Abstract
Platelets play an essential role in hemostasis and thrombosis. We performed a genome-wide association study of platelet count in 12,491 participants of the Hispanic Community Health Study/Study of Latinos by using a mixed-model method that accounts for admixture and family relationships. We discovered and replicated associations with five genes (ACTN1, ETV7, GABBR1-MOG, MEF2C, and ZBTB9-BAK1). Our strongest association was with Amerindian-specific variant rs117672662 (p value = 1.16 × 10−28) in ACTN1, a gene implicated in congenital macrothrombocytopenia. rs117672662 exhibited allelic differences in transcriptional activity and protein binding in hematopoietic cells. Our results underscore the value of diverse populations to extend insights into the allelic architecture of complex traits.
Introduction
Platelets are small, anucleate cells derived from megakaryocyte cytoplasm in the bone marrow. Platelet production results from a series of tightly regulated processes that require lineage commitment of hematopoietic stem cells and leads to the proliferation, terminal differentiation, and maturation of megakaryocytic progenitors. Studying the genetic underpinnings of platelet count (PLT) can provide important insight into molecular mechanisms and pathways involved in both normal and abnormal megakaryopoiesis, which could ultimately have clinical implications for the treatment of bleeding or thrombosis in individuals with a low (thrombocytopenia) or high (thrombocytosis) PLT1, 2, 3 or for the relationship between PLT and cardiovascular or autoimmune disorders.4, 5
Circulating PLT in humans normally ranges between 150,000/μl and 400,000/μl. PLT differs by ethnicity, and these ethnic differences do not appear to be explained by environmental factors.6, 7 Family-based studies have estimated that a large component of the variability of PLT is explained by genetic factors (h2 > 0.50).8, 9, 10 To date, approximately 60 PLT-associated genetic variants have been identified through genome-wide association studies (GWASs) in populations of European, Asian, and African descent.2, 5, 11, 12, 13, 14
Hispanic and/or Latino (Hispanic/Latino) individuals are a highly heterogeneous population with recent admixture among indigenous Amerindian (primarily of South and Central America, Mexico, and the Caribbean islands, hereafter referred to as “Amerindian”), European, and West African ancestral populations. Genetic factors contributing to PLT among Hispanic/Latino populations have not previously been characterized. Notably, certain Mendelian platelet disorders are more common among Hispanic/Latino individuals,15 suggesting the possibility of population-specific genetic contributions to platelet-related phenotypes. To further characterize the role of genetic factors contributing to PLT in Hispanic/Latino populations, we performed a GWAS in 12,491 participants in the Hispanic Community Health Study/Study of Latinos (HCHS/SOL). We sought to identify genetic loci associated with PLT and assess generalization of known loci from other populations to this diverse sample of Hispanic/Latino individuals.
Material and Methods
HCHS/SOL Population
The HCHS/SOL is a community-based cohort study of 16,415 self-identified Hispanic/Latino persons aged 18–74 years and selected from households in predefined census-block groups across four US field centers (in Chicago, Miami, the Bronx, and San Diego). The census-block groups were chosen to provide diversity among cohort participants with regard to socioeconomic status and national origin or background. The HCHS/SOL cohort includes participants who self-identified as having a Hispanic/Latino background; the largest groups are Central American (n = 1,730), Cuban (n = 2,348), Dominican (n = 1,460), Mexican (n = 6,471), Puerto Rican (n = 2,728), and South American (n = 1,068). The sample design and cohort selection have been previously described.16 The HCHS/SOL baseline clinical examination17 occurred between 2008 and 2011 and included comprehensive biological, behavioral, and sociodemographic assessments. This study was approved by the institutional review boards at each field center, where all subjects gave written informed consent.
Measurement of PLT and Exclusion Criteria in HCHS/SOL
PLT was measured in EDTA whole blood with a Sysmex XE-2100 instrument, (Sysmex America) at the University of Minnesota according to national and international standards and procedures. Individuals pregnant at the time of blood draw; those with >5% circulating blasts or immature cells, end-stage renal disease, or any hematologic malignancy; and those undergoing chemotherapy for solid tumors were excluded from our analyses.
Genotyping and Quality Control in HCHS/SOL
Consenting HCHS/SOL subjects were genotyped at Illumina on the HCHS/SOL custom 15041502 B3 array. The custom array comprised the Illumina Omni 2.5M array (HumanOmni2.5-8v.1-1) ancestry-informative markers, known GWAS hits and drug absorption, distribution, metabolism, and excretion (ADME) markers, and additional custom content including ∼150,000 SNPs selected from the CLM (Colombian in Medellin, Colombia), MXL (Mexican Ancestry in Los Angeles, California), and PUR (Puerto Rican in Puerto Rico) samples in the 1000 Genomes phase 1 data to capture a greater amount of Amerindian genetic variation.18
We applied standardized quality-assurance and quality-control (QA/QC) methods19 to generate recommended SNP- and sample-level quality filters. In brief, samples were checked for annotated or genetic sex, gross chromosomal anomalies,20 relatedness21 and population structure,22 missing call rates, batch effects, and duplicate-sample discordance. At the SNP level, checks were performed for Hardy-Weinberg equilibrium, minor allele frequency (MAF), duplicate-probe discordance, Mendelian errors, and missing call rate. These QA/QC procedures yielded a total of 12,803 unique study participants for imputation and downstream association analyses. Of these, 12,491 met specific inclusion criteria related to the study of PLT. A total of 2,232,944 SNPs passed filters for both quality and informativeness (polymorphic and unduplicated) and became candidates for imputation and association testing.
Imputation in HCHS/SOL
Genome-wide imputation was carried out with the full, cosmopolitan 1000 Genomes Project phase 1 reference panel (n = 1,092).23 The HCHS/SOL samples were imputed together with genotyped SNPs passing the quality filter and representing unique genomic positions on the autosomes and non-pseudoautosomal portion of the X chromosome. Genotypes were first pre-phased with SHAPEIT2 (v.2.r644) and then imputed with IMPUTE2 (v.2.3.0).24, 25 Only variants with at least two copies of the minor allele present in any of the four 1000 Genomes continental panels were imputed. In addition to calculating the quality metrics output by IMPUTE2, we also calculated “oevar” (the ratio of the observed variance of imputed dosages to the expected binomial variance) by using the MaCH imputation software.26 We assessed overall imputation quality both by looking at the distribution of imputed quality metrics across the MAF spectrum and by examining results from the IMPUTE2 internal masking experiments. We performed downstream association analyses only on observed variants passing quality filters and all imputed variants (a total of 27,887,661 variants), but we filtered the results on the basis of imputation quality (oevar > 0.3) and MAF > 1%.
Linear Mixed-Effect Model for Association Testing in HCHS/SOL
We analyzed PLT by using linear mixed-effect models (LMMs) to account for the correlations due to genetic relatedness (kinship), shared household, and block group between individuals. The LMM used three independent random effects to model these three sources of dispersion:
where yi is the square-root-transformed platelet value for individual i, xi is a vector of covariate values, α is the corresponding regression parameters, gij is the jth SNP count, where βj is its estimated effect, and bik, bih, and bib are the random effects corresponding to kinship, household, and block group, respectively (independent of each other and the error term εi), of person i. Within the LMM framework, bih (bib) is the same among individuals who live in the same household (block group); for two individuals i and l, the correlation between bik and blk is given by their estimated kinship coefficient.27 The covariates included sex, age, principle components (PCs), recruitment center, smoking, log of sampling weight, and genetic-analysis group (a six-level categorical variable derived from self-identified background). With square-root-transformed PLT, the null-model residuals, given by ei = yi − xiTα − gijβj, were approximately normally distributed and thus compatible with modeling assumptions. We evaluated the goodness of fit of the LMM by using quantile-quantile plots comparing residuals and estimated random effects to a normal distribution and scatter plots describing the relationship between model residuals and covariates.
Ancestry and Relatedness Adjustment in HCHS/SOL
We adjusted analyses for five PCs to prevent spurious association due to population stratification. Analyses accounted for familial relatedness (kinship) by using a random effect with correlation structure specified by pairwise kinship coefficients for preventing inflation of test statistics. The PCs and kinship coefficients were estimated simultaneously with an iterative procedure, alternating between PC-AiR22 (which provides PCs robust to familial relatedness) and PC-Relate21 (which estimates kinship coefficients robust to population structure, admixture, and non-random mating). PC-AiR uses relatedness estimates to identify a mutually unrelated subset of individuals representative of the ancestral diversity of the entire sample, performs PCA on this unrelated subset, and predicts PC values for the remaining individuals. PC-Relate uses PCs to account for genetic similarity due to shared ancestry and provide accurate estimates of kinship coefficients due to familial relatedness. We performed three iterations, each of which used ∼150,000 linkage-disequilibrium (LD)-pruned SNPs.28
SNP-Based Heritability Estimation in HCHS/SOL
Genetic (kinship) and shared environmental (household) effects were estimated from a variance-component analysis that used all genotyped SNPs with MAF > 1% (∼1.7 million) and a subset of 10,093 individuals estimated to be more distant than fourth-degree relatives (i.e., for whom all pairwise kinship coefficient estimates from PC-Relate were less than 2−11/2 ≈ 0.022). Including close relatives in the analysis can lead to inflated heritability estimates as a result of their increased phenotypic correlations due to other factors such as shared environmental effects.29 However, the availability of current household membership data in HCHS/SOL made it possible that the variance-component model could at least partially account for shared environmental effects; therefore, the analysis was repeated with all 12,491 study individuals.
Estimation of SNP Alleles and Allelic Frequencies among Ancestral Populations
We compared allele frequencies of PLT-associated index SNPs across ancestral Hispanic/Latino populations by using data from phase 3 of 1000 Genomes.23 We used the R31 exactci package to calculate exact p values and matching 95% confidence intervals (CIs) for each sub-population from the binomial distribution.30 We also examined whether the derived alleles at our PLT-associated index SNPs were present in other ancestral human or Amerindian populations by using published whole-genome sequence data from Neandertal and Denisovan archaic human samples32, 33 and Papua New Guinea samples.34
Replication of Discovery Loci in Independent Hispanic/Latino Samples
To replicate association findings in Hispanic/Latino samples, we used 1000 Genomes imputed GWAS data available in three additional Hispanics/Latinos samples, including 3,454 from the Women’s Health Initiative (WHI) SNP Health Association Resource (SHARe) project,35 782 from the Multi-Ethnic Study of Atherosclerosis (MESA) cohort,36, 37 and 2,854 from Mount Sinai BioMe Biobank.38 WHI-SHARe and MESA participants were genotyped with the Affymetrix 6.0 chip, and imputation was performed with MaCH.26 BioMe participants were genotyped with the Illumina HumanOmniExpressExome-8 v.1.0 chip, and imputation was performed with IMPUTE224, 25 in 1000 Genomes phase 1 data (March 2012 v.3). Association testing for typed or imputed SNPs was performed by linear regression of square-root-transformed PLT adjusted for age, sex, and PCs.
Meta-analysis and Replication Significance Criteria
Meta-analysis of results from the three replication cohorts for PLT and mean platelet volume (MPV) was performed with the inverse-variance-weighted method implemented in METAL.39 To declare significance for replicated PLT loci, we used Bonferroni correction for the six variants carried forward for a significance threshold of p value < 0.0083.
Admixture Mapping in HCHS/SOL
We implemented a conditional-random-field-based approach, RFMix,40 to infer local ancestry at a set of 236,456 SNPs in common between the HCHS/SOL and reference-panel datasets. We used selected populations from HGDP,41 HapMap 3,42 and 1000 Genomes23 phase 1 to use as a reference panel for detecting European, West African, and Amerindian ancestry. RFMix requires phased data with no missing genotype values. BEAGLE (v.4) was employed for phasing and imputation of sporadic missing genotypes in the HCHS/SOL and reference-panel datasets.43 Admixture mapping is a powerful gene-mapping approach that relies on allele-frequency differences across ancestral populations and the existence of an association between the causal variant and phenotype to identify an association. Using the local-ancestry estimates, we performed a genome-wide admixture-mapping scan by using a LMM43 with a joint test for all three ancestries (European, African, and Amerindian). As a secondary analysis, we performed admixture mapping to test Amerindian against any other ancestry because a priori we were interested in Amerindian ancestry within HCHS/SOL because it has not been well studied in previous admixture-mapping studies. Covariate and ancestry adjustment used in the analyses is described above. The recent history of admixture gives rise to long-range correlation in local-ancestry values across the genome, and thus the critical value for the genome-wide significance level of admixture mapping is substantially lower than that for the genotype test. On the basis of previous simulation results, a nominal p value of 5.7 × 10−5 yielded a genome-wide type I error of 0.05.
Generalization in HCHS/SOL
We performed generalization analysis for PLT-associated SNPs previously reported in GWASs of other populations, including those of European, African, and Japanese ancestry.2, 5, 11, 12, 13 Because all discovery studies, except for that of Kamatani et al.,11 used untransformed PLT as the outcome, we used association results with untransformed PLT for generalization. For SNPs reported in Kamatani et al.,11 we followed their methodology and used the square root of PLT and reported effect sizes in SDs. We performed generalization testing by directional false-discovery rate (FDR) control for the generalization null hypotheses.44 The generalization null hypothesis states that the effect does not exist in both the discovery study and HCHS/SOL and is rejected if there is enough evidence that a SNP affects the outcome, with the same direction of effect, in both the discovery study and HCHS/SOL. We used the number of SNPs tested in the discovery study and the p values for the set of tested SNPs from both the discovery study and HCHS/SOL, and we computed an r value for each of the SNPs to quantify the evidence for generalization. A SNP was generalized if the r value < 0.05.
Functional Annotation of Discovery Loci
We interrogated the PLT-associated loci to determine whether the identified non-coding SNPs and indels and correlated variants (r2 ≥ 0.5, calculated in the HCHS/SOL discovery population) were positioned within predicted regulatory regions, namely enhancers and promoters. These regulatory regions were identified on the basis of the enrichment of various histone-modification and ChIP-seq (chromatin immunoprecipitation followed by sequencing) signals in megakaryocytes.45 A genomic element enriched with the histone H3K4me1 signal was categorized as an enhancer, whereas a genomic element enriched with the histone H3K4me3 signal was categorized as a promoter. SNPs or indels that belonged to either promoter or enhancer categories and overlapped a DNase I hypersensitive site (a general biochemical feature of regulatory regions) in megakaryocytes were prioritized as putatively functional variants. We also reported overlap with ChIP-seq peaks of key megakaryocyte transcription factors and the nearest biologically plausible gene or genes.46 Moreover, because related cell types can share similar regulatory regions, we additionally reported supplementary annotation by using data generated by ENCODE47 on other myeloid lineage cells, including primary erythroblasts, erythroleukemia K562 cells, peripheral-blood-derived erythroblasts (PBDEs), myeloid leukemia (SKNO-1) cells, and human umbilical vein endothelial cells (HUVECs). To provide additional support, we also included overlap with transcription start sites and enhancers identified by an alternate approach in the Fantom5 project. 48 To identify the motifs disrupted by alleles, including ACTN1 (MIM: 102575) variant rs117672662, we utilized HaploReg (v.2)49 and the JASPAR motif database.50
We also included annotations from in silico prediction algorithms including RegulomeDB, the Combined Annotation Dependent Depletion (CADD) score51 (a PHRED-like score indicating deleteriousness of variants and all other substitutions in the genome), GWAVA52 (a score that classifies non-coding variation and uses ENCODE and Roadmap Epigenomic data to prioritize most likely functional variants), and deltaSVM53 (a score that captures how much a variant alters the regulatory potential of the surrounding sequence, particularly in the context of a specific cell type).
eQTL Analysis of American Indians
The eQTL analysis included 1,457 American Indian adults (minimum of 18 years of age) from the urban Phoenix extension of the Family Investigation of Nephropathy and Diabetes; they were examined after they had fasted for ≥8 hr.54 Blood was collected into PAXgene Blood RNA Tubes (Becton Dickinson), and total RNA was isolated with PAXgene Blood miRNA Kits (QIAGEN). Amplification was performed with the Ambion MessageAmp II-Biotin Enhanced aRNA Amplification Kit (Life Technologies), and transcript levels were measured with the Illumina HumanHT-12 v.4 Expression Beadchip according to the manufacturer’s protocol. GenomeStudio software was used for background normalization. Genotyping of rs117672662 was conducted according to the Assays-on-Demand method (Life Technologies). A normalizing transformation of transcription levels was employed in statistical analyses, and association of genotype was analyzed under an additive model with control for age, sex, tribal membership, and European admixture (estimated from 45 markers with large allele-frequency differences between Amerindians and Europeans55).
Cell Culture
THP-1 (ATCC TIB-202) acute monocytic leukemia cells were cultured in RPMI-1640 (Mediatech) supplemented with 10% fetal bovine serum (FBS), and Kasumi-1 (ATCC CRL-2724) acute myeloblastic leukemia cells were cultured in RPMI-1640 supplemented with 20% FBS. The cell cultures were maintained at 37°C with 5% CO2.
Transcriptional Reporter Assays
A 186 bp region (chr14: 69,425,369–69,425,554 according to UCSC Genome Browser hg19) surrounding rs117672662 was amplified with primer pairs 5′-GGTACCGCAGGAAAACATCCACATGA-3′ and 5′-CTCGAGGGAAACAGTGTGGTCAGTCG-3′ (forward) and 5′-CTCGAGGCAGGAAAACATCCACATGA-3′ and 5′- GGTACCGGAAACAGTGTGGTCAGTCG-3′ (reverse) and cloned into the luciferase reporter vector pGL4.23 (Promega) in both orientations with respect to the minimal promoter. The rs117672662 C allele was created with the QuikChange Site-Directed Mutagenesis Kit (Stratagene). Sanger sequencing was used to verify clones for fidelity and genotype. Four verified constructs for each allele in both orientations were transfected in duplicate into THP-1 and Kasumi-1 cells with Renilla control reporter vector (phRL-TK, Promega) with Lipofectamine 3000 (Life Technologies) and incubated for 48 hr. The cells were lysed with Passive Lysis Buffer (Promega), and luciferase activity was measured with the Dual-Luciferase Reporter Assay System (Promega) as previously described.56
Electrophoretic Mobility Shift Assay
Electrophoretic mobility shift assay (EMSA) oligonucleotide probes were designed around the variant rs117672662 (5′-AGAATTAT[T/C]AGCAGAGG-3′) and end labeled with 5′ IRDye 700 (Integrated DNA Technologies). Nuclear protein was extracted with the NE-PER Extraction Kit (ThermoFisher Scientific), and the total nuclear extract was measured with the BCA Protein Assay (ThermoFisher Scientific). All protein-probe binding reactions were incubated for 30 min at room temperature and consisted of the following: 1× binding buffer (10 mM Tris, 50 mM KCl, and 1 mM DTT [pH 7.5]), 1 μg poly(dI-dC), 7 μg nuclear extract, and 200 fmol IRDye-labeled double-stranded oligonucleotide probe in a volume of 20 μl. The competition reactions contained 70-fold excess of unlabeled probe and were incubated with the nuclear extract for 15 min prior to the addition of the IRDye-labeled probe and incubation for another 30 min. In a test for an antibody super-shift, 6 μg of antibody (HOXA5 sc-13199x or GATA1 sc-1234x, Santa Cruz Biotechnologies) was incubated with the nuclear extract for 35 min prior to incubation with the IRDye-labeled probe for 30 min. The probe-protein complexes were resolved with 6% DNA retardation electrophoresis gels (Life Technologies) and visualized with an Odyssey CLx Infrared Imaging System (LI-COR Biosciences).
Results
The characteristics of the 12,491 Hispanic/Latino participants from the HCHS/SOL discovery sample are summarized in Table S1. SNP-based heritability was estimated from a variance-component analysis performed with a subset of 10,093 individuals excluding close familial relatives. Genetic (kinship) effects accounted for 29.4% (95% CI: 22.6%–36.1%) of the variation in PLT, whereas shared environmental (household) effects contributed little (2.6%; 95% CI: 0.0%–6.3%). When the analysis was repeated with all individuals and adjustment for household-membership data, genetic (kinship) effects accounted for 30.8% (95% CI: 25.7%–35.7%), and shared environmental (household) effects accounted for 5.0% (95% CI: 2.1%–7.7%) of PLT variation. Thus, the estimated genetic contribution increased only slightly (from 29.4% to 30.8%) when close relatives were included, and the estimated household contribution was non-zero, suggesting that household membership is a good proxy for the shared environmental effects contributing to PLT in this sample. These heritability estimates are less than what has been reported from family-based studies of PLT (approximately 50%–80%8, 9, 10), which is consistent with previous findings comparing family- and SNP-based heritability estimation.57
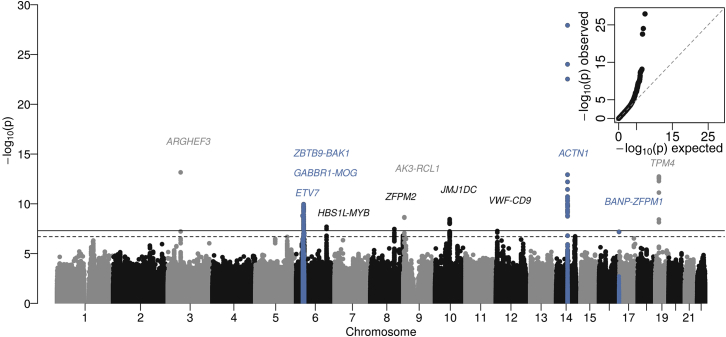
In the HCHS/SOL discovery sample, the genomic inflation factor was 1.046, indicating adequate control of population stratification. Nine loci met the standard significance criteria of p value < 5 × 10−8; three additional loci had p values between 5 × 10−8 and 1 × 10−7 (Table S2). Quantile-quantile and Manhattan plots are shown in Figure 1. For each of the discovery and previously reported PLT loci, we evaluated the extent of LD (calculated HCHS/SOL discovery population) between the index SNP and other nominally significant SNPs in the region (Figures S1A–S1L).
Figure 1.
Manhattan Plot of Discovery Results from HCHS/SOL
The solid line indicates genome-wide significance (p value < 5 × 10−8), and a dashed line indicates suggestive significance (p value < 1 × 10−7). There is an inset quantile-quantile plot of discovery p values. Discovery loci are highlighted in blue, and loci with p values less than the suggestive significance threshold are annotated with the names of the nearest gene(s).
Generalization of PLT Index SNPs from Other Populations to Hispanic/Latino Populations
Of the 12 genome-wide significant or suggestive loci in the HCHS/SOL discovery sample, seven correspond to index SNPs (or LD proxies with r2 ≥ 0.5 calculated in the HCHS/SOL population) previously identified in PLT GWASs of other ancestries (ARHGEF3 [MIM: 612115] rs1354034, TPM4 [MIM: 600317] rs73517714, AK3 [MIM: 609290]-RCL1 [MIM: 611405] rs409801, JMJD1C [MIM: 604503] rs10822155, ZFPM2 [MIM: 603693] rs6993770, HBS1L [MIM: 612450]-MYB [MIM: 189990] rs6934903, and CD9 [MIM: 143030]-VWF [MIM: 613160] rs11064074)2, 5, 12, 13, 14 (Table S2). An eighth significant locus is located in close proximity to BAK1 (MIM: 600516), a gene previously associated with PLT in GWASs from populations of European, African, and Asian descent.2, 5, 11, 12, 14 However, the index SNP (rs62405954) in our Hispanic/Latino discovery sample at the BAK1 locus is distinct from rs210134, the index SNP previously associated with PLT in a GWAS (r2 = 0.06; rs62405954 p value = 1.10 × 10−10, versus 4.6 × 10−7 when adjusted for rs210134; Figure S2).
In order to more comprehensively assess whether previous GWAS PLT SNPs from populations of European, Asian, and African ancestry2, 5, 11, 12, 13, 14 generalize to HCHS/SOL Hispanic/Latino populations, we evaluated all index SNPs in the corresponding ancestral populations (Table S3) by using a directional FDR method58 that rejects the null hypothesis of “no generalization” if there is enough evidence that a SNP is associated with PLT and directionally consistent between the original discovery GWAS and HCHS/SOL. Of the ten SNPs identified previously in populations of African ancestry,12, 13 seven of the SNPs showed evidence of generalization (r value < 0.05) in our Hispanic/Latino sample. Roughly half (27 of 49) of GWAS SNPs identified previously in populations of European ancestry2, 5, 12, 14 generalized to HCHS/SOL. All four SNPs identified previously in a population of Asian ancestry11 generalized to our study. Considering the 55 independent SNPs previously associated with PLT in any population, we found evidence of generalization of 30 (55%) SNPs in the HCHS/SOL population. We hypothesized that some of the SNPs did not generalize as a result of low power. To study this hypothesis, we (1) looked for directional consistency of the effect sizes across our study and previous studies for SNPs that failed to generalize and (2) generated a genetic score for each of the analysis participants by summing all trait-increasing alleles in the SNPs that did not generalize. Out of 25 SNPs that did not generalize, 24 had the same direction of effect in the discovery study and the HCHS/SOL (exact binomial test p value = 1.5 × 10−6). We found a strong association between PLT and the genetic score that we constructed with the non-generalized SNPs (p value = 8.8 × 10−10). Taken together, these tests provide evidence that, indeed, the majority of non-generalized SNPs are associated with PLT.
Discovery of Ancestry-Specific PLT Association Signals in Hispanic/Latino Populations
Four of the 12 loci with significant or suggestive associations in the HCHS/SOL discovery sample (ACTN1 rs117672662, GABBR1 [MIM: 603540]-MOG [MIM: 159465] rs75140056, ETV7 [MIM: 605255] rs9470264, and BANP [MIM: 611564]-ZFPM1 [MIM: 601950] rs80294974) are located within or near genes or genomic regions not previously associated with PLT in GWASs (Table 1). By examining the 55 other genomic regions previously associated with PLT in populations of European, Asian, or African descent, in addition to ZBTB9-BAK1 rs62405954 (described above), we identified one additional signal, MEF2C (MIM: 600662) rs144261491, distinct from the MEF2C European index SNP rs700585 from a GWAS (r2 = 0.009; rs144261491 p value = 3.4 × 10−7, versus 9.0 × 10−7 with adjustment for rs700585; Figure S3).
Table 1.
PLT-Associated Variants from the HCHS/SOL Discovery Analysis
| Nearest Gene (Function) | Chromosomal Position (GRCh37/hg19) | rsID | Coded/Alternative Allele on Plus Strand |
Discovery |
Replication |
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| Coded Allele Frequency | n | Beta (SE) | p Value | n | Beta (SE) | p Value | ||||
| ACTN1 (intronic) | chr14: 69,425,467 | rs117672662 | T/C | 0.94 | 12,491 | 0.604 (0.054) | 1.16 × 10−28 | 7,121 | 0.685 (0.080) | 1.07 × 10−17 |
| ZBTB9-BAK1 (intergenic) | chr6: 33,524,820 | rs62405954 | T/C | 0.86 | 12,491 | −0.239 (0.037) | 1.10 × 10−10 | 7,170 | −0.268 (0.066) | 4.93 × 10−5 |
| GABBR1-MOG (intergenic) | chr6: 29,608,184 | rs75140056 | C/CAT | 0.39 | 12,491 | −0.151 (0.025) | 1.47 × 10−9 | 7,163 | −0.236 (0.041) | 9.82 × 10−9 |
| BANP-ZFPM1 (intergenic) | chr16: 88,376,014 | rs80294974 | G/A | 0.98 | 12,491 | 0.555 (0.103) | 6.60 × 10−8 | 7,060 | −0.082 (0.138) | 0.551 |
| ETV7 (intronic) | chr6: 36,344,980 | rs9470264 | G/A | 0.80 | 12,491 | −0.181 (0.034) | 8.80 × 10−8 | 6,998 | −0.145 (0.048) | 0.00263 |
| MEF2C (intronic) | chr5: 88,133,921 | rs144261491 | C/T | 0.97 | 12,491 | 0.426 (0.083) | 3.35 × 10−7 | 3,982 | 0.624 (0.156) | 6.32 × 10−5 |
Several of the PLT index SNPs discovered in HCHS/SOL had allele frequencies that differed considerably between continental populations represented in 1000 Genomes phase 3 samples23 (Table 2). In particular, ACTN1 rs117672662 and MEF2C rs14426149, associated with decreased PLT, were present at MAFs of ∼7% and ∼4%, respectively, in samples of Amerindian ancestry (Columbians, Mexicans, Peruvians, and Puerto Ricans) but were significantly less common in Asian, European, and African populations.23 Analysis of published archaic genomes32, 33 and genome sequences from New Guinea Papuans34 showed that none of the risk alleles at these six PLT loci appear to be derived from Neandertal, Denisovan, or Australo-Melanesian sequences.
Table 2.
Allele Frequencies of PLT-Associated Variants by 1000 Genomes Continental Populations and Admixed American Sub-populations
| Gene (Function) | Chromosomal Position (GRCh37/hg19) | rsID | Coded/Alternative Allele on Plus Strand | HCHS/SOL Coded Allele Frequency |
Super-population Coded Allele Frequency |
AMR Sub-population Coded Allele Frequency |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AFR | EAS | EUR | SAS | AMR | CLM | MXL | PEL | PUR | |||||
| ACTN1 (intronic)a | chr14: 69,425,467 | rs117672662 | T/C | 0.94 | 1.000 | 1.00 | 0.999 | 0.999 | 0.927 | 0.926 | 0.938 | 0.859 | 0.976 |
| ZBTB9-BAK1 (intergenic) | chr6: 33,524,820 | rs62405954 | T/C | 0.86 | 0.999 | 0.994 | 0.919 | 0.964 | 0.831 | 0.824 | 0.781 | 0.741 | 0.942 |
| GABBR1-MOG (intergenic) | chr6: 29,608,184 | rs75140056 | C/CAT | 0.39 | 0.384 | 0.213 | 0.417 | 0.304 | 0.365 | 0.452 | 0.391 | 0.206 | 0.399 |
| BANP-ZFPM1 (intergenic) | chr16: 88,376,014 | rs80294974 | G/A | 0.98 | 0.998 | 1.000 | 0.963 | 0.993 | 0.991 | 0.995 | 0.992 | 0.994 | 0.986 |
| ETV7 (intronic) | chr6: 36,344,980 | rs9470264 | G/A | 0.80 | 0.825 | 0.877 | 0.992 | 0.995 | 0.765 | 0.809 | 0.656 | 0.612 | 0.918 |
| MEF2C (intronic)a | chr5: 88,133,921 | rs144261491 | C/T | 0.97 | 1.000 | 1.000 | 1.000 | 1.000 | 0.963 | 0.989 | 0.953 | 0.906 | 0.990 |
Abbreviations are as follows: AFR, African; EAS, East Asian; EUR, European; SAS, South Asian; AMR, admixed American; CLM, Colombian from Medellin, Colombia; MXL, Mexican ancestry from Los Angeles, USA; PEL, Peruvians from Lima, Peru; and PUR, Puerto Ricans from Puerto Rico.
For ACTN1 rs117672662 and MEF2C rs144261491, the allele frequencies differ significantly between AMR and AFR, EAS, EUR, and SAS populations.
Admixture mapping based on a joint test of all three local-ancestry estimates in HCHS/SOL confirmed the presence of genome-wide-significant peaks (p value < 5.7 × 10−5) at the BAK1 locus on chromosome 6 and a peak at chromosomal region q13.2, as well as a suggestive peak at the ACTN1 locus on chromosome 14 (Figure S4A). Considering the secondary analysis comparing Amerindian ancestry to all other ancestries, we identified a significant peak at ACTN1 (Figure S4B). Further, there was highly significant concordance between the number of Amerindian ancestral alleles at the ACTN1 locus and the rs117672662 genotype (p value < 2.20 × 10−16; Table S4).
Replication of PLT Loci in Independent Hispanic/Latino Samples
Replication of PLT association findings discovered in HCHS/SOL was carried out in an independent sample of up to 7,170 Hispanic/Latino Americans derived from three multi-ethnic US-based cohorts (WHI [n = 3,534], BioMe Biobank [n = 2,854], and MESA [n = 782]), whose characteristics are described in Table S1. We carried forward six SNPs from the discovery stage for replication: ACTN1 (rs117672662), GABBR1-MOG (rs75140056), ETV7 (rs9470264), BANP-ZFPM1 (rs80294974), ZBTB9-BAK1 (rs62405954), and MEF2C (rs144261491). Of these, five SNPs (all except BANP-ZFPM1 rs80294974) met our pre-specified criteria for replication (p value < 0.05/6 = 0.008; Table 1). BANP-ZFPM1 rs80294974 had a lower frequency (MAF = 2%), so a failure to replicate might be related to limited power to detect this association in the smaller Hispanic/Latino replication sample. The discovery variants and the discovery and generalized variants from HCHS/SOL explain 1.79% and 6.14% of the total variance of PLT, respectively.
In addition to PLT, MPV measurements were available in a subset of 4,041 Hispanic/Latino WHI and BioMe participants in our replication dataset. At four out of five of our replicated loci, the allele associated with lower PLT was also associated with higher MPV (ACTN1 rs117672662 p value = 3.90 × 10−18, GABBR1-MOG rs75140056 p value = 0.003, ETV7 rs9470264 p value = 1.90 × 10−15, and BAK1 rs62405954 p value = 0.03; Table S5).
Functional Annotation and Characterization of the Discovery PLT Loci
At each of the five replicated loci (ACTN1, GABBR1-MOG, ETV7, ZBTB9-BAK1, and MEF2C), we defined the association interval as containing all genotyped or imputed variants (SNPs and indels) in LD (r2 > 0.5) with the index variant. Within each interval, we (1) identified genes and non-coding RNAs and their tissue expression patterns, (2) predicted SNP functionality by using genome-wide epigenomic datasets from megakaryocytes and other blood cell types from the BLUEPRINT,59 ENCODE,47 and FANTOM560 projects (Table S6), and (3) assessed SNP associations with gene expression (eQTL) in whole blood61 (Table S7).
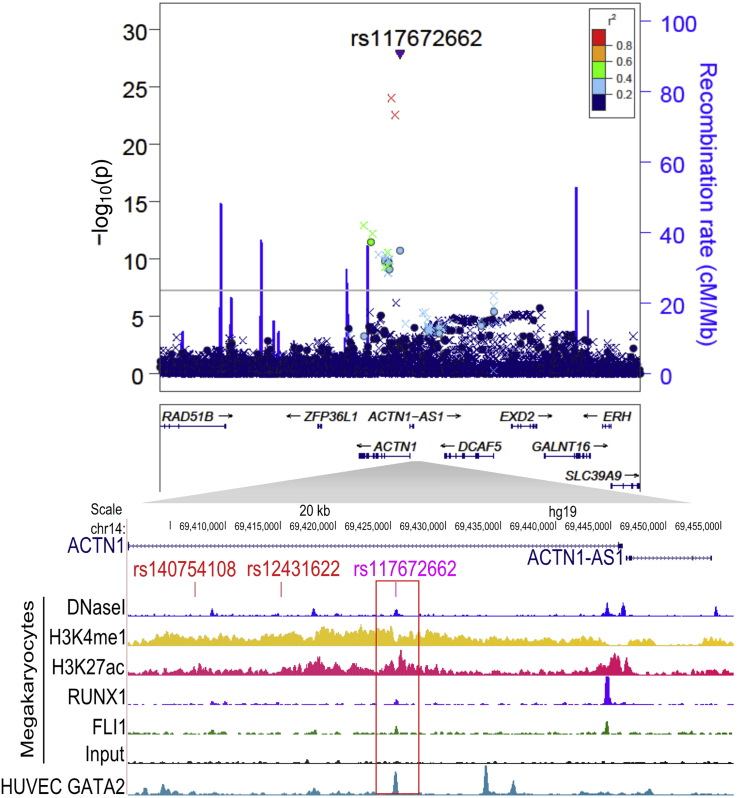
We identified one or more SNPs that overlapped putative megakaryocyte enhancers or promoters and were in LD (r2 ≥ 0.5) with the index SNP at each of the five replicated PLT loci (Table S8). Of particular interest, ACTN1 index SNP rs117672662 lies within a megakaryocyte-specific putative enhancer located within ACTN1 intron 1 (Figure 2). The genomic element harboring rs117672662 overlaps ChIP-seq peaks of key megakaryocyte regulators, including ERG, FLI1, and RUNX1 in SKNO-1 (acute myeloid leukemia) cells, further supporting its role as an enhancer. Furthermore, rs117672662 was predicted to have high regulatory potential according to the cell-type-specific regulatory-motif detection algorithm deltaSVM53 trained on the myelogenous leukemia cell line K562 (score = 9.8; Table S9). The regulatory deltaSVM score for rs117672662 is in the same range as previous predictions for known functional SNPs.53
Figure 2.
Regional Plot of the ACTN1 Locus
The top panel contains a LocusZoom plot of the ACTN1 locus centered on our top Amerindian-specific variant, rs117672662. The LD estimates derived from the HCHS/SOL study samples with respect to rs117672662 and the other variants in the window are color coded according to the scale indicated in the top panel. The imputed SNP, rs117672662, is denoted by a filled triangle, other imputed variants are denoted by an x, and genotyped variants are denoted by a filled circle. Recombination hotspots from HapMap are indicated by the vertical blue lines (see Web Resources). The horizontal line indicates the significance threshold p value ≤ 5 × 10−8. The bottom panel is a UCSC Genome Browser screenshot zoomed in to show rs117672662 and its LD proxies (r2 > 0.8). These variants are aligned with selected signal tracks of megakaryocytes, including DNaseI hypersensitivity, ChIP-seq for enhancer histone modifications H3K4me1 and H3K27ac, ChIP-seq for megakaryocyte transcription factors RUNX1 and Fli1, and the input (no antibody) track. A signal track of ChIP-seq for GATA2, another important megakaryocyte transcription factor in human umbilical vein endothelial cells (HUVECs), is also displayed. The red box is centered on the putative functional SNP rs117672662.
We performed de novo genotyping of the ACTN1 rs117672662 variant in a sample of 1,457 American Indians who were from urban Phoenix and had previously undergone whole-blood transcriptomic analysis. The eQTL analysis did not reveal any significant eQTLs in the region for ACTN1 (C allele: beta [SE] = −0.05 [0.085]; p value = 0.56) or for any other genes in the association interval (Table S10).
Allelic Differences in Enhancer and Protein Binding Activity of ACTN1 rs117672662
To further assess the regulatory properties of ACTN1 variant rs117672662, we performed transcriptional reporter assays in THP-1 monocytic leukemia and Kasumi-1 myeloid leukemia cells. In both cell types, the rs117672662 minor C allele showed higher transcriptional activity than the rs117672662 T allele (Figure S5).
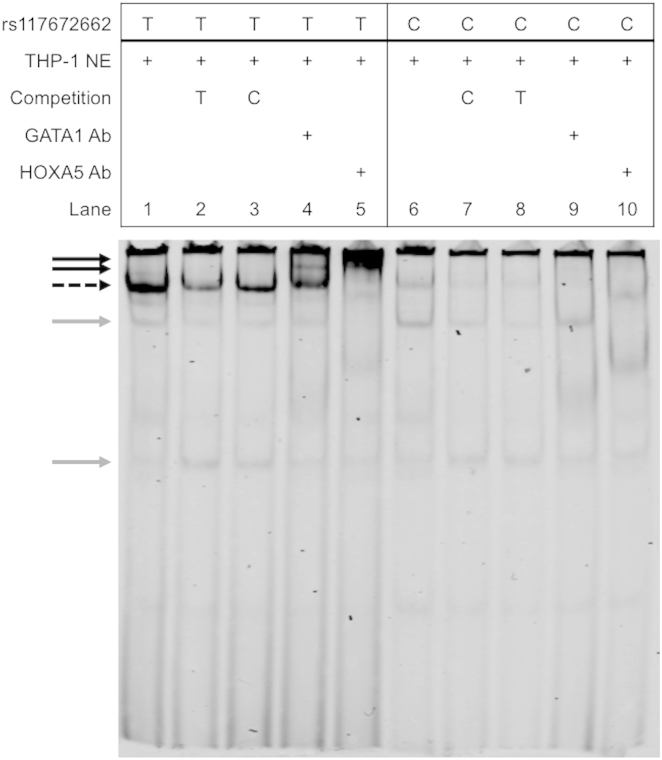
We used THP-1 cells to perform EMSAs and observed that the T allele showed stronger protein binding than the C allele (Figure 3). Including 70-fold excess T allele probe decreased the intensity of the protein-probe band more than including excess C allele probe, supporting allelic differences in specificity of the protein-probe binding (Figure 3 and Figure S6). To characterize the transcription factors binding to the rs1176762662 site, we conducted super-shift assays with antibodies of transcription factors whose DNA binding motifs had been predicted by Haploreg49 to be altered by the variant. An overlapping HOXA5 motif matched better to the T allele, whereas GATA1 and GATA2 motifs matched better to the C allele. With THP-1 cell nuclear lysate, inclusion of HOXA5 antibodies shifted the protein complex bound to the T allele (Figure 3). The addition of GATA1 antibodies showed a partial shift, suggesting that a protein-probe complex might also include GATA1. Taken together, the transcriptional activity and gel-shift assays suggest that a protein complex binding to the T allele contains HOXA5 and might act as a transcriptional repressor of the target gene(s).
Figure 3.
Allelic Differences in Protein Binding at rs117672662
EMSA using oligonucleotide probes containing different alleles at rs117672662 (T allele [lanes 1–5] and C allele [lanes 6–10]). Nuclear extracts from human monocyte THP-1 cells were incubated with IRDye-labeled double-stranded oligonucleotide probe alone (lanes 1 and 6) or with 70-fold excess of unlabeled probe (lanes 2, 3, 7, and 8), GATA1 antibodies (lanes 4 and 9), or HOXA5 antibodies (lanes 5 and 10). The dotted black arrow indicates probe-protein complexes, the solid black arrows indicate probe-protein-antibody complexes, and the gray arrows indicate non-specific probe-protein binding complexes. Further support of the allelic differences is provided in Figure S6.
Discussion
We report the results from a large GWAS of PLT in Hispanic/Latino populations. We identified and replicated associations with three loci including noncoding SNPs in or near ACTN1, ETV7, and GABBR1-MOG and two population-specific variants at previously identified PLT GWAS loci ZBTB9-BAK1 and MEFC2. The ACTN1 and ZBTB9-BAK1 association signals were also detected in a genome-wide scan for local-ancestry admixture. Overall, four of the five PLT association signals (ACTN1, ETV7, MEF2C, and ZBTB9-BAK1) were highly differentiated across populations of European, West African, and Amerindian ancestry. The ACTN1 and MEF2C alleles were found only on an Amerindian ancestral background and therefore could realistically only have been discovered through studies involving Hispanic/Latino or Amerindian populations. We have also demonstrated that approximately 50% of PLT-associated alleles previously identified in European, African American, and Japanese populations generalized to HCHS/SOL populations, suggesting that many of the same regions of the genome are involved in regulation of PLT across global populations.
The identification of a common Amerindian ancestral variant located in a putative enhancer region within intron 1 of ACTN1 adds to previous studies reporting the ACTN1 locus as a source of PLT phenotypic variation. The most likely targets of the rs117672662 variant are either ACTN1 itself or ACTN1-AS1, which is a long non-coding RNA just upstream of ACTN1 and can potentially regulate ACTN1 transcript levels. Missense mutations in ACTN1 have recently been identified in congenital macrothrombocytopenia pedigrees with mild-to-moderate thrombocytopenia, increased MPV, and minimal bleeding manifestations.62, 63 Two subsequent genetic studies (one from Italy and one from the US and Europe) of previously uncharacterized inherited platelet disorders found ACTN1 missense mutations in ∼4%–5% of affected individuals.64, 65 Functional characterization of missense variants in ACTN1 (c.94C>A [p.Gln32Lys] and c.313G>A [p.Val105Ile]) suggest that the variants disrupt the actin cytoskeleton structure and impair megakaryocyte pro-platelet production.63 Although these studies highlight the implications of ACTN1 loss-of-function coding mutations in PLT regulation, transcriptional regulation of ACTN1 is also important for normal megakaryopoiesis.66, 67 In addition to regulating PLT, the actin cytoskeleton is involved in determining platelet size during the final stages of pro-platelet formation from megakaryocytes.68
Consistent with the relationship between ACTN1 mutations and familial macrothrombocytopenia, the minor allele (rs117672662 C allele) of the Amerindian ACTN1 non-coding variant was associated with lower PLT and higher MPV. The rs117672662 C allele displayed increased enhancer activity, whereas the rs117672662 T allele demonstrated a super-shift that could be mediated by HOXA5 and a partial super-shift mediated by GATA1. GATA1 also appeared to bind to the rs117672662 T allele probe. HOXA5 and GATA1 have each been shown to play a role in erythrocyte and megakaryocyte development,69, 70, 71, 72 and both transcription factors are dysregulated in hematopoietic stem cells showing erythroid and megakaryocyte differentiation blockage mediated by HOXA10 overexpression.70 Furthermore, HOXA5 might act as a transcriptional repressor for several genes involved in actin remodeling.73 Despite the evidence for allelic difference in expression and binding of HOXA5 and GATA1, further experiments are needed for elucidating the precise regulatory molecular mechanism by which the ACTN1 rs117672662 C allele alters platelet production and/or PLT.74, 75
Two additional PLT loci and one independent signal in a known locus were identified on chromosome 6 in gene-rich, extended LD regions located about 3.3 Mb centromeric (ETV7), 84 kb telomeric (GABBR1-MOG), and 25 Mb centromeric (ZBTB9-BAK1) to the major histocompatibility complex (MHC) region. The ETV7 index SNP rs9470264 is located within an intron of ETV7, which encodes the Ets family transcription factor TEL-2. ETV7 (or TEL2) is primarily expressed in hematopoietic tissues, including bone marrow and spleen.76, 77 Further, ETV7 is expressed in leukemic cells and appears to be more highly expressed in a subset of leukemic samples,78 pointing to possible roles for this transcription factor in both normal hematopoiesis and oncogenesis. Mutations in a related Ets-encoding gene, ETV6 (MIM: 600618), encoding ubiquitously expressed TEL-1, were recently identified in pedigrees affected by congenital thrombocytopenia,79, 80, 81 and chromosomal translocations involving ETV6 (e.g., ETV6-PDGFRB [MIM: 173410]) are common in hematologic malignancies.78 Despite strong biological plausibility supporting ETV7 in hematopoiesis, in silico functional annotation of the index SNP and LD proxies prioritized a promoter polymorphism in neighboring KCTD20 (MIM: 615932) as the putative functional SNP. Whole-blood eQTL analysis of the ETV7 index SNPs and LD proxies indicated significant differences in KCTD20, but not ETV7, expression (see Table S7), further supporting a role for KCTD20 in regulating PLT at this locus.61
The index variant rs75140056 is located in an intergenic region between MOG and GABBR1 (Figure S1C). Surveying variation in high LD with the index SNP, we prioritized rs29269 as the strongest functional candidate polymorphism on the basis of its position in a putative active GABBR1 promoter in megakaryocytes (Table S8). GABBR1 is a member of the gamma-aminobutyric acid family of inhibitory neurotransmitters and is most notably known for its role in the mammalian CNS. However, a recent study reported differential regulation of GABBR1 in bone-marrow- and fetal-liver-derived megakaryocytes from wild-type mice, thereby suggesting a potential role of GABBR1 in developmental-stage-specific regulation of megakaryopoiesis.82 In addition, several proxy SNPs, including putative functional SNP rs29269, are eQTLs (Table S7) for MHC class I genes (e.g., HLA-F [MIM: 143110] and HLA-G [MIM: 142871]). These observations suggest that variation at this locus might regulate one or more genes.
The index SNP in our Hispanic/Latino discovery sample at the ZBTB9-BAK1 locus (rs62405954) is distinct from rs210134, which was previously associated with PLT in European GWASs. rs62405954 is in LD with rs1002011, which overlaps a DNase hypersensitive site in megakaryocytes and lies within a putative enhancer overlapping the 5′ UTR of VPS52 (MIM: 603443), a gene that encodes an intracellular protein involved in endocytic recycling and is highly expressed in hematopoietic cells of erythroid and megakaryocyte lineages.
In addition, we identified an Amerindian-population-specific variant at MEF2C, a known PLT locus. MEF2C, which encodes a MADS box transcription factor and is differentially expressed at various stages of hematopoiesis, is an important downstream target of stem cell leukemia for lineage-specific megakaryocyte development.83 The Amerindian-ancestry-specific index SNP rs144261491 is distinct from the MEF2C European index SNP rs700585. rs144261491 is in LD with rs200572016 (r2 = 0.8), which lies in a megakaryocyte-specific DNase hypersensitive site in the MEF2C antisense RNA and overlaps several transcription factor binding sites (GATA2, TAL1, and P300) in K562 cells. MEF2C and MEF2C-AS1 are differentially expressed between erythroblasts and megakaryocytes.
In summary, we discovered and replicated three loci associated with PLT in Hispanic/Latino populations, as well as independent signals within two PLT-associated regions previously identified in populations of European descent. Several of these discovered PLT loci are prevalent among populations of Amerindian ancestry but rare or absent among populations of European or African ancestry. Amerindian-specific loci (e.g., SLC16A11 [MIM: 615765]) for metabolic traits (diabetes and glycemic traits) similarly have been identified among other Hispanic/Latino populations.84 Given the role of blood cells in pathogen invasion or defense, population-specific and/or rare variants might be expected to contribute to the regulation of genes relevant to quantitative blood cell phenotypes. The ACTN1 and MEF2C alleles might have arisen by mutation among populations of Amerindian ancestry after the peopling of the Americas85 and persisted as a result of local evolutionary selective pressure or genetic drift. Unlike the SLC16A11 locus,84 our PLT-associated loci did now show evidence of arising from introgression due to admixture with archaic humans. Taken together, our findings emphasize the importance and utility of performing genetic studies in populations with diverse ancestral backgrounds, including Hispanic/Latino populations.
Acknowledgments
We thank the participants and staff of the Hispanic Community Health Study/Study of Latinos (HCHS/SOL). The baseline examination of HCHS/SOL was supported by contracts from the National Heart, Lung, and Blood Institute (NHLBI) to the University of North Carolina (N01-HC65233), University of Miami (N01-HC65234), Albert Einstein College of Medicine (N01-HC65235), Northwestern University (N01-HC65236), and San Diego State University (N01-HC65237). The National Institute on Minority Health and Health Disparities, National Institute on Deafness and Other Communication Disorders, National Institute of Dental and Craniofacial Research (NIDCR), National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK), National Institute of Neurological Disorders and Stroke, and NIH Office of Dietary Supplements additionally contributed funding to HCHS/SOL. The Genetic Analysis Center at the University of Washington was supported by NHLBI and NIDCR contracts (HHSN268201300005C AM03 and MOD03). Additional analysis support was provided by 1R01DK101855-01 and 13GRNT16490017. Genotyping was also supported by National Center for Advancing Translational Sciences UL1TR000124 and NIDDK DK063491 to the Southern California Diabetes Endocrinology Research Center. Additional support for rs117672662 functional studies was provided by R01 DK072193. This research was also supported in part by the Intramural Research Program of the NIDDK, contract no. HHSB268201200054C, and Illumina. S.R.B. was supported by R01-GM110068. We thank Dr. Nick Patterson and the Simons Genome Diversity Project for kindly providing Australo-Melanesian sequence data. The Mount Sinai IPM Biobank Program is supported by the Andrea and Charles Bronfman Philanthropies.
Published: January 21, 2016
Footnotes
Supplemental Data include 6 figures and 12 tables and can be found with this article online at http://dx.doi.org/10.1016/j.ajhg.2015.12.003.
Web Resources
The URLs for data presented herein are as follows:
HapMap recombination hotspots, ftp://ftp.hapmap.org/hapmap/recombination/2008-03_rel22_B36/rates/
OMIM, http://www.omim.org
UCSC Genome Browser, https://genome.ucsc.edu/
Supplemental Data
References
- 1.Freson K., Wijgaerts A., van Geet C. Update on the causes of platelet disorders and functional consequences. Int. J. Lab. Hematol. 2014;36:313–325. doi: 10.1111/ijlh.12213. [DOI] [PubMed] [Google Scholar]
- 2.Gieger C., Radhakrishnan A., Cvejic A., Tang W., Porcu E., Pistis G., Serbanovic-Canic J., Elling U., Goodall A.H., Labrune Y. New gene functions in megakaryopoiesis and platelet formation. Nature. 2011;480:201–208. doi: 10.1038/nature10659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nürnberg S.T., Rendon A., Smethurst P.A., Paul D.S., Voss K., Thon J.N., Lloyd-Jones H., Sambrook J.G., Tijssen M.R., Italiano J.E., Jr., HaemGen Consortium A GWAS sequence variant for platelet volume marks an alternative DNM3 promoter in megakaryocytes near a MEIS1 binding site. Blood. 2012;120:4859–4868. doi: 10.1182/blood-2012-01-401893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Msaouel P., Lam A.P., Gundabolu K., Chrysofakis G., Yu Y., Mantzaris I., Friedman E., Verma A. Abnormal platelet count is an independent predictor of mortality in the elderly and is influenced by ethnicity. Haematologica. 2014;99:930–936. doi: 10.3324/haematol.2013.101949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shameer K., Denny J.C., Ding K., Jouni H., Crosslin D.R., de Andrade M., Chute C.G., Peissig P., Pacheco J.A., Li R. A genome- and phenome-wide association study to identify genetic variants influencing platelet count and volume and their pleiotropic effects. Hum. Genet. 2014;133:95–109. doi: 10.1007/s00439-013-1355-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Segal J.B., Moliterno A.R. Platelet counts differ by sex, ethnicity, and age in the United States. Ann. Epidemiol. 2006;16:123–130. doi: 10.1016/j.annepidem.2005.06.052. [DOI] [PubMed] [Google Scholar]
- 7.Bain B.J. Ethnic and sex differences in the total and differential white cell count and platelet count. J. Clin. Pathol. 1996;49:664–666. doi: 10.1136/jcp.49.8.664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Biino G., Balduini C.L., Casula L., Cavallo P., Vaccargiu S., Parracciani D., Serra D., Portas L., Murgia F., Pirastu M. Analysis of 12,517 inhabitants of a Sardinian geographic isolate reveals that predispositions to thrombocytopenia and thrombocytosis are inherited traits. Haematologica. 2011;96:96–101. doi: 10.3324/haematol.2010.029934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buckley M.F., James J.W., Brown D.E., Whyte G.S., Dean M.G., Chesterman C.N., Donald J.A. A novel approach to the assessment of variations in the human platelet count. Thromb. Haemost. 2000;83:480–484. [PubMed] [Google Scholar]
- 10.Evans D.M., Frazer I.H., Martin N.G. Genetic and environmental causes of variation in basal levels of blood cells. Twin Res. 1999;2:250–257. doi: 10.1375/136905299320565735. [DOI] [PubMed] [Google Scholar]
- 11.Kamatani Y., Matsuda K., Okada Y., Kubo M., Hosono N., Daigo Y., Nakamura Y., Kamatani N. Genome-wide association study of hematological and biochemical traits in a Japanese population. Nat. Genet. 2010;42:210–215. doi: 10.1038/ng.531. [DOI] [PubMed] [Google Scholar]
- 12.Li J., Glessner J.T., Zhang H., Hou C., Wei Z., Bradfield J.P., Mentch F.D., Guo Y., Kim C., Xia Q. GWAS of blood cell traits identifies novel associated loci and epistatic interactions in Caucasian and African-American children. Hum. Mol. Genet. 2013;22:1457–1464. doi: 10.1093/hmg/dds534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qayyum R., Snively B.M., Ziv E., Nalls M.A., Liu Y., Tang W., Yanek L.R., Lange L., Evans M.K., Ganesh S. A meta-analysis and genome-wide association study of platelet count and mean platelet volume in african americans. PLoS Genet. 2012;8:e1002491. doi: 10.1371/journal.pgen.1002491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Soranzo N., Spector T.D., Mangino M., Kühnel B., Rendon A., Teumer A., Willenborg C., Wright B., Chen L., Li M. A genome-wide meta-analysis identifies 22 loci associated with eight hematological parameters in the HaemGen consortium. Nat. Genet. 2009;41:1182–1190. doi: 10.1038/ng.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Witkop C.J., Nuñez Babcock M., Rao G.H., Gaudier F., Summers C.G., Shanahan F., Harmon K.R., Townsend D., Sedano H.O., King R.A. Albinism and Hermansky-Pudlak syndrome in Puerto Rico. Bol. Asoc. Med. P. R. 1990;82:333–339. [PubMed] [Google Scholar]
- 16.Lavange L.M., Kalsbeek W.D., Sorlie P.D., Avilés-Santa L.M., Kaplan R.C., Barnhart J., Liu K., Giachello A., Lee D.J., Ryan J. Sample design and cohort selection in the Hispanic Community Health Study/Study of Latinos. Ann. Epidemiol. 2010;20:642–649. doi: 10.1016/j.annepidem.2010.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sorlie P.D., Avilés-Santa L.M., Wassertheil-Smoller S., Kaplan R.C., Daviglus M.L., Giachello A.L., Schneiderman N., Raij L., Talavera G., Allison M. Design and implementation of the Hispanic Community Health Study/Study of Latinos. Ann. Epidemiol. 2010;20:629–641. doi: 10.1016/j.annepidem.2010.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rosenberg N.A., Li L.M., Ward R., Pritchard J.K. Informativeness of genetic markers for inference of ancestry. Am. J. Hum. Genet. 2003;73:1402–1422. doi: 10.1086/380416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Laurie C.C., Doheny K.F., Mirel D.B., Pugh E.W., Bierut L.J., Bhangale T., Boehm F., Caporaso N.E., Cornelis M.C., Edenberg H.J., GENEVA Investigators Quality control and quality assurance in genotypic data for genome-wide association studies. Genet. Epidemiol. 2010;34:591–602. doi: 10.1002/gepi.20516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laurie C.C., Laurie C.A., Rice K., Doheny K.F., Zelnick L.R., McHugh C.P., Ling H., Hetrick K.N., Pugh E.W., Amos C. Detectable clonal mosaicism from birth to old age and its relationship to cancer. Nat. Genet. 2012;44:642–650. doi: 10.1038/ng.2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Conomos M.P., Reiner A.P., Weir B.S., Thornton T.A. Model-free estimation of recent genetic relatedness. Am. J. Hum. Genet. 2015;98:127–148. doi: 10.1016/j.ajhg.2015.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Conomos M.P., Miller M.B., Thornton T.A. Robust inference of population structure for ancestry prediction and correction of stratification in the presence of relatedness. Genet. Epidemiol. 2015;39:276–293. doi: 10.1002/gepi.21896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abecasis G.R., Auton A., Brooks L.D., DePristo M.A., Durbin R.M., Handsaker R.E., Kang H.M., Marth G.T., McVean G.A., 1000 Genomes Project Consortium An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Howie B., Marchini J., Stephens M. Genotype imputation with thousands of genomes. G3 (Bethesda) 2011;1:457–470. doi: 10.1534/g3.111.001198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Howie B.N., Donnelly P., Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li Y., Willer C.J., Ding J., Scheet P., Abecasis G.R. MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet. Epidemiol. 2010;34:816–834. doi: 10.1002/gepi.20533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Conomos, M.P. (2014). Inferring, estimating and accounting for population and pedigree structure in genetic analyses. PhD thesis (University of Washington).
- 28.Conomos M.P., Laurie C.A., Stilp A.M., Gogarten S.M., McHugh C.P., Nelson S.C., Sofer T., Fernández-Rhodes L., Justice A.E., Graff M. Genetic diversity and association studies in US Hispanic/Latino populations: Applications in the Hispanic Community Health Study/Study of Latinos. Am. J. Hum. Genet. 2015;98:165–184. doi: 10.1016/j.ajhg.2015.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang J., Lee S.H., Goddard M.E., Visscher P.M. GCTA: a tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 2011;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Clopper C.J., Pearson E.S. The Use of Confidence or Fiducial Limits Illustrated in the Case of the Binomial. Biometrika. 1934;26:404–413. [Google Scholar]
- 31.R Core Development Team . R Foundation for Statistical Computing; 2014. R: A language and environment for statistical computing. [Google Scholar]
- 32.Green R.E., Krause J., Briggs A.W., Maricic T., Stenzel U., Kircher M., Patterson N., Li H., Zhai W., Fritz M.H. A draft sequence of the Neandertal genome. Science. 2010;328:710–722. doi: 10.1126/science.1188021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meyer M., Kircher M., Gansauge M.T., Li H., Racimo F., Mallick S., Schraiber J.G., Jay F., Prüfer K., de Filippo C. A high-coverage genome sequence from an archaic Denisovan individual. Science. 2012;338:222–226. doi: 10.1126/science.1224344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Skoglund P., Mallick S., Bortolini M.C., Chennagiri N., Hünemeier T., Petzl-Erler M.L., Salzano F.M., Patterson N., Reich D. Genetic evidence for two founding populations of the Americas. Nature. 2015;525:104–108. doi: 10.1038/nature14895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reiner A.P., Beleza S., Franceschini N., Auer P.L., Robinson J.G., Kooperberg C., Peters U., Tang H. Genome-wide association and population genetic analysis of C-reactive protein in African American and Hispanic American women. Am. J. Hum. Genet. 2012;91:502–512. doi: 10.1016/j.ajhg.2012.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen W., Brehm J.M., Manichaikul A., Cho M.H., Boutaoui N., Yan Q., Burkart K.M., Enright P.L., Rotter J.I., Petersen H. A genome-wide association study of chronic obstructive pulmonary disease in Hispanics. Ann. Am. Thorac. Soc. 2015;12:340–348. doi: 10.1513/AnnalsATS.201408-380OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Manichaikul A., Palmas W., Rodriguez C.J., Peralta C.A., Divers J., Guo X., Chen W.M., Wong Q., Williams K., Kerr K.F. Population structure of Hispanics in the United States: the multi-ethnic study of atherosclerosis. PLoS Genet. 2012;8:e1002640. doi: 10.1371/journal.pgen.1002640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tayo B.O., Teil M., Tong L., Qin H., Khitrov G., Zhang W., Song Q., Gottesman O., Zhu X., Pereira A.C. Genetic background of patients from a university medical center in Manhattan: implications for personalized medicine. PLoS ONE. 2011;6:e19166. doi: 10.1371/journal.pone.0019166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Willer C.J., Li Y., Abecasis G.R. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maples B.K., Gravel S., Kenny E.E., Bustamante C.D. RFMix: a discriminative modeling approach for rapid and robust local-ancestry inference. Am. J. Hum. Genet. 2013;93:278–288. doi: 10.1016/j.ajhg.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cavalli-Sforza L.L. The Human Genome Diversity Project: past, present and future. Nat. Rev. Genet. 2005;6:333–340. doi: 10.1038/nrg1596. [DOI] [PubMed] [Google Scholar]
- 42.Altshuler D.M., Gibbs R.A., Peltonen L., Altshuler D.M., Gibbs R.A., Peltonen L., Dermitzakis E., Schaffner S.F., Yu F., Peltonen L., International HapMap 3 Consortium Integrating common and rare genetic variation in diverse human populations. Nature. 2010;467:52–58. doi: 10.1038/nature09298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Browning S.R., Browning B.L. Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am. J. Hum. Genet. 2007;81:1084–1097. doi: 10.1086/521987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Heller R., Bogomolov M., Benjamini Y. Deciding whether follow-up studies have replicated findings in a preliminary large-scale omics study. Proc. Natl. Acad. Sci. USA. 2014;111:16262–16267. doi: 10.1073/pnas.1314814111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Adams D., Altucci L., Antonarakis S.E., Ballesteros J., Beck S., Bird A., Bock C., Boehm B., Campo E., Caricasole A. BLUEPRINT to decode the epigenetic signature written in blood. Nat. Biotechnol. 2012;30:224–226. doi: 10.1038/nbt.2153. [DOI] [PubMed] [Google Scholar]
- 46.Tijssen M.R., Cvejic A., Joshi A., Hannah R.L., Ferreira R., Forrai A., Bellissimo D.C., Oram S.H., Smethurst P.A., Wilson N.K. Genome-wide analysis of simultaneous GATA1/2, RUNX1, FLI1, and SCL binding in megakaryocytes identifies hematopoietic regulators. Dev. Cell. 2011;20:597–609. doi: 10.1016/j.devcel.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.ENCODE Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Andersson R., Gebhard C., Miguel-Escalada I., Hoof I., Bornholdt J., Boyd M., Chen Y., Zhao X., Schmidl C., Suzuki T., FANTOM Consortium An atlas of active enhancers across human cell types and tissues. Nature. 2014;507:455–461. doi: 10.1038/nature12787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ward L.D., Kellis M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 2012;40:D930–D934. doi: 10.1093/nar/gkr917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sandelin A., Alkema W., Engström P., Wasserman W.W., Lenhard B. JASPAR: an open-access database for eukaryotic transcription factor binding profiles. Nucleic Acids Res. 2004;32:D91–D94. doi: 10.1093/nar/gkh012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kircher M., Witten D.M., Jain P., O’Roak B.J., Cooper G.M., Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 2014;46:310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ritchie G.R., Dunham I., Zeggini E., Flicek P. Functional annotation of noncoding sequence variants. Nat. Methods. 2014;11:294–296. doi: 10.1038/nmeth.2832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lee D., Gorkin D.U., Baker M., Strober B.J., Asoni A.L., McCallion A.S., Beer M.A. A method to predict the impact of regulatory variants from DNA sequence. Nat. Genet. 2015;47:955–961. doi: 10.1038/ng.3331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Knowler W.C., Coresh J., Elston R.C., Freedman B.I., Iyengar S.K., Kimmel P.L., Olson J.M., Plaetke R., Sedor J.R., Seldin M.F., Family Investigation of Nephropathy and Diabetes Research Group The Family Investigation of Nephropathy and Diabetes (FIND): design and methods. J. Diabetes Complications. 2005;19:1–9. doi: 10.1016/j.jdiacomp.2003.12.007. [DOI] [PubMed] [Google Scholar]
- 55.Tian C., Hinds D.A., Shigeta R., Adler S.G., Lee A., Pahl M.V., Silva G., Belmont J.W., Hanson R.L., Knowler W.C. A genomewide single-nucleotide-polymorphism panel for Mexican American admixture mapping. Am. J. Hum. Genet. 2007;80:1014–1023. doi: 10.1086/513522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fogarty M.P., Cannon M.E., Vadlamudi S., Gaulton K.J., Mohlke K.L. Identification of a regulatory variant that binds FOXA1 and FOXA2 at the CDC123/CAMK1D type 2 diabetes GWAS locus. PLoS Genet. 2014;10:e1004633. doi: 10.1371/journal.pgen.1004633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zuk O., Hechter E., Sunyaev S.R., Lander E.S. The mystery of missing heritability: Genetic interactions create phantom heritability. Proc. Natl. Acad. Sci. USA. 2012;109:1193–1198. doi: 10.1073/pnas.1119675109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Heller, R., Bogomolov, M., Benjamini, Y., and Sofer, T. (2015). Testing for replicability in a follow-up study when the primary study hypotheses are two-sided. arXiv, arXiv:1503.02278.
- 59.Martens J.H., Stunnenberg H.G. BLUEPRINT: mapping human blood cell epigenomes. Haematologica. 2013;98:1487–1489. doi: 10.3324/haematol.2013.094243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lizio M., Harshbarger J., Shimoji H., Severin J., Kasukawa T., Sahin S., Abugessaisa I., Fukuda S., Hori F., Ishikawa-Kato S., FANTOM consortium Gateways to the FANTOM5 promoter level mammalian expression atlas. Genome Biol. 2015;16:22. doi: 10.1186/s13059-014-0560-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Westra H.J., Peters M.J., Esko T., Yaghootkar H., Schurmann C., Kettunen J., Christiansen M.W., Fairfax B.P., Schramm K., Powell J.E. Systematic identification of trans eQTLs as putative drivers of known disease associations. Nat. Genet. 2013;45:1238–1243. doi: 10.1038/ng.2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Guéguen P., Rouault K., Chen J.M., Raguénès O., Fichou Y., Hardy E., Gobin E., Pan-Petesch B., Kerbiriou M., Trouvé P. A missense mutation in the alpha-actinin 1 gene (ACTN1) is the cause of autosomal dominant macrothrombocytopenia in a large French family. PLoS ONE. 2013;8:e74728. doi: 10.1371/journal.pone.0074728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kunishima S., Okuno Y., Yoshida K., Shiraishi Y., Sanada M., Muramatsu H., Chiba K., Tanaka H., Miyazaki K., Sakai M. ACTN1 mutations cause congenital macrothrombocytopenia. Am. J. Hum. Genet. 2013;92:431–438. doi: 10.1016/j.ajhg.2013.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bottega R., Marconi C., Faleschini M., Baj G., Cagioni C., Pecci A., Pippucci T., Ramenghi U., Pardini S., Ngu L. ACTN1-related thrombocytopenia: identification of novel families for phenotypic characterization. Blood. 2015;125:869–872. doi: 10.1182/blood-2014-08-594531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Westbury S.K., Turro E., Greene D., Lentaigne C., Kelly A.M., Bariana T.K., Simeoni I., Pillois X., Attwood A., Austin S., BRIDGE-BPD Consortium Human phenotype ontology annotation and cluster analysis to unravel genetic defects in 707 cases with unexplained bleeding and platelet disorders. Genome Med. 2015;7:36. doi: 10.1186/s13073-015-0151-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hamill K.J., Hiroyasu S., Colburn Z.T., Ventrella R.V., Hopkinson S.B., Skalli O., Jones J.C. Alpha actinin-1 regulates cell-matrix adhesion organization in keratinocytes: consequences for skin cell motility. J. Invest. Dermatol. 2015;135:1043–1052. doi: 10.1038/jid.2014.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Raslova H., Kauffmann A., Sekkaï D., Ripoche H., Larbret F., Robert T., Tronik Le Roux D., Kroemer G., Debili N., Dessen P. Interrelation between polyploidization and megakaryocyte differentiation: a gene profiling approach. Blood. 2007;109:3225–3234. doi: 10.1182/blood-2006-07-037838. [DOI] [PubMed] [Google Scholar]
- 68.Thon J.N., Macleod H., Begonja A.J., Zhu J., Lee K.C., Mogilner A., Hartwig J.H., Italiano J.E., Jr. Microtubule and cortical forces determine platelet size during vascular platelet production. Nat. Commun. 2012;3:852. doi: 10.1038/ncomms1838. [DOI] [PubMed] [Google Scholar]
- 69.Alharbi R.A., Pettengell R., Pandha H.S., Morgan R. The role of HOX genes in normal hematopoiesis and acute leukemia. Leukemia. 2013;27:1000–1008. doi: 10.1038/leu.2012.356. [DOI] [PubMed] [Google Scholar]
- 70.Magnusson M., Brun A.C., Miyake N., Larsson J., Ehinger M., Bjornsson J.M., Wutz A., Sigvardsson M., Karlsson S. HOXA10 is a critical regulator for hematopoietic stem cells and erythroid/megakaryocyte development. Blood. 2007;109:3687–3696. doi: 10.1182/blood-2006-10-054676. [DOI] [PubMed] [Google Scholar]
- 71.Pevny L., Simon M.C., Robertson E., Klein W.H., Tsai S.F., D’Agati V., Orkin S.H., Costantini F. Erythroid differentiation in chimaeric mice blocked by a targeted mutation in the gene for transcription factor GATA-1. Nature. 1991;349:257–260. doi: 10.1038/349257a0. [DOI] [PubMed] [Google Scholar]
- 72.Shivdasani R.A., Fujiwara Y., McDevitt M.A., Orkin S.H. A lineage-selective knockout establishes the critical role of transcription factor GATA-1 in megakaryocyte growth and platelet development. EMBO J. 1997;16:3965–3973. doi: 10.1093/emboj/16.13.3965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang C.C., Su K.Y., Chen H.Y., Chang S.Y., Shen C.F., Hsieh C.H., Hong Q.S., Chiang C.C., Chang G.C., Yu S.L., Chen J.J. HOXA5 inhibits metastasis via regulating cytoskeletal remodelling and associates with prolonged survival in non-small-cell lung carcinoma. PLoS ONE. 2015;10:e0124191. doi: 10.1371/journal.pone.0124191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ablain J., Durand E.M., Yang S., Zhou Y., Zon L.I. A CRISPR/Cas9 vector system for tissue-specific gene disruption in zebrafish. Dev. Cell. 2015;32:756–764. doi: 10.1016/j.devcel.2015.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bauer D.E., Kamran S.C., Lessard S., Xu J., Fujiwara Y., Lin C., Shao Z., Canver M.C., Smith E.C., Pinello L. An erythroid enhancer of BCL11A subject to genetic variation determines fetal hemoglobin level. Science. 2013;342:253–257. doi: 10.1126/science.1242088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Potter M.D., Buijs A., Kreider B., van Rompaey L., Grosveld G.C. Identification and characterization of a new human ETS-family transcription factor, TEL2, that is expressed in hematopoietic tissues and can associate with TEL1/ETV6. Blood. 2000;95:3341–3348. [PubMed] [Google Scholar]
- 77.Gu X., Shin B.H., Akbarali Y., Weiss A., Boltax J., Oettgen P., Libermann T.A. Tel-2 is a novel transcriptional repressor related to the Ets factor Tel/ETV-6. J. Biol. Chem. 2001;276:9421–9436. doi: 10.1074/jbc.M010070200. [DOI] [PubMed] [Google Scholar]
- 78.Wang Q., Dong S., Yao H., Wen L., Qiu H., Qin L., Ma L., Chen S. ETV6 mutation in a cohort of 970 patients with hematologic malignancies. Haematologica. 2014;99:e176–e178. doi: 10.3324/haematol.2014.104406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhang M.Y., Churpek J.E., Keel S.B., Walsh T., Lee M.K., Loeb K.R., Gulsuner S., Pritchard C.C., Sanchez-Bonilla M., Delrow J.J. Germline ETV6 mutations in familial thrombocytopenia and hematologic malignancy. Nat. Genet. 2015;47:180–185. doi: 10.1038/ng.3177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Noetzli L., Lo R.W., Lee-Sherick A.B., Callaghan M., Noris P., Savoia A., Rajpurkar M., Jones K., Gowan K., Balduini C.L. Germline mutations in ETV6 are associated with thrombocytopenia, red cell macrocytosis and predisposition to lymphoblastic leukemia. Nat. Genet. 2015;47:535–538. doi: 10.1038/ng.3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Topka S., Vijai J., Walsh M.F., Jacobs L., Maria A., Villano D., Gaddam P., Wu G., McGee R.B., Quinn E. Germline ETV6 Mutations Confer Susceptibility to Acute Lymphoblastic Leukemia and Thrombocytopenia. PLoS Genet. 2015;11:e1005262. doi: 10.1371/journal.pgen.1005262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Woo A.J., Wieland K., Huang H., Akie T.E., Piers T., Kim J., Cantor A.B. Developmental differences in IFN signaling affect GATA1s-induced megakaryocyte hyperproliferation. J. Clin. Invest. 2013 doi: 10.1172/JCI40609. Published online July 1, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gekas C., Rhodes K.E., Gereige L.M., Helgadottir H., Ferrari R., Kurdistani S.K., Montecino-Rodriguez E., Bassel-Duby R., Olson E., Krivtsov A.V. Mef2C is a lineage-restricted target of Scl/Tal1 and regulates megakaryopoiesis and B-cell homeostasis. Blood. 2009;113:3461–3471. doi: 10.1182/blood-2008-07-167577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Williams A.L., Jacobs S.B., Moreno-Macías H., Huerta-Chagoya A., Churchhouse C., Márquez-Luna C., García-Ortíz H., Gómez-Vázquez M.J., Burtt N.P., Aguilar-Salinas C.A., SIGMA Type 2 Diabetes Consortium Sequence variants in SLC16A11 are a common risk factor for type 2 diabetes in Mexico. Nature. 2014;506:97–101. doi: 10.1038/nature12828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Reich D., Patterson N., Campbell D., Tandon A., Mazieres S., Ray N., Parra M.V., Rojas W., Duque C., Mesa N. Reconstructing Native American population history. Nature. 2012;488:370–374. doi: 10.1038/nature11258. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.