks of overall response of plants to low temperature stress still remains unclear. MicroRNAs (miRNAs) are a highly conserved class of endogenous single-stranded small non-coding RNAs that have been clearly shown to serve as negative regulators to modulate plant gene expression at post-transcriptional level by transcript cleavage or translational repression of target genes10,11,12. In recent years, the significant alterations in transcript levels of some miRNAs have been identified in response to cold stress in several plants such as Arabidopsis5,13,14,15, rice16, wheat17, Setaria italica18, Phaseolus vulgaris19, Brachypodium distachyon20, trifoliate orange21, celery21, Camellia sinensis23 and Populus24,25,26,27. Moreover, many predicted and experimentally confirmed targets of cold-responsive miRNAs encode a variety of transcription factors or other regulatory proteins implicated in low temperature response20,21,22,24,26,28. Also, genetic transformation of miRNAs and their targets has been recently demonstrated to alter cold stress tolerance capacity in plants28,29. All of these findings have shown a crucial role of miRNAs in the regulation of gene expression in response of plants to low temperature. Thus, an importance for understanding the initiation and regulation of miRNA gene transcription under low temperature stress.
The biogenesis of miRNAs is complex. Most plant miRNA genes (MIR), located in intergenic regions, are transcribed as independent transcriptional units by RNA polymerase II (Pol II) to produce primary transcripts (pri-miRNA) and then processed into stem-loop structured miRNA precursors (pre-miRNAs) by Dicer-like enzyme 1 (DCL1) in the nucleus10,11,12,30,31,32,33,34. The mature miRNAs, transported to the cytoplasm, are incorporated into the RNA-induced silencing complex (RISC), and then lead to post-transcriptional gene silencing via transcript cleavage or/and translational repression of their target mRNAs by recognizing the base-pairing and interaction with their cognate targets10,11,12,35,36,37,38,39,40,41,42. Although much effort has been focused on elucidating the regulatory function of plant miRNAs, little is known about how MIR genes themselves are regulated. Recently, some studies have shown that plant miRNAs have the class II promoters and may be regulated by a similar mechanism as established for protein-coding genes. The promoters of miRNAs have been predicted in rice by bioinformatic analysis and A. thaliana by 5′ RACE, respectively31,32,43,44,45, indicating the promoter as a crucial control region for the transcription initiation of miRNAs. However, direct evidence for transcriptional regulation of MIR genes by its native promoter is very little to date. Thus, the nature of miRNA promoter remains one of the most interesting open problems in the study of miRNA biogenesis.
Populus suaveolens, a typical freezing-resistant arbor tree of poplar species, can survive under a freezing temperature of approximately −43.5 °C in winter in the distribution of eastern Siberia regions and Great Xing’an Mountain, Northeast of China, and has emerged as a novel ideal model plant to study the freezing resistance mechanism in woody plants25,46,47. Previously, we identified miR475b with a significant down-regulation in P. suaveolens under freezing stress (°C), and revealed that miR475b plays an important role in freezing resistance of P. suaveolens25,27. In this continued study, we report the clone and analysis of miR475b gene and its native promoter from P. suaveolens, and explore the tissue-specific expression pattern of Psu-miR475b promoter. Also, we produce a series of 5′ promoter deletion-GUS reporter constructs, and perform a combination of the histochemical and fluorometric GUS assay and qRT-PCR to functionally characterize a set of regulatory regions and cis-acting elements responsible for the transcriptional activity of Psu-miR475b promoter. Importantly, we comparatively analyze the time-course transcriptional expression profiles of Psu-miR475b and its target genes in Psu-miR475b over-expression transgenic plants controlled by Psu-miR475b-specific promoter or cauliflower mosaic virus 35S (CaMV 35S) constitutive promoter, and investigate the regulatory mechanism of Psu-miR475b promoter for the transcripts of Psu-MIR475b and its targets in the transgenic plants subjected to freezing stress and exogenous hormone treatment. To our knowledge, this is the first report of functional identification and regulatory mechanism of Psu-miR475b promoter governing the transcriptional expressions of Psu-MIR475b and its targets in response to freezing stress.
Results
Cloning and analysis of freezing-responsive Psu-miR475b and its promoter
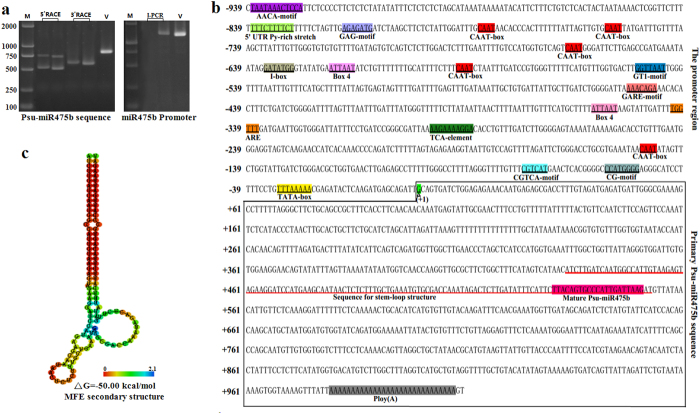
To elucidate the regulatory mechanism of miR475b transcription in response of P. suaveolens to freezing stress, the 1011-bp full-length freezing-responsive pri-miRNA475b with a putative 5′-cap structure and 3′-poly(A) tail (designated as Psu-MIR475b, Accession No. JX262380) was first cloned by 5′ and 3′ RACE from P. suaveolens cDNA (Fig. 1a,b). In order to gain insights into miR475b transcription, the secondary structure of RNA sequences generated from pri-miR475b cDNA was analyzed. We found that miR475b precursor has folding back free energy of −50.00 kcal/mol to form a stable stem-loop structure, and its mature sequence with 21nt length (5′-UUACAGTGCCCATTGATTAAG-3′) located in 3′ arm of stem-loop structure (Fig. 1c). Importantly, based on 5′ end sequence of Psu-MIR475b gene, we used inversion-PCR (IPCR) to obtain full-length (939bp) Psu-miR475b promoter (Accession No. KM288552) from P. suaveolens genomic DNA (Fig. 1a,b).
Figure 1. Cloning and analysis of freezing-responsive Psu-miR475b and its promoter from P. suaveolens.
(a) Amplification of Psu-MIR475b gene (left) and its promoter (right) from P. suaveolens by 5′/3′RACE-PCR and I-PCR, respectively. M means the marker of 2000, and V means the validation for the full-length sequence of miR475b (left) and promoter (right). (b) Analysis of Psu-MIR475b gene sequence and cis-acting elements of promoter. The putative transcriptional start site (TSS) taken as +1 was marked with an upward filled triangle, and the regions from −939 to −1 relative to TSS was for Psu-miR475b promoter. The putative multiple cis-acting elements in Psu-miR475b promoter were underlined and presented with different background. The sequence of primary Psu-miR475b was located in the black box, where mature Psu-miR475b was marked with pink background, and the sequence of stem-loop structure was underlined with red. (c) Predicted stem-loop structure of Psu-miR475b precursor.
Promoter is a crucial control region for transcription initiation of miRNAs. To understand the mechanism of the activation of Psu-MIR475b gene, it is required to locate cis-acting elements within its promoter region. By online programs, we characterized one core promoter element TATA box-like sequence (TTTAAAAA, −32/−25), five CAAT-boxes as common cis-acting elements (−149/−146, −591/−588, −666/−663, −757/−754, −786/−783), five light responsive elements [CG motif (CCATGGGG, −57/−50), Box 4 (ATTAAT, −360/−355, −620/−615), GT1-motif (GGTTAAT, −550/−544), I-box (−635/−629) and GAG-motif (AGAGATG, −817/−811)], four stress-related elements [CGTCA-motif (CGTCA, −75/−71), TCA-element (AAGAAAAGGA, −297/−288), GARE-motif (AAACAGA, −452/−446) and TC-rich repeat (AAACAGA, −687/−678)], two cis-acting regulatory elements [5′UTR Py-rich stretch (TTTCTTTTCT, −838/−829) and ARE (TGGTTT, −342/−339)], and one endosperm-specific regulatory AACA-motif (AATCTAATTT, −590/−581) within Psu-miR475b promoter (Fig. 1b and Table S1). Thus, an enrichment of diverse regulatory cis-acting elements in Psu-miR475b promoter.
Tissue-specific activity for Psu-miR475b promoter in plants
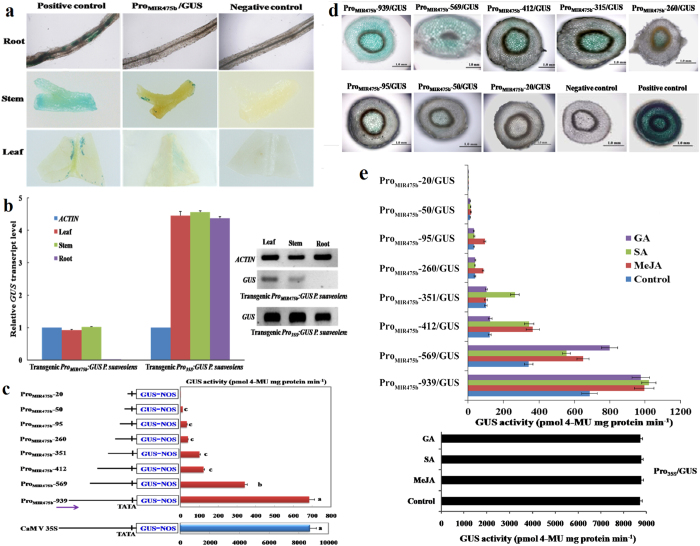
To explore whether Psu-miR475b promoter was similar to those of protein-coding genes, 939 bp full-length Psu-miR475b promoter (ProMIR475b) and CaMV 35S promoter (Pro35S) were respectively fused to GUS reporter gene and transferred into tobacco. We compared GUS activity in different tissues of transgenic ProMIR475b:GUS and Pro35S:GUS tobacco subjected to histochemical GUS staining. The ProMIR475b:GUS plants exhibited GUS expression in the stems and leaves, but no GUS staining was detected in the roots (Fig. 2a). In stark contrast with ProMIR475b:GUS plants, all tested tissues of Pro35S:GUS lines displayed a significant higher GUS expression (Fig. 2a). These results indicate that Psu-miR475b promoter is able to direct GUS gene expression, but differs from CaMV 35S constitutive promoter that served as positive control, directing a stronger expression of GUS.
Figure 2. Characterizations of tissue specificity and multiple cis-regulatory elements for Psu-miR475b promoter.
(a) Tissue-specific activity for Psu-miR475b promoter. The histochemical GUS staining in different tissues of transgenic tobacco shows that Psu-miR475b promoter similar to those of protein-coding genes is able to drive GUS expression but with a tissue-specific manner. GUS staining from CaMV 35S (pBI121 vector) transformant and wild-type tobacco were served as positive and negative controls, respectively. (b) The expression analysis of GUS reporter gene in different tissues of transgenic P. suaveolens plants by RT-PCR and qRT-PCR. The poplar ACTIN gene was used as an endogenous reference gene and its expression level was arbitrarily set to 1.00 for standardization. The means and standard deviations of the relative GUS transcript levels in the respective tissue are shown. (c) 5′ deletion analysis of Psu-miR475b promoter by the fluorometric GUS assay in transgenic tobacco stem. (d) 5′ deletion analysis of Psu-miR475b promoter by histochemical GUS staining in transgenic tobacco stem (as one predominant tissue). (e) GUS activity driven by the Psu-miR475b promoter in stem of transgenic tobacco plants subjected to MeJA, GA or SA. GUS activity from the CaMV 35S (pBI121 vector) transformants served as a comparison. Data are mean and standard deviations of twelve transgenic lines. The numbers below the bars indicate the fold changes of GUS activity. Significance of the changes produced after each treatment was assessed using Student’s t tests (*P < 0.05, **P < 0.01).
Previously, we identified one preferential tissue-specific transcript of Psu-miR475b in the leaves and stems of P. suaveolens27. Thus, it is need to illustrate the tissue-specificity of Psu-miR475b promoter in P. suaveolens to address the regulatory mechanism of Psu-miR475b promoter controlling the transcription of Psu-miR475b. Here, the constructed promoter-GUS chimeric vectors (ProMIR475b:GUS and Pro35S:GUS) were transferred into P. suaveolens for the transcript level assay of GUS gene in different tissues by RT-PCR and qRT-PCR (Fig. 2b). As expected, Psu-miR475b promoter-driving GUS gene was transcribed in the leaves and stems of transgenic P. suaveolens, but no transcript observed for the roots, which was comparable with those observations in transgenic ProMIR475b:GUS tobacco (Fig. 2a). In addition, transgenic P. suaveolens driven by 35S promoter greatly increased GUS activity in all tissues examined (Fig. 2b). Thus, our findings reveal a typical tissue-specific expression pattern for Psu-miR475b promoter in plants.
Characterization of multiple cis-regulatory elements in Psu-miR475b promoter
To understand the regulatory mechanism controlling Psu-MIR475b gene expression by its native promoter, we first sought to determine the functionality of our predicted regulatory regions responsible for the Psu-miR475b promoter activity. Hence, we produced a series of 5′ promoter deletion-GUS constructs, covering different regions from −939 to −1, −569 to −1, −412 to −1, −351 to −1, −260 to −1, −95 to −1, −50 to −1 and −20 to −1 (Figure S1). The multiple transgenic tobacco plants (>12 independent lines) were obtained (Figure S2), and their stems were used as one predominant tissues (Fig. 2a) for the GUS expression analysis. The fluorometric GUS assay clearly demonstrated that compared with full-length promoter (939 bp), the deletion from −939 (relative to TSS) to −570, −569 to −413, −351 to −261, and −95 to −51 caused a significant reduction (about 1.0-, 1.8-, 1.6-, and 1.8-fold, respectively) in GUS activity in transgenic tobacco stems, whereas only 0.2-fold decrease was observed for the deletion from −412 to −352, and −261 to −96 (Fig. 2c). Intriguingly, GUS expression from −50 to −1 was significantly lower than others, while further deletion to −20 abolished the GUS expression (Fig. 2c). These investigations were entirely consistent with our results of histochemical GUS staining in the stems of transgenic tobacco plants (Fig. 2d). Thus, our data indicate that three positive regulatory regions (−939 to −413, −351 to −261 and −95 to −51) are responsible for the basal activity of Psu-miR475b promoter, and one region (−50 to −21) required for transcriptional initiation.
As we were surprised by a cluster of stress-related cis-elements within Psu-miR475b promoter (Fig. 1b and Table S1), we attempted to establish whether or not the basal activity of Psu-miR475b promoter could be affected by the treatments of stress-related stimuli. To achieve this, a series of 5′ promoter deletion-GUS transgenic tobacco was subjected to the treatments of MeJA, SA, ABA and GA, respectively. By using fluorometric GUS assay, an obvious induction of GUS activity was observed in the stems of ProMIR475b-939/GUS and ProMIR475b-569/GUS plants upon treatment by SA, MeJA and GA, but a lower inducible ratio for ProMIR475b-569/GUS plants. In contrast, the GA-inducible expression of GUS gene seemed to disappear in ProMIR475b-412/GUS and ProMIR475b-351/GUS lines, which responded to SA and MeJA treatments. Also, ProMIR475b-260/GUS and ProMIR475b-95/GUS plants exhibited a higher GUS activity in response to MeJA, but not GA and SA. Notably, no inducible expression was shown for ProMIR475b-50/GUS and ProMIR475b-20/GUS plants (Fig. 2e). The present results indicate that the regions from −569 to −413, −351 to −261, and −95 to −51 are respectively required for GA-, SA-, and MeJA-inducible activity of Psu-miR475b promoter. However, no response of GUS activity was observed with or without ABA treatment. It is also interesting to note that Pro35S/GUS lines showed no significant inducible expression under these imposed conditions (Fig. 2e).
Cross-talk between freezing response and stress signaling for ProMIR475b activity regulation
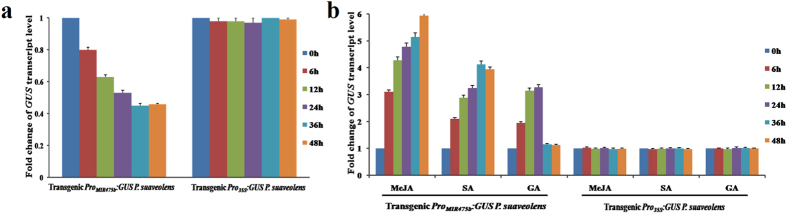
Recently, we identified the significant down-regulation of Psu-miR475b in response of P. suaveolens to 0 °C stress25, which allowed us to explore whether the activity of freezing-responsive Psu-miR475b promoter was specifically regulated by low temperature. Thus, we generated ProMIR475b:GUS and Pro35S:GUS transgenic P. suaveolens (Figure S2), and subjected them to the time-course analysis of GUS transcript level by qRT-PCR after 0 °C treatment for 0–48 h. We observed that during freezing-stress treatment, ProMIR475b-driving GUS expression was significantly decreased from 6 to 48 h, while no expression change was directed by CaMV 35S promoter in transgenic plants (Fig. 3a), indicating that Psu-miR475b promoter activity was specifically down-regulated by freezing stress. In addition, to investigate whether the signaling pathway was involved in the activity regulation of Psu-miR475b promoter, our analysis focuses on identifying the regulatory patterns of Psu-miR475b promoter by MeJA, SA and GA. In the case of treatment with exogenous MeJA, SA or GA for 0–48 h, we found that that in ProMIR475b:GUS plants, GUS expression after MeJA treatment was more sustained and continued to increase over the time points tested, while the relative low transcript of GUS with a peak value at 36 h was detected for SA treatment. By contrast, when exposed to GA, GUS expression first slightly increased and then returned to a normal level after 24 h (Fig. 3b). However, Pro35S:GUS plants showed no inducible expression of GUS by all of the imposed conditions (Fig. 3). These results indicate that Psu-miR475b promoter activity can be induced by SA, MeJA and GA, but with differential time-course regulatory manner respond to different hormones.
Figure 3. The impacts of freezing stress and defense-related stimuli treatments on the activity of Psu-miR475b promoter in transgenic ProMIR475b:GUS and Pro35S:GUS P. suaveolens.
(a) The Psu-miR475b promoter activity is negatively regulated by freezing stress. (b) The Psu-miR475b promoter activity is induced by the applications of exogenous SA, MeJA and GA.
The above investigations prompt us to explore whether freezing response was linked to the hormone signal. Here, we also examine GUS expression driven by Psu-miR475b promoter with serial 5′ deletions under freezing stress for 48 h. Notably, all deletion constructs except ProMIR475b-20 and ProMIR475b-50 showed an obvious decrease of GUS activity when treated with freezing, but no significant difference of GUS expression was observed between ProMIR475b-95 and ProMIR475b-260 or between ProMIR475b-351 and ProMIR475b-412 lines (Figure S3), indicating that the regions from −939 to −413, −351 to −261, and −95 to −51 are required for the regulation of Psu-miR475b promoter activity in response to freezing stress. Impressively, in those regions, MeJA-responsive CGTCA motif (−75/−71), SA-responsive TCA element (−297/−288) and GA-responsive GARE motif (−452/−446) were identified (Fig. 2e). Also, the Psu-miR475b promoter activity was induced by the GA, SA, and MeJA treatments (Fig. 3b). Thus, it could be concluded that a cross-talk between freezing-stress response and hormone signaling may involve in the activity regulation of Psu-miR475b promoter.
ProMIR475b-mediated transcriptions of Psu-miR475b and its targets in transgenic P. suaveolens
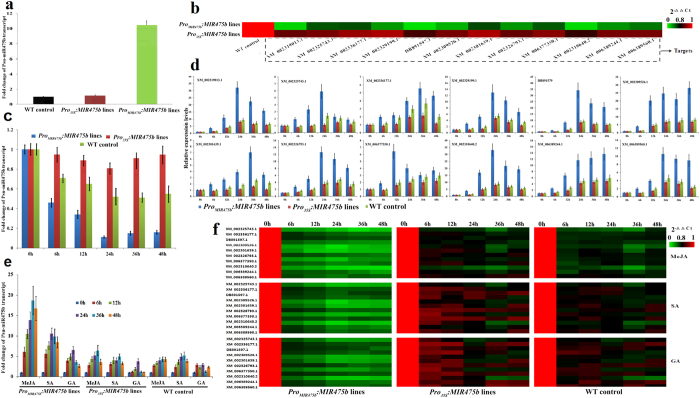
To address the regulatory mechanism of Psu-miR475b-specific promoter controlling Psu-MIR475b expression, we analyzed the time-course transcription pattern of Psu-miR475b by qRT-PCR in Psu-miR475b-overexpressing P. suaveolens under the control of Psu-miR475b promoter or CaMV 35S promoter (Figure S4). Compared with the wild-type (WT) controls, the most striking difference between two types of transgenic lines was observed in ProMIR475b:MIR475b plants with about 6.1-fold increase of Psu-miR475b transcript, which is considerably higher than that (0.3-fold) in Pro35S:MIR475b lines (Fig. 4a), confirming an important contribution of Psu-miR475b promoter to regulate its native gene (Psu-MIR475b) transcription. Recently, we experimentally characterized 12 putative pentatricopeptide repeat protein (PPR) genes (XM_002319013.1, XM_002325743.1, XM_002336177.1, XM_002329199.1, DB891579, XM_002309526.1, XM_002301639.1, XM_002326793.1, XM_006377350.1, XM_002310640.2,XM_006389244.1, XM_006389560.1) as the targets of Psu-miR475b27. To understand how Psu-miR475b regulates the expressions of its targets, and to examine whether Psu-miR475b promoter was involved specifically in the regulation of miR475b-meidiated expression, we further checked the transcript levels of 12 miR475b-targeted PPR genes in transgenic plants. Compared to the WT controls, the transcripts of 12 PPR genes were all markedly reduced (4–9 fold) in ProMIR475b:MIR475b plants, but no significant alteration was observed for Pro35S:MIR475b lines (Fig. 4b). This indicates that the expressions of miR475b-targeted genes in Psu-miR475b-overexpressing plants are specifically regulated by Psu-miR475b promoter. Also, the fact of an inverse correlation between Psu-miR475b (up-regulation) and its targets (down-regulation) (Fig. 4a,b) in transgenic plants showed that the miR475b-targeted transcripts may be cleaved directly by Psu-miR475b, which was confirmed by our previous 5′ RLM- RACE27.
Figure 4. Transcriptional expression analysis of Psu-miR475b and its 12 targets in Psu-miR475b overexpression P. suaveolens by qRT-PCR.
(a) Comparative analysis of Psu-miR475b transcription in transgenic ProMIR475b:MIR475b and Pro35S:MIR475b plants, revealing an important contribution of Psu-miR475b promoter on the up-regulation of its native gene (Psu-MIR475b) transcription. Both miR167e and miR168a-3p were used as inner references. Error bars indicate standard deviations of three technical replicates. (b) Comparative analysis of miR475b-targeted genes in transgenic ProMIR475b:MIR475b and Pro35S:MIR475b plants. The down-regulated expressions of all targets indicate that miR475b-targeted transcripts may be cleaved directly by Psu-miR475b. ACTIN gene was used as reference genes. The relative expression values in heatmap were counted as 2−△△C t, and the wild-type lines were used as the control. (c) The significantly down-regulated transcription of Psu-miR475b by freezing stress. (d) The significantly up-regulated transcription of all 12 targets by freezing stress. (e) The significantly induced transcription of Psu-miR475b by the MeJA, SA and GA treatments, which is tightly correlated with the stronger response of Psu-miR475b promoter to MeJA, SA and GA. (f) The significantly down-regulated transcription of all 12 targets by the MeJA, SA and GA treatments. The finding indicates an important regulatory role of Psu-miR475b promoter for Psu-miR475b-mediated transcriptional repression of its targets responsive to MeJA, SA and GA.
ProMIR475b-mediated transcriptional regulation involved in freezing response and hormone signaling
Considering the fact that freezing stress has shown to confer a negative effect on the Psu-miR475b promoter-directed GUS expression (Fig. 3a), the obtained Psu-miR475b-overexpressing P. suaveolens were also exposed to 0 °C treatment for 0–48 h, and the temporal transcript profiles of Psu-miR475b and its targets were analyzed to reveal the regulatory mechanism of Psu-miR475b promoter for the transcriptional expressions of Psu-MIR475b and its targets in response to freezing stress. We found that the transcript level of Psu-miR475b in ProMIR475b:MIR475b plants was lower after 6 h and greatly decreased with longer stress, while a small decline was detected in Pro35S:MIR475b lines with minimum valve at 24 h, similar to that of the WT controls (Fig. 4c). This finding reveals that Psu-miR475b transcription is altered in response to freezing stress, and a significant down-regulation is specifically driven by its native promoter. Also, the strongly induced expression of all 12 miR475b-targeted genes by freezing was identified in ProMIR475b:MIR475b plants than in both Pro35S:MIR475b plants and WT controls (Fig. 4d), implying that the significant up-regulation of miR475b-targeted genes driven by Psu-miR475b promoter may be involved in freezing-stress response process.
Given that Psu-miR475b promoter activity is induced by MeJA, SA and GA (Fig. 3b), we attempted to address whether the application of these stimuli could trigger the transcriptions of Psu-miR475b and its targets. To this end, the transgenic P. suaveolens of ProMIR475b:MIR475b and Pro35S:MIR475b were respectively treated by MeJA, SA or GA for 0–48 h. Here, Psu-miR475b transcript was up-regulated approximately 17.3-, 9.6- and 5.1-fold in ProMIR475b:MIR475b plants by MeJA, SA and GA treatments for 48 h, peaked at 36, 24 and 24 h respectively, whereas a small induced expression for Psu-MIR475b was detected in both Pro35S:MIR475b lines and WT controls by these treatments (Fig. 4e). These results indicate that an obvious inducible transcription of Psu-miR475b by MeJA, SA and GA is tightly correlated with a stronger response of Psu-miR475b promoter to MeJA, SA and GA. It is also worth noticing that exogenous application of MeJA, SA or GA resulted in a nearly antiparallel transcript pattern of Psu-miR475b (up-regulation) and its targets (down-regulation) in transgenic ProMIR475b:MIR475b plants, but the expressions of all targets remain stable in Pro35S:MIR475b lines under these imposed conditions (Fig. 4e,f).
Discussion
In recent years, bioinformatic analysis has been applied to predict the promoters of miRNAs in Arabidopsis and rice15,31,32,43,44,45, but the experimental cloning and functional identification of MIR promoter was only reported for miR171a, miR172a and miR390a/b in Arabidopsis to data48,49,50. This study presents for the first time a regulatory mechanism of Psu-miR475b promoter governing the transcription of Psu-miR475b and its targets in response of P. suaveolens to freezing stress, where pri-miR475b cDNA and its promoter were firstly cloned from P. suaveolens (Fig. 1a,b). A lower folding back free energy was predicted for Psu-miR475b precursor (Fig. 1c), as reported in Triticum aestivum51, B. distachyon20 and P. tomentosa26. Intriguingly, the primary transcript of Psu-miR475b was capped at the 5′end and polyadenylated at the 3′ end (Fig. 1b), similar to the unique properties of class-II gene transcripts, which has been characterized in A. thaliana MIR genes31,43,45. Moreover, Psu-miR475b has one 21 nt-length mature sequence located in 3′ arm of stem-loop structure, and has uridine (U) as first nucleotide at 5′ end (Fig. 1b,c), which is entirely consistent with P. trichocarpa miR475a/b/c52, also identified as one characteristic feature of miRNAs in plants20,24,31. Together, all our findings reveal that Psu-MIR475b gene is transcribed as a single transcript unit by the RNA pol II mechanism.
The promoter contain essential components for the transcription regulation of MIR gene31,32. TATA-box, as a well-known core motif in the promoters of eukaryotic class-II genes, has been identified in most miRNA promoters of A. thaliana and O. sativa31,32,43,44,45, suggesting that most of plant MIR genes may present the same promoters as the protein-coding genes transcribed by RNA pol II. In this study, our identified one 8-nt TATA box-like sequence of Psu-miR475b promoter within −32 to −25 (Fig. 1c and Table S1) was compatible with those located in protein-coding genes, also correspond to authentic TATA box sequence within the core promoters of plant MIR genes31,32,44. Importantly, the up-regulated GUS was detected in both ProMIR475b:GUS and Pro35S:GUS transgenic tobacco, but the relative lower GUS activity was observed for ProMIR475b:GUS lines (Fig. 2c,d). All our results show indeed that Psu-miR475b promoter may be as pol II promoter, but exhibits a specificity for Psu-miR475b, which support the hypothesis of the differentiation between MIR genes and protein-coding genes45.
Earlier studies in four model species (Caenorhabditis elegans, Homo sapiens, A. thaliana and O. sativa) have revealed many significant conserved motifs in the promoters of MIR genes32. Recently, 11 over-represented cis-elements (AtMYC2, ARF, SORLREP3, G-box, SORLIP1, RY-repeat, LTRE, Evening element, TELO-box, DRE-like and AtMYB2), and 9 under-represented cis-elements (GATA box, LFY motif, T-box, GCC-box, RAV1-B, Bellringer BS3, CArG, HSEs and CCA1) were identified in the promoters of Arabidopsis MIR genes by position weight matrices (PWM)45. In this work, we performed the PWM method to identify a total of 13 cis-acting elements in Psu-miR475b promoter (Fig. 1b and Table S1), among which 11 elements (Box 4, GT1-motif, CGTCA-motif, GAG-motif, CG-motif, TCA-motif, GARE-motif, 5′UTR Py-rich stretch, TC-rich repeat, ARE and AACA-motif) were not previously reported. Our data indicate that these putative cis-regulatory elements may be specific to Psu-miR475b promoter, probably owing to MIR gene promoter with specific cis-acting elements for governing unique transcription of miRNA53. In addition, we present the first 5′ promoter progressive deletion analysis to elucidate the functionality of these potential regulatory cis-elements for the Psu-miR475b promoter activity. The histochemical and fluorometric GUS assay (Fig. 2c,d), combined with our characterizations of cis-acting elements in Psu-miR475b promoter (Fig. 1b and Table S1), suggests that CGTCA motif (involved in MaJA responsiveness), GAR-motif (GA responsiveness), TCA-element (SA responsiveness), TC-rich repeat (stress responsiveness) and 5′UTR Py-rich stretch (conferring high transcription level) may play the key role in the activity regulation of Psu-miR475b promoter, and TATA-box (as core promoter element) function as initiator for initiation of Psu-miR475b transcription. Importantly, the regions from −569 to −413, −351 to −261, and −95 to −51 were respectively required for GA-, SA-, and MeJA-inducible activity of Psu-miR475b promoter (Fig. 2e), also noted in the promoters of some cold-responsive miRNAs (such as miR167/393/408) in A. thaliana14. However, no response of GUS activity to ABA-treatment in transgenic plants may be correlated with the lack of ABA-responsive ABRE motif in the Psu-miR475b promoter (Table S1), as reported in the promoters of some A. thaliana miRNAs14, implying that the activity regulation of Psu-miR475b promoter may be ABA-independent pathway. Together, our deletion analysis suggest a complex regulatory mechanism for the activity of Psu-miR475b promoter controlled by its internal cis-acting elements.
Previously, miR475b has been shown to be down-regulated in response of P. suaveolens to freezing stress27. Our findings on the significant down-regulation of GUS gene by freezing stress in transgenic ProMIR475b:GUS plants (Fig. 3a) and the obvious induced expression by the exogenous SA, MeJA and GA (Fig. 3b) reveal that Psu-miR475b promoter is one low temperature-responsive promoter, and its activity is negatively regulated by freezing stress and positively by the applications of exogenous hormones. This also prompt us to consider that there exists a complex relationship between freezing stress and hormone response signal, and a potential great cross-talk among MeJA, SA and GA for the activity regulation of Psu-miR475b promoter.
Recently, high-throughput sequencing revealed that some cold-induced miRNAs displayed up-regulation in response of T. aestivum to GA, ABA and JA54, suggesting that these miRNAs may involve in an intricate association between the signaling pathways and abiotic stress responses. In this study, the significant down-regulation for Psu-miR475b and up-regulation for its targets were characterized in ProMIR475b:MIR475b transgenic P. suaveolens during freezing stress, while a slight alternation was observed for Pro35S:MIR475b plants (Fig. 4c,d), demonstrating that the transcriptions of Psu-miR475b and its targets driven by its native promoter are specifically altered in response to freezing stress. These results, combined with the finding that the promoter activity of Psu-miR475b is negatively regulated by freezing stress, confirm an involvement of Psu-miR475b promoter in the transcription regulation of Psu-MIR475b and its targets under freezing stress.
Many miRNAs were differentially regulated by exogenous application of plant hormones such as JA, SA, GA and ABA13,54. In this work, compared with Pro35S:MIR475b plants, the significant differential expressions of Psu-miR475b (up-regulation) and its targets (down-regulation) were identified in transgenic ProMIR475b:MIR475b plants by the MeJA, SA and GA treatments (Fig. 4e,f), indicating that the exogenous hormones (MeJA, SA and GA) could regulate the transcripts of Psu-miR475b and its targets, likely though mediating the Psu-miR475b promoter activity. This could be supported by the fact of a higher inducible activity of Psu-miR475b promoter by the treatments of MeJA, SA and GA (Fig. 3b). Thus, we conclude that Psu-miR475b promoter-triggered transcriptions of Psu-MIR475b and its targets may be involved in the complex signaling pathways, mediated by MeJA, SA and GA.
Taken together, our findings that freezing stress-responsive Psu-miR475b and its targets as well as the Psu-miR475b promoter activity are prone to being affected by the treatments of freezing stress and exogenous MeJA, SA or GA, reveal the existence of a great cross-talk between freezing response and stress signaling process. It was suggested that the PPRs may provide a signaling link between mitochondrial electron transport and regulation of stress and hormonal responses in A. thaliana24. Microarray analysis of transcript expression has shown that many genes involved in the biosynthesis and signaling of plant endogenous hormone (such as JA, SA and GA) are down-regulated by cold stress in A. thaliana5. The previously characterized 12 PPRs as the targets for psu-miR475b27, integrated with all our investigations, suggest that the lower activity of Psu-miR475b promoter could be expected mainly due to the smaller biosynthesis capacity of endogenous hormone (MeJA, SA and GA) caused by freezing stress. Thus, the transcriptional expressions of Psu-miR475b and its targets by its native promoter may be involved in a complex signaling pathway and freezing-stressed response. Our findings also evidenced that Psu-miR475b-specific promoter is important determinant for the transcriptional regulation for Psu-miR475b and its targets in response of P. suaveolens to freezing stress.
Methods
Plant materials and growth conditions
P. suaveolens, obtained from Great Xing’an Mountain, Northeast of China, was used as the source material for this study. The plants were propagated by cutting and raised in pots within a controlled environment chamber (photoperiod: 16/8 h light/dark, minimum illumination: 0.2 mM s−1 m−2, day temperature: 20–30 °C) at Beijing Forestry University. Tissue-cultured P. suaveolens plants were raised and synchronized on modified MS medium as our previously described55. The fully developed leaves harvested from the tissue-cultured plantlets were subjected to genetic transformation. Tissue-culture tobacco (Nicotiana tabacum cv. W38) plants were performed on modified MS medium, and the fully developed tobacco leaves were then used for genetic transformation experiments.
Freezing stress treatment of P.suaveolens plantlets
The 2-month-old plantlets with identical growth status were exposed to 0 °C, and the leaves were then harvested at time points 0, 6, 12, 24, 36 and 48 h post-treatment as described previously25. All materials were collected from three individual plants and immediately frozen in liquid nitrogen and stored at −80 °C.
Cloning of pri-miR475b and its promoter and transformation of P. suaveolens
Total RNA was isolated from the tested samples using SV Total RNA Isolation System (Promega, Madison, WI, USA). The synthesis of first strand cDNA, and the 3′- and 5′-RACE of pri-miR475b were performed according to SMARTer RACE cDNA kit (Clontech, USA) illustrate, and then the purified amplification products were sequenced to assemble the full-length sequence of pri-miR475b. 3′- and 5′-RACE outer/inner primer could be seen in Supplementary Table S2. The full-length cDNA sequence (pri-miR475b) was amplified from P. suaveolens genomic DNA by PCR using two gene-specific primers 5′-AGGTAGTCAAGCACCATCACAAA-3′ (forward prime) and 5′-AACCTACAGCATGACCTAGAGGC-3′ (reverse primer), and then cloned into pGM-T Vector, giving pT-MIR475b. The sequence of the amplified DNA fragment was verified by sequencing. A XbaI-SacI fragment from pT-MIR475b containing the MIR475b sequence was then subcloned into the Xba I and Sac I sites of vector pBI121 between the CaMV 35S promoter and the NOS 3′poly (A) signal to generate the 35S:MIR475b construct (named as Pro35S:MIR475b).
For full-length Psu-miR475b promoter from P. suaveolens by IPCR, forward primers were (5′-AATGTCACGGGTAACTAATTCTA-3′(F-1) and 5′-ATAAAGTAAGAATGTCACGGGTA-3′(F-2), and reverse primers were 5′-GCTTTCACCTTCAACAACAAATG-3′(R-1) and 5′-GTAGATGAGATGATTGGGCGAAAA-3′(R-2). To construct the Psu-miR475b-overexpression vector driven by Psu-miR475b promoter, the amplified Psu-MIR475b gene and its promoter were digested with Xba I and Sac I, and joined by ligation with T4 DNA Ligase, followed by PCR amplification to generate Psu-miR475b-promoter/MIR475b construct (ProMIR475b:MIR475b). For a negative control, empty effector plasmid 35S/Em was constructed by the replacement of GUS gene with a native sequence 5′-TCTAGAGGATCCAATTGCTACCGAGCTC-3′ in pBI121. These constructs were first introduced into Agrobacterium tumefaciens strain GV3101 via the freezing-thaw method, and then transferred into P. suaveolens by the leaf disc transformation method.
Promoter-GUS chimeric vector construction and tobacco transformation
A series of 5′ progressive deletions of Psu-miR475b promoter, covering different regions from −939 to −1, −569 to −1, −412 to −1, −351 to −1, −260 to −1, −95 to −1, −50 to −1 and −20 to −1, were respectively generated by PCR using a 5′ sequence of Psu-MIR475b as a template with forward primers containing Hind III restriction site (underlined): 5′-CCCAAGCTTCTAATAAACTCCATTCTCC-3′(f1), 5′-CCCAAGCTTTTTTCATGTTTGGTGA-3′(f2), 5′-CCCAAGCTTTTTGATTTATGGGTTTTT-3′(f3), 5′-CCCAAGCTTTATTGATTTTGGTTTG-3′(f4), 5′-CCCAAGCTTAAAAAGACACCTGTTT-3′(f5), 5′-CCCAAGCTTGCCTTTTAGGGTTTT-3′(f6), 5′-CCCAAGCTTGAGGGCATCCTTTTCCT-3′(f7), 5′-CCCAAGCTTATACTCAAGATGA-3′(f8), and a common reverse primer containing a Xba I restriction site (underlined): 5′-CTCAAGATGAGCAGATTGCTCTAGAGC-3′(r01). Each of the PCR amplified fragments was digested with Xba I and Hind III (Promega, USA) and purified with TIAN-quick Midi Purification Kit (TIANGEN, Beijing, China). They were then fused to the GUS reporter gene of the modified pBI121 vector (Clontech, USA) harboring an Xba I site immediately downstream of the Hind III site, which was previously digested with Xba I and Hind III to release 35S promoter. The resulting vectors, confirmed by DNA sequencing, were respectively named as ProMIR475b-939/GUS, ProMIR475b-569/GUS, ProMIR475b-412/GUS, ProMIR475b-351/GUS, ProMIR475b-260/GUS, ProMIR475b-95/GUS, ProMIR475b-50/GUS and ProMIR475b-20/GUS.
The chimeric vectors for tobacco transformation were performed by the same method of P. suaveolens. The putative transgenic plantlets resistant to kanamycin were further confirmed by PCR, as well as preliminary GUS staining. The verified transgenic tobaccos were then propagated and synchronized from primary transformants in MS medium. One month-old in vitro-grown plantlets were used for subsequent experiments.
Plant treatment
For inducible expression analysis of GUS activity, the in vitro transgenic tobacco plants were sprayed with 200 μM MeJA, 200 μM GA, 5 mM SA, 100 μM ABA, and then incubated at 23 °C for 24 h. To test the effects of the different defense-related stimuli on the activity of Psu-miR475b promoter, the aerial parts between the second and fourth leaves of the in vitro transgenic P. suaveolens plants were sprayed with 200 μM MeJA, 200 μM GA, 5 mM SA or 100 μM ABA solutions, and the poplar samples were then harvested at time points 0, 6, 12, 24, 36 and 48 h post-treatment. All these treatments were described by our previous studies56, and untreated plantlets and plantlets treated with distilled water were used as the controls.
Histochemical and fluorometric GUS assay
For histochemical staining of GUS, fresh tissue samples were dissected from tobacco plants and immediately subjected to the X-Gluc solution56. After overnight incubation at 37 °C, stained samples were bleached with 70% (v/v) ethanol and observed with OLYMPUS BX61 and SZX12 microscopes.
A fluorometric GUS assay was performed as previously described57 and the tobacco stem tissues were ground in liquid nitrogen and homogenized in freshly prepared GUS extraction buffer (50 mM NaH2PO4, pH 7.0, 10 mM EDTA, 0.1% Triton X-100, 0.1% (w/v) sodium laurylsarcosine, 10 mM β-mercaptoethanol). After centrifuging for 15 min at 12,000 rpm at 4 °C, the GUS activity of the supernatant was determined using 4-methylumbelliferyl glucuronide (4-MUG) as a substrate. The fluorescence of the GUS-catalyzed hydrolysis reaction product, 4-methylumbelliferone (4-MU), was measured with the TECAN GENios system. Protein concentration in supernatant was assessed by the Bradford method58, using bovine serum albumin as a standard. GUS activity was normalized to the protein concentration of each supernatant extract and calculated as pmol of 4-MU per milligram of soluble protein per minute.
Gene expression analysis
Genomic DNA was extracted from the mature leaves of P. suaveolens with a Plant Genomic DNA Kit (TIANGEN, Beijing, China). Total RNA was isolated from the tested samples using SV Total RNA Isolation System (Promega, Madison, WI, USA), and treated with RNAse-free DNAse I to eliminate the residual genomic DNA, according to the manufacturer’s instructions (Promega, Madison, WI, USA).
Relative quantification of the expressions for Psu-miR475b and its targets, and GUS gene by quantitative real-time PCR (qRT-PCR) were performed on 7500 Real-Time PCR System, by using MiRcute miRNA SYBR Green Kit (TianGen, Beijing, China) and SuperScriptTM III Platinum® Two-Step qRT-PCR Kit with SYBR® Green (Invitrogen, Carlsbad, CA, USA), respectively. According to the previous reports59,60 and our generated Solexa sequencing (data not shown), miR167e and miR168a-3p were used as inner references for Psu-miR475b, and poplar ACTIN gene as endogenous reference gene for miR475b-targets27. Data were from at least three quantitative PCR replicates per sample and three biological replicates. The specific primers of Psu-miR475b and its target genes, GUS gene and reference gene are shown in Supplementary Table S2.
Prediction of cis-acting elements within Psu-miR475b promoter
The putative cis-elements of Psu-miR475b promoter were identified by online programs including Plant CARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/), PLACE (http://www. dna.affrc.go.jp/ PLACE/ signalup.html), TESS (http://www.cbil.upenn.edu/cgi-bin/tess/tess) and PWM (http://users.soe.ucsc.edu/~kent/improbizer/motifMatcher.html).
Additional Information
How to cite this article: Niu, J. et al. Cross-talk between freezing response and signaling for regulatory transcriptions of MIR475b and its targets by miR475b promoter in Populus suaveolens. Sci. Rep. 6, 20648; doi: 10.1038/srep20648 (2016).
Supplementary Material
Acknowledgments
This research was supported by the National Natural Sciences Foundation of China (No. 31270698).
Footnotes
Author Contributions Experiments were designed by J.N., J.W. and S.L. H.H., Y.C. and R.S. cloned and analyzed freezing-responsive Psu-miR475b and its promoter. J.A. and J.C. participated in transgenic plants. Z.S. and X.L. conducted some sequencing analysis and cis-acting regulatory elements characterization. J.N. and J.W. prepared Figures 1–4 and Figures S1–S4. JN wrote the manuscript with editorial assistance from S.L. All authors have read and approved the manuscript.
References
- Thomashow M. F. Plant cold acclimation: freezing tolerance genes and regulatory mechanisms. Annu Rev Plant Biol 50, 571–599 (1999). [DOI] [PubMed] [Google Scholar]
- Chinnusamy V., Zhu J. & Zhu J. K. Cold stress regulation of gene expression in plants. Trends Plant Sci 12, 444–451 (2007). [DOI] [PubMed] [Google Scholar]
- Fowler S. & Thomashow M. F. Arabidopsis transcriptome profiling indicates that multiple regulatory pathways are activated during cold acclimation in addition to the CBF cold response pathway. Plant Cell 14, 1675–1690 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannah M. A., Heyer A. G. & Hincha D. K. A global survey of gene regulation during cold acclimation in Arabidopsis thaliana. PLoS Genet 1, e26 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee B.-h., Henderson D. A. & Zhu J. K. The Arabidopsis cold-responsive transcriptome and its regulation by ICE1. Plant Cell 17, 3155–3175 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedict C., Geisler M., Trygg J., Huner N. & Hurry V. Consensus by democracy. Using meta-analyses of microarray and genomic data to model the cold acclimation signaling pathway in Arabidopsis. Plant Physiol 141, 1219–1232 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K. & Shinozaki K. Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu Rev Plant Biol 57, 781–803 (2006). [DOI] [PubMed] [Google Scholar]
- Chinnusamy V., Zhu J. K. & Sunkar R. Gene Regulation During Cold Stress Acclimation in Plants in Plant Stress Tolerance, Vol. 639 (ed. Ramanjulu S.) 39–55 (Springer, 2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S. et al. Regulation of the Arabidopsis CBF regulon by a complex low-temperature regulatory network. Plant J 82, 193–207 (2015). [DOI] [PubMed] [Google Scholar]
- Bartel D. P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116, 281–297 (2004). [DOI] [PubMed] [Google Scholar]
- Jones-Rhoades M. W., Bartel D. P. & Bartel B. MicroRNAs and their regulatory roles in plants. Annu Rev Plant Biol 57, 19–53 (2006). [DOI] [PubMed] [Google Scholar]
- Voinnet O. Origin, biogenesis, and activity of plant microRNAs. Cell 136, 669–687 (2009). [DOI] [PubMed] [Google Scholar]
- Sunkar R. & Zhu J. K. Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell 16, 2001–2019 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H. H., Tian X., Li Y. J., Wu C. A. & Zheng C. C. Microarray-based analysis of stress-regulated microRNAs in Arabidopsis thaliana. RNA 14, 836–843 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X. F., Wang G. D., Sutoh K., Zhu J. K. & Zhang W. X. Identification of cold-inducible microRNAs in plants by transcriptome analysis. BBA-Gene Regul Mech 1779, 780–788 (2008). [DOI] [PubMed] [Google Scholar]
- Lv D. K. et al. Profiling of cold-stress-responsive miRNAs in rice by microarrays. Gene 459, 39–47 (2010). [DOI] [PubMed] [Google Scholar]
- Tang Z. H. et al. Uncovering small RNA-mediated responses to cold stress in a wheat thermosensitive genic male-sterile line by deep sequencing. Plant Physiol 159, 721–738 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan Y. et al. Comprehensive genome-wide identification and expression profiling of foxtail millet [Setaria italica (L.)] miRNAs in response to abiotic stress and development of miRNA database. Plant Cell Tissa Org 118, 279–292 (2014). [Google Scholar]
- Arenas-Huertero C. et al. Conserved and novel miRNAs in the legume Phaseolus vulgaris in response to stress. Plant Mol Biol 70, 385–401 (2009). [DOI] [PubMed] [Google Scholar]
- Zhang J. Y., Xu Y. Y., Huan Q. & Chong K. Deep sequencing of Brachypodium small RNAs at the global genome level identifies microRNAs involved in cold stress response. BMC Genomics 10, 449 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X. N., Li X. & Liu J. H. Identification of conserved and novel cold-responsive microRNAs in trifoliate orange (Poncirus trifoliata (L.) Raf.) using high-throughput sequencing. Plant Mol Biol Rep 32, 328–341 (2014). [Google Scholar]
- Jiang Q. et al. High-throughput analysis of small RNAs and characterization of novel microRNAs affected by abiotic stress in a local celery cultivar. Sci Hortic 169, 36–43 (2014). [Google Scholar]
- Zhang Y. et al. Identification and characterization of cold-responsive microRNAs in tea plant (Camellia sinensis) and their targets using high-throughput sequencing and degradome analysis. BMC Plant Biol 14, 271 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S., Sun Y. H. & Chiang V. L. Stress-responsive microRNAs in Populus. Plant J 55, 131–151 (2008). [DOI] [PubMed] [Google Scholar]
- Sun R. Z. et al. Cloning and analysis of the low temperature stress-responsive microRNAs from Populus suaveolens. Genomics Appl Biol 30, 204–211 (2011). [Google Scholar]
- Chen L. et al. Genome-wide identification of cold-responsive and new microRNAs in Populus tomentosa by high-throughput sequencing. Biochem Bioph Res Co 417, 892–896 (2012). [DOI] [PubMed] [Google Scholar]
- Hu H. W. et al. Prediction and expressing analysis of the cold stress-responsive miR475 target genes from Populus suaveolens. Genomics Appl Biol 33, 1–6 (2014). [Google Scholar]
- Hu H. H. et al. Characterization of transcription factor gene SNAC2 conferring cold and salt tolerance in rice. Plant Mol Biol 67, 169–181 (2008). [DOI] [PubMed] [Google Scholar]
- Yang C. H. et al. Overexpression of microRNA319 impacts leaf morphogenesis and leads to enhanced cold tolerance in rice (Oryza sativa L.). Plant Cell Environ 36, 2207–2218 (2013). [DOI] [PubMed] [Google Scholar]
- Lee Y. et al. MicroRNA genes are transcribed by RNA polymerase II. EMBO J 23, 4051–4060 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z. X. et al. Expression of Arabidopsis miRNA genes. Plant Physiol 138, 2145–2154 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X. F., Ruan J. H., Wang G. D. & Zhang W. X. Characterization and identification of microRNA core promoters in four model species. PLoS Comput Biol 3, e37 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozawa M., Miura S. & Nei M. Origins and evolution of microRNA genes in plant species. Genome Biol Evol 4, 230–239 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X. Z., Zhang H. Y. & Li L. Alternative mRNA processing increases the complexity of microRNA-based gene regulation in Arabidopsis. Plant J 70, 421–431 (2012). [DOI] [PubMed] [Google Scholar]
- Llave C., Xie Z. X., Kasschau K. D. & Carrington J. C. Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science 297, 2053–2056 (2002). [DOI] [PubMed] [Google Scholar]
- Vaucheret H., Vazquez F., Crété P. & Bartel D. P. The action of ARGONAUTE1 in the miRNA pathway and its regulation by the miRNA pathway are crucial for plant development. Gene Dev 18, 1187–1197 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodersen P. et al. Widespread translational inhibition by plant miRNAs and siRNAs. Science 320, 1185–1190 (2008). [DOI] [PubMed] [Google Scholar]
- Zhu J. K. Reconstituting plant miRNA biogenesis. P Natl Acad Sci USA 105, 9851–9852 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beauclair L., Yu A. & Bouché N. microRNA-directed cleavage and translational repression of the copper chaperone for superoxide dismutase mRNA in Arabidopsis. Plant J 62, 454–462 (2010). [DOI] [PubMed] [Google Scholar]
- Iwakawa H.-O. & Tomari Y. Molecular insights into microRNA-mediated translational repression in plants. Mol cell 52, 591–601 (2013). [DOI] [PubMed] [Google Scholar]
- Liu Q. K., Wang F. & Axtell M. J. Analysis of complementarity requirements for plant microRNA targeting using a Nicotiana benthamiana quantitative transient assay. Plant Cell 26, 741–753 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B. et al. HOS1 regulates Argonaute1 by promoting transcription of the microRNA gene MIR168b in Arabidopsis. Plant J 81, 861–870 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megraw M. et al. MicroRNA promoter element discovery in Arabidopsis. RNA 12, 1612–1619 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui X., Xu S. M., Mu D. S. & Yang Z. M. Genomic analysis of rice microRNA promoters and clusters. Gene 431, 61–66 (2009). [DOI] [PubMed] [Google Scholar]
- Zhao X., Zhang H. Y. & Li L. Identification and analysis of the proximal promoters of microRNA genes in Arabidopsis. Genomics 101, 187–194 (2013). [DOI] [PubMed] [Google Scholar]
- Lin S. Z., Zhang Z. Y. & Lin Y. Z. Comparison of G6PDH activity and LT50 between P. tomentosa and P. suaveolens during freezing acclimation. Forest Stud China 40 (2003). [Google Scholar]
- Lin S. Z. & Zhang Z. Y. in Studies on molecular biology and freezing-tolerance of poplar 146–150 (Environmental Science Press, Beijing, 2004). [Google Scholar]
- Xue X. Y. et al. Interaction between two timing microRNAs controls trichome distribution in Arabidopsis. PLoS Genet 10, e1004266 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H. J. et al. SHORT VEGETATIVE PHASE (SVP) protein negatively regulates miR172 transcription via direct binding to the pri-miR172a promoter in Arabidopsis. FEBS Letters 586, 2332–2337 (2012). [DOI] [PubMed] [Google Scholar]
- Montgomery T. A. et al. Specificity of ARGONAUTE7-miR390 interaction and dual functionality in TAS3 trans-acting siRNA formation. Cell 133, 128–141 (2008). [DOI] [PubMed] [Google Scholar]
- Yao Y. et al. Cloning and characterization of microRNAs from wheat (Triticum aestivum L.). Genome Biol 8, R96 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S. F. et al. Novel and mechanical stress-responsive microRNAs in Populus trichocarpa that are absent from Arabidopsis. Plant Cell 17, 2186–2203 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naqvi A., Choudhury N. & Haq Q. Small RNA-mediated defensive and adaptive responses in plants. Sust Agri Rev 7, 129–160 (2011). [Google Scholar]
- Pandey R., Joshi G., Bhardwaj A. R., Agarwal M. & Katiyar-Agarwal S. A comprehensive genome-wide study on tissue-specific and abiotic stress-specific miRNAs in Triticum aestivum. PloS One 9, e95800 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y. Z., Lin S. Z. & Zhang Z. Y. Tissue Culture and Rapid Propagation of Populous suaveolens. Plant Physiol Commun 40, 463 (2004). [Google Scholar]
- Lei Y. et al. Cloning and expression of three pathogenesis related protein genes from Populus tomentosa. J Northeast Forest Univ 6, 67–75 (2012). [Google Scholar]
- Jefferson R. A., Kavanagh T. A. & Bevan M. W. GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6, 3901–3907 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72, 248–254 (1976). [DOI] [PubMed] [Google Scholar]
- Lin Y. L. & Lai Z. X. Evaluation of suitable reference genes for normalization of microRNA expression by real-time reverse transcription PCR analysis during longan somatic embryogenesis. Plant Physiol Bioch 66, 20–25 (2013). [DOI] [PubMed] [Google Scholar]
- Feng H. et al. Selection of suitable inner reference genes for relative quantification expression of microRNA in wheat. Plant Physiol Bioch 51, 116–122 (2012). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.