Abstract
The discovery of genetic variation and the assembly of genome sequences are both inextricably linked to advances in DNA-sequencing technology. Short-read massively parallel sequencing has revolutionized our ability to discover genetic variation but is insufficient to generate high-quality genome assemblies or resolve most structural variation. Full resolution of variation is only guaranteed by complete de novo assembly of a genome. Here, we review approaches to genome assembly, the nature of gaps or missing sequences, and biases in the assembly process. We describe the challenges of generating a complete de novo genome assembly using current technologies and the impact that being able to perfectly sequence the genome would have on understanding human disease and evolution. Finally, we summarize recent technological advances that improve both contiguity and accuracy and emphasize the importance of complete de novo assembly as opposed to read mapping as the primary means to understanding the full range of human genetic variation.
Contemporary genetic studies fundamentally require comparing the sequences of individual genomes. The dominant method for this comparison is resequencing, in which random fragments of a genome are obtained and compared to a reference sequence. Such experiments are conducted using instruments for massively parallel sequencing (MPS), in which billions of 100–200-nucleotide sequences may be read by a single instrument in a few days. Although great advances have been made in our knowledge of diversity1, cancer2 and genetic disease3, the genetic information provided by resequencing with current technology is incomplete. There is a lack of sensitivity for detecting small insertions and deletions (indels) and structural variation1,4,5, there is coverage bias against particularly GC-and AT-rich DNA6, the phase of mutations over long ranges must be inferred or imputed as opposed to directly observed, and the architecture of large polymorphic copy number variations is incomplete7–9.
An alternative to resequencing is de novo assembly, in which the entire sequence of two haplotypes is resolved from sequence data without comparison to a reference-genome sequence. Although de novo assembly is, in principle, complete and therefore the ideal for genetic variation discovery, it is still currently impossible to achieve with data generated by typical MPS resequencing projects10. There is evidence that the landscape of sequencing technology is changing in such a way that will ultimately enable more-routine de novo assembly of genomes.
In this Review, we first describe the computational challenges of de novo assembly and review state-of-the-art de novo assembly of human and other mammalian genomes. Next, we discuss the biases involved in detecting sequence variation as a result of incomplete assembly, the implications for biomedicine and the types of variation that may be better accessed with a complete de novo assembly. Finally, we review new approaches coupled with advances in sequencing technology that provide additional information that may be used to resolve de novo assemblies of human genomes.
Strategies and algorithms for assembling genomes
The goal of de novo genome assembly is to determine the sequence of a genome using only randomly sampled sequence fragments, which are typically less than one-millionth the size of a mammalian genome. Most current approaches involve some aspect of a whole-genome shotgun sequencing and assembly (WGSA) strategy, in which random fragments from a genome are sequenced and computationally stitched together to generate sequence contigs and scaffolds11. Under ideal conditions (that is, uniformly high sequence coverage and a genome devoid of repetitive sequences), an assembly may be determined with the simple approach of merging reads with maximal overlap. However, it became clear early on that this method is too simplistic to accurately assemble genomes with complex organizations. Sequence coverage is almost never uniform, and repetitive sequences of varying length, copy number and sequence complicate this process. This makes the correct merging of sequence reads a nearly impossible task in specific regions of the genome (FIG. 1). The key aspects for de novo assembly and repeat resolution are read length, overlap mapping quality and assembly algorithm.
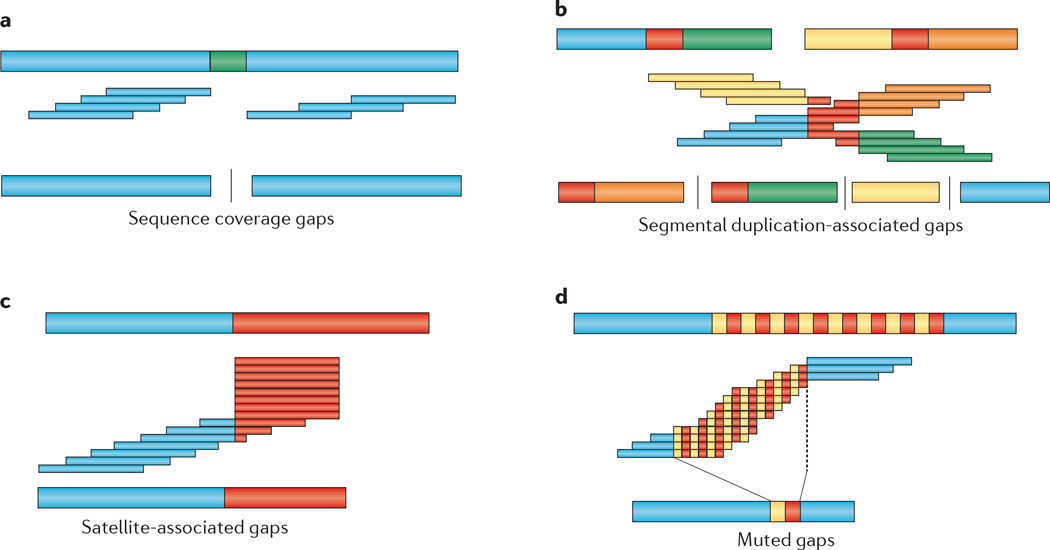
Figure 1. Types of genome assembly gaps.
Abstracted images of genome assemblies are illustrated. The genome architecture being resolved is shown at the top of each figure part as thick bars. Repetitive sequences are shown in red. Read overlaps are illustrated below the genome as thin bars (middle of each figure part), with regions overlapping repeats filled as red. The resulting assembly contigs are shown below (bottom of each figure part). Gaps are shown as vertical bars separating contigs to indicate unresolved sequences. a | The absence or reduction in sequence reads due to potential amplification or sequencing biases creates ‘dropouts’, where the assembled sequence is incomplete. b | Large segmental duplications of high sequence identity (orange and green) make read overlaps ambiguous, leading to multiple gaps flanking segmental duplications. The effect becomes exacerbated if the duplications are structurally polymorphic in a diploid genome. Long-range sequence information is required to resolve the complete sequence. c | Satellite-associated gaps are a special case leading to read ‘pileups’ due to higher-order tandem arrays of repetitive sequence, and they cannot be resolved using paired-end sequence information. These occur primarily in centromeric, acrocentric and telomeric areas of genomes. d | Muted gaps arise when the assembly is contracted relative to the true genome when overlaps are consistent with a smaller representation of the genome. These are often associated with repetitive sequences that cannot be easily amplified and/or are incompatible with cloning and propagation (that is, when they are toxic to Escherichia coli), such as simple tandem repeats.
Early genome assembly strategies
Prior to 2007, the sequence and assembly of mammalian genomes was an expensive and time-consuming operation. Although a few groups initially advocated WGSA for mammalian-genome assembly12, the most-widely used mammalian genomes, human and mouse, were not assembled using this approach. Instead, these assemblies are relatively unique among mammalian genomes in that they were assembled almost entirely using clone-by-clone-based sequencing13. Each genome was divided into roughly 200-kb-long overlapping fragments that were cloned into bacterial artificial chromosomes (BACs) and individually assembled. This offered the advantage that BAC sequences that are repetitive within the context of the entire genome are locally unique, thus making gap-free assembly more tractable. As a result, these genomes have become the benchmark for comparison (FIG. 2a). When the result of a de novo assembly is a sequence per chromosome without gaps and with 99.99% base-pair accuracy, the assembly is considered complete; otherwise, it is considered a draft. In practice, completeness is considered for only euchromatic portions of the genome, and even the most-recent build of the human genome (GRCh38) contains gaps.
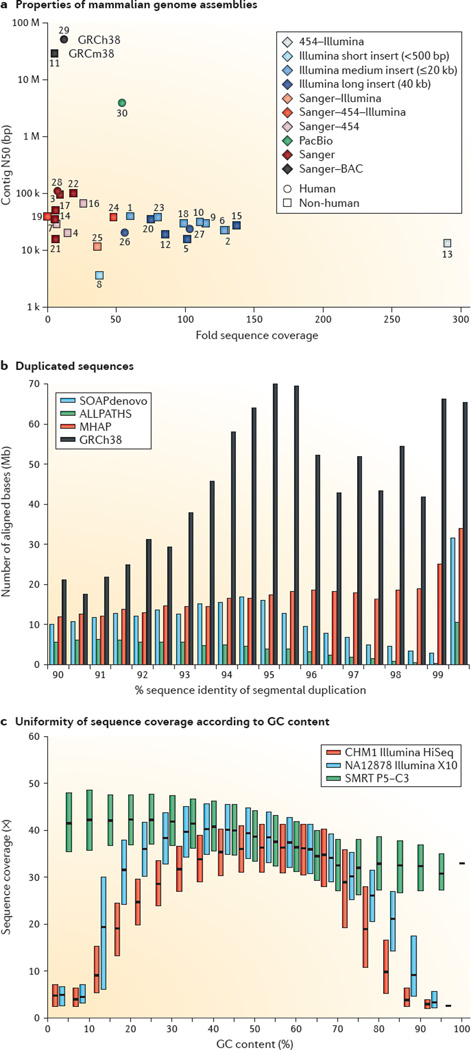
Figure 2. Sequencing and assembly statistics from different platforms.
a | A comparison of sequence coverage versus N50 contig length for 30 mammalian genomes from 25 species deposited into the US National Center for Biotechnology Information (NCBI) genome resource, including 5 human genome assemblies (circles). Colours contrast different sequencing platforms and assembly approaches. GRCh38 (human) and GRCm38 (mouse), generated by Sanger sequencing of bacterial artificial chromosome (BAC) clones represent the highest quality of genome. Genomes are enumerated according to species as follows: 1, Ailuropoda melanoleuca GCA_000004335.1; 2, Bos mutus GCA_000298355.1; 3, Bos taurus GCA_000181335.3; 4, Felis silvestris catus GCA_000687225.1; 5, Ursus maritimus GCA_000687225.1; 6, Balaenoptera acutorostrata GCA_000493695.1; 7, Callithrix jacchus GCA_000004665.1; 8, Daubentonia madagascariensis GCA_000241425.1; 9, Lipotes vexillifer GCA_000442215.1; 10, Pteropus alecto GCA_000325575.1; 11 and 12, Mus musculus GCA_000001635.6; 13, Nasalis larvatus GCA_000772465.1; 14, Nomascus leucogenys GCA_000146795.3; 15, Otolemur garnettii GCA_000181295.3; 16, Pan paniscus GCA_000258655.1; 17, Pan troglodytes GCA_000001515.4; 18, Panthera tigris GCA_000464555.1; 19, Papio anubis GCA_000264685.1; 20, Physeter macrocephalus GCA_000472045.1; 21, Pongo abelii GCF_000001545.4; 22, Rattus norvegicus GCA_000001895.4; 23, Saimiri boliviensis GCA_000235385.1; 24, Tarsius syrichta GCA_000164805.2; 25, Tursiops truncatus GCA_000151865.3; 26–30, Homo sapiens (SOAPdenovo, ALLPATHS, HuRef, GRCh38 and MinHash Alignment Process (MHAP), respectively). b | The amount of duplicated sequence represented in different genome assemblies, as determined by whole-genome assembly comparison (WGAC)107, is shown for SOAPdenovo (YH, GenBank GCA_000004845.2), ALLPATHS (NA12878, GenBank GCA_000185165.1) and MHAP (CHM1, GenBank GCA_000772585), as well as for the human reference genome (GRCh38). None of the de novo assemblies achieves the amount of duplication content resolved by the clone-based GRCh38 assembly, although the resolution of segmental duplication in massively parallel sequencing (MPS)-based assemblies (SOAPdenovo and ALLPATHS) is reduced compared with that of the single-molecule real-time (SMRT) sequence-based assembly MHAP. c | Sequencing read depth is compared to GC composition across the human genome for different platforms: CHM1 Illumina HiSeq (SRP044331), NA12878 Illumina X10 (data from AllSeq) and CHM1 SMRT P5–C3 (SRX533609). (P5–C3 refers to the version of DNA polymerase (P) and chemistry (C) used in the sequencing reaction.) The Illumina bias is decreased in more-modern instruments, whereas the SMRT sequencing coverage is more uniform, with fewer sequence-context gaps. 454, 454 Sequencing; PacBio, Pacific Biosciences.
De novo genome assembly algorithms
Since 2013, the process of de novo assembly of mammalian genomes has shifted from purely WGSA using MPS to assembly with longer sequence reads generated either synthetically or by single-molecule sequencing (SMS). Algorithms for de novo assembly have evolved in concert with these technology improvements. The main algorithmic approaches to de novo assembly — overlap-layout-consensus (OLC), de Bruijn and, more recently, the string graph14 — are each based on a separate theoretical graph framework15. Below, we describe some of the salient features of de novo assembly algorithms and outline specifically their treatment of repeats in the assembly process (FIG. 3).
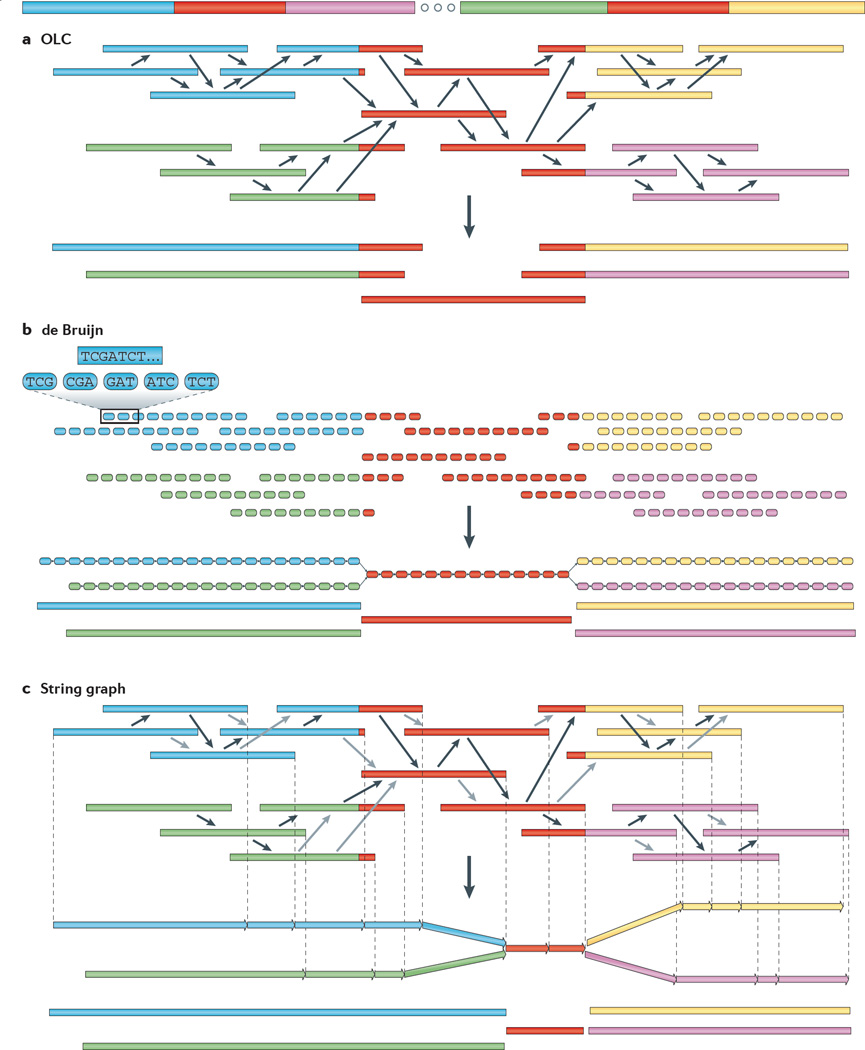
Figure 3. Genome assembly algorithms.
A genome schematic is shown at the top with four unique regions (blue, violet, green and yellow) and two copies of a repeated region (red). Three different strategies for genome assembly are outlined below this schematic. a | Overlap-layout-consensus (OLC). All pairwise alignments (arrows) between reads (solid bars) are detected. Reads are merged into contigs (below the vertical arrow) until a read at a repeat boundary (split colour bar) is detected, leading to a repeat that is unresolved and collapsed into a single copy. b | de Bruijn assembly. Reads are decomposed into overlapping k-mers. An example of the decomposition for k = 3 nucleotides is shown, although in practice k ranges between 31 and 200 nucleotides. Identical k-mers are merged and connected by an edge when appearing adjacently in reads. Contigs are formed by merging chains of k-mers until repeat boundaries are reached. If a k-mer appears in multiple positions (red segment) in the genome, it will fragment assemblies and additional graph operations must be applied to resolve such small repeats. The k-mer approach is ideal for short-read data generated by massively parallel sequencing (MPS). c | String graph. Alignments that may be transitively inferred from all pairwise alignments are removed (grey arrows). A graph is created with a vertex for the endpoint of every read. Edges are created both for each unaligned interval of a read and for each remaining pairwise overlap. Vertices connect edges that correspond to the reads that overlap. When there is allelic variation, alternative paths in the graph are formed. Not shown, but common to all three algorithms, is the use of read pairs to produce the final assembly product.
During OLC assembly, overlaps between all reads are first detected, then contigs are formed by iteratively merging overlapping reads until a read heuristically determined to be at the boundary of a repeat is reached16–18 (FIG. 3a). Repeats shorter than the minimally expected read overlap are often resolved, implying that genome resolution increases with read length. To account for sequencing errors, imprecise read overlaps are allowed, although this procedure may fragment the assembly even when the genomic repeats are nearly identical. The human genome was constructed primarily using OLC algorithms, and notable OLC-based assembly methods include parallel contig assembly program (PCAP)18, Arachne17 and Celera11.
Assembly methods based on de Bruijn graphs begin, somewhat counter-intuitively, by replacing each read with the set of all-overlapping sequences of a shorter, fixed length (FIG. 3b). The length is often denoted k, and the sequences k-mers. Contigs are formed by merging k-mers appearing adjacently in reads halting at k-mers from repeat boundaries (FIG. 3b). This has the cost of requiring highly accurate reads, and it initially discards some of the ability for reads to resolve repeats longer than k bases. It has the benefit of not requiring the storage of pairwise overlaps and having a graph structure representative of the repeat structure of the genome. For these reasons, de Bruijn assembly has been favoured for MPS projects in the ALLPATHS19, SOAPdenovo20 and ABySS21 mammalian next-generation sequencing assembly methods, in which little information is lost using k-mers, given the shorter read lengths.
The string graph14 and the related A-Bruijn graph22 assembly formulations are similar in concept to a de Bruijn graph but have the advantage of not decomposing sequences into k-mers but rather taking the full-length of a sequence read. They are produced based on operations of read overlap (FIG. 3c) and the removal of transitively inferred overlaps. There is an open-source implementation of string graph assembly called FALCON and produced by Pacific Biosciences.
The long sequences and high error rate of SMS reads favour overlap-based approaches over de Bruijn graphs, although it is computationally challenging to detect all pairwise overlaps between SMS reads. Initial approaches using read mapping23 were prohibitively slow, but novel pairwise alignment algorithms24,25 have enabled mammalian de novo assembly with SMS reads using modest computational resources. A final step of consensus calling must be performed for SMS read assembly that incorporates detailed error models of the underlying data, for example with Quiver23 and Nanocorrect26.
Diploid genomes present challenges to de novo assembly when heterozygous structural variation interrupts read overlap consistency. Assembly methods that assume homozygosity will fragment contigs at the boundaries of such structural variants, and methods that are diploid-aware must select only one of the haplotypes for assembly. An alternative representation of an assembly is the reference graph22,27 with a foundation in a repeat graph22, in which every edge represents a sequence and each vertex is at a branching point where alternative haplotypes may be selected or joined back with consensus sequence without structural variation. When available, paired-end sequences may be used to resolve sequences that are not assembled owing to repeats.
Examples of de novo genome assembly approaches
The first human de novo assembly using MPS was largely for proof-of-concept and was highly fragmented with an N50 length of 1.5 kb21. Improvements in MPS-based de novo assemblies have arisen partly from an increase in read lengths, and advances in scaffolding have arisen from an increase in the diversity and lengths of paired-end fragment library sizes (TABLE 1). The common ‘recipe’ for de novo assembly using MPS is similar to the multiple-insert-size design originally proposed for WSGA11. Adapted for Illumina sequencing (Illumina, Inc.), it uses a combination of high coverage reads from a short-insert library (sequences 200–500 bp long), a lower coverage of a medium-insert library (sequences 2 kb, 5 kb and 10 kb in length) and a sparse coverage of long-insert (40 kb in length) fosmid or jumping libraries28. This has been applied to produce draft assemblies of a Yoruban and a Han Chinese individual19,20. The addition of long-insert libraries leads to improved scaffolding and an increase in segmental duplication resolution20 over a previous assembly that used only short-and medium-length inserts. Although additional human genomes have not yet been assembled using this strategy, many mammalian (FIG. 2a) and avian genomes have been generated using MPS for comparative genomics studies29–31.
Table 1.
Human genome assemblies
| ABySS (NA12878) |
SOAPdenovo | SOAPdenovo2 (Yan) |
ALLPATHS-LG (NA12878) |
MHAP (CHM1) |
HuRef | DISCOVAR (NA12878) |
GRCh38 | |
|---|---|---|---|---|---|---|---|---|
|
Accession number |
NA | NA | GCA_ 000004845.2 |
GCA_ 000185165.1 |
GCA_ 000772585 |
GCA_ 000002125.2 |
NA | NA |
| Instrument | Illumina | Illumina | Illumina | Illumina | PacBio | Sanger | Illumina | Sanger |
|
Number of scaffolds* |
2,760,000 | NA | 4,197 | 3,331 | NA | 4,528 | NA | Complete |
|
Scaffold N50 (bp)* |
1,499 | 446,283 | 22,047,463 | 12,140,992 | NA | NA | NA | NA |
|
Number of contigs‡ |
4,348,132 | 3,884,491 | 239,711 | 231,194 | 40,917 | 65,049 | 949,302 | 646 |
|
Contig assembly length (bp) |
NA | NA | 2,851,884,686 | 2,771,622,777 | 3,260,000,000 | 2,782,378,670 | 3,085,280,835 | 3,100,000,000 |
|
Reference coverage (%) |
NA | NA | 87.8 | 81.3 | 89.7 | 89.6 | 90.2 | 100 |
|
Contig N50 (bp)‡ |
870 | 1,050 | 21,011 | 24,024 | 3,945,491 | 111,232 | 178,303 | 57,879,411 |
|
Percentage exons§ |
NA | NA | 89.9 | 79.5 | 93.2 | 93.7 | 95.0 | 100 |
|
Percentage UTRs |
NA | NA | 86.2 | 86.2 | 91.5 | 89.7 | 92.6 | 100 |
|
Sequence coverage |
42 | 71 | 56 | 103.1 | 54 | 7.5 | 69 | NA |
|
Largest clone║ |
210 | 10,000 | 40,000 | 40,000 | NA | 200,000 | 450 | NA |
| Read length¶ | 42 | 75 | 100 | 101 | 4,800 | 540 | 250 | NA |
MHAP, MinHash Alignment Process; NA, not applicable; PacBio, Pacific Biosciences; UTRs, untranslated regions. Each genome assembly was downloaded from GenBank and mapped using BLASR back to GRCh38.
Scaffold count and N50 contig length were calculated from published genome assemblies. The scaffold N50 is not applicable to HuRef, as the assembly is reference-guided, nor to the MHAP and DISCOVAR genome assemblies, because neither produces scaffolds.
Contig counts and N50 were calculated by removing scaffolding information.
We used the coordinates corresponding to the exons from RefSeq and computed the percentage of exons and UTRs identified based on this annotation. ‘Percentage exons’ is the percentage of all 181,147 exons in the reference genome completely contained within a contig, with ‘percentage UTRs’ representing a similar metric for the 97,750 UTRs.
The largest clone denotes the greatest mate-pair span used to generate the assembly, which correlates with resolution of duplicated sequences.
For MHAP and HuRef, the average read length is reported, whereas for ABySS, SOAPdenovo, SOAPdenovo2 and ALLPATHS multiple read sizes are used and the maximum is reported.
More broadly, the set of sequencing technologies and protocols used to assemble the 99 mammalian genomes in GenBank as of February 2015 is diverse: 57 assemblies used only Illumina; 22 were predominantly Sanger-based; 16 mixed Sanger, 454 Sequencing and Illumina technologies; 2 used just 454 Sequencing; 1 used Sequencing by Oligonucleotide Ligation and Detection (SOLiD); and 1 assembly method used Illumina and single-molecule optical mapping32. There is little relationship between sequence coverage and contig N50 for published assemblies (FIG. 2a). There is a similar heterogeneity in assembly methods even within the same sequencing methods; Sanger assemblies have used PCAP18, Arachne17, Celera11 and custom BAC assembly33, whereas genomes assembled predominantly with Illumina reads are roughly balanced between ALLPATHS19 (31 genomes) and SOAPdenovo20 (29 genomes). The quality of most genomes is well below the sequence contiguity and accuracy achieved by the mouse and human clone-ordered reference genomes, albeit at far less cost. Few modern genome assemblies exceed an N50 contig of 100 kb (average = 41 kb). This translates into tens to hundreds of thousands of sequence gaps, most of which correspond to various classes of repeat and MPS biases in GC representation (see above). The average percentage of core eukaryotic genes34 present in these draft genomes is 88%. Despite impressive scaffold N50 lengths, a greater fraction of genomes is not being accurately represented within assemblies (FIG. 2b), gene models and regulatory regions are being missed, and a smaller fraction of genetic variation is being characterized (TABLE 1) relative to clone-based references. As a result, gaps exist within the genomes and arise for various reasons (FIG. 1). We discuss these next in the context of the current human reference genome as well as the draft human genomes generated by MPS.
Types of gaps
Sequence-coverage gaps
Sequencing gaps occur, under the simplest condition, where no sequence reads have been sampled for a particular portion of the genome (FIG. 1a). Prior to the introduction of MPS methods, de novo assembly projects had limited coverage owing to the high cost of deep WGSA33,35,36. With low coverage (less than tenfold sequence coverage), the number of contigs produced in an assembly could be readily estimated using the Lander–Waterman (LW) statistic37. For example, a 5.3-fold sequence coverage from Sanger reads of 540 bp is estimated to result in an assembly with 155,906 gaps, which is roughly consistent with the observation of the low-coverage shotgun sequencing of a human genome with 206,552 gaps36. Such gaps are usually the easiest to remedy with additional coverage or targeted enrichment and sequencing. In principle, genomes sequenced using MPS methods do not suffer from coverage-based gaps, as the probability that a base is in a gap is vanishingly small (P < 1.92−22 at 50× coverage under a Poisson model). Instead, there are platform-dependent sequence compositions that are not well represented, such as the reduced coverage in AT-or GC-rich regions of the genome associated with the MPS platforms6 (FIG. 2c).
Segmental duplication-associated gaps
Over one-third (206/540) of the euchromatic gaps in the human reference genome (GRCh38) are flanked by large, highly identical segmental duplications. Analyses of human genome assemblies over the past 10 years consistently identified large tracts of duplicated DNA as the most common source of gaps, irrespective of the assembly approach or sequencing platform used38. The length (>10 kb), sequence identity (>96%) and structural polymorphism of these regions complicate resolution of this ~3% of the genome. Consider the simple assembly strategy outlined above for a genome that has repeated sequences longer than the length of a read. Most assembly methods iteratively merge overlapping reads into longer sequences called contigs (FIG. 3) and stop merging once the contig is extended to the boundary of a repeat sequence within the genome (FIG. 1b). The assembly process is further complicated by extensive structural polymorphisms, which are enriched more than tenfold in these particular regions1,39,40. Different human haplotypes (that is, allelic variation) have been shown to vary by hundreds of kilobases owing to paralogous sequences that are present in different numbers of copies and in different orientations8,40–42. Therefore, inadvertent assembly of two structurally diverse haplotypes within these regions is more likely than in unique regions of the genome. This results in an assembly that has no biological meaning (the assembly does not represent either of the haplotypes that are actually present) and cannot be deconvoluted until the incorrect joins are broken and the correct haplotype structures resolved41,43. In such regions, a single high-quality reference genome is insufficient, and there have been ongoing efforts to create multiple high-quality alternative reference haplotypes for these regions (FIG. 4A). For example, there are a total of 261 alternative references in the current human reference genome (GRCh38), corresponding primarily to regions of extreme genetic diversity often associated with segmental duplication41,43.
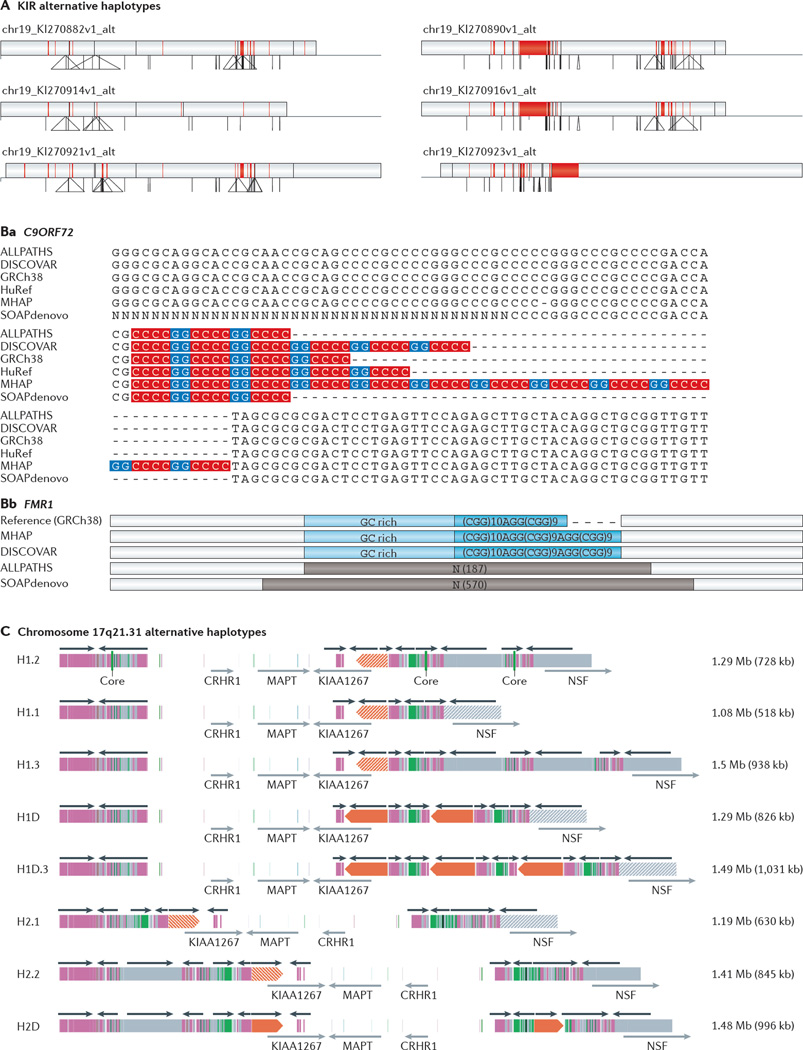
Figure 4. Assembly of complex regions of human genetic variation.
A | Six alternative haplotypes (GRCh38) in the KIR region (chromosome 19q13.42), assembled and sequenced using fosmid clones. The span of each haplotype with respect to the reference genome is denoted by the large rectangle, with the reference genome length being shown as a horizontal line below the rectangle. Deletions (red) are shown within the rectangle and insertions as triangles below, with the base of each triangle representing the length of the insertion. B | A comparison of two GC-rich disease-causing loci, chromosome 9 open reading frame 72 (C9ORF72; which causes frontotemporal dementia (FTD) and amyotrophic lateral sclerosis (ALS)) and fragile X mental retardation 1 (FMR1; which causes fragile X syndrome), in different genome assemblies. The sequence motif associated with the ALS C9ORF72 hexanucleotide repeat (red and blue) is partially resolved in all assemblies except for SOAPdenovo, in which the flanking 3′ region, which contains a divergent repeat motif and interspersed adenine nucleotides, is incomplete (Ba). The FMR1 trinucleotide repeat associated with fragile X syndrome is resolved by the DISCOVAR and MinHash Alignment Process (MHAP) assemblies (Bb). C | Eight different genomic structures associated with direct (H1) and inverted (H2) haplotypes of a gene-rich region on chromosome 17q21.31. The overall length of this region varies from 1.08 Mb to 1.50 Mb owing to variation in segmental duplication content (shown as coloured bars, with the total length given in brackets on the right). The H2D configuration is the only configuration that has large, highly identical duplications in a direct orientation predisposing to deletions that cause Koolen–de Vries syndrome. CRHR1, corticotropin releasing hormone receptor 1; MAPT, microtubule-associated protein tau; NSF, N-ethylmaleimide-sensitive fusion protein. Part C reproduced from REF. 7, Nature Publishing Group.
Satellite-associated gaps
In addition to the gaps associated with gene-rich segmental duplication, other forms of repetitive DNA have been found within gap regions (FIG. 1c). These include short and long runs of tandem repeats designated as short tandem repeats (STRs; also known as microsatellites), variable number of tandem repeats (VNTRs; also known as macrosatellites) and Mb-sized centromeric satellite repeats. These sequences are difficult to assemble because the read overlaps are consistent with different copy numbers of the tandem repeat, making it impossible to determine the exact structure. It is possible to estimate the copy number using read depth, but for complete resolution it is necessary to assemble using reads longer than the total satellite length. For example, 80% of the gaps closed by application of the single-molecule real-time (SMRT) long-read sequencing technology consisted of long-degenerate, low-complexity sequences (5–10 kb long) flanked by GC-rich DNA. Many of these gaps occur within the last 5–10-Mb-long telomeric regions of chromosomal arms. Heterochromatic DNA is an extreme case of repeat-associated gaps, in which short satellite sequences are repeated in tandem hundreds of thousands of times. In addition to it being impossible to resolve the sequence structure of the heterochromatic DNA using standard assembly methods, even length estimates are crude approximations owing to the extensive length heterogeneity within a species. As a result, most centromeric, acrocentric and secondary constrictions of chromosomes are not included in standard genome assemblies. More-recent assemblies (GRCh38) have included decoy satellite sequences as a placeholder to improve mapping44, but this has also resulted in an increase in the number of genome gaps; 212 new centromeric gaps have been added within the decoy sequences, as well as 45 additional gaps adjacent to euchromatic satellite DNA.
Muted gaps
Muted gaps are defined as regions that are inadvertently closed in an assembly but that actually show additional or different sequences in the vast majority of individuals45 (FIG. 1d). Although some muted gaps may theoretically represent a very rare deletion variant in the individual selected for sequence assembly, most arise as a result of errors in the assembly process or the dependence on clone-based sequencing in which the sequences that are toxic to bacteria were deleted during the cloning propagation process5. In the human genome, it has been estimated that there are over 2,600 muted gaps5. These gaps frequently associate with complex repeats, tandem duplications (within a few thousand bases) or long, low-complexity STR sequences, which, as described above, are often collapsed or truncated in the assembly process. Although such gaps are not typically annotated in reference genomes, more-recent assemblies produced by the Genome Reference Consortium46 contain ‘black-tag’ annotations at sites where there are uncertain merges. These sites have been shown to be enriched 200-fold for muted gaps.
Allelic variation gaps
Some regions of a genome also show extraordinary patterns of allelic variation, often reflecting deep evolutionary coalescence (for example, human leukocyte antigen (HLA) or the 17q21.31 polymorphism). Such divergence is frequently missed, especially when diploid genomes are assembled, because a particular haplotype is initially seeded or preferred during the assembly process. In other cases, the similar but not identical allelic haplotypes will appear as repeats to the assembly methods, initiating a contig break at the boundary between similar and diverged sequence. Such regions are not always easily recognized as gaps in the genome (they are a type of muted gap (FIG. 1d) if one haplotype is already resolved). Targeted approaches are usually required to sequence and assemble alternative diverged haplotypes43,47.
Consequences and impact
Accurate assembly of genomes has long been recognized as the key to understanding genetic variation. The more accurate the reference genome, the easier it is to map read data and interpret functional importance. A more-complete reference leads to better annotation, less genotyping error genome-wide1,48 and, concomitantly, a greater likelihood of identifying causal variation associated with human traits. Missing data (in the form of complex genetic variation or sequence gaps) translate into an inability to discover disease-causing mutations and, as a result, contribute to the problem of ‘missing heritability’ (REF. 49). A few recent examples are illustrative of this issue, even with respect to the high calibre of the human reference genome.
Unresolved repeats
The region associated with medullary cystic kidney disease type 1 (MCKD1) was identified in the early 2000s50, but the causative mutation for this straightforward Mendelian disease eluded discovery for several years51. Copy number variant analysis, exome sequencing, targeted capture and characterization of noncoding sequences failed to identify the causal variant. A VNTR region containing repeats of 60 bp in length composed primarily of GC-rich DNA (80%) was associated with a candidate gene encoding mucin 1 (MUC1)51. Southern blot analysis revealed that the VNTR was substantially underrepresented in the human reference genome compared to what had been observed in the normal human population. Targeted cloning, sequencing and de novo assembly was necessary to resolve the sequence structure of this particular region; when the region had been properly assembled, comparisons between individuals with and those without the disease revealed that the disease is associated with a single cytosine insertion within a stretch of seven cytosines (at positions 53–59) in a protein-coding region of MUC1. The resulting frameshift leads to a premature stop codon and hence a truncated mutant MUC1 protein. This truncated MUC1 is defective due to its lack of functional domains and underlies MCKD1 in >60% of families. In this case, both incomplete assembly and short-read mapping biases common to MPS data complicated discovery.
A similar cautionary tale of incomplete genome assembly and variation assessment was revealed for the discovery of the mutations responsible for amyotrophic lateral sclerosis (ALS)52,53 — the most common cause of adult-onset motor neuron disease in the human population. Manual realignment of chromosome flow-sorted sequencing data, as opposed to routine variant-calling algorithms, identified an expanded GGGGCC hexanucleotide repeat located within the first intron of the chromosome 9 open reading frame 72 (C9ORF72) transcript for an estimated 40% of familial ALS cases52,53. The C9ORF72 repeat is probably condensed in the ALLPATHS (NA12878) and HuRef assemblies, but is completely unresolved in the SOAPdenovo assembly (FIG. 4Ba).
GC-rich repetitive sequences are frequently missing or underrepresented from most modern genome assemblies. For example, the CGG trinucleotide sequence within the 5′-untranslated region of the fragile X mental retardation 1 (FMR1) gene is expanded to 200–1,000 copies in individuals with fragile X syndrome54, but the sequence of unaffected individuals, which occurs in 5–55 CGG trinucleotide copies, is missing from assemblies that used reads with lengths of up to 101 bp (ALLPATHS and SOAPdenovo). This locus is resolved in an assembly of NA12878 using 250-bp-long MPS sequences assembled with DISCOVAR. Interestingly, the FMR1 CGG repeat is shown to be further expanded in both the SMS-based MinHash Alignment Process (MHAP) assembly54 and the DISCOVAR Illumina assembly (FIG. 4Bb), suggesting that normal allele lengths, but not pathogenic repeat sizes, can be sequenced and assembled accurately.
In the case of facioscapulohumeral muscular dystrophy55, the genetic basis of the disease was not fully understood until the repeat-rich region was accurately assembled and contraction of the D4Z4 VNTR repeat in conjunction with a point mutation within duplicated DNA was identified in patients with the disease. This particular constellation of genomic features leads to the stable production of a toxic double homeobox 4 (DUX4) transcript and took more than 10 years to deduce, in large part because of difficulties in assembly and in assessing genetic variation in this region of chromosome 4q35.
Unannotated genes
In some cases, functional elements, including entire genes, still await discovery in new human genomes. The discovery of mutations associated with thyrotoxic hypokalaemic periodic catalysis required the cloning and sequencing of inward rectifier potassium channel 18 (KCNJ18; also known as KIR2.6), first by PCR and later in large-insert BAC clones. Variation in this gene, which is ~99% identical to KCNJ12, was originally thought to represent allelic variation of KCNJ12 until the presence of the duplicated copy (KCNJ18) could be definitively proven and mutation of this associated with the phenotype56.
Missing sequence
It is currently estimated that 5–40 Mb of euchromatic sequences are absent from a given human reference genome owing to structural polymorphism57–59 and that an additional 125–150 Mb of gene-rich regions of the genome are inaccessible to standard variation analyses1. High-quality complete de novo sequence assembly would provide access to these regions and the inherent allelic variation within, without having to infer the composition of these regions from structural variation methods. Many of these regions show evidence of association with complex diseases, such as obesity60, lupus61 and infectious diseases62. In the absence of complete sequence structure, the findings remain difficult to interpret or are often mired in controversy because of associations for or against a particular phenotype (for example, C-C motif chemokine 3-like 1 (CCL3L1) and HIV infection63, and α-amylase 1 (AMY1) and obesity60,64). The frequency of recurrent mutation and the size and complexity of structural variation further limits imputation by a flanking-tag single-nucleotide polymorphism1,39. For such regions of the genome, genetic variation discovery is synonymous with high-quality sequence and assembly.
Characterizing normal genetic variation
In addition to disease, the inability to sequence and assemble portions of the genome limits our understanding of normal human genetic variation and evolution. The HLA region is perhaps the best-studied example in which alternative haplotypes differ by as much as 10%-15% between humans, but additional examples of hidden genetic variation, including novel paralogous copies, have emerged as a result of high-quality de novo sequencing and assembly of alternative haplotypes. For example, sequencing of 4 different haplotypes of the chromosome 15q13.3 region harbouring the gene neuronal acetylcholine receptor subunit alpha 7 (CHRNA7) showed that individuals differ by more than 1.5 Mb over this 3-Mb region, primarily with respect to duplicated genes41. These differences have important consequences, such as causing specific haplotypes to be predisposed to, or protected from, recurrent microdeletions that are associated with disease. Some complex structural haplotypes have risen to high allele frequencies in populations, consistent with their role in adaptive evolution. For example, the 17q21.31 inversion polymorphism, haplotype H2 (REFS 7,8) (FIG. 4C), has been associated with increased fecundity and overall global recombination65 but, paradoxically, an increased risk of Koolen–de Vries syndrome66. Recently, novel human-specific duplicate genes — such as SLIT–ROBO Rho GTPase-activating protein 2C (SRGAP2C), which is important for neuronal migration and dendrite density — were discovered as missing from the human reference genome9,67. Local BAC-based assemblies resolved the sequence and structure of three distinct copies of SRGAP2C that emerged during the human lineage of evolution, adding 379,665 bp of new sequence completely absent from the human reference and correcting 559,693 bp of incorrectly mapped sequence. This recently filled gap in SRGAP2C is even more remarkable as it is now considered a potential explanation — along with another human-specific gene, Rho GTPase-activating protein 11B (ARHGAP11B)68 — for the expansion of the human neocortex since humans and australopithecines diverged 2–3 million years ago.
Bioinformatics and technology advances
Recent algorithmic development in de novo assembly of MPS reads has focused on reducing computational space requirements69–71, more-sensitive variant detection72,73, single-cell sequencing74 and metagenomics75. Novel methods for resolving repeat structures with paired ends have only been demonstrated on smaller genomes76. The methods commonly used to produce mammalian assemblies — ABySS, SOAPdenovo and ALLPATHS-LG — have improved assembly results through incremental development19,20 and more-optimized use of library insert sizes, which have improved scaffold N50 statistics. Bioinformatics approaches are intrinsically linked to advances in sequencing technology. Newer approaches that take advantage of longer short-reads (for example, DISCOVAR de novo for 250-bp Illumina reads) hold considerable promise for improving contig lengths and enhancing the discovery of genetic variation72.
Sequencing-based scaffolding methods
More-substantial gains in assembly quality have been made through novel applications of MPS and developments of long-range sequencing technologies. To improve assemblies using existing MPS methods, sequencing techniques have focused on compartmentalizing sequence data into bins based on proximity intervals ranging from 10 kb to 1,000 kb in length77. For example, the ligating adjacent chromatin enables scaffolding in situ (LACHESIS) method78, leverages the observation that most reads sequenced by high-throughput chromatin-interaction sequencing (Hi-C) experiments map to chromatin intervals of ~1 Mb. As a result, this method has been used to significantly enhance the scaffolding of human MPS data at the chromosomal scale. As expected, erroneous scaffolds are highly enriched for segmental duplications (6.4-fold) and simple tandem repeats (2.9-fold). However, Hi-C read counts do not correlate well over shorter genomic distances (<1 Mb), hence contigs are difficult to more-precisely order78, requiring further algorithmic improvement. Thus, local complex repeat architectures cannot be readily resolved at the sequence level with this method, although the method does show promise for phasing79. A complementary technology, contiguity-preserving transposition sequencing (CPT-seq), uses a transposase-mediated barcoding strategy to generate low-coverage (0.05–0.10×) sequencing from midrange (80-kb-long) fragments80 that may be used in scaffolding. However, the sequences read from each fragment are unordered, making it similarly difficult to resolve complicated repeat structures.
Dilution pool sequencing
Another approach shared by several methods is dilution pool sequencing81, in which pools of low-concentration, high-molecular-weight DNA are separately barcoded and sequenced in aggregate; the number of sequences associated with the same barcode is sufficiently low that they do not overlap allelically. Low-level random amplification of DNA from each pool significantly improves haplotype phasing82–84, while greater levels of amplification give sufficient coverage for preassembly of reads from each dilution pool into longer synthetic reads85. This method has been adapted by Illumina as TruSeq and applied to synthetic reads averaging 4.39 kb from Drosophila melanogaster to build an assembly with an N50 of 69.7 kb86. There were 3,524 gaps in this assembly, of which 93% were due to decreased synthetic read coverage attributed to AT-rich regions (29.7%), consistent with PCR bias, and were enriched 2.3-fold for transposable elements and 10.4-fold for simple tandem repeats.
Optical mapping technologies
Complementing the long-range sequencing approaches have been advances in long-range mapping technologies that are enhancing the quality of draft de novo assemblies and highlighting potential inconsistencies and larger forms of structural variation. For example, high-throughput optical mapping technologies87 have now been successfully commercialized that operate by imaging individual DNA molecules at fluorescently labelled restriction sites (such as products created by OpGen, Inc. and BioNano Genomics). Signals are typically read at 1–9 kb intervals, and consensus-assembled optical maps are produced from overlapping restriction patterns of large molecules (>100 kb). These methods have been applied to scaffolding prokaryotic genomes88, discovery of complex structural variation, BAC assembly89 and mammalian genome assembly32. The Irys system, as an example, was recently used to generate optical maps of fragments ≥150 kb to resolve one of the most duplication-rich regions (chromosome 1q21) of the human genome90. These approaches provide accurate scaffolding information for contigs that may be uniquely aligned to the optical map. The methods require the isolation of high-molecular-weight DNA, indicating a resurgence in the need for expertise in pulsed-field gel electrophoresis (PFGE) DNA isolation or the development of new high-molecular-weight DNA isolation methods.
SMS
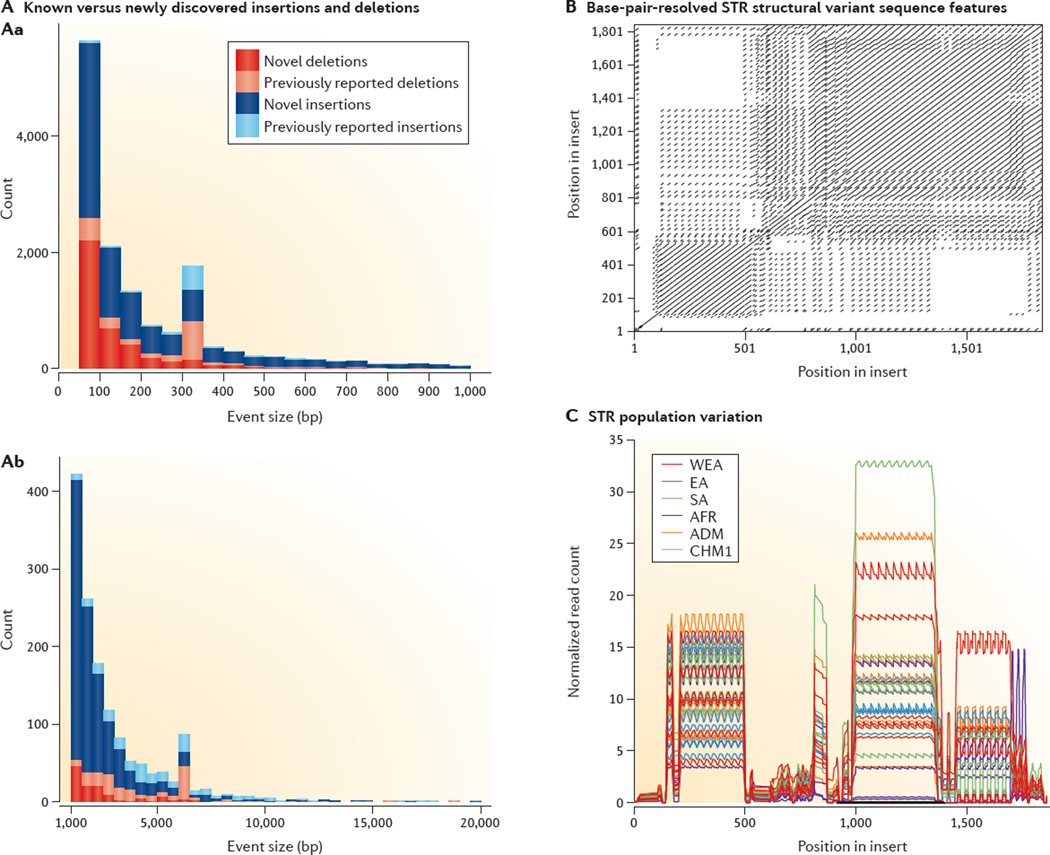
Developments in nanoscale sensing in zero-mode waveguides91 and nanopores92 hold considerable promise for revolutionizing the field of de novo genome assemblies. These technologies have enabled routine sequencing from single molecules without cloning and amplification of DNA. Importantly, the trace signal does not attenuate with read length, unlike in MPS methods that sequence from amplified DNA, allowing read lengths that are 1 to 2 orders of magnitude longer and that have fewer sequencing biases6 (FIG. 2c) and the potential to be limited only by the length of intact DNA after sample preparation. For example, the SMRT P6–C4 chemistry now produces sequence reads of >10 kb from genomic libraries that are >20 kb in length. Sequence error with this technology is more random in nature than it is for MPS, largely due to the small observable signal from nanoscale devices and non-redundant sampling. Mapping SMRT sequencing reads followed by local assembly has revealed that ~50% of the current gaps in the human genome build 37 can now be resolved or reduced5. More importantly, a comparison of one human genome (from a haploid hydatidiform mole, the CHM1 cell line) to the human reference suggests that more than 23,000 structural variants and longer indels, corresponding to 10.6 Mb of sequence, can now be resolved at the single-base-pair level (FIG. 5A,B). In contrast with methods that indirectly detect structural variation with discordant paired-end mapping93,94 and read depth95, base-pair resolution generates alternative reference sequences that allow genotyping analyses using other MPS datasets (FIG. 5C). Remarkably, 85% of insertions and 65% of deletions between 50 bp and 1,000 bp in length are novel, when compared to structural variant maps produced for more than 1,000 genomes sequenced and analysed using MPS technologies4,96. A large, common inversion polymorphism in 15q13.3 mediated by large (>50 kb) inverted duplications97 that is detectable in CHM1 with BAC end mapping was not detected in the SMRT sequence alignments, indicating a limitation of the local alignment approach for detecting large structural variation. This suggests that SMS technologies have the potential to provide a more-comprehensive assessment of the full range of human genetic variation through local alignment, but that the complete assessment of variation will rely on complete de novo assembly.
Figure 5. Human genetic variation detected with local assembly of single molecules.
A | Deletions (red and pink) and insertions (dark and light blue) resolved at base-pair resolution in the genome from the CHM1 cell line through local assembly of the single-molecule real-time (SMRT) reads for events less than 1 kb (Aa) and greater than 1 kb (Ab)5. Copy number variants found in previous studies4,57,108 are in lighter shades, with roughly 85% of events being unique to the CHM1 results. B | An example of a 1.7-kb short tandem repeat (STR) insertion event (represented in a self dot plot) not detected by Illumina resequencing of CHM1 but detected and assembled by SMRT reads. C | This STR insertion contains uniquely identifying 30 bp sequences that, once sequence resolved, may be used to genotype the presence of the insertion in genomes sequenced using Illumina technology. Normalized read depth serves as a proxy for estimating variability of STR length and demonstrates that the STR is highly variable in diverse populations (shown for Western Eurasian (WEA), East Asian (EA), South Asian (SA), African (AFR) and admixed (ADM) individuals). Figure adapted from REF. 5, Nature Publishing Group.
However, current commercially available instruments for SMS (from Oxford Nanopore Technologies and Pacific Biosciences,) have a much higher sequencing error rate than other MPS technologies, such as Illumina (~15%)98,99. Although this can be overcome by higher sequencing coverage (>30×), this translates into a much higher cost per base than other next-generation sequencing methods. Initial applications to assembling SMS reads have been used on human BACs100, bacterial genome assemblies23,101–103 and eukaryotic genome assemblies through hybrid approaches that couple MPS and SMRT sequencing technologies102,104. Efficient methods to detect all pairwise overlaps between reads24,25 have enabled mammalian assembly from SMS reads alone. A draft human genome based solely on de novo assembly of SMRT reads from the CHM1 genome (GenBank accession number GCA_000772585) demonstrates an N50 of 4.5 Mb24, which is 20-fold greater than the initial draft sequence assemblies of the human genome using MPS. This draft closed 51 gaps in the current human reference genome. Analysis of the segmental duplication content of this assembly reveals that the structure of ~200 kb of flanking sequence for 5,543 out of 7,177 segmental duplication regions is resolved, although this is biased towards the resolution of shorter segmental duplication regions, as only 24.8% (41.4 of 167 Mb) of segmentally duplicated bases are resolved. This represents a substantial improvement when compared to some of the early MPS de novo assemblies, in which >90% of duplications were misassembled38. However, the sequences of gaps are enriched tenfold for segmental duplications, indicating the inability to resolve larger segmental duplications with current SMS read lengths. Furthermore, the assembly is based on a haploid sample, and the ability to accurately resolve complex regions in diploid genomes has not yet been demonstrated by long-read technology. This is the critical next step in the application of this technology.
Conclusions and perspective
The ability to sequence genomes more deeply in a rapid and cost-effective manner has not yet translated into improved de novo assemblies of genomes. Although sequencing genomes is routine, their assembly is not. Generating a high-quality genome remains a more-expensive proposition that requires considerably more time and effort to do well. As a result, some of the most genetically diverse and complex regions of the human genome are not understood, and large swathes of structural genetic variation remain undiscovered. If a comprehensive understanding of genetic variation is the key to resolving the missing heritability paradox, cost and sequence coverage are not the only considerations.
Advances in long-read sequencing and single-molecule mapping technologies in the past few years have now made it possible for individual laboratories to consider the possibility of generating high-quality de novo assemblies of new genomes. Using these technologies, efforts are underway to create new vertebrate reference genomes and to produce additional human genomes with quality on par with or exceeding the quality of the human reference (‘gold’ and ‘platinum’ human genome assemblies). Other efforts are focused on applying these technological advances to understand the full range of human genetic variation more completely as standards for clinical sequencing (BOX 1). Efforts are being pursued to assess genetic variation using complete de novo assembly as well as mapping and local re-assembly, as each may be complementary. For example, in the MHAP assembly of CHM1 (REF. 24), 8 gaps in the human reference were closed that were not closed by local assembly5; similarly, local assembly closed 25 gaps not resolved by whole-genome de novo assembly.
Box 1. New human reference genomes and standards for genetic variation.
With the realization that one reference human genome would be insufficient for variation discovery and precision medicine, several initiatives have been launched in the past couple of years to create new human reference genomes and/or sets of highly curated and validated genetic variants for benchmarking of new technologies.
Genome in a Bottle
Genome in a Bottle (GIAB) is a US National Institute of Standards and Technology (NIST) public-private consortium of investigators whose goal is to develop reference standards, methods and datasets for genome analysis. It is using multiple technologies to create standard variant callsets for which the sensitivity and specificity of new variant calling methods developed for clinical sequencing may be evaluated. Data and reference material from one European genome (NA12878) and a father, mother and son Ashkenazim trio (GM24149, GM24143 and GM24385, respectively) have been made available. Additional reference genomes will help tune methods to produce more accurate calls facilitating the implementation of whole-genome sequencing in clinical practice.
Platinum and gold genomes
The goal of this US National Human Genome Research Institute (NHGRI) initiative is to generate new, high-quality reference genomes that are as accurate as (‘gold’), or are more complete and accurate than (‘platinum’), the current human reference genome. Target platinum genomes include two complete hydatidiform moles that are devoid of allelic variation (CHM1 and CHM13) for which draft assemblies are already available using single-molecule real-time (SMRT) technology (for example, GCA_000772585.3 and GCA_001015355.1). In addition, five additional diploid gold genomes have been selected as continental references from Europe, Asia, America and Africa (two samples from Africa). The strategy involves deep genome sequencing using multiple sequence platforms along with targeted bacterial artificial chromosome (BAC)-based sequencing of complex regions to generate haplotype-resolved genome assemblies105.
Human Genome Structural Variation Consortium
The Human Genome Structural Variation Consortium (HGSVC) is a large consortium of investigators (including members of the 1,000 Genomes Project) focused on increasing the sensitivity and specificity of structural variation detection. One of its goals is to deeply characterize the pattern and inheritance of structural variation in two-parent, one-child trios. Using a battery of sequencing and orthogonal mapping technologies, nine genomes are currently being characterized for structural variation, including Yoruban, Puerto Rican and Han Chinese trios. An exhaustive analysis and validation of structural variation in these three families will enhance the development of structural variant discovery and genotyping algorithms106.
Although it is clear that multiple human reference genomes corresponding to different continental or ethnic groups will emerge over the next few years, continued advances in sequencing technology are essential for moving this field forwards. Substantial improvements in sequence read length (>100 kb) as well as reductions in cost and gains in long-read throughput (by a factor of 10) are required before comprehensive and accurate de novo assemblies of human genomes become routine. Until that time, comprehensive de novo assembly of genomes will still depend on hybrid approaches that leverage large-insert clones, mixed whole-genome sequence datasets and long-range sequence data. Nevertheless, the routine and accurate de novo assembly of the complete 6-Gb-long diploid human genome should represent a milestone not only for the genomics community but also for the clinical interpretation of human genetic disease. De novo assembly of human genomes, as opposed to alignment to a reference, promises a more accurate and comprehensive understanding of human genetic variation, consistent with the vision of precision medicine.
Acknowledgments
The authors thank T. Brown for assistance in editing this manuscript. This work was supported, in part, by a US National Institutes of Health grant (2R01HG002385) to E.E.E.. E.E.E. is an investigator of the Howard Hughes Medical Institute.
Glossary
- Resequencing
Characterizing a sample genome and its associated variation by mapping and aligning sequence reads to a reference genome sequence.
- Massively parallel sequencing
(MPS). A general term for a form of DNA sequencing that measures trace signals from millions to hundreds of millions of amplified sequences at once, most frequently referring to sequencing produced by Illumina, Life Technologies and Complete Genomics platforms. Often referred to as next-generation or second-generation sequencing to distinguish it from long-read sequencing approaches (for example, single-molecule sequencing), which are sometimes referred to as third-generation sequencing.
- Structural variation
Large insertion, deletion or inversion differences between homologous chromosomes, or translocation differences involving non-homologous chromosomes. Operationally defined as events >50 bp in size to distinguish from smaller insertion and deletion events.
- Coverage bias
Regions with an excess or deficiency in the number of sequence reads originating as a result of platform differences in sequence chemistry, amplification or cloning.
- Phase
The assignment of genetic variants or alleles to one of two homologous chromosomes.
- De novo assembly
The action of constructing the sequence of a genome from overlapping DNA sequences without guidance from a reference genome.
- Haplotypes
Sets of genetic variants or alleles found on the same chromosome that are inherited together until disrupted by recombination.
- Whole-genome shotgun sequencing and assembly
(WGSA). The reconstruction of a genome from reads redundantly sampled at random, often with the aid of paired-end sequencing.
- Contigs
Continuous (or ‘contiguous’) sequences produced in a de novo assembly, free of any gaps.
- Scaffolds
Sets of ordered and oriented contigs, with the approximate distances between contigs estimated by traversing paired-end sequences that anchor to different contigs. Scaffolds consist of both sequence contigs and gaps.
- Bacterial artificial chromosomes
(BACs). Vectors with an F-plasmid origin of replication used to clonally propagate an organism’s DNA (typically 150–250 kb) by transfection into Escherichia coli.
- Single-molecule sequencing
(SMS). A form of DNA sequencing in which signals are derived from single molecules, frequently referring to sequencing produced by Pacific Biosciences and Oxford Nanopore Technologies platforms.
- Paired-end
Two reads sequenced from opposite ends of the same fragment.
- N50 length
A statistic in genomics defined as the shortest contig at which half the total length of the assembly is made of contigs of that length or greater. It is commonly used as a metric to summarize the contiguity of an assembly.
- Fragment library
A set of DNA fragments of approximately the same length that are paired-end sequenced.
- Segmental duplication
When a sequence is represented two or more times in a genome with high sequence identity and did not arise by retrotransposition. Often defined as paralogous sequences that share ≥90% sequence identity and are ≥1 kb in length.
- Short tandem repeats
(STRs). Tandem repeats in which the individual unit of repetition is less than 10 bp long and varies in length between different individuals in a population.
- Variable number of tandem repeats
(VNTR). Any tandem array of repeated sequence motifs that are highly variable in different individuals of a population. Historically, these were originally used in reference to tandem repeats that varied on the scale of thousands of base pairs over the length of the array.
- Centromeric
Referring to the primary cytogenetic constriction on metaphase chromosomes where the kinetochore forms and spindle fibre attaches during cell division. In humans the centromere is made up primarily of repetitions of higher-order alpha-satellite DNA.
- Heterochromatic DNA
Portions of chromosomes that stain densely, are typically gene poor and are rich in satellite sequences.
- Acrocentric
Relating to a type of chromosome in which the centromere maps very close to the short arm. Acrocentric chromosomes in humans are enriched in beta-satellite and ribosomal DNA sequences, which are repeated as hundreds of copies.
- Secondary constrictions
A cytogenetic term referring to metaphase chromosome constrictions outside the centromere, typically rich in satellites and used to help identify chromosomes.
- Satellite DNA
Highly repetitive DNA composed of thousands to tens of thousands of tandem repeats, usually between 100–300 bp in length, and frequently associated with heterochromatin.
- Muted gaps
Regions that have been incorrectly closed in a genome assembly despite additional sequences being present at these sites in the source genome.
- Coalescence
The genealogy of a region of the genome in which all alleles trace back to a common ancestral sequence.
- Missing heritability
The observation that only a portion of estimated genetic contribution to disease (for example, heritability of a trait from twin studies) can be explained by our current understanding of genetic variation and its transmission properties.
- Exome sequencing
A method for enrichment and targeted sequencing of the protein-coding portions of the genome using massively parallel sequencing.
Footnotes
Competing interests statement
The authors declare competing interests: see Web version for details.
FURTHER INFORMATION
AllSeq: http://allseq.com/x-ten-test-data
FALCON: www.github.com/PacificBiosciences/falcon
GenBank: http://www.ncbi.nlm.nih.gov/genbank/
Genome in a Bottle: https://sites.stanford.edu/abms/giab
US National Center for Biotechnology Information (NCBI) genome resource: http://www.ncbi.nlm.nih.gov/genome
References
- 1.Genomes Project Consortium. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Weinstein JN, et al. The Cancer Genome Atlas Pan-Cancer analysis project. Nat. Genet. 2013;45:1113–1120. doi: 10.1038/ng.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ng SB, et al. Exome sequencing identifies the cause of a mendelian disorder. Nat. Genet. 2010;42:30–35. doi: 10.1038/ng.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mills RE, et al. Mapping copy number variation by population-scale genome sequencing. Nature. 2011;470:59–65. doi: 10.1038/nature09708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chaisson MJP, et al. Resolving the complexity of the human genome using single-molecule sequencing. Nature. 2015;517:608–611. doi: 10.1038/nature13907. Long-read sequencing paired with local assembly reveals structural variation and closes or extends ~50% of the gaps in the reference human genome.
- 6.Ross MG, et al. Characterizing and measuring bias in sequence data. Genome Biol. 2013;14:R51. doi: 10.1186/gb-2013-14-5-r51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Steinberg KM, et al. Structural diversity and African origin of the 17q21.31 inversion polymorphism. Nat. Genet. 2012;44:872–880. doi: 10.1038/ng.2335. High-quality sequencing of the 17q21.31 region reveals a complex haplotype polymorphic region in which certain structural haplotypes predispose for disease.
- 8. Boettger LM, Handsaker RE, Zody MC, McCarroll SA. Structural haplotypes and recent evolution of the human 17q21.31 region. Nat. Genet. 2012;44:881–885. doi: 10.1038/ng.2334. Uses population genetics to infer the architecture and evolutionary history of chromosome 17q21.31 haplotypes. References 7 and 8 show a rapid rise of a particular inverted haplotype in European and Middle Eastern individuals that is consistent with adaptive selection.
- 9. Dennis MY, et al. Evolution of human-specific neural SRGAP2 genes by incomplete segmental duplication. Cell. 2012;149:912–922. doi: 10.1016/j.cell.2012.03.033. Shows that genes potentially responsible for unique aspects of human neuronal development were missing from the reference human genome, highlighting the importance of focusing on obtaining higher-quality reference sequences.
- 10.Motahari AS, Bresler G, Tse DNC. Information theory of DNA shotgun sequencing. IEEE Trans. Inf. Theory. 2013;59:6273–6289. [Google Scholar]
- 11.Myers EW, et al. A whole-genome assembly of Drosophila . Science. 2000;287:2196–2204. doi: 10.1126/science.287.5461.2196. [DOI] [PubMed] [Google Scholar]
- 12.Weber JL, Myers EW. Human whole-genome shotgun sequencing. Genome Res. 1997;7:401–409. doi: 10.1101/gr.7.5.401. [DOI] [PubMed] [Google Scholar]
- 13.Church DM, et al. Lineage-specific biology revealed by a finished genome assembly of the mouse. PLoS Biol. 2009;7:e1000112. doi: 10.1371/journal.pbio.1000112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Myers EW. The fragment assembly string graph. Bioinformatics. 2005;21:i79–ii85. doi: 10.1093/bioinformatics/bti1114. [DOI] [PubMed] [Google Scholar]
- 15. Nagarajan N, Pop M. Sequence assembly demystified. Nat. Rev. Genet. 2013;14:157–167. doi: 10.1038/nrg3367. A review of algorithmic details of fragment assembly.
- 16.Myers EW. Toward simplifying and accurately formulating fragment assembly. J. Comput. Biol. 1995;2:275–290. doi: 10.1089/cmb.1995.2.275. [DOI] [PubMed] [Google Scholar]
- 17.Batzoglou S, et al. ARACHNE: a whole-genome shotgun assembler. Genome Res. 2002;12:177–189. doi: 10.1101/gr.208902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang X, Wang J, Aluru S, Yang S-P, Hillier L. PCAP: a whole-genome assembly program. Genome Res. 2003;13:2164–2170. doi: 10.1101/gr.1390403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gnerre S, et al. High-quality draft assemblies of mammalian genomes from massively parallel sequence data. Proc. Natl Acad. Sci. USA. 2011;108:1513–1518. doi: 10.1073/pnas.1017351108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Luo R, et al. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. GigaScience. 2012;1:18. doi: 10.1186/2047-217X-1-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Simpson JT, et al. ABySS: a parallel assembler for short read sequence data. Genome Res. 2009;19:1117–1123. doi: 10.1101/gr.089532.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pevzner PA, Tang H, Tesler G. De novo repeat classification and fragment assembly. Genome Res. 2004;14:1786–1796. doi: 10.1101/gr.2395204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chin CS, et al. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat. Methods. 2013;10:563–569. doi: 10.1038/nmeth.2474. Describes the method of correcting sequencing error in long SMRT sequences with short SMRT sequences so that they may be assembled using the Celera assembler and consensus called with the Quiver method.
- 24. Berlin K, et al. Assembling large genomes with single-molecule sequencing and locality-sensitive hashing. Nat. Biotechnol. 2015;33:623–630. doi: 10.1038/nbt.3238. Introduces one of the first SMS assemblers. Draft genomes on par with the original human draft sequence may be efficiently assembled with SMS reads.
- 25.Myers G. In: Algorithms in Bioinformatics. Raphael B, Tang J, editors. Springer; 2014. pp. 52–67. [Google Scholar]
- 26.Loman NJ, Quick J, Simpson JT. A complete bacterial genome assembled de novo using only nanopore sequencing data. Nat. Methods. 2015;12:733–735. doi: 10.1038/nmeth.3444. [DOI] [PubMed] [Google Scholar]
- 27. Dilthey A, Cox C, Iqbal Z, Nelson MR, McVean G. Improved genome inference in the MHC using a population reference graph. Nat. Genet. 2015;47:682–688. doi: 10.1038/ng.3257. The first practical study using a graphical representation of the genome to encode the structural diversity of the major histocompatibility complex region.
- 28.Williams LJ, et al. Paired-end sequencing of Fosmid libraries by Illumina. Genome Res. 2012;22:2241–2249. doi: 10.1101/gr.138925.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yim HS, et al. Minke whale genome and aquatic adaptation in cetaceans. Nat. Genet. 2014;46:88–92. doi: 10.1038/ng.2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Parker J, et al. Genome-wide signatures of convergent evolution in echolocating mammals. Nature. 2013;502:228–231. doi: 10.1038/nature12511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang G, et al. Comparative genomics reveals insights into avian genome evolution and adaptation. Science. 2014;346:1311–1320. doi: 10.1126/science.1251385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dong Y, et al. Sequencing and automated whole-genome optical mapping of the genome of a domestic goat (Capra hircus) Nat. Biotechnol. 2013;31:135–141. doi: 10.1038/nbt.2478. [DOI] [PubMed] [Google Scholar]
- 33.Lander ES, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 34.Parra G, Bradnam K, Korf I. CEGMA: a pipeline to accurately annotate core genes in eukaryotic genomes. Bioinformatics. 2007;23:1061–1067. doi: 10.1093/bioinformatics/btm071. [DOI] [PubMed] [Google Scholar]
- 35.Venter JC, et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 36.Istrail S, et al. Whole-genome shotgun assembly and comparison of human genome assemblies. Proc. Natl Acad. Sci. USA. 2004;101:1916–1921. doi: 10.1073/pnas.0307971100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lander ES, Waterman MS. Genomic mapping by fingerprinting random clones: a mathematical analysis. Genomics. 1988;2:231–239. doi: 10.1016/0888-7543(88)90007-9. [DOI] [PubMed] [Google Scholar]
- 38.Alkan C, Sajjadian S, Eichler EE. Limitations of next-generation genome sequence assembly. Nat. Methods. 2011;8:61–65. doi: 10.1038/nmeth.1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sharp AJ, et al. Segmental duplications and copy-number variation in the human genome. Am. J. Hum. Genet. 2005;77:78–88. doi: 10.1086/431652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sudmant PH, et al. Diversity of human copy number variation and multicopy genes. Science. 2010;330:641–646. doi: 10.1126/science.1197005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Antonacci F, et al. Palindromic GOLGA8 core duplicons promote chromosome 15q13.3 microdeletion and evolutionary instability. Nat. Genet. 2014;46:1293–1302. doi: 10.1038/ng.3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pyo CW, et al. Recombinant structures expand and contract inter and intragenic diversification at the KIR locus. BMC Genomics. 2013;14:89. doi: 10.1186/1471-2164-14-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zody MC, et al. Evolutionary toggling of the MAPT 17q21.31 inversion region. Nat. Genet. 2008;40:1076–1083. doi: 10.1038/ng.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Altemose N, Miga KH, Maggioni M, Willard HF. Genomic characterization of large heterochromatic gaps in the human genome assembly. PLoS Comput. Biol. 2014;10:e1003628. doi: 10.1371/journal.pcbi.1003628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Eichler EE. Recent duplication, domain accretion and the dynamic mutation of the human genome. Trends Genet. 2001;17:661–669. doi: 10.1016/s0168-9525(01)02492-1. [DOI] [PubMed] [Google Scholar]
- 46.Church DM, et al. Modernizing reference genome assemblies. PLoS Biol. 2011;9:e1001091. doi: 10.1371/journal.pbio.1001091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Raymond CK, et al. Ancient haplotypes of the HLA Class II region. Genome Res. 2005;15:1250–1257. doi: 10.1101/gr.3554305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li H. Towards better understanding of artifacts in variant calling from high-coverage samples. Bioinformatics. 2014;30:2843–2851. doi: 10.1093/bioinformatics/btu356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Manolio TA, et al. Finding the missing heritability of complex diseases. Nature. 2009;461:747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fuchshuber A, et al. Refinement of the gene locus for autosomal dominant medullary cystic kidney disease type 1 (MCKD1) and construction of a physical and partial transcriptional map of the region. Genomics. 2001;72:278–284. doi: 10.1006/geno.2000.6486. [DOI] [PubMed] [Google Scholar]
- 51.Kirby A, et al. Mutations causing medullary cystic kidney disease type 1 lie in a large VNTR in MUC1 missed by massively parallel sequencing. Nat. Genet. 2013;45:299–303. doi: 10.1038/ng.2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Renton AE, et al. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron. 2011;72:257–268. doi: 10.1016/j.neuron.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.DeJesus-Hernandez M, et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p–linked FTD and ALS. Neuron. 2011;72:245–256. doi: 10.1016/j.neuron.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Eichler EE, et al. Haplotype and interspersion analysis of the FMR1 CGG repeat identifies two different mutational pathways for the origin of the fragile X syndrome. Hum. Mol. Genet. 1996;5:319–330. doi: 10.1093/hmg/5.3.319. [DOI] [PubMed] [Google Scholar]
- 55.Lemmers RJ, et al. Digenic inheritance of an SMCHD1 mutation and an FSHD-permissive D4Z4 allele causes facioscapulohumeral muscular dystrophy type 2. Nat. Genet. 2012;44:1370–1374. doi: 10.1038/ng.2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ryan DP, et al. Mutations in potassium channel Kir2.6 cause susceptibility to thyrotoxic hypokalemic periodic paralysis. Cell. 2010;140:88–98. doi: 10.1016/j.cell.2009.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kidd JM, et al. A human genome structural variation sequencing resource reveals insights into mutational mechanisms. Cell. 2010;143:837–847. doi: 10.1016/j.cell.2010.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Genovese G, et al. Using population admixture to help complete maps of the human genome. Nat. Genet. 2013;45:406–414. doi: 10.1038/ng.2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Li R, et al. Building the sequence map of the human pan-genome. Nat. Biotechnol. 2010;28:57–63. doi: 10.1038/nbt.1596. Describes how the draft assembly of a personal genome using MPS uncovered 19–40 Mb of sequence missing from the reference.
- 60.Falchi M, et al. Low copy number of the salivary amylase gene predisposes to obesity. Nat. Genet. 2014;46:492–497. doi: 10.1038/ng.2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yang Y, et al. Gene copy-number variation and associated polymorphisms of complement component C4 in human systemic lupus erythematosus (SLE): low copy number is a risk factor for and high copy number is a protective factor against SLE susceptibility in European Americans. Am. J. Hum. Genet. 2007;80:1037–1054. doi: 10.1086/518257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shen S, Pyo CW, Vu Q, Wang R, Geraghty DE. The essential detail: the genetics and genomics of the primate immune response. ILAR J. 2013;54:181–195. doi: 10.1093/ilar/ilt043. [DOI] [PubMed] [Google Scholar]
- 63.Hollox EJ, Hoh BP. Human gene copy number variation and infectious disease. Hum. Genet. 2014;133:1217–1233. doi: 10.1007/s00439-014-1457-x. [DOI] [PubMed] [Google Scholar]
- 64.Usher CL, et al. Structural forms of the human amylase locus and their relationships to SNPs, haplotypes and obesity. Nat. Genet. 2015;47:921–925. doi: 10.1038/ng.3340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Stefansson H, et al. A common inversion under selection in Europeans. Nat. Genet. 2005;37:129–137. doi: 10.1038/ng1508. [DOI] [PubMed] [Google Scholar]
- 66.Koolen DA, et al. A new chromosome 17q21.31 microdeletion syndrome associated with a common inversion polymorphism. Nat. Genet. 2006;38:999–1001. doi: 10.1038/ng1853. [DOI] [PubMed] [Google Scholar]
- 67.Charrier C, et al. Inhibition of SRGAP2 function by its human-specific paralogs induces neoteny during spine maturation. Cell. 2012;149:923–935. doi: 10.1016/j.cell.2012.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Florio M, et al. Human-specific gene ARHGAP11B promotes basal progenitor amplification and neocortex expansion. Science. 2015;347:1465–1470. doi: 10.1126/science.aaa1975. [DOI] [PubMed] [Google Scholar]
- 69.Chikhi R, Rizk G. Space-efficient and exact de Bruijn graph representation based on a Bloom filter. Algorithms Mol. Biol. 2013;8:22. doi: 10.1186/1748-7188-8-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Simpson JT, Durbin R. Efficient de novo assembly of large genomes using compressed data structures. Genome Res. 2012;22:549–556. doi: 10.1101/gr.126953.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zimin AV, et al. The MaSuRCA genome assembler. Bioinformatics. 2013;29:2669–2677. doi: 10.1093/bioinformatics/btt476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Weisenfeld NI, et al. Comprehensive variation discovery in single human genomes. Nat. Genet. 2014;46:1350–1355. doi: 10.1038/ng.3121. Shows that MPS deduces more variation than do resequencing methods.
- 73.Li H. Exploring single-sample SNP and INDEL calling with whole-genome de novo assembly. Bioinformatics. 2012;28:1838–1844. doi: 10.1093/bioinformatics/bts280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nurk S, et al. Assembling single-cell genomes and mini-metagenomes from chimeric MDA products. J. Comput. Biol. 2013;20:714–737. doi: 10.1089/cmb.2013.0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Howe AC, et al. Tackling soil diversity with the assembly of large, complex metagenomes. Proc. Natl Acad. Sci. USA. 2014;111:4904–4909. doi: 10.1073/pnas.1402564111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bankevich A, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Adey A, et al. In vitro, long-range sequence information for de novo genome assembly via transposase contiguity. Genome Res. 2014;24:2041–2049. doi: 10.1101/gr.178319.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Burton JN, et al. Chromosome-scale scaffolding of de novo genome assemblies based on chromatin interactions. Nat. Biotechnol. 2013;31:1119–1125. doi: 10.1038/nbt.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Selvaraj S, Dixon JR, Bansal V, Ren B. Whole-genome haplotype reconstruction using proximity-ligation and shotgun sequencing. Nat. Biotechnol. 2013;31:1111–1118. doi: 10.1038/nbt.2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Amini S, et al. Haplotype-resolved whole-genome sequencing by contiguity-preserving transposition and combinatorial indexing. Nat. Genet. 2014;46:1343–1349. doi: 10.1038/ng.3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Snyder MW, Adey A, Kitzman JO, Shendure J. Haplotype-resolved genome sequencing: experimental methods and applications. Nat. Rev. Genet. 2015;16:344–358. doi: 10.1038/nrg3903. [DOI] [PubMed] [Google Scholar]
- 82.Kaper F, et al. Whole-genome haplotyping by dilution, amplification, and sequencing. Proc. Natl Acad. Sci. USA. 2013;110:5552–5557. doi: 10.1073/pnas.1218696110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kuleshov V, et al. Whole-genome haplotyping using long reads and statistical methods. Nat. Biotechnol. 2014;32:261–266. doi: 10.1038/nbt.2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kitzman JO, et al. Haplotype-resolved genome sequencing of a Gujarati Indian individual. Nat. Biotechnol. 2011;29:59–63. doi: 10.1038/nbt.1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Voskoboynik A, et al. The genome sequence of the colonial chordate, Botryllus schlosseri . eLife. 2013;2:e00569. doi: 10.7554/eLife.00569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.McCoy RC, et al. Illumina TruSeq synthetic long-reads empower de novo assembly and resolve complex, highly-repetitive transposable elements. PLoS ONE. 2014;9:e106689. doi: 10.1371/journal.pone.0106689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Schwartz DC, et al. Ordered restriction maps of Saccharomyces cerevisiae chromosomes constructed by optical mapping. Science. 1993;262:110–114. doi: 10.1126/science.8211116. [DOI] [PubMed] [Google Scholar]
- 88.Onmus-Leone F, et al. Enhanced de novo assembly of high throughput pyrosequencing data using whole genome mapping. PLoS ONE. 2013;8:e61762. doi: 10.1371/journal.pone.0061762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lam ET, et al. Genome mapping on nanochannel arrays for structural variation analysis and sequence assembly. Nat. Biotechnol. 2012;30:771–776. doi: 10.1038/nbt.2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.O’Bleness M, et al. Finished sequence and assembly of the DUF1220-rich 1q21 region using a haploid human genome. BMC Genomics. 2014;15:387. doi: 10.1186/1471-2164-15-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Eid J, et al. Real-time DNA sequencing from single polymerase molecules. Science. 2009;323:133–138. doi: 10.1126/science.1162986. [DOI] [PubMed] [Google Scholar]
- 92.Rosenstein JK, Wanunu M, Merchant CA, Drndic M, Shepard KL. Integrated nanopore sensing platform with sub-microsecond temporal resolution. Nat. Methods. 2012;9:487–492. doi: 10.1038/nmeth.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hormozdiari F, Alkan C, Eichler EE, Sahinalp SC. Combinatorial algorithms for structural variation detection in high-throughput sequenced genomes. Genome Res. 2009;19:1270–1278. doi: 10.1101/gr.088633.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Rausch T, et al. DELLY: structural variant discovery by integrated paired-end and split-read analysis. Bioinformatics. 2012;28:i333–i339. doi: 10.1093/bioinformatics/bts378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Handsaker RE, Korn JM, Nemesh J, McCarroll SA. Discovery and genotyping of genome structural polymorphism by sequencing on a population scale. Nat. Genet. 2011;43:269–276. doi: 10.1038/ng.768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.The 1000 Genomes Project Consortium. A global reference for human genetic variation. Nature. 2015 doi: 10.1038/nature15393. http://www.dx.doi.org/10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sharp AJ, et al. A recurrent 15q13.3 microdeletion syndrome associated with mental retardation and seizures. Nat. Genet. 2008;40:322–328. doi: 10.1038/ng.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Chaisson MJ, Tesler G. Mapping single molecule sequencing reads using basic local alignment with successive refinement (BLASR): application and theory. BMC Bioinformatics. 2012;13:238. doi: 10.1186/1471-2105-13-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Quick J, Quinlan AR, Loman NJ. A reference bacterial genome dataset generated on the MinION™ portable single-molecule nanopore sequencer. GigaScience. 2014;3:22. doi: 10.1186/2047-217X-3-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Huddleston J, et al. Reconstructing complex regions of genomes using long-read sequencing technology. Genome Res. 2014;24:688–696. doi: 10.1101/gr.168450.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Bashir A, et al. A hybrid approach for the automated finishing of bacterial genomes. Nat. Biotechnol. 2012;30:701–707. doi: 10.1038/nbt.2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Koren S, et al. Hybrid error correction and de novo assembly of single-molecule sequencing reads. Nat. Biotechnol. 2012;30:693–700. doi: 10.1038/nbt.2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Prjibelski AD, et al. ExSPAnder: a universal repeat resolver for DNA fragment assembly. Bioinformatics. 2014;30:293–301. doi: 10.1093/bioinformatics/btu266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.English AC, et al. Mind the gap: upgrading genomes with Pacific Biosciences RS long-read sequencing technology. PLoS ONE. 2012;7:e47768. doi: 10.1371/journal.pone.0047768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Callaway E. ‘Platinum’ genome takes on disease. Nature. 2014;515:323. doi: 10.1038/515323a. [DOI] [PubMed] [Google Scholar]
- 106.Human Genome Structural Variation Consortium. The phase 3 structural variant dataset. 1000 Genomes. 2015 [online], http://www.1000genomes.org/phase-3-structural-variant-dataset. [Google Scholar]
- 107.Bailey JA, Yavor AM, Massa HF, Trask BJ, Eichler EE. Segmental duplications: organization and impact within the current human genome project assembly. Genome Res. 2001;11:1005–1017. doi: 10.1101/gr.187101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Conrad DF, et al. Origins and functional impact of copy number variation in the human genome. Nature. 2010;464:704–712. doi: 10.1038/nature08516. [DOI] [PMC free article] [PubMed] [Google Scholar]