Abstract
Abiotic stresses such as high temperature, salinity, and drought adversely affect the survival, growth, and reproduction of plants. Plants respond to such unfavorable changes through developmental, physiological, and biochemical ways, and these responses require expression of stress-responsive genes, which are regulated by a network of transcription factors (TFs), including heat stress transcription factors (HSFs). HSFs play a crucial role in plants response to several abiotic stresses by regulating the expression of stress-responsive genes, such as heat shock proteins (Hsps). In this review, we describe the conserved structure of plant HSFs, the identification of HSF gene families from various plant species, their expression profiling under abiotic stress conditions, regulation at different levels and function in abiotic stresses. Despite plant HSFs share highly conserved structure, their remarkable diversification across plants reflects their numerous functions as well as their integration into the complex stress signaling and response networks, which can be employed in crop improvement strategies via biotechnological intervention.
Keywords: plant, heat stress, transcription factors, heat shock proteins, abiotic stress, transcriptional regulation
Introduction
Plants as sessile organisms are routinely confronted by a variety of abiotic or biotic stresses, such as water deficiency, high salt, extreme temperatures, chemical pollutants, oxidative stress, nematodes, herbivores, and pathogens (Al-Whaibi, 2011). Especially, abiotic stress is the primary cause of crop loss worldwide, reducing crop productivity by an estimated 50% annually (Wang et al., 2004). Unlike animals, plants could not change their sites to escape from the unfavorable stresses, but have attained certain adaptations to these rapidly changing stresses during evolution, such as the dominance of sporophyte that encloses the sensitive gametophyte, the presence of leaf epidermis with stomata for gas exchange, the formation of stress resistant dormant organs, and the presence of conducting tissues in long-lived and big plants for long-distance nutrient and water transport (Baniwal et al., 2004; Al-Whaibi, 2011). A network of interconnected cellular stress response systems is a prerequisite for plant survival and productivity (Scharf et al., 2012), and their understanding is important for developing new methods to enhance plant stress tolerance.
A complex stress response network and a wide array of mechanisms for adapting to plants' changing environments at the physiological, biochemical, and molecular levels increase the tolerance to the stresses (Bartels and Sunkar, 2005; Zhou et al., 2009; Nakashima et al., 2012). The phytohormone abscisic acid (ABA) produced under abiotic stress conditions, induces leaf stomata closure and triggers the activation of many stress-related genes, thus playing a key role in responses to abiotic stress factors (Lata and Prasad, 2011). With the molecular techniques such as microarray analysis and large-scale transcriptome analysis, a large array of abiotic stress responsive genes has been identified in plants (Fowler and Thomashow, 2002; Nakashima et al., 2009). These genes not only play a role in the protection of the cells from stress by the production of important enzymes and metabolic proteins (functional proteins) but also in regulating signal transduction and gene expression in the stress response (regulatory proteins; Lata and Prasad, 2011; Nakashima et al., 2012). Among the regulatory proteins, transcription factors (TFs) play a crucial role in the conversion of stress signal perception to stress-responsive gene expression by interacting with cis-acting elements present in the promoter region of various target stress-responsive genes in the signal transduction processes, thus activating signaling cascade whole network of genes that act together in enhancing plant tolerance to the harsh environmental conditions (Akhtar et al., 2012). In plant genomes, ~7% of the coding sequences are assigned to TFs and many of these often belong to large gene families compared with animals and yeasts, such as the heat stress transcription factors (HSFs) family (Baniwal et al., 2004; Udvardi et al., 2007).
Plant HSFs are the terminal components of a signal transduction chain mediating the expression of genes responsive to various abiotic stresses (Nover et al., 2001). Many studies have reported on the central roles of HSFs in various abiotic stresses, including heat stress (HS) (Scharf et al., 2012), however, most analyses of HSFs function in stress responses examine individual stresses, not a combination of abiotic stress factors. In natural conditions, plants are routinely subjected to a combination of different abiotic stresses, such as the combination of drought, heat, and salinity stresses (Sewelam et al., 2014). The response of plants to a combination of different abiotic stresses cannot be directly extrapolated from the response of plants to each of the different stresses applied individually, therefore it is crucial to characterize the acclimation responses of plants to a combination of abiotic stresses and identify multiple stress responsive genes (Mittler, 2006; Colmenero-Flores and Rosales, 2014). Comprehensive characterization of multifunctional HSFs will provide the basis for investigating their functions in plant abiotic stress responses. In this review, the focus will be on the recent progress of the roles of HSFs in abiotic stress responses, with an emphasis on HS. In addition, recent advances in characterization of HSFs regulation will be also discussed.
Structure and classification of plant HSFs
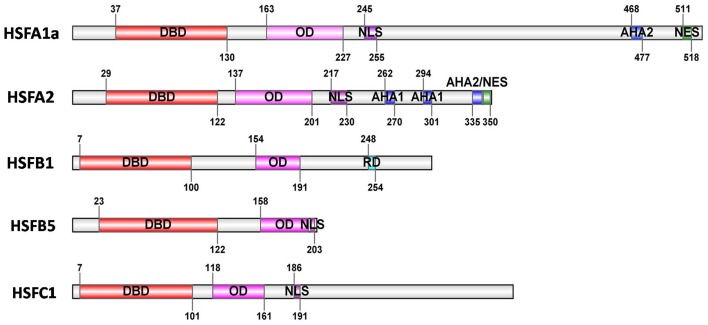
Typically, plant HSF proteins share a well conserved modular structure (Figure 1). The N-terminal DNA binding domain (DBD) is characterized by a central helix-turn-helix motif that specifically binds to the heat stress elements (HSEs) in the target promoters, and subsequently activates the transcription of stress-inducible genes (Baniwal et al., 2004; Sakurai and Enoki, 2010; Scharf et al., 2012). The oligomerization domain (OD) with a bipartite heptad pattern of hydrophobic amino acid residues (HR-A/B region) is connected to the DBD by a flexible linker (Baniwal et al., 2004). Based on the length of the flexible linker region between DBD and HR-A/B regions and the number of amino acid residues inserted into the HR-A/B regions, plant HSFs are classified into three classes, HSFA, B, and C (Nover et al., 2001; Kotak et al., 2004). The HR-A/B regions of HSFBs are compact and similar to all non-plant HSFs, however, members of class HSFA and C have an extended HR-A/B region due to an insertion of 21 (HSFAs) and 7 (HSFCs) amino acid residues between the HR-A and HR-B parts, respectively (Nover et al., 1996; Scharf et al., 2012). The C-terminal activation domains of plant HSFs are characterized by short peptide motifs (AHA motifs), which are crucial for the activator function in many cases (Döring et al., 2000). The AHA motifs formed of aromatic, large hydrophobic, and acidic amino acid residues, are HSFA-specific motifs but not found in class HSFB or C (Döring et al., 2000; Kotak et al., 2004). In addition, nuclear localization signal (NLS) and nuclear export signal (NES) of HSFs function in the assembly of a nuclear import complex built of the target protein and the receptor-mediated export in complex with the NES receptor exportin-α, respectively (Görlich and Kutay, 1999; Heerklotz et al., 2001; Baniwal et al., 2004). Notably, members of class HSFB (except HSFB5) comprise a characteristic tetrapeptide–LFGV–in the C-terminal domain, functioning as repressor domain (RD; Czarnecka-Verner et al., 2000; Ikeda and Ohme-Takagi, 2009; Fragkostefanakis et al., 2015).
Figure 1.
Basic structure of HSFs. The block diagrams represent five tomato HSFs with their conserved functional domains. The conserved domains are identified by Heatster (http://www.cibiv.at/services/hsf/). DBD, DNA binding domain; OD, oligomerization domain (HR-A/B region); NLS, nuclear localization signal; NES, nuclear export signal; AHA, activator motifs; RD, tetrapeptide motif–LFGV–as core of repressor domain. (Adapted from Scharf et al., 2012).
Identification of plant HSF families
Compared with few HSF members in vertebrates (4), Drosophila (1), Caenorhabditis elegans (1), and yeast (one HSF plus three HSF-related proteins; Nover et al., 1996; Nakai, 1999), plant HSF families comprise a large number of HSF members derived from a complex plant-specific superfamily and are present in a wide range of species. In the previous reports, the identification of the HSF family in plants was performed only in few model species such as Arabidopsis, tomato, and rice (Baniwal et al., 2004; Scharf et al., 2012). In recent years, based on the availability of an ever-increasing number of complete plant genomes and EST sequences, a large numbers of HSF families from more than 20 plant species have been identified at genome-wide scale. As shown in Table 1, there are 21 HSF encoding genes in Arabidopsis (Scharf et al., 2012), 24 in tomato (Scharf et al., 2012; Fragkostefanakis et al., 2015), 25 in pepper (Guo et al., 2015), 52 in soybean (Scharf et al., 2012), at least 56 in wheat (Xue et al., 2014), and so on. Compared with the HSF families of soybean, carrot (35 members) and cotton (40 members), the families of Arabidopsis and tomato are considered small. Currently, maximum of HSF genes were identified in wheat and soybean among monocots and eudicots, respectively. The multiplicity of HSFs in plants may be related to the gene duplications and whole-genome duplications at different points of evolution, followed by extensive gene loss (Scharf et al., 2012).
Table 1.
The HSF family in plant species.
| Species | Number of HSF family | References | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HSFA1 | HSFA2 | HSFA3 | HSFA4 | HSFA5 | HSFA6 | HSFA7 | HSFA8 | HSFA9 | HSFB1 | HSFB2 | HSFB3 | HSFB4 | HSFB5 | HSFC1 | HSFC2 | In total | ||
| Arabidopsis thaliana | 4 | 1 | 1 | 2 | 1 | 2 | 2 | 1 | 1 | 1 | 2 | 1 | 1 | 0 | 1 | 0 | 21 | Scharf et al., 2012 |
| Tomato (Solanum lycopersicum) | 4 | 1 | 1 | 3 | 1 | 2 | 1 | 1 | 1 | 1 | 2 | 2 | 2 | 1 | 1 | 0 | 24 | Scharf et al., 2012; Fragkostefanakis et al., 2015 |
| Castor bean (Ricinus communis) | 2 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 2 | 1 | 1 | 0 | 19 | Scharf et al., 2012 |
| Pepper (Capsicum annumm) | 3 | 1 | 1 | 3 | 1 | 3 | 0 | 1 | 4 | 1 | 2 | 2 | 1 | 1 | 1 | 0 | 25 | Guo et al., 2015 |
| Apple (Malus domestica) | 4 | 2 | 3 | 1 | 2 | 0 | 0 | 2 | 2 | 2 | 1 | 2 | 2 | 0 | 2 | 0 | 25 | Giorno et al., 2012 |
| Tea (Camellia sinensis) | 2 | 0 | 1 | 2 | 2 | 1 | 0 | 1 | 0 | 1 | 4 | 0 | 1 | 0 | 1 | 0 | 16 | Liu et al., 2016 |
| Soybean (Glycine max) | 5 | 3 | 4 | 4 | 2 | 3 | 3 | 2 | 2 | 4 | 6 | 2 | 8 | 2 | 2 | 0 | 52 | Scharf et al., 2012 |
| Cotton (Gossypium hirsutum) | 6 | 1 | 1 | 3 | 2 | 2 | 2 | 2 | 3 | 3 | 4 | 1 | 5 | 2 | 3 | 0 | 40 | Wang et al., 2014 |
| Chinese cabbage (Brassica rapa pekinensis) | 8 | 1 | 1 | 1 | 1 | 4 | 2 | 1 | 0 | 2 | 3 | 2 | 2 | 0 | 2 | 0 | 30 | Huang et al., 2015a |
| Poplar (Populus trichocarpa) | 3 | 1 | 1 | 3 | 2 | 2 | 2 | 2 | 1 | 1 | 3 | 2 | 4 | 2 | 1 | 0 | 27 | Scharf et al., 2012 |
| Carrot (Daucus carota) | 2 | 4 | 4 | 8 | 1 | 0 | 5 | 0 | 3 | 2 | 2 | 1 | 2 | 0 | 1 | 0 | 35 | Huang et al., 2015b |
| strawberry (Fragaria vesca) | 2 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 0 | 1 | 0 | 17 | Hu et al., 2015 |
| Willow (Salix suchowensis) | 3 | 1 | 1 | 3 | 1 | 2 | 2 | 2 | 1 | 1 | 2 | 1 | 4 | 2 | 1 | 0 | 27 | Zhang et al., 2015 |
| Chinese white pear (Pyrus bretschneideri) | 3 | 1 | 2 | 4 | 1 | 3 | 2 | 1 | 2 | 2 | 1 | 3 | 1 | 1 | 2 | 0 | 29 | Qiao et al., 2015 |
| Chinese plum (Prunus salicina) | 2 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 0 | 1 | 1 | 1 | 0 | 17 | Qiao et al., 2015 |
| Peach (Amygdalus persica) | 2 | 1 | 1 | 2 | 0 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 0 | 17 | Qiao et al., 2015 |
| European pear (Pyrus communis) | 4 | 3 | 2 | 4 | 1 | 2 | 2 | 2 | 2 | 2 | 3 | 2 | 0 | 2 | 2 | 0 | 33 | Qiao et al., 2015 |
| Maize (Zea mais) | 2 | 3 | 1 | 3 | 1 | 2 | 2 | 2 | 0 | 2 | 4 | 0 | 3 | 0 | 3 | 2 | 30 | Scharf et al., 2012 |
| Rice (Oryza sativa) | 1 | 3 | 1 | 2 | 1 | 2 | 2 | 1 | 0 | 1 | 3 | 0 | 4 | 0 | 2 | 2 | 25 | Scharf et al., 2012 |
| Wheat (Triticum aestivum) | 3 | 9 | 2 | 6 | 2 | 6 | 2 | 3 | 0 | 3 | 5 | 0 | 3 | 0 | 5 | 7 | 56 | Xue et al., 2014 |
| Millet (Sorghum bicolor) | 1 | 3 | 1 | 2 | 1 | 2 | 2 | 1 | 0 | 1 | 3 | 0 | 3 | 0 | 2 | 2 | 24 | Scharf et al., 2012 |
| Brachypodium (Brachypodium distachyon) | 1 | 3 | 1 | 2 | 1 | 2 | 2 | 1 | 0 | 1 | 3 | 0 | 3 | 0 | 2 | 2 | 24 | Scharf et al., 2012 |
Interestingly, among the 25 species listed in Table 1, including 20 eudicots and 5 monocots, members of subclass HSFA9, B3, and B5 were confined to the eudicots but not to the monocots, which emerged presumably after the split of monocots and eudicots. In addition, a variable number of the monocot-specific type HSFC2 genes (2–7 genes) are found in all 5 monocots, not in eudicots, attributing to gene duplications on the monocot lineage. Higher number of class HSFC genes are identified in monocots, such as in wheat, maximum of 5 and 7 genes are assigned into subclass HSFC1 and C2, respectively, which is the most marked difference between monocots and eudicots (Scharf et al., 2012). The large size of the plant HSFs family inevitable complicates the unraveling of their function under stress conditions.
Expression analysis of plant HSF genes
The role of plant HSFs in abiotic stresses, especially in HS, has been recently brought to light (Fragkostefanakis et al., 2015). Although mRNA levels cannot be used to draw immediate conclusions about protein levels, they can point out directions of further investigations (Scharf et al., 2012). Genome-wide expression profiling of plant HSF genes under different abiotic stresses has been investigated extensively in various species. Most plant HSFs are regulated by HS, including up- and down-regulation. Upon HS, the transcript levels of HSFA2 and A6 members became the dominant HSFs in wheat, suggesting an important regulatory role during HS (Xue et al., 2014). Among 23 rice OsHSF genes, 16 OsHSFs were up-regulated by two-folds (log2 value) in response to HS, including 8 genes up-regulated by two-folds only during early heat shock (HS for 10 min) and 8 genes up-regulated at both short (HS for 10 min) and prolong (HS for 30 min) HS treatment, however, OsHSFC1a was noted to be down-regulated by the early HS treatment (Mittal et al., 2009), similarly, many HSF genes from different plant species, such as GhHSF3, 18, 24, 32, 37, and 40 from cotton (Wang et al., 2014), ZmHSF-06, -10, -14, -20, and -21 from maize (Lin et al., 2011), MdHSFA9b and B4a/b from apple (Giorno et al., 2012) showed down-regulation under HS treatment. The expression of Arabidopsis HSFA2 was not detectable in control cell cultures but was detected strongly after HS treatment (Nover et al., 2001), and the similar situation also emerged in the expression profiles of pepper CaHSFA2 (Guo et al., 2015), maize ZmHSF-01 and ZmHSF-04 (HSFA2 group; Lin et al., 2011), apple MdHSFA2a and A2b (Giorno et al., 2012), and tomato SlHSFA2 (Mishra et al., 2002). The HS-dependent translocation of HSFA2 in Arabidopsis (Evrard et al., 2013) and tomato (Chan-Schaminet et al., 2009) and redox-dependent translocation of AtHSFA8 (Giesguth et al., 2015) from the cytosol to nucleus may play central roles in plant HS and oxidative stress responses. In addition, many other abiotic stresses like cold, salinity and drought, and phytohormones such as jasmonic acid (JA), abscisic acid (ABA), salicylic acid (SA), and ethylene (Et) also have been shown to regulate the expression of plant HSF genes (Hu et al., 2015; Huang et al., 2015b; Zhang et al., 2015). The different abiotic stresses and phytohormone signaling pathways are assumed to interact and share some common elements that formed as potential “node” for crosstalk (Akhtar et al., 2012). These plant HSF genes may act as cross-point or node connecting several pathways and simultaneously regulate abiotic and phytohormone signaling pathways.
Plant HSF genes are not only induced by stress response but also by development, cell differentiation, and proliferation. For example, expression of Arabidopsis AtHSFA2 gene increases during the process of callus formation and growth from root explants (Che et al., 2002). In addition, HSFA2 is more highly induced in tomato anther than in the other flower tissues, and further induced under both short and prolonged HS conditions, which is similar to its expression in leaves (Giorno et al., 2010). In rice, the expression of OsHSFA2a gene is highly stimulated by HS particularly in root and shoot tissues as well as during panicle and seed development, while OsHSFA7 and A9 show developing seed-specific expression, in a similar pattern with those of HSFA9 in sunflower and Arabidopsis (Chauhan et al., 2011; Scharf et al., 2012). These studies elaborate the border of conditions that are known to induce plant HSFs expression.
Regulation of plant HSF genes
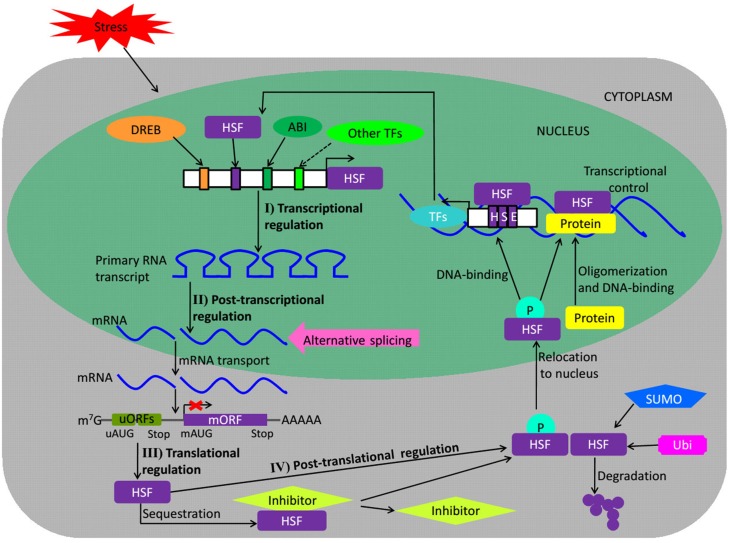
The studies on regulation of plant HSFs mainly focus on four levels including transcriptional, post-transcriptional, translational, and post-translation level (Fragkostefanakis et al., 2015). Transcription is the first step at which activity of a gene can be regulated by binding of specific TFs to the cis-acting elements located on the regulatory region of its promoter (Figure 2). The Arabidopsis AtHSFA1d and A1e binding to the HSE cluster in the 5′-flanking region of AtHSFA2 gene is involved in high light (HL)-inducible HSFA2 expression, activating AtHSFA2 transcription (Nishizawa-Yokoi et al., 2011). Under HS, the Arabidopsis dehydration-responsive element (DRE)-binding protein 2A (DREB2A gene) directly regulates AtHSFA3 transcription via binding the two DRE core elements in the AtHSFA3 promoter (Yoshida et al., 2008). As AtHSFA9 is exclusively expressed in late stages of seed development among the Arabidopsis family of 21 HSFs, a TF may be involved in the regulation of AtHSFA9 expression during seed development. Kotak et al. (2007) reported that ABSCISIC ACID–INSENSITIVE3 (ABI3 gene) could activate the AtHSFA9 promoter based on an RY/Sph motif (8-bp sequence, CATGCATG) as putative seed-related regulatory element in the AtHSFA9 promoter provided an essential binding site for ABI3. Interestingly, unlike Arabidopsis AtHSFA1d and A1e, AtHSFB1 and B2b are transcriptional repressors and negatively regulate the expression of HS-inducible HSFs including not only AtHSFA2 and A7a but also themselves (Ikeda et al., 2011).
Figure 2.
Regulation of HSF proteins. The scheme depicts the regulation of HSFs at different levels during stress. Upstream TFs like DREB, HSF, or ABI may bind to stress-related cis-regulatory elements in the promoter of regulated HSF genes and influence their transcription. Post-transcriptional control of HSFs by alternative splicing may also regulate their expression. The mature mRNAs are again governed during their transport and translation. uORFs regulate HSFs at the translation level. The translated protein may be subjected to activation by phosphorylation or undergo SUMO- and ubiquitin proteasomal-mediated degradation in response to certain environmental cues, other translated HSF proteins may be sequestrated by their inhibitors. Upon their nuclear import, the activated HSF proteins homo- or heterodimerize or bind to promoters of their target genes to control their expression. Broken arrows indicate possible but not firmly demonstrated routes. The red X mark represents translational repression. DREB, dehydration responsive element binding protein; ABI, ABSCISIC ACID–INSENSITIVE protein; TFs, transcription factors; AS, alternate splicing; mRNA, messenger RNA; m7G, cap of mRNA; uORFs, upstream micro open reading frames; mORF, major ORF; uAUG, AUG of uORF; mAUG, AUG of mORF; P, phosphate; SUMO, small ubiquitin-like modifier; Ubi, ubiquitination; HSE, heat stress element. (Adapted from Calkhoven and Ab, 1996; Puranik et al., 2012).
Alternative splicing is a widespread process in eukaryotes that generates two or more different transcripts from the same precursor mRNA molecule by using different splice sites (Guerra et al., 2015). The complex post-transcriptional regulation of HSFs involves alternative splicing during different biological processes (Fragkostefanakis et al., 2015). Alternative splicing induced by HS is observed for AtHSFA2, A4c, A7b, B1, and B2b in Arabidopsis. Arabidopsis AtHSFA2 derives from splicing of the conserved intron in the DBD, and a new heat stress-induced splice variant, AtHSFA2-III encodes a small truncated AtHSFA2 isoform (S-AtHSFA2), which can bind to the TATA box-proximal clusters of HSE in the AtHSFA2 promoter to activate its own transcription, attributing to exon skipping in the intron of the DBD encoding region (Sugio et al., 2009; Liu et al., 2013). The exon skipping pattern of Physcomitrella patens PpHSFA1-1 is similar to that of AtHSFA2, which reveals that heat regulation for alternative splicing evolved early during land colonization of green plants (Chang et al., 2014). The alternative splicing induced by HS is also observed for rice OsHSFA2d, which encodes two main splice variant proteins, OsHSFA2dI localized to the nucleus and OsHSFA2dII localized to the nucleus and cytoplasm, respectively. The transcriptionally inactive spliced form of OsHSFA2d, OsHSFA2dII, is the dominant under normal conditions; however, once the plant suffered from HS, OsHSFA2d is alternatively spliced into the transcriptionally active form, OsHSFA2dI, which participates in the HS response and the unfolded protein response by regulating expression of OsBiP1 (Cheng et al., 2015). Medicago sativa MsHSF1 is composed of four exons and three introns in the primary transcript and generates five splice transcript isoforms, including one spliced transcript MsHSF1b encoding an HSFA1 protein that can specifically bind to the HSEs in vitro and four low-abundant spliced transcripts carrying the premature termination codon (He et al., 2007). These results suggest that the regulation of plant HSFs at post-transcriptional level is diversified.
Recently investigation suggests that the regulation of plant HSFs at translational level is mainly controlled by upstream micro open reading frames (uORFs) in their 5′ untranslated regions (Figure 2; Jorgensen and Dorantes-Acosta, 2012; von Arnim et al., 2014; Fragkostefanakis et al., 2015). However, the information on uORFs of plant HSFs is mainly restricted to Arabidopsis. Zhu et al. (2012) reported that 7 members out of 21 Arabidopsis HSFs have at least one uORF, including AtHSFA1d, A1e, A2, A4a, B1, B2b, and C1, but only for the uORFs of AtHSFB1 and B2b there have been provided experimental evidence. The translation of AtHSFB1 is regulated by uORF2 but not by uORF1, whereas, neither uORFs of AtHSFB2b are involved in regulation of the main ORF translation. The uORF2 represses the translation of AtHSFB1 under normal condition, but the repression is deregulated under HS. The Arabidopsis HSF-like transcription factor TBF1, a major molecular switch for plant growth-to-defense transition, also contains two uORFs in the 5′ untranslated region. Unlike AtHSFB1, both uORFs of TBF1 have inhibitory effects on TBF1 translation, with the effect of uORF2 epistatic to that of uORF1. Both uORFs contain four phenylalanine (Phe) residues, and Phe starvation is shown to alleviate translational repression by the uORFs. Once plants are suffered from pathogen challenge, the uncharged tRNAPhe will temporary increase and the eukaryotic initiation factor 2α (eIF2α) phosphorylation will be triggered, which may facilitate ribosome reattachment to the TBF1 translation start codon downstream of uORFs and release the inhibitory effects of uORFs to initiate TBF1 translation (Jorgensen and Dorantes-Acosta, 2012; Pajerowska-Mukhtar et al., 2012). In general, not only abiotic but also biotic stresses are involved in the translational regulation of plant HSFs controlled by uORFs. However, the mechanism of plant HSFs' translational control via uORFs is still scarce and needs further investigation.
Plant HSFs also undergo intensive post-translational regulation included phosphorylation, ubiquitination, and Small Ubiquitin-like MOdifier (SUMO)-mediated degradation, oligomerization, and interaction with other non-HSF proteins (Figure 2; Scharf et al., 2012; Song et al., 2012). In Arabidopsis, the mitogen-activated protein kinase MAPK6 specifically targets the AtHSFA2, phosphorylates it on T249 and changes its intracellular localization under HS conditions (Evrard et al., 2013); AtHSFA4A interacts with the MAP kinases MPK3 and MPK6 and is phosphorylated in vitro on three distinct sites, and Ser-309 being the major phosphorylation site (Pérez-Salamó et al., 2014). Nishizawa-Yokoi et al. (2010) reported that AtHSFA2 was regulated by the accumulation of polyubiquitinated proteins generated by the inhibition of 26S proteasome and AtHsp90. AtSUMO1 physically interacts with AtHSFA2 at the main SUMOylation site Lys315, leading to the repression of its transcriptional activity and ultimately disrupting the acquired thermotolerance pattern in Arabidopsis (Cohen-Peer et al., 2010). In addition, Arabidopsis FK506-binding proteins (FKBPs), ROF1 (FKBP62), and ROF2 (FKBP65) (Meiri and Breiman, 2009; Meiri et al., 2010), HSF binding protein (AtHSBP; Satyal et al., 1998), and tomato Hsp17.4-II (Port et al., 2004) also act as negative regulators for HSFA2 transcriptional activity. Unfortunately, few active regulation factors involved in HSF regulation are found to date.
Function of plant HSFs in HS stress response
The major objective for agronomic research remains the enhancement of crop productivity under various abiotic stresses (Puranik et al., 2012). Among the major abiotic stresses, HS has an independent mode of action on the physiology and metabolism of plant cells, and has a negative effect on plant growth and development, which may lead to catastrophic loss of crop productivity and result in widespread famine (Bita and Gerats, 2013). To deal with the threat posed by HS, unraveling the independent action and biological consequences is important. Based on the role of central regulators of the HS response (Baniwal et al., 2004), plant HSFs may be used for gene manipulation, contriving tolerance to HS in crops, while characterization of the functional plant HSFs under HS condition is the precondition.
Based on the previous studies, most current information on plant HSFs function under HS condition is derived from HSFA1 and A2 in tomato and Arabidopsis. HSFA1 subfamily is defined as a master regulator of HS responses. Tomato HSFA1a has a unique function as master regulator for acquired thermotolerance, and cannot be replaced by any other HSFs (Mishra et al., 2002). However, no comparable master regulator activity could be identified for any of the four AtHSFA1 (a, b, d, and e) with single or multiple mutants, and the role of master regulator for thermotolerance is shared among the four paralogs due to functional redundancy (Table 2; Liu et al., 2011; Scharf et al., 2012; Fragkostefanakis et al., 2015). Over-expression of soybeans GmHSFA1 can enhance the thermotolerance of transgenic soybeans possibly due to the activation under HS of downstream genes, such as GmHsp70, GmHsp22, and other GmHsps (Table 2; Zhu et al., 2006). Based on its overall sequence (at the protein level) similarity to HSFA1s from other plant species (especially the well-characterized LpHSFA1) and its constitutive expression pattern, GmHSFA1 may be the best candidate of master regulator in soybeans, which needs to be confirmed by an antisense silencing study. HSFA2 has been identified to be the dominant HSF in tomato and Arabidopsis based on its high activator potential for transcription of Hsp genes and the strong accumulation under conditions of long-term HS or repeated cycles of HS and recovery (Mishra et al., 2002; von Koskull-Döring et al., 2007). HSFA2 and A1 form heterodimers resulting in synergistic transcriptional activation of HS genes after HSFA2 is accumulated in the nucleus of cells (Chan-Schaminet et al., 2009). Localization of the tomato HSFA2 protein to the nucleus evidently required interaction with HSFA1, whereas Arabidopsis HSFA2 protein can localize to the nucleus without interacting with the HSFA1 protein (Scharf et al., 1998; Kotak et al., 2004). Over-expression of Arabidopsis HSFA2 in the HSFA1 quadruple knock-out (hsfA1a, b, d, and e) mutant improved the thermotolerance, suggesting that HSFA2 can be active and functional in the absence of HSFA1s in Arabidopsis, and it is tempting to speculate that interactions between HSFA2 and other HSFs may exist in the quadruple knock-out mutants (Liu and Charng, 2013; Fragkostefanakis et al., 2015). Enhanced thermotolerance has also been obtained by ectopic expression of rice HSFA2e and lily HSFA2 in Arabidopsis (Table 2; Yokotani et al., 2008; Xin et al., 2010). In addition to the effects of HSFA1 and A2 members on the thermotolerance level, several other HSFA genes also function in the plant thermotolerance. For example, improved thermotolerance is observed in wheat plants over-expressing wheat TaHSFA6f, which relies on the concerted action of target genes, including TaHsps (TaHSP16.8, TaHSP17, TaHSP17.3, and TaHSP90.1-A1), TaRof1, galactinol synthase, and glutathione-S-transferase (GST; Xue et al., 2015); ectopic expression of tomato HSFA3 and wheat HSF3 in Arabidopsis also enhance its thermotolerance (Li et al., 2013; Zhang et al., 2013).
Table 2.
Overview of plant HSF genotypes and corresponding stress responses.
| Genotype | Gene | Source of gene | Stress responses | References |
|---|---|---|---|---|
| OVER-EXPRESSION | ||||
| AtHSFA1 | Arabidopsis | Increased thermotolerance in transgenic Arabidopsis | Lee et al., 1995 | |
| AtHSFA1b | Arabidopsis | Enhanced water productivity, resistance to drought in transgenic Arabidopsis | Bechtold et al., 2013 | |
| AtHSFA2 | Arabidopsis | Increased themotolerance, salt/osmotic stress tolerance, and enhanced callus growth of transgenic Arabidopsis | Ogawa et al., 2007 | |
| AtHSFA2 | Arabidopsis | Increased tolerance to combined environmental stresses (high-light and heat-shock stresses) in transgenic Arabidopsis | Nishizawa et al., 2006 | |
| AtHSFA2 | Arabidopsis | Enhanced anoxia tolerance in transgenic Arabidopsis | Banti et al., 2010 | |
| AtHSF3 | Arabidopsis | Conferred thermotolerance in transgenic Arabidopsis | Prändl et al., 1998 | |
| AtHSFB1 | Arabidopsis | Repressed expression of HSFA2, HSFA7a, HSFB2b, Hsp15.7CI under moderate heat conditions (28°C) in transgenic Arabidopsis | Ikeda et al., 2011 | |
| AtHSFB2a | Arabidopsis | Reduced biomass production in the early phase of growth and damaged development of female gametophytes in transgenic Arabidopsis | Wunderlich et al., 2014 | |
| LlHSFA1 | Lilium longiflorum | Interaction with LlHSFA2, enhanced thermotolerance in transgenic Arabidopsis | Gong et al., 2014 | |
| LlHSFA2 | Lilium longiflorum | Improved thermotolerance in transgenic Arabidopsis | Xin et al., 2010 | |
| OsHSFA2e | Oryza sativa | Enhanced thermotolerance and tolerance to high-salinity stress in transgenic Arabidopsis | Yokotani et al., 2008 | |
| GmHSFA1 | Glycine max | Enhanced thermotolerance in transgenic soybean | Zhu et al., 2006 | |
| BhHSF1 | Boea hygrometrica | Increased thermotolerance in transgenic Arabidopsis and tobaccos | Zhu et al., 2009 | |
| VpHSF1 | Vitis pseudoreticulata | Reduced the basal thermotolerance, increased acquired thermotolerance, reduced the tolerance to osmotic stress in transgenic tobacco | Peng et al., 2013 | |
| VvHSFA9 | Vitis vinifera | Positive modulation of seed germination and might negatively regulate flowering time of transgenic Arabidopsis | Li et al., 2015 | |
| SlHSFA1 | Solanum lycopersicum | Master regulator of thermotolerance in transgenic tomato | Mishra et al., 2002 | |
| SlHSFA3 | Solanum lycopersicum | Increased thermotolerance and salt hypersensitivity during seed germination in transgenic Arabidopsis | Li et al., 2013 | |
| TaHSF3 | Triticum aestivum | Enhanced tolerance to extreme temperatures in transgenic Arabidopsis | Zhang et al., 2013 | |
| TaHSFA4a | Triticum aestivum | Enhanced Cd tolerance by upregulating metallothionein gene expression in rice plants | Shim et al., 2009 | |
| TaHSFA6f | Triticum aestivum | Improved thermotolerance in transgenic wheat | Xue et al., 2015 | |
| CarHSFB2 | Cicer arietinum | Increased tolerance to drought and heat stress in transgenic Arabidopsis | Ma et al., 2016 | |
| HaHSFA4a and A9 | Helianthus annuus | Synergistic functional effected on tolerance to severe dehydration and to drastic oxidative stress in transgenic tobacco | Personat et al., 2014 | |
| MUTANT | ||||
| AtHSF1 and AtHSF3 | Arabidopsis | No obvious effects on the heat shock response in the individual mutant lines; double mutants were significantly impaired in HS gene expression | Lohmann et al., 2004 | |
| AtHSFA2 | Arabidopsis | The expression of AtHSFA2 was strictly heat stress-dependent and this transcription factor represented a regulator of a subset of stress response genes (Hsp26.5, Hsp25.3, Hsp70b, APX2, RD29A, RD17, GolS1, IPS2, KSC1, ERD7, and ZAT10) in Arabidopsis | Schramm et al., 2006 | |
| AtHSFA2 | Arabidopsis | AtHSFA2 knockout mutant showed an obvious phenotype, and was more sensitive to severe HS than the wild type after long but not short recovery periods. Acquired thermotolerance (AT) decayed faster in the absence of HSFA2. Hsa32 and class I small Hsp were less abundant in the mutant than in the wild type after long recovery. AtHSFA2 sustained the expression of Hsp genes and extended the duration of AT in Arabidopsis | Charng et al., 2007 | |
| AtHSFA2 | Arabidopsis | Heat-dependent acclimation to anoxia was lost in an HSFA2 knockout mutant | Banti et al., 2010 | |
| AtHSFB2a | Arabidopsis | Knockdown of asHSFB2a correlated with an improved biomass production early in vegetative development but with an impaired development of female gametophytes | Wunderlich et al., 2014 | |
| AtHSFA1a/A1b/A1d/ A1e | Arabidopsis | Members of the AtHSFA1 group not only played a pivotal role in HSR but also were involved in growth and development. The basal and acquired thermotolerance capacity was dramatically decreased in the QK mutant but varied in triple KO mutants at different developmental stages. Increased sensitive phenotype of the QK mutant to H2O2, salt and mannitol stresses | Liu et al., 2011 | |
| AtHSFA1a/A1b/A1d/ A1e | Arabidopsis | Constitutive expression of AtHSFA2 rescued the developmental defects of the QK mutant and promoted callus formation in A2QK, but not in A2Wt, after heat treatment. Ectopic expression of AtHSFA2 complemented the defects of QK in tolerance to different heat stress regimes, and to hydrogen peroxide, but not to salt and osmotic stresses, which revealed the overlapping and distinct functions of class A1 and A2 HSFs in Arabidopsis | Liu et al., 2013 | |
| AtHSFA1d and A1e | Arabidopsis | Double knockout mutant significantly suppressed the induction of HSFA2 expression in response to HL and heat shock (HS) stress; HSFA7a, A7b, B1, and B2a were down-regulated compared with those in the wild-type plants under HL stress. The PSII activity of double mutants decreased under HL stress, and double knockout impaired tolerance to HS stress | Nishizawa-Yokoi et al., 2011 | |
| AtHSFB1 and B2b | Arabidopsis | In double mutant plants, the expression of a large number of heat-inducible genes was enhanced in the non-heat condition (23°C) and the plants exhibited slightly higher heat tolerance at 42°C than the wild type; expression of the heat-inducible HSF genes remained consistently higher in mutant than in the wild type under extended heat stress conditions. HSFB1 and B2b appeared to be necessary for the expression of heat stress-inducible heat shock protein genes under heat stress conditions, which was necessary for acquired thermotolerance | Ikeda et al., 2011 | |
| OsHSFA4a | Oryza sativa | Cd tolerance was decreased in rice plants with knocked-down expression of OsHSFA4a | Shim et al., 2009 | |
At, Arabidopsis thaliana; Ll, Lilium longiflorum; Os, Oryza sativa; Gm, Glycine max; Bh, Boea hygrometrica; Vp, Vitis pseudoreticulata; Vv, Vitis vinifera; Sl, Solanum lycopersicum; Ta, Triticum aestivum; Car, Cicer arietinum; Ha, Helianthus annuus; HSR, heat shock response; Wt, wild type; KO, knock-out; QK, quadruple KO; HL, high light; Cd, cadmium; asHSFB2a, a natural long non-coding antisense RNA; APX2, ascorbate peroxidase 2; RD29A and RD17, cold- and drought-regulated genes; GolS1, a galactinol synthase; IPS2, a myo-inositol-1-phosphate synthase; KSC1, a ketoacyl-synthase; ERD7, an ethylene responsive protein; ZAT10, a salt tolerance zinc finger transcription factor.
In contrast to HSFAs, HSFBs have no transcriptional activity on their own due to lack of an activator domain. The HS-induced tomato HSFB1 was suggested to be coactivator of HSFA1a by assembling into an enhanceosome-like complex resulting in the strong synergistic activation of reporter gene expression (Fragkostefanakis et al., 2015). The coactivator function of HSFB1 depends on the recruitment of the plant CREB binding protein (CBP) ortholog histone acetyl transferase HAC1 (von Koskull-Döring et al., 2007). Tomato HSFA1a, A2, and B1 form a triad of functionally interacting HSFs that is responsible for the transcriptional level of HS responsive genes during plant HS response and recovery (Perez et al., 2009; Scharf et al., 2012). However, HSFB1 from Arabidopsis was inactive as coactivator due to the essential histone-like motif GRGKMMK with an invariant Lys residue (underlined) in tomato HSFB1 is replaced by GSRMTETK in Arabidopsis HSFB1 (Bharti et al., 2004). Interestingly, HSFB1 from Arabidopsis is characterized as a repressor of HS-inducible HSFs, such as HSFA2, A7a, B1, and B2b, however, the hsfb1, hsfb2b knockout mutant plants exhibit lower acquired thermotolerance than the wild type. This suggests that HSFB1 and HSFB2b may promote the activity of HSFA1 under HS conditions by repressing Hsps that interfere with the nuclear migration of HSFA1s, an activator of the early HS response (Ikeda et al., 2011). Over-expression of VpHSF1 (a member of class HSFB2 family) from Chinese Wild Vitis pseudoreticulata in tobacco demonstrated that VpHSF1 acted as a negative regulator in basal thermotolerance and a positive regulator in acquired thermotolerance (Peng et al., 2013). The above results indicate striking species-specific deviation in the functional diversification of some members of the HSF family (von Koskull-Döring et al., 2007).
Function of plant HSFs in other abiotic stress responses
Under natural conditions, plants frequently suffer from various abiotic stresses simultaneously; HS is compounded by additional abiotic stresses such as drought and salt stress (Bita and Gerats, 2013). The response of plant cells encountering a single stress condition can not reflect the real conditions in the field (Nishizawa et al., 2006). Gene manipulation of HSFs in plants is a significant approach to ameliorate the effects of combined HS and other abiotic stresses. Characterization of the functional HSFs involved in various abiotic stresses is necessary. The Arabidopsis HSFA1s are involved in response and tolerance to salt, osmotic, and oxidative stresses during seedling establishment (Liu et al., 2011). Especially, Arabidopsis HSFA1b controls a developmental component to drought tolerance and water productivity, however, the effect of HSFA1b over-expression on drought/dehydration tolerance does not involve changes in the expression of DREB2A or many other ABA- or dehydration-responsive genes (Bechtold et al., 2013). Given that Arabidopsis HSFA3 is regulated by DREB2A as part of drought stress signaling pathway (Scharf et al., 2012), it is tempting to speculate that Arabidopsis HSFA1b and A3 involve in different signal pathways to enhance the tolerance to drought stress. In addition, over-expression of chickpea CarHSFB2 in Arabidopsis can increase the transcript levels of some stress-responsive genes (RD22, RD26, and RD29A) at seedling stage under drought stress conditions, thus improving their drought-tolerance (Ma et al., 2016); co-overexpression of sunflower HaHSFA4a and A9 in transgenic tobacco results in synergistic effects on seedling tolerance to severe dehydration and oxidative stress (Personat et al., 2014). As the dominant HSF in thermotolerant cells, HSFA2 also enhances tolerance to various other abiotic stresses, including salt/osmotic stress (Ogawa et al., 2007; Yokotani et al., 2008), anoxia stress (Banti et al., 2010), and combined high-light (HL) and HS stresses (Nishizawa et al., 2006). Unlike the above active regulation factors, tomato SlHSFA3 and V. pseudoreticulata VpHSF1 play negative roles in salt and osmotic stress, respectively (Li et al., 2013; Peng et al., 2013). These results suggest that the complex family of plant HSFs presents a functional diversity under different abiotic stress conditions.
Conclusion and perspectives
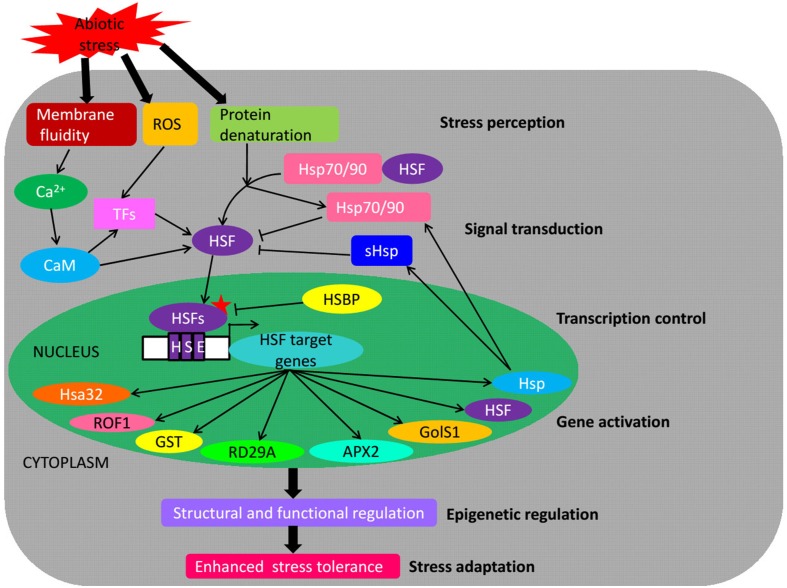
Understanding the molecular mechanisms of plants response to abiotic stresses such as heat, drought and salinity is a prerequisite for the manipulation of plants to improve stress tolerance and productivity. In response to these stresses, many genes are regulated mainly by TFs, and their gene products function in providing stress tolerance to plants (Lata and Prasad, 2011). One such class of the plant TFs is HSF that binds to HSE cis-acting elements in promoters of stress-inducible genes and plays central roles in the acquisition of plant tolerance against abiotic stresses. In this review, we have described the conserved structure of plant HSFs, the HSF gene families from various plant species based on the genome-wide identification, their expression profiling, different regulation levels and function in abiotic stresses. Plant HSF genes are important TFs that regulate the expression of various stress-responsive genes and play a key role in providing tolerance to multifarious abiotic stresses (Figure 3).
Figure 3.
Schematic representation of HSFs as key components in transcriptional regulatory networks during abiotic stress. The scheme integrates both positive (arrows) and negative (bars) regulatory mechanisms. Abiotic stresses provoke a rise of cytoplasmic calcium, ROS accumulation and proteins denaturation inside the cells which convey stress-induced signals to responding genes, directly targeting HSF proteins marked with an asterisk. HSFs induce the activation of various genes playing a central role under abiotic stress conditions, thereby enhancing the abiotic stress tolerance. ROS, reactive oxygen species; CaM, Ca2+–calmodulin; TFs, transcription factors; Hsp, heat shock protein; sHsp, small Hsp; HSE, heat stress element; Hsa32, heat stress- associated 32-kD protein; Rof1, FK506-binding proteins; GST, glutathione-S-transferase; RD29A, drought-regulated gene 29A; APX2, ascorbate peroxidase 2; GolS1, a galactinol synthase; HSBP, HSF binding protein.
HSFs can be employed to engineer transgenic plants with higher tolerance to environmental stresses; however, many important questions should be addressed. The role of HSF genes in plants, especially in important agricultural crops needs a better understanding to minimize their negative effects in transgenic plants. For example, over-expressing VpHSF1 in tobacco not only increased the acquired thermotolerance but also reduced the basal thermotolerance and the tolerance to osmotic stress (Table 2; Peng et al., 2013); over-expression of tomato SlHSFA3 increased thermotolerance of transgenic Arabidopsis, but played a negative role in controlling seed germination under salt stress (Li et al., 2013). Because HSFs and chaperones play the broader role in cellular homeostasis, manipulation of HSFs may disrupt the homeostasis, leading to pleiotropic and undesired effects (Cabello et al., 2014; Fragkostefanakis et al., 2015). Although great progress has been achieved in the characterization of class HSFAs, the biological functions of HSFB and C members, and the HSFs active regulation factors remain to be clarified. Therefore, there is a dire need to understand the exact regulatory mechanisms of all the stress-responsive HSF genes. Most experiments on the role of HSFs in abiotic stress responses are limited to several model plants in laboratory conditions addressing individually abiotic stresses, which cannot represent precisely field conditions. As there is functional divergency between HSF orthologs in different plant species, it is necessary to adjust the research direction of HSFs function from few model plants to a broader variety of plant species, including the desired agricultural crops. In addition, marker-assisted selection can accelerate traditional crop breeding for stress tolerance traits, but decision of HSFs as candidate genes and developing proper functional markers has to be carefully decided due to the implication of HSFs in various developmental and stress response aspects (Fragkostefanakis et al., 2015).
In the future, a combination of advanced high throughput technologies, such as microarray, genomics, and proteomic approaches in various developmental stages and stress conditions will provide us with critical information to elucidate the whole complexity of HSFs integrated abiotic stress responses and different signaling pathways. Further studies are necessary to be focused on the functions of HSFs in agricultural crops under harsh field conditions, the dual (positive or negative) role of HSFs in different stress conditions and establishment of an HSF network in relation to the crosstalk between abiotic stress responses and plant growth, development and metabolism, which may provide practical and biotechnological approaches to improve the crop plants tolerance to extreme environment conditions.
Author contributions
MG, ML, and ZG conceived and designed the paper; MG, JL, XM, and DL collected and analyzed the literature; MG wrote the paper.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (Grant No. 31272163, 31572114), the Shaanxi Agriculture Science and Technology Projects (Grant No. 2014K01-14-01), the Basic Fund for Scientific Research of Northwest A&F University (Grant No. 2452015141), the Opening Fund of Key Laboratory for Crop Biotechnology of Xinjiang Uygur Autonomous Region (Grant No. XJYS0302-2014-03) and the Tang Zhongying Fund for Breeding of Northwest A&F University.
References
- Akhtar M., Jaiswal A., Taj G., Jaiswal J. P., Qureshi M. I., Singh N. K. (2012). DREB1/CBF transcription factors: their structure, function and role in abiotic stress tolerance in plants. J. Genet. 91, 385–395. 10.1007/s12041-012-0201-3 [DOI] [PubMed] [Google Scholar]
- Al-Whaibi M. H. (2011). Plant heat-shock proteins: a mini review. J. King Saud Univ.-Sci. 23, 139–150. 10.1016/j.jksus.2010.06.022 [DOI] [Google Scholar]
- Baniwal S. K., Bharti K., Chan K. Y., Fauth M., Ganguli A., Kotak S., et al. (2004). Heat stress response in plants: a complex game with chaperones and more than twenty heat stress transcription factors. J. Biosci. 29, 471–487. 10.1007/BF02712120 [DOI] [PubMed] [Google Scholar]
- Banti V., Mafessoni F., Loreti E., Alpi A., Perata P. (2010). The heat-inducible transcription factor HsfA2 enhances anoxia tolerance in Arabidopsis. Plant Physiol. 152, 1471–1483. 10.1104/pp.109.149815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartels D., Sunkar R. (2005). Drought and salt tolerance in plants. Crit. Rev. Plant Sci. 24, 23–58. 10.1080/07352680590910410 [DOI] [Google Scholar]
- Bechtold U., Albihlal W. S., Lawson T., Fryer M. J., Sparrow P. A., Richard F., et al. (2013). Arabidopsis HEAT SHOCK TRANSCRIPTION FACTORA1b overexpression enhances water productivity, resistance to drought, and infection. J. Exp. Bot. 64, 3467–3481. 10.1093/jxb/ert185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bharti K., von Koskull-Döring P., Bharti S., Kumar P., Tintschl-Körbitzer A., Treuter E., et al. (2004). Tomato heat stress transcription factor HsfB1 represents a novel type of general transcription coactivator with a histone-like motif interacting with the plant CREB binding protein ortholog HAC1. Plant Cell 16, 1521–1535. 10.1105/tpc.019927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bita C. E., Gerats T. (2013). Plant tolerance to high temperature in a changing environment: scientific fundamentals and production of heat stress-tolerant crops. Front. Plant Sci. 4:273. 10.3389/fpls.2013.00273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabello J. V., Lodeyro A. F., Zurbriggen M. D. (2014). Novel perspectives for the engineering of abiotic stress tolerance in plants. Curr. Opin. Biotechnol. 26, 62–70. 10.1016/j.copbio.2013.09.011 [DOI] [PubMed] [Google Scholar]
- Calkhoven C., Ab G. (1996). Multiple steps in the regulation of transcription-factor level and activity. Biochem. J. 317, 329–342. 10.1042/bj3170329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C. Y., Lin W. D., Tu S. L. (2014). Genome-wide analysis of heat-sensitive alternative splicing in Physcomitrella patens. Plant Physiol. 165, 826–840. 10.1104/pp.113.230540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan-Schaminet K. Y., Baniwal S. K., Bublak D., Nover L., Scharf K. D. (2009). Specific interaction between tomato HsfA1 and HsfA2 creates hetero-oligomeric superactivator complexes for synergistic activation of heat stress gene expression. J. Biol. Chem. 284, 20848–20857. 10.1074/jbc.M109.007336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charng Y. Y., Liu H. C., Liu N. Y., Chi W. T., Wang C. N., Chang S. H., et al. (2007). A heat-inducible transcription factor, HsfA2, is required for extension of acquired thermotolerance in Arabidopsis. Plant Physiol. 143, 251–262. 10.1104/pp.106.091322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauhan H., Khurana N., Agarwal P., Khurana P. (2011). Heat shock factors in rice (Oryza sativa L.): genome-wide expression analysis during reproductive development and abiotic stress. Mol. Genet. Genomics 286, 171–187. 10.1007/s00438-011-0638-8 [DOI] [PubMed] [Google Scholar]
- Che P., Gingerich D. J., Lall S., Howell S. H. (2002). Global and hormone-induced gene expression changes during shoot development in Arabidopsis. Plant Cell 14, 2771–2785. 10.1105/tpc.006668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Q., Zhou Y., Liu Z., Zhang L., Song G., Guo Z., et al. (2015). An alternatively spliced heat shock transcription factor, OsHSFA2dI, functions in the heat stress-induced unfolded protein response in rice. Plant Biol. 17, 419–429. 10.1111/plb.12267 [DOI] [PubMed] [Google Scholar]
- Cohen-Peer R., Schuster S., Meiri D., Breiman A., Avni A. (2010). Sumoylation of Arabidopsis heat shock factor A2 (HsfA2) modifies its activity during acquired thermotholerance. Plant Mol. Biol. 74, 33–45. 10.1007/s11103-010-9652-1 [DOI] [PubMed] [Google Scholar]
- Colmenero-Flores J. M., Rosales M. A. (2014). Interaction between salt and heat stress: when two wrongs make a right. Plant Cell Environ. 37, 1042–1045. 10.1111/pce.12229 [DOI] [PubMed] [Google Scholar]
- Czarnecka-Verner E., Yuan C. X., Scharf K. D., Englich G., Gurley W. B. (2000). Plants contain a novel multi-member class of heat shock factors without transcriptional activator potential. Plant Mol. Biol. 43, 459–471. 10.1023/A:1006448607740 [DOI] [PubMed] [Google Scholar]
- Döring P., Treuter E., Kistner C., Lyck R., Chen A., Nover L. (2000). The role of AHA motifs in the activator function of tomato heat stress transcription factors HsfA1 and HsfA2. Plant Cell 12, 265–278. 10.1105/tpc.12.2.265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evrard A., Kumar M., Lecourieux D., Lucks J., von Koskull-Döring P., Hirt H. (2013). Regulation of the heat stress response in Arabidopsis by MPK6-targeted phosphorylation of the heat stress factor HsfA2. Peer J. 1, e59. 10.7717/peerj.59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler S., Thomashow M. F. (2002). Arabidopsis transcriptome profiling indicates that multiple regulatory pathways are activated during cold acclimation in addition to the CBF cold response pathway. Plant Cell 14, 1675–1690. 10.1105/tpc.003483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fragkostefanakis S., Röth S., Schleiff E., Scharf K. D. (2015). Prospects of engineering thermotolerance in crops through modulation of heat stress transcription factor and heat shock protein networks. Plant Cell Environ. 38, 1881–1895. 10.1111/pce.12396 [DOI] [PubMed] [Google Scholar]
- Giesguth M., Sahm A., Simon S., Dietz K. J. (2015). Redox-dependent translocation of the heat shock transcription factor AtHSFA8 from the cytosol to the nucleus in Arabidopsis thaliana. FEBS Lett. 589, 718–725. 10.1016/j.febslet.2015.01.039 [DOI] [PubMed] [Google Scholar]
- Giorno F., Guerriero G., Baric S., Mariani C. (2012). Heat shock transcriptional factors in Malus domestica: identification, classification and expression analysis. BMC Genomics 13:639. 10.1186/1471-2164-13-639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giorno F., Wolters-Arts M., Grillo S., Scharf K. D., Vriezen W. H., Mariani C. (2010). Developmental and heat stress-regulated expression of HsfA2 and small heat shock proteins in tomato anthers. J. Exp. Bot. 61, 453–462. 10.1093/jxb/erp316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong B., Yi J., Wu J., Sui J., Khan M. A., Wu Z., et al. (2014). LlHSFA1, a novel heat stress transcription factor in lily (Lilium longiflorum), can interact with LlHSFA2 and enhance the thermotolerance of transgenic Arabidopsis thaliana. Plant Cell Rep. 33, 1519–1533. 10.1007/s00299-014-1635-2 [DOI] [PubMed] [Google Scholar]
- Görlich D., Kutay U. (1999). Transport between the cell nucleus and the cytoplasm. Annu. Rev. Cell Dev. Biol. 15, 607–660. 10.1146/annurev.cellbio.15.1.607 [DOI] [PubMed] [Google Scholar]
- Guerra D., Crosatti C., Khoshro H. H., Mastrangelo A. M., Mica E., Mazzucotelli E. (2015). Post-transcriptional and post-translational regulations of drought and heat response in plants: a spider's web of mechanisms. Front Plant Sci. 6:57. 10.3389/fpls.2015.00057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo M., Lu J. P., Zhai Y. F., Chai W. G., Gong Z. H., Lu M. H. (2015). Genome-wide analysis, expression profile of heat shock factor gene family (CaHsfs) and characterisation of CaHsfA2 in pepper (Capsicum annuum L.). BMC Plant Biol. 15:151. 10.1186/s12870-015-0512-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Z. S., Xie R., Zou H. S., Wang Y. Z., Zhu J. B., Yu G. Q. (2007). Structure and alternative splicing of a heat shock transcription factor gene, MsHSF1, in Medicago sativa. Biochem. Biophys. Res. Commun. 364, 1056–1061. 10.1016/j.bbrc.2007.10.131 [DOI] [PubMed] [Google Scholar]
- Heerklotz D., Döring P., Bonzelius F., Winkelhaus S., Nover L. (2001). The balance of nuclear import and export determines the intracellular distribution and function of tomato heat stress transcription factor HsfA2. Mol. Cell. Biol. 21, 1759–1768. 10.1128/MCB.21.5.1759-1768.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y., Han Y. T., Wei W., Li Y. J., Zhang K., Gao Y. R., et al. (2015). Identification, isolation, and expression analysis of heat shock transcription factors in the diploid woodland strawberry Fragaria vesca. Front. Plant Sci. 6:736. 10.3389/fpls.2015.00736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X. Y., Tao P., Li B. Y., Wang W. H., Yue Z. C., Lei J. L., et al. (2015a). Genome-wide identification, classification, and analysis of heat shock transcription factor family in Chinese cabbage (Brassica rapa pekinensis). Genet. Mol. Res. 14, 2189–2204. 10.4238/2015.March.27.5 [DOI] [PubMed] [Google Scholar]
- Huang Y., Li M. Y., Wang F., Xu Z. S., Huang W., Wang G. L., et al. (2015b). Heat shock factors in carrot: genome-wide identification, classification, and expression profiles response to abiotic stress. Mol. Biol. Rep. 42, 893–905. 10.1007/s11033-014-3826-x [DOI] [PubMed] [Google Scholar]
- Ikeda M., Mitsuda N., Ohme-Takagi M. (2011). Arabidopsis HsfB1 and HsfB2b act as repressors of the expression of heat-inducible Hsfs but positively regulate the acquired thermotolerance. Plant Physiol. 157, 1243–1254. 10.1104/pp.111.179036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda M., Ohme-Takagi M. (2009). A novel group of transcriptional repressors in Arabidopsis. Plant Cell Physiol. 50, 970–975. 10.1093/pcp/pcp048 [DOI] [PubMed] [Google Scholar]
- Jorgensen R. A., Dorantes-Acosta A. E. (2012). Conserved peptide upstream open reading frames are associated with regulatory genes in angiosperms. Front. Plant Sci. 3:191. 10.3389/fpls.2012.00191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotak S., Port M., Ganguli A., Bicker F., Koskull−Döring V. (2004). Characterization of C-terminal domains of Arabidopsis heat stress transcription factors (Hsfs) and identification of a new signature combination of plant class A Hsfs with AHA and NES motifs essential for activator function and intracellular localization. Plant J. 39, 98–112. 10.1111/j.1365-313X.2004.02111.x [DOI] [PubMed] [Google Scholar]
- Kotak S., Vierling E., Bäumlein H., von Koskull-Döring P. (2007). A novel transcriptional cascade regulating expression of heat stress proteins during seed development of Arabidopsis. Plant Cell 19, 182–195. 10.1105/tpc.106.048165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lata C., Prasad M. (2011). Role of DREBs in regulation of abiotic stress responses in plants. J. Exp. Bot. 62, 4731–4748. 10.1093/jxb/err210 [DOI] [PubMed] [Google Scholar]
- Lee J. H., Hübel A., Schöffl F. (1995). Derepression of the activity of genetically engineered heat shock factor causes constitutive synthesis of heat shock proteins and increased thermotolerance in transgenic Arabidopsis. Plant J. 8, 603–612. 10.1046/j.1365-313X.1995.8040603.x [DOI] [PubMed] [Google Scholar]
- Li Z., Tian Y., Zhao W., Xu J., Wang L., Peng R., et al. (2015). Functional characterization of a grape heat stress transcription factor VvHsfA9 in transgenic Arabidopsis. Acta Physiol. Plant 37, 1–10. 10.1007/s11738-015-1884-x [DOI] [Google Scholar]
- Li Z., Zhang L., Wang A., Xu X., Li J. (2013). Ectopic overexpression of SlHsfA3, a heat stress transcription factor from tomato, confers increased thermotolerance and salt hypersensitivity in germination in transgenic Arabidopsis. PLoS ONE 8:e54880. 10.1371/journal.pone.0054880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y. X., Jiang H. Y., Chu Z. X., Tang X. L., Zhu S. W., Cheng B. J. (2011). Genome-wide identification, classification and analysis of heat shock transcription factor family in maize. BMC Genomics 12:76. 10.1186/1471-2164-12-76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H. C., Charng Y. Y. (2013). Common and distinct functions of Arabidopsis class A1 and A2 heat shock factors in diverse abiotic stress responses and development. Plant Physiol. 163, 276–290. 10.1104/pp.113.221168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H. C., Liao H. T., Charng Y. Y. (2011). The role of class A1 heat shock factors (HSFA1s) in response to heat and other stresses in Arabidopsis. Plant Cell Environ. 34, 738–751. 10.1111/j.1365-3040.2011.02278.x [DOI] [PubMed] [Google Scholar]
- Liu J., Sun N., Liu M., Liu J., Du B., Wang X., et al. (2013). An autoregulatory loop controlling Arabidopsis HsfA2 expression: role of heat shock-induced alternative splicing. Plant Physiol. 162, 512–521. 10.1104/pp.112.205864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z. W., Wu Z. J., Li X. H., Huang Y., Li H., Wang Y. X., et al. (2016). Identification, classification, and expression profiles of heat shock transcription factors in tea plant (Camellia sinensis) under temperature stress. Gene 576, 52–59. 10.1016/j.gene.2015.09.076 [DOI] [PubMed] [Google Scholar]
- Lohmann C., Eggers-Schumacher G., Wunderlich M., Schöffl F. (2004). Two different heat shock transcription factors regulate immediate early expression of stress genes in Arabidopsis. Mol. Genet. Genomics 271, 11–21. 10.1007/s00438-003-0954-8 [DOI] [PubMed] [Google Scholar]
- Ma H., Wang C., Yang B., Cheng H., Wang Z., Mijiti A., et al. (2016). CarHSFB2, a class B heat shock transcription factor, is involved in different developmental processes and various stress responses in chickpea (Cicer arietinum L.). Plant Mol. Biol. Report. 34, 1–14. 10.1007/s11105-015-0892-8 [DOI] [Google Scholar]
- Meiri D., Breiman A. (2009). Arabidopsis ROF1 (FKBP62) modulates thermotolerance by interacting with HSP90.1 and affecting the accumulation of HsfA2-regulated sHSPs. Plant J. 59, 387–399. 10.1111/j.1365-313X.2009.03878.x [DOI] [PubMed] [Google Scholar]
- Meiri D., Tazat K., Cohen-Peer R., Farchi-Pisanty O., Aviezer-Hagai K., Avni A., et al. (2010). Involvement of Arabidopsis ROF2 (FKBP65) in thermotolerance. Plant Mol. Biol. 72, 191–203. 10.1007/s11103-009-9561-3 [DOI] [PubMed] [Google Scholar]
- Mishra S. K., Tripp J., Winkelhaus S., Tschiersch B., Theres K., Nover L., et al. (2002). In the complex family of heat stress transcription factors, HsfA1 has a unique role as master regulator of thermotolerance in tomato. Genes Dev. 16, 1555–1567. 10.1101/gad.228802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittal D., Chakrabarti S., Sarkar A., Singh A., Grover A. (2009). Heat shock factor gene family in rice: genomic organization and transcript expression profiling in response to high temperature, low temperature and oxidative stresses. Plant Physiol. Biochem. 47, 785–795. 10.1016/j.plaphy.2009.05.003 [DOI] [PubMed] [Google Scholar]
- Mittler R. (2006). Abiotic stress, the field environment and stress combination. Trends Plant Sci. 11, 15–19. 10.1016/j.tplants.2005.11.002 [DOI] [PubMed] [Google Scholar]
- Nakai A. (1999). New aspects in the vertebrate heat shock factor system: Hsf3 and Hsf4. Cell Stress Chaperones 4, 86–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima K., Ito Y., Yamaguchi-Shinozaki K. (2009). Transcriptional regulatory networks in response to abiotic stresses in Arabidopsis and grasses. Plant Physiol. 149, 88–95. 10.1104/pp.108.129791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima K., Takasaki H., Mizoi J., Shinozaki K., Yamaguchi-Shinozaki K. (2012). NAC transcription factors in plant abiotic stress responses. Biochimica et Biophysica Acta 1819, 97–103. 10.1016/j.bbagrm.2011.10.005 [DOI] [PubMed] [Google Scholar]
- Nishizawa A., Yabuta Y., Yoshida E., Maruta T., Yoshimura K., Shigeoka S. (2006). Arabidopsis heat shock transcription factor A2 as a key regulator in response to several types of environmental stress. Plant J. 48, 535–547. 10.1111/j.1365-313X.2006.02889.x [DOI] [PubMed] [Google Scholar]
- Nishizawa-Yokoi A., Nosaka R., Hayashi H., Tainaka H., Maruta T., Tamoi M., et al. (2011). HsfA1d and HsfA1e involved in the transcriptional regulation of HsfA2 function as key regulators for the Hsf signaling network in response to environmental stress. Plant Cell Physiol. 52, 933–945. 10.1093/pcp/pcr045 [DOI] [PubMed] [Google Scholar]
- Nishizawa-Yokoi A., Tainaka H., Yoshida E., Tamoi M., Yabuta Y., Shigeoka S. (2010). The 26S proteasome function and Hsp90 activity involved in the regulation of HsfA2 expression in response to oxidative stress. Plant Cell Physiol. 51, 486–496. 10.1093/pcp/pcq015 [DOI] [PubMed] [Google Scholar]
- Nover L., Bharti K., Döring P., Mishra S. K., Ganguli A., Scharf K. D. (2001). Arabidopsis and the heat stress transcription factor world: how many heat stress transcription factors do we need? Cell Stress Chaperones 6, 177–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nover L., Scharf K. D., Gagliardi D., Vergne P., Czarnecka-Verner E., Gurley W. B. (1996). The Hsf world: classification and properties of plant heat stress transcription factors. Cell Stress Chaperones 1, 215–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa D., Yamaguchi K., Nishiuchi T. (2007). High-level overexpression of the Arabidopsis HsfA2 gene confers not only increased themotolerance but also salt/osmotic stress tolerance and enhanced callus growth. J. Exp. Bot. 58, 3373–3383. 10.1093/jxb/erm184 [DOI] [PubMed] [Google Scholar]
- Pajerowska-Mukhtar K. M., Wang W., Tada Y., Oka N., Tucker C. L., Fonseca J. P., et al. (2012). The HSF-like transcription factor TBF1 is a major molecular switch for plant growth-to-defense transition. Curr. Biol. 22, 103–112. 10.1016/j.cub.2011.12.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng S., Zhu Z., Zhao K., Shi J., Yang Y., He M., et al. (2013). A novel heat shock transcription factor, VpHsf1, from Chinese wild Vitis pseudoreticulata is involved in biotic and abiotic stresses. Plant Mol. Biol. Report. 31, 240–247. 10.1007/s11105-012-0463-1 [DOI] [Google Scholar]
- Perez D. E., Hoyer J. S., Johnson A. I., Moody Z. R., Lopez J., Kaplinsky N. J. (2009). BOBBER1 is a noncanonical Arabidopsis small heat shock protein required for both development and thermotolerance. Plant Physiol. 151, 241–252. 10.1104/pp.109.142125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Salamó I., Papdi C., Rigó G., Zsigmond L., Vilela B., Lumbreras V., et al. (2014). The heat shock factor A4A confers salt tolerance and is regulated by oxidative stress and the mitogen-activated protein kinases MPK3 and MPK6. Plant Physiol. 165, 319–334. 10.1104/pp.114.237891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Personat J. M., Tejedor-Cano J., Prieto-Dapena P., Almoguera C., Jordano J. (2014). Co-overexpression of two Heat Shock Factors results in enhanced seed longevity and in synergistic effects on seedling tolerance to severe dehydration and oxidative stress. BMC Plant Biol. 14:56. 10.1186/1471-2229-14-56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Port M., Tripp J., Zielinski D., Weber C., Heerklotz D., Winkelhaus S., et al. (2004). Role of Hsp17. 4-CII as coregulator and cytoplasmic retention factor of tomato heat stress transcription factor HsfA2. Plant Physiol. 135, 1457–1470. 10.1104/pp.104.042820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prändl R., Hinderhofer K., Eggers-Schumacher G., Schöffl F. (1998). HSF3, a new heat shock factor from Arabidopsis thaliana, derepresses the heat shock response and confers thermotolerance when overexpressed in transgenic plants. Mol Gen. Genet. 258, 269–278. 10.1007/s004380050731 [DOI] [PubMed] [Google Scholar]
- Puranik S., Sahu P. P., Srivastava P. S., Prasad M. (2012). NAC proteins: regulation and role in stress tolerance. Trends Plant Sci. 17, 369–381. 10.1016/j.tplants.2012.02.004 [DOI] [PubMed] [Google Scholar]
- Qiao X., Li M., Li L., Yin H., Wu J., Zhang S. (2015). Genome-wide identification and comparative analysis of the heat shock transcription factor family in Chinese white pear (Pyrus bretschneideri) and five other Rosaceae species. BMC Plant Biol. 15:12. 10.1186/s12870-014-0401-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurai H., Enoki Y. (2010). Novel aspects of heat shock factors: DNA recognition, chromatin modulation and gene expression. FEBS J. 277, 4140–4149. 10.1111/j.1742-4658.2010.07829.x [DOI] [PubMed] [Google Scholar]
- Satyal S. H., Chen D., Fox S. G., Kramer J. M., Morimoto R. I. (1998). Negative regulation of the heat shock transcriptional response by HSBP1. Genes Dev. 12, 1962–1974. 10.1101/gad.12.13.1962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharf K. D., Berberich T., Ebersberger I., Nover L. (2012). The plant heat stress transcription factor (Hsf) family: structure, function and evolution. Biochimica et Biophysica Acta 1819, 104–119. 10.1016/j.bbagrm.2011.10.002 [DOI] [PubMed] [Google Scholar]
- Scharf K. D., Heider H., Höhfeld I., Lyck R., Schmidt E., Nover L. (1998). The tomato Hsf system: HsfA2 needs interaction with HsfA1 for efficient nuclear import and may be localized in cytoplasmic heat stress granules. Mol. Cell. Biol. 18, 2240–2251. 10.1128/MCB.18.4.2240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schramm F., Ganguli A., Kiehlmann E., Englich G., Walch D., von Koskull-Döring P. (2006). The heat stress transcription factor HsfA2 serves as a regulatory amplifier of a subset of genes in the heat stress response in Arabidopsis. Plant Mol. Biol. 60, 759–772. 10.1007/s11103-005-5750-x [DOI] [PubMed] [Google Scholar]
- Sewelam N., Oshima Y., Mitsuda N., Ohme-Takagi M. (2014). A step towards understanding plant responses to multiple environmental stresses: a genome-wide study. Plant Cell Environ. 37, 2024–2035. 10.1111/pce.12274 [DOI] [PubMed] [Google Scholar]
- Shim D., Hwang J. U., Lee J., Lee S., Choi Y., An G., et al. (2009). Orthologs of the class A4 heat shock transcription factor HsfA4a confer cadmium tolerance in wheat and rice. Plant Cell 21, 4031–4043. 10.1105/tpc.109.066902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L., Jiang Y., Zhao H., Hou M. (2012). Acquired thermotolerance in plants. Plant Cell Tissue Organ Cult. 111, 265–276. 10.1007/s11240-012-0198-6 [DOI] [Google Scholar]
- Sugio A., Dreos R., Aparicio F., Maule A. J. (2009). The cytosolic protein response as a subcomponent of the wider heat shock response in Arabidopsis. Plant Cell 21, 642–654. 10.1105/tpc.108.062596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udvardi M. K., Kakar K., Wandrey M., Montanari O., Murray J., Andriankaja A., et al. (2007). Legume transcription factors: global regulators of plant development and response to the environment. Plant Physiol. 144, 538–549. 10.1104/pp.107.098061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Arnim A. G., Jia Q., Vaughn J. N. (2014). Regulation of plant translation by upstream open reading frames. Plant Sci. 214, 1–12. 10.1016/j.plantsci.2013.09.006 [DOI] [PubMed] [Google Scholar]
- von Koskull-Döring P., Scharf K. D., Nover L. (2007). The diversity of plant heat stress transcription factors. Trends Plant Sci. 12, 452–457. 10.1016/j.tplants.2007.08.014 [DOI] [PubMed] [Google Scholar]
- Wang J., Sun N., Deng T., Zhang L., Zuo K. (2014). Genome-wide cloning, identification, classification and functional analysis of cotton heat shock transcription factors in cotton (Gossypium hirsutum). BMC Genomics 15:961. 10.1186/1471-2164-15-961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Vinocur B., Shoseyov O., Altman A. (2004). Role of plant heat-shock proteins and molecular chaperones in the abiotic stress response. Trends Plant Sci. 9, 244–252. 10.1016/j.tplants.2004.03.006 [DOI] [PubMed] [Google Scholar]
- Wunderlich M., Gross-Hardt R., Schöffl F. (2014). Heat shock factor HSFB2a involved in gametophyte development of Arabidopsis thaliana and its expression is controlled by a heat-inducible long non-coding antisense RNA. Plant Mol. Biol. 85, 541–550. 10.1007/s11103-014-0202-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin H., Zhang H., Chen L., Li X., Lian Q., Yuan X., et al. (2010). Cloning and characterization of HsfA2 from Lily (Lilium longiflorum). Plant Cell Rep. 29, 875–885. 10.1007/s00299-010-0873-1 [DOI] [PubMed] [Google Scholar]
- Xue G. P., Drenth J., McIntyre C. L. (2015). TaHsfA6f is a transcriptional activator that regulates a suite of heat stress protection genes in wheat (Triticum aestivum L.) including previously unknown Hsf targets. J. Exp. Bot. 66, 1025–1039. 10.1093/jxb/eru462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue G. P., Sadat S., Drenth J., McIntyre C. L. (2014). The heat shock factor family from Triticum aestivum in response to heat and other major abiotic stresses and their role in regulation of heat shock protein genes. J. Exp. Bot. 65, 539–557. 10.1093/jxb/ert399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokotani N., Ichikawa T., Kondou Y., Matsui M., Hirochika H., Iwabuchi M., et al. (2008). Expression of rice heat stress transcription factor OsHsfA2e enhances tolerance to environmental stresses in transgenic Arabidopsis. Planta 227, 957–967. 10.1007/s00425-007-0670-4 [DOI] [PubMed] [Google Scholar]
- Yoshida T., Sakuma Y., Todaka D., Maruyama K., Qin F., Mizoi J., et al. (2008). Functional analysis of an Arabidopsis heat-shock transcription factor HsfA3 in the transcriptional cascade downstream of the DREB2A stress-regulatory system. Biochem. Biophys. Res. Commun. 368, 515–521. 10.1016/j.bbrc.2008.01.134 [DOI] [PubMed] [Google Scholar]
- Zhang J., Li Y., Jia H. X., Li J. B., Huang J., Lu M. Z., et al. (2015). The heat shock factor gene family in Salix suchowensis: a genome-wide survey and expression profiling during development and abiotic stresses. Front. Plant Sci. 6:748. 10.3389/fpls.2015.00748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., Xu Z. S., Li P., Yang L., Wei Y., Chen M., et al. (2013). Overexpression of TaHSF3 in transgenic Arabidopsis enhances tolerance to extreme temperatures. Plant Mol. Biol. Report. 31, 688–697. 10.1007/s11105-012-0546-z [DOI] [Google Scholar]
- Zhou R., Li B., Liu H., Sun D. (2009). Progress in the participation of Ca2+–calmodulin in heat shock signal transduction. Prog. Nat. Sci. 19, 1201–1208. 10.1016/j.pnsc.2008.12.011 [DOI] [Google Scholar]
- Zhu B., Ye C., Lü H., Chen X., Chai G., Chen J., et al. (2006). Identification and characterization of a novel heat shock transcription factor gene, GmHsfA1, in soybeans (Glycine max). J. Plant Res. 119, 247–256. 10.1007/s10265-006-0267-1 [DOI] [PubMed] [Google Scholar]
- Zhu X., Thalor S. K., Takahashi Y., Berberich T., Kusano T. (2012). An inhibitory effect of the sequence-conserved upstream open-reading frame on the translation of the main open-reading frame of HsfB1 transcripts in Arabidopsis. Plant Cell Environ. 35, 2014–2030. 10.1111/j.1365-3040.2012.02533.x [DOI] [PubMed] [Google Scholar]
- Zhu Y., Wang Z., Jing Y., Wang L., Liu X., Liu Y., et al. (2009). Ectopic over-expression of BhHsf1, a heat shock factor from the resurrection plant Boea hygrometrica, leads to increased thermotolerance and retarded growth in transgenic Arabidopsis and tobacco. Plant Mol. Biol. 71, 451–467. 10.1007/s11103-009-9538-2 [DOI] [PubMed] [Google Scholar]