The interaction between blood cells and disease has continued to grow both more complex and increasingly intriguing. Once thought to be static populations, circulating blood cells are now known to interact and communicate in ways far beyond the singular processes historically attributed to each population. A primary example is the platelet, an anucleate cell with a traditional role in hemostasis and thrombosis. The platelets’ defined biological roles have expanded exponentially over the last decade to include immunity, inflammation and mediation of oncogenesis.1 Platelets, although anucleate, contain a wealth of transcriptomic information. When viewed from the perspective of a large population analysis, platelets demonstrate wide diversity, important patterns, including association with obesity and diabetes, and distinct expression profiles as compared to white cells.2 Platelets' ability to participate in diverse systemic responses has been elucidated by our growing understanding of their contents and the revelation of their capacity to share these contents. Platelets are now known to horizontally transfer RNA, traffic pathogens and regulate physiological and pathophysiological processes far beyond hemostasis.3, 4
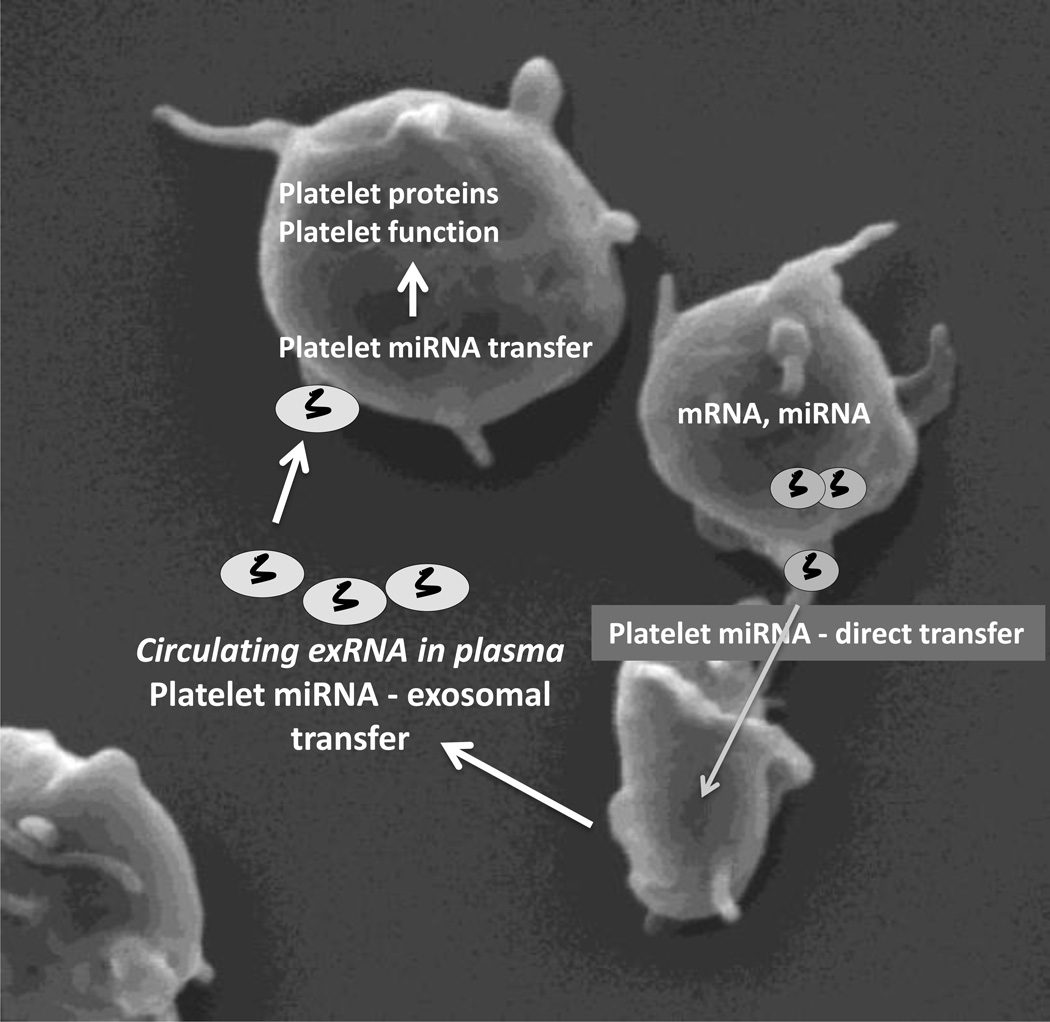
As part of our expanded understanding of platelet transfer processes, we have begun to appreciate the ability of the platelet to transfer transcripts, including mRNA and miRNA, to recipient cells with functional implications (Figure).4, 5 The observation that platelets possess the capacity to transfer cytosolic RNA suggests a new function for platelets in the regulation of vascular homeostasis.4 The processes involved in this horizontal cellular transfer of RNA are not fully understood; however, it is believed to be multifactorial with some contribution of platelet exosome release and uptake by other platelets or vascular cells. Platelet specific exosomes have been investigated and found to be functional, mediating vascular processes. For example, platelets from patients with myocardial infarction exhibit loss of specific miRNAs, and activated platelets shed miRNAs that can regulate endothelial cell gene expression.5
The Connection Between Plasma and Platelet microRNAs.
Platelets transfer microRNA (miRNA) and other coding and noncoding RNAs both through direct transfer and via exosome release where it can be detected in the plasma. The transfer of these miRNAs leads to altered vascular cellular phenotypes including changes in platelet proteins and platelet function (platelet image courtesy of Olga Vitseva).
While there are limited studies available about platelet specific exosomes or platelet specific RNA, the field of plasma-derived RNA or extracellular RNAs (exRNAs) has rapidly and greatly expanded. Although few of these studies specifically characterize the cell of origin of these exosomes, the most abundant microparticle subtype in circulation is platelet-derived, suggesting many exosomes are also platelet-derived. Most plasma exRNA studies have focused primarily on microRNA (miRNA) expression. MicroRNAs are highly conserved, small non-coding RNAs that regulate protein expression by complementary sequence recognition, binding, and translational repression of protein coding mRNA transcripts. Studies of circulating miRNAs have primarily been conducted to gain a greater understanding of disease processes or identify potential biomarkers such as in cardiovascular disease. For example, 852 distinct plasma miRNAs were measured in 83 patients presenting for cardiac catheterization. Eight plasma miRNAs were found to have over 2-fold increased expression in patients with significant coronary disease as compared to those with minimal coronary disease or normal coronary arteries.6 Importantly, cell-specific miRNA profiles are distinct, as demonstrated in patients with different types of myocardial infarction, ischemic heart disease or acute coronary syndromes who displayed unique miRNA distributions amongst their plasma, platelets, and leukocytes.7 Lastly, the largest study to date, from over five thousand people, demonstrated the rich genetic connection of circulating miRNAs with cis-miR-eQTLs that may be associated with complex traits.8
Adding to the exploration of the interaction of circulating exRNAs and platelet processes is the study of Kaudewitz and colleagues examining the association and potential function of plasma miRNAs and platelet activity.9 This group performed sequencing from plasma-derived RNA to identify specific small RNAs including miRNAs and YRNAs. They then examined a small cohort of patients with a history of acute coronary syndromes and recorded platelet function studies and showed that specific exRNAs correlated with platelet function tests. In a secondary larger population, they demonstrate that circulating plasma platelet protein markers associate with specific exRNAs. They then expand the mechanistic relevance of their findings by demonstrating that inhibition of miR-126 in a murine model reduced platelet aggregation and specific platelet function receptor expression.
The findings of this study are consistent with previous investigations demonstrating an association of plasma miRNAs with cardiovascular disease as well as studies demonstrating the relevance of platelet miRNA to platelet function.6, 7, 10 It has been previously shown that platelet miRNAs are able to repress expression of platelet proteins and, consistent with the study by Kaudewitz and colleagues,9 miRNA profiles are associated with and may predict platelet reactivity.10 As suggested by these studies, platelets and their response to antiplatelet therapy may be important to the circulating miRNA pool.11 The strengths of the current study lie in the use of the mechanistic information and the specific insight from the animal model. Although the study by Kaudewitz and colleagues does not demonstrate that miRNA changes in the human populations were specifically derived from platelets, this has been previously shown.12 Specifically, it is known that platelet microparticles, rich in miRNAs, can modify the transcriptome of macrophages and reprogram their function towards a phagocytic phenotype.12 Also consistent with these studies, the importance of platelet activation on the platelet miRNA content has been demonstrated to cause changes in the platelet proteome.13
While the study by Kaudewitz and colleagues provides intriguing translational insights, like many studies before, it does not definitely define a new biomarker of platelet function. A limitation of this and many of the other circulating exRNA investigations is the small populations utilized and the limited numbers of miRNA targets studied.14 The issue with such an approach is that it assumes the miRNAs chosen primarily affect a single direct downstream target and ignores the dogma that a single miRNA can result in abundant expression changes due to the combinatorial effect of multiple gene targets. Whether this assumption is correct will likely depend on the setting. As a single miRNA can target many genes, this cautions that the use of miRNAs as markers of platelet function may be more complex as compared to many classic platelet function assays.
In summary, our understanding of platelet function continues to progress as does our appreciation of their complex regulatory processes. The study by Kaudewitz and colleagues adds to our understanding of the utility of small non-coding RNAs released by platelets into the circulation and how they may reflect platelet processes. They add to these findings by showing functional data linking miRNAs to platelet aggregation. However, as a therapeutic, the use of miRNAs will require thorough analysis examining specificity, toxicity and human utility. As a biomarker of platelet reactivity, the use of plasma-based exRNA measurements is highly intriguing but, as we have learned from the differences between associative studies and large prospective controlled trials,15 extensive clinical data is needed to infer true utility.
Acknowledgments
Sources of Funding
Dr. Freedman is supported by P01-HL085381, RFA-HL-12-008 and RFA-RM-12-013NIH from Common Fund, through the Office of Strategic Coordination/Office of the NIH Director.
Footnotes
Disclosures
None
References
- 1.Clancy L, Freedman JE. The role of circulating platelet transcripts. Journal of thrombosis and haemostasis : JTH. 2015;13(Suppl 1):S33–S39. doi: 10.1111/jth.12922. [DOI] [PubMed] [Google Scholar]
- 2.Freedman JE, Larson MG, Tanriverdi K, O'Donnell CJ, Morin K, Hakanson AS, Vasan RS, Johnson AD, Iafrati MD, Benjamin EJ. Relation of platelet and leukocyte inflammatory transcripts to body mass index in the framingham heart study. Circulation. 2010;122:119–129. doi: 10.1161/CIRCULATIONAHA.109.928192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Koupenova M, Vitseva O, MacKay CR, Beaulieu LM, Benjamin EJ, Mick E, Kurt-Jones EA, Ravid K, Freedman JE. Platelet-tlr7 mediates host survival and platelet count during viral infection in the absence of platelet-dependent thrombosis. Blood. 2014;124:791–802. doi: 10.1182/blood-2013-11-536003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Risitano A, Beaulieu LM, Vitseva O, Freedman JE. Platelets and platelet-like particles mediate intercellular rna transfer. Blood. 2012;119:6288–6295. doi: 10.1182/blood-2011-12-396440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gidlof O, van der Brug M, Ohman J, Gilje P, Olde B, Wahlestedt C, Erlinge D. Platelets activated during myocardial infarction release functional mirna, which can be taken up by endothelial cells and regulate icam1 expression. Blood. 2013;121:3908–3917. S3901–S3926. doi: 10.1182/blood-2012-10-461798. [DOI] [PubMed] [Google Scholar]
- 6.Freedman JE, Ercan B, Morin KM, Liu CT, Tamer L, Ayaz L, Kanadasi M, Cicek D, Seyhan AI, Akilli RE, Camci C, Cengiz B, Oztuzcu S, Tanriverdi K. The distribution of circulating microrna and their relation to coronary disease. F1000 Research. 2012;1:50. doi: 10.12688/f1000research.1-50.v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ward JA, Esa N, Pidikiti R, Freedman JE, Keaney JF, Tanriverdi K, Vitseva O, Ambros V, Lee R, McManus DD. Circulating cell and plasma microrna profiles differ between non-st-segment and st-segment-elevation myocardial infarction. Family medicine & medical science research. 2013;2:108. doi: 10.4172/2327-4972.1000108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huan T, Rong J, Liu C, Zhang X, Tanriverdi K, Joehanes R, Chen BH, Murabito JM, Yao C, Courchesne P, Munson PJ, O'Donnell CJ, Cox N, Johnson AD, Larson MG, Levy D, Freedman JE. Genome-wide identification of microrna expression quantitative trait loci. Nature communications. 2015;6:6601. doi: 10.1038/ncomms7601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kaudewitz D, Skroblin P, Bender LH, Barwari T, Willeit P, Pechlaner R, Sunderland NP, Willeit K, Morton A, Armstrong PC, Chan MV, Lu R, Yin X, Gracio F, Dudek K, Langley S, Zampetaki A, de Rinaldis E, Ye S, Warner TD, Saxena A, Kiechl S, Storey R, Mayr M. Association of micrornas and yrnas with platelet function. Circulation research. 2015 doi: 10.1161/CIRCRESAHA.114.305663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nagalla S, Shaw C, Kong X, Kondkar AA, Edelstein LC, Ma L, Chen J, McKnight GS, Lopez JA, Yang L, Jin Y, Bray MS, Leal SM, Dong JF, Bray PF. Platelet microrna-mrna coexpression profiles correlate with platelet reactivity. Blood. 2011;117:5189–5197. doi: 10.1182/blood-2010-09-299719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Willeit P, Zampetaki A, Dudek K, Kaudewitz D, King A, Kirkby NS, Crosby-Nwaobi R, Prokopi M, Drozdov I, Langley SR, Sivaprasad S, Markus HS, Mitchell JA, Warner TD, Kiechl S, Mayr M. Circulating micrornas as novel biomarkers for platelet activation. Circulation research. 2013;112:595–600. doi: 10.1161/CIRCRESAHA.111.300539. [DOI] [PubMed] [Google Scholar]
- 12.Laffont B, Corduan A, Rousseau M, Duchez AC, Lee CH, Boilard E, Provost P. Platelet microparticles reprogram macrophage gene expression and function. Thrombosis and haemostasis. 2015;115 doi: 10.1160/TH15-05-0389. [DOI] [PubMed] [Google Scholar]
- 13.Cimmino G, Tarallo R, Nassa G, De Filippo MR, Giurato G, Ravo M, Rizzo F, Conte S, Pellegrino G, Cirillo P, Calabro P, Ohman T, Nyman TA, Weisz A, Golino P. Activating stimuli induce platelet microrna modulation and proteome reorganisation. Thrombosis and haemostasis. 2015;114:96–108. doi: 10.1160/TH14-09-0726. [DOI] [PubMed] [Google Scholar]
- 14.Freedman JE, Tanriverdi K. Defining mirna targets: Balancing simplicity with complexity. Circulation. 2013;127:2075–2077. doi: 10.1161/CIRCULATIONAHA.113.003058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Price MJ, Berger PB, Teirstein PS, Tanguay JF, Angiolillo DJ, Spriggs D, Puri S, Robbins M, Garratt KN, Bertrand OF, Stillabower ME, Aragon JR, Kandzari DE, Stinis CT, Lee MS, Manoukian SV, Cannon CP, Schork NJ, Topol EJ. Standard- vs high-dose clopidogrel based on platelet function testing after percutaneous coronary intervention: The gravitas randomized trial. Jama. 2011;305:1097–1105. doi: 10.1001/jama.2011.290. [DOI] [PubMed] [Google Scholar]