Abstract
Advanced age-related macular degeneration (AMD) is the leading cause of blindness in the elderly with limited therapeutic options. Here, we report on a study of >12 million variants including 163,714 directly genotyped, most rare, protein-altering variant. Analyzing 16,144 patients and 17,832 controls, we identify 52 independently associated common and rare variants (P < 5×10–8) distributed across 34 loci. While wet and dry AMD subtypes exhibit predominantly shared genetics, we identify the first signal specific to wet AMD, near MMP9 (difference-P = 4.1×10–10). Very rare coding variants (frequency < 0.1%) in CFH, CFI, and TIMP3 suggest causal roles for these genes, as does a splice variant in SLC16A8. Our results support the hypothesis that rare coding variants can pinpoint causal genes within known genetic loci and illustrate that applying the approach systematically to detect new loci requires extremely large sample sizes.
Advanced age-related macular degeneration (AMD) is a neurodegenerative disease and the leading cause of vision loss among the elderly affecting 5% of those >75 years of age1,2. The disease is characterized by reduced retinal pigment epithelium (RPE) function and photoreceptor loss in the macula. Advanced AMD is classified as wet (choroidal neovascularization, CNV, when accompanied by angiogenesis) or dry AMD (geographic atrophy, GA, when angiogenesis is absent). These advanced stages of disease are typically preceded by clinically asymptomatic earlier stages3. Advanced AMD is estimated to affect 10 million patients worldwide, reaching >150 million for earlier stages4. At present, understanding of disease biology and therapies remains limited5.
Genetic variants can help uncover disease mechanisms and provide entry points into therapy. Analyses of common variation have uncovered numerous risk loci for many complex diseases (see Web Resources) including 21 loci for AMD6-12. However translation into biological insights remains a challenge, since the functional consequences of disease-associated common variants are typically subtle13 and hard to decipher.
With advances in sequencing technology, genetic analyses are gradually extending to rare variants, which often have more obvious functional consequences14,15 and can thus accelerate translation into biological understanding14,16. For example, identifying multiple disease-associated coding variants (particularly knock-out alleles) in the same gene provides strong evidence that disrupting gene function leads to disease17. So far, studies that implicate specific rare variants in complex diseases either rely on special populations8,18,19, on targeted examinations of a few genes7,9-11,20,21, or on genome-wide assessments of relatively modest numbers of individuals22-25. In contrast, systematic analyses of common variation are now available in hundreds of thousands of phenotyped individuals26,27. Thus, there remains considerable uncertainty about the relative role of rare variants in complex disease and about the sample sizes and study designs that will enable systematic identification of these variants16.
Here, we set out to systematically examine common and rare variation of AMD in the International AMD Genomics Consortium (IAMDGC). The preceding largest study of AMD examined ∼2.4 million variants including ∼18,000 imputed or genotyped protein-altering variants using meta-analysis6. Customizing a chip for de novo centralized genotyping, we analyze >12 million variants including 163,714 directly typed protein-altering variants in 43,566 unrelated subjects of predominantly European ancestry. Our study constitutes a detailed simultaneous assessment of common and rare variation in a complex disease and a large sample, setting expectations for other well-powered studies.
Results
The study data and genomic heritability
We gathered advanced AMD cases with GA and/or CNV, intermediate AMD cases, and control subjects across 26 studies (Supplementary Table 1). While recruitment and ascertainment strategies varied (Supplementary Table 2), DNA samples were collected and genotyped centrally. Making maximal use of genotyping technologies, we utilized a chip with (i) the usual genome-wide variant content, (ii) exome content comparable to the exome chip (adding protein-altering variants from across all exons), and a specific customization to add (iii) protein-altering variants detected by our prior sequencing of known AMD loci (see Methods) and (iv) previously observed and predicted variation in TIMP3 and ABCA4, two genes implicated in monogenic retinal dystrophies. After quality control, we retained 439,350 directly typed variants including a grid of 264,655 primarily non-coding (93%) common variants (frequency among controls >1%) and 163,714 protein-altering variants (including 8,290 from known AMD loci), mostly rare (88% with frequency among controls ≤1%). Imputation to the 1000 Genomes reference panel enabled examining a total of 12,023,830 variants (Supplementary Table 3A). Our final data set included a total of 43,566 subjects consisting of 16,144 advanced AMD patients and 17,832 control subjects of European ancestry for our primary analysis, as well as 6,657 Europeans with intermediate disease and 2,933 subjects with Non-European ancestry (Supplementary Table 3B, Supplementary Figure 1).
Altogether, our genotyped markers accounted for 46.7%28 of variability in advanced AMD risk in the European ancestry subjects (95% confidence interval [CI] 44.5% to 48.8%). Regarding AMD subtypes, estimates for CNV (h2 = 44.3%, CI 42.2% to 46.5%) and GA (h2 = 52.3%, CI 47.2% to 57.4%) were similar; bivariate analyses29 showed genetic correlation of 0.85 (CI 0.78 to 0.92) between disease subtypes.
Thirty-Four Susceptibility Loci for AMD
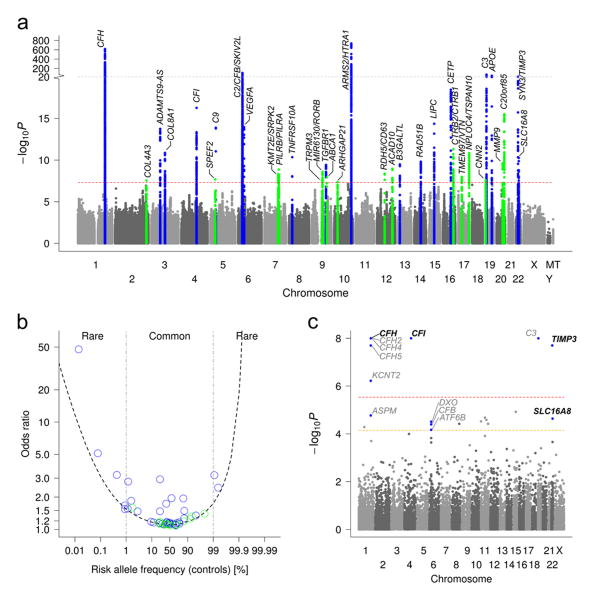
We first conducted a genome-wide single variant analysis of the >12 million genotyped or imputed variants (applying genomic control λ=1.13) comparing the 16,144 advanced AMD patients and 17,832 controls of European ancestry (full results online; see Web resources). We obtained >7000 genome-wide significant variants (P ≤ 5×10–8, Supplementary Figure 2). Sequential forward selection (Supplementary Figure 3) identified 52 independently associated variants at P ≤ 5×10–8 (Supplementary Table 4, Supplementary File 1). These are distributed across 34 locus regions (Figure 1A), each extending across the identified and correlated variants, r2≥0.5, ±500kb (Supplementary Table 5). The 34 loci include 16 loci that reached genome-wide significance for the first time (novel loci, Table 1) and include genes with compelling biology like extra-cellular matrix genes (COL4A3, MMP19, MMP9), an ABC transporter linked to HDL cholesterol (ABCA1), and a key activator in immune function (PILRB). Also included are 18 of the 21 AMD loci that reached genome-wide significance previously6,9 (known loci, Table 1), between-study heterogeneity was low, particularly for the new loci (Supplementary Note 1, Supplementary Table 6, 7).
Figure 1. Genome-wide search reveals 34 loci and genes with rare variant burden for AMD.
(a) We conducted a genome-wide single variant association analysis for >12 million variants in 16,144 advanced AMD patients versus 17,832 controls. Shown is the Manhattan Plot exhibiting P-values for association highlighting novel (P < 5×10–8 for the first time, green) and known (blue) AMD loci (see Table 1). (b) We computed independent effect size (Odds Ratios) of each of the 52 identified variants (Supplementary Table 4). Shown are these effect sizes versus the frequency of the AMD risk increasing allele and a 80% power curve. (c) We conducted a genome-wide gene-based test for disease burden based on the protein-altering variants testing 17,044 RefSeq genes by the variable threshold test51. Shown is the Manhattan Plot with P-values, the red horizontal line indicating genome-wide significance (P ≤ 0.05/17,044 = 2.9×10–6) and the yellow line indicating AMD-locus-wide significance (given 703 genes in the 34 AMD loci, P ≤ 0.05/703 = 7.1×10–5). No gene outside the 34 loci is genome-wide significant; 14 genes are AMD-locus-wide significant (blue), four remain significant after locus-wide conditioning (bold letters, Supplementary Table 11).
Table 1. Thirty-four loci for age-related macular degeneration.
Our genome-wide single-variant association analysis identified 34 loci for advanced AMD with genome-wide significance (P < 5 × 10–8) based on logistic regression in 16,144 cases and 17,832 controls of European ancestry. Shown are P-values and effect sizes (Odds Ratios, OR) for the variant with the smallest P-value per locus (lead variant) and the number of independent signals per locus (see Supplementary Table 4)
| Lead Variant | Chr | Positiona | Major/minor allele | Locus nameb | # Signalsc | MAF | Association | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||||
| Cases | Controls | OR | P | ||||||||
| KNOWN (previously reported with genome-wide significance, P < 5 × 10–8) | |||||||||||
|
| |||||||||||
| rs10922109 | 1 | 196,704,632 | C/A | CFH | 8 | 0.223 | 0.426 | 0.38 | 9.6 × 10–618 | ||
| rs62247658 | 3 | 64,715,155 | T/C | ADAMTS9-AS2 | 1 | 0.466 | 0.433 | 1.14 | 1.8 × 10–14 | ||
| rs140647181 | 3 | 99,180,668 | T/C | COL8A1 | 2 | 0.023 | 0.016 | 1.59 | 1.4 × 10–11 | ||
| rs10033900 | 4 | 110,659,067 | C/T | CFI | 2 | 0.511 | 0.477 | 1.15 | 5.4 × 10–17 | ||
| rs62358361 | 5 | 39,327,888 | G/T | C9 | 1 | 0.016 | 0.009 | 1.80 | 1.3 × 10–14 | ||
| rs116503776 | 6 | 31,930,462 | G/A | C2/CFB/SKIV2L | 4 | 0.090 | 0.148 | 0.57 | 1.2 × 10–103 | ||
| rs943080 | 6 | 43,826,627 | T/C | VEGFA | 1 | 0.465 | 0.497 | 0.88 | 1.1 × 10–14 | ||
| rs79037040 | 8 | 23,082,971 | T/G | TNFRSF10A | 1 | 0.451 | 0.479 | 0.90 | 4.5 × 10–11 | ||
| rs1626340 | 9 | 101,923,372 | G/A | TGFBR1 | 1 | 0.189 | 0.209 | 0.88 | 3.8 × 10–10 | ||
| rs3750846 | 10 | 124,215,565 | T/C | ARMS2/HTRA1 | 1 | 0.436 | 0.208 | 2.81 | 6.5 × 10–735 | ||
| rs9564692 | 13 | 31,821,240 | C/T | B3GALTL | 1 | 0.277 | 0.299 | 0.89 | 3.3 × 10–10 | ||
| rs61985136 | 14 | 68,769,199 | T/C | RAD51B | 2 | 0.360 | 0.384 | 0.90 | 1.6 × 10–10 | ||
| rs2043085 | 15 | 58,680,954 | T/C | LIPC | 2 | 0.350 | 0.381 | 0.87 | 4.3 × 10–15 | ||
| rs5817082 | 16 | 56,997,349 | C/CA | CETP | 2 | 0.232 | 0.264 | 0.84 | 3.6 × 10–19 | ||
| rs2230199 | 19 | 6,718,387 | C/G | C3 | 3 | 0.266 | 0.208 | 1.43 | 3.8 × 10–69 | ||
| rs429358 | 19 | 45,411,941 | T/C | APOE | 2 | 0.099 | 0.135 | 0.70 | 2.4 × 10–42 | ||
| rs5754227 | 22 | 33,105,817 | T/C | SYN3/TIMP3 | 1 | 0.109 | 0.137 | 0.77 | 1.1 × 10–24 | ||
| rs8135665 | 22 | 38,476,276 | C/T | SLC16A8 | 1 | 0.217 | 0.195 | 1.14 | 5.5 × 10–11 | ||
|
| |||||||||||
| NOVEL (reported with genome-wide significance, P < 5 × 10–8, for the first time) | |||||||||||
|
| |||||||||||
| rs11884770 | 2 | 228,086,920 | C/T | COL4A3 | 1 | 0.258 | 0.278 | 0.90 | 2.9 × 10–8 | ||
| rs114092250 | 5 | 35,494,448 | G/A | PRLR/SPEF2 | 1 | 0.016 | 0.022 | 0.70 | 2.1 × 10–8 | ||
| rs7803454 | 7 | 99,991,548 | C/T | PILRB/PILRA | 1 | 0.209 | 0.190 | 1.13 | 4.8 × 10–9 | ||
| rs1142 | 7 | 104,756,326 | C/T | KMT2E/SRPK2 | 1 | 0.370 | 0.346 | 1.11 | 1.4 × 10–9 | ||
| rs71507014 | 9 | 73,438,605 | GC/G | TRPM3 | 1 | 0.427 | 0.405 | 1.10 | 3.0 × 10–8 | ||
| rs10781182 | 9 | 76,617,720 | G/T | MIR6130/RORB | 1 | 0.328 | 0.306 | 1.11 | 2.6 × 10–9 | ||
| rs2740488 | 9 | 107,661,742 | A/C | ABCA1 | 1 | 0.255 | 0.275 | 0.90 | 1.2 × 10–8 | ||
| rs12357257 | 10 | 24,999,593 | G/A | ARHGAP21 | 1 | 0.243 | 0.223 | 1.11 | 4.4 × 10–8 | ||
| rs3138141 | 12 | 56,115,778 | C/A | RDH5/CD63 | 1 | 0.222 | 0.207 | 1.16 | 4.3 × 10–9 | ||
| rs61941274 | 12 | 112,132,610 | G/A | ACAD10 | 1 | 0.024 | 0.018 | 1.51 | 1.1 × 10–9 | ||
| rs72802342 | 16 | 75,234,872 | C/A | CTRB2/CTRB1 | 1 | 0.067 | 0.080 | 0.79 | 5.0 × 10–12 | ||
| rs11080055 | 17 | 26,649,724 | C/A | TMEM97/VTN | 1 | 0.463 | 0.486 | 0.91 | 1.0 × 10–8 | ||
| rs6565597 | 17 | 79,526,821 | C/T | NPLOC4/TSPAN10 | 1 | 0.400 | 0.381 | 1.13 | 1.5 × 10–11 | ||
| rs67538026 | 19 | 1,031,438 | C/T | CNN2 | 1 | 0.460 | 0.498 | 0.90 | 2.6 × 10–8 | ||
| rs142450006 | 20 | 44,614,991 | TTTTC/T | MMP9 | 1 | 0.124 | 0.141 | 0.85 | 2.4 × 10–10 | ||
| rs201459901 | 20 | 56,653,724 | T/TA | C20orf85 | 1 | 0.054 | 0.070 | 0.76 | 3.1 × 10–16 | ||
Chr = Chromosome; MAF = minor allele frequency; OR = Odds Ratio
Chromosomal position is given based on NCBI RefSeq hg19;
The locus name is a label of the region using the nearest gene(s), but does not necessarily state the responsible gene;
number of independent variants in this locus; hg19 = human genome reference assembly (version 19)
Most associated variants are common (45 out of 52) with fully conditioned odds ratios (OR) from 1.1 to 2.9 (Figure 1B, Supplementary Table 4) with two interacting variants (Supplementary Note 2). We also observed seven rare variants with frequencies between 0.01% and 1% and ORs between 1.5 and 47.6 (Figure 1B, Supplementary Table 4). All of these variants were also rare in Non-European ancestries (Supplementary Table 8, extended association results on Non-European in Supplementary File 2). All seven rare variants are located in/near complement genes: four previously described non-synonymous (CFH:Arg1210Cys, CFI:Gly119Arg, C9:Pro167Ser, C3:Lys155Gln)7-11; three others (CFH: rs148553336, rs191281603, rs35292876) described here for the first time including two with the rare allele decreasing the disease risk. . To ensure validity of our results, we verified associations of lead variants in sensitivity analyses that relied on alternate association tests, adjusted for age, gender, or ten ancestry principal components, or were restricted to population-based controls or controls ≥ 50 years of age (data not shown). Altogether, our genome-wide single variant analysis nearly doubles the number of AMD loci and variants.
Prioritizing variants within 52 association signals
It is often challenging to translate common variant association signals into mechanistic understanding of biology; two key challenges are (i) variants with similar signals because of linkage disequilibrium and (ii) subtle functional consequences. Without narrowing lists of candidate variants, follow-up functional experiments are complicated. To prioritize among nearby variants, we computed each variant's ability to explain the observed signal and derived, for each of the 52 signals, the smallest set of variants that included the causal variant with 95% probability 30,31. The 52 credible sets each included from 1 to >100 variants (total of 1,345 variants, Supplementary File 3). Twenty-seven (of 52) sets were small with ≤10 variants (19 with ≤5 variants, Supplementary Table 9); seven sets included only one variant. Among the 205 variants with >5% probability of being causal, we observe 11 protein-altering (all non-synonymous) variants (versus 2 expected assuming 1% protein-altering variants, P for enrichment = 8.7×10–6, Supplementary Table 10). We recognize that the analysis has limitations [for example, when causal variants when the signal is due to a combination of multiple variants, as in the counter example in Supplementary Figure 4].
Rare Variant Association Signals
Analysis of rare variants that alter peptide sequences (non-synonymous), truncate proteins (premature stop), or affect RNA splicing (splice site) can help to identify causal mechanisms – particularly when multiple associated variants reside in the same gene16,32. We examined the cumulative effect of rare protein-altering variants in each ancestry group. Genome-wide, no signal was detected with P ≤ 0.05/17,044 = 2.9×10–6 outside the 34 AMD loci (Figure 1C). Within the 34 loci, we found 14 genes with significant disease burden (P < 0.05/703 genes = 7.1×10–5, Supplementary Table 11). To eliminate settings where a rare variant burden finding is a linkage disequilibrium shadow of a nearby common variant, we re-evaluated each burden signal conditioning on nearby single variants (from Supplementary Table 4). Four of the 14 genes retained P < 0.05/703 = 7.1×10–5 in this analysis (CFH, CFI, TIMP3, SLC16A8; conditioned P = 1.2×10–6, 1.0×10–8, 9.0×10–8, or 3.1×10–6, respectively, Table 2). Sensitivity analyses provide similar (excluding previously sequenced subjects) and extended results (prioritizing variants with high predicted functionality, Supplementary Note 3, Supplementary Table 12).
Table 2. Four genes with a significant rare variant burden within the 34 AMD loci independent from other identified variants.
We computed a gene-based burden test of rare protein-altering variants comparing 16,144 advanced AMD cases and 17,832 controls. Shown are P-values from the variable threshold test (up to 100 million permutations) and Odds Ratios from the collapsed burden test, both adjusted for the other identified variants in the respective locus (locus-wide conditioning). Four genes (among the 703 genes in the 34 AMD locus regions) showed a significant (P < 0.05/703 = 7.1 × 10–5) burden. Details about the corresponding rare variants underlying the observed burden can be found in Supplementary File 4. Results for the 14 genes that show significant burden within the 34 AMD loci without locus-wide conditioning are shown in Supplementary Table 11. Rare variants were defined here as variants with minor allele frequency in cases and controls < 1% in each of the ancestries, European, Asian, and African.
| Gene | Optimal Threshold for Rare Variants Count (%) | Number of Variants below Optimal RAC | Summed Rare Allele Count (Frequency [%]) | Pa | Odds Ratio | |
|---|---|---|---|---|---|---|
|
| ||||||
| Total (Exome Chip Base + Custom) | Cases N = 16,144 | Controls N = 17,832 | ||||
| CFH | 10 (0.015%) | 37 (9+28) | 88 (0.273%) | 38 (0.107%) | 1.2 × 10–6 | 2.94 |
| CFI | 46 (0.068%) | 43 (17+26) | 213 (0.660%) | 82 (0.230%) | 1.0 × 10–8 | 2.95 |
| TIMP3 | 14 (0.021%) | 9 (1+8) | 29 (0.0898%) | 1 (0.00280%) | 9.0 × 10–8 | 31.21 |
| SLC16A8 | 648 (0.954%) | 9 (7+2) | 487 (1.51%) | 392 (1.10%) | 3.1 × 10–6 | 1.40 |
RAC = rare allele count;
P-values are from the variable threshold test conditioned on other identified variants in the locus (locus-wide conditioned).
Several interesting patterns emerge, many of which we owe to our chip design. First, three of the four rare variant burden signals (CFH, CFI, TIMP3) are due to variants with frequency <0.1%, all genotyped (Supplementary File 4). Many human genetic studies have used frequency thresholds of 1% to 5% as a working definition of “rare”, but our data suggests that trait associated variants with clear function may often be much rarer – necessitating very large sample sizes for analysis. In two genes (CFH, CFI), the rare burden was detected because we enriched arrays with variants from previous sequencing of AMD loci10 (54 of 80 variants). The burden findings in CFH (new, Supplementary Note 4) and CFI9 together with variants CFH:Arg1210Cys and CFI:Gly119Arg7,9, corroborate a causal role for these genes in AMD etiology.
The third signal (TIMP3) was in a gene previously associated with Sorsby's fundus dystrophy, a rare monogenic disease with early onset at <45 years of age but with clinical presentation strikingly similar to AMD33,34. Because the majority of Sorsby's alleles disrupt cysteine-cysteine bonds in TIMP3, we arrayed all possible cysteine disrupting sites together with other previously described Sorsby's risk alleles 33,34. The nine rarest TIMP3 variants were cumulatively associated with >30-fold increased risk of disease. TIMP3 resides in an established AMD locus5,35 targeted in previous sequencing efforts32,35 that were too small to evaluate rare variation on this scale (1 variant in 17,832 controls versus 29 variants in 16,144 cases). Interestingly, although Sorsby-associated TIMP3 variants typically occur in exon 5, four of the unpaired cysteine residues we observed map to other exons – perhaps because unpaired cysteines in different locations impair protein folding in different ways. AMD cases with these rare TIMP3 risk alleles still exhibited higher counts of AMD risk alleles across the genome than controls, suggesting that TIMP3 is not a monogenic cause of AMD but contributes to disease together with alleles at the other risk loci. Our finding illustrates a locus where complex and monogenic disorders arise from variation in the same gene, similar to MC4R and POMC in obesity36 or UMOD in kidney function37. In a similar approach, we analyzed 146 rare protein-altering variants in ABCA4, a gene underlying Stargardt disease38, but found no association (P=0.97).
The rare variant burden signal in SLC16A8 was primarily driven by a putative splice variant (c.214+1G>C, rs77968014, minor allele frequency among controls, CAF = 0.81%, OR = 1.5, imputed with R2=0.87, Supplementary File 4). This is not a burden from multiple rare variants, but a single variant emerging as significant due to the reduced multiple testing from gene-wide testing (single variant association P = 9.1×10-6, conditioned on rs8135665 P = 1.3×10-6). This variant is interesting as it is predicted to disrupt processing of the encoded transcript (as +1 G variant, Human Splicing Finder 3.0). SLC16A8 encodes a cell membrane transporter, involved in transport of pyruvate, lactate and related compounds across cell membranes39. This class of proteins mediates the acidity level in the outer retinal segments, and SLC16A8 gene knock-out animals have changes in visual function and scotopic electroretinograms, but not overt retinal pathology40. Interestingly, a progressive loss of SLC16A8 expression in eyes affected with GA was reported with increasing severity of disease41. In summary, our chip design and our large data set enabled us not only to detect interesting features of AMD genetics, but also to provide guidance for future investigations on rare variants.
From Disease Loci to Biological Insights
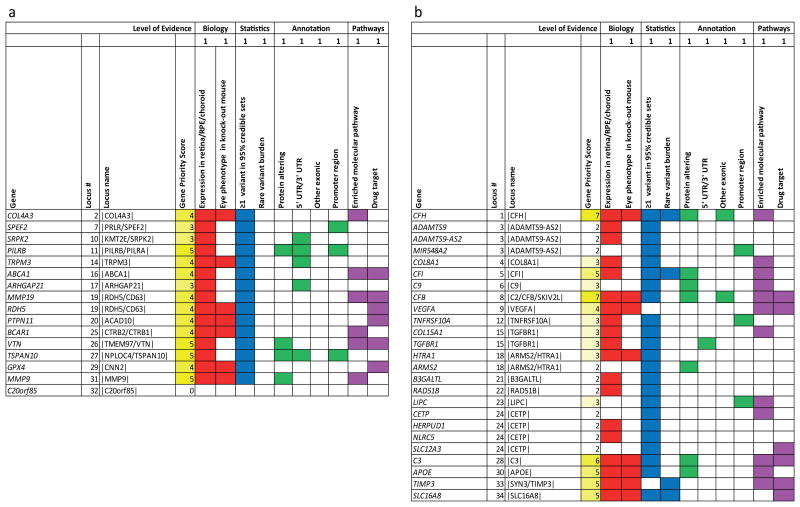
Many analyses can further narrow the list of candidate genes in our loci. We annotated the 368 genes closest to our 52 association signals (index variant and proxies, r2 ≥0.5, ±100kb, Supplementary File 5), noting among these the genes those that contained associated credible set variants (Supplementary File 3) or a rare variant burden (Table 2) – these are the highest priority candidates, consistent with previous analysis of putative cis-regulatory variants42. We further checked whether genes were expressed in retina (82.6% of genes) or RPE/choroid (86.4%, Supplementary File 6). We sought relevant eye phenotypes in genetically modified mice (observed in 32 of the 368 queried genes, Supplementary File 7). We tagged genes in biological pathways enriched across loci, such as the alternative complement pathway, HDL transport, and extracellular matrix organization and assembly (Supplementary Table 13) – highlighting genes that connect multiple pathways (COL4A3/COL4A4, ABCA1, MMP9, and VTN). We also highlighted genes that were approved or experimental drug targets (31 of the 368 queried, Supplementary File 8). Finally, we prioritized genes where at least one of the credible set variants (Supplementary File 3) was protein-altering or located in a putative functional region (promoter, 3′/5′ UTR).
All this information is summarized in the gene priority score table (Supplementary File 9, Supplementary Note 5, Supplementary Table 14), which uses a simple customizable scoring scheme to assign priority: the scheme using equal weights for each column assigns highest scores (Figure 2A, Supplementary Table 15) to genes such as master regulators of immune function (PILRB), matrix metalloproteinase genes (MMP9, MMP19), genes involved in in lipid metabolism (ABCA1, GPX4), an inhibitor of the complement cascade (VTN), another collagen gene (COL4A3), a gene causing a developmental monogenic disorder (PTPN11), and a retinol dehydrogenase (RDH5). Six of these are current drug targets (ABCA1, MMP19, RDH5, PTPN11, VTN, GPX4). In the known AMD loci, the highest scores per locus included the usual suspects (CFH, CFI, CFB, C3, and APOE) as well as TIMP3 and SLC16A8 (Figure 2B). This summary of evidence is not amenable to formal statistical enrichment testing, but may help prioritize genes for follow-up functional experiments.
Figure 2. Genes with top priority based on biological and statistical evidence combined.
We queried 368 genes in the 34 narrow AMD regions (index and proxies, r2 ≥0.5, ±100kb) for biological (red; expression in retina/RPE/choroid, Supplementary File 6; ocular mouse phenotype, Supplementary File 7), statistical, (blue; ≥1 credible set variant in gene ±50 kb, Supplementary File 3; rare variant burden, Table 2), putative functional (green; ≥ 1 credible set variant in gene ±50 kb being protein-altering, 5′/3′ UTR, other exonic, or putative promoter, Supplementary File 3), and molecular (magenta; enriched molecular pathway, drug target) evidence. We here focus on the gene(s) with the highest gene priority score (GPS) per locus (full list of genes in Supplementary File 9). Shown are (a) the 16 genes with highest GPS in the 15 novel AMD loci (one novel locus without any gene), and (b) the 25 genes with highest GPS in the 18 known AMD loci. Colored fields indicate yes and GPS counts number of colored fields per row.
Commonalities and differences of advanced AMD subtypes
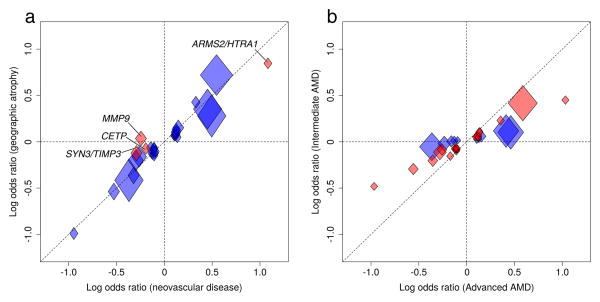
Previously identified risk variants all contribute to the two advanced AMD subtypes, CNV and GA. We compared association signals between our 10,749 cases with CNV and 3,235 cases with GA. Four of the 34 lead variants show significant difference (Pdiff < 0.05/34 = 0.00147) between disease subtypes (in the loci ARMS2/HTRA1, CETP, MMP9, SYN3/TIMP3, Figure 3A, Supplementary Table 16). Variant rs42450006 upstream of MMP9 was the only one that was specific to one subtype, being exclusively associated with CNV (frequency in controls = 14.1%; ORCNV = 0.78 vs. ORGA = 1.04; Pdiff = 4.1×10–10), but not with GA (PGA=0.39, Supplementary Note 6). The MMP9 signal for neovascular disease fits well with prior evidence: upregulation of MMP9 appears to induce neovascularization43 and interacts with VEGF signaling in the RPE44. VEGF currently provides an effective therapy for patients with CNV, but the struggle to keep vision continues. Beyond confirming a shared genetic predisposition of the two subtypes, our data identifies – for the first time – one variant that is specific to one subtype.
Figure 3. Comparison of advanced AMD subtypes and intermediate versus advanced AMD.
We compared associations of the 34 lead variants across different AMD phenotypes. Shown are effect sizes (log Odds Ratio) per minor allele in controls as well as 95% confidence intervals (widths and heights of diamonds). (a) Comparison of neovascular disease (10,749 CNV cases vs. 17,832 controls) and GA (3,235 GA cases vs. 17,832 controls) identified four variants (in loci MMP9, ARMS2/HTRA1, CETP, and SYN3/TIMP3) with significantly different association comparing CNV with GA (Pdiff < 0.05/34, marked in red, see also Supplementary Table 16). (b) Comparison of intermediate AMD (6,657 cases vs. 17,832 controls) with advanced AMD (16,144 cases vs. 17,832 controls) identifies 24 variants with nominally significant (P < 0.05, marked in red) association with intermediate AMD (Pbinomial = 4.8 × 10–24), all of which have the same effect direction and less extreme effect sizes compared to advanced AMD (Supplementary Table 17).
Commonalities and differences of advanced and early AMD
We evaluated our association signals in 6,657 individuals with intermediate AMD, defined as having more than five macular drusen greater than 63μm and/or pigmentary changes in the RPE. Examining all genotyped variants28, we found a correlation of 0.78, indicating substantial overlap between genetic determinants of advanced and intermediate AMD (95% CI 0.69 to 0.87). Among our 34 index variants, 24 showed nominally significant association (Pintermediate ≤ 0.05) with intermediate AMD (2 expected, Pbinomial = 4.8×10–24); all had ORs in the same direction but smaller in magnitude (Figure 3B, Supplementary Table 17). The other 10 variants showed no association with intermediate AMD (Pintermediate > 0.05), despite sufficient power (Supplementary Table 18). Interestingly, these 10 variants point to 7 extra-cellular matrix genes (COL15A1, COL8A1, MMP9, PCOLCE, MMP19, CTRB1/2, ITGA7, Supplementary Table 19), based on which one may hypothesize that the extra-cellular matrix points to a disease subtype without early stage manifestation or with extremely rapid progression. If confirmed, a group of rapidly progressing patients or without early symptoms might eventually derive maximum benefit from genetic diagnosis and future preventive therapies.
An Accounting of AMD Genetics
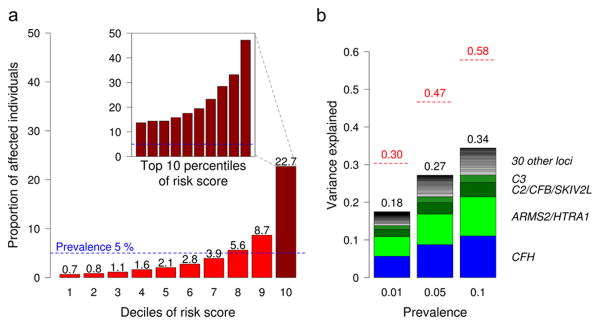
To account for progress made here in understanding AMD genetics, we estimated the proportion of disease risk explained by our 52 independent variants and compared it to our initial estimates of heritability obtained by examining all genotyped variants. We computed a weighted risk score of the 52 variants45 and modeled a population risk score distribution (see Materials and Methods). Individuals in the highest decile of genetic risk have a 44-fold increased risk of developing advanced AMD compared to the lowest decile; of these, 22.7% are predicted to have AMD in an elderly general population above 75 years of age with ∼5% disease prevalence (Figure 4A, Supplementary Table 20). Altogether, the 52 variants explain 27.2% of disease variability (Figure 4B, also highlighting results based on other prevalence assumptions), including a 1.4% contribution from rare variants. The 52 identified variants thus explain more than half of the genomic heritability; the balance might be attributed to additional variation not studied here, or to genetic interaction with environmental factors such as smoking, diet or sunlight exposure.
Figure 4. Variance explained and absolute risk of disease based on the 52 identified variants.
(a) Absolute disease risk (=proportion of affected) by genetic risk score intervals (deciles and top 10 percentiles in embedded bar plot) based on our cases-control-data weighted to model a general population with 5% disease prevalence (see also Supplementary Table 20). (b) Shown is disease liability explained by the 52 identified variants (bars) compared to the genomic heritability based on all genotyped variants (red lines) assuming disease prevalence of 1%, 5%, or 10%, respectively.
Discussion
We set out to improve our understanding of rare and common genetic variation for macular degeneration biology, to guide the development of therapeutic interventions and facilitate early diagnosis, monitoring and prevention of disease. We systematically examine rare variation (through direct genotyping) and common variation (through genotyping and imputation) for AMD in a study designed to discover >80% of associated protein-altering variants with an allele frequency of >0.1% and >3-fold increased disease risk (or >0.5% frequency and >1.8-fold increased disease risk). Our study provides a simultaneous assessment of common and rare variation enabling us to understand the relative roles of rare and common variants and the scientific insights to be gained from rare variation.
Rare protein-altering variants are an attractive target for genetic studies because most of these variants are expected to damage gene function. Furthermore, observing that many rare variants in a gene are, together, associated with a change in disease risk strongly suggests that the gene is causally implicated in disease biology and – further – suggests the consequences of mimicking or blocking gene action using a drug. Our study demonstrates that when rare variants are systematically assessed genome-wide, significant signals can be assigned to single rare variants as well as to rare variant burden in individual genes.
Our study also demonstrates the challenges of these analyses. For three of the genes where we identified a rare variant burden, the accumulated evidence was spread across very rare variants with frequencies <0.1% in controls. Most of these variants derived from sequencing AMD patients. This emphasizes the value of a hybrid approach with direct targeted sequencing of patient samples for variant discovery, followed by genotyping in larger samples for association analysis. Another conclusion is about required sample sizes: although such rare variants are expected to exist in nearly all genes, no rare variant burden was observed in most of the 34 loci we studied. For these loci, identifying causal mechanisms through the study of rare protein-altering variants will require a combination of more sequencing and even larger sample sizes. While our findings of rare variant burden are predominantly from targeted enrichment, the knowledge about effect sizes and frequencies of contributing variants illustrates that applying the approach genome-wide to detect new loci requires extremely large sample sizes. In our view, a recent estimate that sequencing of 25,000 patients will be needed to identify genes where rare variants have a substantial impact on disease risk is likely to be optimistic, particularly given the fact that effect sizes for AMD risk alleles appear to be larger than for many other complex traits 16.
In addition to corroborating previous reports of rare variants that disrupt genes in the complement pathway and lead to large increases in disease risk, our study also includes two unexpected rare variant findings. First, we show that a putative splice variant in SLC16A8 can greatly increase the risk of age-related macular degeneration – providing strong evidence that the gene is directly involved in disease biology. SLC16A8 is a lactate transporter expressed39 specifically by the RPE; a deficit of lactate transport results in acidification of the retina and photoreceptor dysfunction in Slc16a8 knock-out mice40. Second, we show a >30-fold excess of rare TIMP3 mutations among putative cases of macular degeneration. TIMP3 is an especially attractive candidate that has been the subject of previous, underpowered, genetic association studies.
While it has been hypothesized that studies of rare and low frequency genetic variants will greatly increase the proportion of genetic risk that can be explained, our results don't support this. Our study and others successfully identify many low frequency disease risk alleles, and these provide clues about disease biology, but our results also show that common variants make a much larger contribution to disease risk. Common variants also suggest interesting leads and pathways for future analysis (Supplementary Table 15, Figure 2A), including attractive candidates such as immune regulators (PILRB), genes implicated in mouse ocular phenotypes (MMP9, MMP19, COL4A3, PTPN11, GPX4, and RDH5), and proven drug targets (ABCA1, MMP19, RDH5, PTPN11, VTN, GPX4). In a literature search, we identified no previous candidate gene association studies targeting our novel loci, although several model organism, cellular, and functional studies evaluated potential links between genes in these loci and AMD (highlights of this search in Supplementary Table 15) and a few loci were nominally associated and proposed as candidates in prior genome-wide searches 46,47. As richer functional annotations of the genome48 become available in diverse cell types, systematic assessment of overlap between these and our loci should clarify disease biology.
Our study also suggests additional important observations. While our results show that the majority of genetic risk is shared between GA and CNV, we also identify – for the first time – a variant that is specific to one advanced AMD subtype: a genetic variant near MMP9 is specific to CNV, a candidate gene also supported by prior gene expression analyses in the Bruch's membrane of patients with neovascular disease49. Future efforts extending to longitudinal data might help improve the dissection of pure CNV and pure GA and their genetic make-up even further. If substantiated, the fact that nearly all disease associated variants modulate risk of both CNV and GA has potentially significant therapeutic consequences. It implies that individuals at high risk of CNV are also at high risk of GA. This suggests that therapeutic strategies which mitigate CNV but not GA will only provide temporary relief to patients – who are likely to remain at high risk of developing GA and may still require future interventions to prevent it.
Therefore, our findings have several important implications for future studies of rare variation in human complex traits. First, they clearly emphasize the need for very large sample sizes in population studies: the functionally most interesting variants we identify have frequencies in the range of 0.01 – 1.0% and, despite their strong impact on disease risk, could only be implicated using 10,000s of individuals. Second, they illustrate the value of hybrid approaches, where sequencing is used to detect interesting variants and custom arrays and imputation are used to examine these variants in very large samples. Since all the large effect rare variants we identify reside in or near GWAS loci, as with most complex trait associated rare variants 7-11,20,21,23,50, focused studies around GWAS loci may continue to be a cost-effective compromise. Third, our analysis of cysteine variants in TIMP3 illustrates not only the potential for targeted variant discovery but the critical need to understand the consequences of rare variants when analyzing them together. While very large samples will be needed, our results also show that the effort to extend genetic studies to rare variants is worthwhile as these variants can pinpoint causal genes and advance our understanding of disease biology.
Online Methods
Study data and phenotype
In the International AMD Genomics Consortium (IAMDGC), we gathered 26 studies with each including (i) advanced AMD cases with GA and/or CNV in at least one eye and age at first diagnosis ≥ 50 years, (ii) intermediate AMD cases with pigmentary changes in the RPE or more than five macular drusen greater than 63μm and age at first diagnosis ≥ 50 years, or (iii) controls without known advanced or intermediate AMD. Recruitment and ascertainment strategies varied by study (Supplementary Tables 1 and 2, Supplementary Note 7). All groups collected data according to the Declaration of Helsinki principles. Study participants provided informed consent and protocols were reviewed and approved by local ethics committees.
DNA and chip design
We gathered DNA samples of more than 50,000 individuals. Groups with very limited amounts of available DNA contributed aliquots after whole-genome amplification (8% of subjects).
We utilized a custom-modified HumanCoreExome array by Illumina, Inc., which includes (i) tagging variants across the genome (genome chip content) and (ii) a catalogue of protein-altering variants (exome chip content). Our customization of the array included three additional tiers to enrich for variants from 22 AMD loci implicated by our previous genome-wide association analysis6 based on 19 index variants with genome-wide significance, 3 with consistent effect direction in the replication stage and 4×10–7 ≤ P ≤ 2×10–6) by selecting (iii) tagging variants (pair-wise tagging r2 < 0.8) from Phase I 1000G/HapMap52,53 common variants (minor allele frequency, MAF, ≥ 1 % in European or East Asian individuals) using Tagger implemented in Haploview54 within ±100kb of the 22 index variants expanded to cover all correlated variants (r2 [EUR] > 0.5) and the complete gene (transcript ±1 kb), (iv) protein-altering variants within 500 kb of the 22 index variants as identified from public general population data bases (dbSNP55, the NHLBI Exome Sequencing Project56, the Phase I 1000 Genomes Project, see Web Resources), and (v) protein-altering variants within the 500 kb of the 22 index variants identified by re-sequencing AMD case-control study data (targeted re-sequencing of 2,335 AMD cases and 789 controls10,57 and whole-genome sequencing 60 AMD cases and 60 controls; G. Abecasis and A. Swaroop). The customization further included (vi) the 1,000 top independent (> 2 Mb distant) variants from the previous analysis and additional 100 top variants from each the previous CNV only and the previous GA only analysis, (vii) and 375 variants in ABCA4, including known variants causing Stargardt disease58, benign variants, and those of unknown significance, as well as 10 known and 44 predicted cysteine mutations in TIMP3, motivated by the known variants causing Sorsby's fundus dystrophy33,34 (also B. Weber, personal communication).
Annotation
Variant identifiers were based on NCBI dbSNP v137. Chromosomal position and functional annotation of the variant was based on the NCBI Reference Sequence Human Genome Build 19 (RefSeq hg19)59 and SeattleSeq Annotation 13860 (see Web Resources). We particularly focus on protein-altering variants including non-synonymous coding variants (missense, stop loss, in-frame insertion/deletion, frameshift, premature stop codon) and splice sites. We converted the description of splice site variants to HGVS nomenclature using Mutalyzer version 2.0.beta-3361 (see Web Resources).
Genotypes
We genotyped all subjects centrally at the Center for Inherited Diseases Research (CIDR), Johns Hopkins University School of Medicine, Baltimore, MD, USA. From the 569,645 genotyped variants, our quality control excluded poorly genotyped variants as evidenced by genotype call rates < 98.5% (5.8%), deviations from Hardy-Weinberg equilibrium with P < 10–6 (0.34%), variants that mapped at multiple genome locations (0.25%) or variants failing other criteria, resulting in 521,950 (91.6%) variants passing all quality criteria. After excluding monomorphic variants (15.8%), we yielded 264,655 common variants distributed across autosomes, sex chromosomes, and mitochondria, as well as 163,714 directly genotyped protein-altering variants including 8,290 from previously implicated AMD loci (Supplementary Table 3A). For these variants, genotype call rates averaged 99.9% (99.1% for subjects with amplified DNA).
We phased the autosomal and X-chromosomal genotype data using SHAPEIT (200 states, 2.5 Mb windows)62, then imputed genotypes based on the 1000 Genomes Project63 reference panel (1000G Phase I, version 3, SHAPEIT2 Reference) using MINIMAC64 (reference-based 2.5 Mb chunks, 500 kb buffer regions). We then merged study variants that were excluded during imputation (not found in the reference panel) back into the final data set. We excluded common variants (CAF ≥ 1%) with bad imputation quality, R2 < 0.3, and adopted a more stringent exclusion criterion for rare variants (CAF < 1%), R2 < 0.8, for the initial identification of lead variants. This yielded a total of 12,023,830 genotyped (439,350) or imputed (11,584,480) quality-controlled variants (Supplementary Table 3A).
Analyzed subjects
Using the genomic information for subject-level quality control, we excluded duplicated and related individuals (kinship coefficient (x003D5) ≥ 0.0884, i.e. 3rd degree relatives or closer)65, subjects with discrepancies between reported gender and sex chromosomal information or with atypical sex chromosome configurations66, or subjects with genotyping call rates < 98.5%; we derived ancestry based on the first two principal components using autosomal genotyped variants together with genotype information of the samples from the Human Genome Diversity Project (HGDP)67. Our final data set contained 43,566 successfully genotyped unrelated subjects including 16,144 advanced AMD cases and 17,832 controls of European ancestry, 6,657 intermediate AMD cases of European ancestry, and 2,933 subjects (advanced AMD or controls) of Asian or African ancestries (Supplementary Table 3B).
Genomic heritability and genomic correlation
Combined contribution of genotyped variants to disease was evaluated using a variance-component based heritability analysis68. This analysis used genotypes to build a similarity matrix, summarizing the overall genetic kinship between each pair of individuals, and then examined the correspondence between genetic and phenotypic similarity. We estimated the explained variance on all genotyped, autosomal variants using restricted maximum likelihood (REML) analysis implemented in GCTA28 (see Web Resources). We jointly estimated the contributions of rare (MAF in controls < 1 %) and common (MAF in controls ≥ 1%) genotyped variants by first separately calculating their genetic relationship matrices before adding both to the model. Obtained estimates of variance explained were transformed from the observed scale to the liability scale assuming various levels of disease prevalence68.
We estimated the genomic correlation between disease sub-phenotypes using bivariate REML analyses implemented in GCTA and only included common (MAF in controls ≥ 1%) genotyped variants 29. We compared 10,749 cases with CNV versus 3,325 cases with GA (excluding the 2,070 cases with mixed CNV and GA) and we compared 6,657 intermediate AMD cases with 16,144 advanced AMD cases. For both analyses, we used the control subjects as reference and avoided shared controls between traits by randomly splitting the 17,832 unrelated European control individuals into two sub-samples of 8,916 individuals.
Genome-wide single variant association analysis
Single-variant association tests analyzing the 16,144 advanced AMD cases and 17,832 controls of European ancestry were based on the Firth bias-corrected likelihood ratio test69, which is recommended for genetic association studies that include rare variants70, as implemented in EPACTS (see Web Resources). Analyses were adjusted for two principal components and source of DNA (whole-blood or whole-genome amplified DNA). Allele dosages of the imputed data were utilized, Sensitivity analyses were conducted to evaluate the influence of alternative association tests, alternative covariate adjustment including age or sex, or up to 10 principal components instead of two, as well as the influence of restricting to population-based controls, or to controls aged 50 years or older. Genomic control correction71 was used to account for potential population stratification using all genotyped variants with minor allele count ≥ 20 outside of 20 previously described AMD loci6,9. As usual for genome-wide association studies, we considered P-values ≤ 5 × 10–8 as genome-wide significant.
To identify independently associated variants, we adopted a sequential forward selection approach: We first computed single variant association for each of the > 12 million variants. Then we selected the variant with the smallest P-value and its flanking ±5 Mb region, repeating the process until no genome-wide significant variant (P ≤ 5 × 10–8) was left yielding a number of 10 Mb regions. Within each of these large regions, we re-analyzed each variant conditioning on the top variant, and repeated this process by adding the previously identified genome-wide significant variant(s) within the respective 10 Mb region. This yielded one or more independently associated genome-wide significant variant(s) per 10 Mb region.
A locus region was defined by a genome-wide significant variant and its correlated variants (r2≥ 0.5) ± 500kb; overlapping locus regions were merged to one locus, so some loci contained more than one index variant (details in Supplementary Figure 3).
In order to derive independent effect sizes (log odds ratios) for all identified variants, we computed a fully conditioned logistic regression model including all identified variants.
Bayesian approach to prioritize variants
n order to summarize the statistical evidence of a variant for its association strength, we computed the Bayes factor for each variant, which is a measure of the strength of the association that is comparable irrespective of variant frequency or study sample size. It provides the probability of the genotype configuration at a variant (in cases and controls) under the alternative hypothesis (association) divided by the probability of the genotype configuration under the null hypothesis (no association). It is computed using the association results per variant 72. The posterior probability of each variant is then computed as the Bayes factor relative to the sum of all variants' Bayes factors across one locus region and can be thought of as the relative strength of evidence in favor of each SNP studied in the respective region. This assumes that there is one causal variant per region and that the causal variant is in the analyzed data set.
Expanding to loci with multiple association signals and thus a single alleged causal variant per signal, we used the association results per SNP obtained by conditioning on the other independent variants at that locus for computing the Bayes factor.
We derived 95% credible sets of variants per signal, which is the minimal set of variants, for which the sum of the posterior probabilities accumulates beyond 95%. This approach was recommended for fine-mapping of association signals and for prioritizing variants73. Assuming that there is only one causal variant in an association signal and that the causal variant is contained among the analyzed variants, such a credible set of variants contains the causal variant with 95% probability.
We annotated functionality of the variants in each of the 95% credible sets (see above).
Gene-based burden analysis
Single variant analyses have limited power to depict rare variants with association. Gene-based burden tests evaluating accumulated association from multiple rare variants per gene have been shown to complement such analyses and improve power to detect a burden of disease. We computed the burden of disease using the variable threshold test51 as implemented in EPACTS. These analysis assume that all variants in a gene either increase or decrease disease risk. When variants with opposite directions of effect reside in the same gene, power will be reduced. An analysis with SKAT and SKAT-O, which both allow for variants with opposite directions of effect to reside in the same gene, did not identify additional signals (data not shown).
We focused this analysis on protein-altering variants, since we assumed that the other (not protein-altering) variants would outnumber these predicted deleterious variants by far and would thus dilute a disease burden from the deleterious variants. Assuming a negative selection against such deleterious variants that cause their frequency to be low across ancestries, we restricted our rare variant definition to variants with MAF < 1% (cases and controls combined) in each of our ancestry groups (African, Asian, and European). We utilized the genotypes of these rare protein-altering variants if genotyped directly, or rounded imputed allele dosages to the next best genotype if imputed; imputed variants were restricted to those of highest imputation quality (RSQ >= 0.8).
We assessed statistical significance by adaptive permutation testing with variable thresholds (up to 100 million permutations; minimal P-value = 1 × 10–8)51. When rare variants appear on a haplotype associated with disease through a common variant allele already identified for AMD, the rare variant burden would depict a mere shadow of the already identified variant. Therefore, we repeated the variable threshold test conditioned on the variant(s) identified in the respective locus by single variant analysis (locus-wide conditioning), to unravel a gene-based burden of rare variants independent of risk variants identified in single variants tests.
First, we searched for rare variant disease burden genome-wide applying a genome-wide Bonferroni-corrected significance threshold of 0.05 / 17,044 = 2.9 × 10–6 (17,044 genes genome-wide with at least 1 variant included in the analysis, i.e. with ≥ 1 rare protein-altering variant). In a second view on this, we focused on our 34 identified AMD loci and here applied a significance threshold based on the 703 genes overlapping with the locus regions (P < 0.05 / 703 = 7.1 × 10–5). Odds ratio estimates of the burden were derived by logistic regression using the Wald test on the collapsed burden. As there was an overlap of the sequenced subjects with the chip data subjects, we conducted a sensitivity analysis for the burden test excluding overlapping subjects (see Supplementary Note 8).
Follow-up queries for genes underneath the association signals
In order to derive information for all genes underneath our 52 identified association signals (spread across the 34 AMD loci), we built a gene list containing all genes that overlapped with a more narrow definition of locus regions: We have been using a particularly comprehensive definition of the locus region during the signal identification step (index variants and proxies, r2 ≥ 0.5, ±500kb), to avoid far-reaching linkage disequilibrium that may generate shadow signals (particularly in the light of strong associations in the CFH, C3, C2/CFI, and ARMS2/HTRA1 loci) and to optimally differentiate independent signals within locus. We have also used this wide locus region definition for the rare variant burden test again to fully correct for independent signals in the respective wider locus regions and to be conservative in the multiple testing corrections for the AMD-locus-wide burden test search. However, this wide definition is less adequate when prioritizing genes around the identified signals under the assumption that most protein-altering or regulating variants exert their effects in cis42. We thus focused the gene list for further queries to a more narrow locus region definition (index variants and proxies, r2 ≥ 0.5, ± 100kb) and yield 368 overlapping RefSeq genes (Supplementary File 5).
Gene expression
For the 368 genes in our gene list (see above), we sought to obtain gene expression in relevant tissues, retina, RPE, and choroid, in two independent data sets (see details in Supplementary Note 9). A consensus rating of gene expression observed in the two labs was derived as follows: Expression of a gene in one set of tissues (retina or RPE/choroid) was inferred, if both labs detected expression in the respective set of tissues; if at least one of the labs did not observe expression, the gene was considered as not expressed; gene expression of all other genes (one lab observing expression and the other with missing, or both labs with missing data) was regarded as missing.
Mouse model phenotypes
For the 368 genes in our gene list, we queried the Mouse Genome Informatics (MGI)74 and the International Mouse Phenotyping Consortium (IPMC)75 data bases (see Web Resources), and manually curated results by information from published literature. We determined whether a gene exhibited a relevant eye-phenotype (i.e. retina, RPE, or choroid phenotypes) in established genetic mouse models (knock-out, knock-in, or trans-genic mice).
Enrichment for molecular pathways
For the 368 overlapping genes, we performed functional enrichment analysis using INRICH76 with default settings unless stated otherwise (see Supplementary Note 10). Target intervals of this analysis were the narrow AMD locus regions (index variants and proxies, r2 ≥ 0.5, ± 100kb, Supplementary Table 5). Since there is no consensus approach to pathway analysis, we queried multiple data bases: (i) Kyoto Encyclopedia of Genes and Genomes (KEGG)77, (ii) Reactome78, and (iii) Gene Ontology (GO) Consortium79 (see Web Resources). For example, while KEGG is a manually curated database on metabolic pathways, GO also includes automatic annotations and more comprehensive set of cellular processes and molecular functions.
Drug pathways and targets
In order to derive information on whether the product of a gene among the 368 genes in our gene list was a direct drug target, we searched the DrugBank database (Version 4.1) which contains 4,207 drug targets (= genes) and 7,740 drugs 80(see Web Resources).
Explained variability in disease liability
Based on the 52 identified AMD variants, we estimated the explained proportion of disease liability explained by these variants (see Web Resources)81 using the log Odds Ratio estimates from the model including all 52 identified variants (fully conditioned) to derive independent effect sizes. We compared this proportion explained by the 52 variants with the earlier derived genomic heritability based on all genotyped variants (see above).
Genetic risk score and relative and absolute genetic risk of AMD
For each individual, we computed a genetic risk score (GRS) as the effect size weighted sum of the AMD risk increasing alleles for all 52 independent variants divided by the sum of all effect sizes. To derive a a realistic genetic risk score distribution, we modeled a general population based on our case-control data, which requires an assumption on the prevalence of advanced AMD(see Supplementary Note 11). For this modeled general population, we derived the GRS distribution and its deciles. For the weighting, the log Odds Ratios for each of the 52 variants were derived from the fully adjusted model (including all 52 variants) to assure independence of effect sizes.
We derived relative risk estimates (as Odds Ratios) for each GRS decile with the first decile as reference. This relative risk estimate is independent of the prevalence except that the decile to form the genetic risk groups used the GRS distribution as expected in a general population (which requires a prevalence assumption). We also computed absolute risk estimates per GRS decile as the proportion of advanced AMD cases applying the weights and prevalence assumptions as described above.
Supplementary Material
Acknowledgments
We thank all participants of all the studies included for enabling this research by their participation to these studies. Computer resources for this project have been provided by the High Performance Computing Centers of the University of Michigan and the University of Regensburg. Group-specific acknowledgements can be found in the Supplementary Note. The Center for Inherited Diseases Research (CIDR) Program contract number is HHSN268201200008I. This and the main consortium work were predominantly funded by 1X01HG006934-01 to G.R.A. and R01 EY022310 to J.L.H.
Web Resources
Full GWAS results: http://amdgenetics.org/
The following Web Resources have been utilized:
GWAS catalog http://www.ebi.ac.uk/gwas/home),
Exome Variant Server, NHLBI GO Exome Sequencing Project:
http://evs.gs.washington.edu/EVS/
EPACTS: http://www.sph.umich.edu/csg/kang/epacts/index.html
SHAPEIT: https://mathgen.stats.ox.ac.uk/genetics_software/shapeit/shapeit.html
MINIMAC: http://genome.sph.umich.edu/wiki/Minimac
1000 Genomes Reference Panel:
http://www.sph.umich.edu/csg/abecasis/MACH/download/1000G.2013-09.html
The Human Genome Diversity Project data:
http://genome.sph.umich.edu/wiki/LASER and http://www.hagsc.org/hgdp
SeattleSeq: http://snp.gs.washington.edu/SeattleSeqAnnotation138/index.jsp
Mutalyzer: https://mutalyzer.nl
NCBI Reference Sequence (RefSeq, downloaded December, 2012):
http://www.ncbi.nlm.nih.gov/refseq/
Human Splicing Finder 3.0: http://www.umd.be/HSF3/index.html
PubMed (retrieved November 11, 2014): http://www.pubmed.org
Mouse Genome Informatics (MGI) databases: http://www.informatics.jax.org
International Mouse Phenotyping Consortium Database: https://www.mousephenotype.org
INRICH: http://atgu.mgh.harvard.edu/inrich/
KEGG: Kyoto Encyclopedia of Genes and Genomes (KEGG): http://www.genome.jp/kegg/
MSigDB database v4.0: http://www.broadinstitute.org/gsea/index.jsp
Reactome (downloaded January 12th, 2015): http://www.reactome.org
Gene Ontology (GO) Consortium (downloaded January 12th, 2015): http://geneontology.org
DrugBank (downloaded June 4, 2014): http://www.drugbank.ca
GCTA: http://www.complextraitgenomics.com/software/gcta/
Variance explained by genetic variants: https://sites.google.com/site/honcheongso/software/varexp
Author Contributions
Clinical Ascertainment, Contribution of Samples, Study Coordination, and Data Analysis: Gonçalo R. Abecasis, Anita Agarwal, Jeeyun Ahn, Rando Allikmets, Isabelle Audo, Paul N. Baird, Elisa Bala, Mustapha Benchaboune, Hélène Blanché, John Blangero, Frédéric Blon, Alexis Boleda, Caroline Brandl, Kari E. Branham, Murray H. Brilliant, Kathryn P. Burdon, Melinda S. Cain, Peter Campochiaro, Albert Caramoy, Daniel Chen, David Cho, Itay Chowers, Ian J. Constable, Jamie E. Craig, Angela Cree, Christine Curcio, Margaret DeAngelis, Jean-François Deleuze, Anneke I. den Hollander, Bal Dhillon, Lebriz Ersoy, Lindsay A. Farrer, Sascha Fauser, Henry Ferreyra, Ken Flagg, Johanna R. Foerster, Lars G. Fritsche, Linn Gieser, Bamini Gopinath, Michael B. Gorin, Srinivas Goverdhan, Robyn H. Guymer, Shira Hagbi-Levi, Stephanie A. Hagstrom, Jonathan L. Haines, Janette Hall, Michael A. Hauser, Caroline Hayward, Scott J. Hebbring, John R. Heckenlively, Iris M. Heid, Alex W. Hewitt, Joshua D. Hoffman, Frank G. Holz, Carel B. Hoyng, David J. Hunter, Timothy Isaacs, Sudha K. Iyengar, Matthew P. Johnson, Nicholas Katsanis, Jane Khan, Ivana K. Kim, Terrie E. Kitchner, Caroline C. W. Klaver, Barbara E. K. Klein, Michael L. Klein, Ronald Klein, Jaclyn L. Kovach, Alan M. Kwong, Stewart Lake, Thomas Langmann, Reneé Laux, Yara T. E. Lechanteur, Kristine E. Lee, Thierry Léveillard, Mingyao Li, Helena Hai Liang, Gerald Liew, Danni Lin, Andrew Lotery, Hongrong Luo, David A. Mackey, Guanping Mao, Tammy M. Martin, Ian L. McAllister, J. Allie McGrath, Joanna E. Merriam, John C. Merriam, Stacy M. Meuer, Paul Mitchell, Saddek Mohand-Saïd, Anthony T. Moore, Emily L. Moore, Chelsea E. Myers, Anton Orlin, Mohammad I. Othman, Hong Ouyang, Kyu Hyung Park, Neal S. Peachey, Margaret A. Pericak-Vance, Eric A. Postel, Christina Rennie, Andrea J. Richardson, Guenther Rudolph, José-Alain Sahel, Nicole T. M. Saksens, Debra A. Schaumberg, Tina Schick, Hendrik P. N. Scholl, Stephen G. Schwartz, William K. Scott, Sebanti Sengupta, Humma Shahid, Giuliana Silvestri, R. Theodore Smith, Eric Souied, Emmanuelle Souzeau, Dwight Stambolian, Zhiguang Su, Anand Swaroop, Ava G. Tan, Barbara Truitt, Evangelia E. Tsironi, Cornelia M. van Duijn, Claudia N. von Strachwitz, Brendan J. Vote, Jie Jin Wang, Bernhard H. F. Weber, Daniel E. Weeks, Cindy Wen, Armin Wolf, Zhenglin Yang, John R. W. Yates, Donald Zack, Kang Zhang
Phenotype Committee: Ivana K. Kim (lead), Sudha K. Iyengar (lead), Margaret DeAngelis (lead), Gabriëlle H. S. Buitendijk, Emily Y. Chew, Itay Chowers, Anneke I. den Hollander, Sascha Fauser, Michael B. Gorin, Jonathan L. Haines, Iris M. Heid, Alex W. Hewitt, Caroline C. W. Klaver, Barbara E. K. Klein, Michael L. Klein, Ronald Klein, Thierry Léveillard, Andrew Lotery, Kyu Hyung Park, Jie Jin Wang, Kang Zhang
Data Analysis
Team 1: Quality control of data: Jennifer L. Bragg-Gresham, Margaret DeAngelis, Lars G. Fritsche, Mathias Gorski, Wilmar Igl, Ivana K. Kim
Team 2: single variant analysis: Lars G. Fritsche (lead), Iris M. Heid (lead), Gonçalo R. Abecasis (lead), Wilmar Igl (lead), Jennifer L. Bragg-Gresham, Gabriëlle H. S. Buitendijk, Valentina Cipriani, Margaret DeAngelis, Mathias Gorski, Felix Grassmann, Michelle Grunin, Jonathan L. Haines, Robert P. Igo Jr., Sudha K. Iyengar, Caroline C. W. Klaver, Matthias Olden, Klaus Stark, Xiaowei Zhan
Team 3: pathway and rare variant burden analysis: Lars G. Fritsche (lead), Jessica N. Cooke Bailey (lead), Matthew Schu (lead), Gonçalo R. Abecasis, Milam A. Brantley Jr., Matthew Brooks, Gabriëlle H. S. Buitendijk, Monique D. Courtenay, Margaret DeAngelis, Eiko K. de Jong, Anneke I. den Hollander, Lindsay A. Farrer, Felix Grassmann, Jonathan L. Haines, Iris M. Heid, Joshua D. Hoffman, Wilmar Igl, Robert P. Igo Jr., Sudha K. Iyengar, Yingda Jiang, Margaux A. Morrison, Matthias Olden, Margaret A. Pericak-Vance, Rebecca J. Sardell, William K. Scott, Klaus Stark, Anand Swaroop, Bernhard H. F. Weber, Daniel E. Weeks, Xiaowei Zhan
Team 4: analysis of non-SNP variation: Robert P. Igo Jr. (lead), Sudha K. Iyengar (lead), Paul N. Baird (lead), Gonçalo R. Abecasis, Monique D. Courtenay, Lars G. Fritsche, Jonathan L. Haines
Team 5: functional data analysis: Dwight Stambolian (lead), Bernhard H. F. Weber (lead), Margaret DeAngelis (lead), Sudha K. Iyengar (lead), Valentina Cipriani, Jessica N. Cooke Bailey, Monique D. Courtenay, Eiko K. de Jong, Anneke I. den Hollander, Sascha Fauser, Lars G. Fritsche, Felix Grassmann, Jonathan L. Haines, Caroline Hayward, Iris M. Heid, Wilmar Igl, Denise J. Morgan, Margaux A. Morrison, Rinki Ratnapriya, Chloe M. Stanton, Anand Swaroop, Xiaowei Zhan
Design of Overall Experiment
Gonçalo R. Abecasis, Margaret DeAngelis, Lars G. Fritsche, Jonathan L. Haines, Iris M. Heid, Sudha K. Iyengar, Margaret A. Pericak-Vance, Bernhard H. F. Weber
Genotyping and QC
Kimberly F. Doheny (lead), Jane Romm (lead), Lars G. Fritsche (lead), Mathias Gorski (lead), Gonçalo R. Abecasis, Jennifer L. Bragg-Gresham, Monique D. Courtenay, Felix Grassmann, Jonathan L. Haines, Iris M. Heid, Joshua D. Hoffman, Wilmar Igl, Matthias Olden, Xiaowei Zhan
Writing Team
Lars G. Fritsche (lead), Iris M. Heid (lead), Gonçalo R. Abecasis, Jessica N. Cooke Bailey, Margaret DeAngelis, Jonathan L. Haines, Wilmar Igl, Sudha K. Iyengar, Ivana K. Kim, Dwight Stambolian, Bernhard H. F. Weber
Critical review of manuscript
Gonçalo R. Abecasis, Rando Allikmets, Paul N. Baird, Murray H. Brilliant, Itay Chowers, Jessica N. Cooke Bailey, Margaret DeAngelis, Sascha Fauser, Anneke I. den Hollander, Lindsay A. Farrer, Lars G. Fritsche, Michael B. Gorin, Stephanie A. Hagstrom, Jonathan L. Haines, Caroline Hayward, Iris M. Heid, Alex W. Hewitt, Wilmar Igl, Sudha K. Iyengar, Ivana K. Kim, Caroline C. W. Klaver, Barbara E. K. Klein, Michael L. Klein, Ronald Klein, Thierry Léveillard, Andrew Lotery, Paul Mitchell, Anthony T. Moore, Kyu Hyung Park, Neal S. Peachey, Margaret A. Pericak-Vance, Debra A. Schaumberg, Dwight Stambolian, Anand Swaroop, Jie Jin Wang, Bernhard H. F. Weber, Daniel E. Weeks, John R. W. Yates, Kang Zhang
Steering Committee of IAMDGC consortium
Anand Swaroop, Gonçalo R. Abecasis, Alex W. Hewitt, Murray H. Brilliant, Kang Zhang, Bernhard H. F. Weber, Iris M. Heid, Margaret DeAngelis, Lindsay A. Farrer, Kyu Hyung Park, Ivana K. Kim, Dwight Stambolian, Thierry Léveillard, Andrew Lotery, Itay Chowers, Sudha K. Iyengar, Stephanie A. Hagstrom, Neal S. Peachey, Barbara E. K. Klein, Ronald Klein, Debra A. Schaumberg, Margaret A. Pericak-Vance, Paul Mitchell, Jie Jin Wang, Rando Allikmets, Anthony T. Moore, John R. W. Yates, Jonathan L. Haines, Sascha Fauser, Anneke I. den Hollander, Paul N. Baird, Michael L. Klein, Michael B. Gorin, Daniel E. Weeks, Caroline Hayward, Caroline C. W. Klaver
Senior Executive Committee of IAMDGC consortium
Gonçalo R. Abecasis, Margaret DeAngelis, Jonathan L. Haines, Sudha K. Iyengar, Margaret A. Pericak-Vance, Bernhard H. F. Weber
Accession Code
Data permitted for sharing by respective Institutional Review Boards, and/summary statistics reported in the paper will be archived in the database of Genotypes and Phenotypes (dbGaP) under accession phs001039.v1.p1. Full GWAS summary statistics are available at http://amdgenetics.org/.
Conflicts of Interest
Inventor status for patents held by University of Pittsburgh regarding the 10q26 AMD susceptibility locus (DEW, MBG). VC, ATM and JRW are co-inventors or beneficiaries of patents related to genetic discoveries in AMD. IC serves as a consultant for Novartis, Bayer, Allergan, and Lycored. Royalties for AMD patents held by the University of Regensburg (LGF, BHFW), Royalties for AMD patents held by the University of Michigan (GRA, AS, MIO, KEB), Scientific Advisory Board for Regeneron Genetics Center (GRA). PM holds a consultant position for Bayer Inc. and Novartis Inc. AL has acted as a consultant to Bayer, Allergan, Roche and Novartis Pharmaceuticals. SGS has acted as a consultant to Alimera and Bausch + Lomb and has received writing fees from Vindico.
References
- 1.Smith W, et al. Risk factors for age-related macular degeneration: Pooled findings from three continents. Ophthalmology. 2001;108:697–704. doi: 10.1016/s0161-6420(00)00580-7. [DOI] [PubMed] [Google Scholar]
- 2.Chakravarthy U, Evans J, Rosenfeld PJ. Age related macular degeneration. BMJ. 2010;340:c981. doi: 10.1136/bmj.c981. [DOI] [PubMed] [Google Scholar]
- 3.Ferris FL, et al. A simplified severity scale for age-related macular degeneration: AREDS Report No. 18. Arch Ophthalmol. 2005;123:1570–4. doi: 10.1001/archopht.123.11.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wong WL, et al. Global prevalence of age-related macular degeneration and disease burden projection for 2020 and 2040: a systematic review and meta-analysis. Lancet Glob Health. 2014;2:e106–16. doi: 10.1016/S2214-109X(13)70145-1. [DOI] [PubMed] [Google Scholar]
- 5.Fritsche LG, et al. Age-related macular degeneration: genetics and biology coming together. Annu Rev Genomics Hum Genet. 2014;15:151–71. doi: 10.1146/annurev-genom-090413-025610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fritsche LG, et al. Seven new loci associated with age-related macular degeneration. Nat Genet. 2013;45:433–9. 439e1–2. doi: 10.1038/ng.2578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Raychaudhuri S, et al. A rare penetrant mutation in CFH confers high risk of age-related macular degeneration. Nat Genet. 2011;43:1232–6. doi: 10.1038/ng.976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Helgason H, et al. A rare nonsynonymous sequence variant in C3 is associated with high risk of age-related macular degeneration. Nat Genet. 2013;45:1371–4. doi: 10.1038/ng.2740. [DOI] [PubMed] [Google Scholar]
- 9.Seddon JM, et al. Rare variants in CFI, C3 and C9 are associated with high risk of advanced age-related macular degeneration. Nat Genet. 2013;45:1366–70. doi: 10.1038/ng.2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhan X, et al. Identification of a rare coding variant in complement 3 associated with age-related macular degeneration. Nat Genet. 2013;45:1375–9. doi: 10.1038/ng.2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van de Ven JP, et al. A functional variant in the CFI gene confers a high risk of age-related macular degeneration. Nat Genet. 2013;45:813–7. doi: 10.1038/ng.2640. [DOI] [PubMed] [Google Scholar]
- 12.Arakawa S, et al. Genome-wide association study identifies two susceptibility loci for exudative age-related macular degeneration in the Japanese population. Nat Genet. 2011;43:1001–4. doi: 10.1038/ng.938. [DOI] [PubMed] [Google Scholar]
- 13.Gibson G. Rare and common variants: twenty arguments. Nat Rev Genet. 2011;13:135–45. doi: 10.1038/nrg3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Do R, Kathiresan S, Abecasis GR. Exome sequencing and complex disease: practical aspects of rare variant association studies. Hum Mol Genet. 2012;21:R1–9. doi: 10.1093/hmg/dds387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nelson MR, et al. An abundance of rare functional variants in 202 drug target genes sequenced in 14,002 people. Science. 2012;337:100–4. doi: 10.1126/science.1217876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zuk O, et al. Searching for missing heritability: designing rare variant association studies. Proc Natl Acad Sci U S A. 2014;111:E455–64. doi: 10.1073/pnas.1322563111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vogelstein B, et al. Cancer genome landscapes. Science. 2013;339:1546–58. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Styrkarsdottir U, et al. Severe osteoarthritis of the hand associates with common variants within the ALDH1A2 gene and with rare variants at 1p31. Nat Genet. 2014;46:498–502. doi: 10.1038/ng.2957. [DOI] [PubMed] [Google Scholar]
- 19.Styrkarsdottir U, et al. Nonsense mutation in the LGR4 gene is associated with several human diseases and other traits. Nature. 2013;497:517–20. doi: 10.1038/nature12124. [DOI] [PubMed] [Google Scholar]
- 20.Rivas MA, et al. Deep resequencing of GWAS loci identifies independent rare variants associated with inflammatory bowel disease. Nat Genet. 2011;43:1066–73. doi: 10.1038/ng.952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Flannick J, et al. Loss-of-function mutations in SLC30A8 protect against type 2 diabetes. Nat Genet. 2014;46:357–63. doi: 10.1038/ng.2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cruchaga C, et al. Rare coding variants in the phospholipase D3 gene confer risk for Alzheimer's disease. Nature. 2014;505:550–4. doi: 10.1038/nature12825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Do R, et al. Exome sequencing identifies rare LDLR and APOA5 alleles conferring risk for myocardial infarction. Nature. 2015;518:102–6. doi: 10.1038/nature13917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lange LA, et al. Whole-exome sequencing identifies rare and low-frequency coding variants associated with LDL cholesterol. Am J Hum Genet. 2014;94:233–45. doi: 10.1016/j.ajhg.2014.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Walters RG, et al. A new highly penetrant form of obesity due to deletions on chromosome 16p11.2. Nature. 2010;463:671–5. doi: 10.1038/nature08727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Locke AE, et al. Genetic studies of body mass index yield new insights for obesity biology. Nature. 2015;518:197–206. doi: 10.1038/nature14177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shungin D, et al. New genetic loci link adipose and insulin biology to body fat distribution. Nature. 2015;518:187–96. doi: 10.1038/nature14132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet. 2011;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee SH, Yang J, Goddard ME, Visscher PM, Wray NR. Estimation of pleiotropy between complex diseases using single-nucleotide polymorphism-derived genomic relationships and restricted maximum likelihood. Bioinformatics. 2012;28:2540–2. doi: 10.1093/bioinformatics/bts474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wellcome Trust Case Control, C et al. Bayesian refinement of association signals for 14 loci in 3 common diseases. Nat Genet. 2012;44:1294–301. doi: 10.1038/ng.2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wen X. Bayesian model selection in complex linear systems, as illustrated in genetic association studies. Biometrics. 2014;70:73–83. doi: 10.1111/biom.12112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nejentsev S, Walker N, Riches D, Egholm M, Todd JA. Rare variants of IFIH1, a gene implicated in antiviral responses, protect against type 1 diabetes. Science. 2009;324:387–9. doi: 10.1126/science.1167728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sorsby A, Mason ME. A fundus dystrophy with unusual features. Br J Ophthalmol. 1949;33:67–97. doi: 10.1136/bjo.33.2.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weber BH, Vogt G, Wolz W, Ives EJ, Ewing CC. Sorsby's fundus dystrophy is genetically linked to chromosome 22q13-qter. Nat Genet. 1994;7:158–61. doi: 10.1038/ng0694-158. [DOI] [PubMed] [Google Scholar]
- 35.Abecasis GR, et al. Age-related macular degeneration: a high-resolution genome scan for susceptibility loci in a population enriched for late-stage disease. Am J Hum Genet. 2004;74:482–94. doi: 10.1086/382786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Speliotes EK, et al. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat Genet. 2010;42:937–48. doi: 10.1038/ng.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kottgen A, et al. New loci associated with kidney function and chronic kidney disease. Nat Genet. 2010;42:376–84. doi: 10.1038/ng.568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Allikmets R, et al. Mutation of the Stargardt disease gene (ABCR) in age-related macular degeneration. Science. 1997;277:1805–7. doi: 10.1126/science.277.5333.1805. [DOI] [PubMed] [Google Scholar]
- 39.Halestrap AP. The SLC16 gene family - structure, role and regulation in health and disease. Mol Aspects Med. 2013;34:337–49. doi: 10.1016/j.mam.2012.05.003. [DOI] [PubMed] [Google Scholar]
- 40.Daniele LL, Sauer B, Gallagher SM, Pugh EN, Jr, Philp NJ. Altered visual function in monocarboxylate transporter 3 (Slc16a8) knockout mice. Am J Physiol Cell Physiol. 2008;295:C451–7. doi: 10.1152/ajpcell.00124.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shoshan V, MacLennan DH, Wood DS. A proton gradient controls a calcium-release channel in sarcoplasmic reticulum. Proc Natl Acad Sci U S A. 1981;78:4828–32. doi: 10.1073/pnas.78.8.4828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stranger BE, et al. Patterns of cis regulatory variation in diverse human populations. PLoS Genet. 2012;8:e1002639. doi: 10.1371/journal.pgen.1002639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lambert C, et al. Gene expression pattern of cells from inflamed and normal areas of osteoarthritis synovial membrane. Arthritis Rheumatol. 2014;66:960–8. doi: 10.1002/art.38315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hollborn M, et al. Positive feedback regulation between MMP-9 and VEGF in human RPE cells. Invest Ophthalmol Vis Sci. 2007;48:4360–7. doi: 10.1167/iovs.06-1234. [DOI] [PubMed] [Google Scholar]
- 45.Rudnicka AR, et al. Age and gender variations in age-related macular degeneration prevalence in populations of European ancestry: a meta-analysis. Ophthalmology. 2012;119:571–80. doi: 10.1016/j.ophtha.2011.09.027. [DOI] [PubMed] [Google Scholar]
- 46.Chen W, et al. Genetic variants near TIMP3 and high-density lipoprotein-associated loci influence susceptibility to age-related macular degeneration. Proc Natl Acad Sci U S A. 2010;107:7401–6. doi: 10.1073/pnas.0912702107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Logue MW, et al. A search for age-related macular degeneration risk variants in Alzheimer disease genes and pathways. Neurobiol Aging. 2014;35:1510.e7–18. doi: 10.1016/j.neurobiolaging.2013.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.The Encode Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hussain AA, Lee Y, Zhang JJ, Marshall J. Disturbed matrix metalloproteinase activity of Bruch's membrane in age-related macular degeneration. Invest Ophthalmol Vis Sci. 2011;52:4459–66. doi: 10.1167/iovs.10-6678. [DOI] [PubMed] [Google Scholar]
- 50.Johansen CT, et al. Excess of rare variants in genes identified by genome-wide association study of hypertriglyceridemia. Nat Genet. 2010;42:684–7. doi: 10.1038/ng.628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Price AL, et al. Pooled association tests for rare variants in exon-resequencing studies. Am J Hum Genet. 2010;86:832–8. doi: 10.1016/j.ajhg.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.The International Hapmap Consortium et al. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–61. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.1000 Genomes Project Consortium et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–5. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 55.Sherry ST, et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–11. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tennessen JA, et al. Evolution and functional impact of rare coding variation from deep sequencing of human exomes. Science. 2012;337:64–9. doi: 10.1126/science.1219240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Age-Related Eye Disease Study Research, G. A randomized, placebo-controlled, clinical trial of high-dose supplementation with vitamins C and E and beta carotene for age-related cataract and vision loss: AREDS report no. 9. Arch Ophthalmol. 2001;119:1439–52. doi: 10.1001/archopht.119.10.1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fritsche LG, et al. A subgroup of age-related macular degeneration is associated with mono-allelic sequence variants in the ABCA4 gene. Invest Ophthalmol Vis Sci. 2012;53:2112–8. doi: 10.1167/iovs.11-8785. [DOI] [PubMed] [Google Scholar]
- 59.Pruitt KD, et al. RefSeq: an update on mammalian reference sequences. Nucleic Acids Res. 2014;42:D756–63. doi: 10.1093/nar/gkt1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ng SB, et al. Targeted capture and massively parallel sequencing of 12 human exomes. Nature. 2009;461:272–6. doi: 10.1038/nature08250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wildeman M, van Ophuizen E, den Dunnen JT, Taschner PE. Improving sequence variant descriptions in mutation databases and literature using the Mutalyzer sequence variation nomenclature checker. Hum Mutat. 2008;29:6–13. doi: 10.1002/humu.20654. [DOI] [PubMed] [Google Scholar]
- 62.Delaneau O, Marchini J, Zagury JF. A linear complexity phasing method for thousands of genomes. Nat Methods. 2012;9:179–81. doi: 10.1038/nmeth.1785. [DOI] [PubMed] [Google Scholar]
- 63.Ristau T, et al. Allergy is a protective factor against age-related macular degeneration. Invest Ophthalmol Vis Sci. 2014;55:210–4. doi: 10.1167/iovs.13-13248. [DOI] [PubMed] [Google Scholar]
- 64.Howie B, Fuchsberger C, Stephens M, Marchini J, Abecasis GR. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat Genet. 2012;44:955–9. doi: 10.1038/ng.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Manichaikul A, et al. Robust relationship inference in genome-wide association studies. Bioinformatics. 2010;26:2867–73. doi: 10.1093/bioinformatics/btq559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Turner S, et al. Quality control procedures for genome-wide association studies. Curr Protoc Hum Genet. 2011;Chapter 1:19. doi: 10.1002/0471142905.hg0119s68. Unit1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cavalli-Sforza LL. The Human Genome Diversity Project: past, present and future. Nat Rev Genet. 2005;6:333–40. doi: 10.1038/nrg1596. [DOI] [PubMed] [Google Scholar]
- 68.Lee SH, Wray NR, Goddard ME, Visscher PM. Estimating missing heritability for disease from genome-wide association studies. Am J Hum Genet. 2011;88:294–305. doi: 10.1016/j.ajhg.2011.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Firth D. Bias reduction of maximum likelihood estimates. Biometrika. 1993;80:27–38. [Google Scholar]
- 70.Ma C, Blackwell T, Boehnke M, Scott LJ, Go TDi. Recommended joint and meta-analysis strategies for case-control association testing of single low-count variants. Genet Epidemiol. 2013;37:539–50. doi: 10.1002/gepi.21742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Devlin B, Roeder K. Genomic control for association studies. Biometrics. 1999;55:997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- 72.Stephens M, Balding DJ. Bayesian statistical methods for genetic association studies. Nat Rev Genet. 2009;10:681–90. doi: 10.1038/nrg2615. [DOI] [PubMed] [Google Scholar]
- 73.Wellcome Trust Case Control Consortium et al. Bayesian refinement of association signals for 14 loci in 3 common diseases. Nat Genet. 2012;44:1294–301. doi: 10.1038/ng.2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Blake JA, Bult CJ, Eppig JT, Kadin JA, Richardson JE. The Mouse Genome Database: integration of and access to knowledge about the laboratory mouse. Nucleic Acids Res. 2014;42:D810–7. doi: 10.1093/nar/gkt1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Brown SD, Moore MW. Towards an encyclopaedia of mammalian gene function: the International Mouse Phenotyping Consortium. Dis Model Mech. 2012;5:289–92. doi: 10.1242/dmm.009878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lee PH, O'Dushlaine C, Thomas B, Purcell SM. INRICH: interval-based enrichment analysis for genome-wide association studies. Bioinformatics. 2012;28:1797–9. doi: 10.1093/bioinformatics/bts191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Croft D, et al. The Reactome pathway knowledgebase. Nucleic Acids Res. 2014;42:D472–7. doi: 10.1093/nar/gkt1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ashburner M, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–9. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Law V, et al. DrugBank 4.0: shedding new light on drug metabolism. Nucleic Acids Res. 2014;42:D1091–7. doi: 10.1093/nar/gkt1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.So HC, Gui AH, Cherny SS, Sham PC. Evaluating the heritability explained by known susceptibility variants: a survey of ten complex diseases. Genet Epidemiol. 2011;35:310–7. doi: 10.1002/gepi.20579. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.