Abstract
Solid tumors house an assortment of complex and dynamically changing microenvironments in which signaling events between multiple cell types are known to play a critical role in tumor progression, invasion, and metastasis. To deepen our understanding of this biology, it is desirable to accurately model these structures in vitro for basic studies and for drug screening; however, current systems fall short of mimicking the complex organization of cells and matrix in vivo. Here we demonstrate the generation of spatially-organized 3D hydrogels of cells and matrix produced from a simple concentric flow device in a single step. Multiple cell types are pre-seeded in different spatial domains such as concentric regions of vessel-like tubular structures to reproducibly establish heterotypic cellular environments in 3D. Using macrophages and breast adenocarcinoma cells as an example of a paracrine loop that regulates metastasis, we explored the effects of clinical drug treatments and observed a dose-dependent modulation of cellular migration. This versatile and tunable approach for tissue fabrication will enable a means to study a wide range of microenvironments and may provide a clinically-viable solution for personalized assessment of patient response to therapeutics.
Keywords: tissue engineering, microfluidics, patterned hydrogel, tumor microenvironment
Anti-cancer drugs are typically assayed on tumor cell lines grown on tissue culture plastic with efficacy measured by growth inhibition or cell death. However, tumor progression in vivo is mediated by dynamic microenvironments where spatiotemporal control of signaling between diverse cell populations is responsible for growth and dissemination.[1,2] Metastasis of breast cancer, in particular, is partially regulated by a paracrine loop between tumor cells (TC) and macrophages (Mϕ) in the primary tumor.[3,4] This interaction enhances the motility of both cells and primes the TC to intravasate into the bloodstream, thus playing a key initiating event in disease progression. This heterotypic cell interaction pair has been directly observed in vivo using intravital microscopy[5] and in vitro using a variety of 2D and 3D culture platforms.[6,7] The development of therapeutic regimens that target heterotypic interactions preceding metastasis is an emerging area for development in cancer therapy. However, there is a deficit of in vitro systems that generate reproducible tissue morphology for a quantitative assessment of heterotypic signaling suitable for therapeutic development.
Compared to traditional 2D culture in a petri dish, 3D culture allows more accurate replication of natural tissue and matrix organization.[8–10] In vitro models developed for drug screening have demonstrated differences in cell proliferation, morphology, and drug response for 3D compared to 2D systems.[11,12] Microfluidic devices provide a means to organize 3D microenvironments such as cysts and tubules, which mimic the basic building blocks of epithelial tissue and allow high-surface-area interfaces between chemically or biologically distinct domains of tissue.[9] Kang et al. developed a process in which they vary chemical composition and topography as a fiber is extruded[13] and Onoe et al. have pioneered a hydrodynamically-focusing method for generating cell-encapsulated fibers on a large scale.[7,14] However, single channel fibers are limited in geometry, and rely on post-processing methods to achieve geometric variability and structural control.
In this report, we demonstrate a versatile approach to multi-domain tissue mimetics by extruding peptide-conjugated alginate fibers under controlled flow rates to modulate flapping instabilities.[15] Controlling the fiber arrangement in a single fluidic extrusion step allows integration of multiple cell types in distinct and controllable spatial domains. We demonstrate the scope of this approach for modeling tissue-mimetic interactions in vitro by filling the inner channel of the fiber with macrophages and incorporating tumor cells in the surrounding peptide-modified alginate. We choose to use a human breast adenocarcinoma (MDA-MB-231) cell type and a mouse macrophage (RAW 264.7) cell type as a model system as this cell pair has been demonstrated to interact in vitro and in vivo through a paracrine loop6,16. We characterize the 3D segregation of this cell pair over time and show how pharmacological inhibitors of migration or TC-Mϕ signaling disrupt the normal spatiotemporal organization.
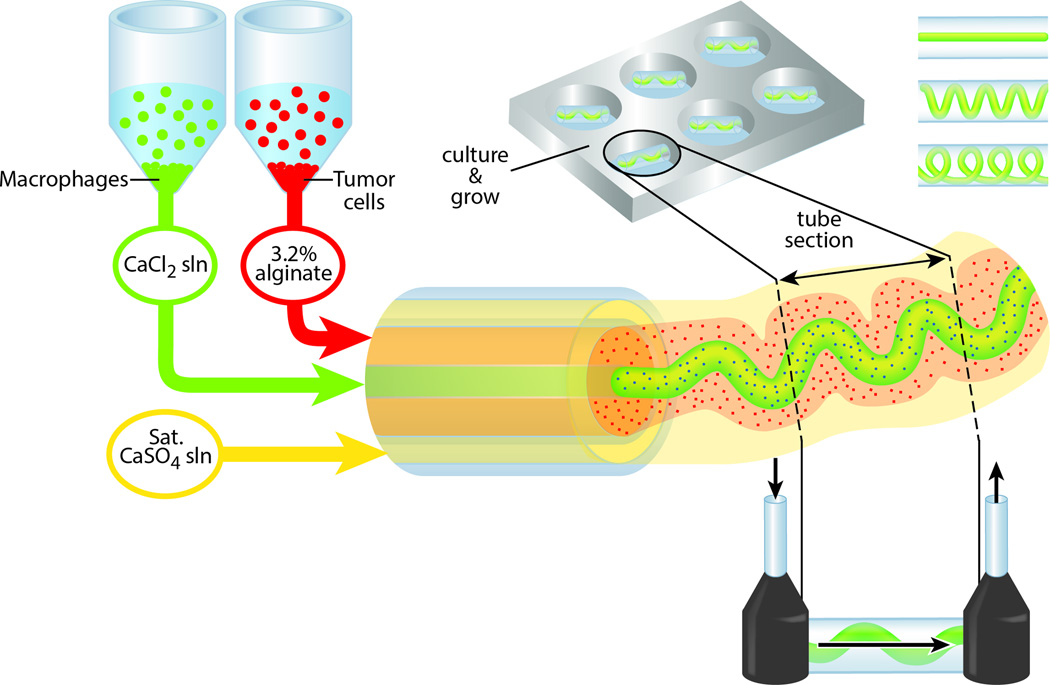
The strategy we present to make high-throughput co-cultured alginate fibers in a single step is illustrated in Figure 1. First, human breast adenocarcinoma (MDA-MB-231; hereafter TC) and mouse macrophage (RAW 264.7; hereafter Mϕ) cells are labeled with CellTracker and then mixed into pre-prepared 3.2% weight alginate and 0.046 g/mL CaCl2 in Dulbecco’s Modified Eagles’ Media respectively. We conjugate the pentapeptide sequence Tyr-Ile-Gly-Ser-Arg (YIGSR) with EDC/NHS to the alginate to support cell-adhesion (Supplementary Figure 1).[17] After being placed in syringe pumps, the solutions are extruded in the microfluidic device in the desired geometry as illustrated in Figure 2, and collected in 45 mg/mL CaCl2 aqueous solution. These fibers are then cut into pieces suitable for mounting as flowable tissue culture chips (Supplementary Figure 2), or used in 96-well plates for long term culture.
Figure 1.
Schematic of dual-cultured hydrogel fiber production.
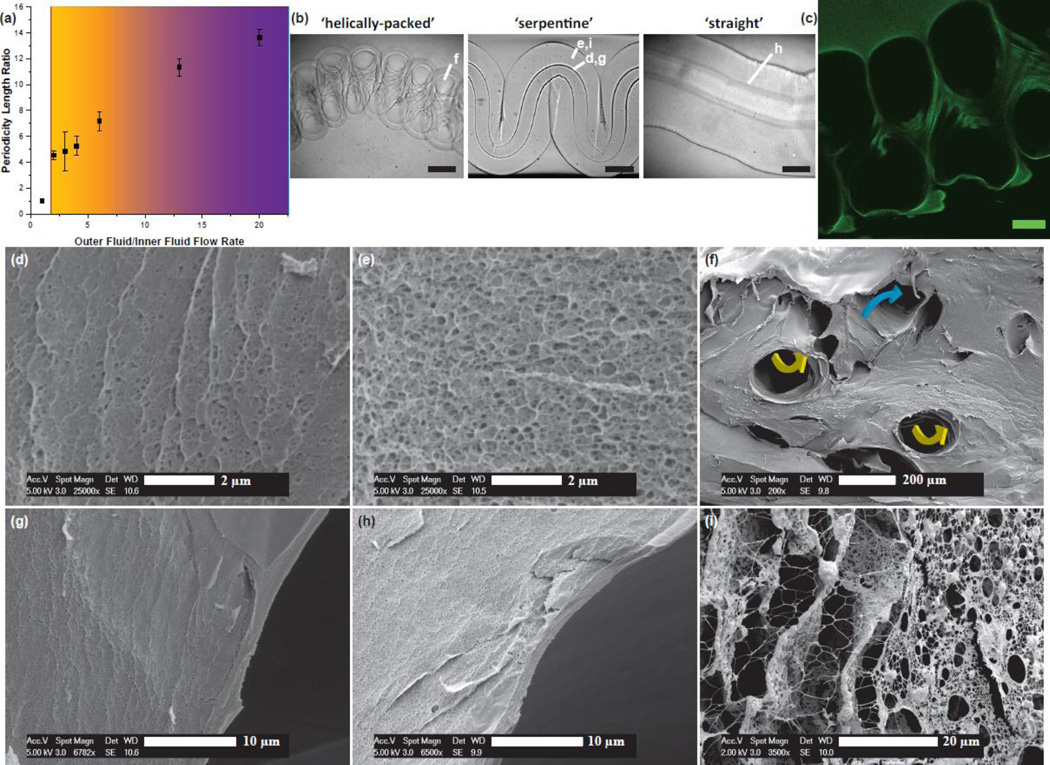
Figure 2.
Fiber architecture characterization and analysis. a, Architecture and pattern amplitude spacing dependence on outer fluid/inner fluid ratio. b, Optical images of select patterned fiber structures with 200 µm scale bars. c, Confocal fluorescence image slice of covalently-conjugated fluorescein to a ‘helically-packed’ fiber with a 50 µm scale bar. (d–i) Calcium-Alginate fibers prior to culturing with cells. d, Close-up ESEM image of tightly-cross-linked inner channel membrane indicating the smaller, partially collapsed pores of the Calcium-Alginate matrix. e, ESEM of the bulk aspect of the alginate fiber matrix. f, Diagonal slice of a patterned alginate fiber showing intersecting inner channel segment with yellow arrows indicating directionality of the vasculature coming out of the plane, and blue arrows indicating a direction into the plane. g, ESEM of patterned fiber close to the opening of the inner channel. h, Macro image of straight fiber near inner channel opening. i, Calcium-Alginate network freeze-dried after culturing with macrophages for four days (control media sample)
Just as vertically extruded soft-serve ice cream twists into swirls when the end is seated in an ice-cream cone, hydrodynamically-focused alginate fibers are manipulated by an analogous push-back force that packs the extruded material into specific hierarchical conformations. Due to the shear-thinning nature of the alginate solution as it is extruded,[18] the solution increases in its ability to bend and twist to accommodate for the stiffness exerted from the gelled, downstream middle fluid. By running the middle fluid at significantly higher volumetric fluid flow rates compared to the outer fluid, the middle fluid tends to pack the extra volume by flapping back and forth in periodic arrangements.[15,19] Based on results shown in Figure 2a, decreases in the outer fluid/inner fluid volumetric flow rates result in tighter packing and form a single concentric co-flow device. We are able to demonstrate ‘straight’, ‘serpentine’, and ‘helically-packed’ architectures by simply changing the flow rate and thus the periodicity. However, this phenomena was only observed at very high flow rates (middle fluid = 20 mL/hr). Using this strategy, we are able to tune the flapping frequency on-the-fly and create hollow-channel fibers with multiple types of patterns on a continuously hollow calcium alginate hydrogel strand (see SI Movie 1). Because sodium alginate is shear-thinning, we use a slower gelating outer fluid (sat. CaSO4) to maintain temporal phase separation behavior until the gelator (45 mg/mL CaCl2) in the inner channel diffuses radially throughout the fiber to ‘lock’ the structures into their respective architecture.
To characterize the geometric structure and porosity of our hollow channels, we use a combination of high speed video recording (Figure 2b), confocal fluorescence microscopy of the hydrogel covalently conjugated to fluorescein (Figure 2c), dynamic mechanical analysis (Supplementary Figure 3), and environmental scanning electron microscopy (ESEM) (Figure 2d–i). ESEM of the freeze-dried alginate, though an indirect measurement of tru porosity in a solvated gel, demonstrates that the relative pore size between samples is homogenous within the bulk structure (Figure 2e) with the exception of an approximately 2 micron crust of tightly-cross-linked hydrogel that surrounds every hollow inner channel (Figure 2g–h). We note that this crust could prove advantageous for applications where several levels of spatial cellular organization are desired (e.g. endothelial perfusion on the channel wall). Inner channel periodicity does not affect cross-linking or porosity in the bulk. Significant structural changes of the hydrogel fibers suggestive of remodeling is seen after 4 days culture with macrophages (Figure 2i).[20] We find that over 90% of encapsulated cells were viable within the first week of culture (Figure 3c), with no significant changes in viability over 3 weeks. Tumor cells in vivo will attract macrophages that secrete epidermal growth factor (EGF) to enhance the metastatic phenotype, thereby priming the tumor cells to intravasate into the vasculature. This paracrine interaction is proposed as a central event mediating metastasis.[3,4,6] We hypothesized that our alginate hydrogels would provide a simple model system of TC-Mϕ co-culture for optimizing pharmacological compounds that disrupt this clinically relevant interaction. To test this hypothesis, we supplemented our co-culture with Gefitinib (GEF), an epidermal growth factor receptor (EGFR) inhibitor,[21] zoledronic acid (ZA), a bisphosphonate that targets osteoclasts and macrophage cells, and a Rac1 inhibitor (RAC) as a broad spectrum modulator of cell migration.[22] We used CellTracker™-labeling and confocal fluorescence microscopy to quantify the co-localization of the tumor cells and macrophages in our co-culture over time (Supplementary Figure 4). At Day 0, the fiber samples are very distinct and the macrophages are exclusively located in the hollow channels of the fibers (Figure 3a). However, after four days of incubation in media or vehicle control, the macrophages became interspersed amongst the entire calcium-alginate hydrogel with a high degree of co-localization with tumor cells. When the co-cultures are treated with GEF, ZA, and RAC, there is a distinct impairment of migration and co-localization with the majority of macrophages remaining in the channel interior.
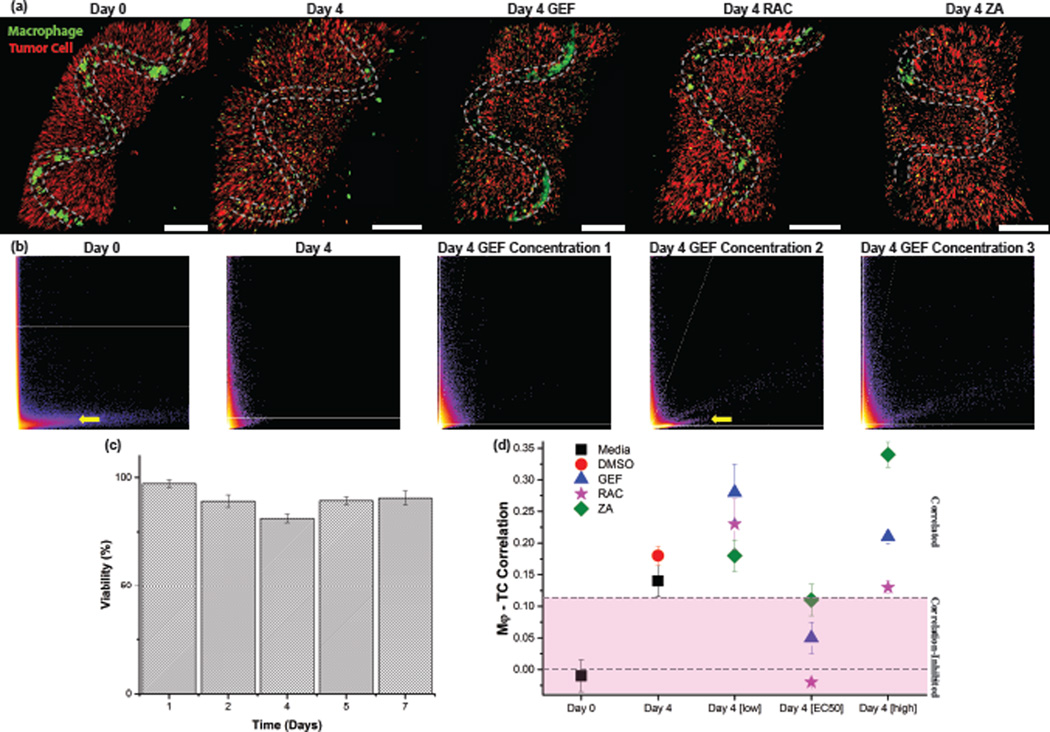
Figure 3.
Co-localization and inhibition analysis of heterotypic co-cultures of macrophage-tumor cells, a, 3-D reconstructions of macrophages (Mϕs) in green and tumor cells (231) in red stained with CellTracker, and the location of the hollow inner channel indicated in dotted grey lines. Scale bar is 400µm. b, Co-localization scatter plots indicating a trend toward correlated pixel values between CellTracker fluorescent channels, which can be partially reversed upon incubation at certain drug concentrations. The yellow arrow indicates the presence of anti-correlated pixel intensities between the two channels. c, Cell viability of co-culture of macrophage and tumor cells in patterned alginate fibers incubated in media over the course of seven days. d, Plot of the calculated Pearson Correlation factors for the three drugs at three concentrations at low, medium[EC50], or high concentrations for three fiber samples averaged. Dashed lines are for visual distinction of zero value correlation and that of the controls.
We quantitated the co-localization of cell-specific CellTracker™ fluorescence signal using the Coloc2 plugin of ImageJ Fiji.[23] At Day 0 when the two cell types are localized in distinct regions, there is a strong anti-correlated band indicated by the yellow arrow in Figure 3b, which indicates a negative calculated correlation factor. A negative correlation factor corresponds to two separate populations of cells, while a high correlation factor corresponds to a well-mixed heterogeneous cell population (Supplemental Figure 7). We use this calculated Pearson correlation as a metric of how well dispersed our two cell populations are, and as a semi-quantitative method to track migration and intermixing of different cell types. After 4 days, when significant numbers of macrophages migrate within the alginate matrix, we see a disappearance of this anti-correlation band and an increase in the calculated correlation factor (Figure 3b). When our cultures were treated with drug concentrations at the approximate EC50 of all three inhibitors (50 µM for ZA and Rac1; 50 nM for GEF), we see the calculated correlation factor decrease. In particular, the inhibitor of Rac1 which is expected to impair cell migration between channels, leads to a return to anti-correlation comparable to initial seeding (Day 0) (Figure 3d). Supplementation of the cultures with drugs well below or above the EC50 fails to abrogate the co-localization.
Next we explored how the ratio of macrophages to tumor cells vary over time in our system (Supplementary Figure 5). There is an increase in the Mϕ/TC ratio from Day 0 to Day 4 in all of the samples, presumable because macrophage growth rates are nearly double that of the TCs.[24,25] By moving from the ‘straight’ fibers to the ‘serpentine’ patterns, the Mϕ/TC ratio increases, which correlates with our fast camera observations. Comparing ‘straight’ fibers with ‘serpentine’ wave-like fibers, we see that the straight fiber samples consistently show higher correlation factors and thus more migration of macrophages to the alginate than their patterned counterparts (Figure 4a). This trend is in contrast to the ratio of Mϕ/TC where the straight fibers result in a 20% decrease in the fraction of macrophages to tumor cells. This is presumably because of the lower volume of the inner channel in the ‘straight’ compared to ‘serpentine’ fibers.
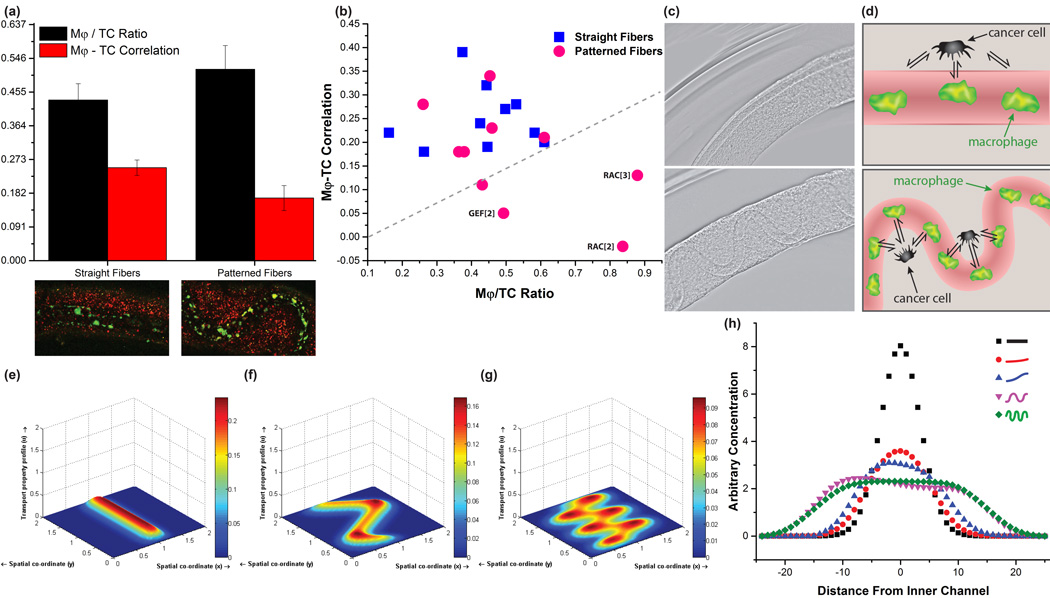
Figure 4.
The influence of geometry on macrophage-tumor cell signaling. a, Comparison of averaged macrophage (Mφ) / tumor cell (TC) ratios to Mφ–TC correlation factors calculated for straight and patterned fibers respectively after drug and media exposure, Mϕ (green), TC (red). T-tests demonstrate statistical significance between the two groups of straight and patterned fibers with a maximum p-value of 0.05 among pharmacological conditions.29 b, Plot relating the Mφ–TC correlation with the Mφ/TC ratio, with labels and a line drawn to illustrate the best performing conditions inhibiting Mφ migration. c, Straight and patterned hollow alginate structures formed in our devices with 200 µm scale bars. d, Cartoon illustrations comparing how the geometric spatiality of cells may affect their signaling in naturally-occurring architectures and model systems. (e–g), Simulations of 2D anisotropic diffusion through Finite Difference Method for channels of e, zero f, one and g, two periodic patterns. h, Cross-sectional plots of simulated diffusion away from inner channels for increasingly periodic patterns.
When we compare the TC-Mϕ correlation to the Mϕ/TC ratio for our fibers treated with pharmacological inhibitors, we can readily identify drug treatments that show the highest inhibition of migration and co-localization (Figure 4b; GEF and RAC). We hypothesize that the different behavior of cells in the ‘straight’ fibers versus the ‘serpentine’ fibers may be on account of increased interactions, not only due to the increase in macrophages, but also from the directionality of the signaling (see Figure 4c). By simulating the diffusion of chemical signaling that may take place from the macrophage positioning within the fiber, it is evident that the patterning periodicity plays a more dominant role in how the diffusant disperses within a fiber than the initial chemical signal concentration (Figure e–h). To supplement the model, we injected Cy3-conjugated streptavidin through the hollow channel and find increased diffusion out of the channel for the serpentine architecture compared to the straight fibers (Supplementary Figure 6). In living systems, structures develop during normal morphogenesis and pathological processes to adopt a breadth of curvilinear and fractal-like forms (e.g. blood vessels, respiratory buds, mammary ducts), where diffusional distances and spatial positioning of cells is critical for function. Our tissue-mimetic fibers may better emulate the non-linear architecture in living systems[26], indicating that this technique may find broad applicability in fabricating model tumor architectures for therapeutic development.
We present a twist on traditional microfluidic concentric flow spinning methods using fiber packing minimization to produce a variety of structures in a single simple device. The ability to quickly tune the packing of vascularized alginate multi-cell tissue scaffolds may lend itself for use as a model system to study other heterotypic interactions. Indeed, we believe this system will prove particularly useful for modeling metastasis because the vessel architecture can be tuned on-the-fly. Not only are the scaffolds easily manufactured, they also offer tremendous potential as model systems for high-throughput screening of drug efficacy, as well as flow-able and vascularized lab-on-a-fiber platforms. We imagine that this packing is not limited to the gelation of calcium-alginate hydrogel fibers but is applicable to a wide range of experimentation required for fast-patterning vasculature in the future.
Experimental Section
Co-culture of adenocarcinoma and macrophage cells within the fibers
MDA-MB-231 human adenocarcinoma cells (ATCC) and RAW 264.7 mouse macrophage cells (ATCC) were cultured in Dulbecco’s modified Eagle’s medium (DMEM, Fisher) supplemented with 10% fetal bovine serum (Life Technologies) and 1% penicillin/streptomycin (p/s), media changed every 2 days and passaged at ~80% confluency using 0.05% Trypsin:EDTA (Life Technologies). CellTracker Green CMFDA dye and CellTracker CM-Dil dye (Life Technologies) were used to label Mϕ and TCs respectively according to manufacturer’s instructions. We dispersed labeled TCs in the alginate solution at a concentration of 5×106 cells/mL. Labeled Mϕ cells were dispersed in CaCl2 solution at a concentration of 4.5×107 cells/mL. After fiber generation, fibers were cut into approximately 20 mm sections and stored in 24-well cell culture plates containing media and pharmacological drugs. For pharmacological inhibition studies, we used Gefitinib (G-4408, LC Labs) at 10, 50, and 100 nM, Zoledronic acid (Cayman Chemical) at 10, 50, 100 µM, and Rac1 Inhibitor II (CAS 1090893-12-1, Calbiochem) at 10, 50, 100 µM concentrations. A vehicle control of 2% DMSO in cell culture media was also used. The cells were incubated at 37°C, 5% CO2 environment, with media changes every 2 days.
Covalently-conjugating alginate fibers
Sodium alginate (71238 Sigma, 1.5 g) was dissolved overnight stirring in 150 mL of PBS at room temperature. EDC (E1769 Sigma, 0.597 g) and sulfo-NHS (56485 Sigma, 0.418 g) were added and stirred for 5 minutes, followed by addition of YIGSR (T7154 Sigma) and stirred for 24 at RT under nitrogen. The solution was dialyzed in Millipore-filtered water for 5 days and lyophilized for 8 days. Conjugation was calculated to be 7% from 1H NMR in D2O (Supplemental Figure 2). To image the fiber patterns in the absence of cellular additives, the 406 mg sections of sodium alginate were added to a solution of 1-ethyl-3-(-3-dimethylaminopropyl) carbodiimide (EDC, 10.5 mg), sulfo-N-hydroxysuccinimide (NHS, 4.83 mg), in 7 mL of PBS and stirred for 5 minutes at room temperature. Then fluoresceinamine (201626 Sigma, 1.4 mg) was added and stirred for 24 hours. The fibers were then washed three times with PBS and fluorescently imaged. These procedures was adapted from Mooney et al.27
Concentric glass capillary microfluidic device manufacture
Glass capillary tubes from Vitrocom were purchased with inner diameters of 100 µm, 700 µm, and 2000 µm. They were glued in a concentric pattern with Loctite 5 Minute Epoxy and washed several times with water and isopropanol.
Producing straight, serpentine, helically-packed architectures
A 3.2% weight solution of sodium alginate was left to gently stir at 3°C for 5 days in PBS for the middle fluid, a 45 mg / mL solution of CaCl2 in media was prepared for the inner fluid, and a saturated solution of CaSO4 in PBS for the outer fluid. The solutions were extruded from Harvard Apparatus PhD 2000 syringe pumps and collected in a bath of inner fluid solution without cells.
Environmental scanning electron microscopy of alginate fibers
After fiber production in the absence of cellular additives, the sections of hydrogel were cut into 20mm sections with 10% ethanol cryo-protectorant and submerged in pressurized liquid nitrogen for 10 minutes, fractured, and then immediately lyophilized in a LABCONCO Freezone 4.5 Liter Freeze Dry system for 36 hours. The fiber sections were then sputter-coated with ~80nm of Au/Pd for imaging.
Cell count and viability assay
Cell viability was measured every day for 7 days. A 20 mm section of cell fiber was collected in a centrifuge tube and suspended in 2 mL of 0.5M ETA solution for 30 minutes at 37 °C to dissolve the alginate followed by the addition of 100 µL 0.05% Trypsin and incubation at 37°C for an additional 5 minutes to form single cell suspension. The solution was centrifuged for 5 min at 300 rcf and the resulting cell pellet was re-suspended in 1mL of fresh cell culture media. A 1:1 mixture of cell suspension and 0.4% Trypan blue solution (Life Technologies) was prepared and counted with a hemocytometer to determine the number of live (unstained) compared to dead (blue stained) cells. 3 counts were averaged for each day.
Cellular migration and correlation analysis
Confocal image stacks of fiber samples were opened in ImageJ Fiji using the Coloc2 plugin with threshold values consistently set throughout samples for the green (Mϕ channels) and the orange (TC channels). The outputs of the plugin are displayed as Pearson Correlation Factors above the threshold values, and the 2-D pixel intensity correlation plot. At least three duplicate samples of each independent experiment were analyzed and ‘t-Test: Two-Sample Assuming Unequal Variances ’ analysis was performed using Microsoft Excel.
Fluorescence imaging of co-cultured fibers
After 4 days in culture, sectioned cell fibers were fixed in 4% paraformaldehyde for 20 minutes and permeabilized with 0.1% Trition X-100 for 30 minutes before blocking with 1% bovine serum albumin (BSA, Sigma) for 1 hour. Cell nucleii labeling was performed in 5% goat serum containing 1% BSA with 4',6-diamidino-2-phenylindole (DAPI, 1:5000 dilution) for 1 hour at room temperature. Fluorescence images were taken on a Zeiss 710 multiphoton confocal microscope.
Simulation of Diffusion
The diffusion equation through finite element analysis was simulated and solved implicitly for a grid mesh of 40 by 50 in 2D, using the central difference as the spatial derivatives. For each initial condition in Figure 4(e–h), the total amount of initial concentration and point sources was kept constant to illustrate the ideal case of comparing the diffusion profiles of different inner channel geometries while keeping simulated number of macrophages constant. Increasing point sources in relation to length of simulated inner channel did not produce significant differences in the diffusion profile shape. The code from the MATLAB FileExchange was adopted and repurposed to accommodate more geometrically relevant models.28
Supplementary Material
Acknowledgments
JMG was funded at UIUC from National Science Foundation Grant 0965918 IGERT: Training the Next Generation of Researchers in Cellular and Molecular Mechanics and BioNanotechnology. This research was supported in part by American Cancer Society Illinois Division Grant #281225. AMS acknowledges support from the NIH (R00CA153914). We thank the Beckman ITG facilities, particularly Dr. Robinson, IGB imaging facilities, Micro and Nanotechnology Laboratory facilities, and Dorothy Loudermilk for artistic guidance and support.
Footnotes
Supporting Information is available from the Wiley Online Library or from the author.
References
- 1.Quail DF, Joyce JA. Microenvironmental regulation of tumor progression and metastasis. Nat Med. 2013;19(11):1423–1437. doi: 10.1038/nm.3394. Available at: http://dx.doi.org/10.1038/nm.3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xu X, Farach-Carson MC, Jia X. Three-dimensional in vitro tumor models for cancer research and drug evaluation. Biotechnol Adv. 2014;32(7):1256–1268. doi: 10.1016/j.biotechadv.2014.07.009. doi: http://dx.doi.org/10.1016/j.biotechadv.2014.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Condeelis J, Singer RH, Segall JE. The great escape: when cancer cells hijack the genes for chemotaxis and motility. Annu Rev Cell Dev Biol. 2005;21:695–718. doi: 10.1146/annurev.cellbio.21.122303.120306. [DOI] [PubMed] [Google Scholar]
- 4.Condeelis J, Pollard JW. Macrophages: obligate partners for tumor cell migration, invasion, and metastasis. Cell. 2006;124(2):263–266. doi: 10.1016/j.cell.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 5.Entenberg D, Wyckoff J, Gligorijevic B, et al. Setup and use of a two-laser multiphoton microscope for multichannel intravital fluorescence imaging. Nat Protoc. 2011;6(10):1500–1520. doi: 10.1038/nprot.2011.376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goswami S, Sahai E, Wyckoff JB, et al. Macrophages promote the invasion of breast carcinoma cells via a colony-stimulating factor-1/epidermal growth factor paracrine loop. Cancer Res. 2005;65(12):5278–5283. doi: 10.1158/0008-5472.CAN-04-1853. [DOI] [PubMed] [Google Scholar]
- 7.Miller JS, Stevens KR, Yang MT, et al. Rapid casting of patterned vascular networks for perfusable engineered three-dimensional tissues. Nat Mater. 2012;11(9):768–774. doi: 10.1038/nmat3357. Available at: http://dx.doi.org/10.1038/nmat3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kilian KA, Bugarija B, Lahn BT, Mrksich M. Geometric cues for directing the differentiation of mesenchymal stem cells. Proc Natl Acad Sci U S A. 2010;107(11):4872–4877. doi: 10.1073/pnas.0903269107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.O’Brien LE, Zegers MMP, Mostov KE. Opinion: Building epithelial architecture: insights from three-dimensional culture models. Nat Rev Mol Cell Biol. 2002;3(7):531–537. doi: 10.1038/nrm859. [DOI] [PubMed] [Google Scholar]
- 10.Shamir ER, Ewald AJ. Three-dimensional organotypic culture: experimental models of mammalian biology and disease. Nat Rev Mol Cell Biol. 2014;15(10):647–664. doi: 10.1038/nrm3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Breslin S, O’Driscoll L. Three-dimensional cell culture: the missing link in drug discovery. Drug Discov Today. 2013;18(5–6):240–249. doi: 10.1016/j.drudis.2012.10.003. [DOI] [PubMed] [Google Scholar]
- 12.Hickman JA, Graeser R, de Hoogt R, et al. Three-dimensional models of cancer for pharmacology and cancer cell biology: capturing tumor complexity in vitro/ex vivo. Biotechnol J. 2014;9(9):1115–1128. doi: 10.1002/biot.201300492. [DOI] [PubMed] [Google Scholar]
- 13.Kang E, Jeong GS, Choi YY, Lee KH, Khademhosseini A, Lee S-H. Digitally tunable physicochemical coding of material composition and topography in continuous microfibres. Nat Mater. 2011;10(11):877–883. doi: 10.1038/nmat3108. [DOI] [PubMed] [Google Scholar]
- 14.Onoe H, Okitsu T, Itou A, et al. Metre-long cell-laden microfibres exhibit tissue morphologies and functions. Nat Mater. 2013;12(6):584–590. doi: 10.1038/nmat3606. [DOI] [PubMed] [Google Scholar]
- 15.Hsu C, Kuang J, Sun J. Flapping Instability of Vertically Impinging Turbulent Plane Jets in Shallow Water. J Eng Mech. 2001;127(5):411–420. [Google Scholar]
- 16.Wyckoff J, Wang W, Lin EY, et al. A paracrine loop between tumor cells and macrophages is required for tumor cell migration in mammary tumors. Cancer Res. 2004;64(19):7022–7029. doi: 10.1158/0008-5472.CAN-04-1449. [DOI] [PubMed] [Google Scholar]
- 17.Massia SP, Rao SS, Hubbell JA. Covalently immobilized laminin peptide Tyr-Ile-Gly-Ser-Arg (YIGSR) supports cell spreading and co-localization of the 67-kilodalton laminin receptor with alpha-actinin and vinculin. J Biol Chem. 1993;268(11):8053–8059. Available at: http://www.jbc.org/content/268/11/8053.abstract. [PubMed] [Google Scholar]
- 18.Becker TA, Kipke DR. Flow properties of liquid calcium alginate polymer injected through medical microcatheters for endovascular embolization. J Biomed Mater Res. 2002;61(4):533–540. doi: 10.1002/jbm.10202. [DOI] [PubMed] [Google Scholar]
- 19.Shelley MJ, Zhang J. Flapping and Bending Bodies Interacting with Fluid Flows. Annu Rev Fluid Mech. 2011;43(1):449–465. [Google Scholar]
- 20.Wiesner C, Le-Cabec V, El Azzouzi K, Maridonneau-Parini I, Linder S. Podosomes in space. Cell Adh Migr. 2014;8(3):179–191. doi: 10.4161/cam.28116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mok TS, Wu Y-L, Thongprasert S, et al. Gefitinib or Carboplatin–Paclitaxel in Pulmonary Adenocarcinoma. N Engl J Med. 2009;361(10):947–957. doi: 10.1056/NEJMoa0810699. [DOI] [PubMed] [Google Scholar]
- 22.Wheeler AP, Wells CM, Smith SD, et al. Rac1 and Rac2 regulate macrophage morphology but are not essential for migration. J Cell Sci. 2006;119(13):2749–2757. doi: 10.1242/jcs.03024. [DOI] [PubMed] [Google Scholar]
- 23.Schindelin J, Arganda-Carreras I, Frise E, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9(7):676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhuang JC, Wogan GN. Growth and viability of macrophages continuously stimulated to produce nitric oxide. Proc Natl Acad Sci U S A. 1997;94(22):11875–11880. doi: 10.1073/pnas.94.22.11875. Available at: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC23642/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Watanabe N, Okochi E, Mochizuki M, Sugimura T, Ushijima T. The Presence of Single Nucleotide Instability in Human Breast Cancer Cell Lines. Cancer Res. 2001;61(21):7739–7742. Available at: http://cancerres.aacrjournals.org/content/61/21/7739.abstract. [PubMed] [Google Scholar]
- 26.Goldsmith D, Ritz E, Covic A. Vascular calcification: A stiff challenge for the nephrologist. Kidney Int. 2004;66(4):1315–1333. doi: 10.1111/j.1523-1755.2004.00895.x. Available at: http://dx.doi.org/10.1111/j.1523-1755.2004.00895.x. [DOI] [PubMed] [Google Scholar]
- 27.Rowley JA, Madlambayan G, Mooney DJ. Alginate hydrogels as synthetic extracellular matrix materials. Biomaterials. 1999;20:45–53. doi: 10.1016/s0142-9612(98)00107-0. [DOI] [PubMed] [Google Scholar]
- 28.Shankar S. Diffusion in 1D and 2D. 2012 [Google Scholar]
- 29.Snedecor G, Cochran W. Statistical Methods. 8th. Iowa State University Press; 1989. [Google Scholar]
- 30.Kawarada H, Hirai A, Odani H, Lida T, Nakajima A. Structure characterization of alginate and conformational behaviors of various alkali-metal alginates in solution. Polym Bull. 1990;24(5):551–557. [Google Scholar]
- 31.Guo L, Wang W, Chen Z, Zhou R, Liu Y, Yuan Z. Promotion of microvasculature formation in alginate composite hydrogels by an immobilized peptide GYIGSRG. Sci China Chem. 2012;55(9):1781–1787. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.