Abstract
Long-term vanilla monocropping often results in the occurrence of vanilla Fusarium wilt disease, seriously affecting its production all over the world. In the present study, vanilla exhibited significantly less Fusarium wilt disease in the soil of a long-term continuously cropped black pepper orchard. The entire fungal communities of bulk and rhizosphere soils between the black pepper-vanilla system (i.e., vanilla cropped in the soil of a continuously cropped black pepper orchard) and vanilla monoculture system were compared through the deep pyrosequencing. The results showed that the black pepper-vanilla system revealed a significantly higher fungal diversity than the vanilla monoculture system in both bulk and rhizosphere soils. The UniFrac-weighted PCoA analysis revealed significant differences in bulk soil fungal community structures between the two cropping systems, and fungal community structures were seriously affected by the vanilla root system. In summary, the black pepper-vanilla system harbored a lower abundance of Fusarium oxysporum in the vanilla rhizosphere soil and increased the putatively plant-beneficial fungal groups such as Trichoderma and Penicillium genus, which could explain the healthy growth of vanilla in the soil of the long-term continuously cropped black pepper field. Thus, cropping vanilla in the soil of continuously cropped black pepper fields for maintaining the vanilla industry is executable and meaningful as an agro-ecological system.
Keywords: continuous cropping, black pepper, soil fungal communities, Miseq sequencing, vanilla healthy growth
Introduction
Vanilla (Vanilla planifolia), a herbaceous perennial vine with high economic value, has been widely cropped in tropical and subtropical regions (Minoo et al., 2008). However, the long-term monoculture of this crop often results in the occurrence of soil-borne Fusarium wilt disease, seriously affecting its production worldwide in vanilla-cropping regions (Jayasekhar et al., 2008; Pinaria et al., 2010; Xiong et al., 2015b) and leading to significant economic losses over the last decade. Fungicides and biological control agents have been traditionally suggested as integrated control strategies for vanilla Fusarium wilt disease (Tombe and Sitepu, 1986; Sandheep et al., 2012); however, these methods are usually environmentally unfriendly or inefficient. Thus, exploring an effective method for controlling the vanilla Fusarium wilt disease is extremely important for maintaining the vanilla production. Meanwhile, in tropical China, multiple continuous cropping fields for other tropical crops, such as black pepper and banana, have suffered serious successive cropping obstacles and were given up for growing the same crops (Wang et al., 2013; Xiong et al., 2015a). Because of these associated problems, farmers naturally grow different crops in these fields. By some chance, after our field investigation, an interesting phenomenon was always observed where vanilla with the lowest Fusarium wilt disease incidence (DI) grew in the continuously cropped black pepper field. The causes of the disease decline might be very complex, such as improved soil physical and chemical properties and land management practices (Hilton et al., 2013; Navarro-Noya et al., 2013). However, the detailed mechanisms involved in the healthy vanilla growth associated with the soil microflora variation under the soil of long-term continuously cropped black pepper fields remain unclear.
Exploring continuously cropped field soil for other crop growth is meaningful and sustainable to agro-ecological systems. Meanwhile, to our limited knowledge, few studies have focused on the long-term continuously cropped soil supporting other crop growth; thus, how the variation in soil microbiota under long-term continuously cropped soil could support other crop growth is even less well understood. Soil microorganisms play critical roles in regulating soil fertility, global nutrient cycling, and plant health (Fierer et al., 2012), which might be directly linked to the maintenance of plant health in agro-systems. Within soil ecosystems, the immediate surroundings of the plant root, i.e., the rhizosphere, is a dynamic interface supporting the exchange of resources between plants and their associated soil environment (Peiffer et al., 2013). Rhizosphere microbiota, considered as the second genome of the plant, are significantly influenced by plant roots (Philippot et al., 2013). The main source of microbial communities in the rhizosphere is the adjacent root-free soil, called the bulk soil; hence, the changes brought about in the communities of the bulk soil will have an effect on the assembly and the final composition of rhizosphere communities (Mendes et al., 2014).
The development of high-throughput sequencing, particularly Illumina MiSeq sequencing (Metzker, 2010; Shokralla et al., 2012), offers a powerful strategy for uncovering the complex and diverse soil microbial communities with high throughput, high accuracy, and considerably lower cost. The internal transcribed spacers (ITS1) region has been widely used in the analysis of soil fungal communities (Xu et al., 2012; Lu et al., 2013). The functional diversity of soil fungi and their capacity to colonize diverse microhabitats can influence pathogen levels and play a significant role in improving plant health (Penton et al., 2014). Given that vanilla Fusarium wilt disease is caused by a fungal pathogen, exploring the fungal community involved in the healthy growth of vanilla in black pepper-vanilla agro-ecosystems is quite important.
Thus, in this study, we hypothesize that long-term continuous cropping black pepper orchards harbored a unique soil fungal community associated with healthy vanilla growth. To test this hypothesis, we used pot experiments to evaluate the persistent ability of the soil of long-term continuously cropped black pepper fields to support vanilla healthy growth; and fungal community of the bulk and rhizosphere soils in the black pepper-vanilla and vanilla monoculture systems was accessed by the Illumina MiSeq sequencing.
Materials and methods
No specific permits were required for the described field studies. The locations are not protected. The field studied did not involve endangered or protected species.
Experiment descriptions
The experimental site is located at the Spice and Beverage Research Institute, Wanning City, Hainan Province, China (110°19′E-110°22′E, 18°72′N-18°76′N). It is an area with a tropical monsoon climate, a mean annual temperature of 24.5°C and a mean annual precipitation of 2201 mm. The experimental soil was collected in April 2013 from the 20-years continuously cropped black pepper orchard. The soil was mixed thoroughly and transferred to the greenhouse with an average temperature of 30°C and an average humidity of 72% at the Spice and Beverage Research Institute. Meanwhile, the soil ~200 m away from the black pepper orchard collected from the 21-year continuously cropped vanilla orchard showing serious vanilla Fusarium wilt disease (Xiong et al., 2015b) was considered a control. Both the black pepper and vanilla orchard soils are sandy loam in texture and developed from the same parent material. The experiment was performed using a randomized complete block design in three replicates, where each block had six pots for each treatment, and each pot contained 15 kg soil with three seedling vanillas. The agronomic management and fertilization regime were uniform during the next 18 months (April, 2013 to October, 2014). Vanilla Fusarium wilt disease was monitored immediately after the seedlings were transplanted into the pots based on the observation of typical wilt symptoms. The DI was calculated as the percentage of infected plants among the total number of plants (Wei et al., 2011). It is worth noting that we also used continuously cropped banana and coffee orchards soil to cultivate vanilla in pots. We found the continuously cropped black pepper soil showed the lowest vanilla Fusarium wilt disease and the highest plant biomass (vanilla shoot dry weight). Hence, we got the two vanilla cropping regimes, i.e., black pepper-vanilla system and vanilla monoculture system for the subsequent research.
Soil sample collection and DNA extraction
After removing the vanilla plants from the pots, the bulk soil samples obtained for each replicate from the black pepper-vanilla and vanilla monoculture systems were referred to as “BB” and “VB,” respectively. All six bulk soil samples were passed through a 2 mm sieve, thoroughly homogenized and divided into 2 subsamples: one was air-dried for a soil characteristic analysis according to our previous methods (Xiong et al., 2015b), and the remainder was stored at −80°C for DNA extraction. For the vanilla rhizosphere soil samples, six vanilla plants were randomly selected from each replicate in the black pepper-vanilla and vanilla monoculture systems, the roots were vigorously shaken to dislodge the loosely adhering soil, and the soil remaining attached to the root system was considered to be rhizosphere soil. The rhizosphere soil was collected using the following protocol: the roots were cut into pieces of ~1 cm length and carefully mixed, 20 g of roots were pooled into a 500 mL vol. flask containing 200 mL ddH2O and washed on a shaking platform for 20 min at 180 rpm, the washing buffer was subjected to centrifugation (10,000 g, 10 min), and then the resulting pellet was obtained and defined as the rhizosphere soil. The rhizosphere soil from the black pepper-vanilla system and vanilla monoculture system are referred to as “BR” and “VR,” respectively. All six rhizosphere soil samples were stored at −80°C for DNA extraction.
Total DNA was extracted from the 12 soil samples using a MoBioPowerSoil™ DNA Isolation Kit (Mo Bio Laboratories Inc., Carlsbad, CA, USA) according to the manufacturer's instructions. The genomic DNA concentration and purity were measured using NanoDrop ND-2000 (NanoDrop Technologies, Wilmington, DE) spectrophotometry.
PCR amplification and deep pyrosequencing
The fungi-specific primers ITS1F (CTTGGTCATTTAGAGGAAGTAA) (Gardes and Bruns, 1993) and ITS2 (GCTGCGTTCTTCATCGATGC) (White et al., 1990) were selected to target the ITS1 region. These primer pairs were modified for pyrosequencing by adding the forward Illumina Nextera adapter, a two-base-pair “linker” sequence, and a unique 7-bp barcode sequence to the 5′ end of the forward primer and the appropriate reverse Illumina Nextera adapter and linker sequence at the 5′ end of the reverse primer. PCR amplification was performed in a 25 μl reaction: 2.5 μl of 10 × reaction buffer, 10 μM of each primer, 2.5 mM dNTPs, 40 ng of template, and 0.625 units of Takara Pyrobest (Takara Biotechnology Co., Ltd., Japan). Amplifications were performed with the following temperature regime: 4 min of initial denaturation at 94°C, followed by 35 cycles of denaturation (94°C for 30 s), annealing (50°C for 45 s), extension (72°C for 1 min), and a final extension at 72°C for 7 min. The PCR products were purified using a PCR Purification Kit (Axygen Bio, USA). Then, paired-end sequencing was performed on an Illumina MiSeq sequencer at Personal Biotechnology Co., Ltd (Shanghai, China).
Quantification of the Fusarium oxysporum and fungal abundances
Real-time quantitative polymerase chain reaction (qPCR) was performed according to Chen et al. (2014) for quantifying the soil Fusarium oxysporum and fungi abundances using the SYBR Premix Ex Taq Kit on the ABI PRISM 7500 Real Time PCR System (Applied Biosystems, Germany). The 20 μl reaction mixture contained 10 μl of the Premix Ex Taq™ (2 ×) (Takara), 0.4 μl of each primer (10 μM), 0.4 μl of ROX Reference Dye II (50 ×), 2 μl of template DNA, and 6.8 μl of ddH2O. The specific primer set of F. oxysporum and soil fungi was AFP308R (CGAATT AACGCGAGTCCCAAC)/ITS1F (CTTGGTCATTTAGAGGAAGTAA) (Lievens et al., 2006) and ITS2 (GCTGCGTTCTTCATCGATGC)/ITS1F (CTTGGTCATTTA GAGGAAGTAA), respectively. The thermal conditions were set as follows: 30 s at 95°C for initial denaturation, 40 cycles of 5 s at 95°C, and 34 s at 60°C. The standard curve was obtained using a 10-fold dilution series of plasmid DNA containing a fragment of the ITS region of F. oxysporum and ITS1 gene from the F. oxysporum f. sp. vanillae and soil samples, respectively. All amplifications were performed in triplicate. The specificity of the products was confirmed by a melting curve analysis and agarose gel electrophoresis. The copy numbers were log10-transformed to normalize the values prior to statistical analysis.
Pyrosequencing data analysis
After removing the adaptors and primer sequences, the raw sequences were assembled for each sample according to the unique barcode using QIIME (Caporaso et al., 2010). The split sequences for each sample were merged using FLASH V1.2.7 (Magoč and Salzberg, 2011), and low-quality sequences were then discarded using QIIME. The sequences retained for each sample were processed following the established UPARSE pipeline (Edgar, 2013). Briefly, the sequences with a quality score lower than 0.5 or a length shorter than 200 bp were removed. After discarding the singletons, the remaining reads were assigned to OTUs with a threshold of 97% identity level. Then, the chimera removal processes were performed. Finally, the fungal representative OTUs were classified using the UNITE database (Kõljalg et al., 2013).
The diversity within each individual sample was estimated using non-parametric Shannon diversity indices. Shannoneven was used to measure the evenness of each sample (Schloss et al., 2009). A principal coordinate analysis (PCoA) based on weighted UniFrac metric matrices was performed to explore the differences in fungal community structures among all of the soil samples (Lozupone et al., 2006). A permutational multivariate analysis of variance (Anderson, 2001) was performed to assess the effect of the cropping regime, soil compartment, and their interactions on the fungal community structure (abundance of OTUs and genus) using the adonis function of the R vegan package with 999 permutations.
Statistical analyses
The soil physicochemical characteristics and vanilla Fusarium wilt DI between the black pepper-vanilla and vanilla monoculture systems were compared using Student's t-test. For other parameters in our study, one-way analyses of variance (ANOVA) with Turkey's HSD multiple range test were performed for multiple comparisons. All of the statistical analyses were performed using SPSS v20.0 (SPSS Inc., USA).
Sequence accession numbers
The sequence data have been deposited in the NCBI Sequence Read Archive (SRA) database with the accession number SRP062990.
Results
Soil physical and chemical properties in the black pepper-vanilla and vanilla monoculture systems
The results of soil physical and chemical properties are summarized in Table S1. When compared with the vanilla monoculture system, black pepper-vanilla system presented a significantly (P < 0.05) higher available N content. In contrast, the vanilla monoculture system revealed higher soil pH and the contents of organic matter and available P.
Fusarium wilt DI and fungal abundance in the two vanilla cropping systems
As shown in Table 1, the black pepper-vanilla system significantly reduced vanilla Fusarium wilt DI to 15.56%, whereas the value was over 60% in the vanilla monoculture system. The qPCR results showed that the ITS copies of F. oxysporum were significantly lower in the black pepper-vanilla system compared with those from the vanilla monoculture system in both bulk and rhizosphere soils (Table 1). Strikingly, the F. oxysporum populations significantly increased from the bulk soil to the vanilla rhizosphere soil in both the black pepper-vanilla and vanilla monoculture systems. In addition, the fungal ITS gene copy numbers in bulk soil showed no significant difference between the black pepper-vanilla and vanilla monoculture systems (Table S2). Meanwhile, the fungal ITS gene copy numbers in the rhizosphere soil samples from the black pepper-vanilla system (8.15 × 109 copies g−1 soil) were significantly higher than those from the vanilla monoculture system (2.89 × 109 copies g−1 soil).
Table 1.
Vanilla Fusarium wilt disease incidence and pathogen abundance.
| Cropping regime | Soil compartment | Disease incidence (%) | Fusarium RA | F. oxysporum RA | Log10F. oxysporum ITS copies g−1 soil |
|---|---|---|---|---|---|
| Black pepper-vanilla system | Bulk soil (BB) | 15.56 ± 3.85 b | 6.79±2.23 b | 5.51±2.75 b | 4.80±0.08 d |
| Rhizosphere soil (BR) | 10.36±1.31 b | 8.66±1.15 b | 5.60±0.15 b | ||
| Vanilla monoculture system | Bulk soil (VB) | 62.22 ± 10.08 a | 10.23±0.82 b | 5.59±0.37 b | 5.13±0.13 c |
| Rhizosphere soil (VR) | 26.18±7.54 a | 22.89±6.80 a | 6.18±0.09 a |
Values are means ± standard deviation (n = 3). RA, Relative abundance.
Means followed by the same letter for a given factor are not significantly different (P < 0.05; Turkey's HSD test where there are more than two treatment levels and Student's t-test where there are two treatment levels).
Overall diversity of fungal community
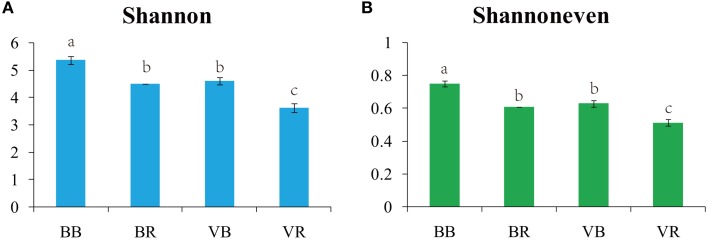
After quality filtering, the pyrosequencing-based analysis of the fungal ITS1 genes resulted in the recovery of 1,260,032 high-quality sequences across the 12 samples (Table S3). The coverage from all samples was above 99%, indicating that the sequencing reads were sufficient for this analysis (Table S3). In both the bulk and rhizosphere soils, the black pepper-vanilla system had a significantly higher fungal diversity (Shannon) and evenness (Shannoneven) values than the vanilla monoculture system (Figure 1). In addition, in both the black pepper-vanilla and vanilla monoculture systems, the fungal community diversity and evenness significantly decreased from the bulk soil to the vanilla rhizosphere soil.
Figure 1.
Shannon diversity indices (A) and Shannoneven indices (B) for the bulk soil and vanilla rhizosphere soil of the black pepper-vanilla and vanilla monoculture systems. Bars represent the standard deviation of the three replicates, and different letters above the bars indicate a significant difference at the 0.05 probability level according to the Turkey's HSD test. “BB” and “VB” represent the bulk soil from the black pepper-vanilla system and the vanilla monoculture system, respectively. “BR” and “VR” represent the rhizosphere soil from the black pepper-vanilla system and the vanilla monoculture system, respectively.
Fungal community composition
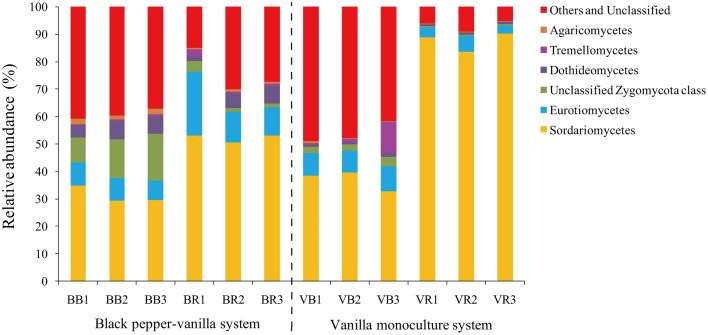
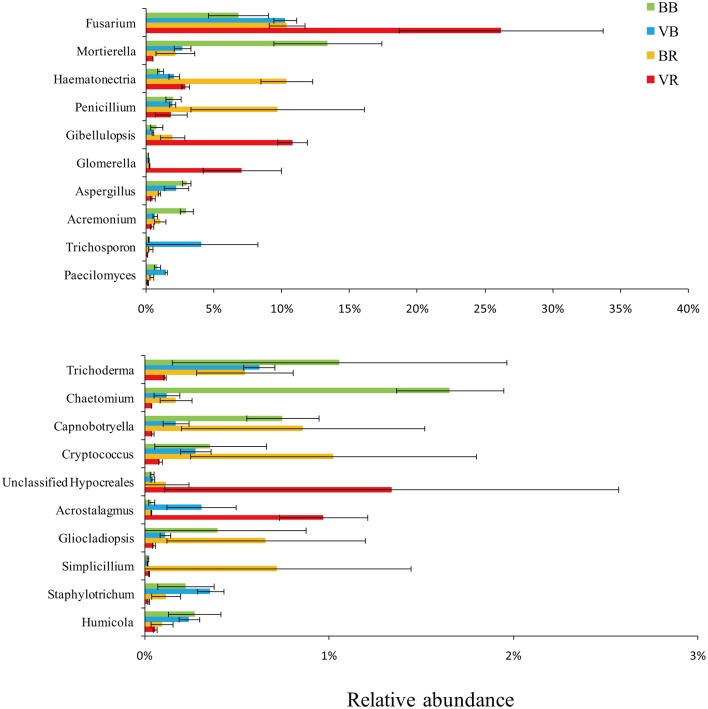
To verify the differences observed in the fungal communities from the black pepper-vanilla and vanilla monoculture systems, the relative abundances (RA) of the different classes and genera from the bulk and rhizosphere soils were compared (Figures 2, 3). In the present study, fungal OTUs across the 12 soil samples were observed predominantly from the six classes (Sordariomycetes, Eurotiomycetes, unclassified Zygomycota class, Dothideomycetes, Tremellomycetes, and Agaricomycetes), accounting for 71.10% of the total fungal sequences. Compared with bulk soil, the relative abundance of the class Sordariomycetes in the vanilla rhizosphere soil significantly increased in both the black pepper-vanilla and vanilla monoculture systems. At the genus level, in the bulk soil, compared with the vanilla monoculture system, the black pepper-vanilla system had a higher relative abundance of Mortierella, Aspergillus, Acremonium, and Chaetomium. As for the rhizosphere soil, the Fusarium genus was significantly more abundant in the vanilla monoculture system than in the black pepper-vanilla system; moreover, the relative abundance of F. oxysporum (OTU level) exhibited a similar trend (Table 1). In addition, the relative abundances of the genera Haematonectria, Trichoderma, and Penicillium were significantly higher in the black pepper-vanilla system with a lower Gibellulopsis abundance.
Figure 2.
Relative abundances of the main fungal classes in the bulk soil and the vanilla rhizosphere soil of the black pepper-vanilla and vanilla monoculture systems. The “Others and Unclassified” comprised the unclassified and low-abundance classes (RA < 0.1%). “BB” and “VB” represent the bulk soil from the black pepper-vanilla system and the vanilla monoculture system, respectively. “BR” and “VR” represent the rhizosphere soil from the black pepper-vanilla system and the vanilla monoculture system, respectively.
Figure 3.
Relative abundances of the top 20 fungal genera in bulk soil and vanilla rhizosphere soil of the black pepper-vanilla and vanilla monoculture systems. Bars represent the standard deviation of the three replicates. “BB” and “VB” represent the bulk soil from the black pepper-vanilla system and the vanilla monoculture system, respectively. “BR” and “VR” represent the rhizosphere soil from the black pepper-vanilla system and the vanilla monoculture system, respectively.
Fungal community structure
A permutational multivariate analysis of variance confirmed that the cropping regime, soil compartment, and their interactions were significant factors of variation for the fungal community structure in terms of both the relative abundance of OTUs and relative abundance of genera (Table 2).
Table 2.
PERMANOVA analysis.
| Source | Df | Abundance of Genera | Abundance of OTUs | ||
|---|---|---|---|---|---|
| Sums of sqs | Pseudo-F | Sums of sqs | Pseudo-F | ||
| Cropping regime (CR) | 1 | 0.18 | 11.79*** | 0.42 | 7.64*** |
| Soil compartment (SC) | 1 | 0.25 | 15.80*** | 0.40 | 7.30*** |
| CR × SC | 1 | 0.09 | 5.54*** | 0.20 | 3.68*** |
| Residuals | 8 | 0.13 | 0.19 | 0.44 | 0.30 |
Indicate significant correlations (P < 0.001).
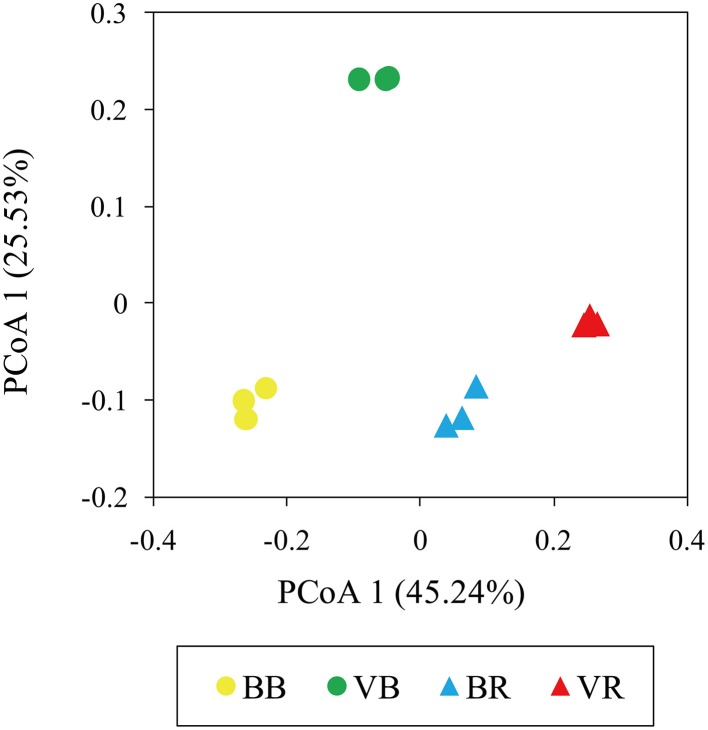
To further compare the variations in fungal community structure between the black pepper-vanilla and vanilla monoculture systems samples, a UniFrac-weighted PCoA was employed. As shown in Figure 4, the bulk soil samples from the black pepper-vanilla system were clearly separated from the vanilla monoculture system, suggesting strong differences in fungal community structures between the different crop regime systems. In addition, the fungal communities in the rhizosphere soils from the two vanilla cropping systems were close together, suggesting that fungal community structures were seriously affected by the vanilla root system.
Figure 4.
UniFrac-weighted principle coordinate analysis of fungal community structures in the bulk soil and vanilla rhizosphere soil of the black pepper-vanilla and vanilla monoculture systems. “BB” and “VB” represent the bulk soil from the black pepper-vanilla system and the vanilla monoculture system, respectively. “BR” and “VR” represent the rhizosphere soil from the black pepper-vanilla system and the vanilla monoculture system, respectively.
Discussion
Obstacles to the continuous cropping of vanilla have always been observed on Hainan Island (Xiong et al., 2015b). In the present study, pot experiments confirmed that long-term continuously cropped black pepper orchard soil showed significantly lower vanilla Fusarium wilt disease, implying that crop rotation is an effective management practice to reduce soil-borne plant disease in agro-systems (Wang et al., 2015). In addition, it will also help us to take advantage of the large area of black pepper continuous cropping soil in tropical China (Zu et al., 2014; Xiong et al., 2015a).
In this study, the black pepper-vanilla system had no effect on the fungal population abundance in the bulk soil. However, alpha diversity estimates of the fungal communities revealed that the black pepper-vanilla system had a significantly higher fungal diversity and evenness than the vanilla monoculture soil (Figure 1). The possible reasons are as follows: residues of black pepper decomposed in the soil and different root exudates could provide more available nutrient to soil microbes, thus improving species richness, heterogeneity, and diversity of the fungal community (Xuan et al., 2011). Furthermore, we could find a negative relationship between the soil fungal diversity and vanilla Fusarium wilt disease, which could support the idea that microbial diversity is a key factor in controlling pathogen invasion (van Elsas et al., 2012). In addition, in the present study, the fungal community diversity significantly decreased from the bulk soil to the vanilla rhizosphere soil in both the black pepper-vanilla and vanilla monoculture systems; as explained by Mendes et al. (2014) plants can select a constant rhizosphere community from highly contrasting reservoirs of bulk soil communities.
The UniFrac-weighted PCoA analysis revealed significant variations in the bulk soil of fungal community structures between the black pepper-vanilla and vanilla monoculture systems (Figure 4). Our results agreed with the findings of Wang et al. (2015) where the crop regime system was the major determinant factor for microbial community structures. The significant variations in bulk soil community structures among the different cropping systems might be attributed to significant differentiations in soil physicochemical characteristics (Table S1), as soil physicochemical properties have significant impacts on microbial community structures (Lauber et al., 2008). Compared with the vanilla monoculture system, black pepper-vanilla system revealed a significantly higher available N content, as nitrogen play a pivotal role in plant growth and might indirectly enhance plant disease suppressiveness (Hayat et al., 2010). In addition, the UniFrac-weighted PCoA analysis suggested fungal community structures were also seriously affected by the vanilla root system, which was consistent with the many previous studies that plant play a key role in shaping the microbial community structures in the rhizosphere (Philippot et al., 2013; Edwards et al., 2015).
The black pepper-vanilla system was shown to have a significant effect on the fungal community compositions in both bulk and rhizosphere soils. Sordariomycetes was the most abundant fungal class (Figure 2), which was generally consistent with the many early studies that found Sordariomycetes to be the most common fungal class in different agricultural systems (Chen et al., 2012; Li et al., 2014). Compared with bulk soil, the abundance of Sordariomycetes significantly increased in vanilla rhizosphere soil in both the black pepper-vanilla and vanilla monoculture systems, as reported by Zhang et al. (2006) who found that members of the Sordariomycetes are ubiquitous in virtually all ecosystems as pathogens and endophytes of plants.
Deeper taxonomic analyses were performed to explore the fungal community compositions of rhizosphere soil in the black pepper-vanilla system associated with vanilla growth. The black pepper-vanilla system showed significantly lower Fusarium and F. oxysporum abundance in the vanilla rhizosphere soil, which could be the most important reason for significantly lower vanilla Fusarium wilt disease in the black pepper-vanilla system (Pinaria et al., 2010). The F. oxysporum abundance is lower in the black pepper-vanilla system might be because that continuous cropping black pepper soil had not previously been used to cultivate vanilla, however, if vanilla is continuously cropped in that soil which could also increase the pathogen load eventually (Xiong et al., 2015b). In addition, in present study, the F. oxysporum populations significantly accumulated from the bulk soil to the vanilla rhizosphere soil in both the black pepper-vanilla and vanilla monoculture systems. Synthesized from the above results, we conclude that iterative crop rotation might be necessary to interrupt the accumulation of F. oxysporum abundance to suppress vanilla Fusarium wilt disease.
Some putatively plant-beneficial fungal groups, such as the genera Trichoderma and Penicillium, increased in the vanilla rhizosphere soil under the black pepper-vanilla system. Trichoderma spp. are known to have an effective antagonistic effect against vanilla Fusarium wilt disease (Jayasekhar et al., 2008; Vijayan et al., 2009). Penicillium is also a famous biocontrol agent for the biological control of Fusarium wilt disease (Larena et al., 2003); however, this has not yet been reported in vanilla systems. Moreover, Haematonectria was the most abundant genus, accounting for 10.33% of the total fungal genera in vanilla rhizosphere soil in the black pepper-vanilla system, which could occupy the rhizosphere niche to avoid pathogen invasion (Qiu et al., 2013). Combining the other variations in fungal genera in the black pepper-vanilla and vanilla monoculture systems and the complex interactions among these microorganisms could explain the status of vanilla Fusarium wilt disease in agro-ecosystems.
In conclusion, compared with the vanilla monoculture system, black pepper-vanilla system harbored a significantly lower abundance of F. oxysporum in vanilla rhizosphere soil, increased the putatively plant-beneficial fungal groups and the fungal diversity, which could explain the decrease in vanilla Fusarium wilt disease in the soil of the long-term continuously cropped black pepper orchard. These results suggested that sustainable agricultural management regime, such as crop rotation might be a meaningful strategy to prevent vanilla Fusarium wilt disease occurrence and will be our future research focus.
Author contributions
Conceived and designed the experiments: W. Xiong, QZ, RL, HW, QS. Performed the experiments: W. Xiong, QZ, CX, W. Xun, JZ. Analyzed the data: W. Xiong, QZ, CX, JZ. Contributed reagents/materials/analysis tools: W. Xun, RL, HW, QS. Wrote the manuscript: W. Xiong, RL, HW, QS.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the National Key Basic Research Program of China (2015CB150500), the National Natural Science Foundation of China (31572212), the Chinese Ministry of Science and Technology (2013AA102802), the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD), 111 project (B12009), and the Agricultural Ministry of China (201103004).
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2016.00117
References
- Anderson M. J. (2001). A new method for non-parametric multivariate analysis of variance. Austral Ecol. 26, 32–46. 10.1111/j.1442-9993.2001.01070.pp.x [DOI] [Google Scholar]
- Caporaso J. G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F. D., Costello E. K., et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336. 10.1038/nmeth.f.303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Xun W., Sun L., Zhang N., Shen Q., Zhang R. (2014). Effect of different long-term fertilization regimes on the viral community in an agricultural soil of Southern China. Eur. J. Soil Biol. 62, 121–126. 10.1016/j.ejsobi.2014.03.006 [DOI] [Google Scholar]
- Chen M., Li X., Yang Q., Chi X., Pan L., Chen N., et al. (2012). Soil eukaryotic microorganism succession as affected by continuous cropping of peanut-pathogenic and beneficial fungi were selected. PLoS ONE 7:e40659. 10.1371/journal.pone.0040659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R. C. (2013). UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 10, 996–998. 10.1038/nmeth.2604 [DOI] [PubMed] [Google Scholar]
- Edwards J., Johnson C., Santos-Medellín C., Lurie E., Podishetty N. K., Bhatnagar S., et al. (2015). Structure, variation, and assembly of the root-associated microbiomes of rice. Proc. Natl. Acad. Sci. U.S.A. 112, E911–E920. 10.1073/pnas.1414592112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fierer N., Leff J. W., Adams B. J., Nielsen U. N., Bates S. T., Lauber C. L., et al. (2012). Cross-biome metagenomic analyses of soil microbial communities and their functional attributes. Proc. Natl. Acad. Sci. U.S.A. 109, 21390–21395. 10.1073/pnas.1215210110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardes M., Bruns T. D. (1993). ITS primers with enhanced specificity for basidiomycetes-application to the identification of mycorrhizae and rusts. Mol. Ecol. 2, 113–118. 10.1111/j.1365-294X.1993.tb00005.x [DOI] [PubMed] [Google Scholar]
- Hayat R., Ali S., Amara U., Khalid R., Ahmed I. (2010). Soil beneficial bacteria and their role in plant growth promotion: a review. Ann. Microbiol. 60, 579–598. 10.1007/s13213-010-0117-1 [DOI] [Google Scholar]
- Hilton S., Bennett A. J., Keane G., Bending G. D., Chandler D., Stobart R., et al. (2013). Impact of shortened crop rotation of oilseed rape on soil and rhizosphere microbial diversity in relation to yield decline. PLoS ONE 8:e59859. 10.1371/journal.pone.0059859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayasekhar M., Manonmani K., Justin C. G. L. (2008). Development of integrated biocontrol strategy for the management of stem rots disease (Fusarium oxysporum f. sp. vanillae) of Vanilla. Agric. Sci. Dig. Abstr. 28, 109–111. Available online at: http://www.indianjournals.com/ijor.aspx?target=ijor:asd&volume=28&issue=2&article=008 [Google Scholar]
- Kõljalg U., Nilsson R. H., Abarenkov K., Tedersoo L., Taylor A. F. S., Bahram M., et al. (2013). Towards a unified paradigm for sequence-based identification of fungi. Mol. Ecol. 22, 5271–5277. 10.1111/mec.12481 [DOI] [PubMed] [Google Scholar]
- Larena I., Melgarejo P., De Cal A. (2003). Drying of Conidia of Penicillium oxalicum, a Biological Control Agent against Fusarium Wilt of Tomato. J. Phytopathol. 151, 600–606. 10.1046/j.0931-1785.2003.00772.x [DOI] [PubMed] [Google Scholar]
- Lauber C. L., Strickland M. S., Bradford M. A., Fierer N. (2008). The influence of soil properties on the structure of bacterial and fungal communities across land-use types. Soil Biol. Biochem. 40, 2407–2415. 10.1016/j.soilbio.2008.05.021 [DOI] [Google Scholar]
- Li X., Ding C., Zhang T., Wang X. (2014). Fungal pathogen accumulation at the expense of plant-beneficial fungi as a consequence of consecutive peanut monoculturing. Soil Biol. Biochem. 72, 11–18. 10.1016/j.soilbio.2014.01.019 [DOI] [Google Scholar]
- Lievens B., Claes L., Vanachter A. C. R. C., Cammue B. P. A., Thomma B. P. H. J. (2006). Detecting single nucleotide polymorphisms using DNA arrays for plant pathogen diagnosis. FEMS Microbiol. Lett. 255, 129–139. 10.1111/j.1574-6968.2005.00074.x [DOI] [PubMed] [Google Scholar]
- Lozupone C., Hamady M., Knight R. (2006). UniFrac–an online tool for comparing microbial community diversity in a phylogenetic context. BMC Bioinformatics 7:371. 10.1186/1471-2105-7-371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L., Yin S., Liu X., Zhang W., Gu T., Shen Q., et al. (2013). Fungal networks in yield-invigorating and -debilitating soils induced by prolonged potato monoculture. Soil Biol. Biochem. 65, 186–194. 10.1016/j.soilbio.2013.05.025 [DOI] [Google Scholar]
- Magoč T., Salzberg S. L. (2011). FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27, 2957–2963. 10.1093/bioinformatics/btr507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendes L. W., Kuramae E. E., Navarrete A. A., van Veen J. A., Tsai S. M. (2014). Taxonomical and functional microbial community selection in soybean rhizosphere. ISME J. 8, 1577–1587. 10.1038/ismej.2014.17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzker M. L. (2010). Sequencing technologies—the next generation. Nat. Rev. Genet. 11, 31–46. 10.1038/nrg2626 [DOI] [PubMed] [Google Scholar]
- Minoo D., Jayakumar V. N., Veena S. S., Vimala J., Basha A., Saji K. V., et al. (2008). Genetic variations and interrelationships in Vanilla planifolia and few related species as expressed by RAPD polymorphism. Genet. Resour. Crop Evol. 55, 459–470. 10.1007/s10722-007-9252-3 [DOI] [Google Scholar]
- Navarro-Noya Y. E., Gómez-Acata S., Montoya-Ciriaco N., Rojas-Valdez A., Suárez-Arriaga M. C., Valenzuela-Encinas C., et al. (2013). Relative impacts of tillage, residue management and crop-rotation on soil bacterial communities in a semi-arid agroecosystem. Soil Biol. Biochem. 65, 86–95. 10.1016/j.soilbio.2013.05.009 [DOI] [Google Scholar]
- Peiffer J. A., Spor A., Koren O., Jin Z., Tringe S. G., Dangl J. L., et al. (2013). Diversity and heritability of the maize rhizosphere microbiome under field conditions. Proc. Natl. Acad. Sci. U.S.A. 110, 6548–6553. 10.1073/pnas.1302837110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penton C. R., Gupta V. V., Tiedje J. M., Neate S. M., Ophel-Keller K., Gillings M., et al. (2014). Fungal community structure in disease suppressive soils assessed by 28S LSU gene sequencing. PLoS ONE 9:e93893. 10.1371/journal.pone.0093893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippot L., Raaijmakers J. M., Lemanceau P., van der Putten W. H. (2013). Going back to the roots: the microbial ecology of the rhizosphere. Nat. Rev. Microbiol. 11, 789–799. 10.1038/nrmicro3109 [DOI] [PubMed] [Google Scholar]
- Pinaria A. G., Liew E. C. Y., Burgess L. W. (2010). Fusarium species associated with vanilla stem rot in Indonesia. Australas. Plant Pathol. 39, 176–183. 10.1071/AP09079 [DOI] [Google Scholar]
- Qiu M., Li S., Zhou X., Cui X., Vivanco J. M., Zhang N., et al. (2013). De-coupling of root–microbiome associations followed by antagonist inoculation improves rhizosphere soil suppressiveness. Biol. Fertil. Soils 50, 217–224. 10.1007/s00374-013-0835-1 [DOI] [Google Scholar]
- Sandheep A. R., Aju K., Asok Ans Jisha M. S. (2012). Biocontrol of Fusarium wilt of vanilla (Vanilla planifolia) using combined inoculation of Trichoderma sp. and Pseudomonas sp. Int. J. Pharma Bio. Sci. 3, 706–716. Available online at: http://www.ijpbs.net/vol-3/issue-3/bio/82.pdf [Google Scholar]
- Schloss P. D., Westcott S. L., Ryabin T., Hall J. R., Hartmann M., Hollister E. B., et al. (2009). Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75, 7537–7541. 10.1128/AEM.01541-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shokralla S., Spall J. L., Gibson J. F., Hajibabaei M. (2012). Next-generation sequencing technologies for environmental DNA research. Mol. Ecol. 21, 1794–1805. 10.1111/j.1365-294X.2012.05538.x [DOI] [PubMed] [Google Scholar]
- Tombe M., Sitepu D. (1986). The control of stem rot of vanilla with fungicides. Ed. Khusus Penelit. Tanam. Rempah Dan Obat 11, 43–47. [Google Scholar]
- van Elsas J. D., Chiurazzi M., Mallon C. A., Elhottovâ D., Krištùfek V., Salles J. F. (2012). Microbial diversity determines the invasion of soil by a bacterial pathogen. Proc. Natl. Acad. Sci. U.S.A. 109, 1159–1164. 10.1073/pnas.1109326109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijayan A. K., Thomas J., Dhanapal K., Thomas J. (2009). Management of rot diseases of vanilla (Vanilla planifolia Andrews) using bioagents. J. Plant. Crops 37, 229–231. Available online at: http://www.cabdirect.org/abstracts/20113073569.html;jsessionid=F7D69B86DF6B31BA1BF18AEDAD901E98 [Google Scholar]
- Wang B., Li R., Ruan Y., Ou Y., Zhao Y., Shen Q. (2015). Pineapple–banana rotation reduced the amount of Fusarium oxysporum more than maize–banana rotation mainly through modulating fungal communities. Soil Biol. Biochem. 86, 77–86. 10.1016/j.soilbio.2015.02.021 [DOI] [Google Scholar]
- Wang B., Yuan J., Zhang J., Shen Z., Zhang M., Li R., et al. (2013). Effects of novel bioorganic fertilizer produced by Bacillus amyloliquefaciens W19 on antagonism of Fusarium wilt of banana. Biol. Fertil. Soils 49, 435–446. 10.1007/s00374-012-0739-5 [DOI] [Google Scholar]
- Wei Z., Yang X., Yin S., Shen Q., Ran W., Xu Y. (2011). Efficacy of Bacillus-fortified organic fertiliser in controlling bacterial wilt of tomato in the field. Appl. Soil Ecol. 48, 152–159. 10.1016/j.apsoil.2011.03.013 [DOI] [Google Scholar]
- White T. J., Bruns T., Lee S., Taylor J. W., others (1990). Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protoc. Guide Methods Appl. 18, 315–322. 10.1016/b978-0-12-372180-8.50042-1 [DOI] [Google Scholar]
- Xiong W., Li Z., Liu H., Xue C., Zhang R., Wu H., et al. (2015a). The effect of long-term continuous cropping of black pepper on soil bacterial communities as determined by 454 pyrosequencing. PLoS ONE 10:e0136946. 10.1371/journal.pone.0136946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong W., Zhao Q., Zhao J., Xun W., Li R., Zhang R., et al. (2015b). Different continuous cropping spans significantly affect microbial community membership and structure in a vanilla-grown soil as revealed by deep pyrosequencing. Microb. Ecol. 70, 209–218. 10.1007/s00248-014-0516-0 [DOI] [PubMed] [Google Scholar]
- Xu L., Ravnskov S., Larsen J., Nilsson R. H., Nicolaisen M. (2012). Soil fungal community structure along a soil health gradient in pea fields examined using deep amplicon sequencing. Soil Biol. Biochem. 46, 26–32. 10.1016/j.soilbio.2011.11.010 [DOI] [Google Scholar]
- Xuan D. T., Guong V. T., Rosling A., Alström S., Chai B., Högberg N. (2011). Different crop rotation systems as drivers of change in soil bacterial community structure and yield of rice, Oryza sativa. Biol. Fertil. Soils 48, 217–225. 10.1007/s00374-011-0618-5 [DOI] [Google Scholar]
- Zhang N., Castlebury L. A., Miller A. N., Huhndorf S. M., Schoch C. L., Seifert K. A., et al. (2006). An overview of the systematics of the Sordariomycetes based on a four-gene phylogeny. Mycologia 98, 1076–1087. 10.3852/mycologia.98.6.1076 [DOI] [PubMed] [Google Scholar]
- Zu C., Li Z., Yang J., Yu H., Sun Y., Tang H., et al. (2014). Acid soil is associated with reduced yield, root growth and nutrient uptake in black pepper (Piper nigrum L.). Agric. Sci. 5, 466–473. 10.4236/as.2014.55047 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.