Abstract
Congenital disorders of glycosylation (CDGs) are disorders of abnormal protein glycosylation that affect multiple organ systems. Because most CDGs have been described in only a few individuals, our understanding of the associated phenotypes and the mechanisms of individual survival are limited. In the process of studying two siblings, aged 6 and 11 years, with MOGS-CDG and biallelic MOGS (mannosyl-oligosaccharide glucosidase) mutations (GenBank: NM_006302.2; c.[65C>A; 329G>A] p.[Ala22Glu; Arg110His]; c.[370C>T] p.[Gln124∗]), we noted that their survival was much longer than the previous report of MOGS-CDG, in a child who died at 74 days of age. Upon mutation analysis, we detected multiple MOGS genotypes including wild-type alleles in their cultured fibroblast and peripheral blood DNA. Further analysis of DNA from cultured fibroblasts of six individuals with compound heterozygous mutations of PMM2 (PMM2-CDG), MPI (MPI-CDG), ALG3 (ALG3-CDG), ALG12 (ALG12-CDG), DPAGT1 (DPAGT1-CDG), and ALG1 (ALG1-CDG) also identified multiple genotypes including wild-type alleles for each. Droplet digital PCR showed a ratio of nearly 1:1 wild-type to mutant alleles for most, but not all, mutations. This suggests that mitotic recombination contributes to the survival and the variable expressivity of individuals with compound heterozygous CDGs. This also provides an explanation for prior observations of a reduced frequency of homozygous mutations and might contribute to increased levels of residual enzyme activity in cultured fibroblasts of individuals with MPI- and PMM2-CDGs.
Main Text
Protein glycosylation takes place in the cytoplasm, endoplasmic reticulum, and Golgi apparatus. Throughout this process, branching linkages of various monosaccharides are added to the amide group of asparagine (N-linked) or the hydroxyl group of serine or threonine (O-linked) (see GeneReviews in Web Resources). Since the first description of “Congenital Disorders of Glycosylation (CDGs)” in 1980, more than 100 different CDGs have been described affecting protein and lipid glycosylation.1, 2, 3, 4 Nearly 60 different disorders in N-linked glycosylation have been reported to date.4 CDGs are defined as either type I defects, involving improper assembly or transfer of dolichol-linked glycans to proteins in the ER, or type II defects, resulting from incorrect modification of the protein-attached glycans in the Golgi.5
The CDGs cause multi-system disease, have a wide range of clinical presentations, and manifest variable expressivity.6 The pleiotropism of the CDGs can be partially attributed to the fact that ∼50% of human proteins are glycosylated and that some glycosylation events are tissue specific.7 A peculiar observation of the CDGs is that very few individuals have homozygous mutations compared to compound heterozygous mutations.8, 9, 10, 11, 12, 13 Combined with biochemical assays for residual enzyme activity and studies of animal models, this paucity of homozygous mutations has been interpreted to suggest that a genotype conveying residual catalytic activity is required for survival.10, 12, 14, 15, 16 It has been proposed that homozygous mutations are either lethal (due to null alleles) or result in a subclinical phenotype (due to hypomorphic alleles), attributed to a wide tolerance of enzymatic activity as reported in unaffected individuals.9, 16 Indeed, Helander et al. have reported two siblings with subclinical presentation of homozygous MPI-CDG due to a variant also found in compound heterozygous MPI-CDG-affected individuals.17 Another unexplained observation among the CDGs is that cultured skin fibroblasts derived from individuals with PMM2-CDG (OMIM: 212065) have high residual enzymatic activity.9
To better understand factors contributing to the prolonged survival of individuals with CDGs, we studied two siblings diagnosed with MOGS-CDG (OMIM: 606056). In contrast to the previously reported individual who died at 74 days of age,18 these individuals have survived into late childhood. A 6-year-old female (individual A) and her 11-year-old brother (individual B), both of European descent, were admitted to the National Institutes of Health Clinical Center under the NHGRI IRB-approved protocol 76-HG-0238, “Diagnosis and Treatment of Patients with Inborn Errors of Metabolism or Other Genetic Disorders.” Written, informed consent was also obtained for both individuals. Both children demonstrated dysmorphic facial features, severe intellectual disability, optic atrophy, mild generalized cerebral atrophy and cerebellar atrophy with thin corpus callosum, hypotonia, persistent leukocystosis, and hypogammaglobinemia. A description of the abnormal immunological phenotype of these individuals has been previously published.19
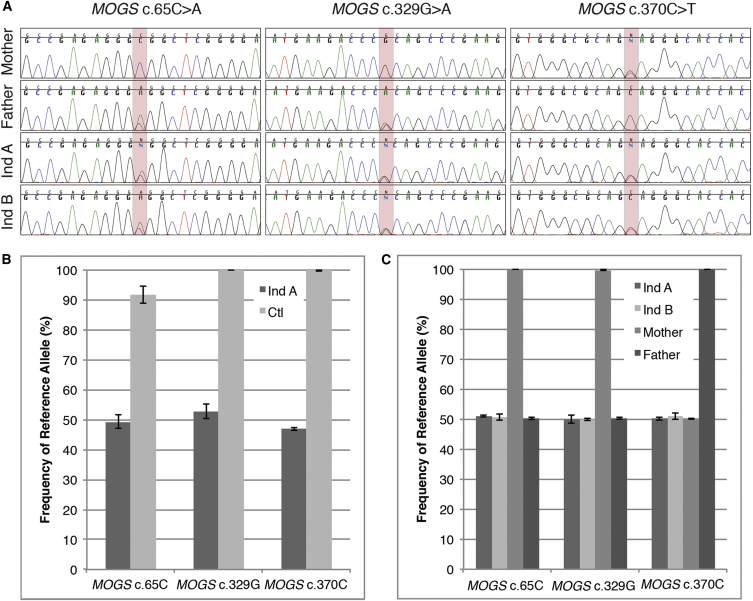
Sanger DNA sequencing of PCR-amplified mannosyl-oligosaccharide glucosidase (MOGS [OMIM: 601336]) coding exons identified mutations (GenBank: NM_006302.2; c.[65C>A; 329G>A];[370C>T]) (Figure 1A). The accession numbers for these SNVs are dbSNP: rs753961807 (c.65C>A) and rs587777323 (c.370C>T) and ClinVar: SCV000256097 (c.329G>A).
Figure 1.
MOGS SNV Analysis in Individuals A and B
(A) Sanger sequencing of MOGS in the parents and individuals A and B reveal two paternally inherited variants at c.65C>A and c.329G>A and one maternally inherited variant at c.370C>T.
(B) Analysis of SNV reference allele frequency by digital PCR in the fibroblasts from MOGS-CDG individual A (Ind A) shows no significant loss of alleles at the three inherited SNVs in vitro.
(C) Digital PCR analysis of SNV alleles in blood-derived DNA shows no loss of alleles in either MOGS-CDG-affected individual (Ind A and Ind B) nor their parents (Mother and Father) in vivo.
Error bars indicate SD.
The paternal mutations encode p.Ala22Glu and p.Arg110His, alterations of two conserved amino acids. The maternal mutation encodes p.Gln124∗. The paternally inherited single-nucleotide variants (SNVs) are predicted by in silico analysis to alter mRNA splicing and protein stability, respectively. The maternally inherited SNV is expected to trigger nonsense-mediated decay of the mRNA. At this time, unlike for PMM2 and MPI activity, there is no established method for measuring MOGS enzymatic activity, making it difficult to examine the enzymatic function of these mutations.
In light of these in silico predictions, we sought to better characterize the MOGS mRNA expressed in fibroblasts from individuals A and B. We amplified, cloned, and sequenced individual cDNA clones. This identified allelic combinations of SNVs absent in the parents (data not shown). To delineate the frequency and origin of these non-parental genotypes, we amplified and sequenced >100 clones of the region of genomic DNA encompassing the inherited SNVs (Table 1). Approximately 60% of the clones for both individuals had recombinant, non-parental genotypes (Table 1); 7% and 13% had a wild-type genotype for individuals A and B, respectively. To determine whether non-parental genotypes also occurred in vivo, we analyzed ≥100 clones and >3,500 PacBio amplicons derived from the blood DNA of both MOGS-CDG-affected individuals. We found 6% and 18% non-parental genotypes for individual A and 19% and 13% non-parental genotypes for individual B (Table 1).
Table 1.
Single-Allele Sequencing Results of MOGS-CDG-Affected Individuals
| Sample | MOGS c.65C>A | MOGS c.329G>A | MOGS c.370C>T | Ind. A, Fibroblast (n = 130) | Ind. B, Fibroblast (n = 126) | Ind. A, Blooda(n = 100) | Ind. B, Blooda(n = 108) | Ind. A, Bloodb(n = 3,896) | Ind.B, Bloodb(n = 5,305) |
|---|---|---|---|---|---|---|---|---|---|
| Genotype and Frequency | A | A | T | 7.69% | 3.97% | 4% | 6.48% | 3.26% | 3.05% |
| C∗ | G∗ | C∗ | 6.92% | 13.49% | 0 | 6.48% | 3.82% | 2.53% | |
| A | G∗ | C∗ | 19.23% | 13.49% | 0 | 0 | 2.62% | 2.21% | |
| C∗ | A | C∗ | 10.00% | 7.14% | 2% | 2.78% | 3.80% | 3.02% | |
| C∗ | A | T | 3.85% | 6.35% | 0 | 0.93% | 1.13% | 0.60% | |
| A | G∗ | T | 11.54% | 14.29% | 0 | 2.78% | 3.52% | 1.79% | |
| A | A | C∗ | 27.69% | 15.87% | 45% | 50.93% | 69.99% | 83.03% | |
| C∗ | G∗ | T | 7.69% | 25.40% | 49% | 29.63% | 11.86% | 3.77% | |
| Total non-parental clonesa/readsb | 59.23% | 58.73% | 6% | 19.44% | 18.15% | 13.20% | |||
Single-allele cloning and sequencing and PacBio amplicon sequencing reveal non-parental genotypes in the fibroblast- and blood-derived DNA from individuals (Ind.) A and B. The total number (n) of alleles sequenced is indicated for each sample type. The allele genotype at each of the three variant sites is shown in the first three columns; reference alleles are indicated with an asterisk. Each row represents a single allele and the allelic percentage among the sequenced population from fibroblast or blood genomic DNA is shown in the adjacent columns as indicated. The last two rows of allele genotypes represent the inherited, parental genotypes.
Single-allele cloning and sequencing.
PacBio amplicon sequencing.
The observed genotypes suggest a rearrangement of genetic information during mitosis, which can be triggered by DNA double-strand breaks (DSBs) and subsequent homology-directed repair.20 Mitotic recombination of chromosomes has been well characterized in model organisms and can result in either reciprocal or non-reciprocal transfer of genetic information between chromosomes.21 In the case of reciprocal recombination, a chromosomal crossover event is resolved with no loss of genetic information; this is the mechanism underlying meiotic recombination. Non-reciprocal recombination results in the loss of genetic information, usually from the damaged allele during DSB repair. The result is a loss of heterozygosity at the repair locus such that both alleles carry the donor or template genotype, e.g., gene conversion.20 Reciprocal recombination inherently requires crossing-over and the number of crossing-over events between chromatids will determine the amount of genetic material that is exchanged. A single cross-over event will result in a complete allelic switch between chromatids from the site of cross-over through the distal arm of the chromosome. Double cross-over events will limit the region of recombination to a shorter segment of the chromosome.22
To test whether reciprocal or nonreciprocal mitotic recombination processes are responsible for the non-parental genotypes observed in cultured fibroblasts and blood from the MOGS-CDG-affected individuals, we used the RainDrop digital PCR system and custom TaqMan genotyping assays to test the frequency of the inherited SNVs (sequences in Table S1). By employing digital PCR, we were able to determine the genotype at the SNV of interest on thousands of individual alleles from fibroblast or blood DNA. This allowed for more accurate quantification of the reference and variant allele frequency versus the relative signal quantification that could be achieved using real-time PCR with these same genotyping assays. Analysis of the fibroblasts derived from individual A showed no significant loss of alleles in vitro (p > .9 by chi-square analysis). Likewise, analyses of blood-derived DNA showed no skewing in the frequency of the three SNVs for either MOGS-CDG-affected individual (Figure 1C), supporting a reciprocal mitotic intragenic recombination process as the origin of the non-parental genotypes in vivo.
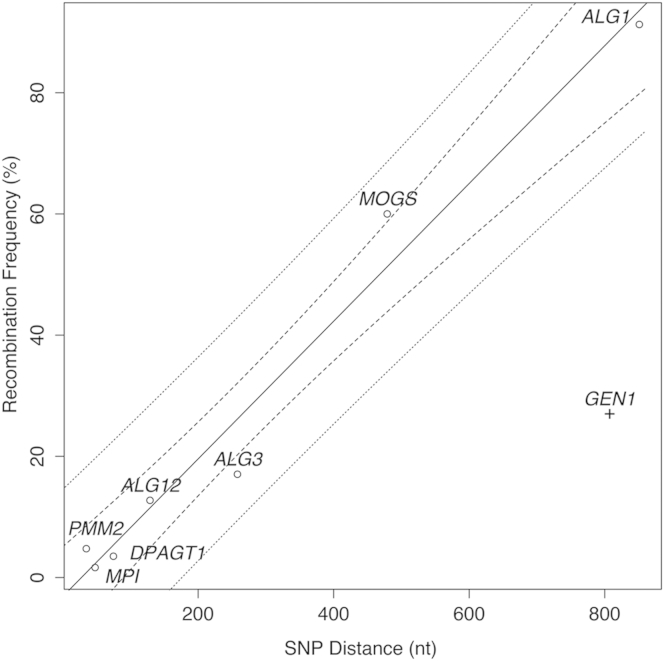
Based on these observations, we proposed that mitotic intragenic recombination accounts for the reported paucity of homozygous versus compound heterozygous mutations associated with CDGs and is a mechanism for individual survival. Blood-derived DNA was unavailable, so we tested this using genomic DNA isolated from cultured fibroblast lines derived from individuals with compound heterozygous mutations of PMM2 (PMM2-CDG [OMIM: 212065]), MPI (MPI-CDG [OMIM: 602579]), ALG3 (ALG3-CDG [OMIM: 601110]), ALG12 (ALG12-CDG [OMIM: 607143]), DPAGT1 (DPAGT1-CDG [OMIM: 608093]), and ALG1 (ALG1-CDG [OMIM: 608540]) (Table 2). Characterization of >100 clones from each cell line detected alleles with non-parental genotypes (Table 2); Sanger sequencing results from representative clones are shown with chromatograms in Figure S1 and FASTA sequences in Table S2. The frequency of non-parental genotypes ranged from approximately 1.5% to 90%, with a strong correlation between the frequency of recombinant genotypes and the chromosomal distance between the variants (R2 = 0.97, Figure 2). In contrast, using Pacific Biosciences (PacBio) amplicon sequencing, recombination analysis of compound heterozygous SNVs at the GEN1 (OMIM: 612449) locus in cultured fibroblasts from an unaffected individual showed only 27% recombination across 800 nucleotides (Table 3; NIH Intramural Sequencing Center, primer sequences in Table S1).23 This recombination frequency is well outside of the 95% prediction interval calculated from the CDG linear model (Figure 2).
Table 2.
Single-Allele Sequencing Results of Additional CDGs
| Disease (Mutations), Accession | Genotype | Category | Percentage | ||
|---|---|---|---|---|---|
| c.395 | c.430 | (n = 105) | |||
| PMM2-CDG (PMM2: c.[395T>C]; [430T>C]), dbSNP: rs150719105, rs80338702 | C | C | mut | 2.86 | |
| T∗ | T∗ | WT | 1.9 | ||
| C | T∗ | het | 46.67 | ||
| T∗ | C | het | 48.57 | ||
| Total non-parental | 4.76 | ||||
| c.1205 | c.1253 | (n = 121) | |||
| MPI-CDG (MPI: c.[1205A>G];[1253G>A]), ClinVar: SCV000256094, SCV000256095 | G | A | mut | 0 | |
| A∗ | G∗ | WT | 1.65 | ||
| A∗ | A | het | 48.76 | ||
| G | G∗ | het | 49.59 | ||
| Total non-parental | 1.65 | ||||
| c.211 | c.470 | (n = 135) | |||
| ALG3-CDG (ALG3: c.[211T>C];[470T>A]), dbSNP: rs119103237, rs119103238 | C | A | mut | 9.63 | |
| T∗ | T∗ | WT | 7.41 | ||
| T∗ | A | het | 49.63 | ||
| C | T∗ | het | 33.33 | ||
| Total non-parental | 17.04 | ||||
| c.301 | c.430 | (n = 102) | |||
| ALG12-CDG (ALG12: c.[301G>A];[430G>A]), dbSNP: rs121907933, rs121907932 | A | A | mut | 2.94 | |
| G∗ | G∗ | WT | 9.8 | ||
| G∗ | A | het | 50 | ||
| A | G∗ | het | 37.25 | ||
| Total non-parental | 12.74 | ||||
| c.509 | c.584 | (n = 115) | |||
| DPAGT1-CDG (DPAGT1: c.[509A>G];[584C>G]), dbSNP: rs28934876; ClinVar: SCV000256096 | G | G | mut | 2.61 | |
| A∗ | C∗ | WT | 0.87 | ||
| A∗ | G | het | 54.78 | ||
| G | C∗ | het | 41.74 | ||
| Total non-parental | 3.51 | ||||
| c.1037 | c.1187+1 | (n = 103) | |||
| ALG1-CDG (ALG1: c.[1037C>G];[1187+1G>A]), dbSNP: rs398124348, rs374928784 | G | A | mut | 4.85 | |
| C∗ | G∗ | WT | 86.41 | ||
| C∗ | A | het | 2.91 | ||
| G | G∗ | het | 5.83 | ||
| Total non-parental | 91.26 | ||||
Single-allele cloning and sequencing reveals non-parental genotypes in genomic DNA from cultured fibroblasts of multiple CDG-affected individuals. Allele genotypes are shown in the first two columns; the reference alleles are indicated by an asterisk. Each row represents a single allele and the frequency of this allele. SNV accession numbers are provided in respective order for each variant.
Figure 2.
Recombination Frequency Increases with Distance
Recombination frequency (percentage) in fibroblasts is plotted versus distance (nucleotides, nt). CDG samples are marked with an open circle and the control sample with a plus sign. The solid line represents a linear model for the recombination versus distance for the CDGs (y = 0.1137∗x-3.1628, R2 = 0.9743). Dashed lines represent the 95% confidence interval and dotted lines the 95% prediction interval.
Table 3.
Single-Allele Sequencing Results of an Unaffected Individual
| Sample (Variants), Accession | Genotype | Category | Percentage | |||
|---|---|---|---|---|---|---|
| c.1638 | c.2445 | (n = 1,269) | ||||
| Control blood (GEN1: c.[1638T>A];[2445C>T]), dbSNP: rs61762986, rs148781334 | A | T | mut | 11.98 | ||
| T∗ | C∗ | WT | 13.95 | |||
| T∗ | T | het | 36.17 | |||
| A | T∗ | het | 37.90 | |||
| Total non-parental | 25.93 | |||||
| c.1638 | c.2445 | (n = 1,115) | ||||
| Control fibroblasts (GEN1: c.[1638T>A];[2445C>T]), dbSNP: rs61762986, rs148781334 | A | T | mut | 15.52 | ||
| T∗ | C∗ | WT | 12.11 | |||
| T∗ | T | het | 33.72 | |||
| A | T∗ | het | 38.65 | |||
| Total non-parental | 27.62 | |||||
Sequence analysis via PacBio amplicon sequencing of single alleles in a healthy individual detects a low level of mitotic recombination in blood and fibroblasts. Allele genotypes are shown in the first two columns; the reference alleles are indicated by an asterisk. Each row represents a single allele and the frequency of this allele in the sequenced population. SNV accession numbers are provided in respective order for each variant.
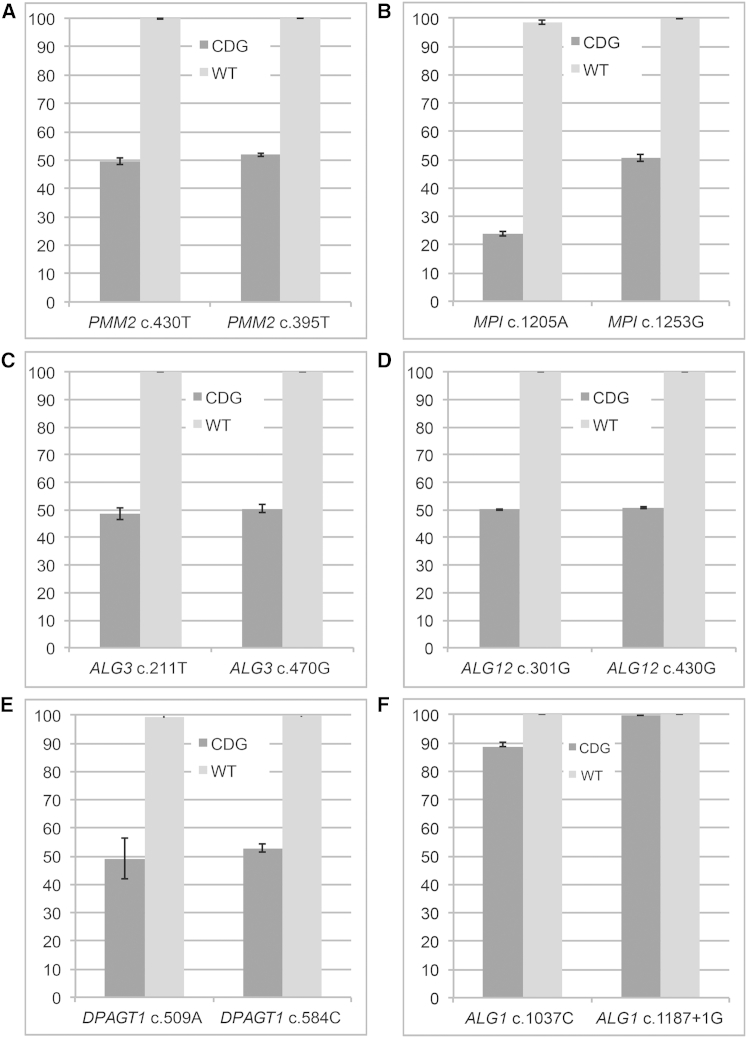
Digital PCR analysis of SNV frequency in the other CDG cell lines also detected evidence for both reciprocal and nonreciprocal mitotic recombination processes. We did not observe a decrease in SNV frequency among the PMM2-, ALG3-, ALG12-, or DPAGT1-CDG cell lines (Table 2 and Figures 3B–3G), whereas we did observe a loss of both pathogenic SNVs for the ALG1-CDG cell line (Figure 3G) and loss of one reference SNV (MPI c.1205A) for the MPI-CDG cell line (Figure 3C). The loss of both pathogenic variants in the ALG1-CDG cell line suggests either nonreciprocal mitotic recombination generating two wild-type alleles or reciprocal mitotic recombination generating a wild-type allele and subsequent loss of heterozygosity. In total, these results provide evidence that increased rates of mitotic intragenic recombination, reciprocal and nonreciprocal, occur in CDG tissues and are a potential mechanism for survival and variability in individuals with CDGs.
Figure 3.
Digital PCR Quantification of SNV Frequencies in Cultured Fibroblasts of CDG-Affected Individuals
The frequency of the reference SNV is shown for unaffected control cells (“WT”) versus primary fibroblast lines from individuals with CDG-causing mutations (“CDG”) in PMM2 (A), MPI (B), ALG3 (C), ALG12 (D), DPAGT1 (E), and ALG1 (F). Error bars indicate SD.
The above observations suggest either that the mutations and genes involved in these CDGs reside at recombination hotspots or that there is selection of cells with increased levels of functional enzyme. The distribution of the studied genes around the genome and the absence of known recombination hotspots at those locations suggested to us that the studied CDG genes do not reside at recombination hotspots. Additionally, because the recombination frequency across these genes and samples had a linear correlation with distance between the heterozygous mutations, we concluded that it was more likely that the recombination rate across each of these genes was comparable rather than that each pair of mutations resided within intragenic recombination hotspots with comparable recombination rates.24
These findings also suggest that, as has been observed for Bloom syndrome (OMIM: 210900) and WRN helicase (OMIM: 604611)-deficient cells,25, 26 recombination and the consequent mosaicism contribute to the variable expressivity of CDGs along with the nature of the mutation.9, 11, 25, 26, 27 Similarly, there are several reports of somatic and revertant mosaicism in human disease, including ichthyosis with confetti (OMIM: 6091650),28 tyrosinemia type I (OMIM: 276700), severe combined immunodeficiency (autosomal [OMIM: 102700] and X-linked [OMIM: 300400]), Wiskott-Aldrich syndrome (OMIM: 301000), epidermolysis bullosa (OMIM: 148066), and Fanconi anemia (OMIM: 607139 and 227645) (reviewed by Hirschorn).29 Among these disorders, there is evidence of gene conversion and somatic recombination as underlying mechanisms for reversion, indicating that our observations in the CDGs are not isolated but reflect an important, albeit rare, phenomenon in human physiology. Mitotic recombination has also been reported both in vivo and in vitro in a mouse model heterozygous for an Aprt mutant allele, further supporting a model whereby selective pressure from disease can lead to increased abundance of recombinant products in the absence of DNA replication defects.30 If an in vivo recombination event occurs more frequently or earlier in life, there is an opportunity for cells expressing wild-type protein to outcompete cells expressing mutant protein and ameliorate the phenotype, as has been observed in Kindler syndrome (OMIM: 173650), a human skin fragility disorder,31 and giving the appearance of an increased mitotic recombination rate.
Here we have provided evidence of mitotic intragenic recombination in compound heterozygous CDGs. This suggests that recombination between compound heterozygous alleles of glycosylation genes causes reversion to a wild-type allele and that survival or growth advantage of cells with the wild-type allele ameliorates the phenotype of the CDGs. A model of CDG survival attributed to mitotic intragenic recombination within somatic cells is further supported by residual enzymatic activity in CDG individual cells8, 9 and the small number of reported homozygous mutations among the CDGs.12, 13
Acknowledgments
H.H.F. is supported by DKR0199551 and The Rocket Fund.
Published: January 21, 2016
Footnotes
Supplemental Data include one figure and two tables and can be found with this article online at http://dx.doi.org/10.1016/j.ajhg.2015.12.007.
Contributor Information
Megan S. Kane, Email: megan.kane@nih.gov.
William A. Gahl, Email: gahlw@helix.nih.gov.
Accession Numbers
The accession numbers for the previously unreported CDG variants studied in this paper are ClinVar: SCV000256094 (MPI; c.1205A>G), SCV000256095 (MPI; c.1253G>A), SCV000256096 (DPAGT1; c.584C>G), and SCV000256097 (c.329G>A).
Web Resources
The URLs for data presented herein are as follows:
GeneReviews, Sparks, S.E., and Krasnewich, D.M. (1993). Congenital Disorders of Glycosylation Overview, http://www.ncbi.nlm.nih.gov/books/NBK1332/
OMIM, http://www.omim.org/
Supplemental Data
Representative sequencing reads of genomic DNA spanning the two indicated SNPs for each allelic combination observed in the CDG fibroblast cell lines in Table 2. Each sequence represents a single clone bearing either the inherited genotype (Ref/Var or Var/Ref) or a non-parental, recombinant genotype (Var/Var or Ref/Ref). Images of the chromatogram traces in the area immediately flanking these SNVs are shown in Figure S1.
References
- 1.Freeze H.H. Understanding human glycosylation disorders: biochemistry leads the charge. J. Biol. Chem. 2013;288:6936–6945. doi: 10.1074/jbc.R112.429274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jaeken J., Matthijs G. Congenital disorders of glycosylation: a rapidly expanding disease family. Annu. Rev. Genomics Hum. Genet. 2007;8:261–278. doi: 10.1146/annurev.genom.8.080706.092327. [DOI] [PubMed] [Google Scholar]
- 3.Morava E., Wosik H., Kárteszi J., Guillard M., Adamowicz M., Sykut-Cegielska J., Hadzsiev K., Wevers R.A., Lefeber D.J. Congenital disorder of glycosylation type Ix: review of clinical spectrum and diagnostic steps. J. Inherit. Metab. Dis. 2008;31:450–456. doi: 10.1007/s10545-008-0822-0. [DOI] [PubMed] [Google Scholar]
- 4.Freeze H.H., Chong J.X., Bamshad M.J., Ng B.G. Solving glycosylation disorders: fundamental approaches reveal complicated pathways. Am. J. Hum. Genet. 2014;94:161–175. doi: 10.1016/j.ajhg.2013.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Leroy J.G. Congenital disorders of N-glycosylation including diseases associated with O- as well as N-glycosylation defects. Pediatr. Res. 2006;60:643–656. doi: 10.1203/01.pdr.0000246802.57692.ea. [DOI] [PubMed] [Google Scholar]
- 6.Schachter H., Freeze H.H. Glycosylation diseases: quo vadis? Biochim. Biophys. Acta. 2009;1792:925–930. doi: 10.1016/j.bbadis.2008.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wolfe L.A., Krasnewich D. Congenital disorders of glycosylation and intellectual disability. Dev. Disabil. Res. Rev. 2013;17:211–225. doi: 10.1002/ddrr.1115. [DOI] [PubMed] [Google Scholar]
- 8.Briones P., Vilaseca M.A., Schollen E., Ferrer I., Maties M., Busquets C., Artuch R., Gort L., Marco M., van Schaftingen E. Biochemical and molecular studies in 26 Spanish patients with congenital disorder of glycosylation type Ia. J. Inherit. Metab. Dis. 2002;25:635–646. doi: 10.1023/a:1022825113506. [DOI] [PubMed] [Google Scholar]
- 9.Grünewald S., Schollen E., Van Schaftingen E., Jaeken J., Matthijs G. High residual activity of PMM2 in patients’ fibroblasts: possible pitfall in the diagnosis of CDG-Ia (phosphomannomutase deficiency) Am. J. Hum. Genet. 2001;68:347–354. doi: 10.1086/318199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vuillaumier-Barrot S., Hetet G., Barnier A., Dupré T., Cuer M., de Lonlay P., Cormier-Daire V., Durand G., Grandchamp B., Seta N. Identification of four novel PMM2 mutations in congenital disorders of glycosylation (CDG) Ia French patients. J. Med. Genet. 2000;37:579–580. doi: 10.1136/jmg.37.8.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dercksen M., Crutchley A.C., Honey E.M., Lippert M.M., Matthijs G., Mienie L.J., Schuman H.C., Vorster B.C., Jaeken J. ALG6-CDG in South Africa: genotype-phenotype description of five novel patients. JIMD Rep. 2013;8:17–23. doi: 10.1007/8904_2012_150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kjaergaard S., Skovby F., Schwartz M. Absence of homozygosity for predominant mutations in PMM2 in Danish patients with carbohydrate-deficient glycoprotein syndrome type 1. Eur. J. Hum. Genet. 1998;6:331–336. doi: 10.1038/sj.ejhg.5200194. [DOI] [PubMed] [Google Scholar]
- 13.Matthijs G., Schollen E., Van Schaftingen E., Cassiman J.J., Jaeken J. Lack of homozygotes for the most frequent disease allele in carbohydrate-deficient glycoprotein syndrome type 1A. Am. J. Hum. Genet. 1998;62:542–550. doi: 10.1086/301763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schneider A., Thiel C., Rindermann J., DeRossi C., Popovici D., Hoffmann G.F., Gröne H.J., Körner C. Successful prenatal mannose treatment for congenital disorder of glycosylation-Ia in mice. Nat. Med. 2012;18:71–73. doi: 10.1038/nm.2548. [DOI] [PubMed] [Google Scholar]
- 15.Matthijs G., Schollen E., Bjursell C., Erlandson A., Freeze H., Imtiaz F., Kjaergaard S., Martinsson T., Schwartz M., Seta N. Mutations in PMM2 that cause congenital disorders of glycosylation, type Ia (CDG-Ia) Hum. Mutat. 2000;16:386–394. doi: 10.1002/1098-1004(200011)16:5<386::AID-HUMU2>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 16.Casado M., O’Callaghan M.M., Montero R., Pérez-Cerda C., Pérez B., Briones P., Quintana E., Muchart J., Aracil A., Pineda M., Artuch R. Mild clinical and biochemical phenotype in two patients with PMM2-CDG (congenital disorder of glycosylation Ia) Cerebellum. 2012;11:557–563. doi: 10.1007/s12311-011-0313-y. [DOI] [PubMed] [Google Scholar]
- 17.Helander A., Jaeken J., Matthijs G., Eggertsen G. Asymptomatic phosphomannose isomerase deficiency (MPI-CDG) initially mistaken for excessive alcohol consumption. Clin. Chim. Acta. 2014;431:15–18. doi: 10.1016/j.cca.2014.01.018. [DOI] [PubMed] [Google Scholar]
- 18.Gerwig G.J., Bause E., Nuytinck L.K., Vliegenthart J.F., Breuer W., Kamerling J.P., Espeel M.F., Martin J.J., Chan N.W., Dacremont G.A., De Praeter C.M., De Paepe A.M., Van Coster R.N. A novel disorder caused by defective biosynthesis of N-linked oligosaccharides due to glucosidase I deficiency. Am. J. Hum. Genet. 2000;66:1744–1756. doi: 10.1086/302948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sadat M.A., Moir S., Chun T.W., Lusso P., Kaplan G., Wolfe L., Memoli M.J., He M., Vega H., Kim L.J.Y. Glycosylation, hypogammaglobulinemia, and resistance to viral infections. N. Engl. J. Med. 2014;370:1615–1625. doi: 10.1056/NEJMoa1302846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen J.M., Cooper D.N., Chuzhanova N., Férec C., Patrinos G.P. Gene conversion: mechanisms, evolution and human disease. Nat. Rev. Genet. 2007;8:762–775. doi: 10.1038/nrg2193. [DOI] [PubMed] [Google Scholar]
- 21.Symington L.S., Rothstein R., Lisby M. Mechanisms and regulation of mitotic recombination in Saccharomyces cerevisiae. Genetics. 2014;198:795–835. doi: 10.1534/genetics.114.166140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Strachan T., Read A.P. Garland Science/Taylor & Francis Group; New York: 2004. Human Molecular Genetics. [Google Scholar]
- 23.Carneiro M.O., Russ C., Ross M.G., Gabriel S.B., Nusbaum C., DePristo M.A. Pacific biosciences sequencing technology for genotyping and variation discovery in human data. BMC Genomics. 2012;13:375. doi: 10.1186/1471-2164-13-375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.de Silva E., Kelley L.A., Stumpf M.P. The extent and importance of intragenic recombination. Hum. Genomics. 2004;1:410–420. doi: 10.1186/1479-7364-1-6-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ellis N.A., Lennon D.J., Proytcheva M., Alhadeff B., Henderson E.E., German J. Somatic intragenic recombination within the mutated locus BLM can correct the high sister-chromatid exchange phenotype of Bloom syndrome cells. Am. J. Hum. Genet. 1995;57:1019–1027. [PMC free article] [PubMed] [Google Scholar]
- 26.Rahn J.J., Lowery M.P., Della-Coletta L., Adair G.M., Nairn R.S. Depletion of Werner helicase results in mitotic hyperrecombination and pleiotropic homologous and nonhomologous recombination phenotypes. Mech. Ageing Dev. 2010;131:562–573. doi: 10.1016/j.mad.2010.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Iqbal Z., Shahzad M., Vissers L.E., van Scherpenzeel M., Gilissen C., Razzaq A., Zahoor M.Y., Khan S.N., Kleefstra T., Veltman J.A. A compound heterozygous mutation in DPAGT1 results in a congenital disorder of glycosylation with a relatively mild phenotype. Eur. J. Hum. Genet. 2013;21:844–849. doi: 10.1038/ejhg.2012.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Choate K.A., Lu Y., Zhou J., Elias P.M., Zaidi S., Paller A.S., Farhi A., Nelson-Williams C., Crumrine D., Milstone L.M., Lifton R.P. Frequent somatic reversion of KRT1 mutations in ichthyosis with confetti. J. Clin. Invest. 2015;125:1703–1707. doi: 10.1172/JCI64415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hirschhorn R. In vivo reversion to normal of inherited mutations in humans. J. Med. Genet. 2003;40:721–728. doi: 10.1136/jmg.40.10.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shao C., Deng L., Henegariu O., Liang L., Raikwar N., Sahota A., Stambrook P.J., Tischfield J.A. Mitotic recombination produces the majority of recessive fibroblast variants in heterozygous mice. Proc. Natl. Acad. Sci. USA. 1999;96:9230–9235. doi: 10.1073/pnas.96.16.9230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kiritsi D., He Y., Pasmooij A.M., Onder M., Happle R., Jonkman M.F., Bruckner-Tuderman L., Has C. Revertant mosaicism in a human skin fragility disorder results from slipped mispairing and mitotic recombination. J. Clin. Invest. 2012;122:1742–1746. doi: 10.1172/JCI61976. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Representative sequencing reads of genomic DNA spanning the two indicated SNPs for each allelic combination observed in the CDG fibroblast cell lines in Table 2. Each sequence represents a single clone bearing either the inherited genotype (Ref/Var or Var/Ref) or a non-parental, recombinant genotype (Var/Var or Ref/Ref). Images of the chromatogram traces in the area immediately flanking these SNVs are shown in Figure S1.