Abstract
Congenital disorders of glycosylation (CDGs) form a genetically and clinically heterogeneous group of diseases with aberrant protein glycosylation as a hallmark. A subgroup of CDGs can be attributed to disturbed Golgi homeostasis. However, identification of pathogenic variants is seriously complicated by the large number of proteins involved. As part of a strategy to identify human homologs of yeast proteins that are known to be involved in Golgi homeostasis, we identified uncharacterized transmembrane protein 199 (TMEM199, previously called C17orf32) as a human homolog of yeast V-ATPase assembly factor Vph2p (also known as Vma12p). Subsequently, we analyzed raw exome-sequencing data from families affected by genetically unsolved CDGs and identified four individuals with different mutations in TMEM199. The adolescent individuals presented with a mild phenotype of hepatic steatosis, elevated aminotransferases and alkaline phosphatase, and hypercholesterolemia, as well as low serum ceruloplasmin. Affected individuals showed abnormal N- and mucin-type O-glycosylation, and mass spectrometry indicated reduced incorporation of galactose and sialic acid, as seen in other Golgi homeostasis defects. Metabolic labeling of sialic acids in fibroblasts confirmed deficient Golgi glycosylation, which was restored by lentiviral transduction with wild-type TMEM199. V5-tagged TMEM199 localized with ERGIC and COPI markers in HeLa cells, and electron microscopy of a liver biopsy showed dilated organelles suggestive of the endoplasmic reticulum and Golgi apparatus. In conclusion, we have identified TMEM199 as a protein involved in Golgi homeostasis and show that TMEM199 deficiency results in a hepatic phenotype with abnormal glycosylation.
Keywords: Golgi homeostasis, COPI vesicular transport, hypercholesterolemia, Congenital Disorders of Glycosylation, alkaline phosphatase, elevated aminotransferases, V-ATPase assembly, Vph2p, TMEM199 deficiency
Main Text
The introduction of exome sequencing in genetics has had an enormous impact on the identification of Mendelian disorders.1 However, genetic variants can appear to be falsely positive, and a considerable percentage of cases still remains genetically unsolved.2, 3 The availability of functional data improves candidate gene selection in the case of well-known biochemical pathways.4 However, this is more complicated for complex pathways with many unknown proteins, such as the secretory pathway, which is composed of the endoplasmic reticulum (ER), the Golgi apparatus, and secretory vesicles. Multiple monogenic disorders have been associated with abnormal membrane trafficking, resulting in diverse clinical phenotypes.5 Protein glycosylation also occurs in the secretory pathway: assembly and quality control of the glycoprotein occurs in the ER and further glycan modification in the Golgi.6 Congenital disorders of glycosylation (CDGs) form a group of >100 monogenic diseases affecting glycosylation.7 In a subgroup of CDGs, abnormal glycosylation of serum proteins is caused by a disturbance of Golgi homeostasis. Examples are abnormal retrograde Golgi transport of glycosyltransferases (conserved oligomeric Golgi [COG] complex defects), abnormal ion transport (TMEM165 deficiency [MIM: 614727]), and abnormal Golgi pH regulation (ATP6V0A2 deficiency [MIM: 219200]).8, 9, 10, 11, 12 ATP6V0A2 is a subunit of the vacuolar H+-ATPase (V-ATPase), the proton pump responsible for acidification of the secretory pathway, among other functions.13
Identification of more genes involved in this emerging group of Golgi homeostasis defects will generate biological insights into organelle homeostasis and vesicular transport. However, this is challenging given the large number of proteins involved, many of these with unknown functions. We set out to identify human Golgi-related proteins by using a bioinformatics search for human homologs of yeast proteins with a known role in ER-to-Golgi transport and/or Golgi glycosylation, including the V-ATPase. Yeast-to-human comparative genomics via a profile-based method, Position-Specific Iterated (PSI)-BLAST,14 revealed transmembrane protein 199 (TMEM199, also known as C17orf32 [GenBank: NP_689677.1]) as a human homolog for yeast Vph2p (also known as Vma12p [GenBank: NP_012803]). Both proteins were identified as each other’s best hit in the third iteration with an E-value of 3e-08 and a reciprocal E-value of 6e-18, suggesting that both proteins are orthologs.15 Vph2p localizes to the ER and has a confirmed role as a V-ATPase assembly factor.16, 17 Subsequently, we searched for TMEM199 mutations in individuals with genetically unsolved Golgi glycosylation defects.
DNA of individuals F1-II2, F1-II3 and F2-II2 was isolated from fibroblasts and subjected to whole-exome sequencing as described.18 All individuals, or their legal representatives, participating in this study provided informed consent, and all biological materials were obtained in accordance with the Declaration of Helsinki.
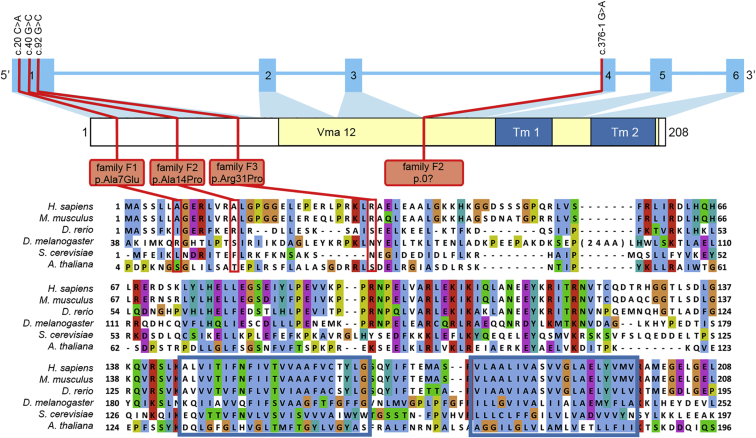
Our in-house bioinformatics pipeline was used to annotate called variants and indels.19 A frequency of >0.2% in an in-house database of over 1,300 exomes was set for exclusion of variants. We searched for non-synonymous variants located in exons and canonical splice sites. As quality criteria, we only included variants called more than five times and with a frequency of more than 20% for heterozygous variants and 80% for homozygous variants. According to a model of autosomal-recessive inheritance, no obvious candidate genes could be identified. For brothers F1-II2 and F1-II3 from family F1, previous homozygosity mapping and targeted chip-based gene sequencing of all genes in the largest homozygous intervals did not reveal a clear candidate gene either. Inspection of the raw exome-sequencing data of both brothers was carried out with the Integrative Genomics Viewer (IGV, v.2.3.14, Broad Institute). Zooming in on all TMEM199 exons (GenBank: NM_152464.2) revealed a missense mutation, c.20C>A (p.Ala7Glu), in exon 1 (Figure 1). This variant had a sequence depth of three reads in one sibling and no coverage for the other affected sibling. Sanger sequencing confirmed the missense mutation to be homozygous in DNA from both affected siblings and heterozygous in both parents. The mutation was absent from a healthy sister, thereby showing complete segregation of the mutation in the family, in agreement with autosomal-recessive inheritance (Figure 2A).
Figure 1.
TMEM199 Domain Structure, Genetic Variants, and Conservation
Schematic representation of the intron-exon structure of TMEM199 and the encoded protein. The mutations and substitutions within the different families are indicated by red lines. The yellow region indicates the conserved Vma12 domain ([Pfam: PF11712] amino acids 78–203). The two predicted transmembrane regions (Tm1 and Tm2) are indicated by a blue box.
Figure 2.
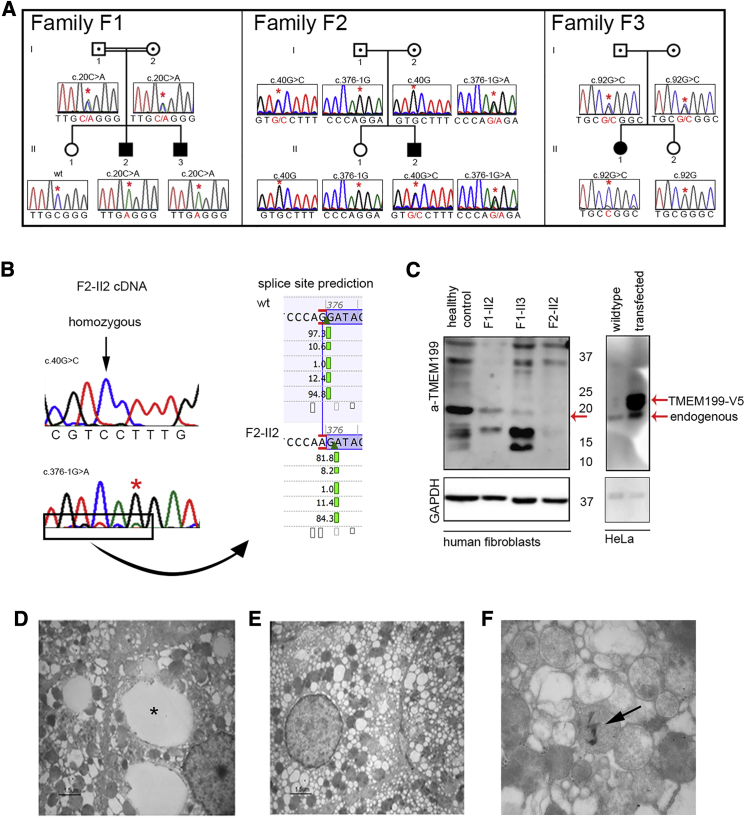
TMEM199 Mutations, TMEM199 Levels, and Liver Ultrastructure
(A) Pedigrees and sequence profiles of TMEM199 mutations in families F1 to F3.
(B) Sequencing of TMEM199 cDNA from individual F2-II2 showed homozygosity for the c.40G>C missense mutation. The insert shows the splice site prediction according to Alamut v.2.5.1 (Interactive Biosoftware), indicating a new splice site one nucleotide downstream.
(C) Western blot analysis of TMEM199 levels was performed on fibroblasts from healthy control individuals and affected individuals with anti-TMEM199 antibody (1:250, Sigma, #HPA027051) (left). TMEM199-V5 was overexpressed in HeLa cells and the Western blot incubated with anti-TMEM199 antibody (right). As a protein loading control, the same blot was incubated with mouse anti-GAPDH antibody (1:2,500, Calbiochem, #CB1001).
(D–F) Transmission electron microscopy of liver tissue from individual F2-II2.
(D) Focally, lipid vacuoles were present in the paranuclear area (asterisk).
(E) Enlarged hepatocytes show diffuse and severe vacuolization most likely derived from rough ER, smooth ER, and/or Golgi apparatus.
(F) Mitochondria featured fragmented cristae and an altered inner matrix appearing as granulo-filamentous material. Sometimes electron-dense crystal-like structures were included in the mitochondria (arrow).
For individual F2-II2, standard filtering of the whole-exome sequencing data indicated a heterozygous variant in the splice acceptor site of TMEM199 exon 4 (c.376-1G>A) (Figure 1). The raw exome-sequencing data revealed an uncalled heterozygous missense variant in exon 1 (c.40G>C [p.Ala14Pro]). Sequencing of parental DNA showed heterozygosity for the c.40G>C mutation in the father and heterozygosity for the c.376-1G>A mutation in the mother. A healthy sister did not show any of these mutations (Figure 2A). To further study the effect of the splice site mutation, we extracted mRNA from skin fibroblasts, synthesized cDNA, and sequenced it by Sanger sequencing. The resulting cDNA sequence showed the c.40G>C variant in homozygous form (Figure 2B), suggesting nonsense mediated decay of mRNA transcribed from the allele with the splice site mutation. Splice site prediction software (Alamut Visual v.2.5.1, Interactive Biosoftware) predicted the mutation to induce a stop codon 14 positions downstream by skipping one nucleotide (Figure 2B).
Sanger sequencing of TMEM199 in a cohort of individuals with genetically unsolved Golgi glycosylation disorders revealed an additional individual in family F3 with a homozygous missense mutation in TMEM199 exon 1 (c.92G>C [p.Arg31Pro]). This variant was heterozygous in both parents (Figures 1 and 2A).
Western blot analysis of TMEM199 was performed in fibroblasts of affected individuals from families F1 and F2, and reduced protein levels were observed (Figure 2C, left). Specificity was demonstrated by detection of TMEM199-V5 with an anti-TMEM199 antibody after overexpression of a TMEM199-V5 construct in HeLa cells, showing a slightly higher molecular weight than endogenous TMEM199 (Figure 2C, right).
TMEM199 (UCSC Genome Browser [GRCh37/hg19], chr17:26,684,687–26,689,089) contains six exons and encodes a protein of 208 amino acids, which includes a conserved Vma12 domain (Pfam: PF11712) at the C terminus. The Vma12 domain is widespread among eukaryotes, including Arabidopsis thaliana, and includes two transmembrane helices as predicted by the CCTOP server (see Figure 1 and Web Resources). Interestingly, three of four mutations alter residues in the N terminus, which seems to be conserved only in higher eukaryotes. The missense mutations were variably predicted to be pathogenic by different prediction programs (Table 1). The Exome Aggregation Consortium (ExAC, see Web Resources) database showed an absence of all variants except the c.92G>C variant, which had a very low frequency (5.002e-05).
Table 1.
Genotypes and Phenotypes of Families Affected by TMEM199 Deficiency
| Family F1 | Family F2 | Family F3 | |
|---|---|---|---|
| Individuals (y.o.b.) |
F1-II2 (1990), F1-II3 (1998) |
F2-II2 (1974) |
F3-II1 (1992) |
|
Genotype | |||
| Genomic change (chr17) | g.26684713C>A | g.26684733G>C / g.26687551G>A | g.26684785G>C |
| cDNA change | c.20C>A | c.40G>C / c.376-1G>A | c. 92G>C |
| Zygosity | homozygous | compound heterozygous | homozygous |
| Protein change | p.Ala7Glu | p.Ala14Pro / p.0? | p.Arg31Pro |
| Allele frequency (ExAC) | 0 | 0 / 0 | 5.002e-05 |
| PhyloP | 0.29 | 3.19 / NA | 1.09 |
| SIFT | not tolerated | tolerated / NA | tolerated |
| PolyPhen-2 (score) | possibly damaging (0.609) | probably damaging (0.990) / NA | probably damaging (0.991) |
| MutationTaster | polymorphism | disease causing / NA | disease causing |
| Grantham distance |
107 |
27 / NA |
103 |
|
Phenotype | |||
| Elevated aminotransferases | + | + | + |
| Elevated alkaline phosphatase | ++ | ++ | ++ |
| Elevated cholesterol and LDL-C | + | + | + |
| Steatosis | + | + | n.d. |
| Low ceruloplasmin | + | + | + |
| Glycosylation profile | abnormal N- and O-glycosylation | abnormal N-glycosylation | abnormal N- and O-glycosylation |
Abbreviations are as follows: y.o.b., year of birth; LDL-C, low-density lipoprotein cholesterol; NA, not applicable; n.d., not determined.
Individuals with mutations in TMEM199 (Table 1) showed a mild clinical presentation with hypercholesterolemia and elevated low-density lipoprotein cholesterol (LDL-C), elevated alkaline phosphatase (ALP, bone-derived) and aminotransferases (ATs, consisting of aspartate aminotransferase [AST] and alanine aminotransferase [ALT]). Liver biopsy showed lipid degeneration and mild steatosis with minimal fibrosis. In addition, slight abnormalities of copper metabolism were noted. However, sequencing of ATP7B (MIM: 606882) for a diagnosis of Wilson disease (WD [MIM: 277900]) failed to detect a mutation. Total cholesterol levels were in between those of control individuals and individuals with familial hypercholesterolemia (FH [MIM: 143890]). Inspection of the raw exome-sequencing data from three individuals revealed no mutations in genes known to be associated with FH (LDLR [MIM: 606945], APOB [MIM: 107730], PCSK9 [MIM: 607786]), although coverage was very poor for exon 1 of these three genes and for exon 10 of PCSK9. Total cholesterol levels of the parents were not measured.
The birth and early childhood of brothers F1-II2 and F1-II3, born 1990 and 1998, respectively, to Greek consanguineous parents, was unremarkable. During a routine blood test at fourteen and six years of age, respectively, elevated serum ATs and ALP were noticed (F1-II2: ALT 54 U/l [normal range, 0–50 U/l], AST 73 U/l [normal range, 0–50 U/l], ALP 745 U/l [normal range, < 360 U/l]; F1-II3: ALT 210 U/l, AST 246 U/l, ALP 1162 U/l). Gamma-glutamyl transferase (gGT) and bilirubin levels were normal. A liver ultrasound was normal and liver biopsy showed steatosis. Their clinical appearance was unremarkable, and their growth and psychomotor development were normal. Isoelectric focusing (IEF) of transferrin (Tf) showed a type 2 CDG profile. Further biochemical analysis showed low serum ceruloplasmin for individual F1-II2 (11.5 mg/dl [normal range, 15–60 mg/dl]) and low-normal ceruloplasmin for individual F1-II3 (15.6 mg/dl). On suspicion of WD, a slightly elevated 24 hr urinary copper/creatinine ratio was found (86.9 and 72.8 μg/g creatinine [normal range, < 60 μg/g creatinine]). However, Kayser-Fleischer rings were absent, and genetic testing for ATP7B found no mutations, therefore a diagnosis of WD was discarded. Additionally, both siblings had elevated levels of total cholesterol (F1-II2: 253 mg/dl [normal range, 120–200 mg/dl]; F1-II3: 336 mg/dl) and LDL-C (F1-II2: 188 mg/dl [normal range, 50–130 mg/dl]; F1-II3: 277 mg/dl). Several coagulation factors were slightly decreased (antithrombin-III, factor XI, factor XIII, and protein S), but the international normalized ratio was normal. During subsequent years, ATs, ALP, and total cholesterol fluctuated from normal to approximately 1.5 times the upper level of normal. During their last visit at the ages of 23 and 15 years, respectively, ALT and AST were within the normal range, but ALP was still slightly elevated. Lipid levels of individual F1-II2 had normalized, but for individual F1-II3, hypercholesterolemia persisted. Ceruloplasmin levels remained low for both individuals.
Individual F2-II2 is a 41-year-old male with a clinical history of unexplained elevation of ATs and ALP. His clinical presentation has been described before.20 During routine pre-operative investigations for inguinal hernia surgery at the age of 11, incidentally, elevated ATs and ALP were found (ALT 190 U/l, AST 153 U/l, and ALP 718 U/l). At age 31, after 21 years of unexplained elevated ATs and ALP, a liver biopsy showed steatosis and minimal fibrosis with a hepatic dry copper weight of 90 μg/g (normal range, <40 μg/g; for WD, >250 μg/g). Serum ceruloplasmin was decreased (8.8 mg/dl) and urinary copper levels were normal. Kayser-Fleischer rings were absent. Based on these parameters, WD was discarded as a diagnosis. Additionally, hypercholesterolemia (204–337 mg/dl) and elevated LDL-C (182–226 mg/dl) were present. Over the course of years, ATs regressed to normal (ALT 46–159 U/l, AST 40–110 U/l), but high ALP persisted (365–557 U/l). Metabolic screening showed a type 2 CDG pattern.
Individual F3-II1 is a 23-year-old female born to Greek parents originating from the same small town. Her neonatal period was unremarkable. She was first seen by a clinician at the age of three months with hypotonia, which persisted during the following years. At the age of one year old, the diagnosis of benign infantile hypotonia was made. During early childhood, she also suffered from psychomotor disability. Brain MRI at the ages of four years and eleven years was normal. Due to attention deficit, carbamazepine was started at the age of six years but was discontinued after two years due to elevation of her liver enzymes, thought to be drug induced. Over the course of years, ATs and ALP remained elevated (ALT 36–172 U/l, AST 31–119 U/l, ALP 132–1,528 U/l) with low-normal ceruloplasmin (17–24 mg/dl). The lipid spectrum was normal until the age of five years, but cholesterol (231–257 mg/dl) and LDL-C (161–171 mg/dl) increased when she got older. At the age of 13 years, metabolic screening revealed abnormal N- and O-glycosylation, in agreement with a diagnosis of type 2 CDG. Subunit analysis of the COG complex failed to detect mutations. Unfortunately, she was lost for follow-up until 2014. In 2014 she was doing well and her ALT, AST, and ALP were only slightly elevated (58 U/l, 52 U/l, and 132 U/l). Serum ceruloplasmin was low (18 mg/dl) and hypercholesterolemia persisted.
We performed ultrastructural analysis of liver tissue from individual F2-II2, directly fixed with 2.5% glutaraldehyde in 0.1 M phosphate buffer. Hepatocytes were enlarged with cytoplasmic degeneration. Severe vacuolization was observed, as well as the presence of large lipid vacuoles, in agreement with optical microscopy analysis (Figure 2D). A marked dilation and vesiculation of the Golgi and/or ER was seen (Figure 2E). In addition, mitochondria featured fragmented cristae, an altered inner matrix appearing as a granulo-filamentous material, and sometimes included electron-dense crystal-like structures (Figure 2F).
All individuals were identified during routine metabolic diagnostics with abnormal glycosylation of serum Tf in CDG screening. Tf IEF showed increased disialo- and trisialotransferrin (see Figure 3A and Table S1 for quantification and ranges), indicating abnormal N-glycosylation. In addition, IEF of serum apolipoprotein C-III (ApoC-III) for analysis of mucin-type O-glycosylation showed an abnormal ApoC-III-1 profile for families F1 and F3 (see Figure 3B and Table S2 for quantification and ranges).
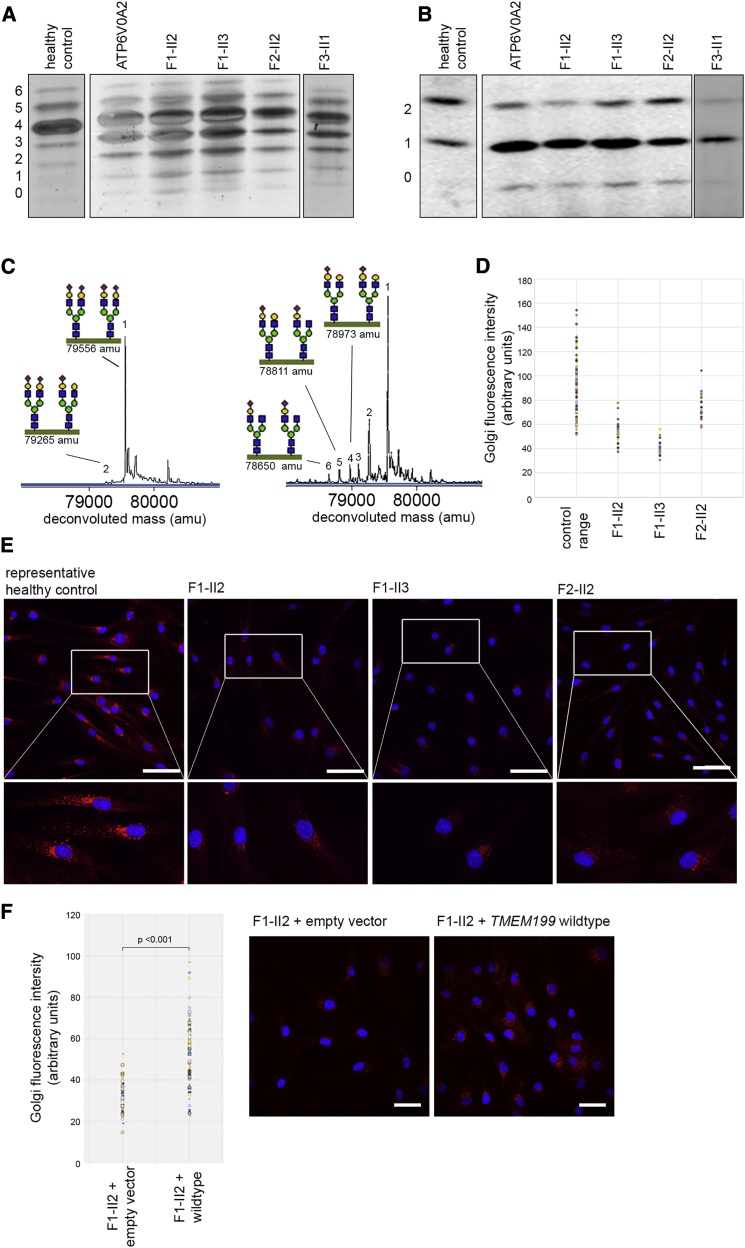
Figure 3.
Glycosylation Abnormalities in TMEM199-Deficient Individuals
(A) Glycosylation screening by IEF of serum Tf. The accompanying numbers represent the total number of sialic acids in the different protein isoforms. Representative N-glycosylation profiles from a healthy control individual, an ATP6V0A2-deficient individual, and TMEM199-deficient individuals are shown.
(B) Glycosylation screening by IEF of serum ApoC-III. ApoC-III has one mucin-type O-linked glycan with one or two sialic acids. For normal ranges and quantification of Tf-IEF and ApoC-III-IEF, see Tables S1 and S2.
(C) The nanochip-C8 QTOF mass spectrum of a healthy control individual (left spectrum) and individual F1-II2 (right spectrum) are shown. Peak 1 (79,556 amu) represents the intact Tf protein with two attached complete glycans. Any subsequent loss of sialic acid and/or galactose can be calculated based on the mass difference with the main peak (i.e., loss of one sialic acid [purple diamond, peak 2]). For glycan structural annotation of peaks 1 to 6, see Table S3.
(D–E) Fibroblasts from healthy control individuals and individuals F1-II2, F1-II3, and F2-II2 were incubated with ManNAl for metabolic labeling of sialic acids. The fluorescent signal indicates incorporation of sialic acids in glycoconjugates. Three healthy control fibroblasts were pooled from experiments performed twice. Scale bars indicate 75 μm.
(F) Fibroblasts of individual F1-II2 were transduced with an empty vector or TMEM199 wild-type construct and incubated with ManNAl, similarly to (D) and (E). Scale bars indicate 50 μm.
To gain further insight into the aberrant N-glycan structures, we performed nanochip-C8 QTOF mass spectrometry of the intact Tf protein, which showed an accumulation of incompletely synthesized glycans lacking galactose and sialic acid for individual F1-II2, in comparison to the spectrum of a healthy individual (Figure 3C, peaks 2–6). Mass spectrometry of total plasma N-glycans showed a similar pattern, with an increase of glycans lacking galactose and sialic acid (Figure S1). Individuals with ATP6V0A2 deficiency show very similar glycosylation abnormalities of Tf, apoC-III, and total plasma N-glycans (Figures 3A and 3B).11
Next, we labeled sialic acids with alkyne-tagged synthetic sugar analogs, as reported before.21 These sialic acid precursors (alkyne-tagged mannosamine [ManNAl]) are incorporated into nascent glycoproteins by the cell, allowing specific detection with fluorescently labeled azides. The immunofluorescent intensity of the azides is a marker for Golgi glycosylation efficiency. As in previous studies, the main pool of labeled glycoproteins located to the Golgi apparatus in healthy control fibroblasts (Figure 3E).21 Quantification of the immunofluorescence signal with TISGolgi, an ImageJ plugin, showed a clear reduction of absolute Golgi fluorescence intensity in individuals F1-II2, F1-II3, and F2-II2 (Figures 3D and 3E). Subsequently, fibroblasts from individual F1-II2 were complemented by transduction with either a pLenti6.2-GFP-V5 (mock) or a pLenti6.2-TMEM199-V5 (wild-type TMEM199) vector. As shown in Figure 3F, the overall Golgi fluorescence intensity increased after transduction with wild-type TMEM199 but not with the mock vector. Together, these data confirm a generalized defect in Golgi processing of protein-linked glycans due to deficiency of TMEM199.
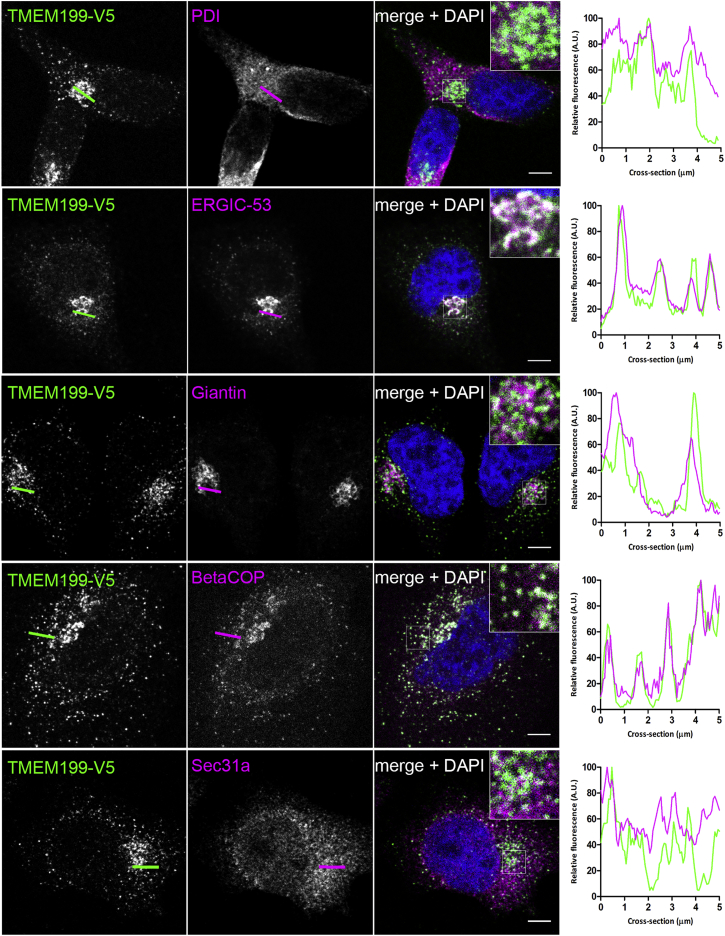
Our study indicated TMEM199 as the human homolog of yeast V-ATPase assembly factor Vph2p. In yeast, V-ATPase assembly factors Vph2p, Vma21p, Pkr1p, and Vma22p are localized to the ER.17 To define the subcellular location of TMEM199, a pLenti6.2-TMEM199-V5 plasmid for expression of C-terminally V5-tagged proteins was transiently overexpressed in HeLa cells. Imaging was done with a Leica SP8 (Leica Microsystems) confocal microscope with 60× water immersion 1.2 NA objective. Figure 4 shows clear co-localization of V5-tagged TMEM199 with ER-Golgi intermediate compartment (ERGIC) and coat protein complex I (COPI) vesicle markers. In addition, partial co-localization was seen with giantin, a Golgi marker, but not with markers for ER or COPII vesicles. On the basis of these results, we conclude that TMEM199 predominantly localizes to the ER-to-Golgi region, but not to the ER.
Figure 4.
Localization of TMEM199
TMEM199 cDNA, compatible with Gateway Technology (Life Technologies), was purchased from Harvard Medical School. HeLa cells were transfected with FuGENE HD Transfection Reagent (Promega), grown on coverslips coated with poly-L-Lysine (Sigma), and incubated overnight. After permeabilization and fixation, cells were stained with fluorescently labeled antibodies against V5 (green in merge) and organelle markers (magenta in merge). Shown are representative cells for the V5 antibody, organelle markers, and a merge with DAPI stain (blue), including a 3-fold magnification. Co-localization is indicated by white in the merged channel. The graphs show the fluorescence intensity profiles along the cross-sections indicated. Scale bars represent 5 μm. Organelle markers used are as follows: anti-V5-Tag (dilution 1:200, Life Technologies, #R960-25), anti-PDI (1:500, Abcam, #ab3672), anti-ERGIC-53 (1:200, Sigma-Aldrich, #E1031), anti-Giantin (1:1,000, BioLegend, #prb-114c), anti- BetaCOP (1:1,000, Abcam, #ab2899), and anti-Sec31a (1:500, Sigma-Aldrich, #HPA005457).
In parallel with identification of TMEM199 mutations, we identified mutations in CCDC115 in a cohort of individuals with abnormal Golgi glycosylation and a storage-disease-like clinical phenotype including hepatosplenomegaly, neurological symptoms, elevated ATs, alkaline phosphatase, and hypercholesterolemia (see the related paper in this issue of AJHG).22 CCDC115 was predicted to be homologous to yeast V-ATPase assembly factor Vma22p. In yeast, Vph2p and Vma22p are localized to the ER and cooperatively stabilize the V0 domain during early assembly of the V-ATPase in the ER.16, 17 The human V-ATPase proton pump consists of 14 subunits divided between the membrane-bound V0 domain and a cytosolic V1 domain.13 It participates in the acidification of the secretory pathway or the extracellular matrix in specialized cells. However, it has numerous additional functions and associations with other cellular mechanisms and pathways.13 A few human disorders involving defects in V-ATPase proteins are known, including X-linked myopathy with excessive autophagy due to mutations in VMA21 (MIM: 310440),23 renal tubular acidosis with deafness due to mutations in the kidney-specific isoforms ATP6V1B1 (MIM: 267300) or ATP6V0A4 (MIM: 602722),24, 25, 26 Zimmerman-Laband syndrome 2 due to mutations in ATP6V1B2 (MIM: 616455),27 osteopetrosis due to mutations in the osteoclast-specific V0a3 subunit (MIM: 259700) and cutis laxa due to mutations in ATP6V0A2.24, 28, 29 ATP6V0A2 encodes the a2 subunit of the V0 domain, which localizes the V-ATPase complex to the Golgi apparatus, and is the only core subunit thus far associated with abnormal glycosylation.11, 30 The glycosylation profile seen in ATP6V0A2 deficiency is similar to that of TMEM199 deficiency; however, the clinical symptoms are clearly different. In addition, a clear effect of TMEM199 mutations was observed on organelle ultrastructure, different from ATP6V0A2 deficiency.12
At the moment, it is difficult to explain the broad heterogeneity in clinical phenotypes among the different V-ATPase associated diseases. Even mutations in TMEM199 and CCDC115, whose gene products are probably interacting in humans, result in different phenotypes. It seems plausible to attribute these differences to different functions of the V-ATPase. On the basis of our experiments, we propose a role for TMEM199 in Golgi homeostasis. However, it remains to be investigated whether this function is mediated through deficient proton transport via an effect on the V-ATPase or whether TMEM199 exerts additional roles in ER-to-Golgi transport.
We have shown that prioritization of exome data based on functional knowledge of abnormal protein glycosylation can successfully elucidate previously unknown Golgi homeostasis defects. Protein interaction maps can be a useful tool for further identification of novel candidate proteins. During our studies, a protein interaction map of Drosophila melanogaster identified interactions between CG7071 (the fruit fly TMEM199) and multiple V-ATPase subunits.31 Recently, an ongoing effort to map the human interactome reported similar protein-protein interactions in humans (accessed via BioGRID, see Web Resources). Such interactomes will be of great benefit for disease-gene prioritization in cohorts of families with genetically unsolved cases of type 2 CDGs.
In conclusion, we used comparative genomics to uncover TMEM199 as a protein involved in Golgi homeostasis, most likely with at least a partial role in V-ATPase functioning. TMEM199 deficiency should be considered in individuals with unexplained mildly elevated serum ATs, elevated ALP, hepatic steatosis, hypercholesterolemia, and low serum ceruloplasmin. We suggest screening for abnormal glycosylation in individuals with these symptoms.
Acknowledgments
We would like to thank the CDG-affected families for their participation in this study. We would also like to thank the group of Dr. Y. Guerardel and Prof. C. Biot for their generous donation of ManNAl. This work was financially supported by grants from the Institute of Genetic and Metabolic Disease (to D.J.L., R.J.R., and J.A.V.), the Dutch Organization for Scientific Research (ZONMW Medium Investment Grant 40-00506-98-9001 and VIDI Grant 91713359 to D.J.L. and VENI grant to A.G.H.), the Metakids foundation (J.C.J., M.v.S., J.P.H.D., and D.J.L.), the AMC graduate school Ph.D scholarship (M.A.W.v.d.B.), the Dr. Karel-Lodewijk Verleysen Award (J.C.J. and J.P.H.D.), the French National Agency (ANR SOLV-CDG to F.F.), and by grant ERARE11-135 of the ERA-Net for Research Programs on Rare Diseases Joint Transnational Call 2011 (EURO-CDG).
Published: January 28, 2016
Footnotes
Supplemental Data include one figure and three tables and can be found with this article online at http://dx.doi.org/10.1016/j.ajhg.2015.12.011.
Accession Numbers
The accession numbers for the pathogenic variants c.92G>C, c.20C>A, and c.40G>C; 376-1G>A reported in this paper are ClinVar: SCV000257475, SCV000257473, and SCV000257476, respectively.
Web Resources
The URLs for data presented herein are as follows:
BioGRID, http://thebiogrid.org
Constrained Consensus TOPology prediction server, http://cctop.enzim.ttk.mta.hu/
ExAC Browser, http://exac.broadinstitute.org
GenBank, http://www.ncbi.nlm.nih.gov/genbank/
Harvard PlasmID database, https://plasmid.med.harvard.edu/PLASMID/Home.jsp
ImageJ, http://imagej.nih.gov/ij
Jalview, http://www.jalview.org
MutationTaster, http://www.mutationtaster.org/
OMIM, http://www.omim.org/
PolyPhen-2, genetics.bwh.harvard.edu/pph2/
SIFT, http://sift.jcvi.org
TISGolgi, tisbio.wix.com/tisbio
UCSC Human Genome Browser, http://genome-euro.ucsc.edu/index.html
Supplemental Data
References
- 1.Yang Y., Muzny D.M., Reid J.G., Bainbridge M.N., Willis A., Ward P.A., Braxton A., Beuten J., Xia F., Niu Z. Clinical whole-exome sequencing for the diagnosis of mendelian disorders. N. Engl. J. Med. 2013;369:1502–1511. doi: 10.1056/NEJMoa1306555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.MacArthur D.G., Manolio T.A., Dimmock D.P., Rehm H.L., Shendure J., Abecasis G.R., Adams D.R., Altman R.B., Antonarakis S.E., Ashley E.A. Guidelines for investigating causality of sequence variants in human disease. Nature. 2014;508:469–476. doi: 10.1038/nature13127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Javed A., Agrawal S., Ng P.C. Phen-Gen: combining phenotype and genotype to analyze rare disorders. Nat. Methods. 2014;11:935–937. doi: 10.1038/nmeth.3046. [DOI] [PubMed] [Google Scholar]
- 4.Timal S., Hoischen A., Lehle L., Adamowicz M., Huijben K., Sykut-Cegielska J., Paprocka J., Jamroz E., van Spronsen F.J., Körner C. Gene identification in the congenital disorders of glycosylation type I by whole-exome sequencing. Hum. Mol. Genet. 2012;21:4151–4161. doi: 10.1093/hmg/dds123. [DOI] [PubMed] [Google Scholar]
- 5.De Matteis M.A., Luini A. Mendelian disorders of membrane trafficking. N. Engl. J. Med. 2011;365:927–938. doi: 10.1056/NEJMra0910494. [DOI] [PubMed] [Google Scholar]
- 6.Ohtsubo K., Marth J.D. Glycosylation in cellular mechanisms of health and disease. Cell. 2006;126:855–867. doi: 10.1016/j.cell.2006.08.019. [DOI] [PubMed] [Google Scholar]
- 7.Freeze H.H., Chong J.X., Bamshad M.J., Ng B.G. Solving glycosylation disorders: fundamental approaches reveal complicated pathways. Am. J. Hum. Genet. 2014;94:161–175. doi: 10.1016/j.ajhg.2013.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Foulquier F., Amyere M., Jaeken J., Zeevaert R., Schollen E., Race V., Bammens R., Morelle W., Rosnoblet C., Legrand D. TMEM165 deficiency causes a congenital disorder of glycosylation. Am. J. Hum. Genet. 2012;91:15–26. doi: 10.1016/j.ajhg.2012.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rosnoblet C., Legrand D., Demaegd D., Hacine-Gherbi H., de Bettignies G., Bammens R., Borrego C., Duvet S., Morsomme P., Matthijs G., Foulquier F. Impact of disease-causing mutations on TMEM165 subcellular localization, a recently identified protein involved in CDG-II. Hum. Mol. Genet. 2013;22:2914–2928. doi: 10.1093/hmg/ddt146. [DOI] [PubMed] [Google Scholar]
- 10.Miller V.J., Ungar D. Re’COG’nition at the Golgi. Traffic. 2012;13:891–897. doi: 10.1111/j.1600-0854.2012.01338.x. [DOI] [PubMed] [Google Scholar]
- 11.Kornak U., Reynders E., Dimopoulou A., van Reeuwijk J., Fischer B., Rajab A., Budde B., Nürnberg P., Foulquier F., Lefeber D., ARCL Debré-type Study Group Impaired glycosylation and cutis laxa caused by mutations in the vesicular H+-ATPase subunit ATP6V0A2. Nat. Genet. 2008;40:32–34. doi: 10.1038/ng.2007.45. [DOI] [PubMed] [Google Scholar]
- 12.Hucthagowder V., Morava E., Kornak U., Lefeber D.J., Fischer B., Dimopoulou A., Aldinger A., Choi J., Davis E.C., Abuelo D.N. Loss-of-function mutations in ATP6V0A2 impair vesicular trafficking, tropoelastin secretion and cell survival. Hum. Mol. Genet. 2009;18:2149–2165. doi: 10.1093/hmg/ddp148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marshansky V., Rubinstein J.L., Grüber G. Eukaryotic V-ATPase: novel structural findings and functional insights. Biochim. Biophys. Acta. 2014;1837:857–879. doi: 10.1016/j.bbabio.2014.01.018. [DOI] [PubMed] [Google Scholar]
- 14.Altschul S.F., Madden T.L., Schäffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Szklarczyk R., Wanschers B.F., Cuypers T.D., Esseling J.J., Riemersma M., van den Brand M.A., Gloerich J., Lasonder E., van den Heuvel L.P., Nijtmans L.G., Huynen M.A. Iterative orthology prediction uncovers new mitochondrial proteins and identifies C12orf62 as the human ortholog of COX14, a protein involved in the assembly of cytochrome c oxidase. Genome Biol. 2012;13:R12. doi: 10.1186/gb-2012-13-2-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Graham L.A., Hill K.J., Stevens T.H. Assembly of the yeast vacuolar H+-ATPase occurs in the endoplasmic reticulum and requires a Vma12p/Vma22p assembly complex. J. Cell Biol. 1998;142:39–49. doi: 10.1083/jcb.142.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Forgac M. Vacuolar ATPases: rotary proton pumps in physiology and pathophysiology. Nat. Rev. Mol. Cell Biol. 2007;8:917–929. doi: 10.1038/nrm2272. [DOI] [PubMed] [Google Scholar]
- 18.Stránecký V., Hoischen A., Hartmannová H., Zaki M.S., Chaudhary A., Zudaire E., Nosková L., Barešová V., Přistoupilová A., Hodaňová K. Mutations in ANTXR1 cause GAPO syndrome. Am. J. Hum. Genet. 2013;92:792–799. doi: 10.1016/j.ajhg.2013.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vissers L.E., de Ligt J., Gilissen C., Janssen I., Steehouwer M., de Vries P., van Lier B., Arts P., Wieskamp N., del Rosario M. A de novo paradigm for mental retardation. Nat. Genet. 2010;42:1109–1112. doi: 10.1038/ng.712. [DOI] [PubMed] [Google Scholar]
- 20.Calvo P.L., Pagliardini S., Baldi M., Pucci A., Sturiale L., Garozzo D., Vinciguerra T., Barbera C., Jaeken J. Long-standing mild hypertransaminasaemia caused by congenital disorder of glycosylation (CDG) type IIx. J. Inherit. Metab. Dis. 2008;31(Suppl 2):S437–S440. doi: 10.1007/s10545-008-1004-9. [DOI] [PubMed] [Google Scholar]
- 21.Vanbeselaere J., Vicogne D., Matthijs G., Biot C., Foulquier F., Guerardel Y. Alkynyl monosaccharide analogues as a tool for evaluating Golgi glycosylation efficiency: application to Congenital Disorders of Glycosylation (CDG) Chem. Commun. (Camb.) 2013;49:11293–11295. doi: 10.1039/c3cc45914d. [DOI] [PubMed] [Google Scholar]
- 22.Jansen J.C., Cirak S., Van Scherpenzeel M., Timal S., Reunert J., Rust S., Perez B., Vicogne D., Krawitz P., Wada Y. CCDC115 deficiency causes a disorder of Golgi homeostasis characterized by abnormal protein glycosylation. Am. J. Hum. Genet. 2016;98:309–321. doi: 10.1016/j.ajhg.2015.12.010. this issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ramachandran N., Munteanu I., Wang P., Ruggieri A., Rilstone J.J., Israelian N., Naranian T., Paroutis P., Guo R., Ren Z.P. VMA21 deficiency prevents vacuolar ATPase assembly and causes autophagic vacuolar myopathy. Acta Neuropathol. 2013;125:439–457. doi: 10.1007/s00401-012-1073-6. [DOI] [PubMed] [Google Scholar]
- 24.Borthwick K.J., Kandemir N., Topaloglu R., Kornak U., Bakkaloglu A., Yordam N., Ozen S., Mocan H., Shah G.N., Sly W.S., Karet F.E. A phenocopy of CAII deficiency: a novel genetic explanation for inherited infantile osteopetrosis with distal renal tubular acidosis. J. Med. Genet. 2003;40:115–121. doi: 10.1136/jmg.40.2.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Karet F.E., Finberg K.E., Nelson R.D., Nayir A., Mocan H., Sanjad S.A., Rodriguez-Soriano J., Santos F., Cremers C.W., Di Pietro A. Mutations in the gene encoding B1 subunit of H+-ATPase cause renal tubular acidosis with sensorineural deafness. Nat. Genet. 1999;21:84–90. doi: 10.1038/5022. [DOI] [PubMed] [Google Scholar]
- 26.Smith A.N., Skaug J., Choate K.A., Nayir A., Bakkaloglu A., Ozen S., Hulton S.A., Sanjad S.A., Al-Sabban E.A., Lifton R.P. Mutations in ATP6N1B, encoding a new kidney vacuolar proton pump 116-kD subunit, cause recessive distal renal tubular acidosis with preserved hearing. Nat. Genet. 2000;26:71–75. doi: 10.1038/79208. [DOI] [PubMed] [Google Scholar]
- 27.Kortüm F., Caputo V., Bauer C.K., Stella L., Ciolfi A., Alawi M., Bocchinfuso G., Flex E., Paolacci S., Dentici M.L. Mutations in KCNH1 and ATP6V1B2 cause Zimmermann-Laband syndrome. Nat. Genet. 2015;47:661–667. doi: 10.1038/ng.3282. [DOI] [PubMed] [Google Scholar]
- 28.Frattini A., Orchard P.J., Sobacchi C., Giliani S., Abinun M., Mattsson J.P., Keeling D.J., Andersson A.K., Wallbrandt P., Zecca L. Defects in TCIRG1 subunit of the vacuolar proton pump are responsible for a subset of human autosomal recessive osteopetrosis. Nat. Genet. 2000;25:343–346. doi: 10.1038/77131. [DOI] [PubMed] [Google Scholar]
- 29.Li Y.P., Chen W., Liang Y., Li E., Stashenko P. Atp6i-deficient mice exhibit severe osteopetrosis due to loss of osteoclast-mediated extracellular acidification. Nat. Genet. 1999;23:447–451. doi: 10.1038/70563. [DOI] [PubMed] [Google Scholar]
- 30.Guillard M., Dimopoulou A., Fischer B., Morava E., Lefeber D.J., Kornak U., Wevers R.A. Vacuolar H+-ATPase meets glycosylation in patients with cutis laxa. Biochim. Biophys. Acta. 2009;1792:903–914. doi: 10.1016/j.bbadis.2008.12.009. [DOI] [PubMed] [Google Scholar]
- 31.Guruharsha K.G., Rual J.F., Zhai B., Mintseris J., Vaidya P., Vaidya N., Beekman C., Wong C., Rhee D.Y., Cenaj O. A protein complex network of Drosophila melanogaster. Cell. 2011;147:690–703. doi: 10.1016/j.cell.2011.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.