Abstract
Expanding on our prior studies with cord blood T-cells, we hypothesized that primary AML-reactive autologous T-cells could be generated ex vivo under immunomodulatory conditions. We purified AML and T-cells from 8 newly diagnosed high-risk patients. After 2 weeks expansion, T-cells were stimulated with IFN-γ treated autologous AML weekly X 3, IL-15 and agonistic anti-CD28 antibody. CTL and ELISpot assays tested functionality; RT-qPCR tested AML and T-cell gene expression profiles. Based on combined positive ELIspot and CTL assays, T-cells reactive against AML were generated in 5/8 patients. Treg proportion declined post-co-cultures in reactive T-cell samples. AML-reactive T-cells displayed an activated gene expression profile. “Resistant” AML blasts displayed genes associated with immunosuppressive MDSC. We discuss our approach to creating primary AML-reactive autologous T-cell and limitations that require further work. Our study provides a platform for future research targeting on generating autologous leukemia reactive T-cells.
Keywords: acute leukemia, adoptive immunotherapy, autologous CTLs, cellular therapy, MDSC, Tregs, AML, T-cells, cytotoxic
INTRODUCTION
Primary leukemia cells can evade immune responses by a variety of mechanisms 1 posing a number of obstacles in generating leukemia-specific cytotoxic T-lymphocytes (CTLs). Various methods have been explored to augment functional capacity of T cells to identify and eliminate leukemic cells 2–13. Many of these methods necessitate either genetic manipulation of T cells or targeting a previously identified “tumor-associated” antigen (TAA). However, most TAAs are not exclusively restricted to tumor cells and many non-malignant hematopoietic cells also share these antigens. On the other hand, TAAs may not be present in all tumors belonging from a common class 14,15. Genetically modified T cells exert their cytotoxic activity against a specific molecule that is generally universally present in malignant T cells of a class, but also in few normal hematopoietic cells. This approach is promising for certain B cell lymphoid malignancies 16, but it has not yet been widely adopted against myeloid malignancies. Alternative new strategies in CTL design with high specificity towards myeloid malignancies and other cancers are required where simultaneous reactivity towards multiple CTL epitopes may be similar to what has been observed with viral lysates 17,18 and DC-tumor fusion vaccines19. Importantly, successful novel approaches would be logistically relatively straightforward to foster wide scale implementation.
In previous studies from our laboratory, we showed that ex vivo expansion of cord blood (CB) T cells with anti-CD3/CD28 co-stimulatory beads and IL-2 plus IL-7 not only lead to their expansion, but also promoted their functional maturation and preserved a polyclonal T-Cell Receptor (TCR) repertoire 20–22. Importantly, subsequent co-cultures of the expanded CBT against myeloid (U937) or lymphoid (IM9) leukemia cell lines generated tumor-specific CTLs from naïve CBT 21,22. With these promising results, we hypothesized that T cells specifically reactive against primary patient acute myeloid leukemia (AML) blasts might also be generated from autologous blood with additional immune-stimulatory modifications during culture conditions.
Here we report on the feasibility and challenges of a novel methodology to generate autologous AML-specific peripheral blood reactive T cells in a significant fraction of tested individuals. First, we overcame barriers to address the low frequency and hyporeactivity of T cells in peripheral blood (PB) during hyperleukocytosis. Then, we implemented steps to render AML blasts immunogenic with our ex vivo culture conditions providing T cell activation signals I, II and III in parallel. To better understand the biological features of the system, we closely monitored both AML targets and T cell responders. We report on differences in the kinetics of regulatory T cells (Tregs) in AML patients during their initial presentation and identify significant differences in the immunophenotypic and genetic profiles of T cells between 5 reactive and 3 non-reactive T cell samples. Finally, we show that the AML blasts in 3 samples with non-reactive T cells (termed “resistant AML”) possessed myeloid derived suppressor cells (MDSC)-like gene expression profile compared with the AML blasts from 5 reactive T cell samples (termed “non-resistant AML”).
MATERIALS AND METHODS
Specimens
Peripheral blood mononuclear cells (PBMCs) were originally collected during leukapheresis of patients with newly diagnosed AML presenting with hyperleukocytosis admitted to the University of Pittsburgh Medical Center Cancer Institute. These PBMCs were cryopreserved and banked in an acute leukemia tissue repository in the Health Sciences Tissue Bank under an Institutional review board (IRB)-approved protocol with written informed consent of patients. We obtained eight cryopreserved PBMCs from this biorepository, which was accessible to other investigators yielding variable cell numbers available to for this study. The median pre-thaw total nucleated cell count was 97.9 (range, 2.14 – 133) × 106 in eight samples.
T cell purification and expansion
As shown in figure 1, T cells were enriched from thawed PBMCs with the immunomagnetic separation method using anti-CD3/CD28 beads (ClinExVivo™, Invitrogen, Sammamish, WA) and cultured with 100 U/mL of IL-2 (Proleukin; Novartis, East Hanover, NJ) and 10ng/ml of IL-7 (R&D Systems) for two weeks (+/− 2 days) in an antigen independent way as previously described 20,22 with the aim to generate sufficient numbers of T cells for AML co-cultures and possibly reverse their hyporeactive/anergic state. Immunophenotypic characterization of T cells was performed on FACSCanto II® multicolor flow cytometer (BD Biosciences) using BD Multitest™ reagents (CD3 FITC, CD8 PE, CD45 PerCP, CD4 APC) and BD Trucount™ absolute count tubes (all from BD Biosciences) to determine the purity of extraction and to quantify the subpopulations of T cells, as per manufacturer’s recommendations 23. T cells were assessed for viability manually using Trypan blue staining. Purified T cells were initially expanded in 96-well round bottom plates and later transferred to 24-well flat bottom plates at a concentration of 0.75 ×106 cell/ml using X-Vivo-15 media (Lonza) containing 5% pooled human serum, 2mM L-glutamine, 100 U/mL each of penicillin, streptomycin and amphotericin (Sigma-Aldrich), with anti-CD3/CD28 beads (beads: T cell ratio of 3:1), 100U/ml of IL-2 and 10ng/ml of IL-7 for 2 weeks as previously reported 20,22.
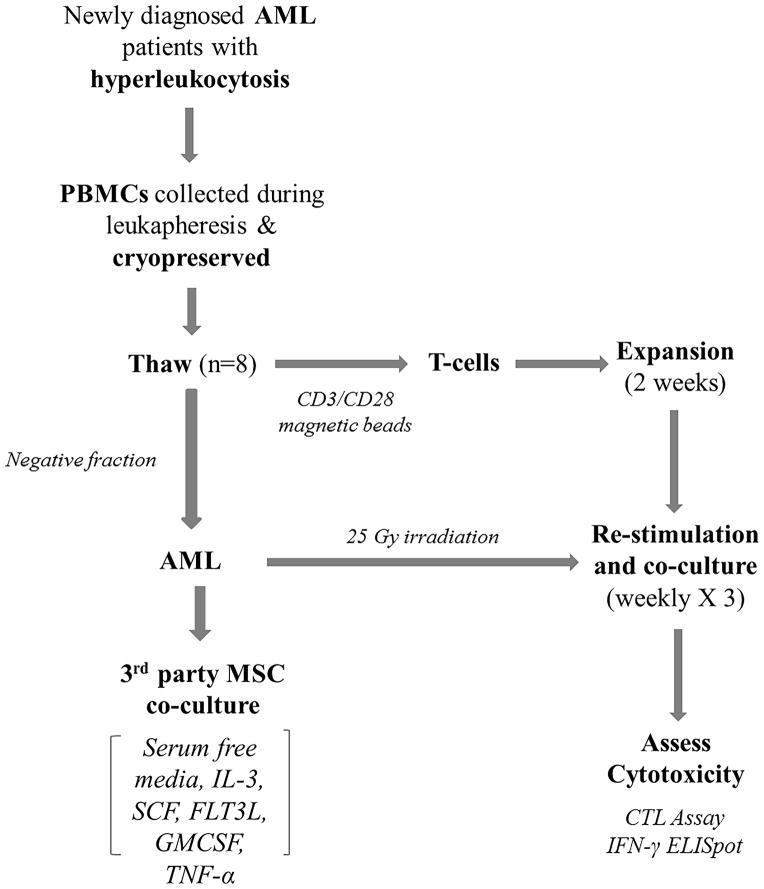
Figure 1. Study schema demonstrating the process of reactive T cell generation.
PBMCs were obtained from patients with AML presenting with hyperleukocytosis by leukapheresis. T cells were expanded from thawed samples (n=8) with anti-CD3/CD28 beads in culture media as we previously reported 22. The “negative fraction” obtained after T cell extraction contained primarily AML blasts, was cultured on a layer of bone marrow derived MSCs in serum-free media and cytokine cocktail. After 2 weeks of T cell expansion, autologous AML blasts were used for co-cultures of expanded T cells for 3 weeks. At the end of 3 weeks, cytotoxicity was assessed using EuTDA assay and IFN-γ ELISpot assay. T cell activation and inhibitory molecules were analyzed before and after co-cultures using multi-color flow cytometry and q-PCR gene expression. Tregs were analyzed using flow cytometry. q-PCR gene expression studies were used to assess MDSC-like phenotype in AML blasts.
Primary AML blasts culture
The “negative fraction” obtained after T cell purification, which constituted primarily of AML blasts, was cultured over a supporting layer of mesenchymal stromal cells (MSCs). The MSCs were isolated from a bone marrow aspirate of a patient with AML in remission and cultured as reported 24,25. Once 80% or greater confluence was achieved, as detected by inverted phase microscopy, MSCs were trypsinized, washed and cultured in 24-well flat bottom plates at a concentration of 5 × 104 cells/well in Dulbecco’s Modified Eagle Medium (DMEM) media containing 20% heat-inactivated pooled human AB serum. Culture medium was changed every 3–4 days until >75–80% confluence was attained, at which time AML blasts were co-cultured over the MSC layer in clinical grade StemSpan™ serum-free expansion media (STEMCELL Technologies) at a concentration of 1 × 106 cells/ml and a cytokine cocktail of 100 ng/ml each of recombinant human IL-3, SCF and FLT3L and 100 U/ml each of GMCSF and IL-4 (all from BioLegend Inc., San Diego, CA) 26–28. The medium and cytokines were replenished every 3–4 days. Two days prior to the planned co-cultures of T cells with autologous AML blasts, 500U/ml of IFN-γ (R & D Systems) was added to the AML culture to enhance their self-antigen presenting potential to T cells. With this protocol, which was applied for the first three samples, we observed gradually declining viability of AML cells for UPIN # 2 and UPIN# 3 in culture. Therefore, we modified the protocol for the remaining 5 samples, where cryopreserved PBMCs were thawed and T cells were purified as described above, but the negative fraction (predominantly AML blasts) was cryopreserved immediately. Two days prior to the planned co-culture with T cells, the cryopreserved AML rich negative fractions were thawed and cultured in serum-free media with MSCs and the same cytokine support and IFN-γ as described above.
In vitro co-cultures and generation of AML-specific reactive T cells
On day 14 (± 2 days) of T cell expansion, the anti-CD3/CD28 beads were removed using magnetic particle concentrator (Dynal MPC-2; Dynal/Invitrogen) and the cells were washed. Simultaneously, cultured autologous AML blasts were also removed from culture, washed and γ-irradiated with single fraction of 26.25Gy over the course of 269 seconds (Compagnie Oris Industrie S.A., France). AML blasts were then co-cultured with expanded autologous T cells at a varying effector: target ratio of 1:1 to 5:1 depending upon the number of AML blasts available. Cultures were maintained in X-Vivo 15 medium (Lonza) supplemented with 5% FCS (Life Technologies, Invitrogen), 2mM L-glutamine, 100 U/mL each of penicillin, streptomycin and amphotericin (Sigma-Aldrich), 100ng/ml of purified anti-human CD28 monoclonal antibody (BD Biosciences) and 5ng/ml IL-15 (R&D Systems). Cultures were re-stimulated with autologous γ-irradiated AML blasts weekly for a total duration of three weeks, at the end of which functional, phenotypic and gene expression assessment was performed.
Protocol amendment
We observed a progressive decline in the number of T cells during the first two weeks of co-cultures in UPIN # 4. Therefore, low dose of IL-2 (10 U/ml) was added during the beginning of the last week of co-cultures in UPIN # 4, which rescued the cells. The protocol was thereafter modified to include low-dose IL-2 (10 U/ml) and IL-12 (10 ng/ml) starting from the first week of co-cultures for the rest of the experiments (UPIN # 5–8).
Assessment of reactive T cell generation
Enzyme-Linked ImmunoSpot (ELISpot) assay
IFN-γ ELISpot assay was performed at the end of third week of co-culture to quantitate the frequency of AML-specific IFN-γ secreting reactive T cells 29. Patients’ own AML blasts served as ‘relevant’ stimulators (S) while U937 cell line served as irrelevant, non-specific targets.
Absence of targets in the culture medium was used as negative control and CD3/CD28 beads were added for positive control. Fifty thousand responder (R) T cells were tested at a R:S ratio of 1:1 to 5:1, depending upon the number of AML blasts available. After 24 hours of incubation, the plates were developed and dried overnight in dark at room temperature. The plates were read and analyzed for the number of spot forming cells (SFCs) in CTL ImmunoSpot® plate reader and CTL ImunoSpot® Software version 5.0.3. (C.T.L., Shaker Heights, OH).
DELFIA® EuTDA cytotoxicity assay
In conjunction with the ELISpot assay, non-radioactive Europium TDA (EuTDA) assay was employed to assess specific cytotoxicity 22. At the end of 3rd week of co-cultures, effectors (viable cells) were harvested to assess cytotoxicity against fresh cultured autologous, wild type AML blasts (without IFN-γ exposure) and two “irrelevant” targets – U937 (myeloid leukemia cell line) loaded with DELFIA® BATDA reagent (Perkin-Elmer) at effector: target ratios of 40:1, 20:1, 10:1 and 5:1 in triplicates, as described previously 22. Cytotoxicity was measured by DELFIA® EuTDA cytotoxicity assay on a Victor 2 microplate reader (Perkin-Elmer) as described 22 and as suggested by the manufacturer 30. Specific release percentage was calculated as [experimental release (counts) - spontaneous release (counts)]/[maximum release (counts) - spontaneous release (counts)] X 100. Spontaneous release percentage was calculated as [spontaneous release (counts) - background (counts)]/[maximum release (counts) - background (counts)] X 100. Of note, standardization of the CTL assay for primary AML blasts was difficult despite testing different loading times, incubation temperatures and washing techniques as suggested by the manufacturer. This was attributed to the inter-sample heterogeneity and relative frailty of previously cryopreserved AML blasts, which led to high background and spontaneous release in a majority of samples.
Immunophenotypic characterization of T cell
We performed eight-color flow cytometric analysis on a FACSCanto II® multicolor flow cytometer (BD Biosciences) to characterize various co-stimulatory and co-inhibitory receptors on T cells (CD3, CD8, CD137, HLA-DR, CTLA-4, PD-1, NKG2D, TCR-γδ) at the end of third week of co-culture. In addition, surface and intracellular immunophenotypic characterization of Treg cells (CD4+CD25brightFoxP3+) was performed before and after co-culture, as previously described 20.
Gene expression profiling of T cells and AML blasts
Genomic DNA was extracted from purified T cells and AML blasts using Trizol method (Life technologies) 31–33. Purification of RNA and real-time quantitative reverse transcriptase Polymerase Chain Reaction analysis (RT-qPCR) to generate cDNA was performed on T cells before and after repeated re-stimulation with autologous AML blasts to detect changes in their genetic profile following in-vitro stimulation period. Similarly, RNA purification and RT-qPCR analysis was performed on AML blasts to detect expression profile of genes associated with MDSC. qPCR analyses were performed on StepOnePlus™ system (Life Technologies) using the Power SYBR® Green RT-PCR reagents and the Power SYBR® Green PCR master mix (Life Technologies) following manufacturer’s recommendations 31. The CT values obtained from the PCR machine were imported into Microsoft Excel 2010 and the analysis of gene expression was done using the 2−ΔΔCt method 34.
A panel of genes analyzed for T cells was chosen based on their known or potential role in T cell co-stimulation or co-inhibition 35. Genes with previously defined role in MDSC generation 36 were chosen for expression analysis on AML blasts. The primers were designed using the NCBI primer blast 15 and target specificity of each primer was confirmed by activating search only for primer pairs specific to the intended PCR template in Homo sapiens using the Refseq mRNA database. This feature enables specificity cross-check not only for forward-reverse primer pairs, but also for forward-forward and reverse-reverse primer pairs 15. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as the house-keeping gene for AML blasts, while five housekeeping genes were selected for T cell gene analysis, including GAPDH, IPO8 (Importin 8), RPL13A (Ribosomal Protein L13a), SDHA (Succinate dehydrogenase complex, subunit A) and TBP (TATA box binding protein) 37. For internal standardization of gene expression, the BestKeeperC© software 38 was used to determine the most reliable and stable housekeeping gene(s) using repeated pair-wise correlation analysis. A “BestKeeper index©” was then calculated based on the geometric average of the most stable housekeeping genes (RPL13A and TBP), which served as the housekeeper for analysis 38.
Statistical Analysis
One-way ANOVA with Bonferroni correction for multiple testing was performed (a) to compare the differences in ELISpot SFCs against AML and U-937 cells and the controls, and (b) to compare the percentage specific release against AML and U-937 cells observed in the CTL assays at various effector: target ratios. Paired sample student t-test with unequal variances was performed to compare the differences in (a) proportion of Treg cells and (b) T cell gene expression occurring pre- and post-co-cultures in reactive and non-reactive T cell samples. Student t-tests with unequal variances were performed to compare the differences in (a) proportion of Treg cells and T cell co-signaling receptors (determined by the FACS analysis) between reactive and non-reactive T cell samples, and (b) AML gene expression differences between resistant and the non-resistant samples. All analyses were two-sided and an alpha value less than 5% was selected as a threshold for statistical significance.
RESULTS
Autologous T cells can be expanded ex vivo from AML patients during hyperleukocytosis
We tested 8 consecutive cryopreserved PBMC samples which were obtained during leukapheresis of newly diagnosed patients with AML presenting with hyperleukocytosis. The immunophenotypic, cytogenetic and molecular characteristics of these AML samples are depicted in table 1. T cell purification and expansion was performed using CD3/CD28 magnetic beads, IL-2 and IL-7 for two weeks (± 2 days). Despite low percentage of T cells extracted from thawed PBMCs (mean CD3+ cells 2.04%, range 0.4 – 7.8%), we were able to expand them in all samples. However, the expansion was inferior as compared to the expansion of healthy T cells seen in our previous experiments 20,22. As shown in figure 2 and table 2, an average 4.98 (range 1.1 – 5.6) fold expansion was achieved at the end of 14-day expansion and a mean (range) fold-expansion of 4.34 (0.9–6.9), 4.8 (0.7–11.6) and 2.69 (1.03 – 3.9) was attained at the end of each successive week respectively during three weeks of co-cultures. Nevertheless, with 5 weeks combined, this translated to median incremental T cell expansion of 279-fold over the starting numbers. There was no effect of IL-12 on T cell expansion. Similarly, the variable effector: target ratio used during co-culture phase did not impact the degree of T cell expansion achieved (data not shown).
Table 1.
Immunophenotype and cytogenetic/molecular characteristics of AML samples
| AML1 | AML2 | AML3 | AML4 | AML5 | AML6 | AML7 | AML8 | |
|---|---|---|---|---|---|---|---|---|
| PB Blasts | 19% | 74% | 18% | 92% | 25% | * | 83% | 79% |
| BM Blasts | 34% | 76% | 34% | 90% | 43% | * | 79% | 65% |
| CD34 | + | − | + | − | − | − | + | − |
| CD33 | + | + | + | + | + | + | + | + |
| CD13 | + | + | + | Partial | + | + | + | |
| CD117 | + | − | + | Partial | + | − | + | − |
| CD45 | Dim + | Dim + | Dim + | Dim + | Dim + | + | Dim + | Dim + |
| HLA-DR | + | + | + | + | + | + | + | |
| Cytogenetics/Molecular | Complex | FLT3 +ve | Complex | t(9:11), Complex | NPM1 +ve | Diploid | inv(16) | Complex |
missing
Figure 2. Mean (SEM) fold expansion of autologous T cells.
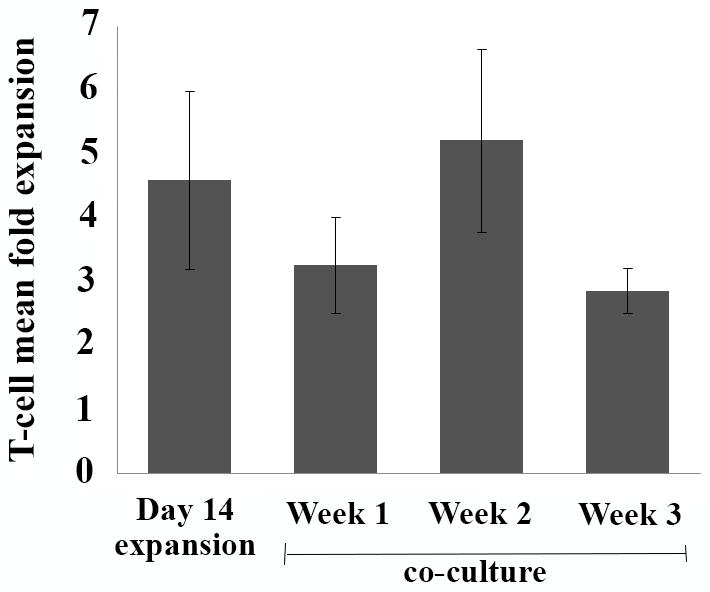
Figure depicts mean (SEM) fold expansion at the end of 14-day expansion period and with each subsequent week of co-cultures.
Table 2.
Total T cell counts (x 106) in eight samples starting from day 1 (post purification) till the end of third week of co-culture with autologous AML blasts.
| Post purification | Day 14 of expansion | Start of co-culture week 1* | End of week 1 of co-culture | Start of co-culture week 2* | End of week 2 of co-culture | Start of co-culture week 3* | End of week 3 of co-culture | |
|---|---|---|---|---|---|---|---|---|
| UPIN 1 | 0.9 | 5 | 2 | 4 | 4 | 44 | 4 | 16 |
| UPIN 2 | 1 | 1.12 | 0.9 | 1.8 | 1.6 | 3.3 | 3 | 10 |
| UPIN 3 | 2.1 | 8.8 | 1.5 | 3.75 | 1 | 11.6 | 3.2 | 7 |
| UPIN 4 | 2.4 | 12.28 | 1 | 0.9 | 0.9 | 0.7 | 0.7 | 2.15 |
| UPIN 5 | 3.7 | 42 | 2.5 | 5.6 | 5.6 | 32.58 | 25 | 70.74 |
| UPIN 6 | 5.2 | 15 | 3.5 | 12.25 | 5 | 19.9 | 7.5 | 29.9 |
| UPIN 7 | 1.34 | 5.93 | 5 | 34.6 | 7.5 | 31 | 10 | 10.26 |
| UPIN 8 | 2.5 | 5.2 | 5 | 30 | 8 | 18.4 | 8 | 19 |
| Mean | 2.3925 | 11.91625 | 2.675 | 11.6125 | 4.2 | 20.185 | 7.675 | 20.63125 |
| Median | 2.25 | 7.365 | 2.25 | 4.8 | 4.5 | 19.15 | 5.75 | 13.13 |
Only a proportion of T cells were used during each week of co-culture, depending upon the number of AML blasts available.
ELISpot assay identifies primary AML- reactive T cells
Ex vivo expanded T cells without antigenic exposure were stimulated for three consecutive weeks with autologous IFN-γ treated AML blasts while receiving co-stimulatory and survival signals from CD28 agonist antibody, IL-15 and low dose (10Uml) IL-2. IFN-γ ELISpot assay was performed at the end of third week of stimulation. Unmodified wild type AML blasts, obtained from the same blood draw as the T cells, were used as specific targets, while U937, a 3rd party AML cell line, served as third party targets to assess for non-specific reactivity.
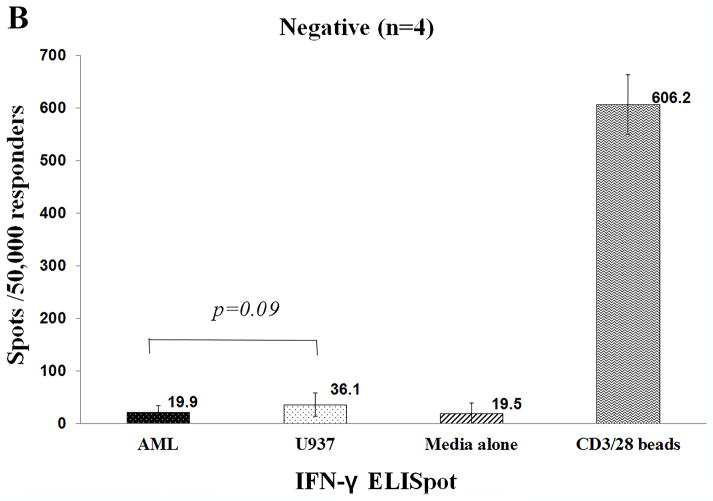
IFN-γ ELISpot assay determined the number of IFN-γ SFCs against AML blasts as compared with irrelevant targets (U937). T cells from 4 patient samples produced significantly higher number of SFCs against AML blasts as compared to the U-937 cells (mean 233 vs. 60 spots/50,000 responders, p=0.006) [Figure 3a]. In contrast, as shown in figure 3b, IFN-γ production was similar against AML blasts, U-937 and negative controls in the remaining four patients. The number of SFCs against AML blasts were significantly higher in the ‘positive’ samples (n=4) as compared to that in the ‘negative’ samples (n=4), [mean 233 vs. 19.9 spots/50,000 responders, p-value 0.0013]. T cells from UPIN#7 also exhibited non-specific activity against 3rd party AML (U937) (mean 211 spots/50,000 responders) cells, although the relevant response against AML blasts was significantly higher (mean 426 spots/50,000 responders), p-value 0.0046. In contrast to the CTL assay where UPIN#1 demonstrated specific cytotoxicity against AML, IFN-γ SFCs were not detectable. Supplemental table 1 depicts the number of SFCs observed in individual experiments against AML, U-937, negative control and positive control.
Figure 3. IFN-γ ELISpot assay performed at the end of third week of co-cultures.
Figure shows the number of IFN-γ spot forming cells/50,000 responders (>95% CD3+ T cells) against autologous AML blasts, irrelevant target (U937), negative control (media alone) and positive control (anti-CD3/CD28 beads). Figure 3A depicts the 4 AML-reactive samples and figure 3B presents the cohort of 4 AML non-reactive samples.
Cytotoxicity of primary AML- reactive T cells was difficult to ascertain
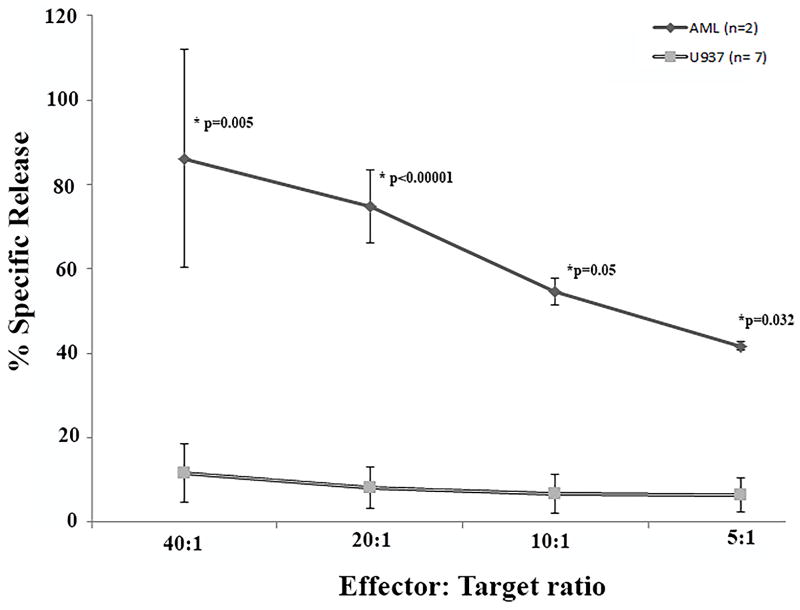
In parallel with the IFN-γ ELISpot assay, we attempted EuDTA cytotoxicity assay. As mentioned above, standardization of patient derived AML blasts was challenging. As a result, only 2/8 samples (UPIN # 1 and 5) yielded results which were deemed evaluable – spontaneous release less than 30%. Significantly, both CTL cultures from these samples demonstrated significantly higher specific cytotoxicity against AML (mean 86%, 75%, 55% and 42% at 40:1, 20:1, 10:1 and 5:1 E:T ratios respectively) but not against U937 cells (Figure 4).
Figure 4. The EuTDA cytotoxicity assay performed at the end of third week of co-cultures.
Ex vivo expanded T cells were re-stimulated with autologous irradiated AML blasts weekly X 3. At the end of week 3, cytotoxicity was assessed using EuTDA assay using labeled autologous AML blasts and irrelevant target (U937 cell line), as described. The X-axis demonstrates mean (SEM) percentage specific release against autologous AML blasts (solid line) and non-specific targets – U937 (grey line) at effector: target ratios of 40:1, 20:1, 10:1 and 5:1 (Y-axis). P-values compare specific release against AML vs U-937.
Combining CTL assay and ELISpot assay results
Based on the EuTDA cytotoxicity assay (UPIN# 1 and 5) and the IFN-γ ELISpot (UPIN# 3, 4, 5 and 7), overall five samples were defined permissive for AML “reactive” T cells while the other 3 cultures yielded “non-reactive” T cells. Three of the five reactive T cell samples were generated from UPIN #1–4 (original protocol) and two from UPIN #5–8 (amended protocol) [Table 5]. Importantly, we did not observe non-specific T cell reactivity in 7/8 samples. Although UPIN # 7 demonstrated some reactivity against the U937 myeloid leukemia cell line in addition to primary AML blasts, yet the numbers of SFCs detected by IFN-γ ELISpot assay were significantly higher against AML blasts than U937 cells, suggesting higher specificity against the relevant target. Taken together these results suggest favorable specificity of the expanded T cells for potential adoptive autologous T cell immunotherapy in patients with AML.
Table 5.
Combined results of ELISpot and CTL assays in 8 samples.
| Cytokines used | IFN-γ ELISpot | EuTDA CTL assay | |
|---|---|---|---|
| UPIN 1 | IL-2, IL-7 (expansion) IL-15, CD28* (co-culture) |
Negative | Positive |
| UPIN 2 | IL-2, IL-7 (expansion) IL-15, CD28* (co-culture) |
Negative | NE |
| UPIN 3 | IL-2, IL-7 (expansion) IL-15, CD28* (co-culture) |
Positive | NE |
| UPIN 4 | IL-2, IL-7 (expansion) IL-15, CD28* (co-culture). IL-2 added during week 3 of co-culture |
Positive | NE |
| UPIN 5 | IL-2, IL-7 (expansion) IL-15, CD28*, IL-2 and IL-12 (co-culture). |
Positive | Positive |
| UPIN 6 | IL-2, IL-7 (expansion) IL-15, CD28*, IL-2 and IL-12 (co-culture). |
Negative | NE |
| UPIN 7 | IL-2, IL-7 (expansion) IL-15, CD28*, IL-2 and IL-12 (co-culture). |
Positive | NE |
| UPIN 8 | IL-2, IL-7 (expansion) IL-15, CD28*, IL-2 and IL-12 (co-culture). |
Negative | NE |
purified anti-human CD28 monoclonal antibody;
NE, non-evaluable
The proportions of Treg cells differ remarkably between reactive and non-reactive T cell samples
Surface and intracellular staining was performed to detect the proportion of T cells exhibiting Treg phenotype (CD4+CD25brightFoxP3+) among total CD4+ population before and after co-cultures with autologous AML blasts. The goal was (a) to detect changes in proportion of Treg cells with in vitro co-cultures (post – vs. pre-‘co-cultures’) and (b) to compare the proportion of Treg cells at the end of co-cultures in reactive versus non-reactive T cell samples.
After 2 weeks of stimulation with AML blasts, we observed a significant decline in the proportion of Treg cells amongst ‘reactive T-cell’ samples as compared to the pre-culture level (56% to 24%, p= 0.046, n=4) [Figure 5]; while in the one ‘non-reactive’ T-cell sample, the proportion increased from 66.7% to 89% from pre to post-‘co-culture’. Regrettably, the limited number of T cells available for the other ‘non-reactive’ T cell samples precluded assessment of their pre-co-culture Treg cell numbers. Notably, the mean proportion of Treg cells at the end of co-culture appeared higher in the ‘non-reactive’ T cell samples (n=3) as compared to the ‘reactive’ T cell samples (n=5), although the results did not attain statistical significance [47.3% (range 22.1 – 89%) vs. 22.4% (range 0 – 93.7%), p=0.18].
Figure 5.
Percentage of CD4+CD25brightFoxP3+Tregs (% of total CD4 population) before and after co-cultures in AML-reactive cultures (n=4).
Significant differences in T cell gene expression exist between ‘reactive’ and ‘non-reactive’ T cell cultures
Gene expression of T cells was examined using qPCR analyses before and after co-culture with autologous AML with a goal to identify signature changes in the expression of various T cell co-signaling molecules in both ‘reactive’ and ‘non-reactive’ T cell cultures.
T cells expanded over 3 weeks with AML stimulation in the ‘reactive’ T cell cohort (n=5) displayed significant up-regulation of several proteins known to be associated with T cell activation, including 4-1BB, HVEM, LIGHT, PRKC-α and PRKC-θ compared to the pre-culture levels. Significantly, these markers of T cell activation were not up-regulated in the ‘non-reactive’ co-cultures (n=3). In fact, activation markers such as PRKC-α, lymphotoxin-α and lymphotoxin-β were down-regulated, while 2-B4, which is a potent inhibitory marker, was up-regulated in non-reactive T cell samples (n=3). No other differences in noted in the expression of co-inhibitory molecules CTLA4, PD1, BTLA, LAG3, TIGIT or CD160 in either ‘reactive’ or ‘non-reactive’ T cells. [Table 3]
Table 3.
Gene expression of T cell co-signaling receptors in reactive and non-reactive T cells post co-culture with autologous AML blasts compared to pre co-culture expression.
| ‘Reactive T cells’ (n=5) | ‘Non-reactive T cells’ (n=3) | |||||
|---|---|---|---|---|---|---|
| Gene | ΔΔ Ct (Post - Pre) (mean, SEM) | Fold change (mean, SEM) | P-value | ΔΔ Ct (Post - Pre) (mean, SEM) | Fold change (mean, SEM) | P-value |
| 4-1BB | −3.17 (0.76) | 14 (7.7) | 0.025 | 1.98 (1.04) | 0.39 (0.22) | 0.19 |
| HVEM | −2.43 (0.61) | 7.3 (3.7) | 0.028 | 0.14 (1.65) | 1.57 (1.28) | 0.95 |
| LIGHT | −3.62 (0.73) | 17.3 (7.3) | 0.016 | 1.78 (1.84) | 1.1 (0.98) | 0.44 |
| PRKC-α | −2.03 (0.47) | 4.6 (1.1) | 0.023 | 1.89 (0.36) | 0.29 (0.08) | 0.034 |
| PRKC-θ | −3.36 (0.59) | 13.7 (6.7) | 0.01 | 0.25 (0.59) | 0.99 (0.41) | 0.71 |
| LAIR1 | −3.81 (0.42) | 16.2 (5.6) | 0.003 | −1.35 (2.20) | 17.15 (16.5) | 0.60 |
| PP2A | −2.40 (0.57) | 6.7 (2.6) | 0.025 | 0.49 (1.57) | 1.89 (1.52) | 0.79 |
| LTA-α | −1.18 (0.78) | 3.61 (2.11) | 0.23 | 2.69 (0.18) | 0.16 (0.02) | 0.043 |
| LTA-β | −0.93 (0.63) | 2.49 (0.99) | 0.24 | 2.24 (0.47) | 0.23 (0.08) | 0.042 |
| IL-17 | −2.66 | 6.31 | - | 5.37 (1.23) | 0.04 (0.02) | 0.048 |
| 2B4 | −1.53 (1.14) | 4.98 (1.82) | 0.27 | −3.48 (0.11) | 11.2 (0.9) | 0.02 |
| CTLA4 | 1.58 (1.68) | 1.22 (1.11) | 0.45 | 1.88 (0.6) | 0.31 (0.1) | 0.09 |
| PD1 | −1.87 (2.54) | 22.37 (19.59) | 0.54 | 2.85 (3.76) | 0.94 (0.93) | 1 |
| BTLA | −0.62 (1.71) | 12.65 (12.1) | 0.74 | −0.28 (2.71) | 4.05 (3.87) | 0.43 |
| LAG3 | 2.12 (2.26) | 1.27 (0.88) | 0.42 | −0.52 (1.2) | 2.25 (0.98) | 0.7 |
| CD160 | 0.48 (1.16) | 1.51 (0.76) | 0.71 | 0.26 (0.18) | 0.84 (0.11) | 0.39 |
| TIGIT | 0.25 (0.85) | 1.20 (0.41) | 0.78 | 3.52 (1.12) | 0.13 (0.06) | 0.09 |
Table showing relative fold change (mean, SEM) in gene expression as assessed by RT q-PCR of various T cell co-signaling molecules after co-cultures with autologous AML blasts compared to their pre-co-cultures expression. The left panel shows reactive T cell samples (n=5); right panel shows same genes in non-reactive T cell samples (n=3).
AML blasts resistant to autologous T cell reactivity demonstrated up-regulation of genes associated with T cell-inhibitory myeloid derived suppressor cell (MDSC) phenotype
In parallel with the T cell studies, we also analyzed primary AML blasts for gene expression associated with MDSC phenotype (n=8) testing the hypothesis that ‘resistant’ AML samples (defined as samples against which reactive T cell generation was unsuccessful) could possess an MDSC-like profile compared with the ‘non-resistant’ AML samples. Indeed, significant up-regulation of down-stream markers of MDSC generation was identifiable in the ‘resistant’ (n=3) as compared to the ‘non-resistant’ (n=5) samples. These included JAK-1, JAK-2, JAK-3, S100A8, S100A9 and c-myc. No differences were noted in the expression of Arg1, IDO2, iNOS, ROS, TGFβ or PDL1 [Table 4].
Table 4.
Gene expression results in AML – Resistant vs. Non-resistant AML samples
| Gene | ΔΔ Ct (mean, SEM) | 95% CI | P-value | Relative fold change | |
|---|---|---|---|---|---|
| JAK1 | −4.63 (1.98) | −9.48 | 0.21 | 0.0579 | 24.83 |
| JAK2 | −5.38 (0.94) | −7.67 | −3.08 | 0.0012 | 41.52 |
| JAK3 | −5.90 (2.17) | −12.81 | 1.01 | 0.0726 | 59.77 |
| S100A8 | −7.16 (2.66) | −14.01 | −0.32 | 0.0432 | 143.27 |
| S100A9 | −8.31 (2.75) | −15.04 | −1.59 | 0.0233 | 318.37 |
| c-myc | −2.78 (0.59) | −4.24 | −1.33 | 0.0034 | 6.89 |
| iNOS | −2.02 (3.7) | −12.23 | 8.19 | 0.6124 | 4.05 |
| ROS | −5.92 (−) | NE | NE | NE | 60.75 |
| Arg1 | 2.81 (4.54) | −16.74 | 22.35 | 0.6 | 2.4 |
| IDO2 | −0.29 (2.79) | −7.13 | 6.55 | 0.92 | 1.22 |
| TGFβ | −7.03 (3.91) | −17.9 | 3.83 | 0.15 | 130.9 |
| PDL1 | 1.43 (3.89) | −9.38 | 12.24 | 0.73 | 0.37 |
Table showing expression of various genes associated with downstream markers of generation, activation and/or proliferation of myeloid derived suppressor cells (MDSCs) in resistant AML (n=3) compared to non-resistant samples (n=5).
DISCUSSION
In these studies we demonstrate the proof of principle for the feasibility to generate autologous AML- reactive T cells in vitro from the peripheral blood of more than half of newly diagnosed AML patients presenting with hyperleukocytosis. To the best of our knowledge, this is the first promising strategy in untreated, freshly diagnosed AML patients with hyperleukocytosis which is well known poor prognostic feature 39–41. This therapeutically novel scenario was attainable by specific immunomodulatory steps. First, AML cells were rendered presumably more immunogenic following IFN-γ pre-treatment. Heightened recognition of autologous AML in T cells was supported with defined but limited co-stimulatory and survival signals (agonist anti-CD28, IL-15) in the culture medium to elicit AML-reactive autologous T cell responses. If we take together the independent results of EuTDA CTL and IFN-γ ELISpot assays, we were able to induce AML-specific autologous T cell responses from 5 out of 8 cryopreserved patient samples. These findings are particularly relevant as biological foundation for future immunotherapy developments because T cells of AML patients are reported to be exhausted and/or anergic 42,43. Our ability to create reactive T cells in some patients suggests that the noted T cell dysfunction can be reversed in a significant fraction of AML patients by creating immune-stimulatory culture conditions.
The expansion of T cells in our samples was inferior as compared to that attained in our previous experiments using healthy CBT cells against immunogenic and immortalized B cell lymphoma 20–22. Nonetheless, we were able to achieve an average expansion of greater than 250-fold by the end of third week of cultures. Furthermore, the presented culture conditions favored highly specific autologous wild type AML-specific responses in all but one sample, where there was detectable IFN-γ-SFC response against myeloid leukemia cell line (U937) in parallel with an even more robust reactivity against the autologous AML blasts. We used clinical grade serum-free media to culture AML blasts to reduce the risk of generating non-specific immune responses against human serum proteins. This could be important to minimize the risk of auto-immune responses with autologous CTL immunotherapy. Importantly, for the sake of simplicity and generalizability we used whole leukemia cell to stimulate autologous T cells rather than targeting a specific TAA. T cells will recognize presumably a larger range of AML associated epitopes and could offer a more broad and widespread applicability in future developments. Similar generalized approach has previously been shown to be promising in the clinical setting 44 and has several unique advantages compared to the use of TAAs 45.
Beyond exploring the feasibility of autologous reactive T cell generation, in parallel experiments we investigated potential mechanisms which may impair and prevent an effective autologous T cell immune response. We found that the proportion of Treg cells was very high at start, but decreased after co-cultures in the reactive T cell samples. Owing to the limited number of T cells available for analysis in non-reactive T cell samples, we were unable to perform pre- versus post-culture analysis in all samples. Therefore, no firm conclusion could be drawn for the non-reactive T cell samples. We also found significant differences in the expression of several co-signaling molecules in T cells contrasting reactive and non-reactive T cell cultures. Specifically, many of the molecules associated with T cell activation (4-1BB, 46–48 HVEM, LIGHT, 49–52 PRKC-α and PRKC-θ 53,54) were up-regulated in ‘reactive’ T cells after 3 week of culture, while the ‘non-reactive’ T cells exhibited down-regulation of many genes associated with T cell activation (PRKC-α, LTA-α and LTA-β 52,55), paired with up-regulation of 2B4, a potent co-inhibitory molecule associated with exhausted T cells 56–58. The expression of co-inhibitory molecules - CTLA4, PD1, BTLA, LAG3, TIGIT and CD160, and the expression of Th1-associated cytokines - IL-2 and IFN-γ were similar among ‘reactive’ and ‘non-reactive’ T cell. The differential gene expression observed in reactive versus non-reactive T cell samples suggests that several T cell activation pathways are either impeded or the inhibitory pathways are activated by certain mechanisms in resistant AML samples.
These T cell alterations may reflect the influence of immunosuppressive MDSC-like AML blasts. Compared to AML blasts that were effectively recognized by reactive T cells, JAK1, JAK2, JAK3, S100A8 and S100A9 were all up-regulated in ‘resistant AML’ samples; although no differences in noted in the expression of PDL1, Arg1, IDO2, iNOS or ROS between ‘resistant’ and ‘non-resistant’ AML. Regardless of the factors that lead to the generation of MDSCs and their mechanism of suppression, the final common pathway in their activation and expansion is reported to involve the JAK-STAT protein family members, which in turn lead to nuclear expression of c-myc and up-regulation of S100A8 and S100A9 59–63.
Despite several novelties described above, there are limitations of our study. First, as mentioned above, we encountered obstacles to perform CTL assay on all primary AML blast samples. The EuTDA CTL assay is critically dependent on membrane integrity and labelling efficacy of the targets. Not only it is challenging to maintain primary AML blasts in cultures, the resultant CTL assay might be uninterpretable due to the poor labeling and subsequent porous nature of ‘friable’ AML blasts. Recognizing this, we performed IFN-γ ELISpot in parallel assays, where the targets, primary patient blasts, need no manipulation/labeling 64. In future studies, ancillary tests such as assessment of CD107 using flow cytometry could be helpful. The addition of IL-12 in UPIN# 5–8 neither altered expression of Th1-associated cytokines, nor had an impact on overall T cell responses, suggesting that IL-12 may not provide critical signals and could be omitted in future studies. Our study lacks additional functional and immunophenotypic characterization of MDSC-like AML blasts, which is of great interest and should be evaluated in future studies. While we were able to demonstrate minimal or no cross-reactivity towards 3rd party AML cell line, due to the small volume of cryopreserved samples and significantly reduced presence of normal hemaotopietic cells, we did not have autologous ‘normal’ cells as controls to demonstrate specificity of T cells against AML. Nevertheless, these T cells did not act as promiscuous cytokine induced killer cells. Lastly, we recognize the limitation of small sample size. To improve upon these limitations, we are pursuing further refinements in manufacturing to build on the proof-of-principle of the experiments described above. In patients with high numbers of circulating AML blasts requiring leukapheresis, collection of fresh PBMC from leukapheresed product (rather than cryopreserved samples) should be considered. On the other hand, in AML patients who do not require leukapheresis, collection of AML blasts from bone marrow aspiration and T cells from peripheral blood could yield optimal cell numbers. The impact of checkpoint inhibitors will be experimentally tested to determine if they enhance or not the numerical and/or functional aspects of T leukemia reactive T cells in clinical grade product development and to test if these agents compromise target specificity or not.
To the best of our knowledge, no study has reported on successful generation of autologous reactive T cells specific against AML from untreated patients with hyperleukocytosis. This is a state which portents particularly poor prognosis 39–41. Our study provides a foundation for future studies which should include larger number of samples and incorporate means to overcome shortcomings of this study.
CONCLUSION
In sum, we demonstrate the feasibility to expand primary patient derived AML-specific autologous reactive T cells from peripheral blood in 5/8 patients during their initial presentation. Despite the known “dysfunctional state” of T cells in AML patients, we were able to expand T cells from all eight patients. The proportion of Treg cells decreased significantly after co-cultures in the ‘reactive’ T cell samples. Several genes associated with T cell activation, such as 4-1BB, PRKC-α, PRKC- θ, HVEM and LIGHT, were up-regulated in reactive T cells. On the other hand, ‘non-reactive’ T cells had down-regulation of T cell activation molecules including PRKC-α, LTA-α and LTA-β in parallel with up-regulation of an inhibitory molecule, 2B4. Other co-inhibitory molecules, including CTLA4, PD1, BTLA, LAG3 and CD160 had similar expression among both ‘reactive’ and ‘non-reactive’ T cell, suggesting that inhibition of T cells may be directly mediated by AML cells. Indeed, ‘resistant AML’ samples displayed gene expression that closely matches what has been reported with immunosuppressive myeloid derived suppressor cells 36. Our results should spur further investigation to extend these findings. This biological feasibility study could provide the foundation to further technical developments to generate personalized leukemia-specific CTLs for patients without overtly complex maneuvers. Just as importantly, if confirmed in larger datasets the differences noted between AML blasts and the contrast between ‘reactive’ and the ‘non-reactive’ T cells could become valuable tools towards building risk models for immunotherapy.
Supplementary Material
Acknowledgments
Part of this work was supported by a grant from the National Cancer Institute to P. Szabolcs (1R01CA132110).
Abbreviations
- AML
Acute myeloid leukemia
- Arg1
Arginase 1
- BTLA
B- and T-lymphocyte attenuator
- CBT
Cord blood T cells
- CTL
Cytotoxic T cells
- CTLA4
Cytotoxic T-lymphocyte-associated protein 4
- FACS
Fluorescence-Activated Cell Sorting
- FLT3
Fms-like tyrosine kinase 3 ligand
- GAPDH
Glyceraldehyde 3-phosphate dehydrogenase
- GM-CSF
Granulocyte-macrophage colony-stimulating factor
- HVEM
Herpes virus entry mediator
- IDO2
Indoleamine 2,3-dioxygenase 2
- IFN
Interferon
- IL
Interleukin
- iNOS
Inducible nitric oxide synthase
- IPO8
Importin-8
- JAK
Janus kinase
- LAG3
Lymphocyte-activation gene 3
- LAIR-1
Leukocyte associated Ig-like receptor-1
- LIGHT
Lymphotoxin related inducible ligand that competes for glycoprotein D binding to HVEM on T cells
- MDSC
Myeloid derived suppressor cells
- MSC
Mesenchymal stromal cells
- PB
Peripheral blood
- PBMC
Peripheral blood mononuclear cells
- PD1
Programmed cell death protein 1
- PDL1
Programmed death-ligand 1
- PP2A
Serine/threonine protein phosphatase 2A
- PRKC
Protein Kinase C
- ROS
Reactive oxygen species
- RPL13A
Ribosomal Protein L13a
- RT-qPCR
Reverse Transcriptase quantitative Polymerase Chain Reaction
- SCF
Stem cell factor
- SDHA
Succinate dehydrogenase complex, subunit A
- STAT
Signal Transducer and Activator of Transcription
- TAA
tumor-associated antigen
- TBP
TATA box binding protein
- TCR
T cell receptor
- TGFb
Transforming growth factor beta 1
- TIGIT
T cell immunoreceptor with Ig and ITIM domains
- Treg
Regulatory T cells
Footnotes
Disclosures: The authors have no financial disclosures or conflicts of interest to declare.
References
- 1.Teague RM, Kline J. Immune evasion in acute myeloid leukemia: current concepts and future directions. J Immunother Cancer. 2013;1(13) doi: 10.1186/2051-1426-1-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Weber G, Gerdemann U, Caruana I, et al. Generation of multi-leukemia antigen-specific T cells to enhance the graft-versus-leukemia effect after allogeneic stem cell transplant. Leukemia. 2013;27(7):1538–1547. doi: 10.1038/leu.2013.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grupp SA, Kalos M, Barrett D, et al. Chimeric antigen receptor-modified T cells for acute lymphoid leukemia. N Engl J Med. 2013;368(16):1509–1518. doi: 10.1056/NEJMoa1215134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maude SL, Frey N, Shaw PA, et al. Chimeric antigen receptor T cells for sustained remissions in leukemia. N Engl J Med. 2014;371(16):1507–1517. doi: 10.1056/NEJMoa1407222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Amrolia PJ, Reid SD, Gao L, et al. Allorestricted cytotoxic T cells specific for human CD45 show potent antileukemic activity. Blood. 2003;101(3):1007–1014. doi: 10.1182/blood-2002-02-0525. [DOI] [PubMed] [Google Scholar]
- 6.Molldrem J, Dermime S, Parker K, et al. Targeted T-cell therapy for human leukemia: cytotoxic T lymphocytes specific for a peptide derived from proteinase 3 preferentially lyse human myeloid leukemia cells. Blood. 1996;88(7):2450–2457. [PubMed] [Google Scholar]
- 7.Molldrem JJ, Lee PP, Wang C, Champlin RE, Davis MM. A PR1-human leukocyte antigen-A2 tetramer can be used to isolate low-frequency cytotoxic T lymphocytes from healthy donors that selectively lyse chronic myelogenous leukemia. Cancer Res. 1999;59(11):2675–2681. [PubMed] [Google Scholar]
- 8.Ohminami H, Yasukawa M, Fujita S. HLA class I-restricted lysis of leukemia cells by a CD8(+) cytotoxic T-lymphocyte clone specific for WT1 peptide. Blood. 2000;95(1):286–293. [PubMed] [Google Scholar]
- 9.Oka Y, Elisseeva OA, Tsuboi A, et al. Human cytotoxic T-lymphocyte responses specific for peptides of the wild-type Wilms’ tumor gene (WT1) product. Immunogenetics. 2000;51(2):99–107. doi: 10.1007/s002510050018. [DOI] [PubMed] [Google Scholar]
- 10.Scheibenbogen C, Letsch A, Thiel E, et al. CD8 T-cell responses to Wilms tumor gene product WT1 and proteinase 3 in patients with acute myeloid leukemia. Blood. 2002;100(6):2132–2137. doi: 10.1182/blood-2002-01-0163. [DOI] [PubMed] [Google Scholar]
- 11.Harrison BD, Adams JA, Briggs M, Brereton ML, Yin JA. Stimulation of autologous proliferative and cytotoxic T-cell responses by “leukemic dendritic cells” derived from blast cells in acute myeloid leukemia. Blood. 2001;97(9):2764–2771. doi: 10.1182/blood.v97.9.2764. [DOI] [PubMed] [Google Scholar]
- 12.Ahmad SM, Larsen SK, Svane IM, Andersen MH. Harnessing PD-L1-specific cytotoxic T cells for anti-leukemia immunotherapy to defeat mechanisms of immune escape mediated by the PD-1 pathway. Leukemia. 2014;28(1):236–238. doi: 10.1038/leu.2013.261. [DOI] [PubMed] [Google Scholar]
- 13.Mutis T, Verdijk R, Schrama E, Esendam B, Brand A, Goulmy E. Feasibility of immunotherapy of relapsed leukemia with ex vivo-generated cytotoxic T lymphocytes specific for hematopoietic system-restricted minor histocompatibility antigens. Blood. 1999;93(7):2336–2341. [PubMed] [Google Scholar]
- 14.Greiner J, Dohner H, Schmitt M. Cancer vaccines for patients with acute myeloid leukemia--definition of leukemia-associated antigens and current clinical protocols targeting these antigens. Haematologica. 2006;91(12):1653–1661. [PubMed] [Google Scholar]
- 15.Anguille S, Van Tendeloo VF, Berneman ZN. Leukemia-associated antigens and their relevance to the immunotherapy of acute myeloid leukemia. Leukemia. 2012;26(10):2186–2196. doi: 10.1038/leu.2012.145. [DOI] [PubMed] [Google Scholar]
- 16.Maus MV, Grupp SA, Porter DL, June CH. Antibody-modified T cells: CARs take the front seat for hematologic malignancies. Blood. 2014;123(17):2625–2635. doi: 10.1182/blood-2013-11-492231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Peggs K, Verfuerth S, Mackinnon S. Induction of cytomegalovirus (CMV)-specific T-cell responses using dendritic cells pulsed with CMV antigen: a novel culture system free of live CMV virions. Blood. 2001;97(4):994–1000. doi: 10.1182/blood.v97.4.994. [DOI] [PubMed] [Google Scholar]
- 18.Foster AE, Bradstock KF, Sili U, Marangolo M, Rooney CM, Gottlieb DJ. A comparison of gene transfer and antigen-loaded dendritic cells for the generation of CD4+ and CD8+ cytomegalovirus-specific T cells in HLA-A2+ and HLA-A2− donors. Biol Blood Marrow Transplant. 2004;10(11):761–771. doi: 10.1016/j.bbmt.2004.05.011. [DOI] [PubMed] [Google Scholar]
- 19.Pyzer AR, Avigan DE, Rosenblatt J. Clinical trials of dendritic cell-based cancer vaccines in hematologic malignancies. Hum Vaccin Immunother. 2014;10(11):3125–3131. doi: 10.4161/21645515.2014.982993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mazur MA, Davis CC, Szabolcs P. Ex vivo expansion and Th1/Tc1 maturation of umbilical cord blood T cells by CD3/CD28 costimulation. Biol Blood Marrow Transplant. 2008;14(10):1190–1196. doi: 10.1016/j.bbmt.2008.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Antony J, Chen X, Szabolcs P. IL-15 Induced Polyclonal CTL Generated From Expanded CBT Cells Against Leukemia Cell Lines Constitutes IFN-γ Producing Cells and TCRγδ Cells. ASH Annual Meeting; December 8, 2012.2012. [Google Scholar]
- 22.Davis CC, Marti LC, Sempowski GD, Jeyaraj DA, Szabolcs P. Interleukin-7 permits Th1/Tc1 maturation and promotes ex vivo expansion of cord blood T cells: a critical step toward adoptive immunotherapy after cord blood transplantation. Cancer Res. 2010;70(13):5249–5258. doi: 10.1158/0008-5472.CAN-09-2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.bdbiosciences.com. BD Multitest™. 2012 available at http://www.bdbiosciences.com/external_files/Doc_Recon_2.0/is/tds/23-3600.pdf.
- 24.Jung S, Panchalingam KM, Rosenberg L, Behie LA. Ex vivo expansion of human mesenchymal stem cells in defined serum-free media. Stem Cells Int. 2012;2012:123030. doi: 10.1155/2012/123030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang HM, Zhang LS. Influence of human bone marrow mesenchymal stem cells on proliferation of chronic myeloid leukemia cells. Ai Zheng. 2009;28(1):29–32. [PubMed] [Google Scholar]
- 26.Glenjen NI, Hatfield K, Bruserud O. Coculture of native human acute myelogenous leukemia blasts with fibroblasts and osteoblasts results in an increase of vascular endothelial growth factor levels. Eur J Haematol. 2005;74(1):24–34. doi: 10.1111/j.1600-0609.2004.00333.x. [DOI] [PubMed] [Google Scholar]
- 27.Ito S, Barrett AJ, Dutra A, et al. Long term maintenance of myeloid leukemic stem cells cultured with unrelated human mesenchymal stromal cells. Stem Cell Res. 2015;14(1):95–104. doi: 10.1016/j.scr.2014.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Choudhury BA, Liang JC, Thomas EK, et al. Dendritic cells derived in vitro from acute myelogenous leukemia cells stimulate autologous, antileukemic T-cell responses. Blood. 1999;93(3):780–786. [PubMed] [Google Scholar]
- 29.Savoldo B, Huls MH, Liu Z, et al. Autologous Epstein-Barr virus (EBV)-specific cytotoxic T cells for the treatment of persistent active EBV infection. Blood. 2002;100(12):4059–4066. doi: 10.1182/blood-2002-01-0039. [DOI] [PubMed] [Google Scholar]
- 30.PerkinElmer. A new simplified, gentle cell-labelling method for non-radioactive cytotoxicity assays. Available at http://www.perkinelmer.com/CMSResources/Images/44-73050APP_DELFIACytotoxicity.pdf.
- 31.LifeTechnologies. Trizol Reagent. Available online at https://tools.lifetechnologies.com/content/sfs/manuals/trizol_reagent.pdf.
- 32.Chomczynski P. A reagent for the single-step simultaneous isolation of RNA, DNA and proteins from cell and tissue samples. Biotechniques. 1993;15(3):532–534. 536–537. [PubMed] [Google Scholar]
- 33.Hummon AB, Lim SR, Difilippantonio MJ, Ried T. Isolation and solubilization of proteins after TRIzol extraction of RNA and DNA from patient material following prolonged storage. Biotechniques. 2007;42(4):467–470. 472. doi: 10.2144/000112401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 35.Chen L, Flies DB. Molecular mechanisms of T cell co-stimulation and co-inhibition. Nat Rev Immunol. 2013;13(4):227–242. doi: 10.1038/nri3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gabrilovich DI, Nagaraj S. Myeloid-derived suppressor cells as regulators of the immune system. Nat Rev Immunol. 2009;9(3):162–174. doi: 10.1038/nri2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ledderose C, Heyn J, Limbeck E, Kreth S. Selection of reliable reference genes for quantitative real-time PCR in human T cells and neutrophils. BMC Res Notes. 2011;4:427. doi: 10.1186/1756-0500-4-427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pfaffl MW, Tichopad A, Prgomet C, Neuvians TP. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper--Excel-based tool using pair-wise correlations. Biotechnol Lett. 2004;26(6):509–515. doi: 10.1023/b:bile.0000019559.84305.47. [DOI] [PubMed] [Google Scholar]
- 39.de Jonge HJ, Valk PJ, de Bont ES, et al. Prognostic impact of white blood cell count in intermediate risk acute myeloid leukemia: relevance of mutated NPM1 and FLT3-ITD. Haematologica. 2011;96(9):1310–1317. doi: 10.3324/haematol.2011.040592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sung L, Aplenc R, Alonzo TA, Gerbing RB, Gamis AS, Group AP. Predictors and short-term outcomes of hyperleukocytosis in children with acute myeloid leukemia: a report from the Children’s Oncology Group. Haematologica. 2012;97(11):1770–1773. doi: 10.3324/haematol.2012.065490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ventura GJ, Hester JP, Smith TL, Keating MJ. Acute myeloblastic leukemia with hyperleukocytosis: risk factors for early mortality in induction. Am J Hematol. 1988;27(1):34–37. doi: 10.1002/ajh.2830270109. [DOI] [PubMed] [Google Scholar]
- 42.Narita M, Takahashi M, Liu A, et al. Leukemia blast-induced T-cell anergy demonstrated by leukemia-derived dendritic cells in acute myelogenous leukemia. Exp Hematol. 2001;29(6):709–719. doi: 10.1016/s0301-472x(01)00636-1. [DOI] [PubMed] [Google Scholar]
- 43.Le Dieu R, Taussig DC, Ramsay AG, et al. Peripheral blood T cells in acute myeloid leukemia (AML) patients at diagnosis have abnormal phenotype and genotype and form defective immune synapses with AML blasts. Blood. 2009;114(18):3909–3916. doi: 10.1182/blood-2009-02-206946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang WG, Liu SH, Cao XM, et al. A phase-I clinical trial of active immunotherapy for acute leukemia using inactivated autologous leukemia cells mixed with IL-2, GM-CSF, and IL-6. Leuk Res. 2005;29(1):3–9. doi: 10.1016/j.leukres.2004.04.015. [DOI] [PubMed] [Google Scholar]
- 45.Smits EL, Lee C, Hardwick N, et al. Clinical evaluation of cellular immunotherapy in acute myeloid leukaemia. Cancer Immunol Immunother. 2011;60(6):757–769. doi: 10.1007/s00262-011-1022-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Croft M. Costimulation of T cells by OX40, 4-1BB, and CD27. Cytokine Growth Factor Rev. 2003;14(3–4):265–273. doi: 10.1016/s1359-6101(03)00025-x. [DOI] [PubMed] [Google Scholar]
- 47.Vinay DS, Kwon BS. Role of 4-1BB in immune responses. Semin Immunol. 1998;10(6):481–489. doi: 10.1006/smim.1998.0157. [DOI] [PubMed] [Google Scholar]
- 48.Watts TH. TNF/TNFR family members in costimulation of T cell responses. Annu Rev Immunol. 2005;23:23–68. doi: 10.1146/annurev.immunol.23.021704.115839. [DOI] [PubMed] [Google Scholar]
- 49.Tamada K, Shimozaki K, Chapoval AI, et al. LIGHT, a TNF-like molecule, costimulates T cell proliferation and is required for dendritic cell-mediated allogeneic T cell response. J Immunol. 2000;164(8):4105–4110. doi: 10.4049/jimmunol.164.8.4105. [DOI] [PubMed] [Google Scholar]
- 50.Chattopadhyay K, Lazar-Molnar E, Yan Q, et al. Sequence, structure, function, immunity: structural genomics of costimulation. Immunol Rev. 2009;229(1):356–386. doi: 10.1111/j.1600-065X.2009.00778.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brandt CS, Baratin M, Yi EC, et al. The B7 family member B7-H6 is a tumor cell ligand for the activating natural killer cell receptor NKp30 in humans. J Exp Med. 2009;206(7):1495–1503. doi: 10.1084/jem.20090681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schneider K, Potter KG, Ware CF. Lymphotoxin and LIGHT signaling pathways and target genes. Immunol Rev. 2004;202:49–66. doi: 10.1111/j.0105-2896.2004.00206.x. [DOI] [PubMed] [Google Scholar]
- 53.Isakov N, Altman A. Protein kinase C(theta) in T cell activation. Annu Rev Immunol. 2002;20:761–794. doi: 10.1146/annurev.immunol.20.100301.064807. [DOI] [PubMed] [Google Scholar]
- 54.Dustin ML. PKC-theta: hitting the bull’s eye. Nat Immunol. 2011;12(11):1031–1032. doi: 10.1038/ni.2141. [DOI] [PubMed] [Google Scholar]
- 55.Nagy B, Ferrer A, Larramendy ML, et al. Lymphotoxin beta expression is high in chronic lymphocytic leukemia but low in small lymphocytic lymphoma: a quantitative real-time reverse transcriptase polymerase chain reaction analysis. Haematologica. 2003;88(6):654–658. [PubMed] [Google Scholar]
- 56.Riches JC, Davies JK, McClanahan F, et al. T cells from CLL patients exhibit features of T-cell exhaustion but retain capacity for cytokine production. Blood. 2013;121(9):1612–1621. doi: 10.1182/blood-2012-09-457531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yang B, Wang X, Jiang J, Cheng X. Involvement of CD244 in regulating CD4+ T cell immunity in patients with active tuberculosis. PLoS One. 2013;8(4):e63261. doi: 10.1371/journal.pone.0063261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bengsch B, Seigel B, Ruhl M, et al. Coexpression of PD-1, 2B4, CD160 and KLRG1 on exhausted HCV-specific CD8+ T cells is linked to antigen recognition and T cell differentiation. PLoS Pathog. 2010;6(6):e1000947. doi: 10.1371/journal.ppat.1000947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sinha P, Okoro C, Foell D, Freeze HH, Ostrand-Rosenberg S, Srikrishna G. Proinflammatory S100 proteins regulate the accumulation of myeloid-derived suppressor cells. J Immunol. 2008;181(7):4666–4675. doi: 10.4049/jimmunol.181.7.4666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nefedova Y, Nagaraj S, Rosenbauer A, Muro-Cacho C, Sebti SM, Gabrilovich DI. Regulation of dendritic cell differentiation and antitumor immune response in cancer by pharmacologic-selective inhibition of the janus-activated kinase 2/signal transducers and activators of transcription 3 pathway. Cancer Res. 2005;65(20):9525–9535. doi: 10.1158/0008-5472.CAN-05-0529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nefedova Y, Huang M, Kusmartsev S, et al. Hyperactivation of STAT3 is involved in abnormal differentiation of dendritic cells in cancer. J Immunol. 2004;172(1):464–474. doi: 10.4049/jimmunol.172.1.464. [DOI] [PubMed] [Google Scholar]
- 62.Foell D, Wittkowski H, Vogl T, Roth J. S100 proteins expressed in phagocytes: a novel group of damage-associated molecular pattern molecules. J Leukoc Biol. 2007;81(1):28–37. doi: 10.1189/jlb.0306170. [DOI] [PubMed] [Google Scholar]
- 63.Cheng P, Corzo CA, Luetteke N, et al. Inhibition of dendritic cell differentiation and accumulation of myeloid-derived suppressor cells in cancer is regulated by S100A9 protein. J Exp Med. 2008;205(10):2235–2249. doi: 10.1084/jem.20080132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Malyguine AM, Strobl S, Dunham K, Shurin MR, Sayers TJ. ELISPOT Assay for Monitoring Cytotoxic T Lymphocytes (CTL) Activity in Cancer Vaccine Clinical Trials. Cells. 2012;1(2):111–126. doi: 10.3390/cells1020111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.