Abstract
The analysis of ethical, legal, and social implications (ELSI) associated with genetics (“genethics”) has focused on traditional concerns in bioethics, such as privacy and informed consent. The analysis of ELSI associated with neuroscience (“neuroethics”) has focused on concerns related to personhood, such as free will or cognitive enhancement. With neurogenomics coming of age, this is an appropriate time to attend to the set of novel concerns that arises when we consider the confluence of these two lines of research. I call this area of ethics inquiry “neurogenethics”, map out the problem space, and highlight future areas of inquiry related to genome editing and gene therapy, optogenetics and memory manipulation, and genomic identity and online communities.
1. Introduction
I previously introduced the term “neurogenethics” to characterize an emerging field from its parent disciplines, genethics and neuroethics (Canli, 2015). These parent disciplines share a common concern for traditional bioethics themes such as informed consent, subject safety, data security, and incidental findings. Yet, each also adopted unique concerns, such as the ethical, legal, and social implications (ELSI) of changes to the germline in genethics, and questions regarding personhood, consciousness or free will in neuroethics (Illes et al., 2007, Roskies, 2007). I suggested that a unique set of new ELSI themes emerges when the study of genetics is applied to the study of the brain, because of its privileged status among all organs as the generator of behavior and our sense of Self and Identity. With neurogenomics coming of age, these themes continue to gain in prominence.
2. Themes in neurogenethics
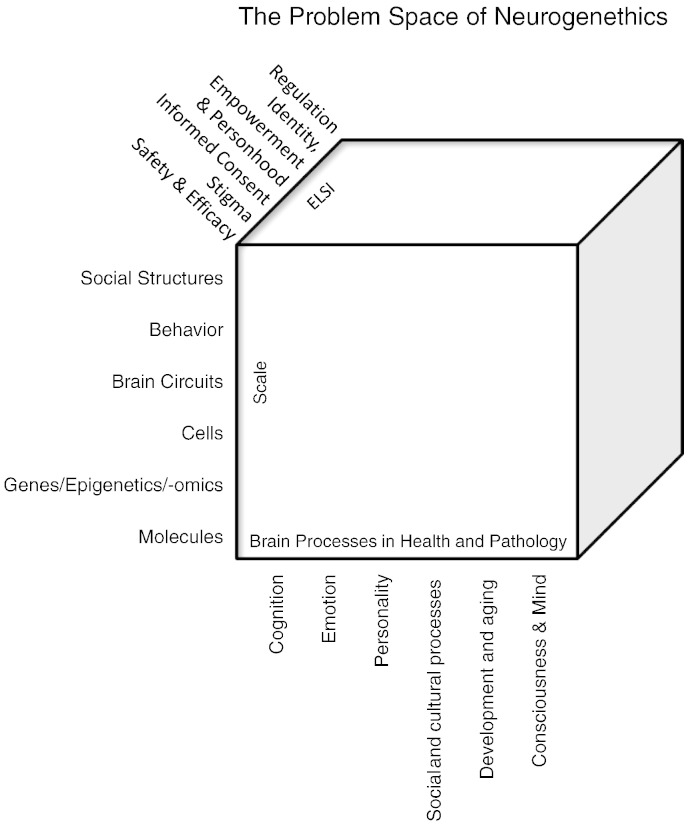
One way to systematically map out what kinds of themes may be unique to neurogenethics is to map out a “problem space”. I have chosen a three-dimensional space organized along scale, brain processes, and ethical-legal-social-implications (Fig. 1, reprinted from Canli, 2015).
Fig. 1.
The problem space of neurogenethics.
Illustrated are three dimensions of the problem space of neurogenetics: scale, brain processes, and ELSI.
Along the dimension of scale, themes can emerge from the level of single molecules to brain circuits, the behaviors they give rise to, and the social structures that emerge from the interactions of large sets of individuals. Along the dimension of brain processes, themes may be related to specific functions, from the narrow and granular (e.g., specific cognitive processes such as memory) to the broad and holistic (e.g., processes related to consciousness and mind). Along the dimension of ELSI, one can again conceptualize a continuum ranging from the narrow and granular (e.g., subject safety) to the broad and holistic (e.g., questions related to regulatory controls of the state versus individual empowerment). I have discussed examples of neurogenethics themes within different quadrants of this problem space elsewhere (Canli, 2015). In the following three sections, I will present new or updated discussions on three specific case studies: genome editing, optogenetics, and genomic identity.
3. Case study 1: genome editing and gene therapy
At the molecular level of scale, new tools in genome editing will likely come into the field of view of neurogenethics, as these tools will be applied to neurodegenerative diseases and behavior. Efforts to accomplish site-specific manipulation of the genome have been catalyzed by the 2012 development of a programmable, RNA-guided DNA endonuclease, the CRISPR–Cas9 system (Jinek et al., 2012). Bacteria such as Escherichia coli use clustered regularly interspaced palindromic repeats (CRISPRs) (Ishino et al., 1987) in a complex with CRISPR-associated (Cas) proteins to fight viral infection (Brouns et al., 2008). (For a brief but detailed review of the history of CRISPR–Cas, see Doudna and Charpentier (2014).) In 2012, Jinek and colleagues then engineered CRISPR into a complex with Cas9 (CRISPR-associated gene 9) (Barrangou et al., 2007, Deltcheva et al., 2011) in a way that a single guide RNA (sgRNA) could direct Cas9 to any DNA sequence of choice (Jinek et al., 2012). The simplicity of the programmable RNA accelerated applications to the point that in less than two years, 1300 publications had used this approach (Doudna and Charpentier, 2014, Maggio and Goncalves, 2015, Xiao-Jie et al., 2015, Pelletier et al., 2015, Hsu et al., 2014).
A recent innovation was the development of “multiplexed” CRISPR-Cas9 for human applications (Konermann et al., 2015). These investigators compiled a library of sgRNAs to target all 70,290 human protein-coding mRNA sequences in the RefSeq database and engineered a CRISPR–Cas9 complex that could activate ten genes simultaneously. With this set of tools, any human protein-coding gene (and sets of up to 10 genes) can be targeted for modification.
Another significant innovation was the application of the CRISPR toolkit to the epigenome (Hilton et al., 2015). These investigators fused a nuclease-null dCas9 protein to the catalytic core of the human acetyltransferase p300, to catalyze acetylation of histone H3 lysine 27 at directed target sites. This tool allows for the targeted transcriptional activation from promoters and enhancers, and regulation of gene expression through epigenomic acetylation. Manipulations of additional epigenomic regulatory mechanisms using the CRISPR–Cas platform are likely to follow.
One application of genome editing by CRISPR–Cas9 will be in gene therapy (Lombardo and Naldini, 2014, Meissner et al., 2014, Wang et al., 2015, Wang and Gao, 2014, Kennedy and Cullen, 2015). Indeed, preclinical studies have already demonstrated its potential utility, such as in HIV-1 provirus (Hu et al., 2014, Ebina et al., 2013) and X-linked severe combined immunodeficiency (Genovese et al., 2014). Of relevance in the context of neurogenomics are potential applications to basic brain functions or neurodegenerative diseases. For example, one study (Swiech et al., 2015) used a sgRNA designed to target the MeCP2 (methyl CpG binding protein 2) gene, which is implicated in Rett syndrome (Chahrour and Zoghbi, 2007). The sgRNA and a Cas9 were packaged into two separate adeno-associated viral (AAV) vectors and injected into the dentate gyrus (DG) of the hippocampus of mice. Following behavioral training, this intervention produced impairments in contextual fear memory that was specific to the learned context and unimpaired in other cognitive or affective tests.
One significant limitation of the CRISPR–Cas9 system in gene therapy has been the challenge of its in vivo delivery, which relies on adeno-associated virus (AAV) vectors. Recombinant AAV (rAAV) vectors have a good safety record, even when injected into the brain (Klein et al., 2002, Tenenbaum et al., 2003, McCown, 2011, Bowers et al., 2011, High and Aubourg, 2011, Weinberg et al., 2013). However, these AAVs can usually only package small genes, approximately 4.5 kb; yet, the Cas9 derived from Streptococcus pyogenes (SpCas9) itself is already 4.2 kb in size. This makes it technically challenging to package both the SpCas9 and the sgRNA into a single AAV vector and leave room for further modifications. This limitation was recently overcome by Zhang's laboratory, which analyzed over 600 Cas9 orthologues to find a smaller Cas9 enzyme for in vivo delivery (Ran et al., 2015). They discovered that Cas9 derived from Staphylococcus aureus (SaCas9), which is more than 1 kb shorter than SpCas9, effectively targeted the intended cholesterol regulatory gene Pcsk9 in the mouse liver, as measured by reduced serum Pcsk9 and total cholesterol levels. Toxicity and off-target analyses also suggested the utility of this novel Cas9 targeting system.
Human applications of gene therapy directed at the brain are currently limited, because they are invasive (injection of the AAV vector directly into the brain). Clinical studies are therefore limited to patients in very advanced stages of neurodegenerative diseases with no further options, as reviewed by Chtarto et al. (2013). The ethical implications of such trials were discussed by Lowenstein (2008), who noted that ethical/safety considerations stand in opposition to therapeutic considerations: the former limit clinical trial participation to patients at very advanced stages, while excluding those at early stages who could potentially most benefit from therapeutic intervention.
I add another potential concern: off-target behavioral effects. The literature of CRISPR–Cas applications is replete with assessments of its off-target effects, reassuring us that the technology reaches its intended targets with minimal modifications to unintended genomic sites. To date, there are very few behavioral studies to determine whether modifications of single, let alone multiplexed, genes produce only changes in targeted behaviors. This could become a significant concern, as this technology will be used to correct genetic deficits related to neurodegenerative diseases or other behavioral deficits. To address this concern, there should be extensive future behavioral studies to examine not only modifications to targeted behaviors but also “off-target” behaviors. It may be impractical to accomplish this in any single study, although individual studies could still be designed to examine off-target behaviors that share common circuitry. For example, a CRISPR–Cas study designed to target the hippocampus and memory functions could also examine behavior related to spatial navigation, which is also subserved by this region. Null results of studies designed to test off-target behavioral effects should be as publishable as the original positive results reporting on “on-target” effects.
4. Case study 2: optogenetics and memory manipulation
One neurogenethics theme straddling the molecular and cellular/circuit level of scale is the manipulation of memory using optogenetics. The basic idea behind the optogenetics approach is to insert light-sensitive molecules called opsins into genetically defined cells of the central nervous system (Boyden et al., 2005). By using different kinds of opsins and wavelengths, and different viral delivery vectors, investigators can select specific types of cells to be excited or inhibited with millisecond resolution. Optogenetics studies have begun to demonstrate how memories can be activated, implanted, or inhibited.
One application of this approach demonstrated activation of a previous memory (Liu et al., 2012). Mice were exposed to a mild electric footshock while placed in one environment (context A) and neurons activated during this experience were labeled with an excitatory opsin called channelrhodopsin-2 (ChR2). When these mice were placed in a novel environment (which elicits exploratory behavior under normal conditions) and the neurons labeled in context A re-activated, these animals exhibited freezing behavior, an index of fear. Thus, reactivation of those neurons that had encoded a fearful memory (footshock exposure in context A) was sufficient to reinstate the memory trace and its accompanying behavior.
A follow-up study by this group demonstrated implantation of a false memory (Ramirez et al., 2013). In a first step, mice were placed into a ‘safe’ environment (context A, no footshock), while neurons encoding the experience were labeled with ChR2. The animals were then placed into a novel environment (context B), in which they did receive footshock at the same time that neurons encoding context A were re-activated. When the animals were placed back into context A, they exhibited fearful behavior despite the fact that they had never experienced foot shock in that environment. Thus, reactivation of the memory of context A while the animals were exposed to context B had been associated with footshock exposure in context B, generating a false memory.
Just as activation of critical neurons can activate a memory, inhibition of neurons can impair its formation. For example, one group investigated the role of the medial prefrontal cortex (mPFC) in episodic memory (Bero et al., 2014). They transfected excitatory cells in the mPFC with an inhibitory opsin (halorhodopsin eNpHR3.0) to allow for inhibition of this region during contextual fear conditioning, which impaired long-term memory, as indexed by reduced freezing behavior in the same context 28 days later.
Although optogenetics in its current form is invasive – requiring injection of viral vectors into the brain and/or transgenic manipulations – human applications are widely anticipated for a range of diseases including neuropsychiatric conditions (Tourino et al., 2013, Huang et al., 2013). A potentially significant step towards non-invasive applications was accomplished by Tsien's group (Lin et al., 2013) who engineered a novel channelrhodopsin (ChR) called red-activatable channelrhodopsin (ReaChR) that is optimally excited by wavelengths that can penetrate the intact skull. In awake mice, a red light shone through the intact skull could activate ReaChR expressing cells in the vibrissa motor cortex to drive vibrissa movements.
In addition to “off-target effects” (discussed in the previous section), another ethical consideration concerns the nature of memory manipulations. In particular, the removal of painful memories has been subject to a vigorous juxtaposition of viewpoints by the President's Council on Bioethics (2010) and Kolber (2010). The President's Council on Bioethics articulated several concerns about memory blunting or removal. Among them was a concern that such a procedure would compromise “our own truthful identities” (p. 91). Kolber, in response, questioned the premise of truthful identity, noting that memories are not verbatim transcripts of experienced events but rather the product of synthesis and reconstruction. Another concern voiced by the Council was that memory dampening would break the link to moral responsibility, such that an amnesic perpetrator could not be held accountable, and hence there could be neither justice nor opportunity for forgiveness. Kolber responded that individuals are indeed held responsible for their actions if they fail to remember. In his arguments, Kolber stressed personal choice, by which one's deliberate decision to either pursue memory dampening or leave it intact would increase one's responsibility.
A striking aspect of ethics debates on memory modifications is that they tend to be void of data, instead reflecting stakeholders' belief systems. Yet, the choices that people would make about modifications of their memories are empirically accessible, and should be explored in future neurogenethics studies.
5. Case study 3: genomic identity and online communities
An emerging neurogenethics theme at the level of social structures is genomic identity, catalyzed by the availability of direct-to-consumer (DTC) commercial genomics services. Small studies of early adopters showed that they were motivated by an interest in health-related information and their individual risk factors (McGowan et al., 2010) or genealogy (Su et al., 2011).
Surveys of medical and genetics experts questioned consumers' ability to interpret genomic information and expected them to overreact to perceived diagnostic results (Hunter et al., 2008, Cho, 2009, McGuire et al., 2009). The data on actual consumer behavior show that customers are more sophisticated, as they take an active role in processing their genomic information, acting on it deliberately, with the input of additional information or health care consultation. A survey of 1048 DTC genomics customers found that consumers did not solely rely on commercial test results, but sought additional information (48%) and discussed their results with a healthcare professional (28%); in 9% of cases, these consultations led to follow-up lab testing.
A recent survey of 998 consumers examined changes in consumers' genetics knowledge and self-efficacy following personal genomic testing (PGT) (Carere et al., 2015). Participants completed survey questions designed to measure genetics knowledge and genetics self-efficacy at baseline and 6 months after receiving genomics results. Whereas genetics knowledge scores were near the ceiling and unchanged across both time points (8.15/9 and 8.25/9, respectively), participants' mean self-efficacy score dropped significantly from 29.1/35 at baseline to 27.7/35 at 6 months (P < 0.0001). On the other hand, the reduction in self-efficacy correlated positively with health-care provider consultation (P = 0.0042), and perceived control over one's health (P < 0.0001). The authors concluded that “[l]owered genetics self-efficacy following PGT may reflect an appropriate reevaluation by consumers in response to receiving complex genetic information”. However, in the absence of baseline data on consumers' intent to solicit health-care provider consultation, this conclusion is speculative.
Does consumers' consultation of health care providers automatically translate to better-informed medical decision-making? A survey of 130 genetic counselors and 38 clinical geneticists in Australia and New Zealand found that only 7% felt confident in accurately interpreting and explaining the results of DTC genomics results (Brett et al., 2012). A systematic review of the literature found “low level of awareness and experience of direct-to-consumer testing in health professionals” (Goldsmith et al., 2013). A survey of 382 primary care physicians and internal medicine providers found that 85% felt unprepared to answer patient questions (Powell et al., 2012). McGowan and colleagues conducted qualitative in-depth interviews with 18 clinicians providing genomic risk assessment services in partnership with the DTC companies DNA Direct and Navigenics (McGowan et al., 2014). These interviews revealed that clinicians' knowledge was based on information provided by these commercial entities “without the ability to critically evaluate the knowledge or assess risks”.
What about the relationship of DTC-genomics regulation versus consumer access to their own genomic data? On the one hand, consumers want access to their genomic data without governmental oversight (66%), although they do want either governmental (73%) or nongovernmental (84%) monitoring of the scientific claims made by DTC genomic service companies (Bollinger et al., 2013).
On the other hand, the regulatory appetite of governmental agencies moved front-and-center in the case of 23andMe and the Food and Drug Administration (FDA), which in November 2013 stopped the company from offering consumers health-related genetic tests. The FDA claimed that the company's Saliva Collection Kit and Personal Genome Service (PGS) constituted a “device within the meaning of section 201(h) of the FD&C Act, 21 U.S.C. 321(h), because it is intended for use in the diagnosis of disease or other conditions or in the cure, mitigation, treatment, or prevention of disease, or is intended to affect the structure or function of the body” (FDA, 2013). These claims were examined by Green and Farahany (2014) who agreed with the FDA that there was a need for transparency in the accuracy of reported data, but also were critical of over-regulation. In the absence of any evidence that consumers respond to genomic health information inappropriately that would cause them physical harm, they found that preventing consumers access to their genomic information constitutes a social harm, in opposition to a “historical trend of patient empowerment that brought informed-consent laws, access to medical records and now direct access to electronic personal health data” (p. 287).
So far, the discussion of DTC genomics has not touched on any aspects relevant to neuroscience. The missing element is the potential that genomic information can have in shaping our sense of Self, as it pertains to behavioral traits, mental illness, or neuropathology. As neurogenomics is coming of age, deeper insights about the relation between genome, brain, and behavior will emerge. I think that the emergence of DTC genomics services may disrupt the traditional model of research that will generate these insights. One of the most exciting and disruptive aspects of DTC genomics is the changing role of the research subject, as illustrated by the approach taken by 23andMe.
In 2009, 23andMe began promoting a “do-it-yourself revolution”, in which the company invited its customers to contribute their genomics data to large-scale datasets. By 2014, 23andMe claimed 500,000 research participants (23andMe, 2014). Data generated from this dataset led to peer-reviewed publications on Parkinson's disease (Do et al., 2011), a replication study of 22 physical traits as well as novel trait associations (Eriksson et al., 2010), and a replication study of another 180 known genetic associations (Tung et al., 2011).
Perhaps more importantly, 23andMe consumers have online tools to share their genomic results with others, and have begun to form online social networks organized around genomic identities. The social and ethical implications of such networks were discussed by Lee and Crawley (2009a,b). One concern they raised was that consumers may be unaware that even pooled genomic datasets can be breached to identify individuals (Homer et al., 2008). On the other hand, the existence of such networks could be used to study how information flows across them, with ramifications for social and ethical considerations such as informed consent, which is currently designed for individual consumers/research participants, but may need to be amended when other parties are affected by sharing genomic information.
I identified three areas of future neurogenethics examination (Canli, 2015). The first is how the change in research participant agency, from passive subject to active consumer, may affect research priorities, design, and interpretation. The second is how the emergence of online social networks based on genomic identity may alter offline behavior, such as social support for Alzheimer's patients and their caregivers. The third area of future neurogenethics examination is global mental health. Given that online social networks have no national boundaries, how will information exchange among individuals who identify themselves by genomic identity change cross-cultural views on mental health, treatment, and stigma? The answers to these and related questions may impact our very sense of Self, as individuals and as a community of Selves.
6. Concluding thoughts
As neurogenomics comes of age, new ethical, legal, and social implications emerge. The pace of technological innovation is breathtaking, as illustrated by the recency of most references in this article. And whereas the applications of these new technologies are, for the most part, eagerly anticipated — the “known unknowns”, their larger implications outside the laboratory or clinic are harder to gauge. Are these the “unknown unknowns”? Perhaps not quite. But even though neurogenethics now moves forward guided by the roadmap of neurogenomics, we will know that the field has come of age itself when it can begin to ask questions that will guide future neurogenomics research.
References
- Canli T. Neurogenethics. In: Canli T., editor. The Oxford Handbook of Molecular Psychology. Oxford University Press; New York, Oxford: 2015. [Google Scholar]
- Illes J. Prospects for prediction: ethics analysis of neuroimaging in Alzheimer's disease. Ann. N. Y. Acad. Sci. 2007;1097:278–295. doi: 10.1196/annals.1379.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roskies A.L. Neuroethics beyond genethics. Despite the overlap between the ethics of neuroscience and genetics, there are important areas where the two diverge. EMBO Rep. 2007;8:S52–S56. doi: 10.1038/sj.embor.7401009. (Spec No) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337(6096):816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishino Y. Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. J. Bacteriol. 1987;169(12):5429–5433. doi: 10.1128/jb.169.12.5429-5433.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouns S.J. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008;321(5891):960–964. doi: 10.1126/science.1159689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doudna J.A., Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR–Cas9. Science. 2014;346(6213):1258096. doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- Barrangou R. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315(5819):1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- Deltcheva E. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011;471(7340):602–607. doi: 10.1038/nature09886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maggio I., Goncalves M.A. Genome editing at the crossroads of delivery, specificity, and fidelity. Trends Biotechnol. 2015 doi: 10.1016/j.tibtech.2015.02.011. [DOI] [PubMed] [Google Scholar]
- Xiao-Jie L. CRISPR–Cas9: a new and promising player in gene therapy. J. Med. Genet. 2015;52(5):289–296. doi: 10.1136/jmedgenet-2014-102968. [DOI] [PubMed] [Google Scholar]
- Pelletier S., Gingras S., Green D.R. Mouse genome engineering via CRISPR–Cas9 for study of immune function. Immunity. 2015;42(1):18–27. doi: 10.1016/j.immuni.2015.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu P.D., Lander E.S., Zhang F. Development and applications of CRISPR–Cas9 for genome engineering. Cell. 2014;157(6):1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konermann S. Genome-scale transcriptional activation by an engineered CRISPR–Cas9 complex. Nature. 2015;517(7536):583–588. doi: 10.1038/nature14136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilton I.B. Epigenome editing by a CRISPR–Cas9-based acetyltransferase activates genes from promoters and enhancers. Nat. Biotechnol. 2015 doi: 10.1038/nbt.3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombardo A., Naldini L. Genome editing: a tool for research and therapy: targeted genome editing hits the clinic. Nat. Med. 2014;20(10):1101–1103. doi: 10.1038/nm.3721. [DOI] [PubMed] [Google Scholar]
- Meissner T.B. Genome editing for human gene therapy. Methods Enzymol. 2014;546:273–295. doi: 10.1016/B978-0-12-801185-0.00013-1. [DOI] [PubMed] [Google Scholar]
- Wang L. Regenerative medicine: targeted genome editing in vivo. Cell Res. 2015;25(3):271–272. doi: 10.1038/cr.2015.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Gao G. State-of-the-art human gene therapy: part II. Gene therapy strategies and clinical applications. Discov. Med. 2014;18(98):151–161. [PMC free article] [PubMed] [Google Scholar]
- Kennedy E.M., Cullen B.R. Bacterial CRISPR/Cas DNA endonucleases: a revolutionary technology that could dramatically impact viral research and treatment. Virology. 2015 doi: 10.1016/j.virol.2015.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W. RNA-directed gene editing specifically eradicates latent and prevents new HIV-1 infection. Proc. Natl. Acad. Sci. U. S. A. 2014;111(31):11461–11466. doi: 10.1073/pnas.1405186111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebina H. Harnessing the CRISPR/Cas9 system to disrupt latent HIV-1 provirus. Sci. Rep. 2013;3:2510. doi: 10.1038/srep02510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genovese P. Targeted genome editing in human repopulating haematopoietic stem cells. Nature. 2014;510(7504):235–240. doi: 10.1038/nature13420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiech L. In vivo interrogation of gene function in the mammalian brain using CRISPR–Cas9. Nat. Biotechnol. 2015;33(1):102–106. doi: 10.1038/nbt.3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chahrour M., Zoghbi H.Y. The story of Rett syndrome: from clinic to neurobiology. Neuron. 2007;56(3):422–437. doi: 10.1016/j.neuron.2007.10.001. [DOI] [PubMed] [Google Scholar]
- Klein R.L. Dose and promoter effects of adeno-associated viral vector for green fluorescent protein expression in the rat brain. Exp. Neurol. 2002;176(1):66–74. doi: 10.1006/exnr.2002.7942. [DOI] [PubMed] [Google Scholar]
- Tenenbaum L., Lehtonen E., Monahan P.E. Evaluation of risks related to the use of adeno-associated virus-based vectors. Curr. Gene Ther. 2003;3(6):545–565. doi: 10.2174/1566523034578131. [DOI] [PubMed] [Google Scholar]
- McCown T.J. Adeno-associated virus (AAV) vectors in the CNS. Curr. Gene Ther. 2011;11(3):181–188. doi: 10.2174/156652311795684759. [DOI] [PubMed] [Google Scholar]
- Bowers W.J., Breakefield X.O., Sena-Esteves M. Genetic therapy for the nervous system. Hum. Mol. Genet. 2011;20(R1):R28–R41. doi: 10.1093/hmg/ddr110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- High K.A., Aubourg P. rAAV human trial experience. Methods Mol. Biol. 2011;807:429–457. doi: 10.1007/978-1-61779-370-7_18. [DOI] [PubMed] [Google Scholar]
- Weinberg M.S., Samulski R.J., McCown T.J. Adeno-associated virus (AAV) gene therapy for neurological disease. Neuropharmacology. 2013;69:82–88. doi: 10.1016/j.neuropharm.2012.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran F.A. In vivo genome editing using Staphylococcus aureus Cas9. Nature. 2015;520(7546):186–191. doi: 10.1038/nature14299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chtarto A. A next step in adeno-associated virus-mediated gene therapy for neurological diseases: regulation and targeting. Br. J. Clin. Pharmacol. 2013;76(2):217–232. doi: 10.1111/bcp.12065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowenstein P.R. Clinical trials in gene therapy: ethics of informed consent and the future of experimental medicine. Curr. Opin. Mol. Ther. 2008;10(5):428–430. [PMC free article] [PubMed] [Google Scholar]
- Boyden E.S. Millisecond-timescale, genetically targeted optical control of neural activity. Nat. Neurosci. 2005;8(9):1263–1268. doi: 10.1038/nn1525. [DOI] [PubMed] [Google Scholar]
- Liu X. Optogenetic stimulation of a hippocampal engram activates fear memory recall. Nature. 2012;484(7394):381–385. doi: 10.1038/nature11028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez S. Creating a false memory in the hippocampus. Science. 2013;341(6144):387–391. doi: 10.1126/science.1239073. [DOI] [PubMed] [Google Scholar]
- Bero A.W. Early remodeling of the neocortex upon episodic memory encoding. Proc. Natl. Acad. Sci. U. S. A. 2014;111(32):11852–11857. doi: 10.1073/pnas.1408378111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tourino C., Eban-Rothschild A., de Lecea L. Optogenetics in psychiatric diseases. Curr. Opin. Neurobiol. 2013;23(3):430–435. doi: 10.1016/j.conb.2013.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang F., Tang B., Jiang H. Optogenetic investigation of neuropsychiatric diseases. Int. J. Neurosci. 2013;123(1):7–16. doi: 10.3109/00207454.2012.728651. [DOI] [PubMed] [Google Scholar]
- Lin J.Y. ReaChR: a red-shifted variant of channelrhodopsin enables deep transcranial optogenetic excitation. Nat. Neurosci. 2013;16(10):1499–1508. doi: 10.1038/nn.3502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- President's Council on Bioethics . Memory blunting: ethical analysis. In: Farah M.J., editor. Neuroethics: An Introduction With Readings. MIT Press; Cambridge, MA: 2010. [Google Scholar]
- Kolber A.J. Ethical implications of memory dampening. In: Farah M.J., editor. Neuroethics: An Introduction With Readings. MIT Press; Cambridge, MA: 2010. [Google Scholar]
- McGowan M.L., Fishman J.R., Lambrix M.A. Personal genomics and individual identities: motivations and moral imperatives of early users. New Genet. Soc. 2010;29(3):261–290. doi: 10.1080/14636778.2010.507485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Y., Howard H.C., Borry P. Users' motivations to purchase direct-to-consumer genome-wide testing: an exploratory study of personal stories. J. Community Genet. 2011;2(3):135–146. doi: 10.1007/s12687-011-0048-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter D.J., Khoury M.J., Drazen J.M. Letting the genome out of the bottle—will we get our wish? N. Engl. J. Med. 2008;358(2):105–107. doi: 10.1056/NEJMp0708162. [DOI] [PubMed] [Google Scholar]
- Cho M.K. Translating genomics into the clinic: moving to the post-Mendelian world. Genome Med. 2009;1(1):7. doi: 10.1186/gm7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuire A.L. Social networkers' attitudes toward direct-to-consumer personal genome testing. Am. J. Bioeth. 2009;9(6–7):3–10. doi: 10.1080/15265160902928209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carere D.A. Consumers report lower confidence in their genetics knowledge following direct-to-consumer personal genomic testing. Genet. Med. 2015 doi: 10.1038/gim.2015.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brett G.R. An exploration of genetic health professionals' experience with direct-to-consumer genetic testing in their clinical practice. Eur. J. Hum. Genet. 2012;20(8):825–830. doi: 10.1038/ejhg.2012.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldsmith L. Direct-to-consumer genomic testing from the perspective of the health professional: a systematic review of the literature. J. Community Genet. 2013;4(2):169–180. doi: 10.1007/s12687-012-0135-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell K.P. Educational needs of primary care physicians regarding direct-to-consumer genetic testing. J. Genet. Couns. 2012;21(3):469–478. doi: 10.1007/s10897-011-9471-9. [DOI] [PubMed] [Google Scholar]
- McGowan M.L. Gatekeepers or intermediaries? The role of clinicians in commercial genomic testing. PLoS One. 2014;9(9):e108484. doi: 10.1371/journal.pone.0108484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollinger J.M., Green R.C., Kaufman D. Attitudes about regulation among direct-to-consumer genetic testing customers. Genet. Test. Mol. Biomarkers. 2013;17(5):424–428. doi: 10.1089/gtmb.2012.0453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FDA . 2013. Warning Letter From 11/22/2013. (4/1/2014) [Google Scholar]
- Green R.C., Farahany N.A. Regulation: the FDA is overcautious on consumer genomics. Nature. 2014;505(7483):286–287. doi: 10.1038/505286a. [DOI] [PubMed] [Google Scholar]
- 23andMe . 2014. The Future of Genetics in People's Lives. (3/21/2014) [Google Scholar]
- Do C.B. Web-based genome-wide association study identifies two novel loci and a substantial genetic component for Parkinson's disease. PLoS Genet. 2011;7(6):e1002141. doi: 10.1371/journal.pgen.1002141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson N. Web-based, participant-driven studies yield novel genetic associations for common traits. PLoS Genet. 2010;6(6):e1000993. doi: 10.1371/journal.pgen.1000993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tung J.Y. Efficient replication of over 180 genetic associations with self-reported medical data. PLoS One. 2011;6(8):e23473. doi: 10.1371/journal.pone.0023473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.S., Crawley L. Research 2.0: social networking and direct-to-consumer (DTC) genomics. Am. J. Bioeth. 2009;9(6–7):35–44. doi: 10.1080/15265160902874452. [DOI] [PubMed] [Google Scholar]
- Lee S.S., Crawley L. Response to open peer commentaries on “Research 2.0: social networking and direct-to-consumer personal genomics”. Am. J. Bioeth. 2009;9(6–7):W1–W3. doi: 10.1080/15265160902967009. [DOI] [PubMed] [Google Scholar]
- Homer N. Resolving individuals contributing trace amounts of DNA to highly complex mixtures using high-density SNP genotyping microarrays. PLoS Genet. 2008;4(8):e1000167. doi: 10.1371/journal.pgen.1000167. [DOI] [PMC free article] [PubMed] [Google Scholar]