Abstract
Comprehensive identification of somatic structural variations (SVs) and understanding their mutational mechanisms in cancer might contribute to understanding biological differences and help to identify new therapeutic targets. Unfortunately, characterization of complex SVs across the whole genome and the mutational mechanisms underlying esophageal squamous cell carcinoma (ESCC) is largely unclear. To define a comprehensive catalog of somatic SVs, affected target genes, and their underlying mechanisms in ESCC, we re-analyzed whole-genome sequencing (WGS) data from 31 ESCCs using Meerkat algorithm to predict somatic SVs and Patchwork to determine copy-number changes. We found deletions and translocations with NHEJ and alt-EJ signature as the dominant SV types, and 16% of deletions were complex deletions. SVs frequently led to disruption of cancer-associated genes (e.g., CDKN2A and NOTCH1) with different mutational mechanisms. Moreover, chromothripsis, kataegis, and breakage-fusion-bridge (BFB) were identified as contributing to locally mis-arranged chromosomes that occurred in 55% of ESCCs. These genomic catastrophes led to amplification of oncogene through chromothripsis-derived double-minute chromosome formation (e.g., FGFR1 and LETM2) or BFB-affected chromosomes (e.g., CCND1, EGFR, ERBB2, MMPs, and MYC), with approximately 30% of ESCCs harboring BFB-derived CCND1 amplification. Furthermore, analyses of copy-number alterations reveal high frequency of whole-genome duplication (WGD) and recurrent focal amplification of CDCA7 that might act as a potential oncogene in ESCC. Our findings reveal molecular defects such as chromothripsis and BFB in malignant transformation of ESCCs and demonstrate diverse models of SVs-derived target genes in ESCCs. These genome-wide SV profiles and their underlying mechanisms provide preventive, diagnostic, and therapeutic implications for ESCCs.
Keywords: ESCC, structural variation, chromothripsis, breakage-fusion-bridge, whole-genome duplication
Introduction
Cancer genomes harbor various somatic forms of genetic alterations spanning from nucleotide-level alterations (e.g., point mutations and small insertions/deletions) to large chromosomal events (e.g., structural variations and copy-number changes), some of which can contribute to tumor development.1 Specially, genomic structural variation (SV) is a hallmark of cancer.1 The fraction of the genome affected by SVs is comparatively larger than that accounted for by SNPs, indicating significant consequences of SVs on phenotypic variation.2 The main types of mechanisms known to cause SVs in human cancer include homologous recombination, nonreplicative nonhomologous repair, and replication-based mechanisms.3 Generally, homologous recombination can occur by non-allelic homologous recombination (NAHR), and deficiency in homologous recombination is implicated as a major source of cancer genome instability.4 In addition, SVs, especially aberrant ligation of double-strand DNA breaks (DSBs), can arise, mostly due to exposure to external DNA-damaging agents, through non-homologous end-joining (NHEJ) or alternative end joining (alt-EJ) mechanisms.5 For complex rearrangements, the mechanisms for repairing DNA replication errors such as fork stalling and template switching (FoSTeS) or microhomology-mediated break-induced repair (MMBIR) have been described.6 Recently, single catastrophic events causing genomic shattering followed by incorrect re-joining of the fragmented DNA, termed chromothripsis, is receiving greater attention as a major mechanism generating complex SVs in human cancer.7
It is well known that SVs have implications in treatment and prediction of individual’s outcome because genome-scale rearrangements can play an unappreciated role in cancer through their ability to move blocks of adjacent genes simultaneously or form gene fusion, leading to concurrent oncogenic events.1 Comprehensive investigation in many types of tumor shows that breakpoints directly generate an oncogenic element that can be used as a therapeutic target, such as driver fusion transcript of EML4-ALK in a subset of non-small-cell lung cancer (NSCLC) that respond to the kinase inhibitor crizotinib and FGFR3-TACC3 fusions in glioblastoma, bladder cancer, lung squamous cell, and head and neck squamous cell carcinoma (HNSCC) that can benefit from targeted FGFR kinase inhibition.1, 8, 9 In addition, gene amplification, a selective copy-number increase of genomic segments through DNA rearrangements, is a clinically important form of genome instability in cancer, because gene amplification causes advanced tumors and acquired therapy resistance.10, 11 Thus, a better understanding of the underlying mechanisms of oncogenic events driven by SVs is important for identification of molecular targets for diagnosis, prognosis, and treatment guidance.
Continuous DNA breaks and rearrangements through chromothripsis, chromoplexy, or a breakage-fusion-bridge (BFB) cycle have been implicated as underlying mechanisms for gene amplification or fusion in human cancer.12 A BFB cycle, a series of chromosome breaks and duplications that generate multiple copy-number states and are assumed to derive from events occurring over many rounds of cell division, has been shown to occur in many malignant solid tumors, including HNSCC and esophageal adenocarcinoma (EAC).13, 14 In contrast to the conventional clusters of complex rearrangements, chromothripsis, despite the large number of rearrangements, exists in only two copy-number states with many transitions between these two states.15 For chromothripsis, the affected chromosome (or regions from one or a few chromosome arms) is somehow fragmented and then stitched together, most likely by NHEJ.15 The segments that are not incorporated into the derivative chromosome are either lost, yielding the low copy-number state, or incorporated into a double-minute (DM) chromosome.7, 15 Chromothripsis has been reported in 2%–5% of diverse cancer entities, with higher frequency in bone cancer (25%) and medulloblastoma (36%).3, 14 In parallel, another mutation mechanism, kataegis, has been identified as distinguishing mutational patterns that often co-occur with large-scale rearrangements.16 Unlike chromothripsis, which refers to an oncogenic mechanism operating on a global level and occurring in one or several chromosomes, kataegis has been found to operate locally, generating large numbers of mutations (or hotspots of hypermutations) in small regions of the genome.16 On the other hand, similar to chromothripsis, kataegis most likely causes a large number of substitution mutations to occur in a region of the genome at one time rather than accumulating in a step-wise fashion.17 Kataegis is remarkably common, occurring, for example, at a rate of 13/21 in breast cancer genomes.16
Massively parallel sequencing strategies offer the potential to carry out genome-wide screening for point mutations, copy-number alterations (CNAs), and rearrangements on a single platform.18 We and others recently reported genomic sequencing analyses of ESCCs, which nominated cancer-associated genes driven by point mutations.19, 20, 21, 22 However, at the level of genome structure, somatic SVs and their underlying mechanisms are largely unknown; the driving forces behind SVs have been less well characterized than those for single-nucleotide alterations in ESCC. In this study, we re-analyzed whole-genome sequencing (WGS) data of 31 ESCCs to characterize SVs and their underlying mechanisms and to identify target genes affected by SVs in ESCC. Our findings revealed different mutational mechanisms for the formation of amplification of cancer-associated genes in ESCC.
Material and Methods
Ethics Approval
This study was approved by the Ethics Committee of Shanxi Medical University (Approval No. 2009029) and the Ethics Committee of Henan Cancer Hospital (Approval No. 2009xjs12). All samples were obtained before treatment according to the guidelines of the local ethics committees, and written informed consents were received from all participants.
Data Processing
The WGS data of a total of 31 paired tumors and matched normal tissues have been deposited at the European Genome-phenome Archive (EGA).19, 22 Raw data were filtered with SOAPnuke (v.1.4.1) to remove sequencing adapters and low-quality reads. High-quality reads were aligned to the NCBI human reference genome (hg19) by BWA (v.0.5.9) with default parameters. Picard (v.1.54) was used to mark duplicates and followed by Genome Analysis Toolkit (v.1.0.6076, GATK IndelRealigner) to improve alignment accuracy. The final BAM file stores all reads and calibrated qualities along with their alignments to the genome. For interesting SVs with fewer numbers of supporting reads, we further inspected IGV and checked the split read alignment (in the .sr/ folder) to verify their accuracy.
Structural Variations Detection
Identification of somatic structural variations (SVs) from short read data is challenging. Meerkat algorithm makes it possible to predict both germline and somatic SVs directly from short read data, focusing on complex events.23 Importantly, Yang et al. verified the accuracy of Meerkat by applying it to two HapMap genomes (NA18507 and NA12878) that were sequenced at high coverage on the Illumina platform and for which complex deletions have been previously reported.23, 24 Also, 48 out of randomly selected 49 (98%) events identified via Meerkat algorithm can be validated by PCR.23 Therefore, the Meerkat algorithm can provide a more comprehensive spectrum of mechanisms of SVs in a genome and is more reliable to detect SVs. In this study, we applied Meerkat (0.185) algorithm with suggested parameters to 31 ESCC genomes to predict somatic SVs and breakpoints as described.23 In brief, we mapped reads against the human reference genome (hg19) to find soft-clipped and unmapped reads (reads that mapped in an unexpected way) and re-mapped them to identify discordant read pairs. Then, we extracted the split reads (20 bp from both ends) to search for reads that cover the candidate breakpoints and refined precise breakpoints by local alignments. Mutational mechanisms were predicted based on homology and sequencing features at the breakpoints. Somatic SVs were generated by filtering out germline events and other artifacts. We used the following criteria to remove artifacts: (1) a large number (thousands or tens of thousands) of somatic SVs in one tumor sample; (2) a dominant event type; (3) the SVs evenly distribute across all chromosomes; (4) if the dominant events are intra-chromosome, they are very uniform in size (usually several hundreds bp or at kb level). The samples that meet these criteria failed our quality-control steps and were discarded from further analysis. Only high confidence calls were used in downstream analysis.
Locally Arranged Genome
To assess the randomness of SVs on chromosomes, we used a goodness-of-fit test against the expected distribution proposed by Campbell et al. with a significant threshold < 0.0001.25 To assess the significance of SV enrichment on chromosomes, we required the number of locally arranged genomes to be more than 50 and clustered chromosomes to have a high SVs mutation rate per Mb exceeding three times the length of the interquartile range from the 75th percentile of the chromosome counts for each tumor.26
Breakage-Fusion-Bridge Detection
We detected BFB events based on the evidence of fold-back inversions and telomere loss.27 Inversions meeting the following criteria were defined as fold-back inversion. (1) Inversion is a single inversion (invers_f or invers_r) detected by Meerkat, which means there is no reciprocal partner of inversion. (2) Inversion must demarcate a copy-number change that we make comparison of reads depth between inverted-amplified and normal space region (the region between breakpoints of fold-back inversion), and the result with q < 0.0001 is defined as significance. (3) The two ends of breakpoints of fold-back inversion must be separated by <20 kb.
Chromothripsis Inference
To infer chromothripsis in ESCCs, we adapted criteria proposed by Campbell et al.25 This analysis is based on ruling out the stepwise rearrangements and required at least ten changes in segmental copy number involving two or three distinct copy-number states on a single chromosome. (1) We manually inspected copy-number profiles for each case for regularity of oscillating copy-number states. ESCC-16T, in which copy number oscillates between two and three and has more than ten transitions, was selected for inclusion. (2) We found statistical evidence (p < 0.001) for breakpoints clustering on chromosomes 3, 8, and 10 of ESCC-16T. (3) In the case of ESCC-16T, due to loss of one haplotype (chromosome 8q) and chromothripsis occurring in amplified haplotype, we could detect allelic imbalance change instead of loss and retention of heterozygosity. (4) For ESCC-16T, chromosome 8q had three copy numbers of amplified haplotype, making it difficult to entirely eliminate the possibility of rearrangements arising from two haplotypes of the sample type. However, the minor copy number always remains one, indicating high possibility of arrangements affecting a specific haplotype. (5) We found statistical evidence of the randomness of fragment joins and segment order. (6) For ESCC-16T, derivate chromosome 8 is difficult to infer the ability to walk the derivative chromosome owing to the loss of some rearrangements.
Copy-Number Alterations
Patchwork was used to determine copy-number alterations (CNAs) across 31 ESCCs.28 First, it fixed windows of 200 bp in the human reference genome, and each window was thought to be a marker. Then, it estimated the log2 ratio between tumor and normal read depth for each window. The log2 ratio of adjacent 50 windows were merged to smooth the data. The merged windows (markers) were further segmented by CBS. After the program combined the allele frequency of germline single-nucleotide variants, absolute copy number for each segments were given. Of 31 ESCC genomes, 19 clearly had clusters of normalized coverage between different copies. For these 19 tumors, we estimated the ploidy, tumor content, and absolute copy number. To identify potential copy-number targets, we combined 31 of WGS data and 123 of comparative genomic hybridization analysis (CGH) data and applied modified GISTIC method to the combined data.19, 22, 29 The amplification or deletion peaks with G-score > 0.1 that corresponds to p < 0.05 and q < 0.05 were defined as significant.
Kataegis
We defined kataegis based on five stringent hallmarks described by Nik-Zainal et al.:16 (1) presence of heavily mutated genomic regions (“macrocluster”) consisting of a few hundred base pairs (“microcluster”) separated by tens of unmutated kilobases; (2) mutation clusters generally colocalized with structural variation breakpoints; (3) mutations that are all of the same type in a long genomic region, and switched to different mutation classes in other regions; (4) within the microcluster region, most mutations being derived from the same parental chromosome; and (5) most substitutions within the hypermutated region being characterized by C>T transitions in TpCpX trinucleotides.
PCR-Sanger Sequencing Validation
For validation of TRAPPC9-CLVS1 or EIF3E-RAD51B fusion transcript, we performed RT-PCR and Sanger sequencing assays on purified tumor and matched normal cells from ESCC-16T or ESCC-19T, respectively. Total RNA (1 μg) from purified tumor and matched normal cells was used for RT-PCR with the SuperScript III First-Strand system (Invitrogen), according to the manufacturer’s instructions. The primers used were designed against exon 18 of TRAPPC9 (MIM: 611966) forward (5′-CGGAATTCACCCTGGAAGCTGTCCTG-3′) and exon 4 of CLVS1 (MIM: 611292) reverse (5′-CCCTCGAGCTGCAACCCTTCAATGGC-3′) or against exon 1 of EIF3E (MIM: 602210) forward (5′-CGGAATTCATGGCGGAGTACG-3′) and exon 5 of RAD51B (MIM: 602948) reverse (5′-CCCTCGAGCTTTCAGCACTAAATG-3′). PCR product for TRAPP9-CLVS1 (334 bp) or EIF3E-RAD51B (243 bp) was analyzed by agarose gel electrophoresis. Amplified PCR products were gel purified and then sequenced via the Sanger method.
Fluorescence In Situ Hybridization Analysis
Frozen tumor and matched normal tissues of interesting ESCC cases were cut a cryostat at 4 μm thickness, fixed in cold acetic acid/methanol for 5 min at 4°C, and dried at room temperature. Slides were stained with Cytocell enumeration probes against interesting genes FGFR1 (MIM: 136350)/CEN8 (Z-2072, Zytovision, German), CCND1 (MIM: 168461)/CEN11 (Z-2071, Zytovision, German), TRAPPC9, CLVS1, EIF3E, and RAD51B, conjugated with FITC or Cy3.5 (Rainbow Scientific). Staining was carried out according to the manufacturer’s protocol. FISH samples were viewed with a fully automated, upright Zeiss Axio-ImagerZ.1 microscope with a 20× objective and DAPI, FITC, and Rhodamine filter cubes. Images were produced using the AxioCam MRm CCD camera and Axiovision v.4.5 software suite. p values were calculated with a two-sample test for equality of proportions with continuity correction.
Real-Time Quantitative PCR
Real-time quantitative PCR (RT-PCR) was performed to quantify the mRNA expression levels of CDCA7 (MIM: 609937), LETM2, FGFR1, or WHSC1L1 (MIM: 607083) using ABI Stepone plus with a SYBR Premix Ex Taq Kit (Takara Bio). GAPDH was used as an endogenous control. Primers for GAPDH (F: 5′-CGGAGTCAACGGATTTGGTCGTAT-3′; R: 5′-AGCCTTCTCCATGGTGGTGAAGAC-3′), CDCA7 (F: 5′-CTTGTCATCAATGCCGTCAG-3′; R: 5′-CAGTTGCAGATTCCTCGACA-3′), LETM2 (F: 5′-GCCCTGGAACACTTAGATCG-3′; R: 5′-TGTTGTCGCAGTTGTTCCTC-3′), FGFR1 (F: 5′-GGCAGCATCAACCACACATA-3′; R: 5′-TCGATGTGCTTTAGCCACTG-3′), and WHSC1L1 (F: 5′-TCGAGAAGAGGCACTGGAAT-3′; R: 5′-GGTGCTGCCCAGTTTTACAT-3′) were used. The detailed protocol was as follows: 95°C for 10 min, 40 cycles of 95°C for 15 s, and 60°C for 1 min, followed by a melting-curve program from 59°C to 95°C with a heating rate of 0.3°C every step and continuous-florescence acquisition. All RT-PCR reactions were completed in triplicate. The relative expression quantification of interesting genes was determined as F = 2−ΔΔCt.
Immunohistochemistry
CDCA7 or LETM2 protein levels in ESCCs were determined by immunohistochemistry with CDCA7 antibody (HPA005565, Sigma) or LETM2 antibody (17180-1-AP, Proteintech). Immunohistochemistry was performed as previously described.22 In brief, sections were incubated with the specific antibody at a 1:40 dilution for 14 hr at 4°C, followed by detection using the PV8000 (Zhongshan) and DAB detection kit (Maixin), producing a dark brown precipitate. Slides were counterstained with hematoxylin. All images were captured at ×100. The cytoplasm H score and the levels of CDCA7 or LETM2 shown by immunohistochemistry were analyzed with Aperio Cytoplasma 2.0 software. Statistic analyses were performed with GraphPad Prism v.6.0 software package. The significance of differences between ESCC and matched normal tissue was determined by paired t test.
Stable CDCA7 Knockdown Clones in ECA109 Cell Line
Vector pLVshRNA-puro was obtained from Addgene and used for CDCA7 knockdown. Two independent shRNAs targeting CDCA7 (5′-CCGGCCGTGACCCTTCCGCATATAACTCGAGTTATATGCGGAAGGGTCACGGTTTTTTG-3′; 5′-CCGGGAGCATCACAGAAGGTATATTCTCGAGAATATACCTTCTGTGATGCTCTTTTTTG-3′) were cloned into pLVshRNA-puro vector (pLV-shRNA1 and pLV-shRNA2). To perform plasmid infections, the ECA109 cells were plated at 40%–50% confluence and incubated at 37°C overnight (16 hr). pLVshRNA-puro vector, pLV-shRNA1, and pLV-shRNA2 were transfected into ECA109 cells using Lipofectamine 2000 reagent (Life Technologies) according to the manufacturer’s instructions. Forty-eight hours after transfection, culture medium was replaced by fresh media containing 2 μg/ml puromycin and subjected to screening stable monoclonies for 3 weeks. During the selection, cells were maintained at culture medium containing 2 μg/ml puromycin. After 3 weeks of selection, approximately 20 monoclonies per dish were selected and transferred into 96-well plate. shRNA knockdown efficiency was determined by RT-PCR and western blotting as described.22
Apoptosis Analysis by Flow Cytrometry
CDCA7 knockdown cells and cells transfected with pLVshRNA-puro vector were labeled with Annexin-FITC/PI Staining Kit (Sangon Biotech) according to the manufacturer’s instruction and analyzed by flow cytometry in BD FACScaliber (BD Bioscience).
RNA Sequencing and Data Analysis
Total RNA was extracted with the RNeasy Mini Kit (QIAGEN) and complementary DNA (cDNA) libraries were synthesized with the TruSeq RNA Sample Preparation Kit v.2 (Illumina). Libraries were sequenced on an Illumina HiSEquation 4000 platform at BGI. Filtering and quality controls were applied according to the standard procedure. The gene expression profiles of CDCA7 knockdown cells versus control cells were compared via gene set enrichment analysis. Differential expression levels (relative RNA counts) between control cells and CDCA7 knockdown cells were considered significantly different with a false discovery rate (FDR) at a threshold of 1%.
Knockdown of LETM2, FGFR1, or WHSC1L1 in ESCC Cell Lines
Three siRNAs targeting LETM2 and one negative control siRNA (NC) (Guangzhou RiboBio) were used to knock down LETM2 in ESCC cell lines (KYSE150 and ECA109). Meanwhile, FGFR1 (siRNA #1: 5′-AGTGGCTTATTAATTCCGATA-3′; siRNA #2: 5′-GCTTGCCAATGGCGGACTCAA-3′; siRNA #3: 5′-GAATGAGTACGGCAGCATCAA-3′) or WHSC1L1 (siRNA #1: 5′-CGAGAGTATAAAGGTCATAAA-3′; siRNA #2: 5′-CCATCATCAATCAGTGTGTAT-3′; siRNA #3: 5′-GCTTCCATTACGATGCACAAA-3′) were knocked down in TE-1 and KYSE150 cells, respectively. To perform infections, the ESCC cells were plated at 40%–50% confluence and incubated at 37°C overnight (16 hr). Cells were transfected with 100 nM (final concentration) siRNA or NC siRNA using Lipofectamine 2000 (Life Technologies) according to manufacturer’s protocols. At 48 hr after transfection, cells were subjected to MTT assay. At 72 hr after transfection, the knockdown efficiency was determined by RT-PCR and western blotting as previously described.22
MTT Assay
5 × 103 cells were seeded in 48-well plates and incubated at normal condition for 24, 48, 72, 96, and 120 hr. Cells were treated with 30 μl of 5 mg/ml of MTT (Invitrogen) solution for 4 hr at 37°C until crystals were formed. MTT solution was removed from each well and 200 μl of DMSO was added to each well to dissolve the crystals. Color intensity was measured by Microplate Reader (Bio-Rad) at 490 nm. Each experiment consisted of five replications and at least three independent experiments were carried out.
Colony Formation Assay
The assay was performed as described previously.22 In brief, cells were seeded at 300–500 cells per well in 6-well plates containing complete DMEM/F12 on day 0 and incubated at 37°C and 5% CO2 for 10 days. On day 10, cells were fixed with 4% polyformaldehyde for 15 min and stained with 1% crystal violet before quantification. The experiments were triplicate and the numbers of colonies containing more than 50 cells were microscopically counted.
Migration and Invasion Assays
Migration and invasion assays were performed in 16-well CIM plates in an xCELLigence RTCA DP system (ACEA Biosciences) using matrigel basement membrane matrix (BD) for real-time cell migration analysis as described previously.22 In brief, 30,000 cells per well were seeded as 5 duplicates in serum-free medium at the upper compartment of the CIM plates coated with or without matrigel. Serum-complemented medium was added to the lower compartment of the chamber, and then we started measurement in xCELLigence RTCA DP system and analyzed the CI (cell index) curves to determine cell invasion activity. For negative controls, we added serum-free medium at both upper and bottom chambers. The cell index representing the amount of migrated cells was calculated with the RTCA Software from ACEA Biosciences. At least three independent experiments were carried out; for each independent experiment, five duplicates were performed for each group.
Immunoblotting
Cells were lysed for 30 min in Triton buffer (1% Triton X-100, 50 mM Tris-HCl [pH 7.6], 150 mM NaCl, 1% sodium deoxycholate, 0.1% SDS) supplemented with protease and phosphatase inhibitors (1 mM PMSF, 2 mM sodium pyrophosphate, 2 mM sodium betaglycerophosphate, 1 mM sodium fluoride, 1 mM sodium orthovanadate, 10 μg/ml leupeptin, and 10 μg/ml aprotinin). Lysates were cleared by centrifugation at 15,000 × g at 4°C for 15 min, and protein concentrations were determined via the Bradford method. 50 μg of protein were separated by SDS-polyacrylamide gel electrophoresis and transferred onto Immobilon-P membranes. Proteins were detected by using anti-LETM2 (Proteintech, 17180-1-AP), anti-FGFR1 (Abcam cat# ab76464; RRID: AB_1523613), anti-WHSC1L1 (Abcam, ab180500), anti-CDCA7 (Abcam cat# ab69609; RRID: AB_1268064), anti-ERK1/2 (Santa Cruz, sc-514302), anti-p-ERK1/2 (Cell Signaling Technology cat# 4376; RRID: AB_331772), anti-AKT1 (Cell Signaling Technology cat# 2967; RRID: AB_331160), and anti-p-AKT1 (Cell Signaling Technology cat# 9018). Antibody binding was detected using horseradish peroxidase-labeled anti-mouse (Sigma) or anti-rabbit (Cell Signaling) antibodies and chemiluminescence was detected with a LAS4000 device (Fuji). Equal protein loading was confirmed with antibodies against GAPDH (Transgen).
Results
Spectrum and Distribution of Somatic SVs across 31 ESCCs
To characterize the mutational spectrum of somatic SVs in ESCC, we applied Meerkat to WGS data of tumors and paired normal tissues from 31 ESCC-affected individuals (Table S1). A total of 5,204 SVs were identified from the 31 ESCC genomes with an average of 168 SVs per tumor, ranging from 10 to 364 (Table S2). Five categories of SVs were observed, including deletions, tandem duplications (TDs), inversions, insertions, and intra- or inter-chromosomal translocations. Among these SVs, the average number of deletions per genome was 58 (ranging from 2 to 191) and make up 35% of SV types. Additionally, about 42% of SVs referred to intra- or inter-chromosomal translocations, with an average of 71 per genome (ranging from 3 to 150). For deletions and intra- or inter-chromosomal translocations, NHEJ and alt-EJ were the dominant mechanisms, with alt-EJ being more abundant in most cases. Moreover, 291 deletions were identified as complex deletions generated by FoSTeS/MMBIR. We noticed that the number of complex deletions were extremely diverse among individuals; some genomes contained a high portion of complex deletions whereas others showed very few (Figure 1A, middle). Besides deletions and translocations, the number of TDs for each genome was remarkably variable, with a range of 5 to 104. We observed no homology at TDs within ESCC genomes, further supporting the underlying mechanism that requires no microhomology or existence of nonhomology-based mechanism to form TDs and complex deletions in tumor cells.24
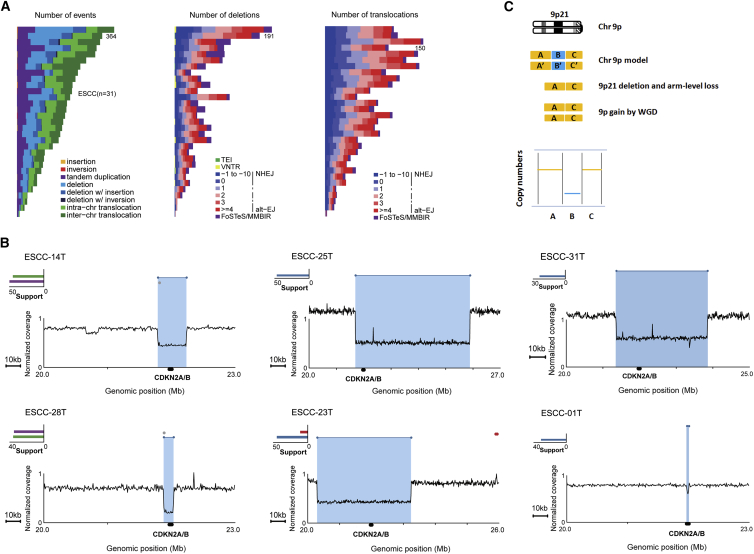
Figure 1.
Spectrum of Somatic SVs across ESCC Genomes and Mutational Mechanisms on CDKN2A in ESCC
(A) Frequencies of SVs (left), deletion (middle), or translocation (right) events and underlying mechanism across 31 ESCC tumors.
(B) Different mutational mechanisms of SVs act on CDKN2A. Representative maps show CDKN2A loss in six ESCCs. A red cluster typically suggests a tandem duplication; a blue cluster typically suggests a deletion; a purple cluster suggests a invers_reverse, and a green cluster suggests a invers_forward.
(C) Model of focal deletion of CDKN2A that occurred before WGD on chromosome 9p in eight ESCC genomes.
Across 31 ESCC genomes, we found that 3,376 SVs occurred in the region of genes and were predicted to directly disrupt sequence of gene such as CDKN2A (MIM: 600160), NOTCH1 (MIM: 190198), NF1 (MIM: 613113), and FANCD2 (MIM: 613984), and 492 genes contained a breakpoint in two or more tumors. Specifically, 29 out of 31 ESCCs harbored CDKN2A deletion; of which, 13 ESCCs had supporting SVs responsible for CDKN2A deletion and 2 out of these 13 genomes demonstrated complex deletions (ESCC-14T and ESCC-28T) (Figures 1B and S1). Notably, all deletions from tumor genomes of these 13 ESCCs were homozygous deletion with both focal deletion and arm-level loss. Furthermore, 8 out of these 13 ESCC genomes had arm-level gain of 9p generated by whole-genome duplication (WGD) (Table S3), and no one had two independent SVs within CDKN2A locus (9p21), suggesting that the focal deletion of CDKN2A happened before WGD in these tumors (Figure 1C). In addition, we also found that NOTCH1 was directly disrupted by TDs in two ESCCs (Figure S2). These results suggested that different mutational mechanisms can act on the same driver (e.g., CDKN2A), and different drivers (e.g., CDKN2A and NOTCH1) might be affected by different mutational mechanisms in ESCC.
SVs tended to be either scattered genome-wide or occurred locally with variable copy numbers across cancer genomes and are more likely to occur in genomic region of fragile sites.30, 31, 32 Across 31 ESCCs, the genomic distribution of SVs was characterized with three features: randomly distributed across chromosomes; clustered in one or more chromosomes; and clustered chromosomes involving SVs accompanied with variable or limited copy numbers. Notably, we observed locally rearranged chromosomes were prevalent in ESCC genomes (17 out of 31 ESCCs) (Table S3). Although the mechanism underlying most of locally rearranged chromosomes remains unknown, it appears that ESCC genomes harboring locally rearranged SVs accompanied with limited copy-number states could be explained as chromothripsis or kataegis (Figure S3). Meanwhile, 21 out of 31 ESCC genomes displayed at least two fold-back inversions in an autosome accompanied with substantial copy-number states, and some of them were likely to be a result of BFB (Table S3).
When analyzing SVs across ESCC genomes, we note that, probably due to the tumor cell purity and ploidy, many of the detected SVs have a smaller number of supporting split reads (Table S2). Additionally, due to a large number of events that were relatively small, we did observe that both breakpoints were in the same gene (Table S4). We further compared the distribution of somatic SVs across a variety of human cancers including breast cancer (BRCA), glioblastoma multiforme (GBM), lung squamous cell carcinoma (LUSC), ovarian serous cystadenocarcinoma (OV), and gastric cancer (GC).23, 33 Consistent with our observation in ESCC, those somatic SVs that had a smaller number of supporting split reads and a high fraction of smaller SVs were also observed in other human cancers (Figures S4A and S4B). Advanced methodology needs to be designed to solve these limitations.
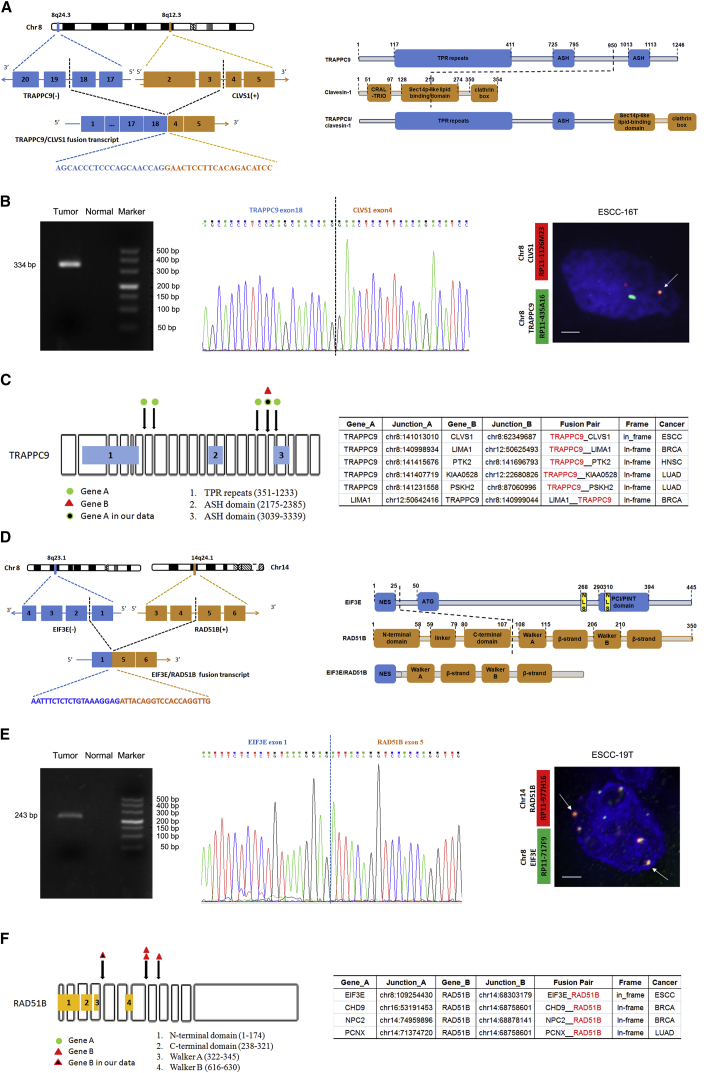
Chromothripsis Leading to High-Level Amplification of FGFR1 and LETM2
It is well known that chromosomes affected by chromothripsis show a characteristic pattern with more than ten transitions oscillating between two and three copy number states on chromosomal arms.7, 15 We further accurately infer the occurrence of chromothripsis by using conceptual criteria proposed by Korbel and Campbell.15 Interestingly, we observed chromothripsis involving chromosome 8 in ESCC-16T (Figure 2A). In addition to general transition between two copy number states, we found a high-level focal amplification (<500 kb, 38,155,351–38,570,827 Mb) rearranged by chromothripsis on chromosome 8p that corresponds to FGFR1 and LETM2 in this tumor. Importantly, no breakpoints were observed within this amplified region, suggesting a strong positive selection of FGFR1 and LETM2 amplifications during ESCC progression/evolution. It was previously shown that a potential by-product of chromothripsis is formation of double-minute chromosomes (DMs) that might harbor oncogenes and have been found in a variety of solid tumors.7, 15 In ESCC-16T, our FISH experiment exhibited multiple scattered FGFR1 signals and two copies of chromosome 8, suggesting that FGFR1 amplification might be due to the formation of DMs (Figure 2A). Moreover, in a second tumor (ESCC-06T), evidence of high-level amplification of this locus harboring FGFR1 was also identified and similarly verified via FISH that showed clustered multiple FGFR1 signals around the centromere of chromosome 8 (Figure 2B), indicating high-level amplification of FGFR1 in ESCC. DMs responsible for FGFR1 amplification were not observed previously in ESCC. Combined with a previous report that FGFR1 was overexpressed in ESCC,20 these findings indicate a oncogenic role of FGFR1 in ESCC. Further functional studies indicated that knockdown of FGFR1 dramatically suppressed cell proliferation, cell migration, and invasion in TE-1 and KYSE150 cells (Figures S5A–S5C). A recent study has demonstrated that focal amplification of the FGFR1 locus on chromosome 8p was associated with cellular dependency on FGFR1 and sensitivity to FGFR inhibitors.34 Consistent with this, a pan-FGFR tyrosine kinase inhibitor has been shown to block tumor proliferation in a subset of NSCLC cell lines with activated FGFR signaling but has no effect on cells that do not activate the pathway.35 Collectively, our results suggest that FGFR1 might be an attractive therapeutic target for ESCC.
Figure 2.
High-Level Amplification of FGFR1 and LETM2 Affected by SVs
(A) The top panel represents different types of SVs indicated by lines with different colors on chromosome 8 in ESCC-16T; the middle panel shows normalized coverage for each window. Zoom-in view of high-level amplification of FGFR1 and LETM2 locus is shown in the bottom panel. FISH analysis demonstrates DM-derived amplification of FGFR1. Scale bar represents 10 μm.
(B) High-level amplification of FGFR1 in ESCC-06T. Top: FGFR1 locus and the high-level amplification region containing MYC gene are shown. Bottom: Zoom-in view of high-level amplification of FGFR1 locus. FISH confirms CNAs by showing FGFR1 amplification as clustered multiple green signals. Scale bar represents 10 μm.
(C) FISH confirms CNAs by showing LETM2 amplification as scattered multiple green signals. Scale bar represents 10 μm.
(D) Immunohistochemical analysis shows LETM2 staining in ESCCs.
(E) LETM2 expression level in multiple ESCC lines determined by RT-PCR and western blotting.
(F) LETM2 knockdown prevents cell proliferation but has no effect on cell migration/invasion as monitored by MTT or in vitro cell migration and invasion assays in KYSE150 and ECA109 cells. Knockdown of LETM2 is demonstrated by immunoblotting; GAPDH was used as loading control.
Data are mean ± SD; each experiment was performed in triplicate. ∗∗p < 0.01.
Additionally, the small circular DNA molecule identified in chromosome 8p of ESCC-16T contains LETM2. FISH analysis further confirmed that LETM2 amplification was due to extra-chromosomal amplification (Figure 2C). Immunohistochemical analysis indicates that LETM2 was upregulated in ESCC tumors and some ESCC cell lines (Figures 2D, 2E, and S6). LETM2 is a mitochondrial gene that is expressed preferentially in spermatocyte to spermatozoon.34 It has been found amplified in breast cancer, lung adenocarcinomas, and squamous cell lung carcinoma.34 However, the function of LETM2 has not been studied in detail. Our result showed that LETM2 knockdown prevented cell proliferation but had no statistical suppression of cell migration and invasion in KYSE150 and ECA109 cells (Figure 2F). Similar trends were observed for WHSC1L1, another potential oncogene located in the 8p12 amplicon (Figures S5D–S5F). Together with genetics observations, these functional analyses strongly implicate these genes as amplification targets in ESCC.
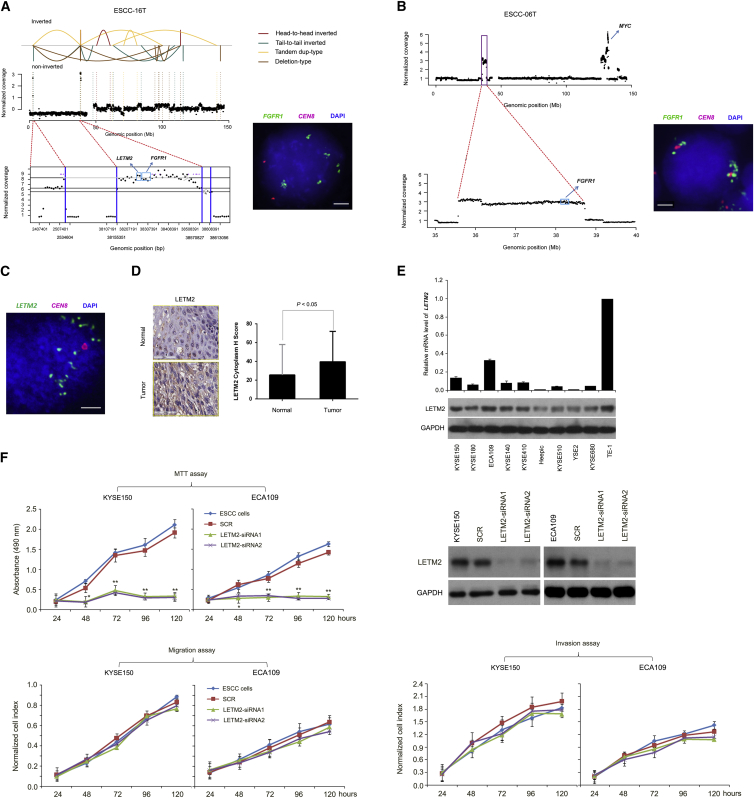
Fusion Genes Caused by Chromosomal Rearrangements
Currently, little is known about the targetable fusion genes underlying ESCC. We therefore screened gene fusion events across 31 ESCC genomes and identified a total of 173 in-frame fusion genes and 231 out-frame fusion genes affected by SVs (Table S4). Notably, in ESCC-16T, the chromothripsis-associated rearrangements led to the formation of putative in-frame fusions involving genes TRAPPC9 at 8q24.3 and CLVS1 at 8q12. This fusion variant was predicted to result in an in-frame fusion of the TRAPPC9 5′ UTR and exon 1–18 with the CLVS1 exon 4–5 and 3′ UTR (Figure 3A). Using primers within exon 18 of TRAPP9 and exon 4 of CLVS1, we confirmed the fusion transcript in purified tumor cells from ESCC-16T (Figure 3B, left and middle). FISH analysis using CLVS1 red probe and TRAPPC9 green probe shows a yellow fusion signal indicative of translocation of TRAPPC9-CLVS1 (Figure 3B, right). In this tumor genome, TRAPPC9 and CLVS1 are adjacent genes on chromosome 8q that are transcribed in opposite directions. TRAPPC9 (trafficking protein particle complex 9) is a 23-exon gene that encodes NIK- and IKK-β-binding protein (NIBP), which activates NF-κB signaling via directly interacting with and activating IKK-β and MAP3K14 kinase.36 TRAPPC9 has been reported correlated with colorectal tumorigenesis and tumor growth and was implicated to be important for lapatinib response in a subgroup of ERBB2-amplified breast cancer.37 CLSV1, also known as CRALBPL, was implicated to be upregulated in hepatocellular carcinoma (HCC) and might be a marker for HCC.38 The function of this fusion transcript in ESCC need to be elucidated in future study.
Figure 3.
Fusion Genes Caused by Chromosomal Rearrangements in ESCC
(A) Details and schematic of the TRAPPC9-CLVS1 fusion transcript caused by complex deletion on chromosome 8 in ESCC-16T.
(B) Validation of the TRAPPC9-CLVS1 fusion transcript via PCR-sanger sequencing (left and middle) and FISH (right).
(C) Left: Junction points of TRAPPC9 in-frame fusions were shown in the transcripts of TRAPPC9 using the bottom symbols. Right: The diversity partners of TRAPPC9 across different types of human cancers.
(D) Details and schematic of the EIF3E-RAD51B fusion transcript caused by interchromosomal translocation between chromosomes 8 and 14 in ESCC-19T.
(E) Validation of the EIF3E-RAD51B fusion transcript via PCR-sanger sequencing (left and middle) and FISH (right).
(F) Left: Junction points of RAD51B in-frame fusions were shown in the transcripts of RAD51B using the bottom symbols. Right: The diversity partners of RAD51B across different types of human cancers.
Another notable inter-chromosome in-frame gene fusion of EIF3E-RAD51B was detected in ESCC-19T. The first exon of EIF3E on chromosome 8, encoding the eukaryotic translation initiation factor 3 subunit, was predicted to join with the last two exons of RAD51B on chromosome 14, a protein that catalyzes repair of DSBs through the process of homologous recombination and are critical for genome stability (Figure 3D). The EIF3E-RAD51B translocation was validated in this tumor by independent PCR sequencing and interphase FISH analyses (Figure 3E). Tumor suppressor or oncogenic effect of EIF3E either through its role as a component of EIF3 translation initiation factor or translation-unrelated function has been reported in various types of human cancer.39 RAD51B, one member of the human RAD51 (MIM: 179617) paralogs, plays a central role in homologous DNA recombination.40 Increased RAD51B protein level has been reported in various cancers, especially gynecological tumors, and linked to uncontrolled recombination, genome instability, tumor recurrence and progression, and increased resistance of tumors to radiotherapy and chemotherapy.40 Interestingly, translocation of RAD51B with other genes has been reported, for example, HMGA2-RAD51B in uterine leiomyoma.41 However, to the best of our knowledge, the EIF3E-RAD51B translocation has not been previously reported in human cancer. Since the N- and C-terminal domains of RAD51B were important to interact with other proteins to catalyze the repair of DNA double-strand breaks, we speculate that the in-frame fusions of EIF3E-RAD51B might cause disruption of EIF3E and RAD51B function, which could result in deregulated homologous recombination or translation initiation, contributing to the tumorigenesis of ESCC.
Recently, Yoshihara et al. analyzed RNA sequencing and DNA copy-number data from 4,366 primary tumor samples and 364 normal samples spanning 13 tumor types.42 To further assess the recurrence of fusion genes identified in ESCCs, we compared our data with the resource of fusion transcripts from Yoshihara’s report.42 We did not find in ESCC recurrent in-frame protein kinase fusions such as FGFR1-TACC3 that was implicated in bladder urothelial carcinoma (BLCA), GBM, HNSCC, low-grade glioma (LGG), and LUSC.42 We then focused on fusions with the same gene fused to multiple different partners. Interestingly, we observed that some in-frame rearrangements were not limited to ESCC but can be detected across cancer at low frequency. For example, TRAPPC9 is paralogous to many oncogenes such as LIMA1 (MIM: 608364), PTK2 (MIM: 600758), PSKH2, and others in BRCA, HNSCC, and lung adenocarcinoma (LUAD) (Figure 3C). RAD51B is a known oncogene and was found to form fusions with various partners (e.g., CHD9, NPC2 [MIM: 601015], PCNX [MIM: 613401]) in BRCA and LUAD (Figure 3F). Moreover, the 3′ partners of TRAPPC9 or EIF3E (e.g., CLVS1, RAD51B) have been reported to be upregulated in human cancers,38, 41 indicating the potential of these fusions to drive carcinogenesis.
Kataegis in ESCC
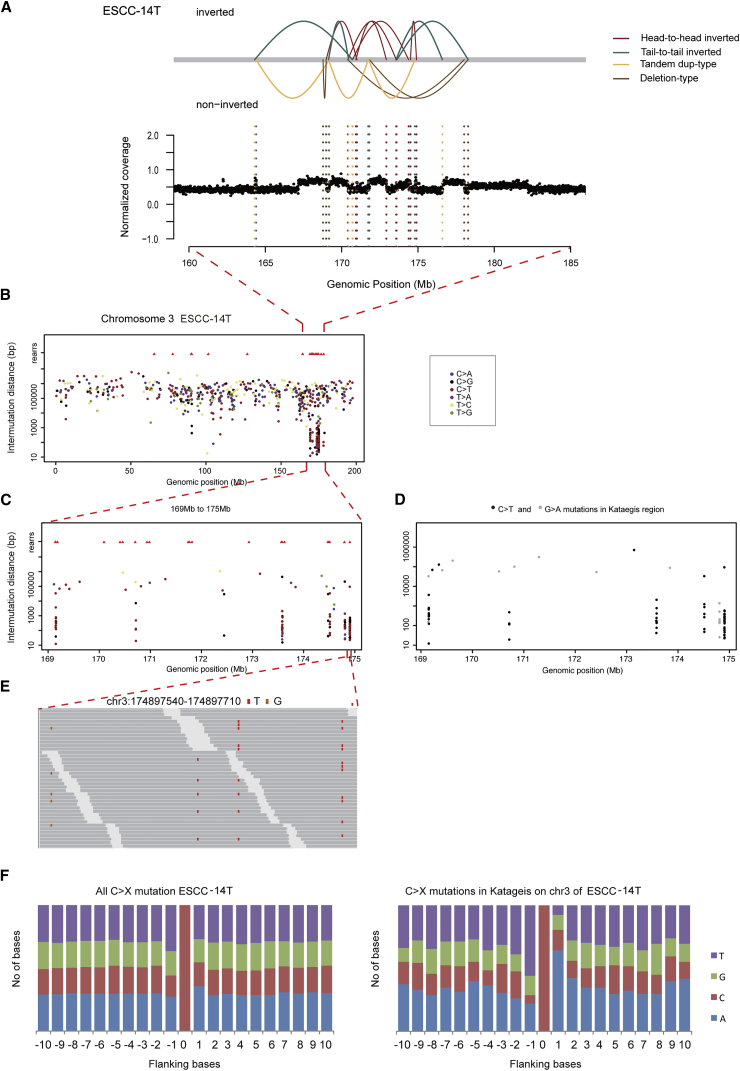
Besides chromothripsis, kataegis also contributes to locally rearranged SVs accompanied with limited copy-number states. Nik-Zainal et al. analyzed the mutational signatures of 21 breast cancers and identified kataegis, a distinct hypermutation phenomenon, in 61% of breast cancers, indicating a direct relevance to tumor initiation and progression.16 To date, there is no implication of kataegis and associated SVs in ESCC. Interestingly, we found locally rearranged variations concentrated in chromosome 3 of ESCC-14T and somatic mutations clustered in the region of 16.9 Mb to 17.5 Mb (Figure 4). Although kataegis was observed in one tumor, perhaps due to the limited sample size, the prevalence of kataegis in other cancer types16, 43 indicates a potential tumorigenic mechanism of kataegis in ESCC development.
Figure 4.
Kataegis on Chromosome 3 in ESCC-14T
(A) SVs observed on chromosome 3 in ESCC-14T. The upper panel represents different types of SVs indicated by lines with different colors; the bottom panel shows normalized coverage for each window.
(B) Each dot of the “rainfall” plots represents a single somatic mutation ordered on the horizontal axis according to its position in human genome. The vertical axis denotes the genomic distance between mutations. The upper red triangles show the position of SV breakpoints.
(C) Highlight of kataegis region (chr3: 169–175 Mb) at increasing resolution to demonstrate microclusters within the macrocluster.
(D) Alternating processivity of kataegis in ESCC-14T. Long regions of C>T mutations are interspersed with regions of G>A mutations.
(E) The processive nature of C>T mutations (IGV image) within region (chr3: 174,897,540–174,897,710 Mb).
(F) Plots of flanking sequence of all C>X mutations and C>X mutations within the regions of kataegis in ESCC-14T.
Breakage-Fusion-Bridge Drives Gene Amplification in ESCC Tumors
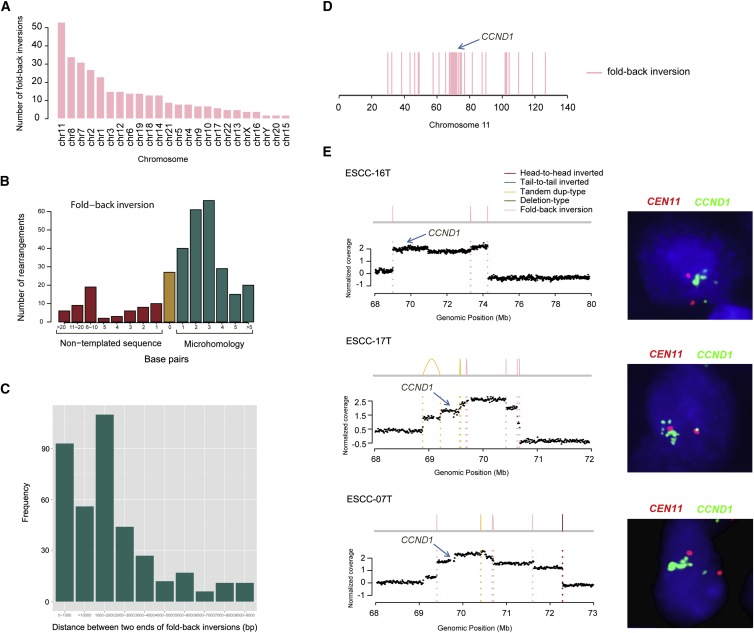
Previous studies from cancer genomes support a BFB event, which is known to begin with telomere loss and is characterized with a class of breakpoints called fold-back inversion.27 Therefore, we used fold-back inversion and telomere loss to infer BFB events for each genome. In total, we obtained 321 fold-back inversions (Table S5A), of which chromosomes 11, 8, and 7 had the most fold-back inversions across 31 ESCC tumors (Figure 5A). Moreover, most of fold-back inversions were mediated by microhomology (Figures 5B and 5C), indicating that homology-mediated fold-back capping of broken ends followed by DNA replication is an underlying mechanism of sister chromatid fusion during BFB cycles in ESCC. In ESCC-11T, five chromosomes (chromosomes 5, 7, 8, 11, and 17) were affected by BFB events (Table S5A). Hence, our large-scale breakpoint analysis of 31 ESCCs exhibited an important role of BFB in tumorigenesis of ESCC.
Figure 5.
BFB Evidence across 31 ESCCs
(A) The number of fold-back inversions across 31 ESCCs.
(B) Sequence length in the breakpoints of fold-back inversion. Patterns of microhomology, non-template sequence, or direct end-joining in the fold-back inversion across 31 ESCCs are shown.
(C) Genomic patterns of fold-back inversions. Histogram showing the distance between the two inverted ends in the set of fold-back inversions.
(D) Distribution of the fold-back inversions on chromosome 11 across 31 ESCCs. Blue arrow indicates a minor cluster around CCND1 locus (11q11.3).
(E) Representative maps show amplification of CCND1 as a result of BFB in three tumors. FISH validation shown on right panel.
Notably, fold-back inversions on chromosome 11 enriched in a minor cluster around CCND1 locus (69,455,873–69,469,242 Mb) at 11q11.3 (Figure 5D). In addition, we observed that 32 chromosomes involving 21 ESCCs displayed at least two inversions and a telomere loss (Table S5B). Of these 21 ESCCs, 10 showed evidence of BFB on chromosome 11, and 9 of them led to a focal amplification of CCND1 showing unbalanced amplified signals (Figures 5E and S7), indicating that the CCND1 amplification was created by BFB cycles in ESCC. Together with the cluster of palindromic junctions, the physical location of the amplicon suggests the BFB cycles as the underlying processes. Additionally, we also found inter-chromosomal SVs enriched in CCND1 locus on chromosome 11 (Figure S8). Amplification of CCND1 has been reported in a variety of tumors and might contribute to tumorigenesis.44 Specifically, CCND1 amplification and overexpression was observed and significantly correlated with lymph node metastasis in ESCC.45 However, the underlying mechanism of CCND1 amplification has not been elucidated. Our results demonstrated that at least two mutational mechanisms, focal amplification via BFB cycles and inter-chromosomal translocations, result in CCND1 amplification in ESCC.
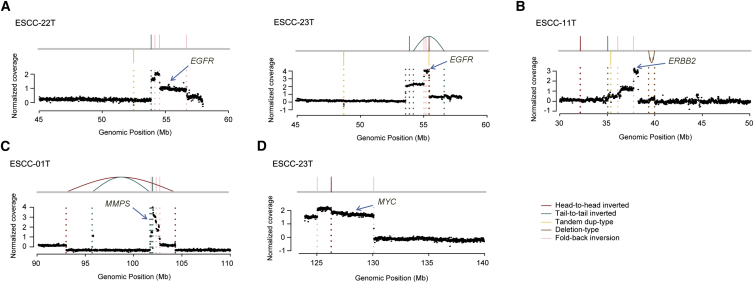
Additionally, we observed that regions amplified by BFB cycles harbor oncogenes such as EGFR (MIM: 131550) (2/31), ERBB2 (MIM: 164870) (1/31), MMPs (1/31), and MYC (MIM: 190080) (1/31) (Figure 6), suggesting that BFB plays an important role in gene amplification in ESCC tumors. In the literature, MYC loci comprise the most significant regions of amplification observed in ESCCs and have been implicated as a reasonable indicator of the accumulation of various activated and inactivated genes involved in carcinogenesis of ESCCs, suggesting deregulation of MYC as a driver event.46 EGFR is an established therapeutic target that is often overexpressed as a consequence of gene amplification in human cancers including ESCCs.47 ERBB2 amplification was observed in breast, esophageal, and other types of cancer and has been a target of anticancer agents.48 MMPs amplification was reported in some human cancers but not ESCC.48 It has been found that regions of DNA gain in cancer rarely coincide with regions of loss and vice versa, suggesting a specialized function for regions characterized by either gain or loss in cancer.49 Therefore, understanding the mechanisms that drive SVs and the gene changes that result from them has significance. WGSs revealed that amplification of MYC, EGFR, and ERBB2 might derive through TDs or chromothripsis-derived DMs.1, 15, 25 However, our data support that BFB events that occurred in ESCCs led to amplification of these genes, which was not proposed previously in ESCC. Therapies targeting these amplified oncogenes would be more practical in ESCC.
Figure 6.
BFB-Derived Gene Amplifications in ESCC
Amplification of other oncogenes caused by BFB events including EGFR (A), ERBB2 (B), MMPs (C), and MYC (D).
Copy-Number Alterations
To investigate copy-number change across ESCC genomes and potential affected genes, we applied Patchwork to determine CNAs based on WGS data of 31 ESCCs and found 19 ESCCs that could be used to determine absolute copy number. Consistently, frequent arm-level changes were observed in ESCC, including frequent copy-number gains of 3q, 5p, 7p, 8q, 12p, 17p, 20p, and 20q and universal deletions affecting 3p, 4p, 4q, 5q, 10p, 13q, and 21q (Figure S9A). Moreover, 19 ESCCs harbored fewer events of copy-number loss than copy-number gain; meanwhile, 70% of loss of heterozygosity (LOH) was copy neutral loss of heterozygosity (CN-LOH) in ESCC. Specifically, we observed frequent CN-LOH on 13q and 17p (Figure S9B). In addition, we found that WGD occurred in 13 out of 19 ESCC genomes. Despite evidence that WGD can result in genetic instability and accelerate oncogenesis,50 the incidence and timing of such events had not been broadly characterized in ESCC. Our results indicate that ESCC tumors have alterations affecting the entire length of chromosome 13q and 17p such as, perhaps, whole chromosome deletion with duplication.
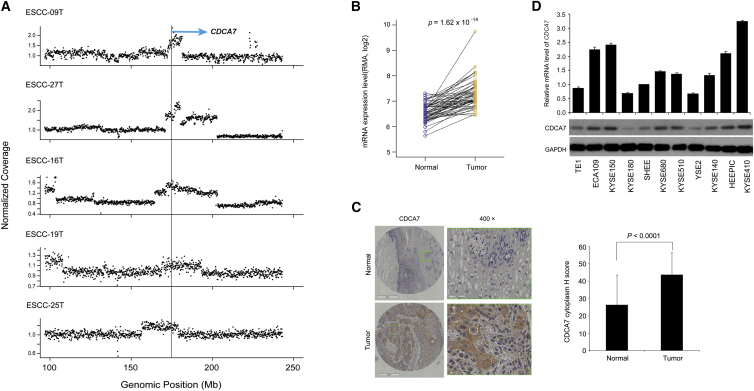
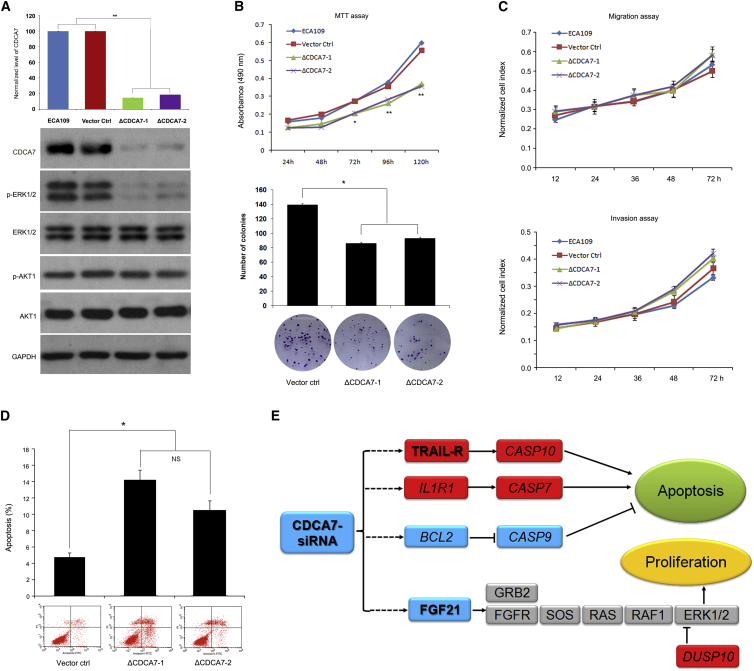
Furthermore, to obtain CNA targets, we applied GISTIC to copy-number profiling from a combination of 31 WGS and 123 CGH data.19, 22 This analysis yielded 11 amplification peaks and 13 deletion peaks, including cancer genes EGFR, CDK6 (MIM: 603368), AKT1 (MIM: 164730), MYC, CCND1, CDKN2A, and others (Table S6). Specifically, we identified a focal amplified region corresponding to CDCA7 in 5 out of 31 ESCC genomes with 2 having high-level copy number (>6 copies; Figure 7A). Moreover, we observed that most of individuals with ESCC tumors showed statistically higher expression level of CDCA7 compared with that of normal tissues as determined by real-time PCR (Figure 7B) and immunohistochemistry analyses (Figures 7C and S10). CDCA7 is a downstream target of MYC and E2F transcription factors and participates in cell cycle progression as a transcriptional regulator of the expression of myriad of target genes.51 Previous transformation studies with cell lines in vitro, analysis of CDCA7 levels in human cancers, and in vivo tumorigenic studies in transgenic mice all support a role for CDCA7 in tumorigenesis.51 However, it has limited implication in ESCC; the mechanism of how CDCA7 is involved in tumorigenesis remains largely unknown. Our result showed that CDCA7 knockdown significantly inhibited cell growth and promoted cell apoptosis but had no differential effect on cell migration and invasion in ESCC cells (Figures 8A–8D), indicating that CDCA7 might involve cell proliferation and apoptosis but not metastasis in ESCC. Moreover, CDCA7 knockdown led to the decrease of phospho-ERK1/2, an essential downstream component of MAPK pathway regulating cell proliferation, whereas no significant effect was seen in AKT pathway (Figure 8A). To further determine the potential targets of CDCA7 in ESCC, we performed RNA-seq of CDCA7 knockdown cells and cells transfected with pLVshRNA-puro vector (used as controls). Indeed, we observed a positive and highly significant enrichment of the expression of cell proliferation or apoptosis-associated target genes, including FGF21 ([MIM: 609436] a MAPK pathway-related gene) and cell-apoptosis-associated genes TRAIL-R, CASP10 (MIM: 601762), IL1R1 (MIM: 147810), CASP7 (MIM: 601761), BCL2 (MIM: 151430), and CASP9 (MIM: 602234). These genes all had outlier expression levels in CDCA7 knockdown cells compared to that of controls (Figure 8E and Table S8). Specifically, a significant decrease of FGF21 in CDCA7 knockdown cells suggests that CDCA7 might regulate cell proliferation via FGF21-ERK1/2 MAPK pathway rather than other pathways in ESCC tumorigenesis (Figure 8E). In addition, CDCA7 knockdown led to the increased expression levels of TRAIL-R, CASP10, IL1R1, and CASP7 and the decreased expression levels of BCL2 and CASP9 (Figure 8E and Table S8), indicating that these genes might be critical for CDCA7 to regulate cell apoptosis. Together with genetic observations, these functional data indicate that CDCA7 might act as an oncogene possibly through deregulation of cell proliferation and apoptosis in ESCC.
Figure 7.
Amplification and Overexpression of CDCA7 in ESCC Tissues
(A) Focal recurrently amplification of CDCA7 shown in five ESCCs. The red line represents genomic position of CDCA7; the dot represents normalized coverage of genomic position (100 kb per window) along chromosome 2.
(B) mRNA expression level of CDCA7 examined from 52 matched normal/tumor ESCC tissues. p value is given by Student’s t test.
(C) Left: Represent images display strongly cytoplasm positivity in ESCC tissues. Right: Expression of CDCA7 was markedly increased in ESCC tissues compared to that of normal esophagus tissue based on judgment of IHC staining intensity.
(D) CDCA7 expression level in multiple ESCC lines determined by RT-PCR and western blotting.
Figure 8.
CDCA7 Deregulates Cell Proliferation and Apoptosis in ESCC
(A) Knockdown of CDCA7 was demonstrated by immunoblotting; GAPDH was used as loading control. Meanwhile, phospho-ERK1/2, ERK1/2, phospho-AKT1, and AKT1 were shown.
(B) Knockdown of CDCA7 significantly prevented cell growth in ECA109 as monitored by MTT (top) and colony formation assays (bottom). ECA109 and cells transfected with pLVshRNA-puro-SCR (scrambled sequence) were used as controls.
(C) Knockdown of CDCA7 shows no significant effect on cell migration and invasion.
(D) Knockdown of CDCA7 significantly promoted cell apoptosis. All data are mean ± SD; each experiment was performed in triplicate. ∗p < 0.05; ∗∗p < 0.01.
(E) Summary of potential CDCA7 target genes involved in cell apoptosis and proliferation in ESCC. The bold characters represent genes with log2 ratio more than 2. Red represents upregulated genes and blue represents downregulated genes.
Detailed information shown in Table S8.
Discussion
In this study, we report a comprehensive description of SVs that characterize ESCC and demonstrate the relative contributions and variability of different mutational mechanisms underlying SVs within ESCC genomes. We found that NHEJ and alt-EJ contribute the most to deletions and translocations. Our findings define a prevalence of locally arranged genomes across 31 ESCC genomes and some of them were delivered by chromothripsis, kataegis, or BFB events. A number of well-known cancer-associated genes (e.g., FGFR1, CDKN2A) and several unreported ESCC-related genes (e.g., LETM2, CDCA7, TRAPPC9-CLVS1, EIF3E-RAD51B) affected by these events were described here. Furthermore, our data provide the potential mechanisms for oncogene amplification or fusion gene formation, which might be critical for tumorigenesis of ESCC.
In studying SVs across ESCC genomes, we observed locally rearranged SVs with either limited (e.g., chromothripsis or kataegis) or substantial (e.g., BFB) copy-number states (Figure S3). In addition to the predominant BFB cycles that were accumulated in a step fashion,27 chromothripsis, a phenomenon in which one or a few chromosomes are shattered into pieces and randomly stitched together in a single event,15 was observed in one ESCC genome, which is consistent with its common rate of 1/40 cancer genomes.11 Moreover, kataegis, most likely co-occuring with large-scale rearrangement in a region of the genome at one time,15 was also observed in an ESCC genome. Combined with the fact that kataegis is remarkably common in human cancers,16, 43 we speculate that it is more likely to have biological significance. However, due to the limited sample size, we observed kataegis in only one ESCC genome. At some point in the future, larger numbers of ESCC genomes at higher resolution will be necessary to create a comprehensive catalog of the significant SVs and define the biological significance of these events in ESCC.
Besides copy-number alterations, translocations in chromothripsis led to gene fusions (e.g., TRAPPC9-CLVS1, EIF3E-RAD51B) (Figure S3). Chromothripsis, occurring as a relatively early tumorigenic event, is thought to represent a driving force of cancer development and progression.7, 15 For example, chromothripsis is implicated as a frequent driver event in uterine leiomyomas, resulting in increased expression of translocated HMGA1 and HMGA2.41 However, distinguishing driver mutations from passenger mutations is challenging. For SVs, recurrence is often used to estimate the likelihood of fusion being a driver; however, because most driver fusions have very low frequency, many studies have small sample sizes (as in the case here), and detection sensitivity might be low, it is hard to define the molecular characteristics of driver fusion.42 In ESCC genomes, we identified two in-frame fusions (TRAPPC9-CLVS1 and EIF3E-RAD51B) via RT-PCR Sanger sequencing and FISH. We did not find the same fusion genes (TRAPPC9-CLVS1 and EIF3E-RAD51B) in other human cancers. This phenomenon also happens in other human cancers. For example, none of predicted fusion events occurred in more than one sample in pancreatic cancer.26 Recent WGSs for structural mutations in cancers showed that most fusion transcripts were singletons unique to individual tumors and not detected in other samples.1 Alternately, we identified additional fusion partners for TRAPPC9 as well as RAD51B. Although these findings have not been validated by functional studies, they illustrate the potential of these fusions to drive carcinogenesis. Further in vitro and in vivo studies are needed to better understand the biological significance of these fusion transcripts in ESCC.
Additionally, BFB events were operative in approximately 68% of ESCCs, indicating that the BFB cycle is an important underlying process for genome instability and gene amplification in ESCC. The BFB events initiate amplification of cancer-associated genes and occur predominantly in early cancer development rather than later stages.25 End-to-end chromosome fusions are often seen in association with telomere erosion and it might be that the dsDNA break initiating BFB repair results from telomere loss. Hence, detecting telomere loss indicative of a BFB event might provide preventive implication for ESCC. However, although we identified BFB-derived amplification of cancer-associated genes, we could not identify candidate target genes from most BFBs because these amplified segments contain many more passenger events. Additionally, a BFB event was defined when a chromosome had at least two inversions and clearly telomere-boundary copy-number loss adjacent to the fold-back inversions.27 It is also possible that some chromosomes without clear telomere-boundary copy-number loss might suffer BFBs. Unfortunately, we could not identify these SV events via current strategies. Existing methods for the detection of SV events show high sensitivity and specificity but still have limitations. In the future, advanced methodology will enable the identification of these events.
We and others previously reported that the two types of esophageal cancers presented different mutational patterns and signatures at the level of SNVs. Specially, a higher frequency of C>G transversions occurred in ESCC than EAC whereas A>C transitions were more frequent in EAC than ESCC.22 A recent combined study of WGSs (22 EACs) and SNP arrays (101 EACs) reported genomic catastrophes that occurred in EAC.14 We then compared the SV events between these two types of esophageal cancers. Evidence of chromothripsis, BFB cycles, and kataegis were reported in both ESCC and EAC. Although TP53, which has been linked to chromothripsis in human cancers,52 was mutated at high frequency in both ESCC and EAC, we found that the frequency of chromothripsis tended to be lower in ESCC than in EAC. Moreover, we note that chromothripsis resulted in DM-derived FGFR1 amplification in ESCC but led to DM-derived MYC amplification in EAC. Otherwise, the high-level amplification of MYC is due to BFB cycles in ESCC, indicating that at least two different mechanisms are responsible for MYC amplification in tumors. Additionally, the genes affected by BFB cycles in these two types of esophageal cancers display differences. BFB cycles are scattered in three genes (KRAS [MIM: 190070], MDM2 [MIM: 164785], and RFC3 [MIM: 600405]) in EAC; in ESCC, they are mainly focused in CCND1 and also scattered in MYC, MMPs, EGFR, and ERBB2 (Figure S11). Unlike ESCCs, EACs arise in a highly genotoxic environment in which the distal esophagus is exposed to high levels of local and systemic injury from reflux of acid, bile, and other gastric contents.53 These findings would suggest that genomic catastrophes, gene activation through chromosomal rearrangements, and telomere integrity might be driving carcinogenesis in esophageal cancer, and the dominant SV type might be different between ESCC and EAC. Further understanding of these events might lead to novel strategies for detection and treatment of esophageal cancers.
Collectively, our findings demonstrated diverse models of SVs contributing to the mutational landscape, with BFB being the most extreme form across ESCC genomes. Besides somatic point mutations and CNAs reported in ESCC previously,19, 20, 21, 22 our findings highlight the oncogenic drives of ESCC through different types of SVs and suggest that complex genomic rearrangements, such as chromothripsis and BFB, are an integral part of mutation mechanisms contributing to ESCC development and should be considered along with simple genomic changes when applying genome-guided treatment strategies. Together with the landscape of point mutations or CNAs described previously, these findings provide a systems explanation for the maintenance of ESCC state. Additional larger panels of ESCC tissues will need to be studied to determine the broader applicability of these results. Currently, identifying SVs is still challenging and remains largely unsolved. Although much effort has focused on candidate genes affected by SVs, most SVs actually occur in non-coding regions.18, 30, 42 As ENCODE project explored potential functions of non-coding sequence,54 more advanced technology is required to characterize those SVs that occurred in non-coding regions and define their contribution in tumorigenesis of ESCC.
Acknowledgments
This work was supported by funding from the National Natural Science Foundation of China (81330063 and 81272189 to Y.C., 81230047 to Q.Z., 81272694 to X.C., 81201956 to J.L., 81402342 to L.Z., and 81502135 to P.K.), the Key Project of Chinese Ministry of Education (NO213005A to Y.C.), the Specialized Research Fund for the Doctoral Program of Higher Education (20121417110001 to Y.C.), a research project supported by Shanxi Scholarship Council of China (2013-053 and 2015-key3 to Y.C.), the Innovative Team in Science & Technology of Shanxi (2013-23 to Y.C.), the Program for the Outstanding Innovative Teams of Higher Learning Institutions of Shanxi (2015-313 to Y.C.), and the 973 National Fundamental Research Program of China (2015CB553904 to Q.Z.).
Published: January 28, 2016
Footnotes
This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/4.0/).
Supplemental Data include 11 figures and 8 tables and can be found with this article online at http://dx.doi.org/10.1016/j.ajhg.2015.12.013.
Contributor Information
Qimin Zhan, Email: zhanqimin@pumc.edu.cn.
Yongping Cui, Email: cuiy0922@yahoo.com.
Accession Numbers
The whole-genome sequencing data of 31 pairs of tumors and matched normal tissues reported in this paper have been deposited to the European Genome-phenome Archive (EGA) under accession numbers EGAS00001001487 and EGAS00001000709.
Web Resources
The URLs for data presented herein are as follows:
European Genome-phenome Archive (EGA), https://www.ebi.ac.uk/ega
OMIM, http://www.omim.org/
Supplemental Data
(A) List of in_frame fusions identified in 31 ESCC genomes.
(B) List of out_frame fusions in 31 ESCC genomes.
References
- 1.Inaki K., Liu E.T. Structural mutations in cancer: mechanistic and functional insights. Trends Genet. 2012;28:550–559. doi: 10.1016/j.tig.2012.07.002. [DOI] [PubMed] [Google Scholar]
- 2.Mills R.E., Walter K., Stewart C., Handsaker R.E., Chen K., Alkan C., Abyzov A., Yoon S.C., Ye K., Cheetham R.K., 1000 Genomes Project Mapping copy number variation by population-scale genome sequencing. Nature. 2011;470:59–65. doi: 10.1038/nature09708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Malhotra A., Lindberg M., Faust G.G., Leibowitz M.L., Clark R.A., Layer R.M., Quinlan A.R., Hall I.M. Breakpoint profiling of 64 cancer genomes reveals numerous complex rearrangements spawned by homology-independent mechanisms. Genome Res. 2013;23:762–776. doi: 10.1101/gr.143677.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hoeijmakers J.H. Genome maintenance mechanisms for preventing cancer. Nature. 2001;411:366–374. doi: 10.1038/35077232. [DOI] [PubMed] [Google Scholar]
- 5.Lee J.A., Carvalho C.M., Lupski J.R. A DNA replication mechanism for generating nonrecurrent rearrangements associated with genomic disorders. Cell. 2007;131:1235–1247. doi: 10.1016/j.cell.2007.11.037. [DOI] [PubMed] [Google Scholar]
- 6.Zhang F., Khajavi M., Connolly A.M., Towne C.F., Batish S.D., Lupski J.R. The DNA replication FoSTeS/MMBIR mechanism can generate genomic, genic and exonic complex rearrangements in humans. Nat. Genet. 2009;41:849–853. doi: 10.1038/ng.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang C.Z., Spektor A., Cornils H., Francis J.M., Jackson E.K., Liu S., Meyerson M., Pellman D. Chromothripsis from DNA damage in micronuclei. Nature. 2015;522:179–184. doi: 10.1038/nature14493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Soda M., Choi Y.L., Enomoto M., Takada S., Yamashita Y., Ishikawa S., Fujiwara S., Watanabe H., Kurashina K., Hatanaka H. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature. 2007;448:561–566. doi: 10.1038/nature05945. [DOI] [PubMed] [Google Scholar]
- 9.Wu Y.M., Su F., Kalyana-Sundaram S., Khazanov N., Ateeq B., Cao X., Lonigro R.J., Vats P., Wang R., Lin S.F. Identification of targetable FGFR gene fusions in diverse cancers. Cancer Discov. 2013;3:636–647. doi: 10.1158/2159-8290.CD-13-0050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hastings P.J., Lupski J.R., Rosenberg S.M., Ira G. Mechanisms of change in gene copy number. Nat. Rev. Genet. 2009;10:551–564. doi: 10.1038/nrg2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu P., Erez A., Nagamani S.C., Dhar S.U., Kołodziejska K.E., Dharmadhikari A.V., Cooper M.L., Wiszniewska J., Zhang F., Withers M.A. Chromosome catastrophes involve replication mechanisms generating complex genomic rearrangements. Cell. 2011;146:889–903. doi: 10.1016/j.cell.2011.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Colnaghi R., Carpenter G., Volker M., O’Driscoll M. The consequences of structural genomic alterations in humans: genomic disorders, genomic instability and cancer. Semin. Cell Dev. Biol. 2011;22:875–885. doi: 10.1016/j.semcdb.2011.07.010. [DOI] [PubMed] [Google Scholar]
- 13.Parikh R.A., White J.S., Huang X., Schoppy D.W., Baysal B.E., Baskaran R., Bakkenist C.J., Saunders W.S., Hsu L.C., Romkes M., Gollin S.M. Loss of distal 11q is associated with DNA repair deficiency and reduced sensitivity to ionizing radiation in head and neck squamous cell carcinoma. Genes Chromosomes Cancer. 2007;46:761–775. doi: 10.1002/gcc.20462. [DOI] [PubMed] [Google Scholar]
- 14.Nones K., Waddell N., Wayte N., Patch A.M., Bailey P., Newell F., Holmes O., Fink J.L., Quinn M.C., Tang Y.H. Genomic catastrophes frequently arise in esophageal adenocarcinoma and drive tumorigenesis. Nat. Commun. 2014;5:5224. doi: 10.1038/ncomms6224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Korbel J.O., Campbell P.J. Criteria for inference of chromothripsis in cancer genomes. Cell. 2013;152:1226–1236. doi: 10.1016/j.cell.2013.02.023. [DOI] [PubMed] [Google Scholar]
- 16.Nik-Zainal S., Alexandrov L.B., Wedge D.C., Van Loo P., Greenman C.D., Raine K., Jones D., Hinton J., Marshall J., Stebbings L.A., Breast Cancer Working Group of the International Cancer Genome Consortium Mutational processes molding the genomes of 21 breast cancers. Cell. 2012;149:979–993. doi: 10.1016/j.cell.2012.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sakofsky C.J., Roberts S.A., Malc E., Mieczkowski P.A., Resnick M.A., Gordenin D.A., Malkova A. Break-induced replication is a source of mutation clusters underlying kataegis. Cell Rep. 2014;7:1640–1648. doi: 10.1016/j.celrep.2014.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Abo R.P., Ducar M., Garcia E.P., Thorner A.R., Rojas-Rudilla V., Lin L., Sholl L.M., Hahn W.C., Meyerson M., Lindeman N.I. BreaKmer: detection of structural variation in targeted massively parallel sequencing data using kmers. Nucleic Acids Res. 2015;43:e19. doi: 10.1093/nar/gku1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Song Y., Li L., Ou Y., Gao Z., Li E., Li X., Zhang W., Wang J., Xu L., Zhou Y. Identification of genomic alterations in oesophageal squamous cell cancer. Nature. 2014;509:91–95. doi: 10.1038/nature13176. [DOI] [PubMed] [Google Scholar]
- 20.Lin D.C., Hao J.J., Nagata Y., Xu L., Shang L., Meng X., Sato Y., Okuno Y., Varela A.M., Ding L.W. Genomic and molecular characterization of esophageal squamous cell carcinoma. Nat. Genet. 2014;46:467–473. doi: 10.1038/ng.2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gao Y.B., Chen Z.L., Li J.G., Hu X.D., Shi X.J., Sun Z.M., Zhang F., Zhao Z.R., Li Z.T., Liu Z.Y. Genetic landscape of esophageal squamous cell carcinoma. Nat. Genet. 2014;46:1097–1102. doi: 10.1038/ng.3076. [DOI] [PubMed] [Google Scholar]
- 22.Zhang L., Zhou Y., Cheng C., Cui H., Cheng L., Kong P., Wang J., Li Y., Chen W., Song B. Genomic analyses reveal mutational signatures and frequently altered genes in esophageal squamous cell carcinoma. Am. J. Hum. Genet. 2015;96:597–611. doi: 10.1016/j.ajhg.2015.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang L., Luquette L.J., Gehlenborg N., Xi R., Haseley P.S., Hsieh C.H., Zhang C., Ren X., Protopopov A., Chin L. Diverse mechanisms of somatic structural variations in human cancer genomes. Cell. 2013;153:919–929. doi: 10.1016/j.cell.2013.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kidd J.M., Sampas N., Antonacci F., Graves T., Fulton R., Hayden H.S., Alkan C., Malig M., Ventura M., Giannuzzi G. Characterization of missing human genome sequences and copy-number polymorphic insertions. Nat. Methods. 2010;7:365–371. doi: 10.1038/nmeth.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Campbell P.J., Yachida S., Mudie L.J., Stephens P.J., Pleasance E.D., Stebbings L.A., Morsberger L.A., Latimer C., McLaren S., Lin M.L. The patterns and dynamics of genomic instability in metastatic pancreatic cancer. Nature. 2010;467:1109–1113. doi: 10.1038/nature09460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Waddell N., Pajic M., Patch A.M., Chang D.K., Kassahn K.S., Bailey P., Johns A.L., Miller D., Nones K., Quek K., Australian Pancreatic Cancer Genome Initiative Whole genomes redefine the mutational landscape of pancreatic cancer. Nature. 2015;518:495–501. doi: 10.1038/nature14169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hermetz K.E., Newman S., Conneely K.N., Martin C.L., Ballif B.C., Shaffer L.G., Cody J.D., Rudd M.K. Large inverted duplications in the human genome form via a fold-back mechanism. PLoS Genet. 2014;10:e1004139. doi: 10.1371/journal.pgen.1004139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mayrhofer M., DiLorenzo S., Isaksson A. Patchwork: allele-specific copy number analysis of whole-genome sequenced tumor tissue. Genome Biol. 2013;14:R24. doi: 10.1186/gb-2013-14-3-r24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mermel C.H., Schumacher S.E., Hill B., Meyerson M.L., Beroukhim R., Getz G. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol. 2011;12:R41. doi: 10.1186/gb-2011-12-4-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Campbell P.J., Stephens P.J., Pleasance E.D., O’Meara S., Li H., Santarius T., Stebbings L.A., Leroy C., Edkins S., Hardy C. Identification of somatically acquired rearrangements in cancer using genome-wide massively parallel paired-end sequencing. Nat. Genet. 2008;40:722–729. doi: 10.1038/ng.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pleasance E.D., Cheetham R.K., Stephens P.J., McBride D.J., Humphray S.J., Greenman C.D., Varela I., Lin M.L., Ordóñez G.R., Bignell G.R. A comprehensive catalogue of somatic mutations from a human cancer genome. Nature. 2010;463:191–196. doi: 10.1038/nature08658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fungtammasan A., Walsh E., Chiaromonte F., Eckert K.A., Makova K.D. A genome-wide analysis of common fragile sites: what features determine chromosomal instability in the human genome? Genome Res. 2012;22:993–1005. doi: 10.1101/gr.134395.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang K., Yuen S.T., Xu J., Lee S.P., Yan H.H., Shi S.T., Siu H.C., Deng S., Chu K.M., Law S. Whole-genome sequencing and comprehensive molecular profiling identify new driver mutations in gastric cancer. Nat. Genet. 2014;46:573–582. doi: 10.1038/ng.2983. [DOI] [PubMed] [Google Scholar]
- 34.Dutt A., Ramos A.H., Hammerman P.S., Mermel C., Cho J., Sharifnia T., Chande A., Tanaka K.E., Stransky N., Greulich H. Inhibitor-sensitive FGFR1 amplification in human non-small cell lung cancer. PLoS ONE. 2011;6:e20351. doi: 10.1371/journal.pone.0020351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marek L., Ware K.E., Fritzsche A., Hercule P., Helton W.R., Smith J.E., McDermott L.A., Coldren C.D., Nemenoff R.A., Merrick D.T. Fibroblast growth factor (FGF) and FGF receptor-mediated autocrine signaling in non-small-cell lung cancer cells. Mol. Pharmacol. 2009;75:196–207. doi: 10.1124/mol.108.049544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hu W.H., Pendergast J.S., Mo X.M., Brambilla R., Bracchi-Ricard V., Li F., Walters W.M., Blits B., He L., Schaal S.M., Bethea J.R. NIBP, a novel NIK and IKK(beta)-binding protein that enhances NF-(kappa)B activation. J. Biol. Chem. 2005;280:29233–29241. doi: 10.1074/jbc.M501670200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schou K.B., Morthorst S.K., Christensen S.T., Pedersen L.B. Identification of conserved, centrosome-targeting ASH domains in TRAPPII complex subunits and TRAPPC8. Cilia. 2014;3:6. doi: 10.1186/2046-2530-3-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhao S., Xu C., Qian H., Lv L., Ji C., Chen C., Zhao X., Zheng D., Gu S., Xie Y., Mao Y. Cellular retinaldehyde-binding protein-like (CRALBPL), a novel human Sec14p-like gene that is upregulated in human hepatocellular carcinomas, may be used as a marker for human hepatocellular carcinomas. DNA Cell Biol. 2008;27:159–163. doi: 10.1089/dna.2007.0634. [DOI] [PubMed] [Google Scholar]
- 39.Gillis L.D., Lewis S.M. Decreased eIF3e/Int6 expression causes epithelial-to-mesenchymal transition in breast epithelial cells. Oncogene. 2013;32:3598–3605. doi: 10.1038/onc.2012.371. [DOI] [PubMed] [Google Scholar]
- 40.Lee P.S., Fang J., Jessop L., Myers T., Raj P., Hu N., Wang C., Taylor P.R., Wang J., Khan J. RAD51B activity and cell cycle regulation in response to DNA damage in breast cancer cell lines. Breast Cancer (Auckl.) 2014;8:135–144. doi: 10.4137/BCBCR.S17766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mehine M., Kaasinen E., Mäkinen N., Katainen R., Kämpjärvi K., Pitkänen E., Heinonen H.R., Bützow R., Kilpivaara O., Kuosmanen A. Characterization of uterine leiomyomas by whole-genome sequencing. N. Engl. J. Med. 2013;369:43–53. doi: 10.1056/NEJMoa1302736. [DOI] [PubMed] [Google Scholar]
- 42.Yoshihara K., Wang Q., Torres-Garcia W., Zheng S., Vegesna R., Kim H., Verhaak R.G. The landscape and therapeutic relevance of cancer-associated transcript fusions. Oncogene. 2015;34:4845–4854. doi: 10.1038/onc.2014.406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Alexandrov L.B., Nik-Zainal S., Wedge D.C., Aparicio S.A., Behjati S., Biankin A.V., Bignell G.R., Bolli N., Borg A., Børresen-Dale A.L., Australian Pancreatic Cancer Genome Initiative. ICGC Breast Cancer Consortium. ICGC MMML-Seq Consortium. ICGC PedBrain Signatures of mutational processes in human cancer. Nature. 2013;500:415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stahl P., Seeschaaf C., Lebok P., Kutup A., Bockhorn M., Izbicki J.R., Bokemeyer C., Simon R., Sauter G., Marx A.H. Heterogeneity of amplification of HER2, EGFR, CCND1 and MYC in gastric cancer. BMC Gastroenterol. 2015;15:7. doi: 10.1186/s12876-015-0231-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ying J., Shan L., Li J., Zhong L., Xue L., Zhao H., Li L., Langford C., Guo L., Qiu T. Genome-wide screening for genetic alterations in esophageal cancer by aCGH identifies 11q13 amplification oncogenes associated with nodal metastasis. PLoS ONE. 2012;7:e39797. doi: 10.1371/journal.pone.0039797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bandla S., Pennathur A., Luketich J.D., Beer D.G., Lin L., Bass A.J., Godfrey T.E., Litle V.R. Comparative genomics of esophageal adenocarcinoma and squamous cell carcinoma. Ann. Thorac. Surg. 2012;93:1101–1106. doi: 10.1016/j.athoracsur.2012.01.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sato F., Kubota Y., Natsuizaka M., Maehara O., Hatanaka Y., Marukawa K., Terashita K., Suda G., Ohnishi S., Shimizu Y. EGFR inhibitors prevent induction of cancer stem-like cells in esophageal squamous cell carcinoma by suppressing epithelial-mesenchymal transition. Cancer Biol. Ther. 2015;16:933–940. doi: 10.1080/15384047.2015.1040959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Matsui A., Ihara T., Suda H., Mikami H., Semba K. Gene amplification: mechanisms and involvement in cancer. Biomol. Concepts. 2013;4:567–582. doi: 10.1515/bmc-2013-0026. [DOI] [PubMed] [Google Scholar]
- 49.Ozery-Flato M., Linhart C., Trakhtenbrot L., Izraeli S., Shamir R. Large-scale analysis of chromosomal aberrations in cancer karyotypes reveals two distinct paths to aneuploidy. Genome Biol. 2011;12:R61. doi: 10.1186/gb-2011-12-6-r61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Carter S.L., Cibulskis K., Helman E., McKenna A., Shen H., Zack T., Laird P.W., Onofrio R.C., Winckler W., Weir B.A. Absolute quantification of somatic DNA alterations in human cancer. Nat. Biotechnol. 2012;30:413–421. doi: 10.1038/nbt.2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Osthus R.C., Karim B., Prescott J.E., Smith B.D., McDevitt M., Huso D.L., Dang C.V. The Myc target gene JPO1/CDCA7 is frequently overexpressed in human tumors and has limited transforming activity in vivo. Cancer Res. 2005;65:5620–5627. doi: 10.1158/0008-5472.CAN-05-0536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rausch T., Jones D.T., Zapatka M., Stütz A.M., Zichner T., Weischenfeldt J., Jäger N., Remke M., Shih D., Northcott P.A. Genome sequencing of pediatric medulloblastoma links catastrophic DNA rearrangements with TP53 mutations. Cell. 2012;148:59–71. doi: 10.1016/j.cell.2011.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Reid B.J., Paulson T.G., Li X. Genetic insights in Barrett’s esophagus and esophageal adenocarcinoma. Gastroenterology. 2015;149:1142–1152.e3. doi: 10.1053/j.gastro.2015.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Consortium E.P., ENCODE Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) List of in_frame fusions identified in 31 ESCC genomes.
(B) List of out_frame fusions in 31 ESCC genomes.