To the Editor: “Collider bias” (also referred to as the “reversal paradox”)1 describes the artificial association created between two exposures (A and B) when a shared outcome (X) is included in the model as a covariate (Figure 1). A recent paper by Aschard et al. described the potential for collider bias when adjusting for heritable covariates in genetic association studies.2 However, in their examples, the authors acknowledged that they could not exclude the possibility of a true biological explanation for the genetic association seen only in the adjusted model. Furthermore, the extent to which this bias could create a completely spurious genetic association, rather than just modify the magnitude of the effect,3 remains unclear.
Figure 1.
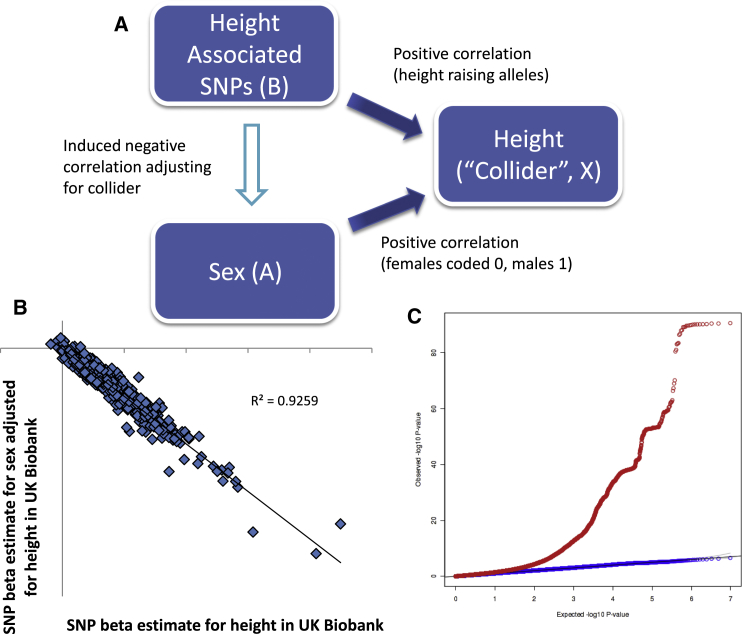
Induced Collider Bias between Genetic Variants, Height, and Sex
(A) Schematic diagram of the scenario in which collider bias can occur between genetic variants, height, and sex.
(B) Spurious autosomal SNP-effect estimates for sex, created by adjusting for height as a covariate, are almost perfectly correlated with SNP-effect estimates for height. In this scenario of collider bias, adjustment for the collider height creates biologically implausible sex associations for the 694 previously identified genome-wide significant autosomal SNPs for height.
(C) A quantile-quantile plot of genome-wide autosomal test statistics for sex ∼ SNP (shown in blue) and sex ∼ SNP + height (shown in red).
We sought to definitively illustrate collider bias by deliberately inducing it to generate a biologically implausible SNP-phenotype association. Both sex (A) and autosomal genetic determinants for adult height (B) have causal effects on height (X), but are themselves implausibly correlated. We theorized that collider bias would induce false-positive associations between (only) autosomal height-associated genetic variants and sex when adjusting for height as a covariate. By applying this model to data from the UK Biobank study, we identified over 200 spurious genome-wide significant associations, illustrating the danger of collider bias in genetic association studies.
Using a sample of 142,630 individuals of white European ancestry from the UK Biobank study,4 we performed a genome-wide association study (GWAS) for sex by using a linear mixed model5 and applying standard quality control metrics.6 As expected, in univariate models (i.e., regression model: sex ∼ SNP), no SNP reached genome-wide significance, and test statistics for 694/697 (3 missing) previously identified height SNPs7 conformed to a null distribution (Pmin = 1.4 × 10−3, p < 0.05 with 27 SNPs, ∼35 SNPs expected by chance). In contrast, when we repeated the analysis, this time including height as a covariate (regression model: sex ∼ SNP + height), 222/694 height SNPs reached genome-wide significance for association with sex. Each height-increasing allele exhibited the expected negative correlation with sex (i.e., lower probability of being male), given the two causal exposures were aligned to be positively associated with height (Figure 1). This was exemplified by the three strongest signals in the reported GWAS meta-analysis of height from the GIANT (genetic investigation of anthropometric traits) consortium: ZBTB38-rs724016 (association with sex: Punadj = 0.05, Padj = 7 × 10−90), GDF5-rs143384 (Punadj = 0.13, Padj = 7 × 10−71), and HMGA2-rs8756 (Punadj = 0.99, Padj = 3 × 10−34). These all showed an apparent robust association with sex only in the height-adjusted model.
Consistent with expectation, among the 694 height-associated SNPs, the beta estimates for sex adjusted for height were almost perfectly correlated with the beta estimate for height adjusted for sex within the UK Biobank study sample (Figure 1). Furthermore, the proportionality between the SNP betas of these two models (after normalizing phenotypes to have the same variance) was roughly equal to the phenotypic correlation (r) of the two exposures (A and B). Outside of the known height loci, there were no SNPs at genome-wide significance for sex adjusted for height that did not exhibit strong evidence for association with height alone (Pmax = 6 × 10−7). These findings are consistent with what might be expected in the presence of collider bias, suggesting that collider bias is only a concern when a SNP is significantly associated with the collider phenotype in a univariate model.
In summary, we have demonstrated that adjusting for causally associated covariates can create apparently highly robust, but actually biologically spurious, associations. The extent of this collider bias is almost perfectly inversely related to the strength of the exposure-collider association. Consideration of causal inference modeling and unadjusted test statistics is therefore of great importance in the design and interpretation of genetic (and non-genetic1) association studies.
Acknowledgments
This work was conducted using the UK Biobank resource. This work was supported by the Medical Research Council (unit programme numbers MC_UU_12015/1 and MC_UU_12015/2).
References
- 1.Tu Y.-K., West R., Ellison G.T.H., Gilthorpe M.S. Why evidence for the fetal origins of adult disease might be a statistical artifact: the “reversal paradox” for the relation between birth weight and blood pressure in later life. Am. J. Epidemiol. 2005;161:27–32. doi: 10.1093/aje/kwi002. [DOI] [PubMed] [Google Scholar]
- 2.Aschard H., Vilhjálmsson B.J., Joshi A.D., Price A.L., Kraft P. Adjusting for heritable covariates can bias effect estimates in genome-wide association studies. Am. J. Hum. Genet. 2015;96:329–339. doi: 10.1016/j.ajhg.2014.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yaghootkar H., Stancáková A., Freathy R.M., Vangipurapu J., Weedon M.N., Xie W., Wood A.R., Ferrannini E., Mari A., Ring S.M. Association analysis of 29,956 individuals confirms that a low-frequency variant at CCND2 halves the risk of type 2 diabetes by enhancing insulin secretion. Diabetes. 2015;64:2279–2285. doi: 10.2337/db14-1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sudlow C., Gallacher J., Allen N., Beral V., Burton P., Danesh J., Downey P., Elliott P., Green J., Landray M. UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 2015;12:e1001779. doi: 10.1371/journal.pmed.1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Loh P.-R., Tucker G., Bulik-Sullivan B.K., Vilhjálmsson B.J., Finucane H.K., Salem R.M., Chasman D.I., Ridker P.M., Neale B.M., Berger B. Efficient Bayesian mixed-model analysis increases association power in large cohorts. Nat. Genet. 2015;47:284–290. doi: 10.1038/ng.3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.UK Biobank. <http://www.ukbiobank.ac.uk/wp-content/uploads/2014/04/UKBiobank_genotyping_QC_documentation-web.pdf>.
- 7.Wood A.R., Esko T., Yang J., Vedantam S., Pers T.H., Gustafsson S., Chu A.Y., Estrada K., Luan J., Kutalik Z., Electronic Medical Records and Genomics (eMEMERGEGE) Consortium. MIGen Consortium. PAGEGE Consortium. LifeLines Cohort Study Defining the role of common variation in the genomic and biological architecture of adult human height. Nat. Genet. 2014;46:1173–1186. doi: 10.1038/ng.3097. [DOI] [PMC free article] [PubMed] [Google Scholar]