Abstract
Nutrigenomics is an emerging science which investigates a certain area of nutrition that uses molecular tools to search access and understand the several responses obtained through a certain diet applied between individual and population groups. The increased need for the use of personalised nutrition in patients is increasing and research is being made on its possible effects. However, research on nutrigenomics and in particular, obesity is still ongoing. Following a current metanalysis on thirty-eight nutrigenomics genes, it seems that a definite association between the genes usually examined in nutrigenomics testing and several diet-related diseases is lacking, even though there is a limited number of studies associating them. In 2014, literature search results in a great number of studies on several polymorphisms. This heterogeneity could only show the way towards new research aims. Nutrigenomics was born due to the need to move from Epidemiology and Physiology to Molecular Biology and Genetics. Currently, there are steps that need to be considered in order for nutrigenomics to be applied: the genes, the gene/protein network, and the strategy towards the determination of the nutrients' influence on gene/protein expression. It is certainly an interesting evolving science with many areas to be investigated further and from different perspectives, as it involves ethics, medicine, genetics and nutrition.
Keywords: Nutrigenomics, Nutrition, Personalised nutrition, Personalised medicine, Science
1. Introduction
1.1. What is nutrigenomics?
Nutrigenomics is an emerging science which investigates a certain area of nutrition that uses molecular tools to search, access and understand the several responses obtained through a certain diet applied between individual and population groups (Sales et al., 2014).
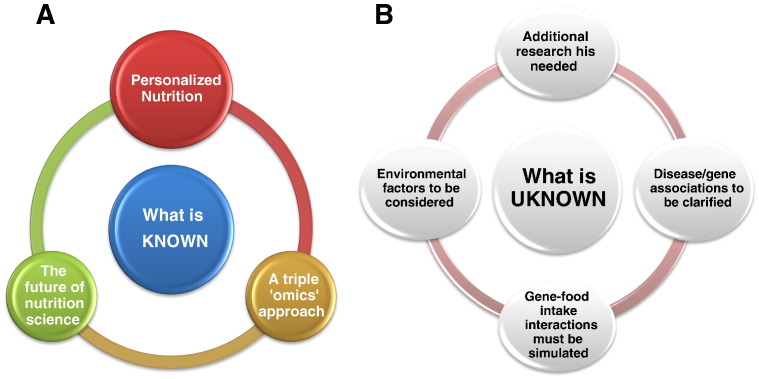
The need for personalised nutrition following the thoughts on the methods and the results of applying personalised medicine to patients was seen after the conclusion of the Human Genome Project. In fact in their review, Sales et al. (2014) show the need for responding to some questions that arose after its conclusion, as nutrigenomics is not investigated thoroughly until now and there are many fields to be researched further. Indeed, the way that gene expression as a response to the metabolic process could influence the health of a person and the interaction between genotype and environment/nutrient as well as the way that this might occur should be investigated in detail (Sales et al., 2014). As depicted in Fig. 1, three ‘omics’ disciplines, transcriptomics, proteomics and metabolomics and the way that these genes–proteins–metabolites could interact and be applied for personalised nutrition, are studied by Nutrigenomics (Affolter et al., 2009). Nutrigenomics application to everyday life would be the future of the Nutrition science and a great series of tools for nutritionists, dietitians, doctors as well as any Health Professional that implicates nutrition therapy for the treatment of disease. It could possibly help into the prevention of diet-related diseases as well as the designing of nutritional strategies and the adverse or beneficial effects of some food or nutrients (Palou, 2007). However, there are some ethical questions that arise whether there is sufficient scientific support at the moment for Nutrigenomics to be applied to everyday life. How could nutrigenomics influence an individual's personalised nutrition? What effects would this science have in terms of public health? Can it already be used? Is there enough scientific background evidence and research for it to be applied? How would the OMICS science help the implementation of the personalised nutrition and is it possible at this stage? Through this article, an investigation of the current situation in nutrigenomics research, as well as the possibilities and chances for studying this science in the future will be discussed in detail.
Fig. 1.
A schematic depiction of what is “known” (A) and what is “unknown” (B) in the field of nutrigenomics, highlighting the findings and challenges that emerge for the field of nutrigenomics.
1.2. Implementing nutrigenomics in real life and in terms of personalised nutrition. Is this possible?
The term “nutrigenomics” was first described in 2001 from Pelegrin (2001) and then, it appears in 2002 in a review by Van Ommen and Stierum (2002). A literature search in PubMed database using ‘nutrigenomics’ as a keyword resulted in 1005 articles since 2001 until present. This search by its own shows a quick route to the future of nutrigenomics and the fields that need to be researched further. Personalised nutrition will be the future in terms of designing and prescribing a diet for individuals based on their genome and their genetic variations. According to a recent survey in Greece on general population (N = 1504) and health professional (N = 87) samples, we have found that there is an increased need for the evolving nutrigenomics science (Pavlidis et al., 2012). At the time of the study, only 11.5% of the respondents were advised to undertake a nutrigenomics test, implying that there is not a great application among nutritionists, medical doctors or health experts. In addition, 23.5% of the respondents were frequently asking their health care providers for nutrigenomics tests, while the surprisingly 80.5% of nutritionists and doctors were willing to recommend a nutrigenomics analysis. However, only 17% have actually done so (Pavlidis et al., 2012). Diseases such as obesity, diabetes, high triglycerides and high cholesterol levels were believed to be associated with the genetic profile of an individual as stated by the beliefs of health professionals and the general public. In particular, 76% of the general public stated that the application of a personalised diet designed according to their genetic profile would be beneficial (Pavlidis et al., 2012).
Nutrigenomics research is still ongoing. A current metanalysis on thirty-eight nutrigenomics genes has shown that there is no definite association between the genes usually examined in nutrigenomics testing and many diet-related diseases, although in some cases there is a limited number of studies associating them (Pavlidis et al., in preparation). Recently, an association between the APOA5 c.-1131C>T and triglyceride and APOA-V levels was reported, when the comparative effect of whole grains, legumes and refined rice was investigated in newly diagnosed patients with diabetes type 2 (Kang et al., 2014). In the same year, Li et al. (2014) investigated the effect of various polymorphisms (rs662799, rs3135506, rs2075291, rs2266788) of the APOA5 gene on triglyceride levels in 1174 Uyghur (mixture of Caucasians and East Asian) subjects. Although a dysregulation of triglycerides levels was evident, the need for further research also became apparent. Additionally, a similar study, showed an age-related association in mice between triglycerides and the APOA5 c.-1131T>C polymorphism, but for those with the TT allele and not the CT or the CC alleles (Kim et al., 2014). These examples show how complex is the study of nutrigenomics in terms of influences, genetic material and genetic predisposition in various populations as well as the environmental factors that could influence their gene-nutrient association. A gene-diet interaction has been found, when two of the most known genes (the FTO and the MC4R) were studied taking into consideration the adherence to the Mediterranean Diet for two polymorphisms (rs9939609 and rs17782313 respectively), although no association to diabetes type 2 was found (Ortega-Azorín et al., 2012). The body weight in children with diabetes could be influenced by the presence of the A allele of the FTO rs9939609 (Luczynski et al., 2014). Another study on Chinese school-aged children, showed an association between rs7206790 and rs11644943 of the FTO gene and obesity (Xu et al., 2014).
An ambitious yet important goal of nutrigenomics is the investigation of the role of metabolic stress and its association to the metabolic syndrome. In this context, nutrigenomics will rather serve a disease prevention role, being complementary to pharmacological approaches. For this, the collection and study of phenotypes combining inflammation, metabolic stress, insulin resistance, and diabetes seem to be necessary (Afman and Muller, 2006). In a molecular context, nutrients can be considered as “signalling molecules” transmitting and translating dietary signals into changes in gene, protein and metabolite expression via the appropriate cellular sensing mechanisms (Wellen and Hotamisligil, 2005). Hence, at a molecular level the question that arises is what is happening in our cells when we eat, when we do not eat, or when we eat too much. On a genomic level, nutrients and in turn, their dietary signals serve as “signatures”. These dietary “signatures” can be precisely linked to the phenotype, in particular when metabolic stress, as well as the early phases of organ-specific insulin resistance occur.
There are nuclear receptors, such as the peroxisome proliferator activator receptor-α (PPARα), which form heterodimers with the retinoid X receptor and bind to specific response elements in the promoter region of genes (Muller and Kersten, 2003). In metabolically active organs (liver, intestine, adipose tissue), they act as nutrient sensors by changing the level of DNA transcription of specific genes in response to nutrient changes. Indicatively, the PPAR group of nuclear receptors acts as nutrient sensors for fatty acids and influences gene expression. There are more than 3000 to 4000 target genes of PPARα that are involved in numerous metabolic processes in the liver; fatty acid oxidation, ketogenesis, gluconeogenesis, amino acid metabolism, cellular proliferation, and the acute-phase response. Fasted PPARα null mice have been shown to suffer from several metabolic defects, such as hypoketonemia, hypothermia, elevated plasma-free fatty acid levels and hypoglycemia (Mandard et al., 2004, Muller and Kersten, 2003). It has been also demonstrated that PPARα directly regulates the expression of genes involved in hepatic gluconeogenesis and glycerol metabolism (Kersten et al., 1999, Muller and Kersten, 2003). Since fatty acids serve as ligands for PPARα, the latter mechanism could explain the stimulatory effect of the elevated plasma-free fatty acids on hepatic gluconeogenesis and glucose output. Despite its important role in the physiological response to food deprivation, the role of PPARα in obesity is less clear, but most likely relevant to our understanding of the obesity-linked pathophysiology of type 2 diabetes (Patsouris et al., 2004a). Visceral obesity is also linked to increased free fatty acid levels (Patsouris et al., 2004b). Noteworthy, these molecules may be recognized by the liver as “hunger” or “in need of glucose” signals, resulting in increased gluconeogenesis in a PPARα-dependent manner, particularly under conditions of hepatic insulin resistance.
The dietary pattern interactions with the common genetic variant of APOC3 (rs5128) have been also investigated in terms of their contribution to metabolic syndrome susceptibility in adults (Tehran Lipid and Glucose study) (Hosseini-Esfahani et al., 2014). The metabolic syndrome risk was found to increase in women with the CC genotype with increasing tertiles of western dietary pattern (WDP) scores compared with women with the CG and GG genotypes, whose risk was decreased with increasing tertiles of WDP scores. Intakes of fast food, salty snacks and soft drinks also showed significant interactions with the rs5128 genotypes in relation to the metabolic syndrome risk (p for interactions < 0.05).
A major focus of nutrigenomics is also on the prevention of chronic diseases that are partly mediated by chronic exposure to certain food components. It is becoming increasingly evident that not all people respond equally to diet. Genetic polymorphisms in apolipoprotein E, fatty acid desaturase, lipoxygenase-5, peroxisome proliferator-activated receptors, apolipoprotein A1, apolipoprotein A2, apolipoprotein A5, and methylenetetrahydrofolate reductase have been associated with cardiovascular disease (Nuno and Heuberger, 2014).
A nice example of nutrigenomics research that allowed the detection of changes in hepatic gene expression patterns in the context of an adaptive response to changes in dietary macronutrient composition refers to the peroxisome proliferator-activated receptor-gamma coactivator-1β (PGC-1β), a co-activator of PPARγ (Lin et al., 2005). In particular, high-fat feeding in mice has been shown to induce hyperlipidemia and atherogenesis and to stimulate PGC-1β expression in the liver. Through molecular studies and microarray analysis, the enhancer effects of PGC-1β on gene transcription (governed by the transcription factors sterol regulatory element binding protein-1 and liver X receptor-α) have been linked to increased lipogenesis and very-low-density lipoprotein excretion. Thus, a mechanism has been proposed by which dietary saturated and transfatty acids can stimulate hyperlipidemia and atherogenesis.
Inflammatory bowel disease refers to both ulcerative colitis and Crohn's disease, two inflammatory disorders of the gastrointestinal tract. Inflammatory bowel disease has a complex aetiology; a genetically determined susceptibility interacting with environmental factors, including nutrients and gut microbiota. So far, genome wide association studies have implicated more than 160 single-nucleotide polymorphisms that relate to disease susceptibility (Ferguson, 2013) Indicatively, Gentschew et al. (2012) have considered genes for selenoproteins as well as low serum selenium jointly (DIO1, DIO2, GPX1, GPX3, SEPHS1, SEPSECS and TXNRD2) as potential Crohn's disease risk factors in a prospective case–control study in Auckland, New Zealand. Crohn's disease patients had significantly lower serum selenium levels as compared with controls. Although 13 out of the 29 SNPs tested showed significant interactions with serum selenium levels on the risk of Crohn's disease, significance remained only for two SNPs in the SEPHS1 gene, and one in the SEPSECS gene, following adjustment for multiple testing. Nevertheless, it is unclear whether low selenium levels are the cause or effect of the disease and hence, a selenoprotein genotype seems to be of importance.
Inflammatory bowel disease is also characterised by low zinc levels. As suggested by Ringstad et al. (1993), low plasma zinc levels could be the outcome of reduced zinc absorption, because of variant genes. At least four isoforms of ZNF365 have been identified (Haritunians et al., 2011). Notably, oral zinc sulfate supplementation has significantly improved intestinal barrier function in a number of patients. Zinc supplementation has been also shown to prevent relapse in those whose intestine did show a response (Ferguson, 2013).
A number of the genetic variants associated with the development of inflammatory bowel disease have been shown to enhance inflammation. In this context, the eicosapentaenoic acid (EPA) and doco-sahexaenoic acid (DHA) – the long chain polyunsaturated omega-3 (n− 3) fatty acids found in oily fish and fish oil supplements – can partly inhibit several aspects of inflammation; adhesion molecule expression, leucocyte chemotaxis and leucocyte–endothelial adhesive interactions (Ferguson, 2013). There is also evidence that vitamin D signalling can directly induce NOD2 expression, implying that vitamin D insufficiency or deficiency plays a causative role in Crohn's disease (Wang et al., 2010). It should be noted that it is likely that this deficiency interacts with polymorphisms of the vitamin D receptor in enhancing inflammatory bowel disease susceptibility (Ferguson, 2013).
Three other conditions, phenylketonuria, celiac disease and lactose intolerance are examples of nutrient and gene interactions. Phenylketonuria (PKU), an inborn error of metabolism, results from the deficiency of hepatic phenylalanine hydroxylase (the enzyme needed to convert phenylalanine to tyrosine). Human phenylalanine hydroxylase gene exhibits two clusters of polymorphic sites (nine in total) at its 5′ and 3′ ends. This genetic variation has been reported to lead to a decrease in the enzyme's activity (DiLella et al., 1986). Excess of serum phenylalanine results in hyperphenylalaninemia as well as several metabolic abnormalities of aromatic amino acids' derivatives. Unless following a recommended diet, individuals with PKU develop postnatal neurological damage (severe mental retardation and seizures) (Surtees and Blau, 2000). In PKU, restriction of phenylalanine and protein is needed as the main part of the nutritional treatment (Sweeney et al., 2012). A genetic variation in the lactase gene leads to an inadequate production of lactase in the small intestine and thus, lactose intolerance (Swallow, 2003). Notably, the disease is present more in some populations than others (Heaney, 2013). Lactose, the primary milk sugar from dairy products cannot be efficiently broken down in individuals with lactose intolerance. Consequently, the dietary recommendation is to limit lactose-containing foods or to use lactase supplements or lactose-free dairy products to prevent gastrointestinal discomfort (Swagerty et al., 2002). Lactose intolerance and PKU are examples that involve a single genetic defect along with a single dietary exposure. On the contrary, celiac disease is characterised by a complex interplay between genetics and nutrients. Celiac disease is a common heritable chronic inflammatory condition of the small intestine that is caused by permanent intolerance to gluten/gliadin (prolamin). Following a non-complete proteolytic digestion, the latter may directly affect intestinal cell structure and functions by modulating gene expression (pro-inflammatory cytokines, adhesion molecules, and enzymes whose gene expression is known to be regulated by NF-κB) and oxidative stress (PPARγ receptor). Thus, gluten should be restricted as part of the nutritional and dietetic management of the disease (Ludvigsson et al., 2014). Furthermore, long chain ω − 3 fatty acids, plant flavonoids and carotenoids have been demonstrated to modulate oxidative stress, gene expression and production of inflammatory mediators. Therefore, their adoption could preserve intestinal barrier integrity, play a protective role against toxicity of gliadin peptides and have a role in nutritional therapy of celiac disease (Ferretti et al., 2012).
In general, this variety of results shows that there are differences in the way that polymorphisms interact with the diseases or conditions as well as the gene–diet interactions that occur. We are still at a starting point of investigating the variety of polymorphisms, their effects and the various interactions (ethnicity, environment, disease/condition, genes, polymorphisms, nutrients).
2. Conclusion
2.1. How can personalised medicine and personalised nutrition be inserted into society?
Currently, personalised medicine and nutrition are not completely applied into the everyday routine of the patients and their carers; doctors, nurses, dietitians or nutritionists. In a recent review, it is described how nutrigenomics should be inserted in public health and how the need for the gene–diet–disease interaction was created in the last years (Neeha and Priyamvadah, 2013). Indeed, nutrigenomics was born due to the need to pass from Epidemiology and Physiology to Molecular Biology and Genetics (Neeha and Priyamvadah, 2013). It is very important to evaluate the genes, the gene/protein network and the method in order to research and check on the influence of the nutrients on gene/protein expression (Neeha and Priyamvadah, 2013). In fact, the difference between personalised nutrition and personalised medicine is that in the first one, genetic data should be related to the optimal diet for a certain genotype in order to reduce the disease risk, while in the latter, genotype data are linked to the risk of developing a disease (Gibney and Walsh, 2013). On the other hand, researchers that are implicated into nutrigenomics should probably think more on the ethical aspects of applying nutrigenomics into everyday life. How could the people include the use of new food products designed and tailored to their ‘needs’ and their ‘genotypes or their genome and their mutations/polymorphisms’? Would they accept the fact that they should search further and also include in their ‘list’ the research results of the test of their genome and what they should or not eat, when going to choose food in the super market? Would a certain ‘medical/nutritional’ therapy exist in terms of, for example, consuming only one specific product designed only for this polymorphism? Would this categorise them as individuals following a ‘specialised diet’? Therefore, the scientific junction between bioethics, nutrigenomics and personalised nutrition in terms of preventing or treating a disease is very important (Patrinos and Prainsack, 2014), in order not only to research the science of nutrigenomics, but also to find ways to implement this in the most ethical way in order for people not to be divided into ‘categories’. Organisations that bridge researchers together from various entities (Patrinos and Prainsack, 2014), could help in this, in order for clinical trials to be designed effectively taking into consideration the ‘ethics’ as well the ‘ways of implementing a research and a new science into the need of the society’.
References
- Affolter M., Raymond F., Kussmann M. Omics in nutrition and health research. In: Mine Y., Miyashita K., Shahidi F., editors. Nutrigenomics and Proteomics in Health and Disease — Food Factors and Gene Interactions. Wiley Blackwell Edition; IA, USA: 2009. pp. 11–29. [Google Scholar]
- Afman L., Muller M. Nutrigenomics: from molecular nutrition to prevention of disease. J. Am. Diet. Assoc. 2006;106:569–576. doi: 10.1016/j.jada.2006.01.001. [DOI] [PubMed] [Google Scholar]
- DiLella A.G., Kwok S.C., Ledley F.D., Marvit J., Woo S.L. Molecular structure and polymorphic map of the human phenylalanine hydroxylase gene. Biochemistry. 1986;25:743–749. doi: 10.1021/bi00352a001. [DOI] [PubMed] [Google Scholar]
- Ferguson L. Nutrigenetics, nutrigenomics and inflammatory bowel diseases. Expert. Rev. Clin. Immunol. 2013;9:717–726. doi: 10.1586/1744666X.2013.824245. [DOI] [PubMed] [Google Scholar]
- Ferretti G., Bacchetti T., Masciangelo S., Saturni L. Celiac disease, inflammation and oxidative damage: a nutrigenetic approach. Nutrients. 2012;4:243–257. doi: 10.3390/nu4040243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentschew L., Bishop K.S., Han D.Y., Morgan A.R., Fraser A.G., Lam W.J., Karunasinghe N., Campbell B., Ferguson L.R. Selenium, selenoprotein genes and Crohn's disease in a case–control population from Auckland, New Zealand. Nutrients. 2012;4:1247–1259. doi: 10.3390/nu4091247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibney M.J., Walsh M.C. The future direction of personalised nutrition: my diet, my phenotype, my genes' Proc. Nutr. Soc. 2013;72(2):219–225. doi: 10.1017/S0029665112003436. [DOI] [PubMed] [Google Scholar]
- Haritunians T., Jones M.R., McGovern D.P., Shih D.Q., Barrett R.J., Derkowski C., Dubinsky M.C., Dutridge D., Fleshner P.R., Ippoliti A., King L., Leshinsky-Silver E., Levine A., Melmed G.Y., Mengesha E., Vasilauskas E.A., Ziaee S., Rotter J.I., Targan S.R., Taylor K.D. Variants in ZNF365 isoform D are associated with Crohn's disease. Gut. 2011;60:1060–1067. doi: 10.1136/gut.2010.227256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heaney R.P. Dairy intake, dietary adequacy, and lactose intolerance. Adv Nutr. 2013;4(2):151–156. doi: 10.3945/an.112.003368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosseini-Esfahani F., Mirmiran P., Daneshpour M.S., Mehrabi Y., Hedayati M., Zarkesh M.M., Azizi F. Western dietary pattern interaction with APOC3 polymorphism in the risk of metabolic syndromes: Tehran Lipid and Glucose study. J. Nutrigenet. Nutrigenomics. 2014;7:105–117. doi: 10.1159/000365445. [DOI] [PubMed] [Google Scholar]
- Kang R., Kim M., Chae J.S., Lee S.-H., Lee J.H. Consumption of whole grains and legumes modulates the genetic effect of the APOA5-1131C variant on changes in triglyceride and apolipoprotein A–V concentrations in patients with impaired fasting glucose or newly diagnosed type 2 diabetes. Trials. 2014;15:100. doi: 10.1186/1745-6215-15-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kersten S., Seydoux J., Peters J.M., Gonzalez F.J., Des-Vvergne B., Wahli W. Peroxisome proliferator-activated receptor alpha mediates the adaptive response to fasting. J. Clin. Invest. 1999;103:1489–1498. doi: 10.1172/JCI6223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M., Chae J.S., Kim M., Lee S.H., Lee J.H. Effects of a 3-year dietary intervention on age-related changes in triglyceride and apolipoprotein A–V levels in patients with impaired fasting glucose or new-onset type 2 diabetes as a function of the APOA5-1131T>C polymorphis. Nutr. J. 2014;13:40. doi: 10.1186/1475-2891-13-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Hu B., Wang Y., Wu D., Jin L., Wang X. Influences of APOA5 variants on plasma triglyceride levels in Uyghur population. PLoS One. 2014;9(10):e110258. doi: 10.1371/journal.pone.0110258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J., Yang R., Tarr P.T., Wu P.H., Handschin C., Li S., Yang W., Pei L., Uldry M., Tontonoz P., Newgard C.B., Spiegelman B.M. Hyperlipidemic effects of dietary saturated fats mediated through PGC-1beta coactivation of SREBP. Cell. 2005;120:261–273. doi: 10.1016/j.cell.2004.11.043. [DOI] [PubMed] [Google Scholar]
- Luczynski W., Fendler W., Ramatowska A., Szypowska A., Szadkowska A., Mlynarski W., Chumiecki M., Jarosz-Chobot P., Chrzanowska J., Noczynska A., Brandt A., Mysliwiec M., Glowinska-Olszewska B., Bernatowicz P., Kowalczuk O., Bossowski A. Polymorphism of the FTO gene influences body weight in children with type 1 diabetes without severe obesity. Int. J. Endocrinol. 2014;Vol. 2014 doi: 10.1155/2014/630712. (Article ID 630712, 5 pages.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludvigsson J.F., Bai J.C., Biagi F., Card T.R., Ciacci C., Ciclitira P.J., Green P.H., Hadjivassiliou M., Holdoway A., van Heel D.A., Kaukinen K., Leffler D.A., Leonard J.N., Lundin K.E., McGough N., Davidson M., Murray J.A., Swift G.L., Walker M.M., Zingone F., Sanders D.S. BSG coeliac disease guidelines development group; British Society of Gastroenterology. Diagnosis and management of adult coeliac disease: guidelines from the British Society of Gastroenterology. Gut. 2014;63(8):1210–1228. doi: 10.1136/gutjnl-2013-306578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandard S., Müller M., Kersten S. Peroxisome proliferator-activated receptor alpha target genes. Cell. Mol. Life Sci. 2004;61:393–416. doi: 10.1007/s00018-003-3216-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller M., Kersten S. Nutrigenomics: goals and strategies. Nat. Rev. Genet. 2003;4:315–322. doi: 10.1038/nrg1047. [DOI] [PubMed] [Google Scholar]
- Neeha V.S., Priyamvadah Kinth P. Nutrigenomics research: a review. J. Food Sci. Technol. 2013;50(3):415–428. doi: 10.1007/s13197-012-0775-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuno N.B., Heuberger R. Nutrigenetic associations with cardiovascular disease. Rev. Cardiovasc. Med. 2014;15:217–225. doi: 10.3909/ricm0658. [DOI] [PubMed] [Google Scholar]
- Ortega-Azorín C., Sorlí J.V., Asensio E.M., Coltell O., Martínez-González M.Á., Salas-Salvadó J., Covas M.I., Arós F., Lapetra J., Serra-Majem L., Gómez-Gracia E., Fiol M., Sáez-Tormo G., Pintó X., Muñoz M.A., Ros E., Ordovás J.M., Estruch R., Corella D. Associations of the FTO rs9939609 and the MC4R rs17782313 polymorphisms with type 2 diabetes are modulated by diet, being higher when adherence to the Mediterranean diet pattern is low. Cardiovasc. Diabetol. 2012;11:137. doi: 10.1186/1475-2840-11-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palou A. From nutrigenomics to personalised nutrition. Gen. Nutr. 2007;2:5–7. doi: 10.1007/s12263-007-0022-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patrinos G.P., Prainsack B. Working towards personalization of medicine: genomics in 2014. Pers. Med. 2014;11(7):611–613. doi: 10.2217/pme.14.79. [DOI] [PubMed] [Google Scholar]
- Patsouris D., Mandard S., Voshol P.J., Escher P., Tan N.S., Havekes L.M., Koenig W., Marz W., Tafuri S., Wahli W., Müller M., Kersten S. PPARalpha governs glycerol metabolism. J. Clin. Invest. 2004;114:94–103. doi: 10.1172/JCI20468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patsouris D., Müller M., Kersten S. Peroxisome proliferator activated receptor ligands for the treatment of insulin resistance. Curr. Opin. Investig. Drugs. 2004;5:1045–1050. [PubMed] [Google Scholar]
- Pavlidis C., Karamitri A., Barakou A., Cooper D.N., Poulas K., Topouzis S., Patrinos G.P. Ascertainment and critical assessment of the views of the general public and healthcare professionals on nutrigenomics in Greece. Pers. Med. 2012;9(2):201–210. doi: 10.2217/pme.12.3. [DOI] [PubMed] [Google Scholar]
- Peregrin T. The new frontier of nutrition science: nutrigenomics. J. Am. Diet. Assoc. 2001;101(11):1306. doi: 10.1016/S0002-8223(01)00309-1. [DOI] [PubMed] [Google Scholar]
- Ringstad J., Kildebo S., Thomassen Y. Serum selenium, copper, and zinc concentrations in Crohn's disease and ulcerative colitis. Scand. J. Gastroenterol. 1993;28:605–608. doi: 10.3109/00365529309096096. [DOI] [PubMed] [Google Scholar]
- Sales N.M.R., Pelegrini P.B., Goerch M.C. Nutrigenomics: definitions and advances of this new science. J. Nutr. Metab. 2014;202759 doi: 10.1155/2014/202759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surtees R., Blau N. The neurochemistry of phenylketonuria. Eur. J. Pediatr. 2000;159(Suppl. 2):S109–S113. doi: 10.1007/pl00014370. [DOI] [PubMed] [Google Scholar]
- Swagerty D.L., Jr., Walling A.D., Klein R.M. Lactose intolerance. Am. Fam. Physician. 2002;65:1845–1850. [PubMed] [Google Scholar]
- Swallow D.M. Genetics of lactase persistence and lactose intolerance. Annu. Rev. Genet. 2003;37:197–219. doi: 10.1146/annurev.genet.37.110801.143820. [DOI] [PubMed] [Google Scholar]
- Sweeney A.L., Roberts R.M., Fletcher J.M. Dietary protein counting as an alternative way of maintaining metabolic control in phenylketonuria. JIMD Rep. 2012;3:131–139. doi: 10.1007/8904_2011_31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Ommen B., Stierum R. Nutrigenomics: exploiting systems biology in the nutrition and health arena. Curr. Opin. Biotechnol. 2002;13(5):517–521. doi: 10.1016/s0958-1669(02)00349-x. [DOI] [PubMed] [Google Scholar]
- Wang T.T., Dabbas B., Laperriere D., Bitton A.J., Soualhine H., Tavera-Mendoza L.E., Dionne S., Servant M.J., Bitton A., Seidman E.G., Mader S., Behr M.A., White J.H. Direct and indirect induction by 1,25-di-hydroxyvitamin D3 of the NOD2/CARD15-defensin beta2 innate immune pathway defective in Crohn disease. J. Biol. Chem. 2010;285(4):2227–2231. doi: 10.1074/jbc.C109.071225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellen K.E., Hotamisligil G.S. Inflammation, stress, and diabetes. J. Clin. Invest. 2005;115:1111–1119. doi: 10.1172/JCI25102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y., Ling J., Yang M., Wang H., Zhang S., Zhang X., Zhu Y. Rs7206790 and rs11644943 in FTO gene are associated with risk of obesity in Chinese school-age population. PLoS One. 2014;9(9):e108050. doi: 10.1371/journal.pone.0108050. (Sep 24) [DOI] [PMC free article] [PubMed] [Google Scholar]