Figure 4. Upregulated mRNA expression levels of COL6A3 in colon cancers revealed by MS and Oncomine analyses.
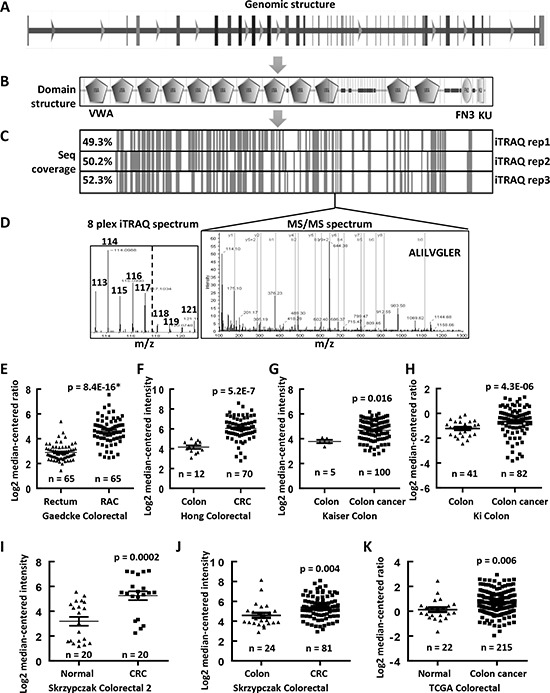
A. The genomic structure of COL6A3 gene contained 44 exons. B. The protein structure of COL6A3, as analyzed by SMART (http://smart.embl.de), contained 12 von Willebrand factor type A domain (VMA), one fibronectin type 3 domain (FN3) and one KU domain. C. The sequence coverage of COL6A3 by MS analysis. The vertical lines indicated the peptides identified in all replicated iTRAQ LC-MS. D. A represented MSMS spectra of a peptide of COL6A3, which was identified in all LC-MS experiments. The iTRAQ spectrum was shown to the left, with a dash line separating the tags labeling the fibroblasts and the colon cancer cell lines. COL6A3 mRNA expression was found to be upregulated in colon cancers comparing with normal colonic tissues revealed by data-mining of gene expression microarray datasets deposited in Oncomine DB (https://www.oncomine.org/) including E. Gaedcke Colorectal [27], F. Hong Colorectal [28], G. Kaiser Colon [29], H. Ki Colon [30], I. Skrzypczak Colorectal 2 [31], J. Skrzypczak Colorectal [31] and K. TCGA Colorectal. Skrzypczak Colorectal 2 contains a subset of selected samples from Skrzypczak Colorectal further treated with micro-dissection. The case number of each category was listed under the dot. The significances are calculated using two-ways of Student's t tests for comparing between normal and tumor samples. The p values marked with an asterisk were calculated using paired Student's t test. A p value < 0.05 was considered as statistically significant. RAC, rectal adenocarcinoma; CRC, colorectal carcinoma.
