Abstract
The dopamine transporter (DAT) is a key regulator of dopaminergic neurotransmission. As such, proper regulation of DAT expression is important to maintain homeostasis, and disruption of DAT expression can lead to neurobehavioral dysfunction. Based on genomic features within the promoter of the DAT gene, there is potential for DAT expression to be regulated through epigenetic mechanisms, including DNA methylation and histone acetylation. However, the relative contribution of these mechanisms to DAT expression has not been empirically determined. Using pharmacologic and genetic approaches, we demonstrate that inhibition of DNA methyltransferase (DNMT) activity increased DAT mRNA approximately 1.5–2 fold. This effect was confirmed by siRNA knockdown of DNMT1. Likewise, the histone deacetylase (HDAC) inhibitors valproate and butyrate also increased DAT mRNA expression, but the response was much more robust with expression increasing over tenfold. Genetic knockdown of HDAC1 by siRNA also increased DAT expression, but not to the extent seen with pharmacological inhibition, suggesting additional isoforms of HDAC or other targets may contribute to the observed effect. Together, these data identify the relative contribution of DNMTs and HDACs in regulating expression. These finding may aid in understanding the mechanistic basis for changes in DAT expression in normal and pathophysiological states.
Keywords: Dopamine transporter, Epigenetic, DNA methyltransferase, Histone deacetylase inhibitor, Histone, Chromatin
Introduction
The neurotransmitter dopamine (DA) mediates essential brain functions including cognition, memory, behavior, coordination of motor output and neuroendocrine modulation. Dysregulation of DA transmission is associated with several neurological diseases and disorders including attention deficit hyperactivity disorder, Parkinson’s disease, Tourette’s syndrome and schizophrenia [1–3]. Dopaminergic neurotransmission is tightly regulated by the dopamine transporter (DAT), a 12 transmembrane domain protein of the Na+/Cl− -dependent family of neurotransmitter transporters [4]. DATs are located at the nerve terminals within the striatum, where they function to terminate dopaminergic transmission through reuptake of DA into the presynaptic nerve terminal [5]. Knockout or knockdown of the DAT gene in animal models results in delayed clearance of DA and down-regulation of dopamine receptors, contributing to several behavioral abnormalities including hyperactivity, cognitive deficits and sleep dysregulation [6–8]. Thus, proper regulation of DAT expression is critical to maintain homeostasis in the dopamine system.
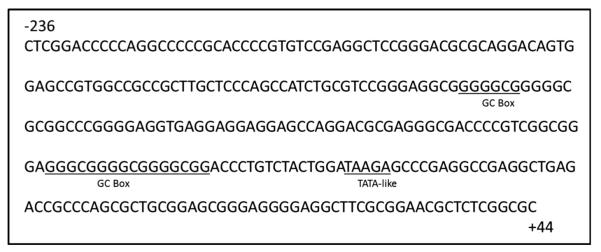
The human DAT gene was cloned more than two decades ago [9, 10]. Localized to chromosome 5p15.3, the gene has a single transcriptional start site [11]. Previous investigators cloned the human DAT promoter and performed functional assays to show that the core promoter is capable of driving reporter gene expression in different cell lines [12, 13]. These studies also defined the minimal essential region of transcription, the core promoter, to be between −236 and +44 relative to the transcription start site. Recent in silico analyses reported a variety of sequence motifs suggesting the potential for the DAT to be regulated by epigenetic mechanisms (Fig. 1), including DNA methylation and histone acetylation [14]. The core promoter lacks a conserved TATA and CAAT box sequence for transcription complex machinery to bind [15], which is generally characteristic of housekeeping genes [16]. Additionally, lacking a TATA box suggests the DAT may be susceptible to regulation by histone acetylation [17]. The core promoter is GC-rich making it possible for regulation through mechanisms involving DNA methylation [18, 19]. These stretches of GC nucleotide-rich segments form about 27 CpG islands, with the human DAT promoter nucleotide sequence comprised of 79 % GCs [20]. CpG islands may also be bound by transcription factors such as SP1, which was shown to contribute to regulation of DAT mRNA expression [21, 22]. However, the relative roles of HDACs and DNMTs in the regulation of DAT mRNA expression have not been established.
Fig. 1.
Features of the human dopamine transporter core promoter demonstrating potential for epigenetic regulation. GC boxes, which are targets of DNMTs, and a TATA-like sequence, which indicates potential for regulation by HDACs, are underlined
To date, these proposed mechanisms of epigenetic regulation have not been evaluated experimentally in a rigorous manner. Using pharmacological and genetic approaches, we sought to determine the relative contribution of DNA methylation and histone acetylation to DAT mRNA expression. The data reveal a modest (~1.5-fold) increase of DAT mRNA expression when DNA metyltransferase enzymes (DNMTs) were inhibited, but a more robust (up to tenfold) increase following pharmacological inhibition of histone deacetylases (HDACs). From a protein standpoint, similar increases in DAT protein were observed following DNMT and HDAC inhibition. These findings demonstrate that DAT expression is sensitive to manipulation of the cellular epigenetic machinery, and may provide insight into potential means to manipulate DAT expression in pathophysiological conditions where the DAT is disrupted.
Materials and Methods
Chemicals
Cell culture supplies, including minimum essential medium (MEM) with Earl’s salts and l-glutamine, nonessential amino acids, sodium pyruvate (100 mM), penicillin/streptomycin, fetal bovine serum (FBS), phosphate-buffered saline (PBS) without calcium and magnesium, and trypsin EDTA (0.05 % trypsin, 0.53 mM EDTA in Hank’s Balanced Salt Solution without sodium bicarbonate, calcium, and magnesium) were purchased from Mediatech (Herndon, VA). Valproic acid sodium salt (VPA), sodium butyrate (NaB), N-phthalyl-l-tryptophan (RG108) and dimethyl sulfoxide (DMSO) were purchased from Sigma-Aldrich (St. Louis, MO).
Cell Culture
SK-N-AS human neuroblastoma cells were purchased from American Type Culture Collection (ATCC, Manassass, VA). Cells were cultured in MEM supplemented with 10 % FBS, 5 mM nonessential amino acids, 5 mM sodium pyruvate, 50 IU/mL penicillin and 50 μg/mL streptomycin. Cells were maintained at 37 °C with 5 % CO2. SK-N-AS cells were selected for these in vitro studies because of their expression of dopaminergic markers, including the DAT [22, 23]. Cells were used between passages 5 and 10 following receipt from ATCC.
Drug Treatment
Cells were plated in supplemented MEM medium at 1 × 106 cells per well of a six well plate 24 h prior to treatment. Stock solutions of RG108 were prepared in DMSO and diluted to working concentrations in media. The final solvent concentration was no greater than 0.1 %, which did not alter the expression of any of the genes of interest compared to control expression. Stock solutions of VPA and NaB were prepared in media. Cells were exposed for 24 h to various concentrations of RG108 (0.25–5 μM), VPA (0.3–5 mM), and NaB (0.5–5 mM) before harvesting and isolation of RNA. Cells were lysed directly in the well with RLT buffer (Qiagen, Valencia, CA), centrifuged for 3 min at 14,000×g, and the supernatant frozen at −80 °C until RNA isolation using the Qiagen RNeasy kit.
siRNA Transfection
SK-N-AS cells were seeded at 1 × 106 per well in 2 mL antibiotic-free medium in a six well plate 24 h prior to transfection. The cells were 50–60 % confluent on the day of transfection. Cells were transfected using Lipofectamine RNAiMAX (Invitrogen, Carlsbad, CA) as per manufacturer instruction. Preliminary experiments determined the optimal siRNA concentration and timecourse of gene knockdown. For these experiments, siRNA (Santa Cruz, CA) was diluted with Opti-MEM Reduced Serum Medium (Invitrogen) to a final concentration of 20 nM per well. Lipofectamine was diluted separately with Opti-MEM. The diluted siRNA and Lipofectamine were mixed together to a final volume of 2 mL and incubated at room temperature for 20 min before adding to cells. A scrambled siRNA sequence was included to control for possible non-specific effects of transfection and was used at a 20 nM per well. 24 h post-transfection, RNA was isolated from cells as described below and knockdown assessed by qPCR.
RNA Isolation and cDNA Synthesis
RNA was isolated from cells using the Qiagen RNeasy Kit per the manufacturer’s instructions, as described previously [23]. RNA concentration was determined using NanoDrop 2000 spectrophotometer (NanoDrop Technologies, Wilmington, DE). A total of 1 μg RNA was used for cDNA synthesis, with the First Strand Synthesis Kit (Invitrogen) as per manufacturer’s instruction. Briefly, 1 μg of RNA, 1 μL random hexamers (50 ng/μL), 1 μL dNTP mix (10 mM) were mixed with DEPC-treated water (10 μL total volume), incubated in the thermocycler for 5 min at 65 °C and then placed on ice for 1 min. Synthesis mix was added to each sample (2 μL 10× RT buffer, 4 μL 25 mM MgCl2, 2 μL 0.1 M DTT, 1 μL RNase OUT) and incubated in the thermocycler at 25 °C for 2 min. SuperScript II RT (1 μL, 50 U/μL) was added to each sample and placed in the thermocycler: 10 min at 25 °C, 50 min at 42 °C, 15 min at 70 °C. The reaction was chilled on ice for 1 min followed by the addition of RNase H (1 μL, 2 U/μL) and thermocycler incubation at 37 °C for 20 min.
Quantitative Real-Time Polymerase Chain Reaction (RT-qPCR)
Reactions were performed in 25 μL total volume using SYBR Green Master Mix (Applied Biosystems, Carlsbad, CA); 1:15 diluted cDNA and 1 μM of forward and reverse primers. Primers were designed using the National Center for Biotechnology Information primer-blast application (http://www.ncbi.nlm.nih.gov/tools/primer-blast/index.cgi): DAT (Accession: NM001044.4; forward 5′-CCATACTGCAAG GTGTGGGC-3′; reverse 5′-CCAGGAGTTGTTGCAGTG GA-3′), DNMT1 (Accession NM 001130823.1; forward 5′-GAGGAAGCTGCTAAGGACTAGTTC-3′; reverse 5′-ACTGCACAATTTGATCACTAAATC-3′), HDAC1 (Accession: NM_004964.2; forward 5′-ACCCGGAGGAAAGTCTGTTA-3′, reverse 5′-GCTTTGTGAGGGCGATAGAT-3′); TATA Binding Protein (Accession: NM_003194.4; forward 5′-TTGTACCGCAGCTGCAAAAT-3′, reverse 5′-CTGAAAATCAGTGCCGTGGT-3′); β-Actin (Accession: NM_001101.3; forward 5′-CTCGCCTTTGCCGATCC-3′, reverse 5′-CATCACGCCCTGGTGC-3′). Thermal cycling conditions of the reaction were: 2 min at 50 °C and 10 min at 95 °C, followed by 40 cycles of 95 °C for 15 s and 1 min at the appropriate annealing temperature for each primer set. Relative gene expression was determined using the SYBR Green technology (Applied Biosystems, Carlsbad, CA) on a ViiA7TM Real-Time PCR system (Applied Biosystems). All samples were run in duplicate with actin and TATA Binding Protein (TBP) expression monitored as reference genes. Data analyzed using actin and TBP gave similar results (data not shown). Therefore, all data presented are based on normalization to TBP expression. All primer sets yielded a single PCR product of designed amplicon size as determined by agarose gel electrophoresis and inspection of the melting curve. Additionally, all primer sets amplified at a similar efficiency, as determined by preliminary studies involving a tenfold dilutional series. Raw data were analyzed using ViiA7 software and relative values were calculated in Microsoft Excel using ΔΔCt method as described by Livak and Schmittgen [24].
Western Immunoblotting
Following 24 h treatment with inhibitors, SK-N-AS cells were lysed with commercial cell lysis buffer (BioVision, Miltipas, CA) containing 0.1 % protease inhibitor cocktail (Sigma-Aldrich, St. Louis, MO). Protein concentrations were determined using the Pierce TM bicinchoninic acid (BCA) assay kit (Thermo Scientific) and 50 μg of protein sample was loaded per lane on a 4–12 % Bis–Tris Polyacrylamide Gel (Invitrogen). Proteins were transferred to a polyvinylidene difluoride (PVDF) membrane and blocked in 7.5 % non-fat milk in 0.1 % Tween 20 and Tris buffered saline (TTBS) for 1 h at room temperature. The membranes were incubated at 4 °C overnight with anti-DAT primary antibody (1:2000; cat #MAB369; Millipore), followed by a 1 h incubation with HRP-conjugated anti-rat secondary antibody (MP Biomedicals)at room temperature. The bound antibody was detected by SuperSignal® West Dura Extended Duration Substrate Kit (Thermo Scientific) and imaged using Alpha Innotech Fluorochem imaging system. The membrane was stripped using Pierce Stripping Buffer (Thermo Scientific, Waltham, MA) and re-probed with anti-tubulin (1:6000; Sigma) antibody to confirm equal protein loading.
Statistical Analysis
All data were analyzed using GraphPad Prism 5.0 Software (GraphPad Software, San Diego, CA). All experiments were performed 3–6 times on different days. Individual experiments were conducted in duplicate or triplicate and averaged. Data are presented as ±SEM and analyzed by ANOVA followed by Dunnett’s test for dose response data and western blot. Student’s t test was used for siRNA data.
Results
Pharmacological Inhibition of DNA Methyltransferases and Histone Deacetylases Increases DAT mRNA Expression
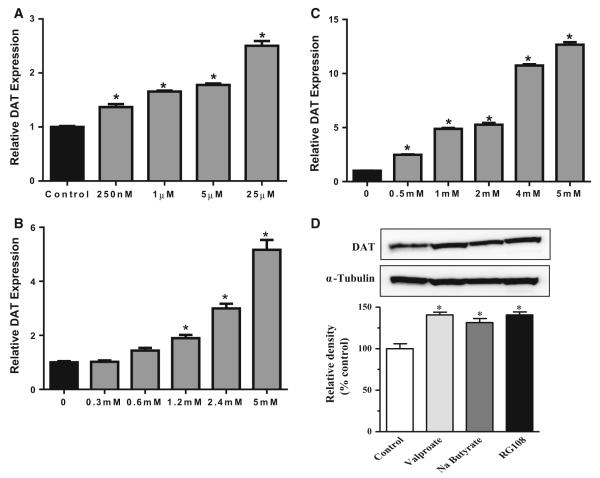
Exposure of cells to the DNMT inhibitor N-phthalyl-l-tryptophan (RG108) caused a significant dose-related increase in DAT mRNA expression (F4,15 = 156.5, p < 0.001), with the highest dose (25 μM) yielding about a 2.5-fold increase in DAT expression (Fig. 2a). To assess the potential for HDAC inhibition to regulate DAT expression, cells were treated with various doses of the HDAC inhibitors valproate and sodium butyrate for 24 h. Valproate treatment caused a significant dose-related increase in DAT mRNA expression (F5,32 = 72.61, p < 0.0001), that peaked at about fivefold at a dose of 5 mM (Fig. 2b). Cells treated with butyrate exhibited significantly increased DAT mRNA (F5,23 = 56.08, p < 0.0001), by up to tenfold at 5 mM (Fig. 2c). At the 25 μM dose of RG108 and the 5 mM dose of both sodium butyrate and valproate, DAT protein levels were significantly increased by about 50 % (F3,11 = 17.68, p = 0.007; Fig. 2d).
Fig. 2.
Pharmacological Inhibition of DNMTs or HDACs increases DAT mRNA expression in human SK-N-AS neuroblastoma cells. DAT mRNA expression following 24 h exposure to a N-phthalyl-l-tryptophan (RG108), b valproate (VPA), or c sodium butyrate (Na butyrate). d Protein levels of DAT are increased following exposure to valproate, sodium butyrate, and RG108. Data represent mean ±SEM, *p ≤ 0.05 compared to control. n = 3–6
siRNA-Mediated Knockdown of DNMT1 and HDAC1 Increases DAT mRNA Expression
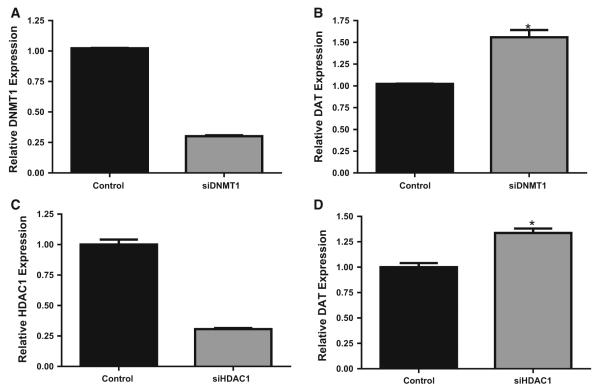
To confirm whether the DNMT inhibitors were acting directly on DNMTs and not other targets, we utilized siRNA knockdown technology to specifically reduce DNMT1 and HDAC1, the most abundant DNMT and HDAC isoforms. qPCR determined that siRNA treatment resulted in 70–85 % knockdown of DNMT1 (Fig. 3a). This was specific, as no effects were observed for the other DNMT isoforms (data not shown). Knockdown of DNMT1 significantly increased DAT mRNA expression by 1.6-fold (t = 6.4, df = 4; p = 0.001; Fig. 3b). Similarly, siRNA targeting HDAC1 significantly reduced HDAC1 mRNA by 60–70 % (Fig. 3c), and was specific to the targeted isoform (data not shown). HDAC1 knockdown resulted in a 1.5-fold increase in relative DAT mRNA expression (t = 5.59, df = 6; p = 0.001; Fig. 3d).
Fig. 3.
Knockdown of DNMT1 and HDAC1 by siRNA increases DAT mRNA expression in human SK-N-AS neuroblastoma cells. a DNMT1 mRNA expression following DNMT1 knockdown, b DAT expression following DNMT1 knockdown, c HDAC1 mRNA expression following HDAC1 knockdown, d DAT expression following HDAC1 knockdown. Data represent mean ±SEM, *p = 0.001 compared to control. n = 3–6
Discussion
In the present study, we used pharmacological inhibitors and siRNA knockdown of DNMTs and HDACs to test the relative contribution of DNA methylation and histone acetylation on the expression of DAT mRNA. The DNMT inhibitor RG108 caused moderate increases in DAT mRNA expression and DAT protein levels. This finding is similar to that observed with tyrosine hydroxylase, where treatment with the DNMT inhibitor 5-aza-2-deoxycytidine increased TH expression [25]. The specific role of DNMT1 was confirmed using siRNA targeting DNMT1.
In contrast to the moderate increases in DAT mRNA following DNMT inhibition or knockdown, we observed up to a tenfold increase in DAT mRNA expression for inhibition of HDACs with sodium butyrate. Our findings confirm previous reports, which observed increased DAT mRNA following in vitro valproate [22] or Trichostatin A treatment [26]. We also extended these findings by using siRNA to specifically target HDAC1, which is a predominant HDAC gene involved in many transcriptional regulation pathways [27, 28]. HDAC1 silencing caused a significant increase in DAT expression, but not to the extent of the pharmacological treatments. This may be because the chemical inhibitors used here target multiple members of the HDAC1 and 2A families. Another possibility is that other families of HDACs may exert a greater effect on the DAT. For example, HDACs 3, 5, and 11 are highly expressed in the substantia nigra and ventral tegmental areas, which contain the dopamine neuron cell bodies [29]. Thus, knockdown of additional HDAC isoforms or a pool of siRNA constructs targeting multiple HDACs may be necessary to reach maximum induction of DAT mRNA.
Because pharmacological alterations in methylation and histone acetylation have a global effect on many genes within cells [30], the effect observed on DAT mRNA expression may be the result of altered expression of transcription factors involved in DAT mRNA regulation. For example, the transcription factors Nurr1 and Pitx3 contribute to the expression of DAT and TH [31, 32] and treatment with DNMT and HDAC inhibitors increase Nurr1 and Pitx3 mRNA expression (Richardson, unpublished data). Moreover, other transcription factors such as Sp1/Sp3, are also involved in the regulation of DAT [33,34]. Indeed, Wang etal. [22] reported that SK-N-AS cells treated with valproate demonstrated a threefold increase in DAT mRNA, which appeared to be mediated by Sp1 and Sp3 transcription factor binding to specific GC-rich regions of the promoter. Our data confirm these findings with valproate and extend them to another another HDAC inhibitor, sodium butyrate. Additionally, we also observed increased DAT mRNA using a DNMT inhibitor and following knockdown of DNMT1. Both HDAC and DNMT inhibitors have significant effects on histone structure and often work in concert to regulate gene expression, suggesting chromatin modifications are likely significant contributors to DAT mRNA expression.
Conclusion
Taken together our data provide mechanistic support for in silico analyses studies which show potential of the DAT to be regulated through epigenetic mechanisms [14, 35]. Although both DNMT and HDAC inhibition altered DAT mRNA expression, the robust effect of HDAC inhibitors suggests a more dominant role of histone acetylation in the regulation of DAT expression compared to DNA methylation. This is consistent with a previous study which showed that the DAT is relatively hypomethylated in primary rat mesencephalic cultures, and thus more resistant to changes in expression due to alterations in promoter methylation [36]. Although future studies are needed to confirm the precise molecular mediators of HDAC and DNMT inhibition on DAT expression and the functional consequences of altered mRNA expresssion, the current findings increase our understanding of the role of epigenetic pathways in DAT gene regulation and provide evidence of pharmacological and genetic means to manipulate DAT expression.
Acknowledgments
This research was supported in part by NIEHS R01ES015991, R01ES021800, P30ES005022, and T32ES007148. The content of this article is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health, which had no role in the design, analysis, or writing of this manuscript.
Footnotes
Conflict of interest The authors declare no conflicts of interest regarding the content of this manuscript.
References
- 1.Haberland N, Hetey L. Studies in postmortem dopamine uptake. II. Alterations of the synaptosomal catecholamine uptake in postmortem brain regions in schizophrenia. J Neural Transm. 1987;68:303–313. doi: 10.1007/BF02098505. [DOI] [PubMed] [Google Scholar]
- 2.Cook EH, Jr, Stein MA, Krasowski MD, Cox NJ, Olkon DM, Kieffer JE, Leventhal BL. Association of attention-deficit disorder and the dopamine transporter gene. Am J Hum Genet. 1995;56:993–998. [PMC free article] [PubMed] [Google Scholar]
- 3.Malison RT, McDougle CJ, van Dyck CH, Scahill L, Baldwin RM, Seibyl JP, Price LH, Leckman JF, Innis RB. [123I]-beta-CIT SPECT imaging of striatal dopamine transporter binding in Tourette’s disorder. Am J Psychiatry. 1995;152:1359–1361. doi: 10.1176/ajp.152.9.1359. [DOI] [PubMed] [Google Scholar]
- 4.Giros B, Caron MG. Molecular characterization of the dopamine transporter. Trends Pharmacol Sci. 1993;14:43–49. doi: 10.1016/0165-6147(93)90029-j. [DOI] [PubMed] [Google Scholar]
- 5.Nirenberg MJ, Vaughan RA, Uhl GR, Kuhar MJ, Pickel VM. The dopamine transporter is localized to dendritic and axonal plasma membranes of nigrostriatal dopaminergic neurons. J Neurosci. 1996;16:436–447. doi: 10.1523/JNEUROSCI.16-02-00436.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jones SR, Gainetdinov RR, Jaber M, Giros B, Wightman RM, Caron MG. Profound neuronal plasticity in response to inactivation of the dopamine transporter. Proc Natl Acad Sci USA. 1998;95:4029–4034. doi: 10.1073/pnas.95.7.4029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gainetdinov RR, Caron MG. Monoamine transporters: from genes to behavior. Annu Rev Pharmacol Toxicol. 2003;43:261–284. doi: 10.1146/annurev.pharmtox.43.050802.112309. [DOI] [PubMed] [Google Scholar]
- 8.Giros B, Jaber M, Jones SR, Wightman RM, Caron MG. Hyperlocomotion and indifference to cocaine and amphetamine in mice lacking the dopamine transporter. Nature. 1996;379:606–612. doi: 10.1038/379606a0. [DOI] [PubMed] [Google Scholar]
- 9.Kilty JE, Lorang D, Amara SG. Cloning and expression of a cocaine-sensitive rat dopamine transporter. Science. 1991;254:578–579. doi: 10.1126/science.1948035. [DOI] [PubMed] [Google Scholar]
- 10.Vandenbergh DJ, Persico AM, Hawkins AL, Griffin CA, Li X, Jabs EW, Uhl GR. Human dopamine transporter gene (DAT1) maps to chromosome 5p15.3 and displays a VNTR. Genomics. 1992;14:1104–1106. doi: 10.1016/s0888-7543(05)80138-7. [DOI] [PubMed] [Google Scholar]
- 11.Vandenbergh DJ, Persico AM, Uhl GR. A human dopamine transporter cDNA predicts reduced glycosylation, displays a novel repetitive element and provides racially-dimorphic TaqI RFLPs. Brain Res Mol Brain Res. 1992;15:161–166. doi: 10.1016/0169-328x(92)90165-8. [DOI] [PubMed] [Google Scholar]
- 12.Kouzmenko AP, Pereira AM, Singh BS. Intronic sequences are involved in neural targeting of human dopamine transporter gene expression. Biochem Biophys Res Commun. 1997;240:807–811. doi: 10.1006/bbrc.1997.7754. [DOI] [PubMed] [Google Scholar]
- 13.Sacchetti P, Brownschidle LA, Granneman JG, Bannon MJ. Characterization of the 5′-flanking region of the human dopamine transporter gene. Brain Res Mol Brain Res. 1999;74:167–174. doi: 10.1016/s0169-328x(99)00275-2. [DOI] [PubMed] [Google Scholar]
- 14.Shumay E, Fowler JS, Volkow ND. Genomic features of the human dopamine transporter gene and its potential epigenetic States: implications for phenotypic diversity. Plos One. 2010;5:e11067. doi: 10.1371/journal.pone.0011067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martinat C, Bacci JJ, Leete T, Kim J, Vanti WB, Newman AH, Cha JH, Gether U, Wang H, Abeliovich A. Cooperative transcription activation by Nurr1 and Pitx3 induces embryonic stem cell maturation to the midbrain dopamine neuron phenotype. Proc Natl Acad Sci USA. 2006;103:2874–2879. doi: 10.1073/pnas.0511153103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blake MC, Jambou RC, Swick AG, Kahn JW, Azizkhan JC. Transcriptional initiation is controlled by upstream GC-box interactions in a TATAA-less promoter. Mol Cell Biol. 1990;10:6632–6641. doi: 10.1128/mcb.10.12.6632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Choi JK, Kim YJ. Epigenetic regulation and the variability of gene expression. Nat Genet. 2008;40:141–147. doi: 10.1038/ng.2007.58. [DOI] [PubMed] [Google Scholar]
- 18.Wang Y, Leung FC. An evaluation of new criteria for CpG islands in the human genome as gene markers. Bioinformatics. 2004;20:1170–1177. doi: 10.1093/bioinformatics/bth059. [DOI] [PubMed] [Google Scholar]
- 19.Hillemacher T, Frieling H, Hartl T, Wilhelm J, Kornhuber J, Bleich S. Promoter specific methylation of the dopamine transporter gene is altered in alcohol dependence and associated with craving. J Psychiatr Res. 2009;43:388–392. doi: 10.1016/j.jpsychires.2008.04.006. [DOI] [PubMed] [Google Scholar]
- 20.Kawarai T, Kawakami H, Yamamura Y, Nakamura S. Structure and organization of the gene encoding human dopamine transporter. Gene. 1997;195:11–18. doi: 10.1016/s0378-1119(97)00131-5. [DOI] [PubMed] [Google Scholar]
- 21.Holler M, Westin G, Jiricny J, Schaffner W. Sp1 transcription factor binds DNA and activates transcription even when the binding site is CpG methylated. Genes Dev. 1988;2:1127–1135. doi: 10.1101/gad.2.9.1127. [DOI] [PubMed] [Google Scholar]
- 22.Wang J, Michelhaugh SK, Bannon MJ. Valproate robustly increases Sp transcription factor-mediated expression of the dopamine transporter gene within dopamine cells. Eur J Neurosci. 2007;25:1982–1986. doi: 10.1111/j.1460-9568.2007.05460.x. [DOI] [PubMed] [Google Scholar]
- 23.Hossain MM, Richardson JR. Mechanism of pyrethroid pesticide-induced apoptosis: role of calpain and the ER stress pathway. Toxicol Sci. 2011;122:512–525. doi: 10.1093/toxsci/kfr111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(–delta delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 25.Aranyi T, Faucheux BA, Khalfallah O, Vodjdani G, Biguet NF, Mallet J, Meloni R. The tissue-specific methylation of the human tyrosine hydroxylase gene reveals new regulatory elements in the first exon. J Neurochem. 2005;94:129–139. doi: 10.1111/j.1471-4159.2005.03173.x. [DOI] [PubMed] [Google Scholar]
- 26.Bence M, Koller J, Sasvari-Szekely M, Keszler G. Transcriptional modulation of monoaminergic neurotransmission genes by the histone deacetylase inhibitor trichostatin A in neuroblastoma cells. J Neural Transm. 2012;119:17–24. doi: 10.1007/s00702-011-0688-4. [DOI] [PubMed] [Google Scholar]
- 27.Ma P, Schultz RM. Histone deacetylase 1 (HDAC1) regulates histone acetylation, development, and gene expression in preimplantation mouse embryos. Dev Biol. 2008;319:110–120. doi: 10.1016/j.ydbio.2008.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Montgomery RL, Davis CA, Potthoff MJ, Haberland M, Fielitz J, Qi X, Hill JA, Richardson JA, Olson EN. Histone deacetylases 1 and 2 redundantly regulate cardiac morphogenesis, growth, and contractility. Genes Dev. 2007;21:1790–1802. doi: 10.1101/gad.1563807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Broide RS, Redwine JM, Aftahi N, Young W, Bloom FE, Winrow CJ. Distribution of histone deacetylases 1–11 in the rat brain. J Mol Neurosci. 2007;31:47–58. doi: 10.1007/BF02686117. [DOI] [PubMed] [Google Scholar]
- 30.Robert MF, Morin S, Beaulieu N, Gauthier F, Chute IC, Barsalou A, MacLeod AR. DNMT1 is required to maintain CpG methylation and aberrant gene silencing in human cancer cells. Nat Genet. 2003;33:61–65. doi: 10.1038/ng1068. [DOI] [PubMed] [Google Scholar]
- 31.Volpicelli F, De Gregorio R, Pulcrano S, Perrone-Capano C, di Porzio U, Bellenchi GC. Direct regulation of Pitx3 expression by Nurr1 in culture and in developing mouse midbrain. Plos One. 2012;7:e30661. doi: 10.1371/journal.pone.0030661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aarnisalo P, Kim CH, Lee JW, Perlmann T. Defining requirements for heterodimerization between the retinoid X receptor and the orphan nuclear receptor Nurr1. J Biol Chem. 2002;277:35118–35123. doi: 10.1074/jbc.M201707200. [DOI] [PubMed] [Google Scholar]
- 33.Wang J, Bannon MJ. Sp1 and Sp3 activate transcription of the human dopamine transporter gene. J Neurochem. 2005;93:474–482. doi: 10.1111/j.1471-4159.2005.03051.x. [DOI] [PubMed] [Google Scholar]
- 34.Hwang DY, Hong S, Jeong JW, Choi S, Kim H, Kim J, Kim KS. Vesicular monoamine transporter 2 and dopamine transporter are molecular targets of Pitx3 in the ventral midbrain dopamine neurons. J Neurochem. 2009;111:1202–1212. doi: 10.1111/j.1471-4159.2009.06404.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhao Y, Xiong N, Liu Y, Zhou Y, Li N, Qing H, Lin Z. Human dopamine transporter gene: differential regulation of 18-kb haplotypes. Pharmacogenomics. 2013;14:1481–1494. doi: 10.2217/pgs.13.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.He XB, Yi SH, Rhee YH, Kim H, Han YM, Lee SH, Lee H, Park CH, Lee YS, Richardson E, Kim BW. Prolonged membrane depolarization enhances midbrain dopamine neuron differentiation via epigenetic histone modifications. Stem Cells. 2011;29:1861–1873. doi: 10.1002/stem.739. [DOI] [PubMed] [Google Scholar]