Abstract
HAK/KUP/KT K+ transporters have been widely associated with K+ transport across membranes in bacteria, fungi, and plants. Indeed some members of the plant HAK/KUP/KT family contribute to root K+ uptake, notably at low external concentrations. Besides such role in acquisition, several studies carried out in Arabidopsis have shown that other members are also involved in developmental processes. With the publication of new plant genomes, a growing interest on plant species other than Arabidopsis has become evident. In order to understand HAK/KUP/KT diversity in these new plant genomes, we discuss the evolutionary trends of 913 HAK/KUP/KT sequences identified in 46 genomes revealing five major groups with an uneven distribution among angiosperms, notably between dicotyledonous and monocotyledonous species. This information evidenced the richness of crop genomes in HAK/KUP/KT transporters and supports their study for unraveling novel physiological roles of such transporters in plants.
Keywords: HAK/KUP/KT, transporter, potassium, phylogeny, angiosperm
Introduction
Potassium is an essential macronutrient for plants, making up to 2–7% of the plant’s total dry weight (Evans and Sorger, 1966; Leigh and Wyn Jones, 1984). It fulfills a number of important functions, such as enzyme activation, neutralization of negative charges and, more specific to plants, the maintenance of cell turgor that leads to plant growth and organ movement (Marschner, 2012). As sessile organisms, plants need to take up K+ from the soil. This is firstly achieved by root epidermal and cortical cells. Then, K+ is loaded in the stele and transported to the shoot and distributed to the leaves (Ahmad and Maathuis, 2014; Wigoda et al., 2014). Potassium short- and long-distance transport involves the movement of K+ through cell membranes, notably the plasma membrane which in many cases occurs against steep concentration gradients (like in the root–soil interface, for instance). In plants, there are five major multi-gene families that encode K+-permeable transport systems: (i) Shaker-like K+ channels, (ii) tandem-pore K+ (TPK) channels, (iii) HAK/KUP/KT transporters, (iv) HKT transporters, and (v) cation-proton antiporters (CPAs; Mäser et al., 2001). They have become the essentials of the K+ transport toolkit during terrestrial plant evolution due to their widespread presence in different land plant lineages (Gomez-Porras et al., 2012).
Here we focus on the HAK/KUP/KT (High-Affinity K+/K+ UPtake/K+ Transporter) transporter family. Plant HAK/KUP/KT transporters were first identified in barley and Arabidopsis (Quintero and Blatt, 1997; Santa-María et al., 1997; Fu and Luan, 1998; Kim et al., 1998) from their homology to bacterial KUP and fungal HAK transporters (Schleyer and Bakker, 1993; Bañuelos et al., 1995). Due to the different acronyms used in these early reports, the composite name of HAK/KUP/KT is widely used to refer to the whole family in plants. Plant HAK/KUP/KT proteins possess 10–15 transmembrane (TM) segments with both N- and C-termini in the intracellular side of the membrane, the latter being much longer (Rubio et al., 2000; Gomez-Porras et al., 2012). They have been widely shown to mediate K+ fluxes when expressed in K+-uptake deficient bacteria or yeast. Moreover, plant HAK/KUP/KT proteins differ in their affinity for K+ and can mediate cation influx as well as efflux (Fu and Luan, 1998; Rubio et al., 2000; Senn et al., 2001; Bañuelos et al., 2002; Garciadeblas et al., 2002; Ahn et al., 2004). Different studies reported that HAK/KUP/KT transporters poorly discriminate between K+, Rb+, and Cs+ and are inhibited by NH4+ (Santa-María et al., 1997; Rubio et al., 2000; Bañuelos et al., 2002; Martínez-Cordero et al., 2004). Plant HAK/KUP/KT proteins exhibit a great diversity in terms of subcellular localization (plasma membrane, tonoplast, or other endomembranes; Bañuelos et al., 2002; Jaquinod et al., 2007; Qi et al., 2008; Osakabe et al., 2013; Rigas et al., 2013) and expression patterns (root meristems, vascular tissues, guard cells, fruits, or specialized organs such as flytraps; Elumalai et al., 2002; Ahn et al., 2004; Vicente-Agullo et al., 2004; Davies et al., 2006; Osakabe et al., 2013; Scherzer et al., 2015).
Regarding their functions, some members of the plant HAK/KUP/KT family contribute to root K+ uptake, notably at low external concentrations (high-affinity range) through active K+ transport (Nieves-Cordones et al., 2014). Such high-affinity K+ transporters are expected to be H+/K+ symporters (Rodriguez-Navarro et al., 1986; Maathuis and Sanders, 1994), but experimental support for this notion is still required. Several studies carried out in Arabidopsis have shown that other members are involved in the regulation of cell size, auxin distribution or osmotic stress adaptation (Very et al., 2014). Such three roles highlight the great importance and role diversity of HAK/KUP/KT transporters in plant physiology besides K+ acquisition.
During the last two decades, research on Arabidopsis has notably accelerated the acquisition of information concerning the molecular and physiological mechanisms around K+ transport and HAK/KUP/KT proteins. This has been possible mainly because of the availability of its genome sequence and the use of T-DNA insertion lines to knock-out gene function. In recent years, genome sequences from many plant species have become available. This, together with the establishment of genome-editing techniques, such as Transcription Activator-Like Effector Nucleases (TALEN) or Clustered Regularly Interspaced Short Palindromic Repeats-Cas system (CRISPR-Cas) opens the door to investigate HAK/KUP/KT gene function in crops much faster (Andersen et al., 2015). It is true that research on crop species can benefit from the information gained in Arabidopsis, but the study of certain physiological processes, such as the development of a fleshy fruit, need to be carried out in appropriate species. In order to orientate further research in HAK/KUP/KT function in crop species, we present a multi-species phylogenetic analysis of plant HAK/KUP/KT proteins (comprising 913 members from 46 sequenced genomes) evidencing the presence of five major clades and remarkable specificities depending on the angiosperm group considered.
HAK/KUP/KT Phylogeny in Angiosperms
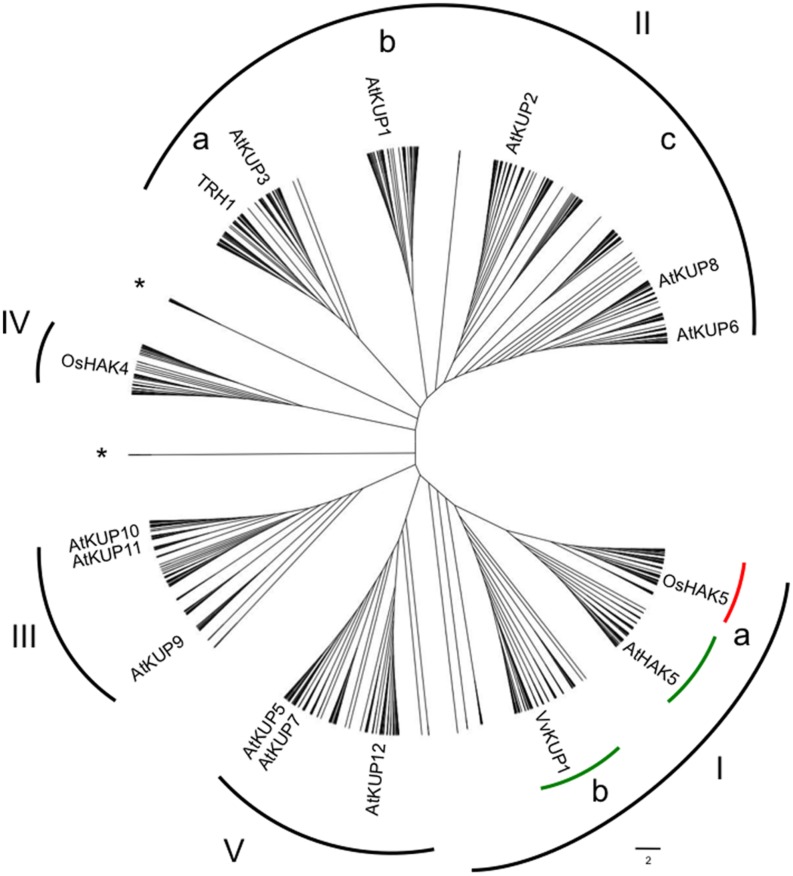
Phylogenetic relationships within the HAK/KUP/KT family have consistently shown the existence of several clades in angiosperm species, but with weak biological support for such distribution (Rubio et al., 2000; Gupta et al., 2008; Gomez-Porras et al., 2012; Very et al., 2014). Since the number of sequenced angiosperm genomes, and thus that of HAK/KUP/KT available sequences, has notably increased in the last years, we wanted to assess the robustness and the species distribution of the different clades. For that purpose, we made an inventory of HAK/KUP/KT protein sequences from 43 angiosperm genomes plus three outgroup species (one gymnosperm, Picea abies, and two primitive non-seed plants Selaginella moellendorffii and Physcomitrella patens; Supplementary Table S1). The phylogenetic tree obtained by maximum likelihood for such sequences revealed five major clades (I to V) where I to IV followed previous numeration (Rubio et al., 2000; Gupta et al., 2008; Gomez-Porras et al., 2012; Very et al., 2014) (Figure 1). Representative HAK/KUP/KT transporters that have been functionally characterized are found throughout the tree. Several subgroups were identified in clade I (Ia and Ib) and in clade II (IIa, IIb, and IIc). Then, we assessed the HAK/KUP/KT sequence distribution in the different analyzed species and the angiosperm orders to which they belong (Table 1). Results from the common ancestor of dicotyledonous and monocotyledonous species, Amborella trichopoda, evidenced the presence of HAK/KUP/KT transporters in that ancestor in all of the aforementioned clades. They also suggested that clade I separation into Ia and Ib occurred at the beginning of the angiosperm lineage since A. trichopoda has Ia and Ib transporters and outgroup sequences belonging to clade I were not placed in any of this two major subclades. It is worth to note that clade Ib only contained sequences from dicotyledonous species, but not from monocotyledonous ones (Figure 1, Table 1). This result suggests that clade Ib disappeared in the monocotyledonous lineage because, as stated before, it was already present in the A. trichopoda genome. Within HAK/KUP/KT transporters from dicotyledonous genomes, different transporter distributions among orders were identified and, in some cases, groups of related species displayed empty clades that are indicative of important events in the evolution of HAK/KUP/KT transporters in dicotyledonous orders. Indeed, in Solanales, clade IIb transporters were not identified, while in Cucurbitales it was the case for clade IV. The analysis of HAK/KUP/KT transporters from Brassicales species provided striking results: clades Ib and IV were absent in HAK/KUP/KT transporters from the Brassicaceae family (eight genomes, including Arabidopsis thaliana) while in Carica papaya (belonging to Brassicales but not to such family) had one transporter belonging to clade Ib and three in clade IV. Thus, a loss of both clades could have taken place during the evolution of Brassicaceae. At the outgroup level, HAK/KUP/KT transporters from Physcomitrella patens were found in clade I, IV, and V whereas 13 sequences from this organism fell apart in two separate branches independent from the five major clades. With respect to Selaginella moellendorffii and Picea abies, we did not observe sequences in clades IIa and IIb in the former and in clade III in the latter.
FIGURE 1.
Phylogenetic tree of the HAK/KUP/KT family in plants containing 913 sequences from 46 fully sequenced angiosperm genomes plus three outgroups (Picea abies, Selaginella moellendorffii, and Physcomitrella patens). Protein sequences fall into five main clades (I to V) where V is a novel clade. Some sub-clades within clade I contain only sequences from dicotyledonous (green lines) or monocotyledonous (red line) species. Asterisks represent outgroup sequences, which did not fall into main clades. Letters depict sub-clades within clades I and II. Representative members within the different clades are shown. Retrieved sequences from public genomic resources were ascribed to the HAK/KUP/KT family by using Orthomcl (http://orthomcl.org/orthomcl/). Protein organization was evaluated with MEME suite website (http://meme-suite.org/). Then, sequences were aligned by MAFFT (http://mafft.cbrc.jp/alignment/server/) and then alignment curation by G-block analysis in Seaview was applied prior to tree building. Tree building was constructed with MEGA6 by maximum-likelihood analysis. The scale bar represents number of substitutions per site. See also Supplementary Table S1.
Table 1.
HAK/KUP/KT gene distribution among angiosperm orders and outrgroups.
| Group | Order | Species | Clade |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Ia | Ib | IIa | IIb | IIc | III | IV | V | Total | |||
| Amborellales | Amborella trichopoda | 1 | 2 | 2 | 1 | 4 | 1 | 2 | 2 | 15 | |
| Dicotyledons | Solanales | Solanum lycopersicum | 2 | 3 | 3 | 0 | 5 | 2 | 2 | 4 | 21 |
| Solanum tuberosum | 2 | 2 | 3 | 0 | 4 | 2 | 0 | 2 | 15 | ||
| Lamiales | Mimulus guttatus | 0 | 3 | 2 | 0 | 4 | 1 | 1 | 2 | 13 | |
| Vitales | Vitis vinifera | 1 | 4 | 2 | 2 | 5 | 1 | 1 | 2 | 18 | |
| Fabales | Cicer arietinum | 1 | 1 | 2 | 2 | 4 | 1 | 1 | 3 | 15 | |
| Glycine max | 3 | 1 | 6 | 3 | 10 | 3 | 1 | 5 | 32 | ||
| Medicago truncatula | 3 | 2 | 2 | 3 | 4 | 3 | 0 | 3 | 20 | ||
| Phaseolus vulgaris | 2 | 0 | 3 | 1 | 6 | 2 | 1 | 3 | 18 | ||
| Rosales | Fragaria vesca | 3 | 1 | 2 | 2 | 4 | 2 | 3 | 0 | 17 | |
| Malus domestica | 3 | 6 | 0 | 3 | 7 | 3 | 2 | 4 | 28 | ||
| Prunus persica | 2 | 1 | 1 | 2 | 4 | 3 | 0 | 2 | 15 | ||
| Cucurbitales | Cucumis melo | 1 | 4 | 1 | 1 | 4 | 2 | 0 | 2 | 15 | |
| Cucumis sativus | 1 | 4 | 1 | 1 | 4 | 2 | 0 | 2 | 15 | ||
| Malpighiales | Jatropha curcas | 1 | 2 | 2 | 1 | 3 | 1 | 1 | 1 | 12 | |
| Linum usitatissimum | 1 | 2 | 2 | 1 | 9 | 5 | 0 | 3 | 23 | ||
| Manihot esculenta | 1 | 5 | 3 | 1 | 7 | 1 | 1 | 2 | 21 | ||
| Populus trichocarpa | 3 | 1 | 3 | 4 | 8 | 4 | 2 | 4 | 29 | ||
| Ricinus communis | 1 | 2 | 2 | 1 | 4 | 1 | 1 | 2 | 14 | ||
| Salix purpurea | 3 | 1 | 3 | 2 | 6 | 4 | 1 | 3 | 23 | ||
| Myrtales | Eucalyptus grandis | 6 | 9 | 2 | 1 | 6 | 3 | 1 | 2 | 30 | |
| Sapindales | Citrus clementina | 1 | 5 | 2 | 1 | 3 | 2 | 1 | 2 | 17 | |
| Citrus sinensis | 1 | 3 | 2 | 2 | 3 | 2 | 1 | 2 | 16 | ||
| Malvales | Gossypium raimondii | 2 | 3 | 3 | 3 | 7 | 2 | 1 | 5 | 26 | |
| Theobroma cacao | 1 | 3 | 2 | 3 | 4 | 1 | 1 | 2 | 17 | ||
| Brassicales | Arabidopsis halleri | 2 | 0 | 2 | 1 | 3 | 3 | 0 | 3 | 14 | |
| Arabidopsis lyrata | 3 | 0 | 2 | 1 | 3 | 3 | 0 | 3 | 15 | ||
| Arabidopsis thaliana | 1 | 0 | 2 | 1 | 3 | 3 | 0 | 3 | 13 | ||
| Boechera stricta | 1 | 0 | 2 | 1 | 3 | 3 | 0 | 3 | 13 | ||
| Brassica rapa | 1 | 0 | 3 | 2 | 4 | 6 | 0 | 3 | 19 | ||
| Capsella grandis | 1 | 0 | 2 | 1 | 3 | 3 | 0 | 3 | 13 | ||
| Capsella rubella | 1 | 0 | 2 | 1 | 4 | 3 | 0 | 3 | 14 | ||
| Eutrema salsugineum | 1 | 0 | 2 | 1 | 3 | 4 | 0 | 3 | 14 | ||
| Carica papaya | 0 | 1 | 1 | 2 | 4 | 2 | 3 | 1 | 14 | ||
| Ranunculales | Aquilegia coerulea | 3 | 0 | 2 | 2 | 3 | 1 | 4 | 2 | 17 | |
| Monocotyledons | Zingiberales | Musa acuminata | 0∗ | 0 | 4 | 3 | 7 | 5 | 1 | 3 | 24 |
| Poales | Brachypodium distachyon | 6 | 0 | 3 | 1 | 7 | 3 | 5 | 2 | 27 | |
| Hordeum vulgare | 5 | 0 | 2 | 0 | 4 | 2 | 0 | 2 | 15 | ||
| Oryza sativa | 8 | 0 | 3 | 1 | 5 | 3 | 4 | 3 | 27 | ||
| Panicum virgatum | 21 | 0 | 6 | 2 | 12 | 6 | 4 | 6 | 57 | ||
| Setaria italica | 12 | 0 | 3 | 1 | 5 | 3 | 3 | 3 | 30 | ||
| Sorghum bicolor | 11 | 0 | 4 | 1 | 5 | 3 | 2 | 4 | 30 | ||
| Zea mays | 9 | 0 | 3 | 1 | 5 | 3 | 3 | 3 | 27 | ||
| Outgroups | Species |
Cluster |
|||||||||
| I | IIa | IIb | IIc | III | IV | V | Total | ||||
| Gymnosperms | Picea abies | 5 | 1 | 1 | 3 | 0 | 1 | 2 | 13 | ||
| Lycopodiophytes | Selaginella moellendorffii | 2 | 0 | 0 | 2 | 4 | 2 | 1 | 11 | ||
| Bryophytes | Physcomitrella patens | 1 | 0 | 0 | 0 | 0 | 2 | 2 | 18∗∗ | ||
Clades where no sequence was found in a given species are shown in red. ∗There is one protein belonging to cluster I but it is located in a different branch from Ia and Ib. ∗∗Thirteen sequences fell apart from the five major clades described for angiosperms.
Despite the number of HAK/KUP/KT transporters whose physiological role has been established is relatively small, some conclusions can be drawn from the present analysis. Transporters involved in root high-affinity K+ uptake both from dicotyledonous or monocotyledonous species fall into clade Ia: HvHAK1, AtHAK5, OsHAK1, CaHAK1, SlHAK5/LeHAK5, and EsHAK5/ThHAK5, for instance (Santa-María et al., 1997; Bañuelos et al., 2002; Martínez-Cordero et al., 2004; Nieves-Cordones et al., 2007; Rubio et al., 2008; Aleman et al., 2009). However, recent work on rice OsHAK5 and OsHAK21, which also belong to clade Ia, showed more specialized functions when compared to the typical high-affinity K+ transporter OsHAK1 (Chen et al., 2015). For instance, OsHAK5 and OsHAK21 were involved in K+ transport to aerial parts during K+ deficiency or salt stress, respectively (Yang et al., 2014; Shen et al., 2015). Since rice and other grasses belonging to the Poaceae family exhibited a higher number of clade Ia HAK/KUP/KT sequences than dicotyledonous genomes (10.29 vs. 1.84), it could be interpreted as a specific diversification of Ia high-affinity K+ transporters in Poaceae species. It is worth to note that disruption of the OsHAK1 gene led to a dramatic decrease in grain yield (Chen et al., 2015), whereas such a phenotype has not been observed in the AtHAK5 KO mutant (Nieves-Cordones and Rubio, unpublished results). It would be interesting to know which is the contribution to grain yield of OsHAK1-like transporters from other cereal species, since they could be good targets to improve food production. Regarding clade Ib HAK/KUP/KT transporters, two reports have provided us with some information about this group. First, VvKUP1/VvHAK1-a from grapevine was shown to be expressed in flowers and grape berry skin, showing its highest expression level in the latter tissue during the pre-veraison stage (Davies et al., 2006). Second, DmHAK5 from Dionaea muscipula (Venus flytrap) contributes to high-affinity K+ uptake in digesting traps (Scherzer et al., 2015). Further characterization of clade Ib transporters will clarify whether they are specialized in transporting K+ in tissues other than roots. Interestingly, recent reports showed that some clade I HAK/KUP/KT transporters, including DmHAK5 (clade Ib), SlHAK5, CaHAK1, and AtHAK5 (clade Ia), are activated by CIPK23-CBL1/9 complexes, which provide novel insights into the regulation of high-affinity K+ transport (Ragel et al., 2015; Scherzer et al., 2015). Moreover, such regulatory network offers a new alternative that could be used to enhance K+ uptake in tomato and pepper plants.
Clade II has been associated in Arabidopsis with developmental processes, especially those which demand turgor-driven cell expansion. In clade IIa, there is AtKUP4/TRH1 (Tiny Root Hairs 1) which contributes to the polar localization of auxin transporters in the root apex that, in turn, establishes auxin gradients necessary for both gravitropic responses and root hair formation (Rigas et al., 2001, 2013; Vicente-Agullo et al., 2004). The first cloned HAK/KUP/KT transporter from Arabidopsis, AtKUP1/KT1, belongs to clade IIb, but no physiological role has been attributed to it so far (Quintero and Blatt, 1997; Fu and Luan, 1998; Kim et al., 1998). In clade IIc, there are AtKUP2/6/8 which have been shown to negatively regulate plant growth and cell size by mediating K+ efflux rather influx (Osakabe et al., 2013). Analysis of an AtKUP2/6/8 triple null mutant also evidenced impaired ABA responses in guard cells and lateral root cells. Phosphorylation of AtKUP6 by OST1 connected osmotic stress adaptation to the regulation of K+ fluxes mediated by HAK/KUP/KT transporters.
With respect the other clades, GhKT1 from cotton (clade III; Gossypium hirsutum) was specifically upregulated during cotton fiber elongation (Ruan et al., 2001). Regarding clade IV transporters, only two have been characterized so far. LjKUP from Lotus japonicus was highly expressed during late nodulation development and complemented K+ uptake deficient bacteria (Desbrosses et al., 2004). On the other hand, PpHAK13 from the outgroup species Physcomitrella patens is a high-affinity Na+ transporter, with low K+ permeability, that was repressed under the presence of high Na+ concentrations (Benito et al., 2012). The latter transporter raises the question whether other plant HAK/KUP/KT transporters are permeable to Na+ at low external concentrations. Finally, belonging to clade V, PpHAK1 from Physcomitrella patens was shown to regulate steady K+ content and plant morphology under non-K+-limiting conditions and to contribute to high-affinity Rb+ and Cs+ uptake during K+ starvation (Garciadeblas et al., 2007).
Besides their physiological roles, subcellular localization of HAK/KUP/KT transporters has been assessed in some cases and it was shown to be quite diverse. Furthermore, there is not a clear relationship between phylogenetic clade to which a transporter belongs and its targeted cell membrane. For instance, several members are targeted to the plasma membrane, such as AtHAK5, OsHAK1, OsHAK21, OsHAK5 (clade I; Qi et al., 2008; Yang et al., 2014; Chen et al., 2015; Shen et al., 2015), AtKUP6 (clade IIc; Osakabe et al., 2013), and LjKUP (clade IV; Desbrosses et al., 2004) while others are targeted to the tonoplast (OsHAK10, clade IIc, and AtKUP5 clade V; Jaquinod et al., 2007; Bañuelos et al., 2002) or endoplasmatic reticulum-like endomembranes (AtKUP4/TRH1; Rigas et al., 2013).
Conclusion and Perspectives
Plant HAK/KUP/KT K+ transporters have been shown to play key roles in plant physiology like K+ acquisition, abiotic stress adaptation and developmental processes. Interestingly, the fact that HAK/KUP/KT transporters are permeable to K+ only explains a part of the phenotypes exhibited by the plants lacking them as it is the case of AtHAK5 or AtKUP4/TRH1, where energization of AtHAK5-mediated K+ uptake or the relationship between auxin distribution and AtKUP4/TRH1 activity deserve further attention. From our analysis, it can be deduced that the contribution of HAK/KUP/KT K+ transporters to plant physiology may substantially differ among species, especially when entire clades are missing in a given group of species as shown. Therefore, Arabidopsis can still be a good model for certain well conserved roles of HAK/KUP/KT K+ transporters, AtHAK5 for example, but research on other species, notably crops, is required: (i) to study transporters belonging to clades Ib or IV (missing in the Brassicaceae family) or clades where significant gene duplication occurred (clade Ia in monocots) and (ii) to investigate physiological aspects which are absent in Arabidopsis (fleshy fruit development, for instance). Besides, some HAK/KUP/KT proteins can be already regarded as interesting candidates for future crop improvement strategies, for example GhKT1 (specifically upregulated during cotton fiber elongation), VvKUP1/VvHAK1-a (highly expressed during the preveraison stage) and OsHAK1 (critical for rice grain yield). In line with this statement, the peach fruit is the organ where more HAK/KUP/KT genes are expressed in this species (Song et al., 2015). Further research on this transporter family will contribute to understanding how we can engineer plants for food and renewable biomass production.
Author Contributions
MN-C, RR, and FR performed the experimental analyses. MN-C wrote the article with inputs from AC, RMR, VM, IG, and FR.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. This work was funded by grant AGL2012-33504 from Ministerio de Economia y Competitividad, Spain. RR is recipient of an FPU predoctoral contract from Ministerio de Educación, Cultura y Deporte, Spain.
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.00127
| List of HAK/KUP/KT sequences used in Figure 1 (Excel file).
References
- Ahmad I., Maathuis F. J. (2014). Cellular and tissue distribution of potassium: physiological relevance, mechanisms and regulation. J. Plant Physiol. 171 708–714. 10.1016/j.jplph.2013.10.016 [DOI] [PubMed] [Google Scholar]
- Ahn S. J., Shin R., Schachtman D. P. (2004). Expression of KT/KUP genes in Arabidopsis and the role of root hairs in K+ uptake. Plant Physiol. 134 1135–1145. 10.1104/pp.103.034660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aleman F., Nieves-Cordones M., Martinez V., Rubio F. (2009). Differential regulation of the HAK5 genes encoding the high-affinity K+ transporters of Thellungiella halophila and Arabidopsis thaliana. Environ. Exp. Bot. 65 263–269. 10.1016/j.envexpbot.2008.09.011 [DOI] [Google Scholar]
- Andersen M. M., Landes X., Xiang W., Anyshchenko A., Falhof J., Østerberg J. T., et al. (2015). Feasibility of new breeding techniques for organic farming. Trends Plant Sci. 20 426–434. 10.1016/j.tplants.2015.04.011 [DOI] [PubMed] [Google Scholar]
- Bañuelos M. A., Garciadeblas B., Cubero B., Rodríguez-Navarro A. (2002). Inventory and functional characterization of the HAK potassium transporters of rice. Plant Physiol. 130 784–795. 10.1104/pp.007781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bañuelos M. A., Klein R. D., Alexander-Bowman S. J., Rodriguez-Navarro A. (1995). A potassium transporter of the yeast Schwanniomyces occidentalis homologous to the Kup system of Escherichia coli has a high concentrative capacity. EMBO J. 14 3021–3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benito B., Garciadeblas B., Rodriguez-Navarro A. (2012). HAK transporters from Physcomitrella patens and Yarrowia lipolytica mediate sodium uptake. Plant Cell Physiol. 53 1117–1123. 10.1093/pcp/pcs056 [DOI] [PubMed] [Google Scholar]
- Chen G., Hu Q., Luo L., Yang T., Zhang S., Hu Y., et al. (2015). Rice potassium transporter OsHAK1 is essential for maintaining potassium-mediated growth and functions in salt tolerance over low and high potassium concentration ranges. Plant Cell Environ. 38 2747–2765. 10.1111/pce.12585 [DOI] [PubMed] [Google Scholar]
- Davies C., Shin R., Liu W., Thomas M. R., Schachtman D. P. (2006). Transporters expressed during grape berry (Vitis vinifera L.) development are associated with an increase in berry size and berry potassium accumulation. J. Exp. Bot. 57 3209–3216. 10.1093/jxb/erl091 [DOI] [PubMed] [Google Scholar]
- Desbrosses G., Kopka C., Ott T., Udvardi M. K. (2004). Lotus japonicus LjKUP is induced late during nodule development and encodes a potassium transporter of the plasma membrane. Mol. Plant Microbe Interact. 17 789–797. 10.1094/MPMI.2004.17.7.789 [DOI] [PubMed] [Google Scholar]
- Elumalai R. P., Nagpal P., Reed J. W. (2002). A mutation in the Arabidopsis KT2/KUP2 potassium transporter gene affects shoot cell expansion. Plant Cell 14 119–131. 10.1105/tpc.010322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans H. J., Sorger G. J. (1966). Role of mineral elements with emphasis on univalent cations. Annu. Rev. Plant Physiol. 17 47–76. 10.1146/annurev.pp.17.060166.000403 [DOI] [Google Scholar]
- Fu H. H., Luan S. (1998). AtKUP1: a dual-affinity K+ transporter from Arabidopsis. Plant Cell 10 63–73. 10.2307/3870629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garciadeblas B., Barrero-Gil J., Benito B., Rodriguez-Navarro A. (2007). Potassium transport systems in the moss Physcomitrella patens: pphak1 plants reveal the complexity of potassium uptake. Plant J. 52 1080–1093. 10.1111/j.1365-313X.2007.03297.x [DOI] [PubMed] [Google Scholar]
- Garciadeblas B., Benito B., Rodriguez-Navarro A. (2002). Molecular cloning and functional expression in bacteria of the potassium transporters CnHAK1 and CnHAK2 of the seagrass Cymodocea nodosa. Plant Mol. Biol. 50 623–633. 10.1023/A:1019951023362 [DOI] [PubMed] [Google Scholar]
- Gomez-Porras J. L., Riaño-Pachón D. M., Benito B., Haro R., Sklodowski K., Rodríguez-Navarro A., et al. (2012). Phylogenetic analysis of K+ transporters in bryophytes, lycophytes, and flowering plants indicates a specialization of vascular plants. Front. Plant Sci. 3:167 10.3389/fpls.2012.00167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta M., Qiu X., Wang L., Xie W., Zhang C., Xiong L., et al. (2008). KT/HAK/KUP potassium transporters gene family and their whole-life cycle expression profile in rice (Oryza sativa). Mol. Genet. Genomics 280 437–452. 10.1007/s00438-008-0377-7 [DOI] [PubMed] [Google Scholar]
- Jaquinod M., Villiers F., Kieffer-Jaquinod S., Hugouvieux V., Bruley C., Garin J., et al. (2007). A proteomics dissection of Arabidopsis thaliana vacuoles isolated from cell culture. Mol. Cell. Proteomics 6 394–412. 10.1074/mcp.M600250-MCP200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E. J., Kwak J. M., Uozumi N., Schroeder J. I. (1998). AtKUP1: an Arabidopsis gene encoding high-affinity potassium transport activity. Plant Cell 10 51–62. 10.2307/3870628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leigh R. A., Wyn Jones R. G. (1984). A hypothesis relating critical potassium concentrations for growth to the distribution and functions of this ion in the plant cell. New Phytol. 97 1–13. 10.1111/j.1469-8137.1984.tb04103.x [DOI] [Google Scholar]
- Maathuis F. J. M., Sanders D. (1994). Mechanism of high-affinity potassium uptake in roots of Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 91 9272–9276. 10.1073/pnas.91.20.9272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marschner P. (2012). Marschner’s Mineral Nutrition of Higher Plants, 3rd Edn. London: Academic Press. [Google Scholar]
- Martínez-Cordero M. A., Martinez V., Rubio F. (2004). Cloning and functional characterization of the high-affinity K+ transporter HAK1 of pepper. Plant Mol. Biol. 56 413–421. 10.1007/s11103-004-3845-4 [DOI] [PubMed] [Google Scholar]
- Mäser P., Thomine S., Schroeder J. I., Ward J. M., Hirschi K., Sze H., et al. (2001). Phylogenetic relationships within cation transporter families of Arabidopsis. Plant Physiol. 126 1646–1667. 10.1104/pp.126.4.1646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieves-Cordones M., Aleman F., Martinez V., Rubio F. (2014). K+ uptake in plant roots. The systems involved, their regulation and parallels in other organisms. J. Plant Physiol. 171 688–695. 10.1016/j.jplph.2013.09.021 [DOI] [PubMed] [Google Scholar]
- Nieves-Cordones M., Martinez-Cordero M. A., Martinez V., Rubio F. (2007). An NH4+-sensitive component dominates high-affinity K+ uptake in tomato plants. Plant Sci. 172 273–280. 10.1016/j.plantsci.2006.09.003 [DOI] [Google Scholar]
- Osakabe Y., Arinaga N., Umezawa T., Katsura S., Nagamachi K., Tanaka H., et al. (2013). Osmotic stress responses and plant growth controlled by potassium transporters in Arabidopsis. Plant Cell 25 609–624. 10.1105/tpc.112.105700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Z., Hampton C. R., Shin R., Barkla B. J., White P. J., Schachtman D. P. (2008). The high affinity K+ transporter AtHAK5 plays a physiological role in planta at very low K+ concentrations and provides a caesium uptake pathway in Arabidopsis. J. Exp. Bot. 59 595–607. 10.1093/jxb/erm330 [DOI] [PubMed] [Google Scholar]
- Quintero F. J., Blatt M. R. (1997). A new family of K+ transporters from Arabidopsis that are conserved across phyla. FEBS Lett. 415 206–211. 10.1016/S0014-5793(97)01125-3 [DOI] [PubMed] [Google Scholar]
- Ragel P., Ródenas R., García-Martín E., Andrés Z., Villalta I., Nieves-Cordones M., et al. (2015). CIPK23 regulates HAK5-mediated high-affinity K+ uptake in Arabidopsis roots. Plant Physiol. 169 2863–2873. 10.1104/pp.15.01401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigas S., Debrosses G., Haralampidis K., Vicente-Agullo F., Feldmann K. A., Grabov A., et al. (2001). TRH1 encodes a potassium transporter required for tip growth in Arabidopsis root hairs. Plant Cell 13 139–151. 10.1105/tpc.13.1.139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigas S., Ditengou F. A., Ljung K., Daras G., Tietz O., Palme K., et al. (2013). Root gravitropism and root hair development constitute coupled developmental responses regulated by auxin homeostasis in the Arabidopsis root apex. New Phytol. 197 1130–1141. 10.1111/nph.12092 [DOI] [PubMed] [Google Scholar]
- Rodriguez-Navarro A., Blatt M. R., Slayman C. L. (1986). A potassium-proton symport in Neurospora crassa. J. Gen. Physiol. 87 649–674. 10.1085/jgp.87.5.649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan Y. L., Llewellyn D. J., Furbank R. T. (2001). The control of single-celled cotton fiber elongation by developmentally reversible gating of plasmodesmata and coordinated expression of sucrose and K+ transporters and expansin. Plant Cell 13 47–60. 10.1105/tpc.13.1.47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio F., Nieves-Cordones M., Aleman F., Martinez V. (2008). Relative contribution of AtHAK5 and AtAKT1 to K+ uptake in the high-affinity range of concentrations. Physiol. Plant. 134 598–608. 10.1111/j.1399-3054.2008.01168.x [DOI] [PubMed] [Google Scholar]
- Rubio F., Santa-Maria G. E., Rodriguez-Navarro A. (2000). Cloning of Arabidopsis and barley cDNAs encoding HAK potassium transporters in root and shoot cells. Physiol. Plant. 109 34–43. 10.1034/j.1399-3054.2000.100106.x [DOI] [Google Scholar]
- Santa-María G. E., Rubio F., Dubcovsky J., Rodriguez-Navarro A. (1997). The HAK1 gene of barley is a member of a large gene family and encodes a high-affinity potassium transporter. Plant Cell 9 2281–2289. 10.2307/3870585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherzer S., Böhm J., Krol E., Shabala L., Kreuzer I., Larisch C., et al. (2015). Calcium sensor kinase activates potassium uptake systems in gland cells of Venus flytraps. Proc. Natl. Acad. Sci. U.S.A. 112 7309–7314. 10.1073/pnas.1507810112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schleyer M., Bakker E. P. (1993). Nucleotide sequence and 3’-end deletion studies indicate that the K(+)-uptake protein kup from Escherichia coli is composed of a hydrophobic core linked to a large and partially essential hydrophilic C terminus. J. Bacteriol. 175 6925–6931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senn M. E., Rubio F., Bañuelos M. A., Rodriguez-Navarro A. (2001). Comparative functional features of plant potassium HvHAK1 and HvHAK2 transporters. J. Biol. Chem. 276 44563–44569. 10.1074/jbc.M108129200 [DOI] [PubMed] [Google Scholar]
- Shen Y., Shen L., Shen Z., Jing W., Ge H., Zhao J., et al. (2015). The potassium transporter OsHAK21 functions in the maintenance of ion homeostasis and tolerance to salt stress in rice. Plant Cell Environ. 38 2766–2779. 10.1111/pce.12586 [DOI] [PubMed] [Google Scholar]
- Song Z. Z., Ma R. J., Yu M. L. (2015). Genome-wide analysis and identification of KT/HAK/KUP potassium transporter gene family in peach (Prunus persica). Genet. Mol. Res. 14 774–787. 10.4238/2015.January.30.21 [DOI] [PubMed] [Google Scholar]
- Very A. A., Nieves-Cordones M., Daly M., Khan I., Fizames C., Sentenac H. (2014). Molecular biology of K+ transport across the plant cell membrane: what do we learn from comparison between plant species? J. Plant Physiol. 171 748–769. 10.1016/j.jplph.2014.01.011 [DOI] [PubMed] [Google Scholar]
- Vicente-Agullo F., Rigas S., Desbrosses G., Dolan L., Hatzopoulos P., Grabov A. (2004). Potassium carrier TRH1 is required for auxin transport in Arabidopsis roots. Plant J. 40 523–535. 10.1111/j.1365-313X.2004.02230.x [DOI] [PubMed] [Google Scholar]
- Wigoda N., Moshelion M., Moran N. (2014). Is the leaf bundle sheath a “smart flux valve” for K+ nutrition? J. Plant Physiol. 171 715–722. 10.1016/j.jplph.2013.12.017 [DOI] [PubMed] [Google Scholar]
- Yang T., Zhang S., Hu Y., Wu F., Hu Q., Chen G., et al. (2014). The role of a potassium transporter OsHAK5 in potassium acquisition and transport from roots to shoots in rice at low potassium supply levels. Plant Physiol. 166 945–959. 10.1104/pp.114.246520 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
| List of HAK/KUP/KT sequences used in Figure 1 (Excel file).