SUMMARY
BYL719, which selectively inhibits the alpha isoform of the phosphatidylinositol 3-kinase (PI3K) catalytic subunit (p110α), is currently in clinical trials for the treatment of solid tumors, especially luminal breast cancers with PIK3CA mutations and/or HER2 amplification. This study reveals that, even among these sensitive cancers, the initial efficacy of p110α inhibition is mitigated by rapid re-accumulation of the PI3K product PIP3 produced by the p110β isoform. Importantly, the reactivation of PI3K mediated by p110β does not invariably restore AKT phosphorylation, demonstrating the limitations of using phospho-AKT as a surrogate to measure PI3K activation. Consistently, we show that the addition of the p110β inhibitor to BYL719 prevents the PIP3 rebound and induces greater antitumor efficacy in HER2-amplified and PIK3CA mutant cancers.
INTRODUCTION
Phosphatidylinositol 3-kinase (PI3K) lipid kinases promote cell growth and survival of many cancer types, including breast cancers that have genetic activation of this pathway in up to 70% of cases (Hernandez-Aya and Gonzalez-Angulo, 2011). The class IA PI3Ks are holoenzymes consisting of a regulatory subunit (p85) bound to one of three possible catalytic subunits, p110α (encoded by PIK3CA), p110β, and p110δ. All class IA PI3Ks are activated by recruitment to phosphotyrosine peptides, such as those on receptor tyrosine kinases (RTKs) and adaptors (e.g., IRS-1), via the SH2 domains of p85. However, p110β is the only member that can also be activated by G protein-coupled receptors (GPCRs). Class I PI3Ks function by phosphorylating the D-3 position of the inositol ring of phosphatidylinositol (4,5)-bi-sphosphate (PI(4,5)P2) to generate the second messenger phos-phatidylinositol (3,4,5)-trisphosphate (PIP3). PIP3 activates its effectors, such as AKT, by recruiting them to the membrane via their pleckstrin homology (PH) domains (Engelman, 2009).
Due to the technical challenges of directly measuring the PI3K product, PIP3, the phosphorylation level of the downstream effector AKT has historically been used to indirectly assess the activity of class I PI3K. Although this approach has proven to be rather useful, it is based on the assumption that this readout is always proportional to PIP3 levels, which may apply to only a limited range of PI3K activation levels and/or in specific cellular contexts. For instance, AKT phosphorylation correlates poorly with PIK3CA mutation status in some types of breast cancer (Stemke-Hale et al., 2008; Vasudevan et al., 2009). Furthermore, the use of AKT phosphorylation as readout of PI3K activity ignores other potentially relevant downstream targets of PI3K. For example, one report demonstrated that PI3K controls cell viability of some PIK3CA mutant breast cancers through SGK3 in an AKT-independent manner (Vasudevan et al., 2009). More recently, it has been demonstrated that in luminal breast cancers PI3K regulates Rac-ERK signaling independently of AKT via the PH-domain containing Rac guanine exchange factor, P-Rex1 (Ebi et al., 2013).
Over the past several years, significant investment has been made into the rational development of both isoform-specific and pan-PI3K inhibitors. However, it is unclear which type of PI3K inhibitor will be most effective among the different subtypes of breast cancer. One potential limitation of pan-PI3K inhibitors is the toxicity that may emerge when all PI3K isoforms are suppressed. Thus, if a single PI3K isoform is driving cell proliferation in a specific cancer, then isoform-specific inhibitors may exhibit a greater therapeutic window, and thus permit more complete inhibition of the critical isoform. Among all class IA members, p110α is the isoform predominantly mutated in cancers (Samuels et al., 2004) and has a prominent role in controlling cell growth in solid malignancies (Bader et al., 2006; Foukas et al., 2006; Zhao et al., 2006). For these reasons, the α-specific inhibitor BYL719 is currently being tested in clinical trials (Juric and Baselga, 2012; Juric et al., 2012). Because preclinical data indicate that breast cancers are particularly sensitive to pan-PI3K pathway inhibitors (Faber et al., 2009; O’Brien et al., 2010), we explored which genetically defined breast cancers were more sensitive to BYL719 and if these tumors were solely dependent on p110α.
RESULTS
Reactivation of PI3K Signaling following Inhibition with α-Selective Inhibitor BYL719 in HER2-Amplified Breast Cancers
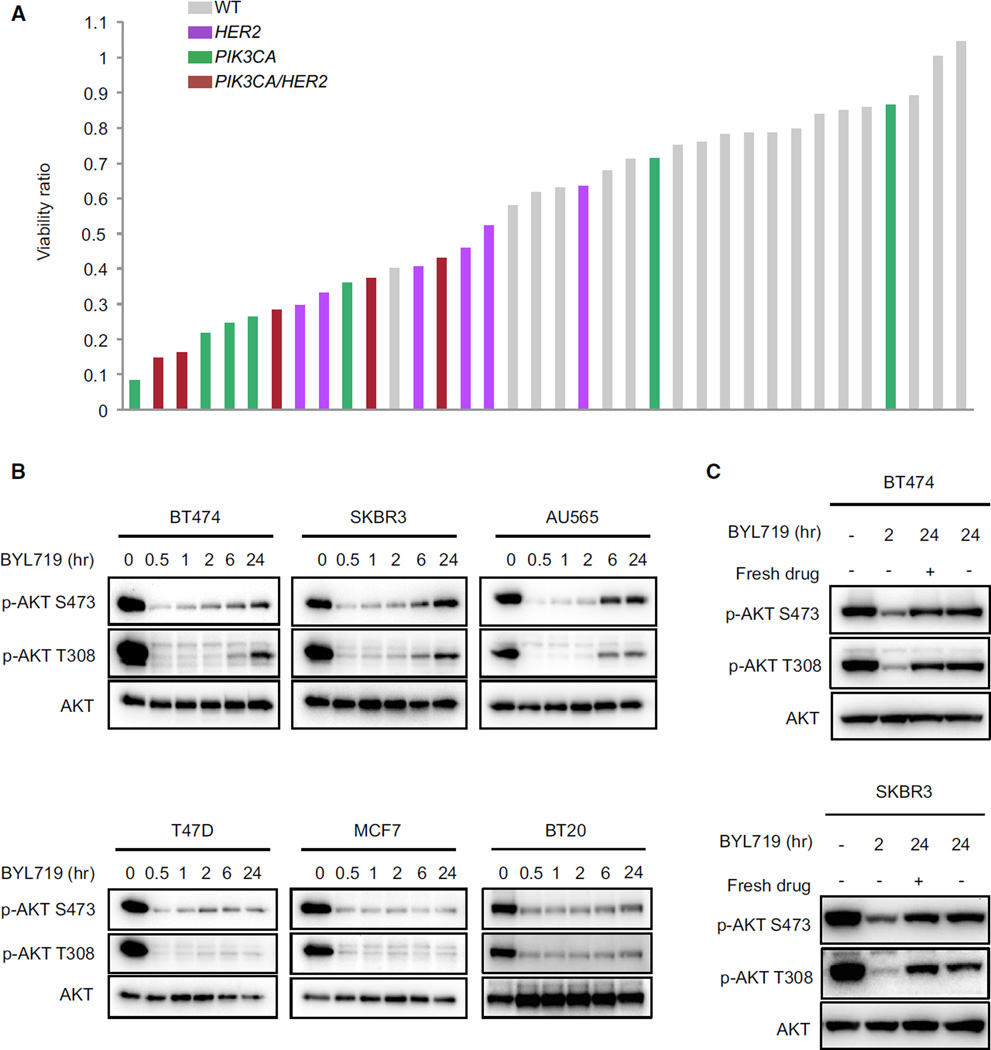
To identify the breast tumors most sensitive to the selective p110α inhibitor, BYL719, we tested the effect of this drug on cell proliferation in a large panel of breast cancer cell lines. As shown in Figure 1A (Table S1 available online), breast cancer cell lines harboring mutations in PIK3CA and/or HER2 amplifications were significantly more sensitive to the antiproliferative effects of BYL719 than breast cancer cells that are wild-type for both genes. These data were further supported by analysis in a larger panel of 321 cancer cell lines (Figure S1A and Table S2) and are in agreement with results of previous reports (Fritsch et al., 2014; Huang et al., 2012). To test whether p110α inhibition alone was sufficient to completely abrogate PI3K–AKT signaling, time-course experiments using BYL719 in multiple PIK3CA hot-spot mutant or HER2-amplified breast cancer cell lines were performed. While BYL719 effectively suppressed AKT over the 24 hr time course in PIK3CA mutant cells (HER2 wild-type), a more pronounced rebound of AKT phosphorylation was observed after 24 hr in the HER2-amplified cells (Figure 1B). This rebound of AKT activation after 24 hr in the HER2-amplified cells is due to a biological effect rather than degradation or efflux of the p110α inhibitor, because similar rebound of AKT signaling occurred despite the addition of fresh drug to the media of cells after 22 hr (Figure 1C). Furthermore, the intracellular concentration of BYL719 was the same at 1 and 24 hr after treatment (Figure S1B). Accordingly, in HER2-amplified cells, AKT signaling completely recovered at longer time points (48 and 72 hr), while it remained more effectively suppressed in PIK3CA mutant cells (Figure S1C). Notably, BYL719 treatment did not change expression levels of the PIP3 phosphatase PTEN (Figure S1D), suggesting that the restoration of AKT signaling is not due to alteration of the phosphatase expression.
Figure 1. Rebound of Phospho-AKT Occurs after 24 hr Treatment with Selective p110α Inhibitor in HER2-Amplified Cell Lines.
(A) Effects of BYL719 on proliferation in a panel of 36 breast cancer cell lines. HER2-amplified versus HER2 WT, Fishers test p = 0.0363; PIK3CA mutant versus PIK3CA WT, Fishers test p = 0.0188.
(B) HER2-amplified (upper) and PIK3CA hot-spot mutant (lower) cell lines were treated with 1 µM BYL719 for the indicated period of time. Lysates were immunoblotted to detect the indicated proteins.
(C) The indicated cell lines were treated with 1 µM BYL719 for indicated period of time (fresh drug was added after 22 hr). Lysates were immunoblotted to detect the indicated proteins.
See also Figure S1 and Tables S1 and S2.
Validation of a Nonradioactive Method to Directly Measure PI3K Activity
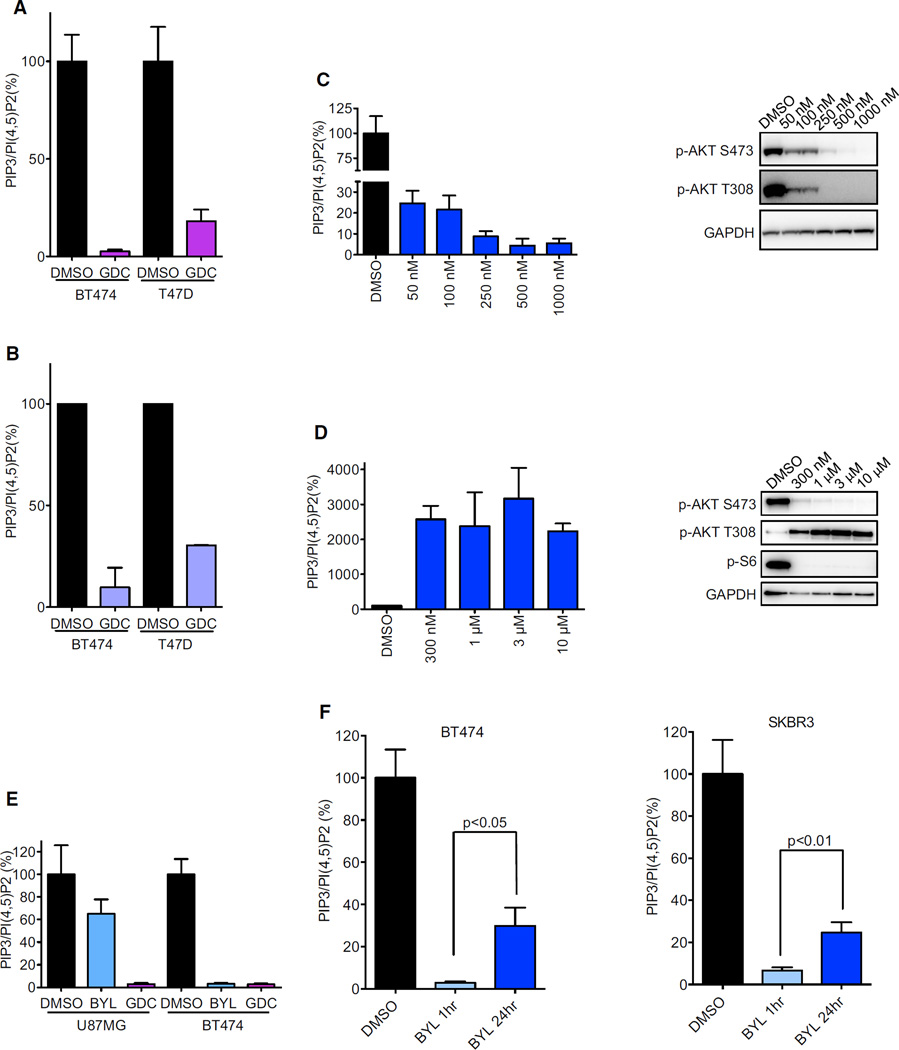
Because AKT activation may also occur via PI3K–independent mechanisms (Guo et al., 2011), we aimed to determine if the reactivation of AKT phosphorylation was due to restored produc-tionof PIP3 despite the presence of BYL719. To directly measure PIP3 levels, an ELISA kit, which has recently been developed to quantify the levels of different phosphoinositides, was utilized. Since PI(4,5)P2 is the most abundant phosphoinositide (Guillou et al., 2007) and its levels are not influenced by PI3K activity (Condliffe et al., 2005), PI(4,5)P2 levels were used as a control for total phosphoinositides, as previously described (Clark et al., 2011; Guillou et al., 2007). PIP3/PI(4,5)P2 ratios were measured after treatment with different inhibitors to test the specificity and reliability of this method under multiple conditions. First, BT474 and T47D cells were treated with the pan-PI3K inhibitor, GDC-0941; as expected, there was potent suppression of the PIP3/PI(4,5)P2 ratio (Figure 2A). Importantly, comparable results and equivalent range of suppression were observed when the PIP3/PI(4,5)P2 ratio was measured with metabolic labeling of cells with [3H] inositol followed by high-performance liquid chromatography (HPLC) (Figure 2B), a classical technique for phosphoinositide measurement (Guillou et al., 2007). To further assess the reliability of this method for PIP3 quantification, T47D cells were treated with varying concentrations of BYL719 for 1 hr. As shown in Figure 2C, this treatment led to a dose-dependent decrease of PIP3 that correlated well with the reduction of AKT phosphorylation. Then, PIP3 levels were determined under a condition in which an increase of PIP3 levels was expected. T47D cells were exposed for 24 hr to different concentrations of AZD8055, an mTORC1/2 catalytic inhibitor. Under these conditions, PIP3 levels dramatically increased, indicating the stimulation of PI3K activity (Figure 2D, left), in accordance with release of the well-established mTORC1 negative feedback loops leading to reactivation of PI3K signaling (O’Reilly et al., 2006; Rodrik-Outmezguine et al., 2011). Accordingly, the mTORC2-dependent AKT phosphorylation on Serine 473 was completely abrogated whereas the PI3K/PDK1-depen-dent AKT phosphorylation on Threonine 308 was induced by derepression of the negative feedback (Figure 2D, right), in agreement with the PIP3 measurements. Finally, we compared PIP3 levels using different PI3K inhibitors. To do so, BT474 and U87MG cells were treated for 1 hr with BYL719 and a pan-PI3K inhibitor GDC-0941. As expected, in BT474 cells, BYL719 was sufficient to downregulate PIP3 levels, while in U87MG cells, which are PTEN null and largely p110β-dependent (Wee et al., 2008), GDC-0941, but not BYL719, suppressed PIP3 levels (Figure 2E). All together, these data demonstrate that this method consistently and reliably quantifies PIP3 levels.
Figure 2. Validation of ELISA Kit to Measure the Ratio of PIP3/PI(4,5)P2.
(A) Phospholipids were isolated from cells treated with DMSO or 1 µM GDC-0941 for 1 hr and relative PIP3 and PI(4,5)P2 levels were quantified by ELISA. Each data point represents the average of three independent experiments, each carried out in triplicate. For normalization, the PIP3/PI(4,5)P2 ratio was set to 100% for DMSO treated cells.
(B) Phospholipids were isolated from cells treated as in (A) and measured with HPLC analysis after metabolic labeling of the cells for 48 hr.
(C and D) T47D cells were treated with indicated concentrations of BYL719 for 1 hr (C) or AZD8055 for 24 hr (D). Phospholipids were isolated and PIP3 and PI(4,5) P2 levels were quantified by ELISA (left) or cell lysates were immunoblotted to detect the indicated proteins (right).
(E) Cells were treated with 1 µM BYL719 or 1 µM GDC-0941 for 1 hr and relative PIP3 and PI(4,5)P2 levels were quantified by ELISA.
(F) Phospholipds were isolated from cells treated with 1 µM BYL719 for indicated time and relative PIP3 and PI(4,5)P2 levels were quantified by ELISA. p < 0.05 by Student’s t test.
All error bars in this figure represent ± SEM. See also Figure S2.
The p110β Isoform Is Responsible for the Reactivation of PIP3 Signaling after Treatment with BYL719 in HER2-Amplified Breast Cancers
Using the ELISA assay, PIP3 levels were measured in HER2-amplified cells following treatment with BYL719 for 1 or 24 hr. In accordance with AKT phosphorylation (Figure 1B), PIP3 levels were suppressed at 1 hr, but were partially restored after 24 hr treatment (Figure 2F), indicating that this reactivation of AKT was due to a rebound in PIP3 despite the continued presence of BYL719. This AKT reactivation diminished the impact of single-agent BYL719 on cell viability as the AKT inhibitor MK2206 improved the efficacy of BYL719 in blocking cell growth and inducing apoptosis (Figures S2A and S2B).
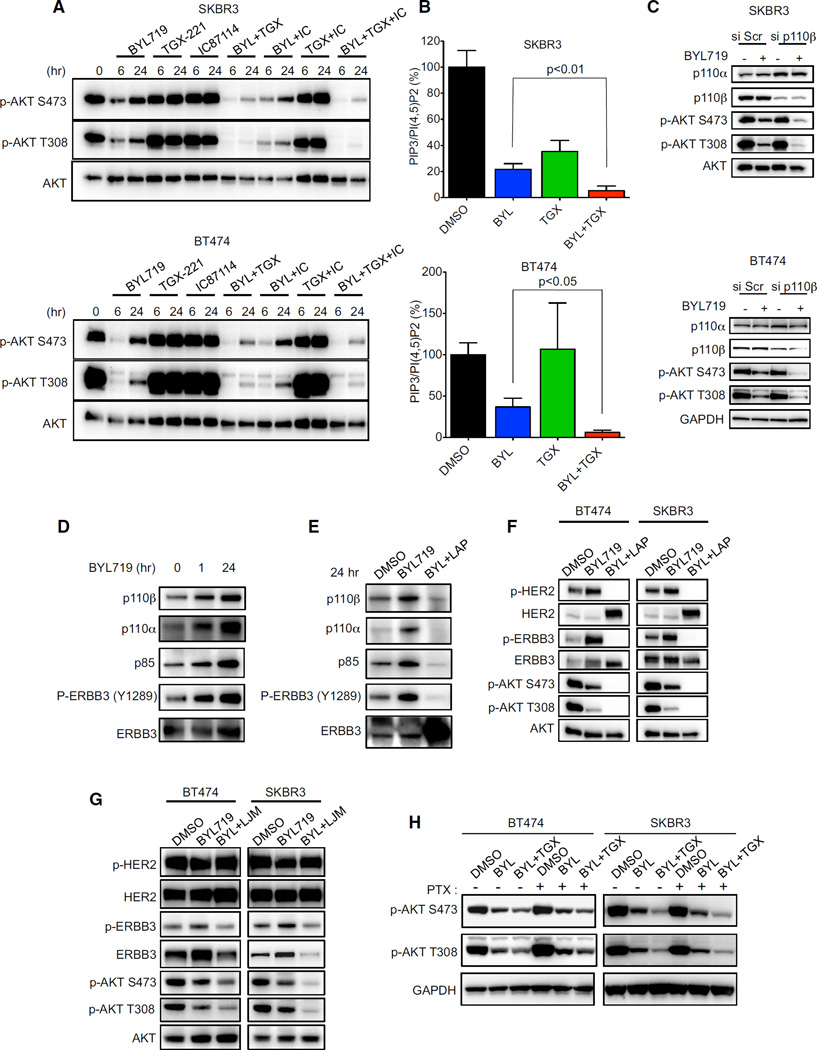
Because BYL719 is a selective p110α inhibitor, we hypothesized that the restored PIP3 levels may be due to activation of another p110 isoform. To investigate if other p110 isoforms were responsible for this rebound of AKT phosphorylation, we utilized p110β and p110δ isoform-selective inhibitors (TGX-221 and IC87114, respectively) at doses previously shown to selectively discriminate among different isoforms (Huang et al., 2012; Kim et al., 2009; Utermark et al., 2012). Specificity was confirmed in the PTEN-deficient PC3 cells, in which the activation of AKT is primarily mediated by p110β, and the HER2-amplified BT474 cells, in which the activation of AKT is primarily mediated by p110α (Figure S3A). The SKBR3 and BT474 cells were treated with different combinations of these isoform-selective inhibitors for 6 and 24 hr (Figure 3A). The combination of BYL719 and TGX-221 effectively mitigated the rebound of AKT phos-phorylation after 24 hr (Figure 3A and Figure S3B). Accordingly, this combination also blocked the re-accumulation of PIP3 (Figure 3B). These results were recapitulated by siRNA-mediated knockdown of p110β in the presence of BYL719 (Figure 3C). Thus, p110β is responsible for the rebound in PIP3 levels and the reaccumulation of AKT phosphorylation in HER2-amplified cancers treated with a p110α-specific inhibitor.
Figure 3. Inhibition of p110β Blocks the Rebound of AKT Phosphorylation and PIP3 Levels in HER2-Amplified Cell Lines.
(A) The indicated HER2-amplified cell lines were treated with p110α inhibitor BYL719 (1 µM), p110β inhibitor TGX-221 (1 µM), and p110δ inhibitor IC87114 (1 µM) or combinations of these inhibitors for the indicated times. Lysates were immunoblotted to detect the indicated proteins.
(B) Cells were treated with BYL719 (1 µM), or TGX-221 (1 µM) or both for 24 hr, phospholipids were isolated from cell lysates and relative PIP3 and PI(4,5)P2 levels were quantified by ELISA. Each data point is the average ± SEM (p < 0.05 by Student’s t test).
(C) Cells were transfected with control (Scr) or p110β-targeted siRNA for 48 hr, followed by treatment with BYL719 (1 µM) for an additional 24 hr. Lysates were immunoblotted to detect the indicated proteins.
(D and E) BT474 cells were treated with 1 µM BYL719 alone for different durations of time (D) or with 1 µM Lapatinib (LAP) for 24 hr (E) and lysates were immunoprecipitated with ERBB3 antibody. Precipitates were analyzed by western blot with the indicated antibodies.
(F) Cells were treated with indicated drugs at 1 µM for 24 hr. Lysates were immunoblotted to detect the indicated proteins.
(G) Cells were treated with 1 µM BYL719 alone or in combination with 10 µg/ml LJM716 (LJM) for 24 hr. Lysates were immunoblotted to detect the indicated proteins.
(H) Cells were treated or not with pertussis toxin (PTX) 100 ng/ml and the indicated drug(s) for 24hr. Lysates were immunoblotted to detect the indicated proteins.
See also Figure S3.
We next aimed to determine molecular mechanisms underlying p110β-induced PIP3 production following initial inhibition of p110α and to identify upstream activators of p110β. ERBB3 is a receptor that heterodimerizes with HER2 and drives PI3K activation in HER2-amplified breast cancer cells (Arteaga and Engelman, 2014). Immunoprecipitation studies revealed increased recruitment of p110β to ERBB3 following 24 hr of treatment with BYL719 (Figure 3D), thus accounting for the induction of p110β-dependent production of PIP3. The increased association between p110β and ERBB3 was likely induced by increased tyrosine phosphorylation and total levels of ERBB3 promoted by PI3K inhibition (Figure 3D), through several feedbacks as previously reported (Chakrabarty et al., 2012; Chandarlapaty et al., 2011). Accordingly, cotreatment with BYL719 and the HER2 inhibitor lapatinib blocked ERBB3 phosphorylation (Figures 3E and 3F). This led to the loss of p110β recruitment (Figure 3E) and the block of the rebound of AKT activation observed 24 hr after treatment with BYL719 alone (Figure 3F). Similarly, siRNA-mediated downregulation of ERBB3 (Figure S3C) or inac-tivation of ERBB3 through LJM716, a specific antibody that blocks ERBB3 activation (Garrett et al., 2013) (Figure 3G), impaired the re-accumulation of AKT phosphorylation. Because p110β can be activated also by GPCRs, cells were treated with pertussis toxin, which inhibits the Gi subunit downstream of GPCRs. This treatment did not block reactivation of AKT (Figure 3H), suggesting that HER2 and ERBB3 have a more prominent role in activating p110β and AKT in HER2-amplified breast cancer cells. Taken together, these data suggest that although inhibition of p110α potently suppresses the PI3K pathway after 1 hr, concomitant inhibition of p110β may be necessary to achieve complete and sustained inhibition of PI3K signaling in HER2-amplified cells.
p110β Activation Mitigates the Growth Arrest and Apoptosis Induced by BYL719 in HER2-Amplified and PIK3CA Mutant Models
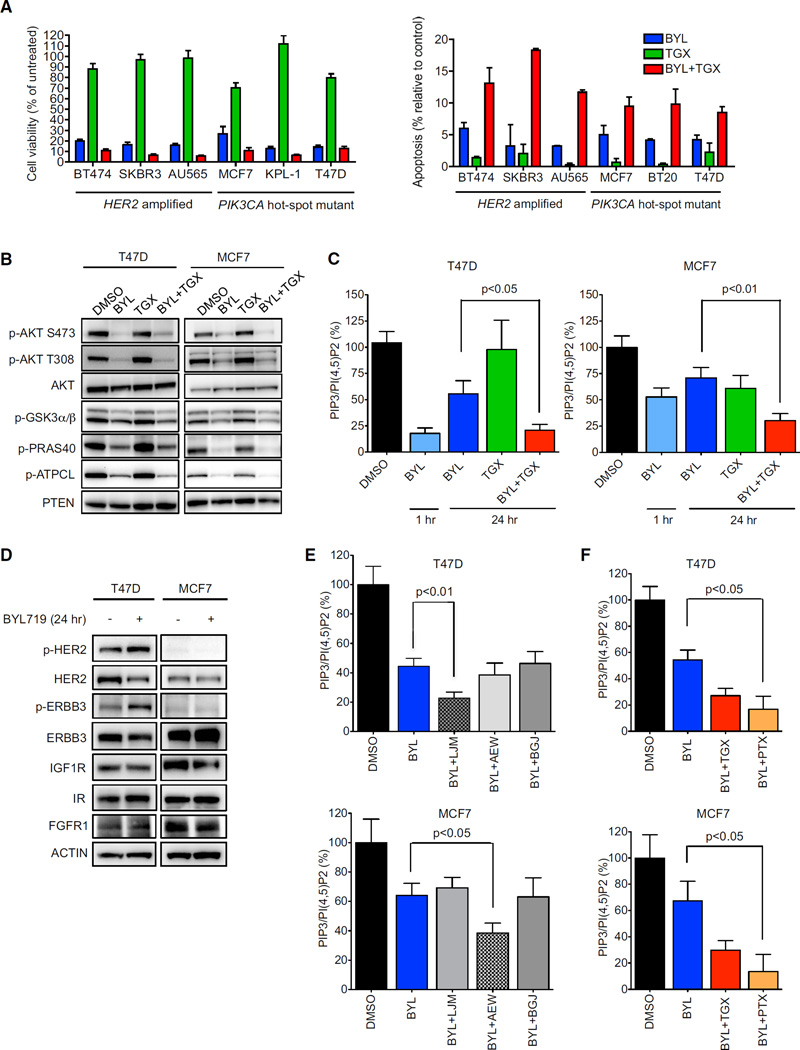
We next determined if the combination of the p110α and β inhibitors demonstrated an additive effect in blocking cell growth and inducing apoptosis in HER2-amplified breast cancers. Long-term cell viability and apoptosis (Figure 4A) assays revealed that dual inhibition of p110α and p110β was more effective than single agent BYL719 in HER2-amplified breast cancers, despite the finding that TGX-221 had no detectable effects on cell viability and growth as a single agent. As a control, we included PIK3CA mutant breast cancers, which had not displayed a substantial rebound of phospho-AKT following treatment of BYL719 (Figure 1B; Figure S1C). To our surprise, combined p110α and p110β inhibition also impaired cell viability to a greater extent than inhibition of p110α alone in two of the three PIK3CA mutant breast cancer cell lines examined. This was initially unexpected because, unlike the HER2-amplified breast cancers, phospho-AKT levels did not substantially rebound in PIK3CA mutant cells for up to 72 hr after treatment with BYL719 (Figure S1C). Accordingly, addition of the p110β inhibitor did not lead to more pronounced suppression of phosphory-lation of AKTor its substratesin these PIK3CA mutant cancer cell lines (Figure 4B). However, measurement of PIP3 levels revealed that BYL719 did not fully suppress PIP3 levels in PIK3CA mutant cells, especially after 24 hr, and combined inhibition of p110α and p110β resulted in stronger reduction of PIP3 levels than BYL719 alone (Figure 4C). Because p110β has been shown to induce phosphorylation of nuclear AKT (Kumar et al., 2011), we determined if inhibition of p110β affected AKT signaling specifically in the nucleus, which may have been missed in assessments of AKT phosphorylation in whole cell lysates (Figure 4B). However, in the PIK3CA mutant cell line MCF7, p110β did not control AKT phosphorylation in either the cytoplasm or the nucleus (Figure S4A). Thus, these findings demonstrate that p110β contributes toPIP3 levels in PIK3CA mutant breast cancer cells, and inhibition of p110β augments the effects of p110α inhibition on cell viability in an AKT-independent manner in these cells.
Figure 4. p110β Contributes to Cell Growth and Survival.
(A) Cells were treated with 1 µM BYL719, 1 µM TGX-221, or both for 14 days and cell viability was determined by Syto60 staining (left) or for 72 hr and the percentage of cells undergoing apoptosis was measured by annexin V positivity (right). Because BT-20 cells did not survive when seeded at low density, they were not assayed for viability.
(B) Cells were treated with DMSO, 1 µM BYL719, 1 µM TGX-221, or the combination for 24 hr and lysates were immunoblotted to detect the indicated proteins.
(C) Cells were treated as (B) and PIP3 and PI(4,5)P2 levels were quantified by ELISA. p < 0.05 by Student’s t test.
(D) Cells were treated with 1 µM BYL719 for 24 hr and lysates were immunoblotted to detect the indicated proteins.
(E) Cells were treated with single agent or combination of BYL719 (1 µM), LJM716 (LJM) (10 µg/ml), NVP-AEW541 (AEW) (1 µM), or NVP-BGJ398 (BGJ) (250 nM) or the indicated combination for 24 hr and PIP3 and PI(4,5)P2 levels were quantified by ELISA. p < 0.05 by Student’s t test.
(F) Cells were treated with BYL719 (1 µM), TGX-221 (1 µM), or pertussis toxin (PTX) (100 ng/ml) or the indicated combination for 24 hr and PIP3 and PI(4,5)P2 levels were quantified by ELISA. p < 0.05 by Student’s t test.
All error bars in this figure represent ± SEM. See also Figure S4.
To identify the mechanism underlying p110β activation in PIK3CA mutant cells, lysates from cells treated for 24 hr with BYL719 were hybridized to p-RTK arrays (Figure S4B). Treatment with p110α inhibitor induced increased phosphorylation of HER2 and ERBB3 in T47D cells whereas no changes were observed in the p-RTK arrays probed with the MCF7 lysates. We also specifically probed for the RTKs previously found to be activated following treatment with PI3K inhibitor (Chakrabarty et al., 2012) (Figure 4D). These results confirmed the induction of HER2 and ERBB3 phosphorylation in T47D cells. However, in MCF7 cells, BYL719 led to lower levels of total insulin-like growth factor 1 receptor (IGF1R) (Figure 4D). We speculated that this might possibly reflect an increase in its phosphorylation and activation. To test this hypothesis, we immunoprecipitated the IGF1R–adaptor IRS1 and assessed its phosphorylation. An increased in tyrosine phosphorylation of IRS1 after BYL719 treatment was observed (Figure S4C), suggesting that BYL719 indeed induced the activation of the IGF1R-dependent signaling pathway in MCF7 cells. To test if HER2/ERBB3 and IGF1R signaling were responsible for the activation of p110β activation in T47D and MCF7 cells, respectively, we measured PIP3 levels after treating both cell lines with BYL719 alone, or in combination with the anti-ERBB3 antibody LJM716, the IGF1R inhibitor NVP-AEW541, or the fibroblast growth factor receptor inhibitor NVP-BGJ398 (Figure 4E). In T47D cells, only the combination of BYL719 and LJM716 blocked the re-accumulation of PIP3 similarly to the combination of p110α and p110β inhibitors, whereas, in MCF7 cells, the rebound of PIP3 was prevented by the combination of BYL719 and AEW541. Consistent with this result, a previous report demonstrated that IGF1R inactivation sensitizes MCF7 cells to PI3K inhibitors (Chakrabarty et al., 2012). These data suggest that p110β activation is also driven by RTKs in PIK3CA mutants. However, pertussis toxin also prevented the rebound of PIP3 after BYL719 treatment (Figure 4F), suggesting that input from both RTKs and GPCRs is required for p110β engagement and activation in PIK3CA mutant models.
Combination of p110α and p110β Inhibition Induces Tumor Regression In Vivo
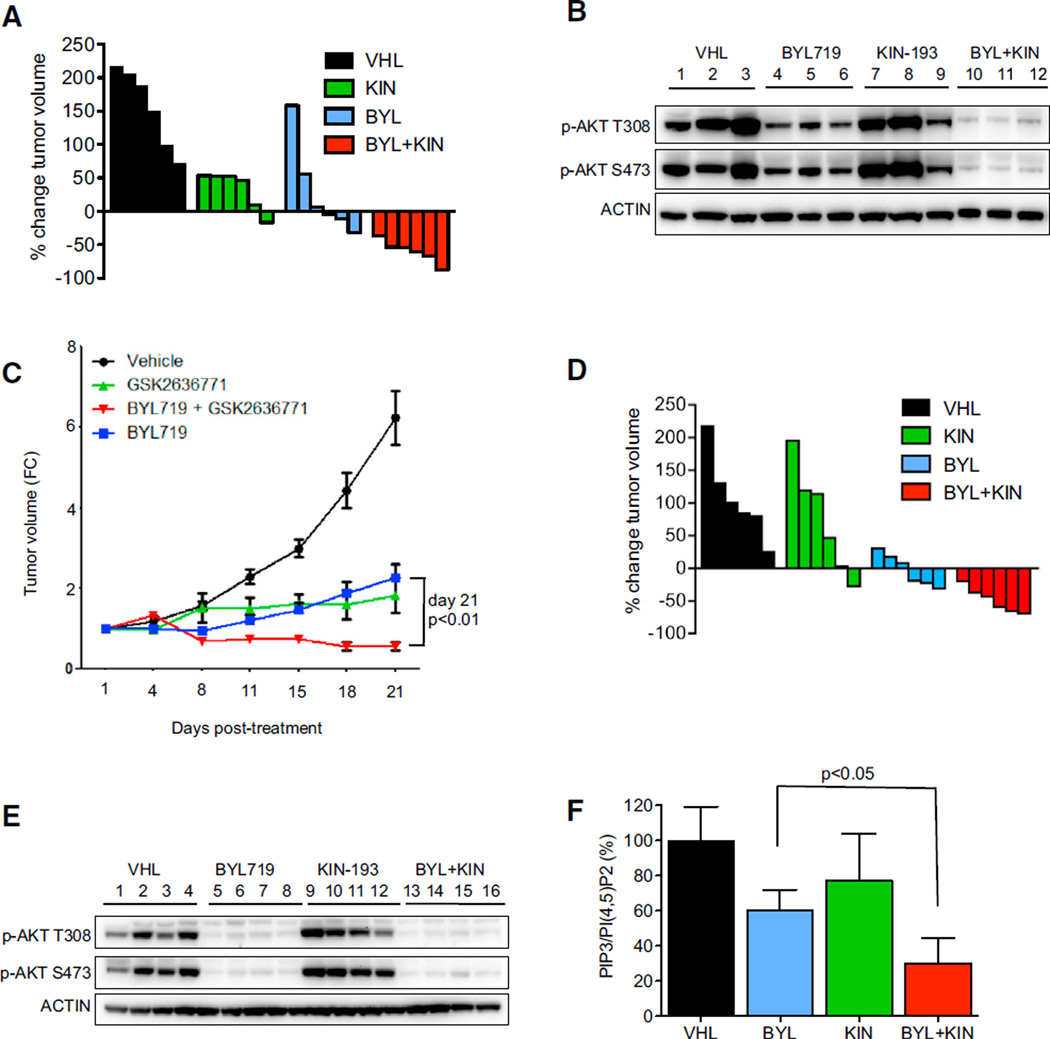
These findings led us to test the efficacy of combined p110α and p110β inhibitors in vivo. Because TGX-221 cannot be used for in vivo studies, we used the structurally related p110β selective inhibitor KIN-193 (Ni et al., 2012). Dose-response assays in vitro confirmed comparable sensitivity of PTEN-deficient LNCaP cells to TGX-221 and KIN-193, as well as a similar lack of potency in BT474 cells (Figure S5A). Whereas BT474 xeno-grafts primarily exhibited static tumor growth when treated with BYL719 alone, tumors regressed when treated with the combination of p110α and p110β inhibitors (Figure 5A and Figure S5B). Pharmacodynamic studies of the drug-treated tumors were consistent with the in vitro results. Both phospho-AKT and PIP3 levels were partially restored following BYL719 administration in HER2-amplified tumors (Figures S5C and S5D). However, the addition of the p110β inhibitor led to greater suppression of AKT phosphorylation than BYL719 treatment alone (Figure 5B). To further assess the activity of combination of p110α and p110β inhibitor, we used a genetically engineered mouse model (GEMM) of HER2-amplified breast cancer. Consistent with the findings in the human xenograft model, both BYL719 and GSK2636771, a structurally unrelated p110β inhibitor currently in clinical trials, slowed tumor growth but only the combination induced tumor shrinkage (Figure 5C).
Figure 5. Combined p110α and p110β Inhibition Leads to Tumor Regression In Vivo.
(A) Waterfall plot showing the percent change in tumor volume (relative to initial volume) for individual BT474 tumors in tumor-bearing mice treated with the vehicle (VHL), BYL719 (BYL) 25 mg/kg per oral gavage daily, KIN-193 (KIN) 20 mg/kg per IP injection twice per day, and combination (BYL/KIN) groups following 21 days of treatment.
(B) Mice harboring BT474 tumors were administered with vehicle (VHL), BYL719 (BYL) 25 mg/kg, KIN-193 (KIN) 20 mg/kg, or the combination of BYL/KIN for 3 days. Mice were killed 12 hr after the last treatment with BYL719 and 2 hr after treatment with KIN-193. For the combination-treated mice, tumors were also collected 12 hr after treatment with BYL719 and 2 hr after KIN-193. Lysates were prepared from tumors and blotted with the indicated antibodies.
(C) Tumor growth assay showing the antitumor effect of BYL719 25 mg/kg per oral gavage daily and GSK2636771 30 mg/kg per oral gavage daily alone or in combination on endogenous NeuT tumors. p < 0.05 by two-way ANOVA. The average tumor volume ± SEM for each cohort is displayed.
(D) Waterfall plot showing the percent change in tumor volume of MCF7 xenografts (relative to initial volume) for individual tumors treated as in (A).
(E) Mice harboring MCF7 were administered as in (B). Mice were killed 2 hr after the last treatment with BYL719 and/or KIN-193. Lysates were prepared and blotted with the indicated antibodies.
(F) Phospholipids were isolated from the same tumors as (E) and PIP3 and PI(4,5)P2 levels were quantified by ELISA. Each data point is the average ± SEM (p < 0.05 by Student’s t test). See also Figure S5.
To validate our in vitro findings in PIK3CA mutant cells, we also tested MCF7 tumor xenografts. MCF7 tumors regressed only with the combination of p110α and p110β inhibitors (Figure 5D and Figure S5E). In contrast to the analyses of the HER2-ampli-fied xenograft tumors, lysates from MCF7 tumors revealed that suppression of AKT phosphorylation was not further increased by the addition of the p110β inhibitor (Figure 5E), consistent with the in vitro findings (Figure 4B). However, PIP3 levels were more effectively suppressed in MCF7 tumors treated with the combination in comparison to BYL719 alone (Figure 5F), supporting the notion that PIP3 may serve as an informative biomarker to more fully assess PI3K inhibition and efficacy in vivo.
DISCUSSION
In this study, we observed that p110α inhibition alone is not sufficient to completely suppress PI3K activity in HER2-amplified and PIK3CA mutant breast cancer cells, even though these cancers are among the most sensitive to single-agent p110α-specific inhibitors. Although p110α is the major isoform responsible for PI3K signaling in these cancers, p110β activation partially recovered PIP3 levels within 24 hr of BYL719 treatment. In HER2-amplified cancers, this led to reactivation of AKT, but in PIK3CA mutant cancers, it did not. However, in both breast cancer models, concomitant inhibition of p110α and p110β induced greater antitumor activity. We speculate that this p110β-dependent rebound of PIP3 requires several hours to manifest for two main reasons: (1) the time required for maximal BYL719-dependent release of the negative feedbacks leading to tyrosine phos-phorylation of PI3K activators (e.g., ERBB3) and subsequent p110β recruitment, and (2) since p110β is a relatively inefficient enzyme in comparison to p110α (Beeton et al., 2000), PIP3 accumulation can only be observed after sufficient time following activation. Although p110β has been identified as the primary PI3K isoform driving PI3K signaling in PTEN null tumors (Jia et al., 2008; Ni et al., 2012), the experiments in this study suggest that p110β activation also promotes the viability of p110α-driven breast cancers following treatment with p110α-specific inhibitors.
In vivo, p110α inhibition blocked tumor growth but did not induce tumor shrinkage in either the HER2-amplified or PIK3CA mutant xenograft models. Interestingly, early reports from clinical trials showed that, even among patients whose cancers harbor PIK3CA mutations, BYL719 did not induce regressions in the majority of cases (Juric and Baselga, 2012; Juric et al., 2012), consistent with our observation that p110α inhibition alone does not cause tumor shrinkage in the PIK3CA mutant MCF7 tumors. Surprisingly, two structurally distinct p110β inhibitors showed an effect as single agents on the growth of the HER2 amplified tumors in vivo (both in BT474 xenograft and GEMM model overexpressing HER2) despite the lack of any antiproliferative effects in vitro. This is consistent with previous reports showing that mice expressing a catalytically inactive isoform of p110β showed slower tumor growth in a HER2-driven model (Ciraolo et al., 2008) and that p110β inhibition causes modest but significant reduction of tumor growth in HER2-induced murine cancer model (Utermark et al., 2012). It is possible that some of the effects observed in vivo with the p110β inhibitor are non-cell-autonomous, or that specific upstream activating mechanisms render p110β more relevant in vivo. Importantly, tumor regressions resulted from concomitant inhibition of p110α and p110β in the GEMM overexpressing HER2, and HER2-amplified and PIK3CA mutant xenograft models. We believe it might be beneficial to combine two isoform-selective inhibitors instead of using a pan-PI3K inhibitor, as the currently available pan-PI3K inhibitors do not equally block all p110 isoforms. For example, the pan-PI3K inhibitor GDC-0941 is equipotent against p110α and p110δ, but displays only modest (10-fold less) activity against p110β (Raynaud et al., 2009). Accordingly, GDC-0941 failed to induce tumor regression in MCF7 xeno-graft (Ebi et al., 2013), whereas a combination of BYL719 and KIN-193 resulted in remarkable tumor shrinkage (Figure 5D). For this reason, the combination of two selective and potent isoform-specific inhibitors may have a better therapeutic window by maximally inhibiting p110α and p110β, while sparing the toxicities associated with inhibiting other PI3K isoforms. The article by Schwartz et al. (2014) in this issue of Cancer Cell demonstrates that p110α inhibitor adds to the efficacy of p110β inhibitor in PTEN-deficient cancers further supporting the potential value of targeting both isoforms also in PTEN-deficient contexts. Together, these findings point to the value of inhibiting both isoforms in cancers because inhibition of only one p110 isoform leads to the activation of the other isoform. Perhaps, future pan-PI3K inhibitors that equally and specifically inhibit p110α and p110β would be similarly advantageous.
Since p110β-dependent PIP3 activation in the BYL719-treated PIK3CA mutant cancers did not lead to a rebound of AKT activation, it remains unknown which PIP3-dependent signals affect cell viability in these cells. We attempted to address this question using an unbiased mass spectrometry approach comparing the phosphopeptides that were differentially modulated by single-agent BYL719 versus the combination of BYL719 and TGX-221 in MCF7 cells. The results reveal that co-inhibition of p110α and p110β leads to sustained downregulation of some intracellular signaling nodes, including downstream effectors of the mTORC1-signaling pathway (data not shown). Interestingly, it has recently been demonstrated that SGK3 activation, which in turn can activate mTORC1-pathway, can be partially triggered by a pool of PIP3 that acts independently of AKT (Bago et al., 2014). However, it remains unknown which of these pathways are most responsible for driving the p110β-mediated effects on cell viability.
The precise mechanism of how p110β is recruited and activated in cancer is still uncertain; however, it has been suggested that it can occur through GPCRs (Dbouk et al., 2012). Indeed, we observed that input from both RTKs and GPCRs is necessary for p110β activation in PIK3CA mutant cells. These findings are consistent with recent data from Backer and colleagues demonstrating that there is synergistic activation of p110β by concomitant binding to both the Gbg subunit and phosphotyrosine peptides (Dbouk et al., 2012). In contrast, the observation that pertussis toxin treatment did not affect p110β-dependent AKT phosphorylation in HER2-amplified cancer cells, whereas lapatinib and LJM716 abolished it, suggests that the input from GPCRs is dispensable for activation of p110β-dependent signaling in these cells. However, it is possible that GPCRs that are not inhibited by pertussis toxin may contribute to activation of p110β in HER2-amplified cells. These findings raise the question why GPCRs may not be necessary to engage p110β in HER2-amplified cells. Since HER2-amplified cells have dramatically increased HER2/ERBB3 activation in comparison to PIK3CA mutant cells, we speculate that the RTK component might be sufficient to induce p110β activation in HER2-amplified cancers. In PIK3CA mutant cells, our findings demonstrate that different RTKs, specifically ERBB3 for T47D and IGF1R for MCF7, work in concert with GPCRs to recruit and activate p110β. Future studies on clinical specimens will be required to identify if RTKs other than HER2/ERBB3 and IGF1R commonly activate p110β signaling in PIK3CA mutant breast cancers following treatment with p110α specific inhibitor. These data may inform combination therapies that might be most effective in PIK3CA mutant breast cancers.
Our data also raise the related question as to why PIP3 levels correlate with AKT phosphorylation in HER2-amplified but not in PIK3CA mutant breast cancer cells. It has been previously reported that specific pools of PIP3 do not correlate with phos-pho-AKT in some cellular contexts (Mandl et al., 2007), and that some PIK3CA mutant breast cancers with low phospho-AKT levels are more reliant on AKT-independent PI3K effectors (Vasudevan et al., 2009). The PH domain of AKT has a relatively low affinity for PIP3 in comparison to other PH domains (Park et al., 2008), thus AKT membrane localization and activation critically depends on elevated and localized PIP3 levels. We speculate that, in HER2-amplified cells, HER2 is highly localized in membrane rafts (Pickl and Ries, 2009), and the recruitment and activation of p110β by HER2/ERBB3 heterodimers following BYL719 treatment produces highly concentrated local pools of PIP3, leading to potent activation AKT. However, in PIK3CA mutants, the receptors (RTKs and GPCRs) responsible for p110β activation may be more diffuse on the membrane, producing less concentrated local pools of PIP3, which are insufficient to activate AKT. In this scenario, proteins with PH domains that have higher affinities for PIP3 than AKT can be activated by the lower concentrations of local PIP3 produced by p110β. Therefore, the mechanism of p110β activation may contribute to the disconnection between PIP3 levels (and thus PI3K activity) and AKT activation in PIK3CA mutant breast cancers.
Importantly, AKT phosphorylation is currently used as surrogate marker to assess PI3K activity both in preclinical and clinical studies. This approach is useful and feasible with available standard techniques; however, measuring only AKT phosphor-ylation may risk potentially missing important information by oversimplifying the PI3K signal transduction pathway. The data in this work support the notion that direct and comprehensive methods are needed to evaluate PI3K activity and investigate its functions. Thus, in this study we evaluated PI3K activity by measuring PIP3 levels, and thereby elucidated the benefit of combining p110α and p110β inhibitors to more efficiently block PI3K–dependent oncogenic signaling in “p110α-driven” breast cancers.
EXPERIMENTAL PROCEDURES
Cell Lines and Reagents
BT474 and MCF7 cells were cultured in Dulbecco’s modified Eagle’s medium with 10% fetal bovine serum (FBS). T47D, SKBR3, and AU565 cells were cultured in RPMI1640 with 10% FBS. KPL-1 and BT-20 cells were cultured in DF12 with 10% FBS. Cell lines were obtained from the Center for Molecular Therapeutics at Massachusetts General Hospital Cancer Center. The following drugs were used: TGX-221 (Selleckchem), Lapatinib (LC laboratories and Ab-mole), GDC-0941 (supplied by the Targeting PI3K in Women’s Cancers Stand Up to Cancer Dream Team), AZD8055 (Selleckchem), IC87114 (Selleckchem), BYL719 and LJM716 (kindly provided by Novartis), KIN-193 (kindly provided by Jean Zhao), GSK2636771 (Abmole), NVP-BGJ398 (Selleckchem), NVP-AEW541 (Selleckchem), and MK2206 (Selleckchem).
Immunoblotting
Antibodies against p-AKT(Ser473), p-AKT(Thr308), p110α, p110β, p-GSK3α/β, p-PRAS40, p-ATP citrate lyase, p-ERBB3, p-HER2, HER2, PARP, ACTIN, IR, IGFR, FGFR1, p-RPS6 (Ser240–244), p-Tyrosine, and PTEN were from Cell Signaling Technology. Antibodies against AKT and ERBB3 were purchased from Santa Cruz Biotechnology. GAPDH, IRS1, and p85 antibodies were from Millipore.
Small Interfering RNA Knockdown
Cells were seeded into six-well plates at density of 1.5 × 105 cells/well. Twenty-four hours later, cells were transfected with ON-TARGETplus SMART-pool siRNA against p110β (Dharmacon), ERBB3 (Ambion), or ON-TARGET plus Nontargeting Pool as negative control using HiPerFect transfection reagent (QIAGEN) according to the manufacturer’s instructions. Transfected cells were cultured at 37°C for 72 hr before analysis.
HPLC Phosphoinositide Analysis
Cells were metabolically labeled with 10 µCi/ml [3H] inositol for 48 hr in inositol-free DMEM supplemented with dialyzed fetal calf serum (GIBCO) and 200 mM L-glutamine. After the cells were labeled, they were treated as indicated and lysed in 1 N HCl. Lipids were extracted in chloroform-methanol (1:1, vol/vol). Phosphoinositides were separated by anion-exchange HPLC (Beckman), detected by a flow scintillation analyzer (Perkin-Elmer), and quantified using ProFSA software (Perkin-Elmer) as described previously (Mandl et al., 2007).
PIP3/PI(4,5)P2 Quantification
Cells were seeded at density of 1 × 107 cells/10 cm dish. After treatment, the media was removed by aspiration and 5 ml of ice-cold 0.5 M TCA was immediately added. Cells were scraped, transferred into a 15 ml tube on ice, and centrifuged at 3,000 revolutions per minute (rpm) for 7 min at 4°C. The pellet was resuspended in 3 ml of 5% tricarboxylic acid/1 µM EDTA, vortexed, and centrifuged at 3,000 rpm for 5 min, the supernatant was discarded, and this washing step was repeated one more time. Afterward, neutral lipids were extracted adding 3 ml of MeOH:CHCl3 (2:1) and continuously vortexing over 10 min at room temperature. Extracts were centrifuged at 3,000 rpm for 5 min, the supernatant was discarded, and this extraction step was repeated one more time. The acidic lipids were extracted adding 2.25 ml MeOH:CHCl3:12 M HCl (80:40:1) with continuous vortexing over 25 min at room temperature. Extracts were centrifuged at 3,000 rpm for 5 min and the supernatant was transferred to a new 15 ml tube; 0.75 ml of CHCl3 and 1.35 ml of 0.1 M HCl were added to the supernatant, vortexed, and centrifuged at 3,000 rpm for 5 min to separate organic and aqueous phases. The organic (lower) phase was collected; 1.45 ml were transferred into new vial for PIP3 measurement and 0.05 ml were transferred into a new vial for PI(4,5)P2 measurement. All samples were dried in a vacuum dryer for 1 hr. PIP3 samples were resuspended in 120 ml of PBS-Tween+3% Protein Stabilizer (provided by the Echelon kit). PI(4,5)P2 samples were re-suspended in 120 µl of PBS+ 0.25% Protein Stabilizer. Samples were sonicated in an ice-water bath for 10 min, vortexed, and spun down before adding totheELISA. All experiments were performed at least three times, each carried out in biological triplicate. Once phospholipids were isolated from cells, PIP3 and PI(4,5)P2 levels were measured using ELISA kits (Echelon, K-2500s and K4500) according to the manufacturer’s instructions.
Apoptosis Analysis
Cells were seeded at ~30%-40% confluence in 6 cm plates. After overnight incubation, media was aspirated and replaced with media with or without indicated drugs. After 72 hr, media was collected. Cells were harvested, rinsed once with PBS, and resuspended in Annexin binding buffer (BD Biosciences). Cells were stained with propidium iodide (BD Biosciences) and Annexin V Cy5 (Biovision) according to the manufacturer’s protocol and analyzed on a LSRII flow cytometer (BD Biosciences).
Viability Assays
Three thousand cells were seeded in quadruplicate in 24 wells and treated with fresh media and drug every 72 hr. At day 14, cells were washed with PBS and fixed in 4% formaldehyde for 20 min. Then, cells were washed twice with dH20, and stained for 1 hr with Syto60 (Invitrogen). The plates were then read by measuring the absorption at 700 nm with the Odyssey Imaging System (LI-COR). For CellTiter-Glo assay (Promega) 5,000 cells were seeded in triplicate in 96 wells and the following day treated with media or drug. At day 3,50 µl of CellTiter-Glo was added to each well and the plate was read on a Centro LB 960 microplate luminometer (Berthold Technologies) according to the Promega protocol.
BYL719 Screening Sensitivity
The assay was performed in 96-well plates, and each treatment was done in triplicate. Every cell line was treated with 2 µM BYL719aswellasDMSOasacon-trol. After 72 hr of drug treatment, cells were stained with the fluorescent dye Syto60. The viability ratio was obtained by [average fluorescence read of 2mMBYL719-treatedcells]/[averagefluorescencereadofDMSO-treatedcells].
Xenograft Mouse Studies
For xenograft experiments, female mice were implanted with estrogen 0.72 mg of 17b–estradiol pellets (Innovative Research of America). A suspension of 15–20 × 106 cells was inoculated subcutaneously into the left flanks of 6- to 8-week-old female athymic nude mice. Tumors were monitored until they reached approximately the average size of 250–400 mm3. At this time, mice were randomized to control and treatment groups (n = 6 per group). BYL719 was dissolved in 0.5% carboxymethyl cellulose sodium salt (Sigma) and administered at 25 mg/kg once a day by oral gavage. KIN-193 was dissolved in 7.5% NMP (Sigma), 40% PEG400 (Sigma), 52.5% dH2O, and given by IP injection twice a day at 20 mg/kg. Tumors were measured twice weekly using calipers. For pharmacodynamic analyses, tumor-bearing mice were administered with drugs or vehicle for 3 days. Tumor tissue was excised and snapfrozen in liquid nitrogen for immunoblotting or phosphoinositide measurement. Virgin female BALB-neuT mice transgenic for the rat-transforming neu oncogene expressed under the control of mouse mammary tumor virus promoter (Ciraolo et al., 2008) were weekly monitored for mammary tumor development by palpation. Once palpable tumors were present, tumor size was measured twice a week using a caliper. Treatments were initiated when a cumulative tumor burden of around 25 mm3 was reached, at which point mice were randomized into four groups of five mice each. GSK2636771 was dissolved on 0.5% metilcellulose and 0.2% Tween 80. Mice were daily treated by oral gavage with GSK2636771 (30 mg/kg), BYL719 (25 mg/kg), or a combination of the two drugs for 21 days. The combo mice received the two treatments 30 min apart. The final group of animals was treated with vehicles of both agents. All mice were killed using CO2 inhalation per institutional guidelines at Massachusetts General Hospital or University of Torino. Experiments were approved by the Institutional Animal Care and Use Committee at Massachusetts General Hospital or University of Torino.
Supplementary Material
Significance.
Clinical development of PI3K inhibitors is underway, and there is emerging enthusiasm for using isoform-specific PI3K inhibitors that may have a greater therapeutic index. We found that breast cancer cells harboring PIK3CA mutations and/or HER2 amplification were among those more sensitive to the selective p110α inhibitor BYL719. However, even among these cell lines, PI3K signaling was rapidly restored after the initial response to BYL719 via the activation of the p110β isoform. Because BYL719 is currently tested in the clinic for PIK3CA mutant breast cancer, this reactivation is critical because it may limit its efficacy in the clinic. Importantly, we showed that the p110β inhibitor was effective in blocking PIP3 rebound and potentiated the antitumor effects of p110α inhibitor in vivo.
Acknowledgments
This study is supported by grants from the NIH R01CA137008 (to J.A.E.) and AIRC, Italy (to E.H.). We thank Targeting PI3K in Women’s Cancers Stand Up to Cancer Dream Team for supplying GDC-0941. We thank Dr. Lucia Rameh for performing the PIP3 measurement using HPLC. We thank Drs. Rosen and Schwartz for critical reading of this manuscript. A.H. and Y.W. are employees and shareholders of Novartis Pharmaceuticals. C.H.B. received research support from Novartis Pharmaceuticals. J.A.E. is a consultant for Novartis and receives research support from Novartis Pharmaceuticals.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information includes Supplemental Experimental Procedures, five figures, and two tables and can be found with this article online at http://dx.doi.org/10.1016/j.ccell.2014.11.007.
REFERENCES
- Arteaga CL, Engelman JA. ERBB receptors: from oncogene discovery to basic science to mechanism-based cancer therapeutics. Cancer Cell. 2014;25:282–303. doi: 10.1016/j.ccr.2014.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bader AG, Kang S, Vogt PK. Cancer-specific mutations in PIK3CA are oncogenic in vivo. Proc. Natl. Acad. Sci. USA. 2006;103:1475–1479. doi: 10.1073/pnas.0510857103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bago R, Malik N, Munson MJ, Prescott AR, Davies P, Sommer E, Shpiro N, Ward R, Cross D, Ganley IG, Alessi DR. Characterization of VPS34-IN1, a selective inhibitor of Vps34, reveals that the phosphatidylinositol 3-phosphate-binding SGK3 protein kinase is a downstream target of class III phosphoinositide 3-kinase. Biochem J. 2014;463:413–427. doi: 10.1042/BJ20140889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beeton CA, Chance EM, Foukas LC, Shepherd PR. Comparison of the kinetic properties of the lipid- and protein-kinase activities of the p110alpha and p110beta catalytic subunits of class-Ia phosphoinositide 3-kinases. Biochem. J. 2000;350:353–359. [PMC free article] [PubMed] [Google Scholar]
- Chakrabarty A, Sánchez V, Kuba MG, Rinehart C, Arteaga CL. Feedback upregulation of HER3 (ErbB3) expression and activity attenuates antitumor effect of PI3K inhibitors. Proc. Natl. Acad.Sci. USA. 2012;109:2718–2723. doi: 10.1073/pnas.1018001108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandarlapaty S, Sawai A, Scaltriti M, Rodrik-Outmezguine V, Grbovic-Huezo O, Serra V, Majumder PK, Baselga J, Rosen N. AKT inhibition relieves feedback suppression of receptor tyrosine kinase expression and activity. Cancer Cell. 2011;19:58–71. doi: 10.1016/j.ccr.2010.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciraolo E, Iezzi M, Marone R, Marengo S, Curcio C, Costa C, Azzolino O, Gonella C, Rubinetto C, Wu H, et al. Phosphoinositide 3-kinase p110beta activity: key role in metabolism and mammary gland cancer but not development. Sci. Signal. 2008;1:ra3. doi: 10.1126/scisignal.1161577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark J, Anderson KE, Juvin V, Smith TS, Karpe F, Wakelam MJ, Stephens LR, Hawkins PT. Quantification of PtdInsP3 molecular species in cells and tissues by mass spectrometry. Nat. Methods. 2011;8:267–272. doi: 10.1038/nmeth.1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condliffe AM, Davidson K, Anderson KE, Ellson CD, Crabbe T, Okkenhaug K, Vanhaesebroeck B, Turner M, Webb L, Wymann MP, et al. Sequential activation of class IB and class IA PI3K is important for the primed respiratory burst of human but not murine neutrophils. Blood. 2005;106:1432–1440. doi: 10.1182/blood-2005-03-0944. [DOI] [PubMed] [Google Scholar]
- Dbouk HA, Vadas O, Shymanets A, Burke JE, Salamon RS, Khalil BD, Barrett MO, Waldo GL, Surve C, Hsueh C, et al. G protein-coupled receptor-mediated activation of p110β by Gbg is required for cellular transformation and invasiveness. Sci. Signal. 2012;5:ra89. doi: 10.1126/scisignal.2003264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebi H, Costa C, Faber AC, Nishtala M, Kotani H, Juric D, Della Pelle P, Song Y, Yano S, Mino-Kenudson M, et al. PI3K regulates MEK/ERK signaling inbreast cancer via the Rac-GEF, P-Rex1. Proc. Natl. Acad. Sci. USA. 2013;110:21124–21129. doi: 10.1073/pnas.1314124110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelman JA. Targeting PI3K signalling in cancer: opportunities, challenges and limitations. Nat. Rev. Cancer. 2009;9:550–562. doi: 10.1038/nrc2664. [DOI] [PubMed] [Google Scholar]
- Faber AC, Li D, Song Y, Liang MC, Yeap BY, Bronson RT, Lifshits E, Chen Z, Maira SM, García-Echeverría C, et al. Differential induction of apoptosis in HER2 and EGFR addicted cancers following PI3K inhibition. Proc. Natl. Acad. Sci. USA. 2009;106:19503–19508. doi: 10.1073/pnas.0905056106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foukas LC, Claret M, Pearce W, Okkenhaug K, Meek S, Peskett E, Sancho S, Smith AJ, Withers DJ, Vanhaesebroeck B. Critical role for the p110alpha phosphoinositide-3-OH kinase in growth and metabolic regulation. Nature. 2006;441:366–370. doi: 10.1038/nature04694. [DOI] [PubMed] [Google Scholar]
- Fritsch C, Huang A, Chatenay-Rivauday C, Schnell C, Reddy A, Liu M, Kauffmann A, Guthy D, Erdmann D, De Pover A, et al. Characterization of the novel and specific PI3Kα inhibitor NVP-BYL719 and development of the patient stratification strategy for clinical trials. Mol. Cancer Ther. 2014;13:1117–1129. doi: 10.1158/1535-7163.MCT-13-0865. [DOI] [PubMed] [Google Scholar]
- Garrett JT, Sutton CR, Kurupi R, Bialucha CU, Ettenberg SA, Collins SD, Sheng Q, Wallweber J, Defazio-Eli L, Arteaga CL. Combination of antibody that inhibits ligand-independent HER3 dimerization and a p110α inhibitor potently blocks PI3K signaling and growth of HER2+ breast cancers. Cancer Res. 2013;73:6013–6023. doi: 10.1158/0008-5472.CAN-13-1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillou H, Stephens LR, Hawkins PT. Quantitative measurement of phosphatidylinositol 3,4,5-trisphosphate. Methods Enzymol. 2007;434:117–130. doi: 10.1016/S0076-6879(07)34007-X. [DOI] [PubMed] [Google Scholar]
- Guo JP, Coppola D, Cheng JQ. IKBKE protein activates Akt independent of phosphatidylinositol 3-kinase/PDK1/mTORC2 and the pleck-strin homology domain to sustain malignant transformation. J. Biol. Chem. 2011;286:37389–37398. doi: 10.1074/jbc.M111.287433. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Hernandez-Aya LF, Gonzalez-Angulo AM. Targeting the phos-phatidylinositol 3-kinase signaling pathway in breast cancer. Oncologist. 2011;16:404–414. doi: 10.1634/theoncologist.2010-0402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang A, Fritsch C, Wilson C, Reddy A, Liu M, Lehar J, Quadt C, Hofmann F, Schlegel R. Single agent activity of PIK3CA inhibitor BYL719 in a broad cancer cell line panel. Cancer research. 2012;72(Supplement 8) [Google Scholar]
- Jia S, Liu Z, Zhang S, Liu P, Zhang L, Lee SH, Zhang J, Signoretti S, Loda M, Roberts TM, Zhao JJ. Essential roles of PI(3)K-p110beta in cell growth, metabolism and tumorigenesis. Nature. 2008;454:776–779. doi: 10.1038/nature07091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juric D, Baselga J. Tumor genetic testing for patient selection in phase I clinical trials: the case of PI3K inhibitors. J. Clin. Oncol. 2012;30:765–766. doi: 10.1200/JCO.2011.39.6390. [DOI] [PubMed] [Google Scholar]
- Juric D, Rodon J, Gonzalez-Angulo AM, Burris HA, Bendell JC, Berlin JD, Middleton MR, Bootle D, Boehm M, Schmitt A, et al. BYL719, a next generation PI3K alpha specific inhibitor: Preliminary safety, PK, and efficacy results from the first-in-human study. Cancer research. 2012;72(Supplement 1) [Google Scholar]
- Kim S, Mangin P, Dangelmaier C, Lillian R, Jackson SP, Daniel JL, Kunapuli SP. Role of phosphoinositide 3-kinase beta in glycopro-tein VI-mediated Akt activation in platelets. J. Biol. Chem. 2009;284:33763–33772. doi: 10.1074/jbc.M109.048553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Redondo-Muñoz J, Perez-García V, Cortes I, Chagoyen M, Carrera AC. Nuclear but not cytosolic phosphoinositide 3-kinase beta has an essential function in cell survival. Mol. Cell. Biol. 2011;31:2122–2133. doi: 10.1128/MCB.01313-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandl A, Sarkes D, Carricaburu V, Jung V, Rameh L. Serum withdrawal-induced accumulation of phosphoinositide 3-kinase lipids in differentiating 3T3-L6 myoblasts: distinct roles for Ship2 and PTEN. Mol. Cell. Biol. 2007;27:8098–8112. doi: 10.1128/MCB.00756-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni J, Liu Q, Xie S, Carlson C, Von T, Vogel K, Riddle S, Benes C, Eck M, Roberts T, et al. Functional characterization of an isoform-selec-tive inhibitor of PI3K–p110beta as a potential anticancer agent. Cancer discovery. 2012;2:425–433. doi: 10.1158/2159-8290.CD-12-0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien C, Wallin JJ, Sampath D, GuhaThakurta D, Savage H, Punnoose EA, Guan J, Berry L, Prior WW, Amler LC, et al. Predictive biomarkers of sensitivity to the phosphatidylinositol 30 kinase inhibitor GDC-0941 in breast cancer preclinical models. Clinical cancer research. 2010;16:3670–3683. doi: 10.1158/1078-0432.CCR-09-2828. [DOI] [PubMed] [Google Scholar]
- O’Reilly KE, Rojo F, She QB, Solit D, Mills GB, Smith D, Lane H, Hofmann F, Hicklin DJ, Ludwig DL, et al. mTOR inhibition induces upstream receptor tyrosine kinase signaling and activates Akt. Cancer Res. 2006;66:1500–1508. doi: 10.1158/0008-5472.CAN-05-2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park WS, Heo WD, Whalen JH, O’Rourke NA, Bryan HM, Meyer T, Teruel MN. Comprehensive identification of PIP3-regulated PH domains from C. elegans to H. sapiens by model prediction and live imaging. Mol. Cell. 2008;30:381–392. doi: 10.1016/j.molcel.2008.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickl M, Ries CH. Comparison of 3D and 2D tumor models reveals enhanced HER2 activation in 3D associated with an increased response to trastuzumab. Oncogene. 2009;28:461–468. doi: 10.1038/onc.2008.394. [DOI] [PubMed] [Google Scholar]
- Raynaud FI, Eccles SA, Patel S, Alix S, Box G, Chuckowree I, Folkes A, Gowan S, De Haven Brandon A, Di Stefano F, et al. Biological properties of potent inhibitors of class I phosphatidylinositide 3-kinases: from PI-103 through PI-540, PI-620 to the oral agent GDC-0941. Mol. Cancer Ther. 2009;8:1725–1738. doi: 10.1158/1535-7163.MCT-08-1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrik-Outmezguine VS, Chandarlapaty S, Pagano NC, Poulikakos PI, Scaltriti M, Moskatel E, Baselga J, Guichard S, Rosen N. mTOR kinase inhibition causes feedback-dependent biphasic regulation of AKT signaling. Cancer discovery. 2011;1:248–259. doi: 10.1158/2159-8290.CD-11-0085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuels Y, Wang Z, Bardelli A, Silliman N, Ptak J, Szabo S, Yan H, Gazdar A, Powell SM, Riggins GJ, et al. High frequency of mutations of the PIK3CA gene in human cancers. Science. 2004;304:554. doi: 10.1126/science.1096502. [DOI] [PubMed] [Google Scholar]
- Schwartz S, Wongvipat J, Trigwell CB, Hancox U, Carver BS, Rodrik-Outmezguine V, Will M, Yellen P, de Stanchina E, Baselga J, et al. Feedback suppression of PI3Kα signaling in PTEN-mutated tumors is relieved by selective inhibition of PI3Kβ. Cancer Cell. 2014;27:109–122. doi: 10.1016/j.ccell.2014.11.008. this issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stemke-Hale K, Gonzalez-Angulo AM, Lluch A, Neve RM, Kuo WL, Davies M, Carey M, Hu Z, Guan Y, Sahin A, et al. An integrative genomic and proteomic analysis of PIK3CA, PTEN, and AKT mutations in breast cancer. Cancer Res. 2008;68:6084–6091. doi: 10.1158/0008-5472.CAN-07-6854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utermark T, Rao T, Cheng H, Wang Q, Lee SH, Wang ZC, Iglehart JD, Roberts TM, Muller WJ, Zhao JJ. The p110α and p110β isoforms of PI3K play divergent roles in mammary gland development and tumorigenesis. Genes Dev. 2012;26:1573–1586. doi: 10.1101/gad.191973.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasudevan KM, Barbie DA, Davies MA, Rabinovsky R, McNear CJ, Kim JJ, Hennessy BT, Tseng H, Pochanard P, Kim SY, et al. AKT-independent signaling downstream of oncogenic PIK3CA mutations in human cancer. Cancer Cell. 2009;16:21–32. doi: 10.1016/j.ccr.2009.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wee S, Wiederschain D, Maira SM, Loo A, Miller C, deBeaumont R, Stegmeier F, Yao YM, Lengauer C. PTEN-deficient cancers depend on PIK3CB. Proc. Natl. Acad. Sci. USA. 2008;105:13057–13062. doi: 10.1073/pnas.0802655105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao JJ, Cheng H, Jia S, Wang L, Gjoerup OV, Mikami A, Roberts TM. The p110alpha isoform of PI3K is essential for proper growth factor signaling and oncogenic transformation. Proc. Natl. Acad. Sci. USA. 2006;103:16296–16300. doi: 10.1073/pnas.0607899103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.