Abstract
Cancer etiology is influenced by alterations in protein synthesis that are not fully understood. In this study, we took a novel approach to investigate the role of the eukaryotic translation initiation factor eIF5A in human cervical cancers, where it is widely overexpressed. eIF5A contains the distinctive amino acid hypusine, which is formed by a posttranslational modification event requiring deoxyhypusine hydroxylase (DOHH), an enzyme that can be inhibited by the drugs ciclopirox and deferiprone. We found that proliferation of cervical cancer cells can be blocked by DOHH inhibition with either of these pharmacologic agents, as well as by RNA interference–mediated silencing of eIF5A, DOHH, or another enzyme in the hypusine pathway. Proteomic and RNA analyses in HeLa cervical cancer cells identified two groups of proteins in addition to eIF5A that were coordinately affected by ciclopirox and deferiprone. Group 1 proteins (Hsp27, NM23, and DJ-1) were downregulated at the translational level, whereas group 2 proteins (TrpRS and PRDX2) were upregulated at the mRNA level. Further investigations confirmed that eIF5A and DOHH are required for Hsp27 expression in cervical cancer cells and for regulation of its key target IκB and hence NF-κB. Our results argue that mature eIF5A controls a translational network of cancer-driving genes, termed the eIF5A regulon, at the levels of mRNA abundance and translation. In coordinating cell proliferation, the eIF5A regulon can be modulated by drugs such as ciclopirox or deferiprone, which might be repositioned to control cancer cell growth.
Introduction
Despite advances in detection and prevention, cervical cancer remains the third most frequently diagnosed female cancer worldwide, with an estimated 275,000 deaths in 2008 (1). For the United States, the National Cancer Institute estimated that more than 12,000 new cases will be diagnosed in 2013, and that every third patient with this diagnosis will die despite state-of-the-art treatment. The identification of novel targets in cancer cells and the analysis of the molecular response to their suppression will promote the rational development of novel therapeutic modalities.
Translation, a key process in the gene expression pathway, is often dysregulated in cancer (2). A strong correlation has been established between cancer and overexpression of the eukaryotic initiation factor 5A (eIF5A), which functions in protein synthesis (3). Humans have 2 eIF5A isoforms: eIF5A1, expressed in many normal tissues, and eIF5A2, which enjoys more limited expression and distribution. Elevated levels of both isoforms characterize a variety of cancers and tumor-derived cell lines, and accumulating evidence links eIF5A to cell proliferation, cancer progression, invasiveness, metastasis, and poor clinical prognosis (3, 4).
Both isoforms carry the amino acid hypusine, which is apparently unique to eIF5A and essential for many (if not all) of its functions (3). Hypusine is formed posttranslationally in sequential reactions catalyzed by 2 dedicated enzymes, deoxyhypusine synthase (DHS) and deoxyhypusine hydroxylase (DOHH; Fig. 1A). The singularity of this pathway presents attractive targets for drug development and cancer therapy (5).
Figure 1.
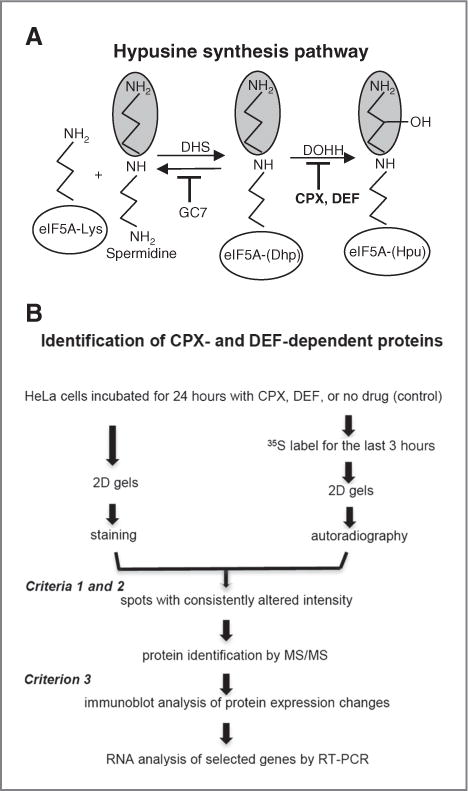
Pathway of eIF5A modification and experimental strategy. A, hypusine formation and inhibition. DHS catalyzes aminobutyl transfer from spermidine onto the ɛ-amino group of lysine-50 of human eIF5A using NAD+ as cofactor, yielding deoxyhypusine (Dhp). DOHH hydroxylates Dhp to hypusine (Hpu) in an Fe(II)-dependent reaction using molecular oxygen. The spermidine analog GC7 (N1-guanyl-1,7-diaminoheptane) inhibits DHS. The drugs ciclopirox (CPX; 6-cyclohexyl-1-hydroxy-4-methyl-(1H)-pyrid-2-one) and deferiprone (DEF; 1,2-dimethyl-3-hydroxypyridin-4-one) inhibit DOHH (19). B, strategy for identification of CPX- and DEF-dependent proteins.
The involvement of DOHH in cell-cycle progression was recognized early (6), and specific inhibitors were characterized (7). Of particular interest are 2 drugs that inhibit DOHH and hypusine formation at clinically relevant concentrations: ciclopirox (CPX), a topical antifungal (8), and deferiprone (DEF), used to treat transfusional iron-overload such as in thalassemia (9). Both drugs block cell proliferation and display antineoplastic potential. Thus, CPX has been shown to inhibit the proliferation of cells in culture (10–12) and of breast cancer and myeloma xenograft growth in mice (11–13). CPX also inhibits angiogenesis and lymphangiogenesis in established culture models (10, 14). DEF inhibits the growth of HeLa cells derived from cervical carcinoma as well as other cancer cell lines (15), and its analog mimosine slows the growth of subcutaneous lung and pancreatic cancer xenografts in mice (16). The DHS substrate analog GC7 (Fig. 1A) also impairs cancer cell growth, for example, of glioblastoma cells (17).
This study addresses the relationship between eIF5A, hypusine and gene expression in cervical cancer, and identifies cellular protein targets of the drugs CPX and DEF. We show that mature, hypusyl-eIF5A1 is highly expressed in proliferating cervical cancer tissue, of both the squamous cell and adenocarcinoma types, and in adenocarcinoma-derived HeLa cells. Cell proliferation was inhibited, and morphologic changes occurred, after treatment with the drugs or eIF5A silencing. We devised a proteomic approach (Fig. 1B) to identify proteins that are regulated by both CPX and DEF using HeLa cells as a model. Five proteins, in addition to mature eIF5A itself, were identified whose synthesis and accumulation were coordinately affected by these drugs. The proteins play key roles in cancer cell proliferation, survival, and metastasis and fall into 2 groups. Group 1 proteins were downregulated by the drugs at the translational level, whereas group 2 proteins were upregulated at the mRNA level. The group 1 protein Hsp27, a molecular chaperone, is a cancer biomarker and potential target for cancer therapy (18). RNA interference experiments showed that hypusyl-eIF5A is required for Hsp27 synthesis and modulates transcription via NF-κB. Our data suggest that mature eIF5A1 regulates the expression of a set of genes, designated the eIF5A regulon, that are required for cell proliferation. Taken together, these findings prompt the design of clinical trials to examine the use of the drugs CPX and DEF and related agents, in anti-cancer therapy.
Materials and Methods
Immunohistochemistry
Cervical cancers were diagnosed by an experienced pathologist as squamous cell carcinoma (12 samples) or adenocarcinoma (9 samples). Tissue sections were stained for eIF5A using NIH-353 antibody (kindly provided by Dr. M.H. Park) and with Ki-67 antibody (Dako) as described previously, in compliance with an Institutional Review Board-approved protocol allowing anonymous use of archival, formalin-fixed, paraffin-embedded biopsy material (5).
Cell culture
Cells from the American Type Culture Collection were maintained as recommended and seeded 1 day before treatment. CPX (Sigma-Aldrich) and DEF (Calbiochem) were freshly dissolved in PBS and added to the medium for 24 to 72 hours.
Immunoblotting
Cells were lysed as described previously (19). Proteins were assayed (DC Protein Assay, BioRad) and 1 to 20 μg samples were resolved by SDS-PAGE. After transfer to polyvinylidene difluoride (PVDF) membranes, blots were probed with primary antibody followed by horseradish peroxidase–conjugated secondary antibody (Jackson Immunoresearch). Rabbit anti-peroxiredoxin 2 antibody was from Upstate Cell Signaling, mouse anti-eIF5A and anti-NM23 from Becton Dickinson (BD; Transduction Laboratories), mouse anti-TrpRS and anti-Hsp27 from Novus Biologicals, rabbit anti-DJ-1 from Cell Signaling Technology, and mouse anti-β-actin and anti-α-tubulin from Sigma. Rabbit anti-DOHH antibody was kindly provided by Dr. Myung Hee Park (NIH/NIDCR, Bethesda, MD). Signal was detected using enhanced chemiluminescence.
RNA interference
For cell growth assays, HeLa cells were transfected with 20 nmol/L siRNA and U2OS cells with 50 nmol/L siRNA using Hiperfect (Qiagen). Cells were harvested at 72 hours posttransfection unless otherwise stated. siRNA sequences (sense/antisense) were as follows: eIF5A1 (Ambion), 5′-GGUCCAUCUGGUUGGUAUUTT/5′-AAUACCAACCAGAUGGACCTT; luciferase (Ambion) as negative control for HeLa cell proliferation assays, 5′-CGUACGCGGAAUACUUCGATT/5′-UCGAAGUAUUCCGCGUACGTT. DHS and DOHH “ON-TARGET plus SMART pool” and nontargeting control siRNAs were from Dharmacon. For luciferase assays, HeLa cells were transfected with 50 nmol/L siRNA using Hiperfect (Qiagen). Cells were re-seeded at 48 hours and co-transfected at 72 hours with the HIV-1 molecular clone pNL4-3 lucE− and pCMV-Renilla plasmid using Jet-PEI (polyplus transfections). Cells were harvested 24 hours later and expression of reporter genes analyzed (19).
Metabolic labeling
HeLa cells were incubated for 21 hours with 30 μmol/L CPX or 200 μmol/L DEF, washed twice with PBS and once with Dulbecco’s Modified Eagle Medium (DMEM) lacking methionine and cysteine, then incubated for 3 hours in the same medium containing 50 μCi/mL of Trans[35S]-label (MP Biomed) with drug as appropriate. Cells were washed with ice-cold PBS before scraping. For measurement of hypusine and deoxyhypusine synthesis, cells were labeled with [3H]-spermidine for 24 hours and analyzed as described previously (20).
Two-dimensional gel electrophoresis and protein identification
HeLa cell extracts were prepared by incubation for 30 minutes at 4°C in lysis buffer (50 mmol/L Tris-HCl, pH 7.5, 150 mmol/L NaCl, 0.5% Nonidet P-40, 1 mmol/L dithiothreitol, 0.1% SDS, 50 mmol/L NaF, 1 mmol/L Na orthovanadate, and protease inhibitor cocktail from Sigma-Aldrich), and analyzed in 2-dimensional (2D) gels (21). Nonradioactive gels were fixed (40% ethanol, 10% acetic acid), stained with SYPRO Ruby (BioRad), and scanned on a Typhoon scanner (GE Healthcare). Radioactive gels were dried under vacuum and exposed to autoradiography film. Spot intensities were compared visually in 3 replicate experiments for each of the analysis using spots with unchanged densities as landmarks. Spots from stained gels were processed for MALDI-TOF-TOF mass spectrometry analysis (Supplementary Table S1; ref. 21).
RNA extraction and real-time PCR
Total RNA was isolated using TRIzol (Invitrogen Life Technologies). cDNA was generated using 2 μg total RNA, random hexamers, and MultiScribe reverse transcriptase (Applied Biosystems). PCR was carried out in an Applied Biosystems 7500 apparatus, using SYBR Green mix (Roche Applied Biosystems) and AmpErase (Applied Biosystems). Primers were: Hsp27, 5′-GACACTCCTGCAGCAGATGCA/5′-GTTCAACTCTTTTGCGTGGCAG; NM23, 5′-CCATTCTTTGCCGGCCTG/5′-GTGAAAAGCAATGTGGTCTG; PRDX2, 5′-GCGCATCGGAAAGCCAGC/5′-CAAGCGTCTGGTCACGTCAG; TrpRS, 5′-GGAGCTGTTCAACAGCATCG/5′-CATTCTCCACAGCATAGCTATA; eIF5A1, 5′-GACTTCCAGCTGATTGGCATCCAG/5′-GCGGGCCTTATTTTGCCATGGCCTTGATTG; DJ-1, 5′-GATGTCATGAGGCGAGCTGGG/5′-CTCCTTCACAGCAGCAGACTC; and β-actin, 5′-AAATCGTGCGTGACATTAAGG/5′-AGCACTGTGTTGGCGTACAG.
Results
eIF5A expression in human cervical carcinoma
eIF5A is overexpressed in a number of malignant tissues including vulvar high-grade intraepithelial neoplasia (VIN), the precursor to vulvar cancer (5). To evaluate expression in cervical cancer, we conducted immunohistochemical studies with NIH-353 antibody, which recognizes mature hypusyl-eIF5A1 (5), and with Ki-67 antibody to identify proliferating cells. NIH-353 immunoreactivity located strictly to Ki-67–positive areas in both major types of cervical cancer, that is, invasive solid tumor cords and nests in squamous cell carcinoma and invasive vacuole-containing glandular sheets of columnar cells in adenocarcinoma (Fig. 2A). These observations indicate that hypusyl-eIF5A1 is highly expressed in the vast majority of cervical tumors.
Figure 2.

eIF5A expression in cervical cancer and in cancer-derived cell lines. A, tissue localization of mature, hypusyl-eIF5A1 (detected by NIH-353; left) and of Ki-67 protein (right) by immunohistochemical staining of squamous cell carcinoma (top; neighboring sections) and adenocarcinoma (bottom; adjacent sections). Bars, 600 μm. NIH-353 strongly stains infiltrating malignant areas and faintly stains glandular structures (arrow) and nonmalignant tissue elements, reproducing the Ki-67 pattern. B, expression of eIF5A in cell lines. Whole-cell extracts (10 μg protein) were analyzed by immunoblotting using antibody NIH-353 against eIF5A with actin as standard. C, as in B, comparing the indicated amounts of HeLa and U2OS protein, standardized with tubulin.
Correspondingly, hypusyl-eIF5A is abundant in HeLa cells, derived from cervical adenocarcinoma. Immunoblotting with NIH-353 antibody revealed varying levels of expression among cancer cell types (Fig. 2B). Comparison with U2OS cells (derived from osteosarcoma) indicates a range of at least 5-fold in proliferating cell lines (Fig. 2C). The high level of mature eIF5A1 observed in HeLa cells corroborates previous observations (20), matching its high expression in cervical cancer biopsies (Fig. 2A).
Mature eIF5A is required for cell growth
RNA silencing was deployed to determine whether eIF5A1 overexpression is directly involved in cell proliferation. Reduction in HeLa cell number became evident 4 days after transfection with siRNA directed against eIF5A1 (si5A) compared with control siRNA (siC; Fig. 3A, top). Inhibition of cell proliferation correlated with the reduction in the eIF5A1 protein level (Fig. 3A, bottom). Notably, both declined more rapidly in U2OS cells that contain less eIF5A (Fig. 3B). Furthermore, depletion of the eIF5A modifying enzyme DOHH (Fig. 1A) also reduced proliferation of HeLa and U2OS cells (Fig. 3A and B). These data indicate that hypusyl-eIF5A is critical for cell proliferation in both of these cancer cell lines.
Figure 3.
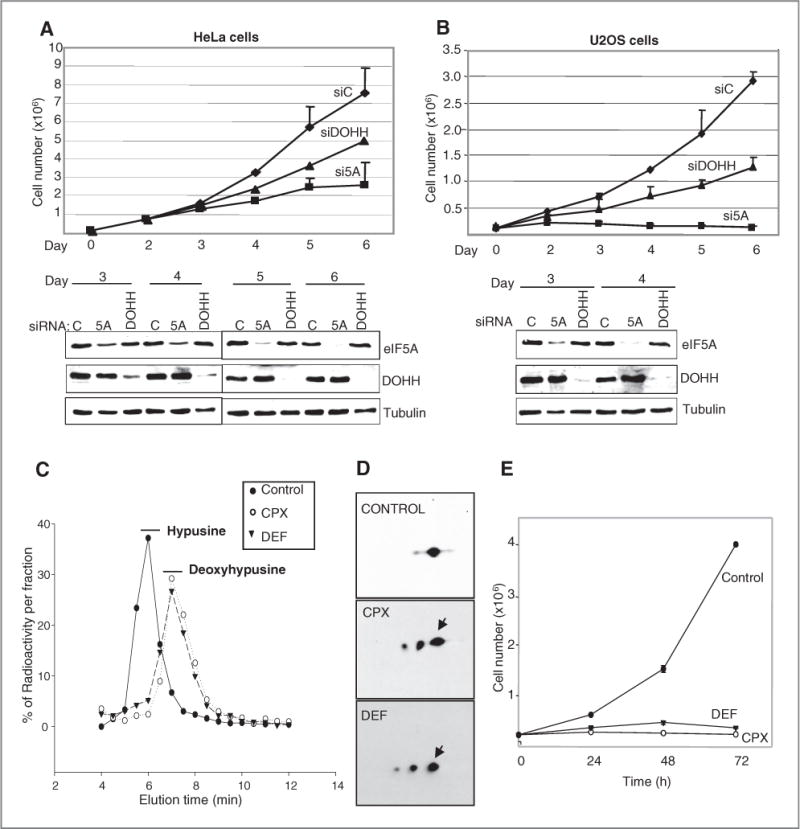
Hypusyl-eIF5A requirement for cell proliferation. A, HeLa cells were transfected with siRNAs directed against eIF5A or DOHH or with control siRNA (siC). Trypan blue-excluding viable cells were counted (top) and cell extract protein (1 μg) was analyzed by immunoblotting (bottom) on the days indicated. eIF5A was detected by eIF5A1 (BD) antibody directed against the protein’s C terminus (independent of hypusine modification). B, as in A, using 10 μg samples of U2OS cell extract protein. C, HeLa cells were incubated with [3H]-spermidine and CPX (30 μmol/L) or DEF (200 μmol/L) as indicated. Acid hydrolysates were fractionated and fractions assayed for radioactivity. Positions of [3H]-hypusine and [3H]-deoxyhypusine are indicated. D, extracts of cells treated with CPX or DEF were resolved in 2D gels. eIF5A was detected using BD antibody (arrows, spots with similar isoelectric points). E, cell proliferation in HeLa cell cultures exposed to the drugs.
CPX and DEF inhibit eIF5A maturation and cell proliferation
We used HeLa cells to study the effects of 2 drugs, CPX and DEF, which inhibit the hydroxylation of deoxyhypusyl-eIF5A by DOHH (Fig. 1A). To monitor the drugs’ action in HeLa cells, we measured hypusine formation by labeling eIF5A with [3H]-spermidine. Inhibitor constants, Ki, were 6.25 μmol/L for CPX and 152 μmol/L for DEF (Supplementary Fig. S1). Pharmacologic concentrations of CPX (30 μmol/L) or DEF (200 μmol/L; refs. 10, 19) suppressed hypusine labeling by >90% with concomitant appearance of deoxyhypusine (Fig. 3C). Immature forms of eIF5A1 accumulated in the presence of the drugs as revealed by 2D gel electrophoresis (Fig. 3D). Without drug treatment, eIF5A was represented by a single predominant spot with a pI of about 5.3, corresponding to hypusyl-eIF5A (22). Treatment with CPX or DEF led to the appearance of 2 spots with more acidic pIs (~5.2 and ~5.1) associated with acetylated forms of deoxyhypusyl-eIF5A (23). Thus, the drugs prevented the maturation of eIF5A and led to the accumulation of its immediate precursor, deoxyhypusyl-eIF5A.
Consistent with their inhibitory action on DOHH, both drugs rapidly inhibited HeLa cell proliferation (Fig. 3E), similar to the effects of eIF5A or DOHH knockdown (Fig. 3A). Drug-treated cells retained >93% of control viability at 24 hours, exhibited minimal apoptosis, and were arrested in the G1 to S-phase of the cell cycle (Supplementary Fig. S2), as reported previously (6, 7, 15, 24). They exhibited morphologic changes and an increase in size (Supplementary Fig. S3), reminiscent of the phenotype of eIF5A-deficient yeast (25, 26).
Modulation of protein expression by CPX and DEF
eIF5A is believed to be required for the translation of a specific class of proteins (24, 25). As CPX and DEF inhibit eIF5A maturation and HeLa cell proliferation, we sought to identify cellular proteins that are downstream targets of the drugs. Our strategy combined the analysis of protein levels and translation in drug-treated and untreated cells using 2D gels (Fig. 1B). Representative gel images are presented (Supplementary Fig. S4), together with sections of stained gels (Fig. 4A) and [35S]-labeled gels (Fig. 4B) containing spots whose intensities changed with drug treatment. Several spots decreased in intensity as expected (e.g., spot 10) but others were observed to increase (e.g., spot 6).
Figure 4.
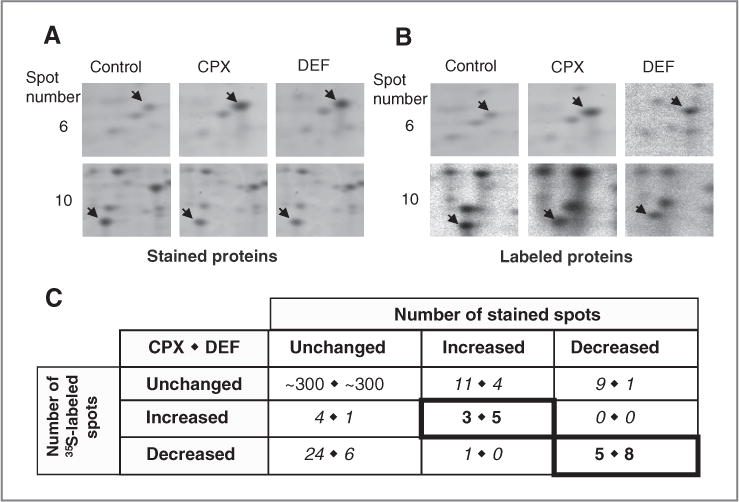
Effects of CPX and DEF on HeLa cell protein synthesis and accumulation. A, extracts of HeLa cells treated with CPX or DEF and control cells were resolved in 2D gels and stained with SYPRO Ruby. Sections show representative differentially expressed stained spots (arrows). B, as in A, except that the cells were labeled with [35S]-methionine and -cysteine and spots were detected by autoradiography. C, enumeration of 2D gel spots that changed after treatment with CPX (left numbers) or DEF (right numbers) as detected by staining (columns) and labeling (rows). Bold-bordered numerals denote concordant spots, which were examined further. Italicized numerals denote discordant spots.
We enumerated the spots in both the staining and labeling analyses that increased, decreased, or were unaltered in intensity after drug treatment (Fig. 4C). The overwhelming preponderance of spots did not change in intensity with CPX or DEF. Among those that did change, attention was focused on proteins whose labeling and accumulation varied in parallel (criterion 1, Fig. 1B). With CPX, 5 spots decreased and 3 spots increased in both types of analysis; with DEF, 8 spots decreased and 5 increased in both types of analysis (highlighted in Fig. 4C).
As the drugs might impinge on cellular pathways in addition to that involving eIF5A, we further limited consideration to spots that were affected in the same way by CPX and DEF (criterion 2). Seven spots varied coordinately with the 2 drugs: 3 increased and 4 decreased. It is notable that all 3 proteins upregulated by CPX and nearly all (4 of 5) of the downregulated proteins were shared with DEF and that most (3 of 5) of the proteins upregulated by DEF and half (4 of 8) of the downregulated proteins were in common. This high degree of concordance lends confidence that the changes reflect a common pharmacologic target and biochemical pathway.
Identification of DOHH-dependent proteins
Proteins whose synthesis was affected by both drugs were eluted from unlabelled gels, analyzed by mass spectrometry, and identified through database searches (Fig. 5A; Supplementary Table S1). Identifications were checked by immunoblotting with specific antibodies (criterion 3, Fig. 1B). All were confirmed except for one upregulated protein identified as 4-hydroxyphenylpyruvate dioxygenase (4-HPD), which could not be detected by immunoblotting (data not shown). Accordingly, 4-HPD is not included in the list of identified proteins and was not pursued further.
Figure 5.

Identification and expression of drug-regulated proteins. A, identity and properties of proteins affected by both CPX and DEF and identified by mass spectrometry. Molecular weights (MW) and isoelectric points (pI) were calculated from protein sequences (www.phosphositeplus.org and www.expasy.org). B–G, gene expression analysis of drug effects in HeLa cells. Top, immunoblot analysis of the downregulated (B–E) and upregulated (F and G) proteins with actin as a control. Bar graphs, protein levels relative to actin were evaluated by immunoblotting (filled bars) and corresponding RNA levels were assayed using RT-PCR (gray bars). Data are means of 3 independent experiments with SD.
In addition to eIF5A1, the downregulated proteins are heat shock protein 27 (Hsp27), DJ-1 (PARK7), and non-metastatic protein 23 (NM23), designated group 1. The confirmed upregulated proteins, designated group 2, are peroxiredoxin 2 (PRDX2) and tryptophanyl-tRNA synthetase (TrpRS).
Differential regulation of gene expression by the drugs
Immunoblotting and real-time (RT)-PCR were carried out to examine the effect of CPX and DEF on the identified proteins and their cognate mRNAs (Fig. 5B–G).
Consistent with the action of the drugs on its posttranslational modification (Fig. 3C and D), eIF5A1 expression was not significantly affected at the total protein level (Fig. 5B, filled bars) or at the RNA level (Fig. 5B, gray bars).
On the other hand, CPX and DEF reduced the expression of Hsp27, NM23, and DJ-1 proteins (Fig. 5C–E, filled bars), in agreement with the 2D gel data. The degree of inhibition was about 50% with CPX and about 30% with DEF. Exceptionally, the DEF-induced reduction in the level of DJ-1 was less, possibly because the drug affects the oxidation status of this protein, resulting in a change that is registered primarily in 2D gels (27). As with eIF5A1, RT-PCR revealed no significant changes in transcript levels (Fig. 5C–E, gray bars), indicating that the downregulation of Group 1 proteins by the drugs is due to translational control.
The drug-induced upregulation of PRDX2 and TrpRS was confirmed by immunoblotting (Fig. 5F and G, filled bars). Increased expression of these proteins was accompanied by increased levels of their transcripts (Fig. 5F and G, gray bars), indicating that CPX and DEF upregulate group 2 proteins at the level of mRNA transcription or stability. Interestingly, the increase in TrpRS RNA markedly exceeded that of the protein, possibly reflecting its secretion as an angiostatic factor (discussed below).
Mature eIF5A modulates Hsp27 levels and NF-κB activity
To document the connection between a drug target and eIF5A modification, we selected Hsp27 for further study. Hsp27 expression is increased in high-grade intraepithelial neoplasia and squamous cell carcinoma of the cervix (28) and several other cancers. It has been directly implicated in tumor progression (29) and radio- and chemoresistance (18). Furthermore, Hsp27 activates signaling via the transcription factor NF-κB by facilitating the turnover of its inhibitor IκB (30).
Our findings with CPX and DEF (Fig. 5C) suggested that hypusyl-eIF5A is required for elevated Hsp27 synthesis and, by extension, for NF-κB–dependent transcription. To examine these inferences, we depleted HeLa cells for eIF5A1 or its modifying enzymes. Immunoblotting showed that siRNA directed against eIF5A1, DHS, or DOHH reduced the level of Hsp27 (Fig. 6A), as with drug treatment (Fig. 5C) and led to concomitant elevation of IκB as predicted (Fig. 6A).
Figure 6.
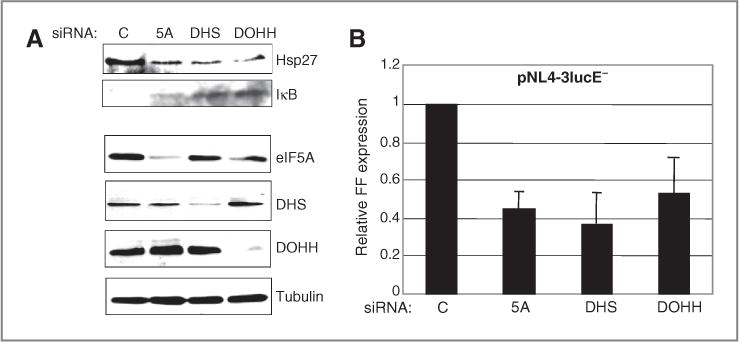
Depletion of mature eIF5A induces IκB and inhibits transcription directed by the HIV-1 promoter. HeLa cells were transfected with siRNAs directed against eIF5A, DHS, or DOHH or control siRNA, then with pCMV-Renilla and the HIV-1 molecular clone pNL4-3lucE− encoding firefly luciferase (FF). A, immunoblot analysis. eIF5A was detected with BD antibody. B, expression of FF luciferase normalized to Renilla luciferase activity with SD.
NF-κB is essential for HIV-1 transcription (31), and CPX and DEF have been shown to inhibit expression from the HIV-1 promoter (19). We therefore depleted HeLa cells for eIF5A1, DHS, or DOHH and then transfected with an HIV-1 molecular clone expressing firefly luciferase. Reporter gene expression was inhibited by 50% to 60% in all cases (Fig. 6B). As IκB is barely detectable in control cells (Fig. 6A), these results indicate that hypusyl-eIF5A plays an important role in regulating Hsp27 levels and NF-κB activity in HeLa cells.
Discussion
The identification of 2 groups of target proteins sensitive to CPX and DEF has implications for the regulation of gene expression by eIF5A and for the exploration of the drugs’ therapeutic potential in cancer.
Robustness of target identification
The cornerstone of our strategy for identifying drug targets was multiplexed 2D gel analysis, and its reliability follows from the stipulation of 3 stringent criteria (Fig. 1B). First, we considered only those proteins that responded in the same way to drug treatment when staining and labeling analyses were compared. Of the spots that increased or decreased in at least one type of analysis, about 25% (21 of 82) satisfied this criterion (Fig. 4C). Second, we demanded coordinate response to CPX and DEF: 7 of the remaining spots satisfied this criterion. Third, after identification by mass spectrometry, we required confirmation by immunoblotting: 6 proteins met this final criterion (Fig. 5A), testifying to the rigor of the overall strategy.
Criterion 1 was evidently an effective filter, but the requirement for consistent behavior probably eliminated genuine drug targets that turn over slowly. Indeed, almost half of the spots with altered intensity (30 of 61) displayed decreased labeling but unchanged staining, hallmarks of proteins with long half-lives. Most of the other discordant spots were unchanged in labeling yet increased (15 of 61) or decreased (10 of 61) in staining, perhaps signifying posttranslational processing. A few (5 of 61) increased in labeling without a change in staining, possibly attributable to a large protein pool size. Reassuringly, only one spot behaved anomalously, increasing in one type of analysis and decreasing in the other.
Criterion 2, which was motivated by the likelihood that drugs can have differential side effects, was also an effective filter. It eliminated 7 spots that displayed drug-specific behavior, potential false-positives that were more prevalent with DEF (6 spots) than CPX (1 spot).
Although robust, it should also be noted that the protocol favors proteins that are relatively abundant, rapidly synthesized, and methionine/cysteine-containing. Some 300 proteins were surveyed in our gels, a sampling of less than 1% of the human proteome, so the target proteins identified almost certainly represent a subset of those affected by CPX and DEF.
Putative eIF5A regulon
Results presented here (Fig. 3C and D) establish that CPX and DEF inhibit DOHH in HeLa cells, leading to a deficit in hypusine and accumulation of deoxyhypusine in eIF5A. Although both drugs chelate iron (8, 9), the lowering of intracellular iron is insufficient for this activity (19, 32). Rather, they appear to fit into the enzyme’s active site and abstract its iron, causing structural collapse (19). While contributions of other pathways cannot be ruled out, the evidence points to eIF5A maturation as the primary target with far-reaching consequences including inhibition of cell proliferation resulting from the depletion of hypusyl-eIF5A and/or the accumulation of deoxyhypusyl-eIF5A (Fig. 3).
The 5 newly identified drug-sensitive proteins (Fig. 5A) are not documented cellular binding partners of eIF5A or its modifying enzymes (33), nor do they uniformly harbor proline-rich translational signals characterized for the bacterial eIF5A homolog EF-P (Supplementary Fig. S5). Rather, they appear to be secondary drug targets regulated by hypusyl-eIF5A, as shown for Hsp27 (Fig. 6), and they fall into 2 groups distinguished by both the direction of the drug response and the level at which it is exerted.
Group 1 targets are downregulated by CPX and DEF apparently at the level of translation, consistent with the function of eIF5A in protein synthesis (34). Depletion of eIF5A in yeast decreases protein synthesis only partially (25, 35), however, implying that it functions as a selective translational enhancer for group 1 mRNAs.
Group 2 targets are upregulated at the RNA level by CPX and DEF, indicating that their transcription or mRNA stability is increased. eIF5A has been implicated in mRNA turnover in yeast (35) and human cells (Hoque and colleagues, unpublished results), suggesting that eIF5A-dependent selective turnover could be responsible for group 2 mRNA regulation.
Coordinate regulation of these proteins has also been observed in proteomic studies of cells treated with other agents. For example, the expression of eIF5A and NM23 is induced and that of PRDX2 and TrpRS is repressed, after p53 activation by mitomycin C (36). We therefore advance a model (Fig. 7) in which hypusyl-eIF5A1 modulates a regulon, or RNA operon (37), that controls cell proliferation and possibly other processes such as the response to oxidative stress. Mechanistically, we envision that eIF5A (or possibly one or more of its target proteins) preferentially binds a subset of cellular mRNAs (38) and recruits additional mRNA-binding proteins that impinge differentially on the 2 groups of gene products, thereby facilitating the translation of group 1 mRNAs and accelerating the turnover of group 2 mRNAs.
Figure 7.
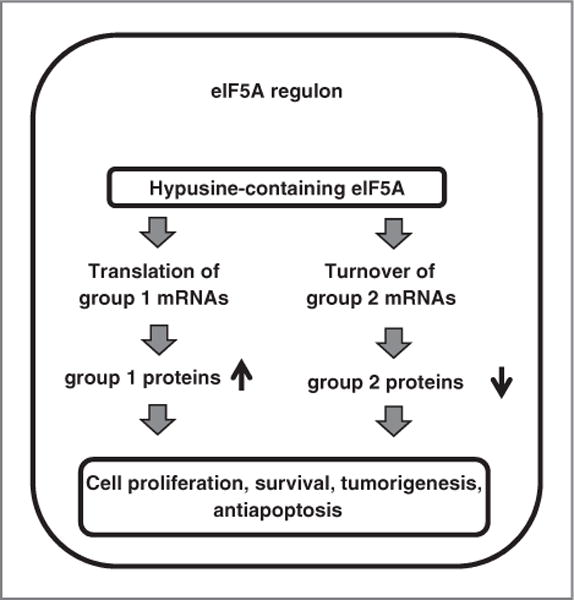
Model for the function of the eIF5A regulon. Mature eIF5A is proposed to affect protein expression differentially by enhancing the translation of group 1 mRNAs, but the turnover of group 2 mRNAs (see text for details).
Oncological implications
Like eIF5A, group 1 proteins are associated with cell proliferation and cancer (39–41). The best characterized is the chaperonin Hsp27, a marker for unfavorable prognosis in many human cancers and a potential therapeutic target (18, 29, 42, 43). Similarly, DJ-1 is highly expressed in a number of human cancers (41). DJ-1 enhances cell survival via activation of NF-κB and its silencing inhibits cell proliferation (41, 44). NM23 catalyzes the phosphorylation of nucleoside diphosphates and its expression is related to cell proliferative activity (45). Although initially considered an inhibitor of metastasis, accumulating data reveal differential expression of NM23 isoforms and indicate a more complex relationship with cancer. Recent work identified NM23 as a predictor of poor survival in pancreatic cancer (46). Furthermore, all three group 1 proteins associate with factors that control cell shape, providing a plausible explanation for the morphologic changes brought about by CPX and DEF, which are possibly related to cell motility changes and metastasis (Supplementary Fig. S4; ref. 41). Thus, the downregulation of Hsp27, DJ-1, and NM23 by CPX and DEF suggests that these drugs could have a therapeutic effect in malignancies characterized by overexpression of group 1 proteins.
Conversely, group 2 proteins can exert negative effects on cell proliferation. High TrpRS expression correlates with lower risk of recurrence and increased survival in patients with colorectal cancer (47). In addition to its canonical intracellular role in tRNA aminoacylation, N-terminally truncated forms of TrpRS are secreted and have anti-angiogenic activity, blocking VEGF-induced cell proliferation and migration (48, 49). Thus, like IFN-γ administration, drug-induced increase of TrpRS is consistent with anti-angiogenic and antiproliferative activity that could discourage tumor growth. PRDX2 is a member of a family of cellular peroxidases that have a complex relationship with tumor formation (50). The growth of breast cancer cells that metastasize to lung was inhibited by PRDX2 knockdown, whereas overexpression in bone metastatic breast cancer cells reduced the skeletal tumor burden and bone destruction (51). This suggests that the enzyme prevents tumor growth in hypoxic conditions, possibly signaled by deoxyhypusyl-eIF5A, although it protects against oxidative stress in oxygen-rich environments. Therefore, upregulation of group 2 proteins by CPX and DEF may restrain the growth of certain tumors.
In summary, we propose that eIF5A coordinates a set of genes at the level of translation (increased) and of mRNA abundance (decreased, possibly by facilitating mRNA decay). These genes define an eIF5A regulon that activates cell proliferation, and loss of eIF5A function has anti-cancer effects. The widely used drugs CPX and DEF interfere with eIF5A posttranslational modification and function, therefore qualify as candidates for exploratory oncologic trials and further drug development. Consistent with this conclusion, oral CPX has shown promise in treating patients with acute myelogenous leukemia in a recent proof-of-concept trial (A.D. Schimmer, personal communication).
Supplementary Material
Acknowledgments
The authors thank Anita Antes for invaluable assistance, Dr Jyoti Mundra for contributions to RNA interference experiments, and the Foundation of UMDNJ for support. Apologies for limited literature citation.
Footnotes
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Disclosure of Potential Conflicts of Interest
D.S. Heller has ownership interest (including patents) with Hanauske-Abel HM, Park MH, Wolf EC, Clement PMJ, Cracchiolo BM, Heller D (2002) Method of detecting proliferating cells by antibodies and their derivatives directed at the hypusine region of eIF-5A. United States Patent pending (NJMS-02-71). H.M. Hanauske-Abel has ownership interest (including patents) from Cornell University owned US patent #6,046,219 in deferiprone. B. Cracchiolo and H.M. Hanauske-Abel have ownership interest (including patents) from Rutgers University and NIH co-owned US patent #7,141,589 in ciclopirox, Rutgers University and NIH co-owned US patent #7,141,589 in ciclopirox (for Dr. Cracchiolo, spouse). No potential conflicts of interest were disclosed by the other authors.
Authors’ Contributions
Conception and design: E. Memin, M. Hoque, H.M. Hanauske-Abel, T. Pe’ery, M.B. Mathews
Development of methodology: E. Memin, M. Hoque, T. Pe’ery, M.B. Mathews
Acquisition of data (provided animals, acquired and managed patients, provided facilities, etc.): E. Memin, M. Hoque, M.R. Jain, D.S. Heller
Analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis): E. Memin, M. Hoque, M.R. Jain, D.S. Heller, H. Li, B. Cracchiolo, H.M. Hanauske-Abel, T. Pe’ery, M.B. Mathews
Writing, review, and/or revision of the manuscript: E. Memin, M. Hoque, M.R. Jain, D.S. Heller, H. Li, B. Cracchiolo, H.M. Hanauske-Abel, T. Pe’ery, M.B. Mathews
Administrative, technical, or material support (i.e., reporting or organizing data, constructing databases): M. Hoque
Study supervision: M. Hoque, H. Li, T. Pe’ery, M.B. Mathews
References
- 1.Jemal A, Center MM, DeSantis C, Ward EM. Global patterns of cancer incidence and mortality rates and trends. Cancer Epidemiol Biomarkers Prev. 2010;19:1893–907. doi: 10.1158/1055-9965.EPI-10-0437. [DOI] [PubMed] [Google Scholar]
- 2.Ruggero D. Translational control in cancer etiology. Cold Spring Harb Perspect Biol. 2013;5:a012336. doi: 10.1101/cshperspect.a012336. pii. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Caraglia M, Park MH, Wolff EC, Marra M, Abbruzzese A. eIF5A isoforms and cancer: two brothers for two functions? Amino Acids. 2011;44:103–9. doi: 10.1007/s00726-011-1182-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shek FH, Fatima S, Lee NP. Implications of the use of eukaryotic translation initiation factor 5A (eIF5A) for prognosis and treatment of hepatocellular carcinoma. Int J Hepatol. 2012;2012:760928. doi: 10.1155/2012/760928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cracchiolo BM, Heller DS, Clement PM, Wolff EC, Park MH, Hanauske-Abel HM. Eukaryotic initiation factor 5A-1 (eIF5A-1) as a diagnostic marker for aberrant proliferation in intraepithelial neoplasia of the vulva. Gynecol Oncol. 2004;94:217–22. doi: 10.1016/j.ygyno.2004.03.018. [DOI] [PubMed] [Google Scholar]
- 6.Hoffman BD, Hanauske-Abel HM, Flint A, Lalande M. A new class of reversible cell cycle inhibitors. Cytometry. 1991;12:26–32. doi: 10.1002/cyto.990120105. [DOI] [PubMed] [Google Scholar]
- 7.Abbruzzese A, Hanauske-Abel HM, Park MH, Henke S, Folk JE. The active site of deoxyhypusyl hydroxylase: use of catecholpeptides and their component chelator and peptide moieties as molecular probes. Biochim Biophys Acta. 1991;1077:159–66. doi: 10.1016/0167-4838(91)90053-3. [DOI] [PubMed] [Google Scholar]
- 8.Subissi A, Monti D, Togni G, Mailland F. Ciclopirox: recent nonclinical and clinical data relevant to its use as a topical antimycotic agent. Drugs. 2010;70:2133–52. doi: 10.2165/11538110-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 9.Neufeld EJ. Update on iron chelators in thalassemia. Hematology Am Soc Hematol Educ Program. 2010;2010:451–5. doi: 10.1182/asheducation-2010.1.451. [DOI] [PubMed] [Google Scholar]
- 10.Clement PM, Hanauske-Abel HM, Wolff EC, Kleinman HK, Park MH. The antifungal drug ciclopirox inhibits deoxyhypusine and proline hydroxylation, endothelial cell growth and angiogenesis in vitro. Int J Cancer. 2002;100:491–8. doi: 10.1002/ijc.10515. [DOI] [PubMed] [Google Scholar]
- 11.Zhou H, Shen T, Luo Y, Liu L, Chen W, Xu B, et al. The antitumor activity of the fungicide ciclopirox. Int J Cancer. 2010;127:2467–77. doi: 10.1002/ijc.25255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eberhard Y, McDermott SP, Wang X, Gronda M, Venugopal A, Wood TE, et al. Chelation of intracellular iron with the antifungal agent ciclopirox olamine induces cell death in leukemia and myeloma cells. Blood. 2009;114:3064–73. doi: 10.1182/blood-2009-03-209965. [DOI] [PubMed] [Google Scholar]
- 13.Kim Y, Schmidt M, Endo T, Lu D, Carson D, Schmidt-Wolf IG. Targeting the Wnt/beta-catenin pathway with the antifungal agent ciclopirox olamine in a murine myeloma model. In Vivo. 2011;25:887–93. [PubMed] [Google Scholar]
- 14.Luo Y, Zhou H, Liu L, Shen T, Chen W, Xu B, et al. The fungicide ciclopirox inhibits lymphatic endothelial cell tube formation by suppressing VEGFR-3-mediated ERK signaling pathway. Oncogene. 2011;30:2098–107. doi: 10.1038/onc.2010.590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Simonart T, Boelaert JR, Mosselmans R, Andrei G, Noel JC, De Clercq E, et al. Antiproliferative and apoptotic effects of iron chelators on human cervical carcinoma cells. Gynecol Oncol. 2002;85:95–102. doi: 10.1006/gyno.2001.6570. [DOI] [PubMed] [Google Scholar]
- 16.Chang HC, Weng CF, Yen MH, Chuang LY, Hung WC. Modulation of cell cycle regulatory protein expression and suppression of tumor growth by mimosine in nude mice. Int J Oncol. 2000;17:659–65. doi: 10.3892/ijo.17.4.659. [DOI] [PubMed] [Google Scholar]
- 17.Preukschas M, Hagel C, Schulte A, Weber K, Lamszus K, Sievert H, et al. Expression of eukaryotic initiation factor 5A and hypusine forming enzymes in glioblastoma patient samples: implications for new targeted therapies. PLoS One. 2012;7:e43468. doi: 10.1371/journal.pone.0043468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zoubeidi A, Gleave M. Small heat shock proteins in cancer therapy and prognosis. Int J Biochem Cell Biol. 2012;44:1646–56. doi: 10.1016/j.biocel.2012.04.010. [DOI] [PubMed] [Google Scholar]
- 19.Hoque M, Hanauske-Abel HM, Palumbo P, Saxena D, D’Alliessi Gandolfi D, Park MH, et al. Inhibition of HIV-1 gene expression by Ciclopirox and Deferiprone, drugs that prevent hypusination of eukaryotic initiation factor 5A. Retrovirology. 2009;6:90. doi: 10.1186/1742-4690-6-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Clement PM, Johansson HE, Wolff EC, Park MH. Differential expression of eIF5A-1 and eIF5A-2 in human cancer cells. Febs J. 2006;273:1102–14. doi: 10.1111/j.1742-4658.2006.05135.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jain MR, Li M, Chen W, Liu T, de Toledo SM, Pandey BN, et al. In vivo space radiation-induced non-targeted responses: late effects on molecular signaling in mitochondria. Curr Mol Pharmacol. 2011;4:106–14. doi: 10.2174/1874467211104020106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Park MH. The identification of an eukaryotic initiation factor 4D precursor in spermidine-depleted Chinese hamster ovary cells. J Biol Chem. 1988;263:7447–9. [PubMed] [Google Scholar]
- 23.Klier H, Csonga R, Joao HC, Eckerskorn C, Auer M, Lottspeich F, et al. Isolation and structural characterization of different isoforms of the hypusine-containing protein eIF-5A from HeLa cells. Biochemistry. 1995;34:14693–702. doi: 10.1021/bi00045a010. [DOI] [PubMed] [Google Scholar]
- 24.Hanauske-Abel HM, Slowinska B, Zagulska S, Wilson RC, Staiano-Coico L, Hanauske AR, et al. Detection of a sub-set of polysomal mRNAs associated with modulation of hypusine formation at the G1-S boundary. Proposal of a role for eIF-5A in onset of DNA replication. FEBS Lett. 1995;366:92–8. doi: 10.1016/0014-5793(95)00493-s. [DOI] [PubMed] [Google Scholar]
- 25.Kang HA, Hershey JW. Effect of initiation factor eIF-5A depletion on protein synthesis and proliferation of Saccharomyces cerevisiae. J Biol Chem. 1994;269:3934–40. [PubMed] [Google Scholar]
- 26.Zanelli CF, Valentini SR. Pkc1 acts through Zds1 and Gic1 to suppress growth and cell polarity defects of a yeast eIF5A mutant. Genetics. 2005;171:1571–81. doi: 10.1534/genetics.105.048082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim YC, Kitaura H, Taira T, Iguchi-Ariga SM, Ariga H. Oxidation of DJ-1-dependent cell transformation through direct binding of DJ-1 to PTEN. Int J Oncol. 2009;35:1331–41. [PubMed] [Google Scholar]
- 28.Ono A, Kumai T, Koizumi H, Nishikawa H, Kobayashi S, Tadokoro M. Overexpression of heat shock protein 27 in squamous cell carcinoma of the uterine cervix: a proteomic analysis using archival formalin-fixed, paraffin-embedded tissues. Hum Pathol. 2009;40:41–9. doi: 10.1016/j.humpath.2008.06.010. [DOI] [PubMed] [Google Scholar]
- 29.Straume O, Shimamura T, Lampa MJ, Carretero J, Oyan AM, Jia D, et al. Suppression of heat shock protein 27 induces long-term dormancy in human breast cancer. Proc Natl Acad Sci U S A. 2012;109:8699–704. doi: 10.1073/pnas.1017909109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Parcellier A, Schmitt E, Gurbuxani S, Seigneurin-Berny D, Pance A, Chantome A, et al. HSP27 is a ubiquitin-binding protein involved in I-kappaBalpha proteasomal degradation. Mol Cell Biol. 2003;23:5790–802. doi: 10.1128/MCB.23.16.5790-5802.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nabel G, Baltimore D. An inducible transcription factor activates expression of human immunodeficiency virus in T cells. Nature. 1987;326:711–3. doi: 10.1038/326711a0. [DOI] [PubMed] [Google Scholar]
- 32.Hanauske-Abel HM, Saxena D, Palumbo PE, Hanauske AR, Luchessi AD, Cambiaghi TD, et al. Drug-induced reactivation of apoptosis abrogates HIV-1 infection. PLoS One. 2013;8:e74414. doi: 10.1371/journal.pone.0074414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sievert H, Venz S, Platas-Barradas O, Dhople VM, Schaletzky M, Nagel CH, et al. Protein-protein-interaction network organization of the hypusine modification system. Mol Cell Proteomics. 2012;11:1289–305. doi: 10.1074/mcp.M112.019059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Henderson A, Hershey JW. The role of eIF5A in protein synthesis. Cell Cycle. 2011;10:3617–8. doi: 10.4161/cc.10.21.17850. [DOI] [PubMed] [Google Scholar]
- 35.Zuk D, Jacobson A. A single amino acid substitution in yeast eIF-5A results in mRNA stabilization. Embo J. 1998;17:2914–25. doi: 10.1093/emboj/17.10.2914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rahman-Roblick R, Roblick UJ, Hellman U, Conrotto P, Liu T, Becker S, et al. p53 targets identified by protein expression profiling. Proc Natl Acad Sci U S A. 2007;104:5401–6. doi: 10.1073/pnas.0700794104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Keene JD. RNA regulons: coordination of post-transcriptional events. Nat Rev Genet. 2007;8:533–43. doi: 10.1038/nrg2111. [DOI] [PubMed] [Google Scholar]
- 38.Xu A, Jao DL, Chen KY. Identification of mRNA that binds to eukaryotic initiation factor 5A by affinity co-purification and differential display. Biochem J. 2004;384:585–90. doi: 10.1042/BJ20041232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vargas-Roig LM, Fanelli MA, Lopez LA, Gago FE, Tello O, Aznar JC, et al. Heat shock proteins and cell proliferation in human breast cancer biopsy samples. Cancer Detect Prev. 1997;21:441–51. [PubMed] [Google Scholar]
- 40.Cipollini G, Berti A, Fiore L, Rainaldi G, Basolo F, Merlo G, et al. Down-regulation of the nm23.h1 gene inhibits cell proliferation. Int J Cancer. 1997;73:297–302. doi: 10.1002/(sici)1097-0215(19971009)73:2<297::aid-ijc22>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 41.He X, Zheng Z, Li J, Ben Q, Liu J, Zhang J, et al. DJ-1 promotes invasion and metastasis of pancreatic cancer cells by activating SRC/ERK/uPA. Carcinogenesis. 2012;33:555–62. doi: 10.1093/carcin/bgs002. [DOI] [PubMed] [Google Scholar]
- 42.Mjahed H, Girodon F, Fontenay M, Garrido C. Heat shock proteins in hematopoietic malignancies. Exp Cell Res. 2012;318:1946–58. doi: 10.1016/j.yexcr.2012.05.012. [DOI] [PubMed] [Google Scholar]
- 43.Schultz CR, Golembieski WA, King DA, Brown SL, Brodie C, Rempel SA. Inhibition of HSP27 alone or in combination with pAKT inhibition as therapeutic approaches to target SPARC-induced glioma cell survival. Mol Cancer. 2012;11:20. doi: 10.1186/1476-4598-11-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shen Z, Jiang Z, Ye D, Xiao B, Zhang X, Guo J. Growth inhibitory effects of DJ-1-small interfering RNA on laryngeal carcinoma Hep-2 cells. Med Oncol. 2011;28:601–7. doi: 10.1007/s12032-010-9474-7. [DOI] [PubMed] [Google Scholar]
- 45.Keim D, Hailat N, Melhem R, Zhu XX, Lascu I, Veron M, et al. Proliferation-related expression of p19/nm23 nucleoside diphosphate kinase. J Clin Invest. 1992;89:919–24. doi: 10.1172/JCI115672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Takadate T, Onogawa T, Fujii K, Motoi F, Mikami S, Fukuda T, et al. Nm23/Nucleoside diphosphate kinase-a as a potent prognostic marker in invasive pancreatic ductal carcinoma identified by proteomic analysis of laser micro-dissected formalin-fixed paraffin-embedded tissue. Clin Proteomics. 2012;9:8. doi: 10.1186/1559-0275-9-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ghanipour A, Jirstrom K, Ponten F, Glimelius B, Pahlman L, Birgisson H. The prognostic significance of tryptophanyl-tRNA synthetase in colorectal cancer. Cancer Epidemiol Biomarkers Prev. 2009;18:2949–56. doi: 10.1158/1055-9965.EPI-09-0456. [DOI] [PubMed] [Google Scholar]
- 48.Kapoor M, Zhou Q, Otero F, Myers CA, Bates A, Belani R, et al. Evidence for annexin II-S100A10 complex and plasmin in mobilization of cytokine activity of human TrpRS. J Biol Chem. 2008;283:2070–7. doi: 10.1074/jbc.M706028200. [DOI] [PubMed] [Google Scholar]
- 49.Wakasugi K, Slike BM, Hood J, Otani A, Ewalt KL, Friedlander M, et al. A human aminoacyl-tRNA synthetase as a regulator of angiogenesis. Proc Natl Acad Sci U S A. 2002;99:173–7. doi: 10.1073/pnas.012602099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Neumann CA, Fang Q. Are peroxiredoxins tumor suppressors? Curr Opin Pharmacol. 2007;7:375–80. doi: 10.1016/j.coph.2007.04.007. [DOI] [PubMed] [Google Scholar]
- 51.Stresing V, Baltziskueta E, Rubio N, Blanco J, Arriba M, Valls J, et al. Peroxiredoxin 2 specifically regulates the oxidative and metabolic stress response of human metastatic breast cancer cells in lungs. Oncogene. 2013;32:724–35. doi: 10.1038/onc.2012.93. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


