Abstract
Many infectious diseases caused by eukaryotic pathogens have a devastating, long-term impact on animal health and welfare. Hundreds of millions of animals are affected by parasitic nematodes of the order Stronglida. Unlocking the molecular biology of representatives of this order, and understanding nematode-host interactions, drug resistance and disease using advanced technologies could lead to entirely new ways of controlling the diseases that they cause. Oesphagostomum dentatum (nodule worm; superfamily Strongyloidea) is an economically important strongylid nematode of swine worldwide. The present article reports recent advances made in biology and animal biotechnology through the draft genome and developmental transcriptome of O. dentatum, in order to support biological research of this and related parasitic nematodes as well as the search for new and improved interventions. This first genome of any member of the Strongyloidea is 443 Mb in size and predicted to encode 25,291 protein-coding genes. Here, we review the dynamics of transcription throughout the life cycle of O. dentatum, describe double-stranded RNA interference (RNAi) machinery and infer molecules involved in development and reproduction, and in inducing or modulating immune responses or disease. The secretome predicted for O. dentatum is particularly rich in peptidases linked to interactions with host tissues and/or feeding activity, and a diverse array of molecules likely involved in immune responses. This research progress provides an important resource for future comparative genomic and molecular biological investigations as well as for biotechnological research toward new anthelmintics, vaccines and diagnostic tests.
Keywords: Biotechnology, Genomics, Transcriptomics, Bioinformatics, Nodule worm disease, Livestock
1. Introduction
Oesophagostomum dentatum (nodule worm) is an economically important parasite of swine; this dioecious nematode belongs to the large order of the Strongylida (strongylids) that infect humans and animals worldwide (Anderson, 2000; Taylor et al., 2007). For instance, more than 1.3 billion people are infected with strongylids, such as Necator americanus and/or Ancylostoma duodenale (hookworms), which feed on blood in the small intestine (Hotez et al., 2013), causing adverse effects on human health, particularly in children. Other strongylids infect livestock and cause substantial production losses due to subclinical infections and clinical disease (Cantacessi et al., 2012; Lichtenfels et al., 1997), with billions of dollars spent annually on treatments to control these worms. In addition to their socioeconomic impact, various strongylid nematodes have developed resistance against the main drug classes commonly used to treat the diseases that they cause (Gilleard, 2006). Therefore, there is a need to work towards new treatment or control methods. This quest should be facilitated by acquiring a deep understanding of the molecular biology and biochemistry of key representatives.
O. dentatum is transmitted orally to the host and has a complex 3-week life cycle (Christensen et al., 1995; Spindler, 1933) (Fig. 1): eggs are excreted in host faeces; the first-stage larva (L1) develops inside the egg to then hatch (within 1 day) and moult through to the second- (L2) and third-stage (L3) larvae within a week; the infective L3s are then ingested by the host, exsheath and, after a histotrophic phase, develop through the fourth-stage larvae (L4s) to dioecious adults, which feed on nutrients in the large intestine. Because of its short life cycle and ability to develop in vitro for weeks through several moults (Daugschies and Watzel, 1999), O. dentatum is a useful model system for profound investigations of fundamental biological processes in nematodes. What has been missing, however, is basic information on the genome and transcriptomes to underpin such explorations.
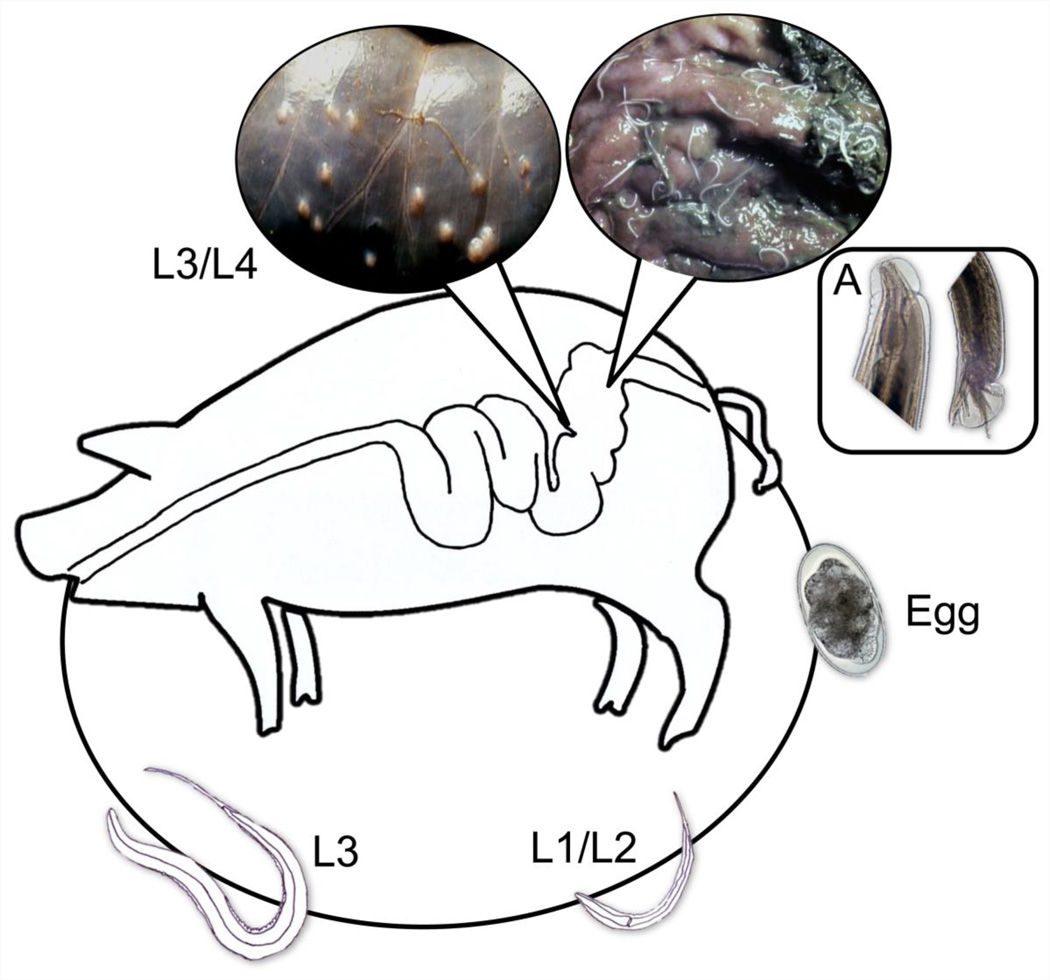
Fig. 1.
Development of Oesophagostomum dentatum. In a 3-week life cycle of the parasite, eggs are excreted in host faeces; the first-stage larva (L1) develops inside the egg to hatch and moult through to the second- (L2) and third-stage (L3) larval stages within a week; the infective L3s are then ingested by the pig host, exsheath and, after a short tissue phase, develop through the fourth-stage larvae (L4) to dioecious adults (A) which both feed on mucus and the contents of the large intestines.
Recent advances in the sequencing of the draft genomes and transcriptomes of selected, blood-feeding strongylid nematodes Haemonchus contortus (barber’s pole worm of sheep) and N. americanus (Schwarz et al., 2013; Tang et al., 2014) now provide a sound basis to tackle related nematodes that do not feed on blood. Here, we report on recent advances made in biology and biotechnology through the draft genome, developmentally-staged transcriptome to substantially enhance our understanding of this pathogen at the molecular level, across all defined life cycle stages, and its relationship with the porcine host. This genome not only delivers an important resource to the scientific community for a wide spectrum of genomic, systematic, biological and epidemiological studies, it also provides a solid foundation for the development of new interventions (drugs, vaccines and diagnostic tests) against O. dentatum and related nematodes of the superfamily Strongyloidea.
2. Genome characteristics and protein-encoding gene sets
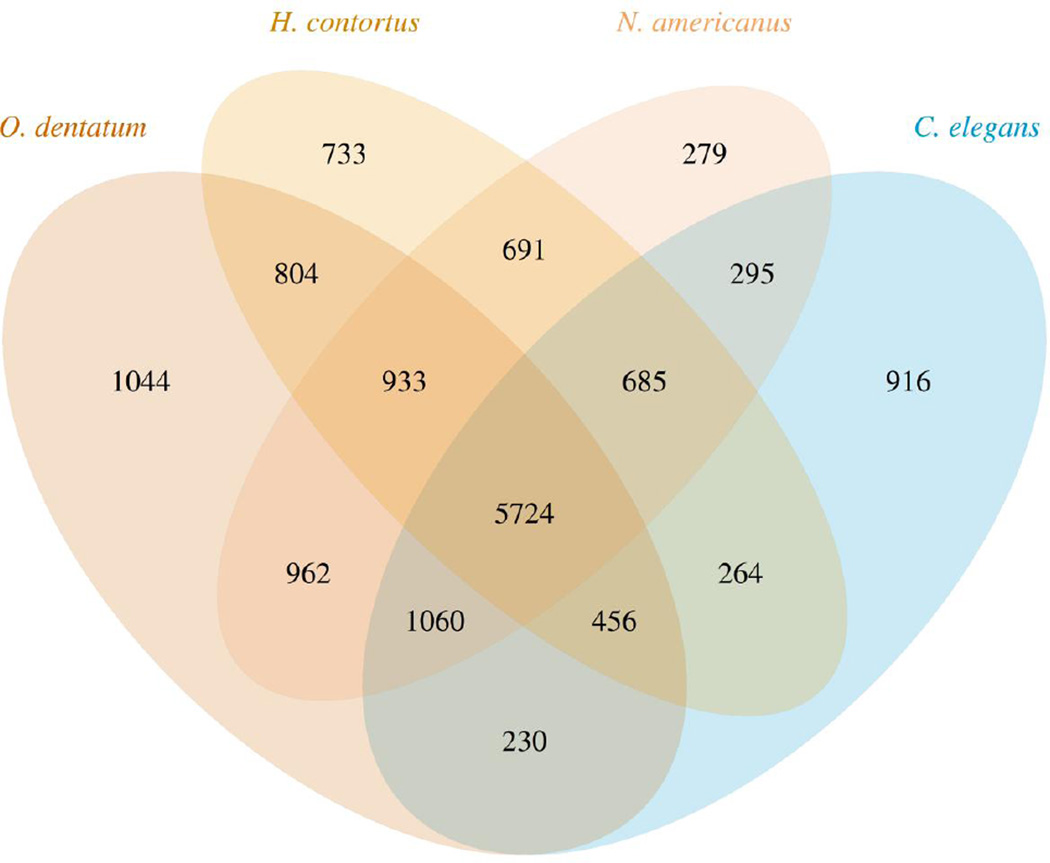
The nuclear genome of O. dentatum (Od-Hann strain) was sequenced, assembled and annotated (Appendix A). The final draft assembly was 443 Mb, half of which was represented by supercontigs of > 34.6 kb. This genome is amongst the largest of any nematode studied to date (Table 1), and the assembly represents most (90%) of the genome (Parra et al., 2007; Parra et al., 2009). The GC content is 41.3%, and the estimated repeat content is 30.9%, equating to 136.7 Mb of the genome. In total, 1,591 repeat families were predicted and annotated (Table 1); 350 transposons, including 137 DNA transposons, 112 LTR retrotransposons and 89 non-LTR retrotransposons were identified amongst these repeat families. The protein-encoding genes predicted (n = 25,291) represent 12.4% of the genome and have an average density of 57 genes per Mb. The GC content of coding sequences is 47.2%. Functional annotation of predicted proteins by sequence comparisons identified 4,540 unique domains (IPR) and 1,354 gene ontology (GO) terms for 62% and 48% of the O. dentatum genes, respectively; KEGG-based annotations were assigned to 56% of the proteins predicted for O. dentatum, which represented 4,171 unique KEGG orthology groups (KOs). Comparison of the gene set of O. dentatum with those of three other nematodes from the same taxonomic order (Blaxter et al., 1998) identified 15,076 orthologous groups (OGs) (Fig. 2); 11,213 of these OGs contained at least one O. dentatum gene (66.6% of all protein-coding genes), 2,699 OGs included genes from at least one other nematode but none from C. elegans, and 1,044 OGs (representing 2,972 genes) were unique to O. dentatum.
Table 1.
Features of the genome of Oesophagostomum dentatum and three other nematode species (Haemonchus contortus, Necator americanus and Caenorhabditis elegans) included for comparative purposes.
| O. dentatum | H. contortus | N. americanus | C. elegans | |
|---|---|---|---|---|
| Estimated genome size (Mega bases) | 443 | 320 | 244 | 100 |
| Assembly statistics | ||||
| Total number of supercontigs (>=1 kb) | 64,258 | 14,419 | 11,713 | 6 |
| Total number of base pairs (bp) in supercontigs | 443,038,381 | 319,640,208 | 244,009,025 | 100,272,607 |
| Number of N50 supercontigs | 3096 | 11,000 | 283 | 3 |
| N50 supercontig length (bp) | 26,460 | 56,328 | 213,095 | 17,500 |
| Number N90 supercontigs | 31,598 | 6,085 | 1,336 | 1 |
| N90 supercontig length (bp) | 2153 | 13,105 | 29,214 | |
| GC content of whole genome (%) | 33% | 42.40% | 40.20% | 35.40% |
| Repetitive sequences (%) | 30.86% | 13.40% | 24% | 21% |
| Protein-coding loci | ||||
| Total number of protein coding genes | 25,291 | 23,610 | 19,151 | 20,517 |
| Mean gene loci footprint (bp) | 2171 | 6167 | 4289 | 3035 |
| Mean number of exons per gene | 5.3 | 7 | 6.4 | 6.1 |
| Mean exon size (bp) | 122 | 139 | 125 | 203 |
| Mean intron size (bp) | 352 | 6 | 5.4 | 5.1 |
N50: number-50% of all nucleotides in the assembly are in 3096 supercontigs, length-50% of the genome is in supercontigs with a minimum length of 26kb; N90: number-90% of all nucleotides in the assembly are within 31,598 supercontigs, length-90% of the genome is in supercontigs with a minimum length of 2153.
(considering space on either strand)/(considering same strand space)
Fig. 2.
A Venn diagram showing the species distribution of constituent genes of orthologous groups obtained for Oesophagostomum dentatum and three other nematode species (Haemonchus contortus, Necator americanus and Caenorhabditis elegans).
Some common drug target entities, including kinases, phosphatases, GTPases, G protein-coupled receptors (GPCRs) and transporters, were annotated. In total, 327 kinases and 445 phosphatases were encoded in the genome of O. dentatum (Table 1). All the major classes of kinases were identified, with tyrosine (TK; n = 76), CAMK (51), CMGC (49) and casein kinases (CK1; 45) being abundantly represented (67.6%). The phosphatases annotated include mainly protein tyrosine (n = 77), serine/threonine (63), dual specificity (56) and histidine (41) phosphatases. In addition, 159 GTPases including 91 small GTPases within the families Rab (n = 68), Rho (42), Arf (9) and Ran (6) were predicted. A small number (n = 12) of large GTPases, including dynamin, GBP and mitofusin (Table 1), were also identified. Examples of salient small GTPase homologs are rab-5, ran-1, sar-1, mig-2 and rheb-1, whose C. elegans orthologs are involved in reproductive, embryonic and/or larval development (www.wormbase.org). In total, 375 GPCRs and 2687 transporters and ion channels were also identified (Table 1), including voltage-gated channels (VICs) and ligand-gated ion channels (LGICs). Ion-channels have been explored previously as targets for nematocides such as monepantel, cydectin and levamisole (Kaminsky et al., 2008; Martin et al., 1998; Prichard et al., 2012). Interestingly, although homologs of acr-23 (the C. elegans monepantel receptor) have been identified in H. contortus and N. americanus (Laing et al., 2013; Schwarz et al., 2013; Tang et al., 2014), none was found in the O. dentatum gene set; acr-23 mutants are resistant to the amino-acetonitrile derivative anthelmintic monepantel (Kaminsky et al., 2008).
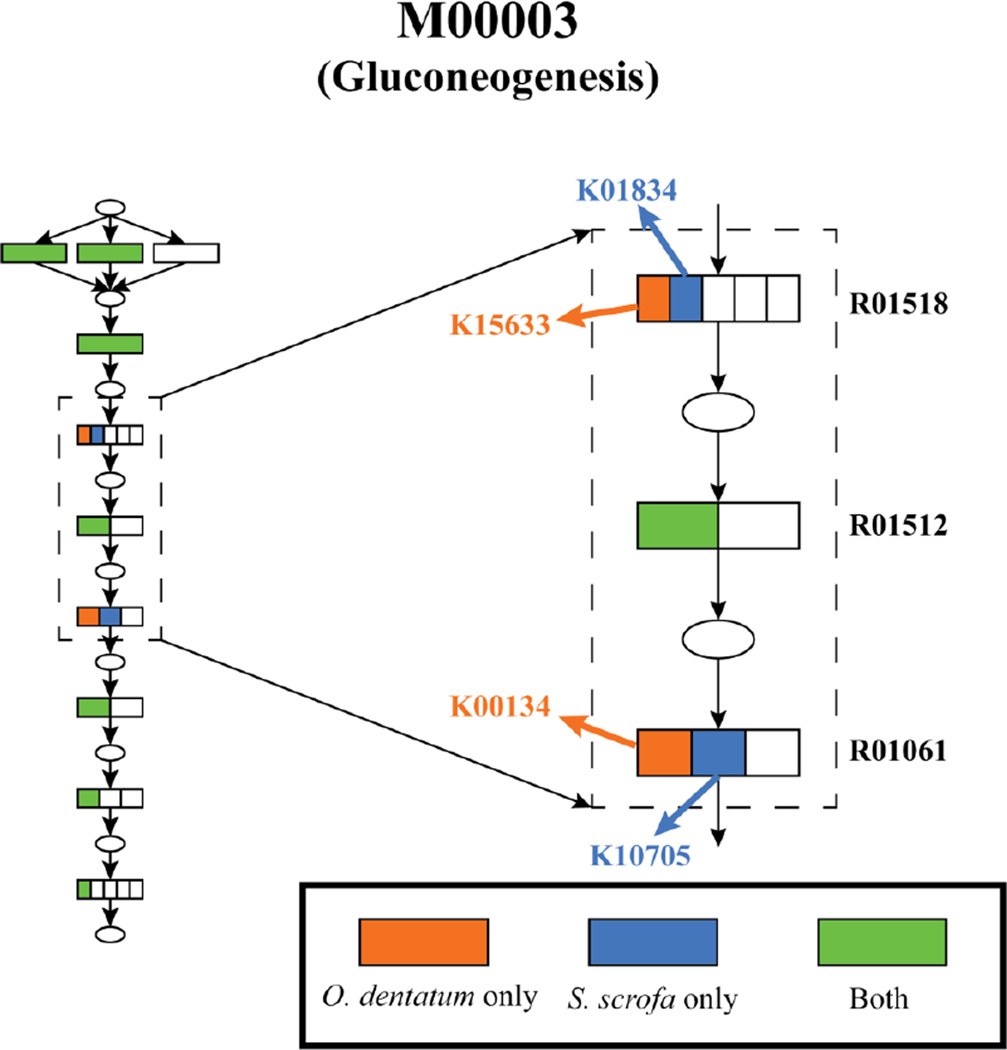
The metabolic potential of O. dentatum and its host, Sus scrofa (pig) were compared, in order to establish differences in pathways that may be exploited therapeutically. Metabolic modules were reconstructed using enzyme annotation. The high confidence set of KEGG orthology annotations resulted in the identification of 3,774 O. dentatum genes associated with 2,850 unique KOs, 295 pathways and 218 modules. The absence of a module, because of a lack of a single enzyme, might be the consequence of the draft nature of the genome or an incorrect functional annotation. Among the modules predicted for O. dentatum, there was no module that could be identified confidently as being absent from its host (i.e. lacking multiple critical enzymes). Among the modules that were present in both the parasite and the host, a search was conducted for cases where the parasite and the host used different enzymes (i.e. KEGG orthology groups) for the same reaction. Overall, four modules were found in which the worm uses enzymes that are not predicted to be encoded in the pig genome (Table 2, Fig. 3). Such enzymes might be assessed as potential drug targets that are selective for the parasite (without an adverse effect on the host).
Table 2.
Metabolic modules identified in Oesophagostomum dentatum and its porcine host, based on differential representation of metabolic KEGG orthology groups (KOs).
| Module ID |
Module description |
Reaction ID |
Description of reaction | KO in O. dentatum only |
Corre- sponding EC ID |
Name of enzyme |
|---|---|---|---|---|---|---|
| M00001 | Glycoloysis (Embden- Meyehof) |
R01518 | D-phosphoglycerate 2,3- phosphomutase |
K15633 | 5.4.2.12 | Cofactor independent phosphoglycerate mutase |
| M00003 | Gluco- neogenesis |
R01518 | D-phosphoglycerate 2,3- phosphomutase |
K15633 | 5.4.2.12 | Cofactor independent phosphoglycerate mutase |
| M00003 | Gluco- neogenesis |
R01061 | D-glyceraldehyde-3- phosphate:NAD+ oxidoreductase |
K00134 | 1.2.1.12 | Glyceraldehyde-3-P- dehydrogenase |
| M00083 | Fatty acid biosynthesis, elongation |
R04533 | (3R)-3-Hydroxybutanoyl- [acyl-carrier protein]:NADP+ oxidoreductase |
K00059 | 1.1.1.100 | Beta-ketoacyl-ACP reductase |
| M00087 | Beta-oxidation | R04738 | (S)-3-Hydroxyhexadecanoyl- CoA hydro-lyase |
K01692 | 4.2.1.17 | Enoyl-CoA hydratase |
ID = identification code.
Fig. 3.
Oesophagostomum dentatum and pig use non-orthologous enzymes for selected, crucial metabolic reactions. An example of non-orthologous enzyme usage in host and parasite is shown. Ellipses represent metabolites; rectangles represent reaction steps in the pathway, with divisions inside the rectangles representing different KEGG orthology groups of enzymes that catalyze that reaction. Reactions phosphoglycerate mutase (R01518) and glyceraldehyde 3-P dehydrogenase (R01061) are critical glycolysis steps that are catalyzed by enzymes belonging to different KEGG orthology groups in the two organisms, as evident by the different KO identification codes (IDs) in the schematic.
3. Transcriptional alterations during development
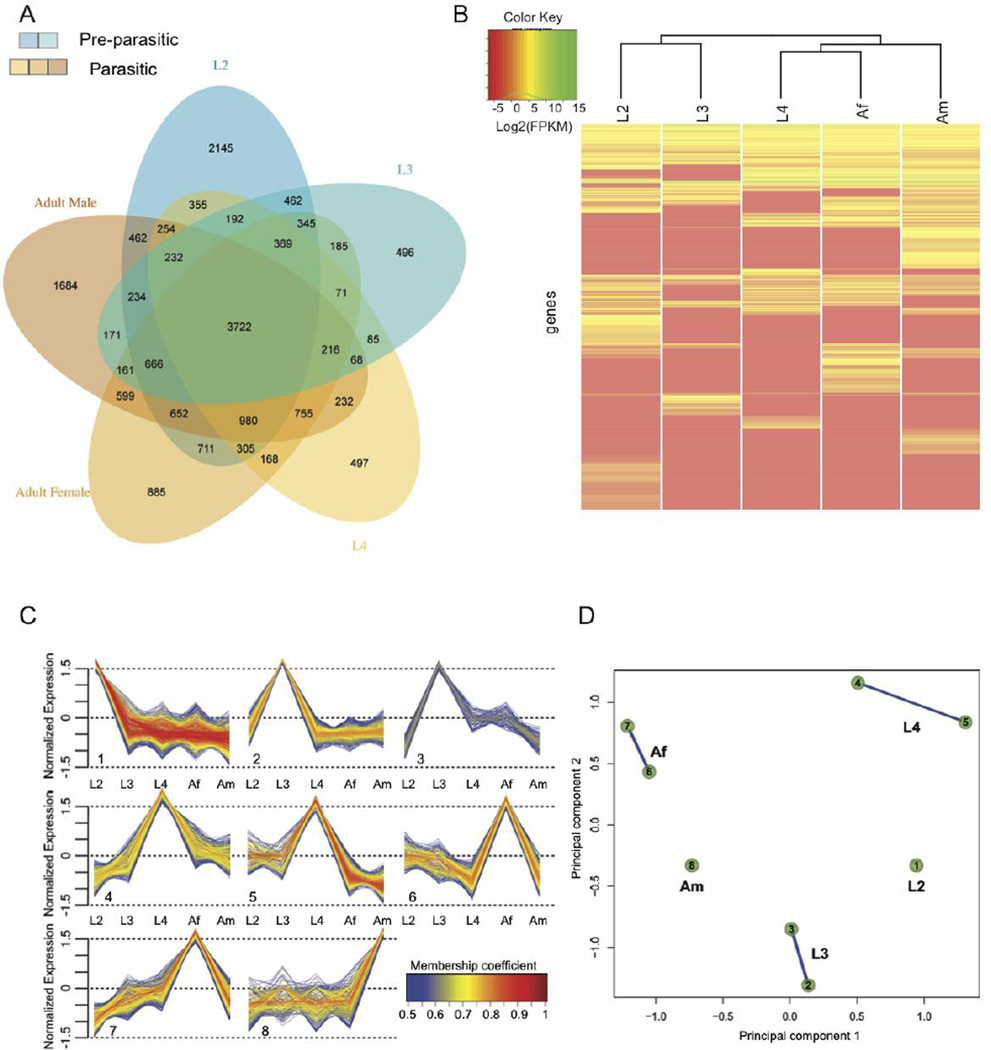
Like other strongylid nematodes, O. dentatum development requires a number of temporally regulated processes (Kotlán, 1948). The basic tissue types are generated during embryogenesis, with each type differentiating at a particular point in the developmental cycle. Essential tissues and organs differentiate in the larval stages (L1 to L4) and develop further to allow a parasitic mode of existence and reproduction in the host animal (L4 to adult). Major growth occurs from L2 to L4, and again to the adult stage. Development occurs in two different locations, namely in the environment (e.g., pasture or ground) for the free-living stages L1 to L3, and in the host for the dioecious L4 and adult stages (Fig. 1). Each of these stages has distinct requirements, in terms of motility, sensory perception, regulation of hormones of the endocrine system and metabolism. The free-living, infective L3 stage persists in the environment until being ingested by the host, in which it then receives a signal (primarily pH, temperature and CO2 changes) to make the transition to the parasitic phase in the gut. Such complex developmental changes in the nematode require tight control and rapid regulation of transcription. An investigation of transcriptional dynamics during the development of O. dentatum from L2 to the adult stage revealed that 18,359 genes (encoding 72.6% of the predicted proteins) were transcribed in at least one stage (Fig. 4A). These include 3,722 genes (20.3%) that are transcribed in all five developmental stages, representing a core set of functions required by the parasite, irrespective of stage. Of all genes, 31.1% (n = 5,707) were transcribed in only one of the five developmental stages studied. Interestingly, no orthologs of most (n = 2,928) of these genes were detected in H. contortus, N. americanus or C. elegans, indicating that they are unique to O. dentatum. This finding is similar to results for Brugia malayi (Li et al., 2012), for which 46.3% of genes encoding stage-specific transcripts represent currently unknown proteins.
Fig. 4.
Transcription throughout the development of Oesophagostomum dentatum. (A) Venn diagram showing genes whose transcription is detected in the developmental stages assayed. Blue shades represent pre-parasitic stages; orange shades represent parasitic stages. (B) Heat map showing transcriptional levels for the genes either specific to a stage or overexpressed in a stage. Based on these, the free-living stages (L2 and L3) cluster separately from the parasitic stages (L4 and Adult stages). (C) Soft clustering of all genes with non-zero FPKM in at least 3 stages was done. Eight clusters associated into 5 different groups based on the extent of shared genes among them, each characterized primarily by having high level of normalized transcription in one of the 5 developmental stages. (D) “Cluster Overlap” panel for gene clusters presented in C. Genes that did not have >0.5 membership coefficient in any of the clusters were not assigned to any clusters and are not shown here.
4. Dynamics of transcription
Clustering based on transcription levels indicated clear differences between free-living and parasitic stages (Fig. 4B). Furthermore, associations of transcription with different stages were identified based on (a) stage-specific transcription profiles of 5,707 genes (Fig. 4A) and (b) membership to one of eight clusters (comprising 4,089 genes), inferred by the fuzzy clustering (Figs. 4C and 4D) of profiles for 9,222 genes which are transcribed in at least three stages. An assessment of functional enrichment of genes associated with the individual developmental stages of O. dentatum (Fig. 4) showed that the free-living L2 stage had the highest number of stage-specific genes (n = 2,145, with an additional 947 genes in cluster 1 (cf. Fig. 4C), which consists of genes transcribed at high levels in L2). In this developmental stage, there was an expansion of the number of transcribed genes linked to lipid absorption and metabolism (e.g., fat-2 and spp-3), cell-cell signaling and hedgehog signaling (daf-6 and ptc-1), ubiquitin-related processes (cyn-4, dre-1 and siah-1), cuticular components (dpy-3, dpy-7, col-2 and col-34), proteolysis (adm-4, asp-1, cpl-1 and cpr-1), responses to oxidative stress, peroxidase, oxygenase and/or oxidoreductase activities (cyp-13A genes, bli-3, mlt-7, phy-2) as well as molecular binding and/or transport (e.g., calm-1, cmd-1, ncs-2 and twk-10). The functions of these genes appear to reflect rapid larval development, adaptation to a free-living environment, stress associated with this adaptation and/or a need to search for microbial food sources under perilous and varied conditions outside of the host animal. For example, L2 has the largest number of transcribed genes (n = 13) encoding peroxidases or superoxide dismutases (compared with 10 in all other stages and three in the adults) (Fig. 5). In the transition to the ensheathed L3 stage, the 63.3% reduction seen in the number of stage-specific genes (in clusters 2 or 3; Fig. 4C) was consistent with this stage being developmentally arrested. The processes enriched were primarily linked to neuropeptide signaling (e.g., flp-9, npr-3 and aexr-1), GPCR signaling (gpa-8 and ntr-1), translation and ribosome-related functions (e.g., eef-1B.1, mrpl-35 and rpl-2), molecular binding and/or transport (asic-1 and vha-2) as well as inositol binding and synthesis (inos-1 and snx-3), likely associated with survival of this free-living stage in the environment and its preparedness to invade the host animal. Following invasion and histotropic development of L3s in the gut wall of the host, the 25.4% increase in the number of L4 stage specific genes (in clusters 4 and 5; Fig. 4C) was linked to processes including molecular binding (e.g., idh-1), lysosome-related proteolysis (e.g., clp-3, lap-1 and nep-22) and cuticular synthesis (collagen genes including sqt-1, col-7, col-8, col-19, col-39 and col-62), likely reflecting substantial growth and development of this stage within the host animal. Two hyaluronidase genes (of ten encoded in the genome) were highly transcribed in the L4 stage; products from these genes catalyze the degradation of hyaluronic acid, an integral part of connective tissue in the host (Csóka et al., 1997; Laurent et al., 1996), and have been reported previously to be developmentally regulated in parasitic nematodes (Rhoads et al., 2000). There is a major increase in the number of genes transcribed specifically in the adult stage of O. dentatum (n = 4,144 genes in clusters 6–8; see Fig. 4C; an increase of 191% in L4); the enrichment related to mRNA processing, RNA binding and processing (e.g., adr-2 and prp-6), tRNA-aminoacylation (e.g., dars-1, ears-1 and lars-1), ATP binding (mrp-2), life-span (hsp-1) and DNA replication-related processes (e.g., rfc-2, rcq-5 and hus-1), protein processing and modification (e.g., air-1, cul-3, gly-13, kin-34, pas-2 and pptr-1), most of which are inferred to be associated with extensive digestive processes, longevity and/or reproduction in the worm within the large intestine of the host animal.
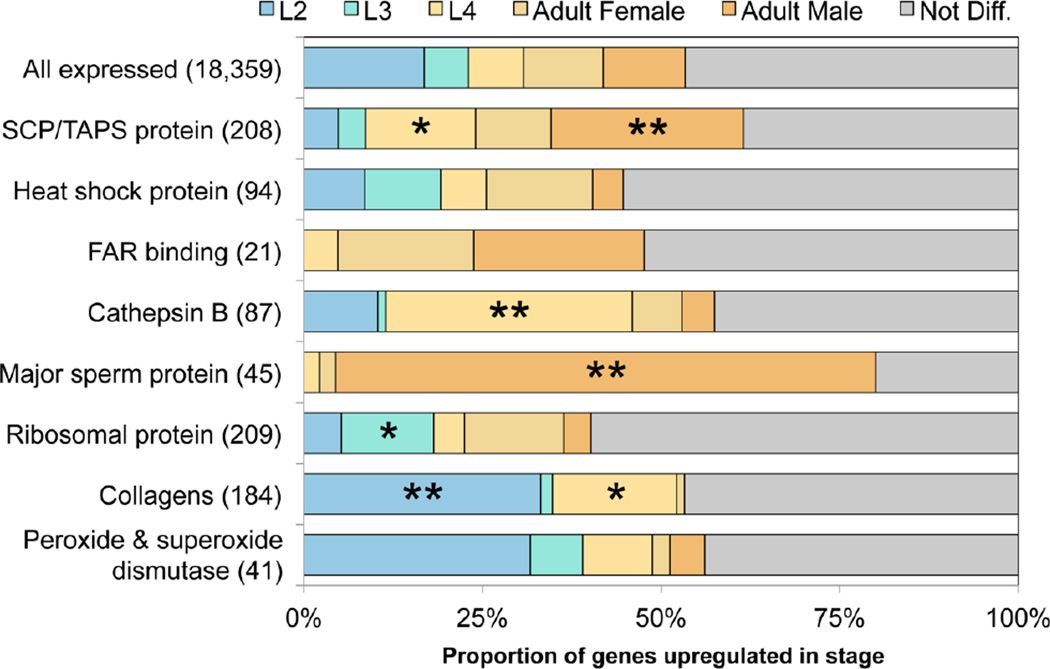
Fig. 5.
Transcription profiles of genes of Oesophagostomum dentatum encoding selected protein families based on developmental stage and parasitism. Genes annotated with certain functions that are either exclusively transcribed in a developmental stage or have high level of normalized transcription in it (i.e. belong to the corresponding expression profile cluster(s); cf. Fig. 4).
During crucial life-cycle transitions (egg to L2, and L3 to L4), which are linked to substantial growth and development (Gasser et al., 2007), the transcription of numerous genes (n = 93) encoding collagens and cuticular proteins was enriched (Fig. 5). Such structural molecules are pivotal for the maintenance of nematode body shape, protection against the external environment and contact with host interface. Of the 184 expressed collagen genes in O. dentatum, 58% (n = 131) were highly transcribed in individual stages, indicating that collagens are expressed in waves coinciding with the four molts (Johnstone and Barry, 1996). This dynamic, development-related transcriptional profile for this relatively large complement of genes (Fig. 5) is consistent with previous studies in which numerous collagen genes were transcribed in molting worms under nutrient-rich conditions, but no such genes were transcribed in developmentally arrested larvae (Mitreva et al., 2004).
In the L4 and/or adult stages of O. dentatum, there was an abundance of peptidase genes (n = 320) with enriched transcription (Fig. 4). Conspicuous were those encoding excretory/secretory (ES) peptidases of various clans, including cysteine peptidases (CA: predominantly C01A], serine peptidases (SA; predominantly S01A) and metallopeptidases (MA; predominantly M12A, but also numerous instances of M13, M10A and M01), which are likely involved in the degradation of cells and digestion of proteins following establishment in the gut, and which may be critical for growth, development and survival of O. dentatum in the porcine host (Gasser et al., 2007; Lant and Derry, 2013) and might represent potential drug targets (Page et al., 2014).
Subsequently, the functions of genes with enriched transcription in parasitic (L4 and adult) stages but not free-living stages of O. dentatum and without orthologs in C. elegans (179 genes) were studied. Such genes were linked to the cellular components such as “extracellular space” and “lysosome” as well as biological processes including “proteolysis”. Specifically, the genes related to these enrichment terms encoded C13 (legumain family), cathepsin B, papain family and pepsin A family type peptidase activities, all of which are recognized to be lysosome components and/or involved in digesting tissues or extracellular matrices, which are all functions that are critical to the feeding and survival of the parasite inside the host animal. For example, 63 of all 87 cathepsin B genes were transcribed exclusively in the parasitic stages, consistent with evidence that these genes are up-regulated in parasitic nematodes (Hartman et al., 2001; Jasmer et al., 2004). In addition, 16 of 21 genes encoding nematode fatty acid retinoid (FAR) binding proteins (IPR008632) were transcribed exclusively in parasitic stages. FAR proteins do not have homologs in mammals and are likely localized to the surface of parasites, placing them in a prime position for interactions with the host animal (Basavaraju et al., 2003; Nisbet et al., 2004). As 84 of the 179 genes enriched in parasitic stages could not be functionally annotated, they may relate to critical processes linked to parasitism.
5. Gender-enriched transcription
Groups of genes were studied that are differentially transcribed between female and male adults of O. dentatum. The transcription of 2,682 and 1,519 genes was enriched in females and males, respectively. A large number of these genes had orthologs in C. elegans, with 80% of the female-specific and 61% of male-specific transcripts with significant sequence identity to genes of C. elegans. Both the female and male gene sets were enriched for molecules linked predominantly to growth and reproduction, and germline, embryonic and genital development (Fig. 4). The female-enriched set had orthologs associated with the germline (e.g., fem-2, let-49 and mes-6), oogenesis or egg laying (e.g., ima-2 and mei-1), embryogenesis (e.g., let-858, mes-6 and rab-6.1), vulva development (e.g., air-1 and rab-8) and/or other reproductive and biological processes in C. elegans. In addition, vitellogenin genes were transcribed exclusively in females. Vitellogenins are egg-yolk precursor proteins expressed in almost all oviparous animals (Byrne et al., 1989); these proteins are abundantly transcribed during egg production and linked to an upregulation of protein synthesis and processing in females (Nisbet et al., 2008). The female-enriched set also contained key ribosomal proteins, heat shock proteins and chaperonins (Fig. 5), likely to be involved in reproductive processes and possibly host-pathogen-interactions (Gillan and Devaney, 2014). Interestingly, a gene (OESDEN_15934) encoding an endonuclease (CRN-1, the C. elegans homolog of FEN-1) likely to be intimately involved in switching the state of cells from DNA replication/repair to DNA degradation during apoptosis (Nisbet et al., 2008; Parrish et al., 2003) was highly transcribed in females (Fig. 5). This information suggests that selected oocytes might undergo apoptosis during oogenesis to ensure normal development of sister oocytes, consistent with evidence for some other ecdysozoans (Andux and Ellis, 2008; Buszczak and Cooley, 2000). Interestingly, transcriptional enrichment in the female also relates to actin and cytoskeletal activity, interpreted to be linked to the contractility in the ovary of O. dentatum, and consistent with the finding of non-striated actin filament networks in the myoepithelial sheath of the proximal ovary of C. elegans - which provides contractile forces essential for the transport of eggs in the uterus and “ovulation” (Ono and Ono, 2014).
Within the male-enriched set are genes encoding 64 kinases and 71 phosphatases with orthologs in C. elegans as well as other proteins linked to spermatogenesis/sperm (e.g., fog-3, msp-3, msp-10, msp-19 and msp-31; ssp-9, ssp-10, ssp-11 and ssp-16), some of which have been identified previously in small-scale molecular studies (Boag et al., 2003; Cottee et al., 2004). For instance, major sperm proteins (MSPs) are molecules that are exclusive to male nematodes (Cottee et al., 2004; Gasser et al., 2007), reflected in the present transcription profiles of the majority of 45 msp genes of O. dentatum (Fig. 5). Interestingly, there are at least 1,508 gender-enriched genes (i.e. 35.9% of all 4,201 sex-enriched genes) in O. dentatum that do not have orthologs in other strongylid nematodes (e.g., H. contortus and N. americanus) or C. elegans, indicating the existence of numerous genes and gene products that are unique to reproductive and/or developmental processes in O. dentatum. Although some genes of O. dentatum were not found to be transcribed in any of the five developmental stages investigated here, they may be involved in developmentally regulated processes within particular (e.g., neural) tissues of the worm. It is possible that low-level transcription in certain cells or tissues might not have been detected by RNA-seq because of a dilution effect caused by the bulk of other transcripts. For instance, lov-1, a gene required for a mating behavior and vulva location by the male (Barr and Sternberg, 1999) is known to be transcribed/expressed in the HOA and HOB neurons of C. elegans (Gasser et al., 2007), but was not detectable in any of the five developmental stages of O. dentatum studied here. Another possible reason for the absence of such transcripts might relate to a lack of RNA-seq data for stages of O. dentatum other than those included in the present study, such as those undergoing exsheathment, moulting, mating or arrested development (hypobiosis) (Ondrovics et al., 2013; Ondrovics et al., 2014) Future studies are warranted to explore transcription during short developmental transition periods.
6. Nematode-host Interactions and immunobiology
Many molecules, particularly ES proteins, are recognized to have critical roles in worm establishment, infection, and immune modulation and/or evasion (Hewitson et al., 2009, Pineda et al., 2014). In O. dentatum, 3,081 of 11,087 genes (27.8%) were inferred to encode ES proteins to be transcribed exclusively in parasitic (L4 and adult) stages, and fewer (2,153; 19.4%) only in free-living stages. These percentages are significantly higher (P=0.031 and P=0.045) than when compared against all genes with specific transcription in parasitic and free-living stages, respectively. These findings suggest that, while most ES proteins are deployed during the lifecycle, a considerable number are specifically associated with particular functions in either the parasitic- or free-living stages of the worm.
A total of 162 genes were predicted to encode ES proteins (Appendix B) likely to have immunomodulatory or immunogenic functions, many of which (46%) were transcribed at high levels in both parasitic stages (L4 and adults); 27% of them are ‘over-transcribed’ in parasitic stages (P<3×10−7 compared with all transcribed genes). Conspicuous were transcripts encoding numerous peptidases, SCP/Tpx-1/Ag5/PR-1/Sc7 (SCP/TAPS), transthyretin-like (TTL) and FAR binding proteins as well as eicosanoids.
A set of 160 genes was inferred to encode ES peptidases in the L4 and adult stages with abundant transcription (Appendix B). These genes encoded mainly cysteine-type (n = 51), serine-type (47) and metallopeptidases (47) as well as some aspartate- (9) and threonine-type (6) peptidases (mainly clans CA, MA and SA). In addition, transcription of genes encoding 75 peptidase inhibitors (primarily aprotinin- and Bombyx subtilisin-like molecules) was assessed. Many secreted peptidases likely to represent the ‘degradome’ and respective inhibitors are known to enable parasitic worms to invade, penetrate tissue barriers and feed (Knox, 2011; McKerrow et al., 2006; Nikolaou and Gasser, 2006; Tang et al., 2014); some of them (e.g., ES-62 in Acanthocheilonema viteae) have been reported to induce or modulate the host’s immune response against the parasite (Hewitson et al., 2008).
A large repertoire of genes encoding SCP/TAPS proteins (also called activation-associated proteins, ASPs) (Datu et al., 2009; Hawdon et al., 1996) was predicted; these molecules are characterized by the presence of SCP-like domains (IPR014044 and/or IPR001283). Of the 284 predicted SCP/TAPS proteins (250 single-, 32 double- and 2 triple-SCP-like domain molecules), 167 were inferred to be ES molecules and 119 were transcribed exclusively in the parasitic stages (L4 and adult stages) as compared with only 19 transcribed exclusively in free-living stages (Appendix B). In total, 179 of the predicted SCP/TAPS proteins did not have orthologs in H. contortus, N. americanus or C. elegans, and only 16 had C. elegans orthologs, similar to recent observations in Necator (Tang et al., 2014). The large number of genes encoding SCP/TAPS proteins in O. dentatum compared with only 34 such genes in C. elegans suggests that many of these proteins are involved in functions specific to O. dentatum, with potential relevance to parasitism and/or disease. Some (n = 66) of the SCP/TAPS proteins predicted were classified as NIFs, 30 of which were predicted to be ES proteins with immunobiological roles (Hewitson et al., 2009). Although NIFs had not been reported previously for O. dentatum, the SCP-1 homolog in Ancylostoma caninum binds the canine integrin CR3 (CD11b/CD18) and inhibits the oxidative burst by neutrophils (Moyle et al., 1994; Rieu et al., 1996). While the functional significance of most SCP/TAPS proteins is still unknown, they deserve detailed curation and investigation, given that they have been explored as vaccine candidates for other nematodes. Although not yet curated, SCP/TAPS genes are expanded in N. americanus compared with some other parasitic nematodes of animals studied to date (Tang et al., 2014). One representative, Na-ASP-2, has been tested in humans as a vaccine candidate (Bethony et al., 2008), but induced allergic responses following natural exposure to hookworm (Diemert et al., 2012). The crystal structure of Na-ASP-2 reveals charge segregation, like that of mammalian chemokines, suggesting that this protein is a ligand or agonist of selected GPCRs (chemokine receptors) (Asojo et al., 2005).
Another set of molecules (n = 81) likely to be involved in host-parasite interactions are TTL proteins, 64 of which were ES proteins and 28 were transcribed only in parasitic stages of O. dentatum (Appendix B). TTLs are relatively conserved (Rahat et al., 2008), and some are enzymes that catalyze the hydrolysis of 5-hydroxyisourate (HIU) to OHCU (in purine metabolism pathway) (Lee et al., 2005; Ramazzina et al., 2006). Some TTLs can bind hormones, such as thyroxine (T4) and vitamin A (Li and Buxbaum, 2011) or enable cell corpse engulfment by binding phosphatidylserine on the surface of apoptotic cells (Wang et al., 2010). TTLs represent a relatively large group of proteins usually specific to nematodes (Jacob et al., 2007; Parkinson et al., 2004), and have been identified previously in O. dentatum (Ondrovics et al., 2014). For instance, the O. dentatum homolog of C. elegans TTL-5 has been linked to L3 exsheathment (Ondrovics et al., 2014). TTL proteins have also been found in the strongylid nematodes A. caninum, H. contortus and Ostertagia ostertagi (Saverwyns et al., 2008, Vercauteren et al., 2003), Brugia malayi (human filarial nematode) (Hewitson et al., 2008) and Heterodera glycines, Meloidogyne incognita, Xiphinema index and Radophilis similis (plant parasitic nematodes) (Furlanetto et al., 2005; Gao et al., 2003; Jacob et al., 2007; McCarter et al., 2004). Although TTLs identified in these studies have been associated with stages relevant to host-parasite interactions, the precise functional roles of these molecules need to be confirmed.
Some of the most important effectors involved in immunity and inflammation are eicosanoids, which are signaling molecules comprising bioactive lipids (including leukotrienes, lipoxins, prostaglandins and thromboxanes) commonly found in some invertebrates (Rowley et al., 2005; Stanley, 2006). They were amongst the first developmentally regulated molecules described for O. dentatum (Daugschies, 1995; 1996; Daugschies and Ruttkowski, 1998). In mammals, they are known to exert complex control over the development of organ systems, such as the immune and nervous systems, but very little is known about them in nematodes (Daugschies and Joachim, 2000). Previous studies have shown that eicosanoids can be identified in homogenates and ES products from L3s and L4s in stage-specific compositions. The genome of C. elegans is not known to encode any orthologs of mammalian COX and LOX (cyclooxygenase and lipoxygenase) (Kosel et al., 2011; Lesa et al., 2003), proteins responsible for the conversion of precursor 20-carbon fatty acids to biologically active forms. However, they have been detected in O. dentatum, for which COX had a stage-specific expression profile (Joachim et al., 2001). Although COX1 or COX2 homologs were not detected in O. dentatum, four genes encoding proteins with LH2 domains (LOX homology domain) were identified. Previously, LOX was reported to have relatively consistent expression throughout development (Joachim et al., 2001); however, adult-specific transcription for 3 of 4 of these LOX genes was recorded. Along with arachidonic acid (AA)-derived pro-inflammatory eicosanoids, LOX is also involved in the production of anti-inflammatory eicosanoids derived from dihomo-γ-linolenic acid (DGLA) and eicosapentanoic acid (EPA). This suggests that particular eicosanoids play a role in suppressing host immune responses, thereby helping the parasite survive and reproduce inside the host. For example, the presence of parasite prostaglandins and leukotrienes in host tissues (such as the intestinal wall) may aid the invasion and establishment of O. dentatum (Daugschies and Joachim, 2000). Although very little is known about lipids in nematodes in general (Wenk, 2006), analyses of esterified fatty acids (FA) from extracts from L3, L4 and adult males and females of O. dentatum and O. quadrispinulatum (a closely related species) demonstrated a relative increase in somatic long-chain FA content during larval development. Cultured larvae appeared to consume polyunsaturated fatty acids including AAs (the precursors of eicosanoids) from the medium (Joachim et al., 2000). AA can be converted to eicosanoids via COX and LOX activities, but also via a cytochrome P450 (CYP450)-dependent pathway (Capdevila et al., 2002; Kosel et al., 2011). In total, 76 CYP450 genes are predicted to be encoded in O. dentatum (Appendix B), including orthologs of cyp-29 and cyp-33, which are C. elegans homologs of mammalian AA-hydroxylases and AA-epoxygenases, respectively, and inferred to be involved in this CYP450-dependent eicosanoid production pathway in O. dentatum. Interestingly, in this parasite, 38 of the genes predicted to encode CYP450 are stage-specifically transcribed in at least one stage; most of them (n = 26) are transcribed exclusively in L2. The present information provides a basis to now investigate in detail the biological, physiological and immunobiological significance of these molecules in O. dentatum.
7. Prediction of essential genes and RNA interference machinery
Essential genes are those that are crucial for life in any organism. Following the characterization of the genomes of C. elegans (C. elegans Sequencing Consortium, 1998) and Drosophila melanogaster (vinegar fly) (Adams et al., 2000), such genes and/or their products were explored on a large scale in these model metazoans (Boutros et al., 2004; Kamath et al., 2003), in order to understand developmental and many other biological, physiological and disease processes. Large-scale screens for essentiality have not been conducted in most parasitic nematodes, such as O. dentatum, mainly because their entire life cycles cannot be maintained in vitro, thus compromising analyses. Nonetheless, using in silico approaches (Lee et al., 2008; Zhong and Sternberg, 2006), it has been possible to infer, indirectly, essential genes in selected worms for targeted experimentation (Chen et al., 2011; Samarasinghe et al., 2011), the characterization of RNAi pathways (Schwarz et al., 2013) and/or the prioritization of anthelmintic targets (e.g., (Jex et al., 2011; Schwarz et al., 2013; Taylor et al., 2013b, Zhu et al., 2015) for genomic-guided drug discovery (Campbell et al., 2011; Taylor et al., 2013a).
This in silico approach was used to predict essential genes and the RNAi machinery of O. dentatum. In total, 2,109 genes were identified that have essential orthologs in C. elegans, which are known to yield lethal phenotypes upon gene knock-down or -knock out (Appendix B). Thereof, 57 genes encode protein homologs involved in the RNAi machinery of C. elegans (Dalzell et al., 2011), with many RNAi effector genes up-regulated in parasitic stages of O. dentatum (i.e. 13 in L4 and adult stages and 5 in L2; Appendix B). Although none of the exportin or mutator RNAi effector genes encoded in C. elegans (xpo-1, xpo-2, xpo-3, mut-2, mut-7 and mut-16) were detected in O. dentatum, seven genes encoded a protein domain (IPR013598) related to that of yeast exportin-1, suggesting that O. dentatum exportins have diverged considerably in sequence from their C. elegans counterparts. Interestingly, according to the present analysis, only two of four RNA-induced silencing complex (RISC) proteins and 11 of 28 argonaute proteins of C. elegans have orthologs in O. dentatum, indicating substantial differences between these two nematodes and unique silencing pathways within each nematode. In spite of these differences and challenges of conducting RNAi in parasitic nematodes (Geldhof et al., 2007; Knox et al., 2007; Lok, 2012), there seems to be a prospect of undertaking targeted functional genomic experiments in O. dentatum, possibly using virus-based transduction, given that this worm can be maintained in vitro through several moults (Daugschies and Watzel, 1999).
8. Conclusions and biotechnological implications
This article provides the first global insight into the molecular biology of the first representative of economically important parasitic nematodes of the superfamily Strongyloidea. The complexity of the O. dentatum genome is consistent with that seen in blood-feeding strongylid nematodes, such as H. contortus and N. americanus (Laing et al., 2013; Schwarz et al., 2013; Tang et al., 2014), and might relate to major sequence variation in non-coding regions within or among individual worms in the populations used for sequencing. A high mutation rate in such regions might partly explain the rapid emergence of anthelmintic resistance in strongylid nematodes (Gilleard, 2006; James et al., 2009; Keiser and Utzinger, 2010).
Some 40% of genes of O. dentatum have homologs in C. elegans, indicating considerable orthology between the two nematodes. However, numerous genes have orthologs exclusively in related parasitic nematodes (~15%) or have no known homolog, and cannot be assigned a function using current in silico methods and publicly available data sets. Such orphan genes and their products are of particular fundamental and applied research interest, and might relate specifically to a parasitic mode of existence, or parasite-specific functions, pathways or processes that might represent new and unique targets for drug design. Although such orphans are challenging to work on, there is considerable scope for exploring (relatively conserved) parasitic nematode- or strongylid-specific molecules using a combination of genomic and proteomic tools, particularly now that the draft genome and transcriptome resources are available for O. dentatum. Studying them will likely depend on available in vitro culture techniques, such that a gene silencing approach can be utilized to attempt to deduce gene/protein functions in the parasite itself. O. dentatum could provide a useful platform for this purpose, given that this nematode can be maintained in culture in vitro for considerably longer periods (at least 4 weeks) than other nematodes of the same order (Strongylida) (usually 1–3 weeks) (cf. Eckert, 1997) and because the parasite can be rectally transplanted, for example from in vitro culture, into the host without the need for surgical intervention (Gasser, Cottee, 2007), providing the prospect for well-controlled in vivo studies of worms, whose genes have been silenced using perturbation techniques. Having defined the RNAi machinery for O. dentatum might assist functional genomic work in selected life cycle stages, in spite of known challenges in strongylid nematodes studied to date (Geldhof et al., 2007; Knox et al., 2007). However, recent work on the flatworm Schistosoma mansoni shows considerable promise for virus-based transduction to deliver microRNA-adapted small hairpin RNAs (shRNAmirs) into the parasite to achieve effective gene silencing with a phenotypic read out in vivo in an experimental host animal (Hagen et al., 2014). This approach could be assessed in O. dentatum and/or related strongylids. In conclusion, the knowledge created here through new genomic resources should facilitate deep explorations of gene and protein functions, such as those involved in developmental and reproductive pathways, as well as comparative genomic studies. Moreover, with anthelmintic resistance problems in strongylid nematodes, designing new interventions based on an improved understanding of the molecular biology of these worms has considerable merit.
Acknowledgments
The authors thank the faculty and staff of the Genome Institute at Washington University who contributed the work described in this article. The genome sequencing and annotation work was funded by US National Institutes of Health grant U54HG003079 to R.K.W. Comparative genome analysis was funded by grants AI081803 and GM097435 to M.M. P.W.S. is an investigator with the Howard Hughes Medical Institute. RBG’s research was supported by the Australian Research Council (SRC), the National Health & Medical Research Council of Australia (NHMRC) and the Victorian Life Sciences Computation Initiative (VR0007).
APPENDICES
Appendix A
Methodology
Materials
O. dentatum (Od-Hann strain) was produced in Landrace-Large White crossbred pigs (3 months of age upon infection) maintained under helminth-free conditions. Pigs were inoculated via oral intubation with 5,000–10,000 infective third-stage larvae (L3s) of O. dentatum. Larvae were produced by copro-culture (27 °C) from the feces from pigs with patent infection (Talvik et al., 1997); first- and second-stage larvae (called L1s and L2s, respectively) were harvested from cultures after 24 h of cultivation, third-stage larvae (L3s) after 7 days; larvae were purified and concentrated using the Baermann funnel technique (Talvik et al., 1997) for 6 h. Larval stages were identified morphologically (Goodey, 1926). L1s and L2s were washed extensively in H2O; L3s were additionally subjected to a small-scale agar gel migration (Talvik et al., 1997). Fourth-stage larvae (L4s) and adults of O. dentatum were collected from the large intestine from infected pigs, following sacrifice by stunning with captive bolt and subsequent exsanguination at 13 days and 1 month, respectively, after infection with L3s. L4s and adults of O. dentatum were washed extensively in physiological saline, and the sexes were separated prior to freezing. All of the developmental stages or sexes of O. dentatum collected were snap-frozen separately in liquid nitrogen and stored at −70 °C until use. Animal ethics approval (no. GZ 68.205/103-II/10b/2008) was granted by the University of Veterinary Medicine, Vienna, Austria.
DNA-sequencing, assembly and analyses
Paired-end whole-genome shotgun libraries (3 kb, 8 kb and 20 kb insert sizes) were constructed from genomic DNA from adult worms of O. dentatum and then sequenced using 454 technology (Roche). After trimming linker sequences, the reads were assembled using the program Newbler (Margulies et al., 2005). A repeat library was generated using Repeatmodeler, and repeats were characterized using the program CENSOR v.4.2.28 (Kohany et al., 2006) against RepBase (release 19.02) (Jurka et al., 2005). Repeats were then masked using RepeatMasker, and protein-coding genes were predicted ab initio using a combination of programs SNAP (Korf, 2004), Fgenesh, AUGUSTUS (Stanke et al., 2008) as well as MAKER (Cantarel et al., 2008) which aligns mRNA, expressed sequence tag (EST) and protein evidence from the same or different species, to aid in gene structure predictions. A high confidence gene set was predicted from the MAKER output, followed by quality assurance (Tang et al., 2014). The following quality indices (QIs) were calculated: (QI1) length of the 5’-UTR; (QI2) fraction of splice sites confirmed by an EST alignment; (QI3) fraction of exons that overlap an EST alignment; (QI4) fraction of exons that overlap EST or protein alignments; (QI5) fraction of splice sites confirmed by a SNAP prediction; (QI6) fraction of exons that overlap a SNAP prediction; (QI7) number of exons in mRNA; (QI8) length of the 3’ UTR; (QI9) length of the protein sequence produced by mRNA. Then, the following decision making steps were followed: (a) genes are screened for overlaps (<10% overlap permitted); (b) If QI2 and QI3 were > 0, or QI4 was > 0, then the gene is retained; (c) each gene was compared against SWISSPROT (Boeckmann et al., 2003) by BLASTp (E-value: <10−6); if a match was found, the gene was kept; (d) genes were compared against the Pfam database (Finn et al., 2006) using RPSBLAST (E-value: <10−3); if a Pfam entry was found, a gene was retained; (e) genes were compared against CDD (Marchler-Bauer et al., 2011) using RPSBLAST (E<10−3; coverage > 40%). If both cut-offs were achieved, genes were retained; (f) if no match was found, then a sequence similarity-based search was conducted against GenesDB from KEGG (Kanehisa et al., 2012), and genes with ≥ 55% identity and a bit score of ≥ 35 were retained. Orthologous groups of molecules were inferred among four nematode species using the program OrthoMCL (Li et al., 2003) using an inflation parameter value of 1.5. C. elegans homologs were identified employing the best identity match to the C. elegans proteome (E-value of 10−5).
RNA-sequencing and analyses
Total RNA was extracted (Wang et al., 2010) separately from the three larvae stages (L2, L3 and L4) and from male and female adults of O. dentatum. The integrity and yield of the RNA were verified using Bioanalyzer 2100 (Agilent Technologies). Each RNA sample (100–500 ng) was treated with Ambion Turbo DNase (Ambion/Applied Biosystems), mRNA purified using the MicroPoly(A) Purist Kit (Ambion/Applied Biosystems), and 1 ng of this RNA used as the template for cDNA library construction using the Ovation® RNA-Seq v.2 kit (NuGEN Technologies Inc.). Non-normalized cDNA was used to construct multiplexed Illumina paired-end small-insert libraries (Illumina Inc, San Diego, CA) using the following approach: (i) 500 ng of cDNA was sheared using a sonicator (Covaris S220) to a size range of 200–400 bp; (ii) PCR was used to enrich for adaptor-ligated fragments and to index libraries; (iii) AMPure paramagnetic beads (Agencourt, Beckman Coulter Genomics) were used for size selection (300–500 bp); (iv) quantitative PCR was used to establish the cluster counts required for sequencing using the GAIIx Illumina platform to produce sequences of 100 bp reads. One lane was run to produce 2 × 101 bp read pairs for each sample, resulting in a total of 6.5–7 Gb of read data.
RNA-seq data were processed using in-house scripts. The program Burrows-Wheeler Aligner (BWA) was used to detect any extraneous (e.g., host or microbial) reads for removal, and DUST (Hancock and Armstrong, 1994) to identify regions of low compositional complexity that were converted into Ns. A script was used to remove reads of < 60 non-Ns. Clean reads were mapped using the program TopHat (Trapnell et al., 2009) against the O. dentatum genome, and the number of fragments mapping to each coding exon was determined using HTSeq-Count (Anders et al., 2014). For each stage and gene, transcription was calculated following normalization for library depth (using the total number of fragments mapped per stage) and gene length, resulting in fragments per kilobase exonic sequence per million fragments mapped (FPKM) values.
Functional annotation
Amino acid sequences predicted from the protein-coding genes were used to detect conserved protein domains using InterProScan (Quevillon et al., 2005; Zdobnov and Apweiler, 2001) employing default settings. InterProScan results were the basis of annotation of several gene families of interest. Other gene families of interest were annotated based on orthologs in other species. Transporter proteins were annotated using homology to proteins in the transporter classification database (TCDB) (Saier et al., 2014), additional ligand-gated ion channels were annotated using other relevant resources (LGICdb) (Donizelli et al., 2006). Peptidases and peptidase inhibitors were annotated using the MEROPS database (Rawlings et al., 2012), and kinases were detected by building an HMM model for kinases using domain models from the Kinomer website (http://www.compbio.dundee.ac.uk/kinomer/allPK.hmm) and defining custom score thresholds per kinase class (Miranda-Saavedra and Barton, 2007), adjusted until an hmmpfam search (HMMER v2.3.2) identified all C. elegans kinases (TK, 5.50E-03; CAMK, 9.60E-07; CK1, 1.10E-02; CMGC, 6.70E-03; AGC, 1.10E-14; STE, 3.40E-03; RGC, 4.80E-05; TKL, 8.70E-03; PDHK, 4.70E-160; PIKK, 1.40E-06; Alpha, 8.50E-66; RIO, 7.50E-10). ES proteins were predicted using the programs SignalP 4.0 (Petersen et al., 2011), Phobius (Kall et al., 2004) and Secretome P (Bendtsen et al., 2004). KEGG orthology annotations was conducted using the program KEGGscan (Wylie et al., 2008) using a P-value cutoff of 10−5; the program KAAS (Moriya et al., 2007) was used to identify a high confidence set of KEGG orthology annotations for metabolic module analysis. The resultant protein-coding gene sequences for O. dentatum and annotations are available for download at Nematode.net (Martin et al., 2015) and at ParaSite within WormBase (Harris et al., 2014) in nucleotide and amino acid formats.
Differential transcription analysis
The Bioconductor package edge R (Robinson et al., 2010) was used for the analysis of RNA-seq data for individual developmental stages and sexes of O. dentatum. Paired-end reads were filtered for quality using Trimmomatic software (Lohse et al., 2012) (parameters: phred64, ILLUMINACLIP:illuminaClipping.fa:2:40:20,LEADING:3, TRAILING:3, SLIDINGWINDOW:4:20, MINLEN:40). Quality-filtered paired-end reads were mapped to the cDNA sequences using BWA (Li and Durbin, 2010). Genes over-transcribed in a specific stage were identified by using transcription profile clusters obtained using the program mfuzz (Kumar and Futschik, 2007). For this, the HTSeq python library (Anders et al., 2014) was used to establish the numbers of reads that mapped to annotated gene loci, which were then converted to FPKM values and clustered. This analysis was conducted for genes (n = 9,222) with FPKM values of >0 in three or more stages using the “mean fill” and “NA mode” settings. Five different sets of clusters were defined, with each set corresponding to high transcription in one of the five developmental stages. For functional enrichment, genes were assigned to a cluster based on their membership value (threshold of 0.5), leaving 4,089 genes. The gene set inferred to be associated with a particular stage was the union of the set of genes over-transcribed in that stage (based on mfuzz) and the set of genes transcribed exclusively in that stage. Functional enrichment in these sets was studied using the program “func_hyper” of the package FUNC (Prufer et al., 2007), using the reference set of all 18,359 genes confirmed by transcription in at least one stage for comparison. To detect putative parasitism-related gene functions, a parasitism-enriched gene-set was obtained by including only those genes that are transcribed in all the parasitic stages (i.e. L4 and the adult stage of both genders), not transcribed in the free-living stages (L2 and L3), and do not share an orthogroup with any C. elegans gene. To find differentially transcribed genes between two stages, the stage-wise read counts were used as input to edgeR. The differential transcription levels were calculated by pairwise comparison of life cycle stages of O. dentatum. An edgeR biological coefficient of variation of 0.1 and a false discovery rate (FDR) ≤ 0.05 were used.
Additional information and accession numbers/codes
Data analyses were conducted in a Unix environment or Microsoft Excel 2011 using standard commands. Statistical analysis was primarily performed using the programming language R, and most other bioinformatics analyses were done using in-house perl scripts. The whole-genome sequence of O. dentatum has been deposited in DDBJ/EMBL/GenBank under BioProject ID PRJNA72579, project accession JOOK00000000. The version described in this paper is the first version JOOK01000000. All short read data have been deposited in the NCBI Short Read Archive under the following accessions: SRR000205-SRR000206-SRR001309-to-SRR001315-SRR360867-to-SRR360880-SRR361450-to-SRR361456-SRR361463-SRR361470-SRR332584-SRR332587-SRR332590.
References
- Anders S, Pyl PT, Huber W. HTSeq - A Python framework to work with high-throughput sequencing data. Bioinformatics. 2014;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendtsen JD, Jensen LJ, Blom N, Von Heijne G, Brunak S. Feature-based prediction of non-classical and leaderless protein secretion. Protein Eng Des Sel. 2004;17:349–356. doi: 10.1093/protein/gzh037. [DOI] [PubMed] [Google Scholar]
- Boeckmann B, Bairoch A, Apweiler R, Blatter MC, Estreicher A, Gasteiger E, et al. The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nuc Acids Res. 2003;31:365–370. doi: 10.1093/nar/gkg095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantarel BL, Korf I, Robb SM, Parra G, Ross E, Moore B, et al. MAKER: an easy-to-use annotation pipeline designed for emerging model organism genomes. Genome Res. 2008;18:188–196. doi: 10.1101/gr.6743907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donizelli M, Djite MA, Le Novere N. LGICdb: a manually curated sequence database after the genomes. Nucleic Acids Res. 2006;34:D267–D269. doi: 10.1093/nar/gkj104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Mistry J, Schuster-Böckler B, Griffiths-Jones S, Hollich V, Lassmann T, et al. Pfam: clans, web tools and services. Nucleic Acids Res. 2006;34:D247–D251. doi: 10.1093/nar/gkj149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodey T. Some stages in the development of Oesophagostomum dentatum from the pig. J Helminthol. 1926;4:191–198. [Google Scholar]
- Hancock JM, Armstrong JS. SIMPLE34: an improved and enhanced implementation for VAX and Sun computers of the SIMPLE algorithm for analysis of clustered repetitive motifs in nucleotide sequences. Computer applications in the biosciences : CABIOS. 1994;10:67–70. doi: 10.1093/bioinformatics/10.1.67. [DOI] [PubMed] [Google Scholar]
- Harris TW, Baran J, Bieri T, Cabunoc A, Chan J, Chen WJ, et al. WormBase 2014: new views of curated biology. Nucleic Acids Res. 2014;42:D789–D793. doi: 10.1093/nar/gkt1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurka J, Kapitonov VV, Pavlicek A, Klonowski P, Kohany O, Walichiewicz J. Repbase Update, a database of eukaryotic repetitive elements. Cytogenet Genome Res. 2005;110:462–467. doi: 10.1159/000084979. [DOI] [PubMed] [Google Scholar]
- Kall L, Krogh A, Sonnhammer EL. A combined transmembrane topology and signal peptide prediction method. J Mol Biol. 2004;338:1027–1036. doi: 10.1016/j.jmb.2004.03.016. [DOI] [PubMed] [Google Scholar]
- Kanehisa M, Goto S, Sato Y, Furumichi M, Tanabe M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 2012;40:D109–D114. doi: 10.1093/nar/gkr988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohany O, Gentles AJ, Hankus L, Jurka J. Annotation, submission and screening of repetitive elements in Repbase: RepbaseSubmitter and Censor. BMC Bioinformatics. 2006;7:474. doi: 10.1186/1471-2105-7-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korf I. Gene finding in novel genomes. BMC Bioinformatics. 2004;5:59. doi: 10.1186/1471-2105-5-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar L, Futschik ME. Mfuzz: a software package for soft clustering of microarray data. Bioinformation. 2007;2:5–7. doi: 10.6026/97320630002005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Stoeckert CJ, Jr, Roos DS. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 2003;13:2178–2189. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohse M, Bolger AM, Nagel A, Fernie AR, Lunn JE, Stitt M, et al. RobiNA: a userfriendly, integrated software solution for RNA-Seq-based transcriptomics. Nucleic Acids Res. 2012;40:W622–W627. doi: 10.1093/nar/gks540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A, Lu S, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, et al. CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 2011;39:D225–D229. doi: 10.1093/nar/gkq1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda-Saavedra D, Barton GJ. Classification and functional annotation of eukaryotic protein kinases. Proteins. 2007;68:893–914. doi: 10.1002/prot.21444. [DOI] [PubMed] [Google Scholar]
- Martin J, Rosa BA, Ozersky P, Hallsworth-Pepin K, Zhang X, Bhonagiri-Palsikar V, et al. Helminth.net: expansions to Nematode.net and an introduction to Trematode.net. Nucleic Acids Res. 2015;43:D698–D706. doi: 10.1093/nar/gku1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriya Y, Itoh M, Okuda S, Yoshizawa AC, Kanehisa M. KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 2007;35:W182–W185. doi: 10.1093/nar/gkm321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nature Methods. 2011;8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- Quevillon E, Silventoinen V, Pillai S, Harte N, Mulder N, Apweiler R, et al. InterProScan: protein domains identifier. Nucleic Acids Res. 2005;33:W116–W120. doi: 10.1093/nar/gki442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlings ND, Barrett AJ, Bateman A. MEROPS: the database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res. 2012;40:D343–D350. doi: 10.1093/nar/gkr987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saier MH, Jr, Reddy VS, Tamang DG, Vastermark A. The transporter classification database. Nucleic Acids Res. 2014;42:D251–D258. doi: 10.1093/nar/gkt1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanke M, Diekhans M, Baertsch R, Haussler D. Using native and syntenically mapped cDNA alignments to improve de novo gene finding. Bioinformatics. 2008;24:637–644. doi: 10.1093/bioinformatics/btn013. [DOI] [PubMed] [Google Scholar]
- Talvik H, Christensen CM, Joachim A, Roepstorff A, Bjorn H, Nansen P. Prepatent periods of different Oesophagostomum spp. isolates in experimentally infected pigs. Parasitology Res. 1997;83:563–568. doi: 10.1007/s004360050298. [DOI] [PubMed] [Google Scholar]
- Tang YT, Gao X, Rosa BA, Abubucker S, Hallsworth-Pepin K, Martin J, et al. Genome of the human hookworm Necator americanus. Nat Genet. 2014;46:261–269. doi: 10.1038/ng.2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Abubucker S, Martin J, Wilson RK, Hawdon J, Mitreva M. Characterizing Ancylostoma caninum transcriptome and exploring nematode parasitic adaptation. BMC Genomics. 2010;11:307. doi: 10.1186/1471-2164-11-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wylie T, Martin J, Abubucker S, Yin Y, Messina D, Wang Z, et al. NemaPath: online exploration of KEGG-based metabolic pathways for nematodes. BMC Genomics. 2008;9:525. doi: 10.1186/1471-2164-9-525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zdobnov EM, Apweiler R. InterProScan - an integration platform for the signature-recognition methods in InterPro. Bioinformatics. 2001;17:847–848. doi: 10.1093/bioinformatics/17.9.847. [DOI] [PubMed] [Google Scholar]
Appendix B
Annotation and Transcription data summary for O. dentatum deduced proteins. Various gene categories of interest have been annotated. Gene transcription level values (FPKM) and stage-associated transcription is indicated (based on pair-wise comparisons and soft clustering)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams MD, Celniker SE, Holt RA, Evans CA, Gocayne JD, Amanatides PG, et al. The genome sequence of Drosophila melanogaster. Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- Anderson RC. Nematode Parasites of Vertebrates: their Development and Transmission. second. CABI; 2000. [Google Scholar]
- Andux S, Ellis RE. Apoptosis maintains oocyte quality in aging Caenorhabditis elegans females. PLoS Genet. 2008;4:e1000295. doi: 10.1371/journal.pgen.1000295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asojo OA, Goud G, Dhar K, Loukas A, Zhan B, Deumic V, et al. X-ray structure of Na-ASP-2, a pathogenesis-related-1 protein from the nematode parasite, Necator americanus and a vaccine antigen for human hookworm infection. J Mol Biol. 2005;346:801–814. doi: 10.1016/j.jmb.2004.12.023. [DOI] [PubMed] [Google Scholar]
- Barr MM, Sternberg PW. A polycystic kidney-disease gene homologue required for male mating behaviour in C. elegans. Nature. 1999;401:386–389. doi: 10.1038/43913. [DOI] [PubMed] [Google Scholar]
- Basavaraju SV, Basavaraju S, Zhan B, Kennedy MW, Liu Y, Hawdon J, et al. Ac-FAR-1, a 20 kDa fatty acid- and retinol-binding protein secreted by adult Ancylostoma caninum hookworms: gene transcription pattern, ligand binding properties and structural characterisation. Mol Biochem Parasitol. 2003;126:63–71. doi: 10.1016/s0166-6851(02)00253-0. [DOI] [PubMed] [Google Scholar]
- Bethony JM, Simon G, Diemert DJ, Parenti D, Desrosiers A, Schuck S, et al. Randomized, placebo-controlled, double-blind trial of the Na-ASP-2 hookworm vaccine in unexposed adults. Vaccine. 2008;26:2408–2417. doi: 10.1016/j.vaccine.2008.02.049. [DOI] [PubMed] [Google Scholar]
- Blaxter ML, De Ley P, Garey JR, Liu LX, Scheldeman P, Vierstraete A, et al. A molecular evolutionary framework for the phylum Nematoda. Nature. 1998;392:71–75. doi: 10.1038/32160. [DOI] [PubMed] [Google Scholar]
- Boag PR, Ren P, Newton SE, Gasser RB. Molecular characterisation of a male-specific serine/threonine phosphatase from Oesophagostomum dentatum (Nematoda: Strongylida), and functional analysis of homologues in Caenorhabditis elegans. Int J Parasitol. 2003;33:313–325. doi: 10.1016/s0020-7519(02)00263-1. [DOI] [PubMed] [Google Scholar]
- Boutros M, Kiger AA, Armknecht S, Kerr K, Hild M, Koch B, et al. Genome-wide RNAi analysis of growth and viability in Drosophila cells. Science. 2004;303:832–835. doi: 10.1126/science.1091266. [DOI] [PubMed] [Google Scholar]
- Buszczak M, Cooley L. Eggs to die for: cell death during Drosophila oogenesis. Cell Death Differ. 2000;7:1071–1074. doi: 10.1038/sj.cdd.4400755. [DOI] [PubMed] [Google Scholar]
- Byrne BM, Gruber M, Ab G. The evolution of egg yolk proteins. Progress in Biophys Mol Biol. 1989;53:33–69. doi: 10.1016/0079-6107(89)90005-9. [DOI] [PubMed] [Google Scholar]
- C. elegansSequencing Consortium. Genome sequence of the nematode C. elegans: a platform for investigating biology. Science. 1998;282:2012–2018. doi: 10.1126/science.282.5396.2012. [DOI] [PubMed] [Google Scholar]
- Campbell BE, Tarleton M, Gordon CP, Sakoff JA, Gilbert J, McCluskey A, et al. Norcantharidin analogues with nematocidal activity in Haemonchus contortus. Bioorganic & Med Chem Lett. 2011;21:3277–3281. doi: 10.1016/j.bmcl.2011.04.031. [DOI] [PubMed] [Google Scholar]
- Cantacessi C, Campbell BE, Gasser RB. Key strongylid nematodes of animals - Impact of next-generation transcriptomics on systems biology and biotechnology. Biotechnol Adv. 2012;30:469–488. doi: 10.1016/j.biotechadv.2011.08.016. [DOI] [PubMed] [Google Scholar]
- Capdevila JH, Harris RC, Falck JR. Microsomal cytochrome P450 and eicosanoid metabolism. Cell Mol Life Sci. 2002;59:780–789. doi: 10.1007/s00018-002-8466-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N, Xu MJ, Nisbet AJ, Huang CQ, Lin RQ, Yuan ZG, et al. Ascaris suum: RNAi mediated silencing of enolase gene expression in infective larvae. Exp Parasitol. 2011;127:142–146. doi: 10.1016/j.exppara.2010.07.019. [DOI] [PubMed] [Google Scholar]
- Christensen CM, Barnes EH, Nansen P, Roepstorff A, Slotved HC. Experimental Oesophagostomum dentatum infection in the pig: worm populations resulting from single infections with three doses of larvae. Int J Parasitol. 1995;25:1491–1498. doi: 10.1016/0020-7519(95)00085-2. [DOI] [PubMed] [Google Scholar]
- Cottee PA, Nisbet AJ, Boag PR, Larsen M, Gasser RB. Characterization of major sperm protein genes and their expression in Oesophagostomum dentatum (Nematoda: Strongylida) Parasitology. 2004;129:479–490. doi: 10.1017/s003118200400561x. [DOI] [PubMed] [Google Scholar]
- Csóka TB, Frost GI, Stern R, Csóka AB. Hyaluronidases in tissue invasion. Invasion Metastasis. 1997;17:297–311. [PubMed] [Google Scholar]
- Dalzell JJ, McVeigh P, Warnock ND, Mitreva M, Bird DM, Abad P, et al. RNAi effector diversity in nematodes. PLoS Neg Trop Dis. 2011;5:e1176. doi: 10.1371/journal.pntd.0001176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datu BJ, Loukas A, Cantacessi C, O'Donoghue P, Gasser RB. Investigation of the regulation of transcriptional changes in Ancylostoma caninum larvae following serum activation, with a focus on the insulin-like signalling pathway. Vet Parasitol. 2009;159:139–148. doi: 10.1016/j.vetpar.2008.10.026. [DOI] [PubMed] [Google Scholar]
- Daugschies A. Oesophagostomum dentatum: population dynamics and synthesis of prostanoids by histotropic stages cultured in vitro. Exp Parasitol. 1995;81:574–583. doi: 10.1006/expr.1995.1151. [DOI] [PubMed] [Google Scholar]
- Daugschies A. Investigations into the production and function of leukotrienes during histotropic development of Oesophagostomum dentatum. Parasitology Res. 1996;82:416–422. doi: 10.1007/s004360050138. [DOI] [PubMed] [Google Scholar]
- Daugschies A, Joachim A. Eicosanoids in parasites and parasitic infections. Adv Parasitol. 2000;46:181–240. doi: 10.1016/s0065-308x(00)46009-4. [DOI] [PubMed] [Google Scholar]
- Daugschies A, Ruttkowski B. Modulation of migration of Oesophagostomum dentatum larvae by inhibitors and products of eicosanoid metabolism. Int J Parasitol. 1998;28:355–362. doi: 10.1016/s0020-7519(97)00153-7. [DOI] [PubMed] [Google Scholar]
- Daugschies A, Watzel C. In vitro development of histotropic larvae of Oesophagostomum dentatum under various conditions of cultivation. Parasitol Res. 1999;85:158–161. doi: 10.1007/s004360050527. [DOI] [PubMed] [Google Scholar]
- Diemert DJ, Pinto AG, Freire J, Jariwala A, Santiago H, Hamilton RG, et al. Generalized urticaria induced by the Na-ASP-2 hookworm vaccine: implications for the development of vaccines against helminths. J Allerg Clin Immunol. 2012;130:169 e6–176 e6. doi: 10.1016/j.jaci.2012.04.027. [DOI] [PubMed] [Google Scholar]
- Eckert J. Alternatives to animal experimentation in parasitology. Vet Parasitol. 1997;71:99–120. doi: 10.1016/s0304-4017(97)00027-7. [DOI] [PubMed] [Google Scholar]
- Furlanetto C, Cardle L, Brown DJF, Jones JT. Analysis of expressed sequence tags from the ectoparasitic nematode Xiphinema index. Nematology. 2005;7:95–104. [Google Scholar]
- Gao B, Allen R, Maier T, Davis EL, Baum TJ, Hussey RS. The parasitome of the phytonematode Heterodera glycines. Mol Plant Microbe In. 2003;16:720–726. doi: 10.1094/MPMI.2003.16.8.720. [DOI] [PubMed] [Google Scholar]
- Gasser RB, Cottee P, Nisbet AJ, Ruttkowski B, Ranganathan S, Joachim A. Oesophagostomum dentatum: potential as a model for genomic studies of strongylid nematodes, with biotechnological prospects. Biotechnol Adv. 2007;25:281–293. doi: 10.1016/j.biotechadv.2007.01.008. [DOI] [PubMed] [Google Scholar]
- Geldhof P, Visser A, Clark D, Saunders G, Britton C, Gilleard J, et al. RNA interference in parasitic helminths: current situation, potential pitfalls and future prospects. Parasitology. 2007;134:609–619. doi: 10.1017/S0031182006002071. [DOI] [PubMed] [Google Scholar]
- Gillan V, Devaney E. Nematode Hsp90: highly conserved but functionally diverse. Parasitology. 2014;141:1203–1215. doi: 10.1017/S0031182014000304. [DOI] [PubMed] [Google Scholar]
- Gilleard JS. Understanding anthelmintic resistance: the need for genomics and genetics. Int J Parasitol. 2006;36:1227–1239. doi: 10.1016/j.ijpara.2006.06.010. [DOI] [PubMed] [Google Scholar]
- Hagen J, Young ND, Every AL, Pagel CN, Schnoeller C, Scheerlinck J-PY, Gasser RB, Kalinna BH. Omega-1 knockdown in Schistosoma mansoni eggs by lentivirus transduction reduces granuloma size in vivo. Nat Commun. 2014;5:5375. doi: 10.1038/ncomms6375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman D, Donald DR, Nikolaou S, Savin KW, Hasse D, Presidente PJ, et al. Analysis of developmentally regulated genes of the parasite Haemonchus contortus. Int J Parasitol. 2001;31:1236–1245. doi: 10.1016/s0020-7519(01)00248-x. [DOI] [PubMed] [Google Scholar]
- Hawdon JM, Jones BF, Hoffman DR, Hotez PJ. Cloning and characterization of Ancylostoma -secreted protein. A novel protein associated with the transition to parasitism by infective hookworm larvae. J Biol Chem. 1996;271:6672–6678. doi: 10.1074/jbc.271.12.6672. [DOI] [PubMed] [Google Scholar]
- Hewitson JP, Grainger JR, Maizels RM. Helminth immunoregulation: the role of parasite secreted proteins in modulating host immunity. Mol Biochem Parasitol. 2009;167:1–11. doi: 10.1016/j.molbiopara.2009.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewitson JP, Harcus YM, Curwen RS, Dowle AA, Atmadja AK, Ashton PD, et al. The secretome of the filarial parasite, Brugia malayi: proteomic profile of adult excretory-secretory products. Mol Biochem Parasitol. 2008;160:8–21. doi: 10.1016/j.molbiopara.2008.02.007. [DOI] [PubMed] [Google Scholar]
- Hotez PJ, Diemert D, Bacon KM, Beaumier C, Bethony JM, Bottazzi ME, et al. The Human Hookworm Vaccine. Vaccine. 2013;31(Suppl 2):B227–B232. doi: 10.1016/j.vaccine.2012.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob J, Vanholme B, Haegeman A, Gheysen G. Four transthyretin-like genes of the migratory plant-parasitic nematode Radopholus similis: members of an extensive nematode-specific family. Gene. 2007;402:9–19. doi: 10.1016/j.gene.2007.07.015. [DOI] [PubMed] [Google Scholar]
- James CE, Hudson AL, Davey MW. Drug resistance mechanisms in helminths: is it survival of the fittest? Trends Parasitol. 2009;25:328–335. doi: 10.1016/j.pt.2009.04.004. [DOI] [PubMed] [Google Scholar]
- Jasmer DP, Mitreva MD, McCarter JP. mRNA sequences for Haemonchus contortus intestinal cathepsin B-like cysteine proteases display an extreme in abundance and diversity compared with other adult mammalian parasitic nematodes. Mol Biochem Parasitol. 2004;137:297–305. doi: 10.1016/j.molbiopara.2004.06.010. [DOI] [PubMed] [Google Scholar]
- Jex AR, Liu S, Li B, Young ND, Hall RS, Li Y, et al. Ascaris suum draft genome. Nature. 2011;479:529–533. doi: 10.1038/nature10553. [DOI] [PubMed] [Google Scholar]
- Joachim A, Ruttkowski B, Daugschies A. Oesophagostomum dentatum: expression patterns of enzymes involved in eicosanoid production. Parasitol Int. 2001;50:211–215. doi: 10.1016/s1383-5769(01)00077-0. [DOI] [PubMed] [Google Scholar]
- Joachim A, Ryll M, Daugschies A. Fatty acid patterns of different stages of Oesophagostomum dentatum and Oesophagostomum quadrispinulatum as revealed by gas chromatography. Int J Parasitol. 2000;30:819–827. doi: 10.1016/s0020-7519(00)00067-9. [DOI] [PubMed] [Google Scholar]
- Johnstone IL, Barry JD. Temporal reiteration of a precise gene expression pattern during nematode development. EMBO J. 1996;15:3633–3639. [PMC free article] [PubMed] [Google Scholar]
- Kamath RS, Fraser AG, Dong Y, Poulin G, Durbin R, Gotta M, et al. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. 2003;421:231–237. doi: 10.1038/nature01278. [DOI] [PubMed] [Google Scholar]
- Kaminsky R, Ducray P, Jung M, Clover R, Rufener L, Bouvier J, et al. A new class of anthelmintics effective against drug-resistant nematodes. Nature. 2008;452:176–180. doi: 10.1038/nature06722. [DOI] [PubMed] [Google Scholar]
- Keiser J, Utzinger J. The drugs we have and the drugs we need against major helminth infections. Adv Parasitol. 2010;73:197–230. doi: 10.1016/S0065-308X(10)73008-6. [DOI] [PubMed] [Google Scholar]
- Knox D. Proteases in blood-feeding nematodes and their potential as vaccine candidates. Adv Exp Med Biol. 2011;712:155–176. doi: 10.1007/978-1-4419-8414-2_10. [DOI] [PubMed] [Google Scholar]
- Knox DP, Geldhof P, Visser A, Britton C. RNA interference in parasitic nematodes of animals: a reality check? Trends Parasitol. 2007;23:105–107. doi: 10.1016/j.pt.2007.01.007. [DOI] [PubMed] [Google Scholar]
- Kosel M, Wild W, Bell A, Rothe M, Lindschau C, Steinberg CE, et al. Eicosanoid formation by a cytochrome P450 isoform expressed in the pharynx of Caenorhabditis elegans. Biochem J. 2011;435:689–700. doi: 10.1042/BJ20101942. [DOI] [PubMed] [Google Scholar]
- Kotlán A. Studies on the life-history and pathological significance of Oesophagostomum spp. of the domestic pig. Acta Vet Hung. 1948:14–30. [Google Scholar]
- Laing R, Kikuchi T, Martinelli A, Tsai IJ, Beech RN, Redman E, et al. The genome and transcriptome of Haemonchus contortus a key model parasite for drug and vaccine discovery. Genome Biol. 2013:14. doi: 10.1186/gb-2013-14-8-r88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lant B, Derry WB. Methods for detection and analysis of apoptosis signaling in the C. elegans germline. Methods. 2013;61:174–182. doi: 10.1016/j.ymeth.2013.04.022. [DOI] [PubMed] [Google Scholar]
- Laurent TC, Laurent UB, Fraser JR. The structure and function of hyaluronan: An overview. Immunol Cell Biol. 1996;74:A1–A7. doi: 10.1038/icb.1996.32. [DOI] [PubMed] [Google Scholar]
- Lee I, Lehner B, Crombie C, Wong W, Fraser AG, Marcotte EM. A single gene network accurately predicts phenotypic effects of gene perturbation in Caenorhabditis elegans. Nat Genet. 2008;40:181–188. doi: 10.1038/ng.2007.70. [DOI] [PubMed] [Google Scholar]
- Lee Y, Lee DH, Kho CW, Lee AY, Jang M, Cho S, et al. Transthyretin-related proteins function to facilitate the hydrolysis of 5-hydroxyisourate, the end product of the uricase reaction. FEBS Lett. 2005;579:4769–4774. doi: 10.1016/j.febslet.2005.07.056. [DOI] [PubMed] [Google Scholar]
- Lesa GM, Palfreyman M, Hall DH, Clandinin MT, Rudolph C, Jorgensen EM, et al. Long chain polyunsaturated fatty acids are required for efficient neurotransmission in C. elegans. J Cell Sci. 2003;116:4965–4975. doi: 10.1242/jcs.00918. [DOI] [PubMed] [Google Scholar]
- Li BW, Wang Z, Rush AC, Mitreva M, Weil GJ. Transcription profiling reveals stage- and function-dependent expression patterns in the filarial nematode Brugia malayi. BMC Genomics. 2012;13:184. doi: 10.1186/1471-2164-13-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Buxbaum JN. Transthyretin and the brain re-visited: is neuronal synthesis of transthyretin protective in Alzheimer's disease? Mol Neurodegener. 2011;6:79. doi: 10.1186/1750-1326-6-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtenfels JR, Hoberg EP, Zarlenga DS. Systematics of gastrointestinal nematodes of domestic ruminants: advances between 1992 and 1995 and proposals for future research. Vet Parasitol. 1997;72:225–238. doi: 10.1016/s0304-4017(97)00099-x. discussion 238-45. [DOI] [PubMed] [Google Scholar]
- Lok JB. Nucleic acid transfection and transgenesis in parasitic nematodes. Parasitology. 2012;139:574–588. doi: 10.1017/S0031182011001387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin RJ, Murray I, Robertson AP, Bjørn H, Sangster N. Anthelmintics and ion-channels: after a puncture, use a patch. Int J Parasitol. 1998;28:849–862. doi: 10.1016/s0020-7519(98)00048-4. [DOI] [PubMed] [Google Scholar]
- McCarter JP, Mitreva MD, Martin J, Dante M, Wylie T, et al. Analysis and functional classification of transcripts from the nematode Meloidogyne incognita. Genome Biol. 2004;4:R26. doi: 10.1186/gb-2003-4-4-r26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKerrow JH, Caffrey C, Kelly B, Loke P, Sajid M. Proteases in parasitic diseases. Annual Rev Pathology. 2006;1:497–536. doi: 10.1146/annurev.pathol.1.110304.100151. [DOI] [PubMed] [Google Scholar]
- Mitreva M, McCarter JP, Martin J, Dante M, Wylie T, Chiapelli B, et al. Comparative genomics of gene expression in the parasitic and free-living nematodes Strongyloides stercoralis and Caenorhabditis elegans. Genome Res. 2004;14:209–220. doi: 10.1101/gr.1524804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyle M, Foster DL, McGrath DE, et al. A hookworm glycoprotein that inhibits neutrophil function is a ligand of the integrin CD11b/CD18. J Bio Chem. 1994;269:10008–10015. [PubMed] [Google Scholar]
- Nikolaou S, Gasser RB. Prospects for exploring molecular developmental processes in Haemonchus contortus. Int J Parasitol. 2006;36:859–868. doi: 10.1016/j.ijpara.2006.04.007. [DOI] [PubMed] [Google Scholar]
- Nisbet AJ, Cottee P, Gasser RB. Molecular biology of reproduction and development in parasitic nematodes: progress and opportunities. Int J Parasitol. 2004;34:125–138. doi: 10.1016/j.ijpara.2003.09.012. [DOI] [PubMed] [Google Scholar]
- Nisbet AJ, Cottee PA, Gasser RB. Genomics of reproduction in nematodes: prospects for parasite intervention? Trends Parasitol. 2008;24:89–95. doi: 10.1016/j.pt.2007.12.001. [DOI] [PubMed] [Google Scholar]
- Ondrovics M, Silbermayr K, Mitreva M, Young ND, Gasser RB, Joachim A. Proteomics elucidates key molecules involved in exsheathment in vitro in Oesophagostomum dentatum. Int J Parasitol. 2014;44:759–764. doi: 10.1016/j.ijpara.2014.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ondrovics M, Silbermayr K, Mitreva M, Young ND, Razzazi-Fazeli E, Gasser RB, et al. Proteomic analysis of Oesophagostomum dentatum (Nematoda) during larval transition, and the effects of hydrolase inhibitors on development. PLoS One. 2013;8:e63955. doi: 10.1371/journal.pone.0063955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono K, Ono S. Two actin-interacting protein 1 isoforms function redundantly in the somatic gonad and are essential for reproduction in Caenorhabditis elegans. Cytoskeleton (Hoboken) 2014;71:36–45. doi: 10.1002/cm.21152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page AP, Stepek G, Winter AD, Pertab D. Enzymology of the nematode cuticle: A potential drug target? Int J Parasitol Drugs Drug Resist. 2014;4:133–141. doi: 10.1016/j.ijpddr.2014.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson J, Mitreva M, Whitton C, Thomson M, Daub J, Martin J, et al. A transcriptomic analysis of the phylum Nematoda. Nat Genet. 2004;36:1259–1267. doi: 10.1038/ng1472. [DOI] [PubMed] [Google Scholar]
- Parra G, Bradnam K, Korf I. CEGMA: a pipeline to accurately annotate core genes in eukaryotic genomes. Bioinformatics. 2007;23:1061–1067. doi: 10.1093/bioinformatics/btm071. [DOI] [PubMed] [Google Scholar]
- Parra G, Bradnam K, Ning Z, Keane T, Korf I. Assessing the gene space in draft genomes. Nucleic Acids Res. 2009;37:289–297. doi: 10.1093/nar/gkn916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrish JZ, Yang C, Shen B, Xue D. CRN-1, a Caenorhabditis elegans FEN-1 homologue, cooperates with CPS-6/EndoG to promote apoptotic DNA degradation. EMBO J. 2003;22:3451–3460. doi: 10.1093/emboj/cdg320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pineda MA, Lumb F, Harnett MM, Harnett W. ES-62, a therapeutic anti-inflammatory agent evolved by the filarial nematode Acanthocheilonema viteae. Mol Biochem Parasitol. 2014;194:1–8. doi: 10.1016/j.molbiopara.2014.03.003. [DOI] [PubMed] [Google Scholar]
- Prichard R, Menez C, Lespine A. Moxidectin and the avermectins: Consanguinity but not identity. Int J Parasitol Drugs Drug Resist. 2012;2:134–153. doi: 10.1016/j.ijpddr.2012.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prufer K, Muetzel B, Do HH, Weiss G, Khaitovich P, Rahm E, et al. FUNC: a package for detecting significant associations between gene sets and ontological annotations. BMC Bioinformatics. 2007;8:41. doi: 10.1186/1471-2105-8-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahat O, Yitzhaky A, Schreiber G. Cluster conservation as a novel tool for studying protein-protein interactions evolution. Proteins. 2008;71:621–630. doi: 10.1002/prot.21749. [DOI] [PubMed] [Google Scholar]
- Ramazzina I, Folli C, Secchi A, Berni R, Percudani R. Completing the uric acid degradation pathway through phylogenetic comparison of whole genomes. Nat Chem Biol. 2006;2:144–148. doi: 10.1038/nchembio768. [DOI] [PubMed] [Google Scholar]
- Rhoads ML, Fetterer RH, Romanowski RD. A developmentally regulated hyaluronidase of Haemonchus contortus. J Parasitol. 2000;86:916–921. doi: 10.1645/0022-3395(2000)086[0916:ADRHOH]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Rieu P, Sugimori T, Griffith DL, Arnaout MA. Solvent-accessible residues on the metal ion-dependent adhesion site face of integrin CR3 mediate its binding to the neutrophil inhibitory factor. J Biol Chem. 1996;271:15858–15861. doi: 10.1074/jbc.271.27.15858. [DOI] [PubMed] [Google Scholar]
- Rowley AF, Vogan CL, Taylor GW, Clare AS. Prostaglandins in non-insectan invertebrates: recent insights and unsolved problems. J Exp Biol. 2005;208:3–14. doi: 10.1242/jeb.01275. [DOI] [PubMed] [Google Scholar]
- Samarasinghe B, Knox DP, Britton C. Factors affecting susceptibility to RNA interference in Haemonchus contortus and in vivo silencing of an H11 aminopeptidase gene. Int J Parasitol. 2011;41:51–59. doi: 10.1016/j.ijpara.2010.07.005. [DOI] [PubMed] [Google Scholar]
- Saverwyns H, Visser A, Van Durme J, Power D, Morgado I, Kennedy MW, et al. Analysis of the transthyretin-like (TTL) gene family in Ostertagia ostertagi--comparison with other strongylid nematodes and Caenorhabditis elegans. Int J Parasitol. 2008;38:1545–1556. doi: 10.1016/j.ijpara.2008.04.004. [DOI] [PubMed] [Google Scholar]
- Schwarz EM, Korhonen PK, Campbell BE, Young ND, Jex AR, Jabbar A, et al. The genome and developmental transcriptome of the strongylid nematode Haemonchus contortus. Genome Biol. 2013;14:R89. doi: 10.1186/gb-2013-14-8-r89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spindler LA. Development of the nodular worm, Oesophagostomum longicaudum, in the pig. J Agr Res. 1933;46:531–542. [Google Scholar]
- Stanley D. Prostaglandins and other eicosanoids in insects: biological significance. Ann Rev Entomol. 2006;51:25–44. doi: 10.1146/annurev.ento.51.110104.151021. [DOI] [PubMed] [Google Scholar]
- Tang YT, Gao X, Rosa BA, Abubucker S, Hallsworth-Pepin K, Martin J, et al. Genome of the human hookworm Necator americanus. Nat Genet. 2014;46:261–269. doi: 10.1038/ng.2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor CM, Martin J, Rao RU, Powell K, Abubucker S, Mitreva M. Using existing drugs as leads for broad spectrum anthelmintics targeting protein kinases. PLoS Pathog. 2013a;9:e1003149. doi: 10.1371/journal.ppat.1003149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor CM, Wang Q, Rosa BA, Huang SC-C, Powell K, Schedl T, et al. Discovery of anthelmintic drug targets and drugs using chokepoints in nematode metabolic pathways. PLoS Pathog. 2013b;9:e1003505. doi: 10.1371/journal.ppat.1003505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor M, Coop R, Wall R. Veterinary Parasitology. third. Oxford Liver Fluke: Blackwell Publishing; 2007. [Google Scholar]
- Vercauteren I, Geldhof P, Peelaers I, Claerebout E, Berx G, Vercruysse J. Identification of excretory-secretory products of larval and adult Ostertagia ostertagi by immunoscreening of cDNA libraries. Mol Biochem Parasitol. 2003;126:201–208. doi: 10.1016/s0166-6851(02)00274-8. [DOI] [PubMed] [Google Scholar]
- Wang X, Li W, Zhao D, Liu B, Shi Y, Chen B, et al. Caenorhabditis elegans transthyretinlike protein TTR-52 mediates recognition of apoptotic cells by the CED-1 phagocyte receptor. Nat Cell Biol. 2010;12:655–664. doi: 10.1038/ncb2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenk MR. Lipidomics of host-pathogen interactions. FEBS letters. 2006;580:5541–5551. doi: 10.1016/j.febslet.2006.07.007. [DOI] [PubMed] [Google Scholar]
- Zhong W, Sternberg PW. Genome-wide prediction of C. elegans genetic interactions. Science. 2006;311:1481–1484. doi: 10.1126/science.1123287. [DOI] [PubMed] [Google Scholar]
- Zhu XQ, Korhonen PK, Cai H, Young ND, Nejsum P, von Samson-Himmelstjerna G, et al. Genetic blueprint of the zoonotic pathogen Toxocara canis. Nat Commun. 2015;6:6145. doi: 10.1038/ncomms7145. [DOI] [PMC free article] [PubMed] [Google Scholar]