Abstract
The GluD2 receptor is a fundamental component of postsynaptic sites in Purkinje neurons, and is required for normal cerebellar function. GluD2 and the closely related GluD1 are classified as members of the ionotropic glutamate receptor (iGluR) superfamily on the basis of sequence similarity, but do not bind l-glutamate. The amino acid neurotransmitter D-Ser is a GluD2 receptor ligand, and endogenous D-Ser signaling through GluD2 has recently been shown to regulate endocytosis of α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid–type iGluRs during synaptic plasticity in the cerebellum, such as long-term depression. Here, we investigate the pharmacology of the orthosteric binding site in GluD2 by examining the activity of analogs of D-Ser and GluN1 glycine site competitive antagonists at GluD2 receptors containing the lurcher mutation (GluD2LC), which promotes spontaneous channel activation. We identify several compounds that modulate GluD2LC, including a halogenated alanine analog as well as the kynurenic acid analog 7-chloro-4-oxo-1H-quinoline-2-carboxylic acid (7-chlorokynurenic acid; 7-CKA). By correlating thermodynamic and structural data for 7-CKA binding to the isolated GluD2 ligand binding domain (GluD2-LBD), we find that binding 7-CKA to GluD2-LBD differs from D-Ser by inducing an intermediate cleft closure of the clamshell-shaped LBD. The GluD2 ligands identified here can potentially serve as a starting point for development of GluD2-selective ligands useful as tools in studies of the signaling role of the GluD2 receptor in the brain.
Introduction
The GluD2 receptor (also known as delta2 or Gluδ2) and its closely related paralogue GluD1 are classified to the ionotropic glutamate receptor (iGluR) superfamily on the basis of sequence similarity to the 16 other known iGluR subunit genes (Araki et al., 1993; Lomeli et al., 1993). Similar to the other iGluR subtypes [i.e., the N-methyl-d-aspartate (NMDA), α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA), and kainate receptors], GluD2 receptors are predominantly localized in the postsynaptic density of excitatory synapses in the central nervous system (CNS). GluD2 was previously designated as an “orphan” iGluR, as no endogenous ligands had been identified that could bind and activate the receptor (Araki et al., 1993; Lomeli et al., 1993). D-Ser and glycine have now been identified as ligands for GluD2 (Naur et al., 2007). X-ray crystal structures of the isolated GluD2 ligand binding domain (GluD2-LBD) show that ligand binding to the clamshell-shaped LBD induces a closed-cleft conformation similar to those observed for NMDA receptor (NMDAR), AMPA receptor, and kainate receptor subtypes (Naur et al., 2007). Ligand-induced closure of the LBD is the initial conformational change that triggers ion channel gating in NMDA, AMPA, and kainate receptors, but ligand binding to the GluD2-LBD does not promote gating of the transmembrane ion channel when GluD2 is expressed as a homomeric tetramer. Thus, by its lack of ionotropic activity, the function of the GluD2 receptor in excitatory transmission appears to be fundamentally different from other iGluRs.
The role of GluD2 in the CNS is most studied in the cerebellum, where GluD2 receptors are expressed in glutamatergic synapses of the Purkinje-type neurons (Hirai et al., 2003; Yuzaki, 2004, 2009). A key role for GluD2 in postsynaptic functions in cerebellar Purkinje neurons, including induction of cerebellar long-term depression (LTD), a form of synaptic plasticity that underlies motor learning, has been demonstrated (Hirai et al., 2003; Yuzaki, 2003). Considerable experimental evidence exists that argues against an ionotropic synaptic signaling function of GluD2. For example, it has been demonstrated that transgenic mice carrying GluD2 mutations in the putative ion channel that abolish ion permeation have a normal phenotype and show intact cerebellar LTD (Kakegawa et al., 2007). Still, endogenous D-Ser binding to GluD2 has been shown to regulate LTD in Purkinje neurons (Kakegawa et al., 2011). This modulation required the intact intracellular C-terminal domain (CTD) of GluD2, which interacts with a range of scaffolding and signaling proteins. It can be speculated that D-Ser binding to the extracellular LBD may induce conformational changes at the CTD that potentially control GluD2 interactions with intracellular effector proteins required for LTD induction, suggesting a possible metabotropic signaling role of GluD2 conceptually akin to the recently identified metabotropic component of ligand-mediated NMDA receptor signaling in hippocampal LTD (Nabavi et al., 2013). In addition to a direct signaling role, the extracellular part of GluD2 binds the protein Cbln1, which is secreted from cerebellar granule cells, and this interaction is essential for synapse integrity between Purkinje cells and cerebellar granule cells in adult mice (Miura et al., 2009; Matsuda et al., 2010; Yuzaki, 2010, 2011; Matsuda and Yuzaki, 2011). These data support the idea that GluD2 also has a structural role in synapse formation (Ito-Ishida et al., 2014).
The realization of an active signaling function of GluD2 prompts development of GluD2-selective ligands that can be used as pharmacological tools to further address the molecular function as well as the cellular mechanisms of the receptor. The lurcher mutant of GluD2 (GluD2LC) exhibits spontaneous ion channel activity in the absence of ligand binding (Zuo et al., 1997; Wollmuth et al., 2000), and a previous study identified several ligands capable of blocking the open ion channel of GluD2LC receptors (Williams et al., 2003). Although D-Ser and glycine do not activate currents at wild-type GluD2, both ligands can inactivate spontaneous activated GluD2LC receptors, showing that ligand binding to the LBD can induce conformational changes within the full-length GluD2 receptor (Naur et al., 2007; Hansen et al., 2009). In the present study, we characterized a series of new compounds at GluD2LC. We identified several compounds with potency and activity similar to D-Ser at GluD2LC, including the kynurenic acid analog 7-chloro-4-oxo-1H-quinoline-2-carboxylic acid (7-CKA). Using X-ray crystallography and isothermal titration calorimetry, we established the structural and thermodynamic basis of 7-CKA binding at the isolated GluD2-LBD.
Materials and Methods
Materials.
Ligands were purchased from Tocris (Ellisville, MO), Ascent Scientific (Weston-super-Mare, UK), Enamine (Monmouth Junction, NJ), BioNet (Cornwall, UK), InterBioScreen (Chernogolovka, Russia), Alinda Chemical (Moscow, Russia), Maybridge (Leicestershire, UK), Chembridge (San Diego, CA), Princeton BioMolecular Research (Princeton, NJ), Matrix Scientific (Columbia, SC), and Sigma-Aldrich (St. Louis, MO).
DNA Constructs, cRNA Synthesis, and Protein Expression.
Rat cDNA for GluD2 (GenBank number U08256) was provided by Dr. J. Boulter (University of California, Los Angeles, CA) and sub-cloned into a modified pCI-neo vector (Promega Corp., Madison, WA) as previously described (Hansen et al., 2009). cRNA was synthesized in vitro using the mMessage mMachine kit (Applied Biosystems, Austin, TX). For recombinant bacterial expression and purification of the LBD from rat GluD2, the GluD2-S1S2 construct was used as previously described (Naur et al., 2007). The concentration of protein was determined using the Bradford protein assay.
Electrophysiology.
cRNA for rat GluD2 (hereafter GluD2) and GluD2LC (alanine to serine mutation; GluD2-A754S) was injected into Xenopus laevis oocytes, as previously described (Hansen et al., 2009). Two-electrode voltage-clamp current recordings were made 48–72 hours post-injection. The recording solution contained (in mM) 90 NaCl, 3 KCl, 10 HEPES, and 0.5 BaCl2 (pH 7.6). Solution exchange was computer controlled through an 8-modular valve positioner (Digital MVP Valve; Hamilton Company, Reno, NV). Voltage and current electrodes were filled with 0.3 and 3.0 M KCl, respectively, and current responses were recorded at a holding potential of −40 to −60 mV. Data acquisition and voltage control were accomplished with a two-electrode voltage-clamp amplifier (OC-725; Warner Instruments, Hamden, CT). During screening of compounds at GluD2LC, the ligand activity was expressed as percentage of the mean response to 1 mM D-Ser (∼4-fold higher than D-Ser EC50). D-Ser was applied at the beginning and end of each protocol. Test compounds were applied at either 100 µM or 1 mM, depending on the solubility and the availability of the compound. In general, concentration-response data were obtained from at least six oocytes from two different frogs. Concentration-response data for individual oocytes were normalized to the maximal response to D-Ser (10 mM) determined in the same recording and fitted by the Hill equation. Fitted EC50 values, Hill slopes, and maximal relative responses (i.e., relative efficacy) from individual oocytes were used to calculate the mean and S.E.M.
Crystallization and Structure Determination.
For crystallization experiments, 7-CKA was added in excess of saturation to a GluD2-LBD protein solution of 6.2 mg/ml in 20 mM HEPES (pH 7.0), 10 mM NaCl, and 1 mM EDTA. This gave rise to crystal formation in the test tube within a few days. Crystals were cryoprotected by transferring 1 µl of protein solution containing crystals onto a cover slide and adding glycerol to a final concentration of 30% (w/v) before flash-cooling in liquid nitrogen. A data set extending to 2.5-Å resolution was obtained at 100 K at the I911-2 beamline at MAX-laboratory (Lund, Sweden) (Table 3). The data were indexed and integrated using the diffraction data-integration program iMosflm (MRC Laboratory of Molecular Biology, Cambridge, UK) and scaled using the program Scala in the CCP4 software package (CCP4, Oxon, UK) (Winn, 2003). The structure was solved by molecular replacement using the program Phaser implemented in CCP4 and the apo structure of GluD2-LBD (PDB entry 2V3T; molecule A) as a search model.
TABLE 3.
Data collection and refinement statistics for GluD2-LBD in complex with 7-CKA
| Space Group | P43212 |
|---|---|
| Unit cell (Å) | a = b = 70.2, c = 134.5 |
| No. per a.u.a | 1 |
| Crystal mosaicity (degrees) | 0.78–0.87 |
| Resolution (Å) | 30–2.5 |
| Total observations | 62,324 |
| Unique observations | 12,222 |
| I/σ(I)b | 13.4 (2.8) |
| Completeness (%)b | 99.6 (100) |
| Rmerge (%)b,c | 7.2 (51.9) |
| Rwork (%)d | 19.2 |
| Rfree (%)e | 23.9 |
| No. of GluD2 residues/7-CKA/chloride/glycerol/water | 258/3/1/1/24 |
| Average B-values protein/7-CKA/chloride/glycerol/water (Å2) | 53/40/71/57/44 |
| Root mean square (rms) bond lengths/angles (Å/degree) | 0.003/0.7 |
| Ramachandran favored/outliers (%)f | 98.0/0.4(Asp504) |
Number of protein molecules per asymmetric unit (a.u).
Values in parentheses are statistics for the highest resolution bin (2.66–2.50 Å).
Rmerge (I) = ∑hkl |Ihkl-<Ihkl>| / ∑hklIhkl,, where Ihkl is the measured intensity of the reflections with indices hkl.
Rwork = Σhkl(||Fo,hkl| − |Fc,hkl||)/|Fo,hkl|, where |Fo,hkl| and |Fc,hkl| are the observed and calculated structure factor amplitudes.
Five percent of the reflections in the data set were set aside for calculation of the Rfree value.
The Ramachandran plot was calculated according to MolProbity (Davis et al., 2007) implemented in Phenix.
Ab initio calculations on 7-CKA were performed using the B3LYP function (Lee et al., 1988; Becke, 1993) to determine geometries and energies. Two tautomeric states of 7-CKA were built and geometry optimizations were performed with the split valence polarization (SVP) basis set (Schäfer et al., 1992). The final energies were determined with the triple zeta valence basis set TZVPP (Weigend et al., 1998). All calculations were performed with Turbomole 6.3 (University of Karlsruhe and Forschungszentrum Karlsruhe GmbH, 1989–2007, TURBOMOLE GmbH, since 2007; available from http://www.turbomole.com) and with continuum conductor-like screening model COSMO (Klamt and Schuurmann, 1993) with a dielectric constant of 80.
The protein structure was subjected to simulated annealing refinement followed by iterative cycles of model building in Coot (MRC Laboratory of Molecular Biology, Cambridge, UK) (Emsley and Cowtan, 2004) and refinement in CNS (Yale University, New Haven, CT) (Brunger et al., 1998) and Phenix (Lawrence Berkeley Laboratory, Berkeley, CA) (Adams et al., 2010) until the ratio between the reliability factor (Rwork) and Free R-Factor (Rfree) Rwork/Rfree converged. For refinement statistics, see Table 3. The atomic coordinates of the GluD2-LBD structure in complex with 7-CKA have been deposited in the Protein Data Bank with accession code 5CC2. Degree of domain closure was calculated using DynDom (Hayward and Berendsen, 1998).
Isothermal Titration Calorimetry.
Isothermal titration calorimetry (ITC) was carried out using a VP-ITC calorimeter from MicroCal, LLC (Northampton, MA) as previously described (Naur et al., 2007). In brief, the calorimeter cell (1.40 ml) was filled with purified GluD2-LBD in ITC buffer (in mM: 100 HEPES, 100 NaCl, 2 KCl, pH 7.5) at 20°C. For D-Ala, 50 injections of 20 mM ligand solutions in ITC buffer were carried out at 3-minute intervals, with 3 μl in the initial injection and 6 μl in the subsequent injections. β-Fluoro-DL-alanine (20 mM in ITC buffer) was titrated with 20 injections of 15 μl into the GluD2-LBD solution at 3-minute intervals (1 mg/ml). 7-CKA (1 mM in ITC buffer) was titrated with 3-minute intervals and the initial injection of 3 μl and the remaining of 10 μl into a 2.5-mg/ml solution of GluD2-LBD. The heat of dilution obtained from injecting the ligand into buffer was subtracted before the fitting process. The data analysis was performed using Origin 7.0 (MicroCal) for ITC by using a single binding-site model. The first data points in the analysis were discarded. The stoichiometry N was fixed to 1 in the fitting process.
Data Analysis.
Data were analyzed with the program GraphPad Prism 6.0 (GraphPad Software, San Diego, CA) and the program Clampfit (Axon Instruments, Union City, CA). Composite concentration-response data were fitted to the Hill equation:
ΔI = ΔImax/(1+10^((log EC50-log[A])* nH))
where ΔImax is the maximum current in response to the ligand, nH denotes the Hill coefficient, [A] is the ligand concentration, and EC50 is the ligand concentration that produces half-maximum current response. The EC50 and nH from the individual oocytes were used to calculate the mean and S.E.M. For graphical presentation, data sets from individual oocytes were normalized to the maximum current response in the same recording, making it possible to calculate the mean and the S.E.M. for each data point. The averaged data points were then fitted to the Hill equation and plotted together with the resulting curve.
Results
Screening of Compounds as GluD2 Ligands.
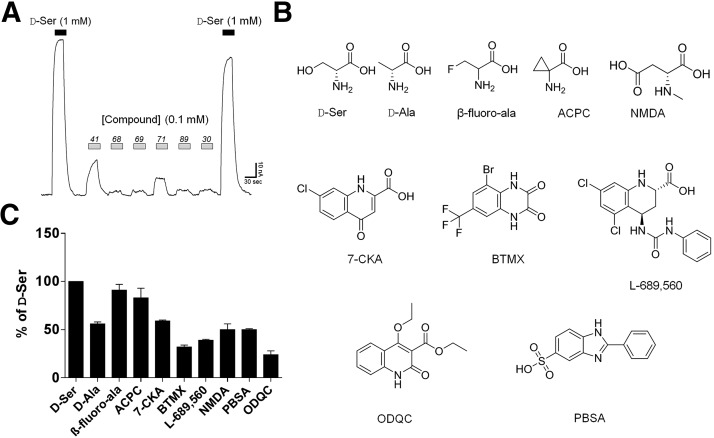
We have previously found that D-Ser, glycine, and D-Ala are capable of binding to GluD2. Although these ligands cannot initiate ion permeation in full-length wild-type GluD2 receptors expressed in heterologous systems, they can inactivate the spontaneous currents generated by GluD2 carrying the lurcher mutation (Naur et al., 2007; Hansen et al., 2009). Thus, we hypothesized that measurement of GluD2LC current can be used to detect novel ligands for the orthosteric binding site in GluD2. We assembled a library of commercially available analogs of D-Ser and other amino acids (Supplemental Tables S1–S4). The GluD2 orthosteric binding site is most similar to the glycine binding site of the NMDAR GluN1 subunit, which also binds D-Ser and glycine. Therefore, we speculated that GluN1 ligands might have affinity for GluD2, and we thus included a range of known GluN1 competitive antagonists and structural analogs hereof in the library (Supplemental Table S4). We initially screened the activity of the library compounds (91 in total) at GluD2LC receptors expressed in Xenopus oocytes using two-electrode voltage-clamp electrophysiology (Fig. 1; Supplemental Tables S1–S4). Compound activity was defined as a change in the spontaneous activity of GluD2LC induced by compound application at concentrations of either 100 µM or 1 mM, depending on the solubility of the compound. No compounds induced an increase in the GluD2LC-mediated spontaneous current (data not shown). Nine compounds were found to inactivate GluD2LC currents by ≥50% of the inactivation produced by a similar concentration of D-Ser (Fig. 1; Supplemental Table S1). None of these compounds were capable of producing a detectable current response at recombinant wild-type GluD2 expressed in Xenopus oocytes (N = 4–8 oocytes for each compound).
Fig. 1.
Identification of novel GluD2 ligands. (A) Representative current traces illustrate the recording protocol used for compound screening at GluD2LC receptors expressed in Xenopus oocytes. Compounds were screened at a concentration of 0.1 or 1 mM, depending on solubility and availability. Numbers refer to compounds in the screening library (see Supplemental Tables S1–S4). (B and C). Chemical structures and summary of activity of compounds identified to have more than 50% activity of D-Ser (1 mM) when applied at a concentration of 1 mM or more than 20% activity when applied at a concentration of 100 µM: D-Ala, β-fluoro-DL-alanine (β-fluoro-Ala), 1-aminocyclopropane carboxylic acid (ACPC), (R)-2-(methylamino)succinic acid (NMDA), 7-CKA, 5-bromo-7-(trifluoromethyl)-1,4-dihydroquinoxaline-2,3-dione (BMTX), (2S,4R)-5,7-dichloro-4-(3-phenylureido)-1,2,3,4-tetrahydroquinoline-2-carboxylic acid (L-689,560), 2-phenyl-1H-benzo[d]imidazole-5-sulfonic acid (PBSA), and ethyl 4-ethoxy-2-oxo-1,2-dihydro-3-quinolinecarboxylate (ODQC).
Concentration-Response Relationship for Novel GluD2 Ligands.
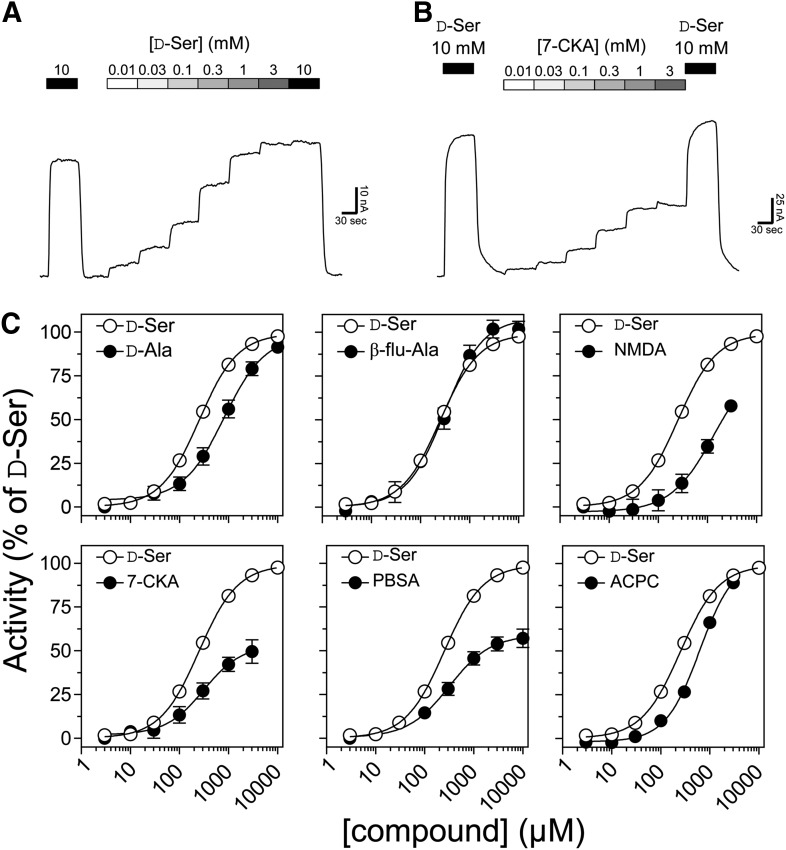
Among the compounds identified to display the highest activity relative to D-Ser for inactivation of GluD2LC current (Fig. 1), we selected compounds representing different chemical classes for generation of full concentration-response data at GluD2LC to determine compound potency (i.e., EC50) and relative efficacy compared with that of D-Ser (Fig. 2). The resulting fitted EC50 values, Hill slopes, and relative efficacies are summarized in Table 1. None of the tested compounds displayed higher potency than D-Ser. The two most potent compounds were the D-Ser analog β-fluoro-DL-alanine and the NMDAR subunit GluN1 competitive antagonist 7-chloro-4-oxo-1H-quinoline-2-carboxylic acid) (7-CKA), which inactivated GluD2LC with potencies close to that of D-Ser. EC50 values for inhibition were 312 and 306 µM for β-fluoro-DL-alanine and 7-CKA, respectively, compared with 250 µM for D-Ser (Fig. 2; Table 1).
Fig. 2.
Concentration-effect relationship for inhibition of GluD2LC currents by selected compounds. Representative current traces for D-Ser (A) and 7-CKA (B) illustrate the recording protocol used for determination of compound potency for inhibition of constitutively active GluD2LC receptors expressed in Xenopus oocytes. (C) Average composite concentration-response curves. Error bars are the S.E.M. and are shown when larger than symbol size. The current responses are normalized to the maximal response produced by D-Ser. ACPC, 1-aminocyclopropane carboxylic acid; BMTX, 5-bromo-7-(trifluoromethyl)-1,4-dihydroquinoxaline-2,3-dione; β-flu-Ala, β-fluoro-DL-alanine; PBSA, 2-phenyl-1H-benzo[d]imidazole-5-sulfonic acid.
TABLE 1.
Compund potency at GluD2LC
| Compound |
EC50a |
Hill Slope |
Efficacy |
|---|---|---|---|
| µM | % D-Ser | ||
| D-Ser | 250 (214–293) | 1.1 (0.9–1.3) | 100 |
| 7-CKA | 312 (236–413) | 1.1 (0.8–1.4) | 53 (47–59) |
| ACPC | >500 | ND | ND |
| β-fluoro-DL-alanine | 306 (270–348) | 1.1 (1.0–1.2) | 108 (103–112) |
| NMDA | >1000 | ND | ND |
| PBSA | 317 (225–448) | 1.0 (0.7–1.3) | 59 (55–63) |
| D-Ala | 805 (686–945) | 1.0 (0.9–1.2) | 98 (92–103) |
ACPC, 1-aminocyclopropane carboxylic acid; PBSA, 2-phenyl-1H-benzo[d]imidazole-5-sulfonic acid.
EC50 values were determined by the nonlinear fitting of concentration-inhibition data collected from 6–10 oocytes (see Materials and Methods). Numbers in parentheses denote the 95% confidence interval for the fitted EC50.
Thermodynamic Characterization of Ligand Binding to the GluD2 Ligand Binding Domain.
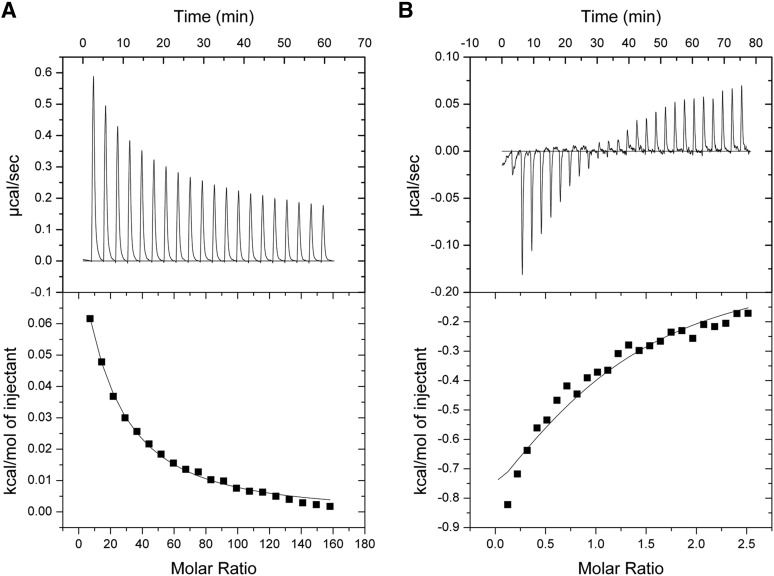
To further study the ligand-binding characteristics of the two most potent new ligands, 7-CKA and analog β-fluoro-DL-alanine, we performed ITC to determine the binding affinity (Ka, ΔG), enthalpy (ΔH), and entropy (ΔS) of ligands for the purified GluD2-LBD (Fig. 3; Table 2). We also included D-Ala in the ITC analysis. ITC previously has been used to study ligand binding to isolated LBDs from the AMPA-type iGluR subunit GluA2 (Kasper et al., 2006; Martinez et al., 2014), GluA4 (Madden et al., 2000), and GluD2 (Naur et al., 2007). Interestingly, 7-CKA binds GluD2-LBD with the highest affinity among compounds characterized by ITC (10-fold higher Ka than D-Ser). Furthermore, 7-CKA has a thermodynamic profile that is very different compared with those of the amino acid ligands β-fluoro-DL-alanine, D-Ser, D-Ala, and glycine (Table 2). Whereas the amino acid ligands elicit a small positive change in enthalpy, and thus their binding is purely entropy-driven, 7-CKA binds with both favorable enthalpy and entropy. Previous ITC analyses of ligand binding to isolated LBDs from AMPA receptors have shown that ligands stabilizing the closed-cleft conformations of the LBD differ from ligands stabilizing an extended cleft conformation with respect to their thermodynamic profile (Kasper et al., 2006; Martinez et al., 2014). On the basis of this, the observed differences in thermodynamic profiles may indicate that 7-CKA has a different binding mode than β-fluoro-DL-alanine, D-Ser, and glycine, and further suggest that 7-CKA may stabilize a distinctly different LBD conformation than the amino acid ligands.
Fig. 3.
Isothermal titration calorimetry studies on GluD2-LBD. A plot of energy transfer rate as a function of molar ratio (top panels) and integrated data after subtraction of the heat of dilution (bottom panels) of β-fluoro-DL-alanine (A) and 7-CKA (B) is shown. The solid lines represent the best-fit curves to the data, using a one-site binding model (Materials and Methods).
TABLE 2.
Thermodynamic characteristics of ligand binding to the isolated GluD2-LBD determined by ITC
| Compound | Kd | ΔG | ΔH | −TΔS |
|---|---|---|---|---|
| µM | kcal/mol | kcal/mol | kcal/mol | |
| Glycinea | 2754 | −3 | 2 | −5 |
| D-Sera | 925 | −4 | 3 | −7 |
| D-Ala | 1919 | −4 | 2 | −5 |
| β-fluoro-DL-alanine | 1519 | −4 | 4 | −7 |
| 7-CKA | 80 | −6 | −2 | −4 |
From Naur et al. (2007).
Structural Analysis of 7-CKA–Bound GluD2-LBD by X-ray Crystallography.
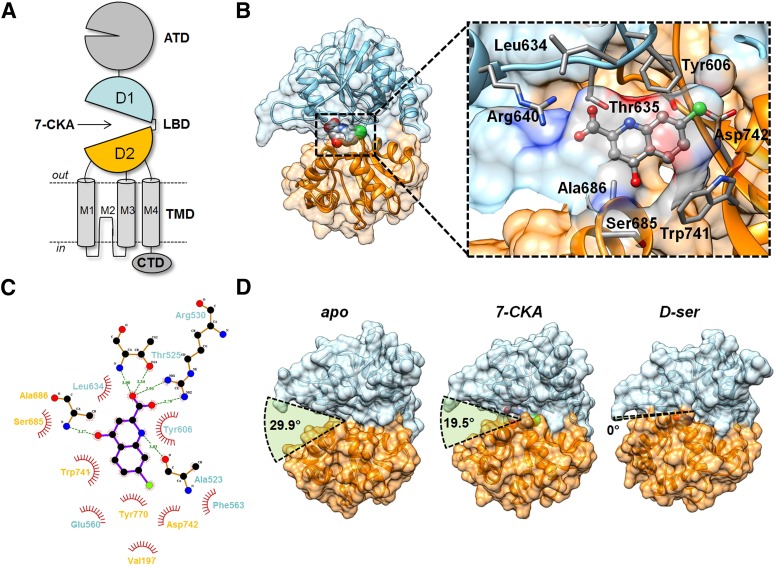
To determine the binding mode of 7-CKA to GluD2 and the molecular details underlying its different thermodynamic profile, we crystallized GluD2-LBD in complex with 7-CKA. The structure of the 7-CKA–bound GluD2-LBD was determined to 2.5 Å resolution and revealed that the complex crystallizes as a monomer with one GluD2 LBD protomer in the asymmetric unit of the crystal (Table 3). The electron density of 7-CKA in the ligand-binding site of the GluD2-LBD is well defined and allowed unambiguous positioning of the ligand and identification of ligand contacts (Fig. 4). The structure showed one molecule of 7-CKA bound within the cleft formed by domains D1 and D2 of the clamshell-shaped LBD structure (Fig. 4, A and B). Similar to D-Ser (Naur et al., 2007), 7-CKA is anchored to the LBD through a bidentate salt bridge formed between the carboxylate group of 7-CKA and the side-chain guanidine group of Arg530 (Fig. 4, B and C). In addition, one carboxylate oxygen atom of 7-CKA forms a hydrogen bond to the backbone nitrogen of Thr525. A hydrogen bond is also seen between the ring nitrogen atom of 7-CKA and the backbone carbonyl of Ala523, suggesting that the ring nitrogen atom is protonated, and that 7-CKA therefore binds in its keto form. To further investigate this, we performed ab initio calculations of the two tautomeric states to determine which state is dominant in solution. The results showed that the keto form had an energy 12.3 kcal/mol lower than the enol form, supporting the proposed hydrogen bond between the ring amine group and the backbone carbonyl of Ala523 (Fig. 4C). The carbonyl oxygen atom of 7-CKA forms contacts to the backbone nitrogen of Ala686 and two water molecules. 7-CKA is surrounded by several hydrophobic residues, of which the ligand forms optimal stacking with Tyr496 (Fig. 4C). In addition, two 7-CKA molecules were found to bind to surface residues of GluD2-LBD (see Supplemental Fig. 1). Compared with the previously reported structure of the GluD2-LBD in complex with D-Ser (Naur et al., 2007), there is a striking difference in the degree of cleft opening between domains D1 and D2 of the GluD2-LBD in the 7-CKA–bound structure (Fig. 4D). Relative to the D-Ser–induced conformation, the D1 and D2 domain opening of GluD2-LBD in complex with 7-CKA is 19.5° more open. Thus, the 7-CKA–induced GluD2-LBD conformation is closer to the apo conformation, which has a domain opening of 29.9° relative to the D-Ser–induced conformation.
Fig. 4.
X-ray structure of GluD2-LBD in complex with 7-CKA. (A) Schematic illustration of the modular architecture of a GluD2 receptor subunit that contains an N-terminal domain (ATD) and a bilobed LBD with D1 and D2 subdomains that are connected to a transmembrane domain (TMD) consisting of three membrane-spanning segments (M1, M3, and M4) and a short membrane re-entrant segment (M2) and a CTD. (B) Ribbon representation of the crystal structure of the isolated GluD2-LBD with 7-CKA bound. The D1 and D2 subdomains are highlighted in orange (D1) and blue (D2). The molecular structure of 7-CKA (shown as stick representation) bound in the cleft between D1 and D2 subdomains is shown as an expanded inset. The binding pocket surface is shown as transparent contour, and key interacting side chains are shown as sticks. The GluD2 amino acid numbering starts with the initiating methionine in the full-length GluD2 sequence that includes the signal peptide. (C) Two-dimensional representation of the binding pocket generated using LigPlot (European Bioinformatics Institute, Cambridge, UK) (Wallace et al., 1995), showing contacts to binding site residues. (D) Domain opening of the GluD2-LBD in the apo (left), 7-CKA (center) and D-Ser (right) bound states. The domain openings are relative to that induced by D-Ser. ATD, amino-terminal domain.
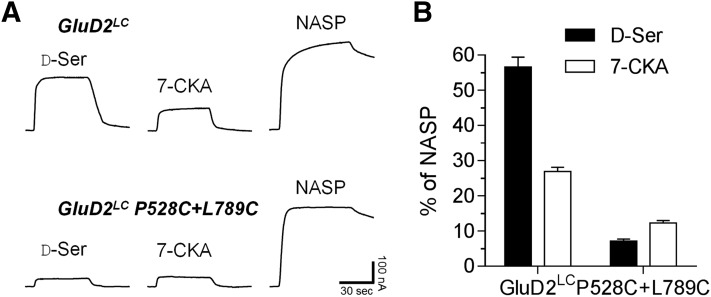
Crystal structures of isolated LBDs from AMPA and NMDA receptor subunits have shown that antagonists unambiguously stabilize LBD conformations with domain opening similar or close to the apo conformation (Pohlsgaard et al., 2011). By contrast, iGluR agonists induce substantial domain closure, which triggers subsequent conformational changes that lead to channel opening (Armstrong and Gouaux, 2000; Pohlsgaard et al., 2011). Comparison of the degree of domain closures in the GluD2-LBD for structures in the apo conformation and with bound 7-CKA or D-Ser suggests that 7-CKA binding at full-length GluD2 induces fewer conformational changes relative to D-Ser, and raises the possibility that 7-CKA is a competitive antagonist at the D-Ser binding site. The GluD2 signaling mechanism has not yet been reconstituted in heterologous expression systems, and at present, it is not possible to directly examine whether ligands have agonist or antagonist activity. So far, direct ligand activity at the wild-type GluD2 receptor can only be studied via effects on the spontaneously active current generated by GluD2LC mutant receptors. Considering the idea of 7-CKA as a potential antagonist of GluD2, it is perhaps surprising that 7-CKA produces similar effect as D-Ser at GluD2LC receptors, e.g., inactivation of the spontaneously active current although with lower efficacy (Fig. 2; Table 1). However, it has been previously shown that reduction of GluD2LC activity by D-Ser results from conformational changes at the LBD dimer interface (Hansen et al., 2009). Interestingly, conformational changes at the LBD dimer interface reflect desensitization of AMPA and kainate receptors (Armstrong et al., 2006; Plested and Mayer, 2009; Daniels et al., 2013; Schauder et al., 2013; Durr et al., 2014; Meyerson et al., 2014), and the extent of desensitization of AMPA receptors is correlated with the degree of domain closure (Jin and Gouaux, 2003). That is, ligands that induce full domain closure desensitize AMPA receptors to a higher extent than ligands that induce less closure. The present finding that 7-CKA inactivates GluD2LC currents less than D-Ser at saturating conditions (Fig. 2C) may suggest that inactivation of GluD2LC channel activity is linked to desensitization and is correlated to the degree of domain closure. However, an alternative mechanism could be that 7-CKA inhibits spontaneous GluD2LC channel opening by stabilizing the LBD in a more open conformation, in contrast to D-Ser, which inhibits by closing the LBD to promote desensitization. To distinguish between these two mechanisms of GluD2LC channel inhibition by 7-CKA, we introduced an engineered disulfide bond at the interface between two GluD2-LBDs (Naur et al., 2007). This engineered disulfide bond (GluD2LC P528+L789C) has been shown to be efficiently formed and to prevent the reduction of GluD2LC activity by D-Ser resulting from conformational changes at the LBD dimer interface (i.e., desensitization) (Hansen et al., 2009). At GluD2LC, 10 mM D-Ser reduced spontaneous channel activity by 57 ± 3% compared with complete channel block by 100 µM 1-naphthyl acetyl spermine, whereas 1 mM 7-CKA inhibited GluD2LC by 27 ± 1% (N = 8) (Fig. 5). GluD2LC P528+L789C receptors with cross-linked dimer interface were inhibited 7 ± 1% by 10 mM D-Ser and 12 ± 1% by 1 mM 7-CKA (N = 6). Thus, cross-linking the GluD2LC dimer interface reduced D-Ser responses by 8.1-fold, whereas 7-CKA responses were modestly reduced by 2.3-fold (Fig. 5), hereby making 7-CKA more efficacious than D-Ser for reducing spontaneous channel activity. This result suggests that 7-CKA responses are mediated by conformational changes that are distinct from those induced by D-Ser binding, consistent with a mechanism by which 7-CKA inhibits spontaneous GluD2LC channel opening by stabilizing the LBD in a more open conformation.
Fig. 5.
Effects of cross-linking the LBD dimer interface in GluD2LC on D-Ser and 7-CKA responses. (A) Representative current traces for responses to 10 mM D-Ser, 1 mM 7-CKA, and 100 µM 1-naphthyl acetyl spermine (NASP) on GluD2LC and GluD2LC P528C+L789C expressed in Xenopus oocytes. The P528C+L789C mutations cross-link the LBD dimer interface in GluD2LC by the formation of a disulfide bond. (B) Bar graph of the mean responses to D-Ser and 7-CKA as a percentage of complete inhibition by the channel blocker NASP. Error bars are the S.E.M. and the data are from six to eight oocytes.
Discussion
The identification of a possible ligand-mediated signaling role of the GluD2 receptor through occupancy of the orthosteric binding site suggests that development of selective ligands for this site could be useful tools for studying the poorly understood cellular and molecular function of GluD2 (Wollmuth et al., 2000; Williams et al., 2003; Hansen et al., 2009; Kakegawa et al., 2011; Orth et al., 2013). Of particular importance is the development of a GluD2-selective antagonist, which could allow further investigation of the role of GluD2 in numerous processes. As a first step toward the development of new GluD2-selective ligands, we have exploited a focused library to identify ligands that constitutively modulate GluD2LC channel activity. This allowed us to identify several new GluD2 ligands, including D-Ser analogs as well as structurally diverse compounds related to known iGluR antagonists. Among the latter group, we focused on the well known NMDA receptor glycine-site competitive antagonist 7-CKA. The molecular mechanism of GluD2 signaling must include conformational changes initiated by D-Ser binding to the LBD, which can be speculated to propagate to structural rearrangement of the CTD. The CTD is linked to an array of intracellular interaction partners that may serve as effector molecules for regulation of processes within the postsynaptic signaling complex that contribute to synaptic plasticity. The crystallographic analysis of the GluD2-LBD shows that 7-CKA stabilizes the domain in a more open conformation relative to D-Ser (Fig. 4). Furthermore, ITC and cross-linking experiments support that 7-CKA induces conformational changes that are distinct from those induced by D-Ser. The observation that 7-CKA stabilizes the LBD in a conformation that is closer to the apo conformation than to the D-Ser–bound conformation may indicate that 7-CKA binding in the intact GluD2 will not induce the receptor conformational changes that underlie D-Ser signaling. In concert, these analyses may suggest that 7-CKA is a competitive antagonist at the D-Ser binding site. Indeed, in their demonstration of D-Ser regulation of cerebellar LTD via GluD2, Kakegawa et al. (2011) found that 7-CKA inhibits LTD induction independently of its activity at NMDARs. However, it should be noted that, until the molecular mechanism of GluD2 signaling is identified, it is not possible to directly test whether effects of ligand binding to the orthosteric site are agonistic or antagonistic. Therefore, the suggestion that 7-CKA is a competitive antagonist at GluD2 is highly speculative.
Among the compounds for which EC50 for GluD2LC inactivation and Kd for binding to the isolated GluD2-LBD were determined, 7-CKA and β-fluoro-DL-alanine were the most potent, with EC50 and Kd in the lower micromolar range (Tables 1 and 2). The results from our initial screening provide some initial insight into the structure-activity relationship that might be of value for future efforts to improve potency. For example, agonist activity at iGluRs in general requires the LBD to adopt a conformation in which the D1 and D2 lobes are near-fully or fully closed around the ligand (Pohlsgaard et al., 2011; Kumar and Mayer, 2013). This may pose specific limits for the size of the agonist structure that depend on the volume of the ligand binding cavity. The structure of D-Ser bound to GluD2-LBD (Naur et al., 2007) shows the ligand binding pocket of GluD2 to be the smallest in volume among all iGluR LBD structures determined so far, with D-Ser completely filling the cavity. The small volume may pose an important size constraint for future design efforts of new GluD2 agonists. Thus, creation of agonists with improved affinity by addition of larger functional groups to the scaffold of the endogenous agonist, as has been found possible for analogs of l-glutamate at AMPA (Vogensen et al., 2011; Juknaite et al., 2012), NMDA (Erreger et al., 2005; Clausen et al., 2008; Hansen et al., 2013), and kainate receptors (Zhou et al., 1997; Juknaite et al., 2012), may prove difficult for GluD2. This is reflected in the present ligand screen where the only D-Ser analogs that preserve activity are those in which the core functional groups are replaced with functionalities of similar size (Fig. 1; Supplemental Table S1). In particular, all substitutions at the α- and β-carbons of D-Ser lead to loss of activity (Supplemental Table S1). Furthermore, replacement of the β-hydroxyl with larger polar substituents rendered the ligand inactive (Supplemental Table S1). The structure of GluD2-LBD in complex with D-Ser (Naur et al., 2007) shows that the hydroxyl group forms a direct interaction with the binding pocket in the form of a hydrogen bond to the hydroxyl group on Tyr543, and the importance of this interaction for compound potency may be illustrated from the decreased EC50 observed for D-Ala and 1-aminocyclopropane carboxylic acid, in which the methyl hydroxyl side chain of D-Ser is removed or replaced by the nonpolar cyclopropane ring (Fig. 2; Table 1).
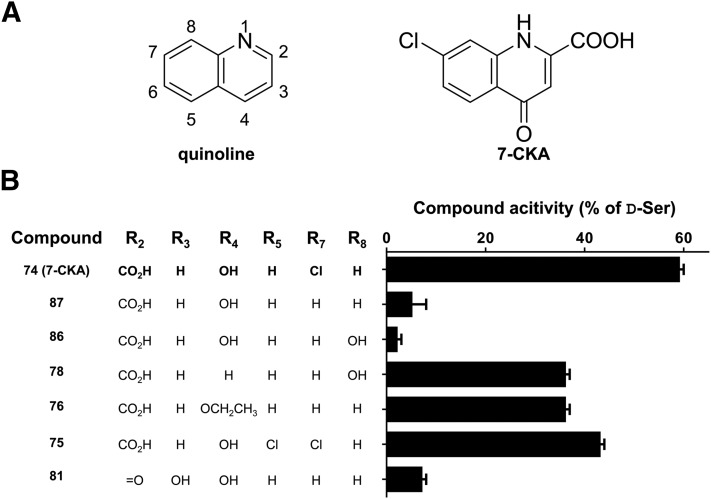
Several compounds containing the quinoline scaffold of 7-CKA were also tested (Supplemental Table S4), allowing some information regarding the 7-CKA pharmacophore to be extracted (Fig. 6). Compound 87 differs from 7-CKA only by the absence of a chlorine atom at the 7-position, and induced 5% inactivation when evaluated at a concentration of 1 mM compared with the 59% inactivation produced by a similar concentration of 7-CKA (Fig. 6), demonstrating a key role of the substituent at the 7-position of the quinoline ring in defining ligand activity. The 7-CKA structure shows the chlorine to be located in a subpocket lined by polar side-chain moieties of Phe563, Asp742, and Tyr770. Future introduction of other polar substituents with hydrogen bond formation properties may be of interest at this position. Interestingly, compounds 78 and 86 also lack chlorine at the 7-position, but have a hydroxyl substituent at the 8-position. Comparison of the activities of these analogs with that of compound 87 shows an interesting pattern with regard to the hydroxyl substituent in the 8-position and ketone in the 4-position: hydroxyl in the 8-position decreases activity when the 4-position contains a ketone (compare compound 87 with compound 86; Fig. 6), but increases activity when the 4-position is unsubstituted. The GluD2-LBD structure with 7-CKA shows that the 4-position ketone forms a hydrogen bond with the backbone nitrogen of Ala686 (Fig. 4C). Ala686 together with Ala523 and Thr525/Arg530 constitute a triangular set of interaction points for the 7-CKA molecule (Fig. 4C). Unlocking this binding mode by removing the 4-position ketone may allow different positioning of the planar quinoline scaffold in the pocket to allow polar substituents in the 8-position to form direct interactions with the binding pocket. Finally, an interesting observation is that compound 76 has activity in the range of 7-CKA (Fig. 6). Compound 76 differs from compound 87 (5% inactivation) only by having an ethoxy substituent instead of ketone in the 4-position. The ethyl moiety to the 4-position oxygen may reach into a hydrophobic subpocket lined by Trp741 (Fig. 6B). It may therefore be worthwhile to explore other alkoxy substituents in the 4-position for improvement of potency.
Fig. 6.
7-CKA structure-activity relationship. Shown is the quinoline scaffold of 7-CKA (A) with ring positions numbered from 1 to 8 along with graphical summary (B) of the activity of analogs with various combinations of ring substituents (R1–R8). Activity measured as compound inhibition of GluD2LC current tested at a compound concentration of 0.1 or 1 mM relative to the inhibition by D-Ser at a similar concentration. Compound numbers refer to the listing in Supplemental Table S4.
In summary, the present identification and characterization of new ligands for the GluD2 receptor constitute a first step toward development of GluD2-selective ligands that could be used as pharmacological tools to explore the role of GluD2 receptors in the CNS. However, further experimentation aimed at providing a more detailed picture of ligand-binding effects on GluD2 receptor function is required. Such studies might focus on understanding the structural changes that are induced by the binding of different ligands in full-length GluD2, or explore functional effects of new disease-associated human mutations that may be identified in GluD2 (Yuan et al., 2015).
Supplementary Material
Acknowledgments
The authors thank the staff at MAX-laboratory for help during data collection.
Abbreviations
- AMPA
α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid
- 7-CKA
7-chloro-4-oxo-1H-quinoline-2-carboxylic acid
- CNS
central nervous system
- CTD
C-terminal domain
- GluD2-LBD
GluD2 ligand binding domain
- GluD2LC
GluD2 receptors containing the lurcher mutation
- iGluR
ionotropic glutamate receptor
- ITC
isothermal titration calorimetry
- LTD
long-term depression
- NMDA
N-methyl-d-aspartate
- NMDAR
NMDA receptor
Authorship Contributions
Participated in research design: Kristensen, Hansen, Kastrup, Traynelis.
Conducted experiments: Kristensen, Hansen, Naur, Olsen, Kurtkaya, Dravid, Kvist, Yi.
Contributed new reagents or analytic tools: Pøhlsgaard, Clausen, Gajhede.
Performed data analysis: Kristensen, Hansen, Yi, Naur, Olsen, Kurtkaya, Dravid, Kastrup, Traynelis.
Wrote or contributed to the writing of the manuscript: Kristensen, Hansen, Olsen, Naur, Clausen Gajhede, Kastrup, Traynelis.
Footnotes
This work was supported by the National Institute of Mental Health [Grant R21-MH062204 to S.F.T.]; National Institute of General Medicine [Grant P20-GM103546 to K.B.H.]; the National Alliance for Research on Schizophrenia and Depression [to S.F.T.]; Alfred Benzon Foundation [grants to A.S.K.. and K.B.H.]; the Danish Medical Research Council [grants to A.S.K., J.S.K., L.O., M.G., and P.N.]; the GluTarget Programme of Excellence at the University of Copenhagen [grant to T.K., J.S.K., and A.S.K.]; the Danish Ministry of Science, Innovation and Higher Education’s EliteForsk Programme [travel grant to T.K.], the Lundbeck Foundation [grant to J.S.K., L.O., M.G., J.P., and P.N.A.]; the Carlsberg Foundation [grant to J.S.K., L.O., and M.G.]; Danscatt [to J.S.K., L.O., M.G., and P.N.A.]; and a University of Copenhagen Drug Research Academy stipend [to P.N.A.].
 This article has supplemental material available at molpharm.aspetjournals.org.
This article has supplemental material available at molpharm.aspetjournals.org.
References
- Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66:213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araki K, Meguro H, Kushiya E, Takayama C, Inoue Y, Mishina M. (1993) Selective expression of the glutamate receptor channel delta 2 subunit in cerebellar Purkinje cells. Biochem Biophys Res Commun 197:1267–1276. [DOI] [PubMed] [Google Scholar]
- Armstrong N, Gouaux E. (2000) Mechanisms for activation and antagonism of an AMPA-sensitive glutamate receptor: crystal structures of the GluR2 ligand binding core. Neuron 28:165–181. [DOI] [PubMed] [Google Scholar]
- Armstrong N, Jasti J, Beich-Frandsen M, Gouaux E. (2006) Measurement of conformational changes accompanying desensitization in an ionotropic glutamate receptor. Cell 127:85–97. [DOI] [PubMed] [Google Scholar]
- Becke AD. (1993) Density‐functional thermochemistry. III. The role of exact exchange. J Chem Phys 98:5648–5652. [Google Scholar]
- Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. (1998) Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr 54:905–921. [DOI] [PubMed] [Google Scholar]
- Clausen RP, Christensen C, Hansen KB, Greenwood JR, Jørgensen L, Micale N, Madsen JC, Nielsen B, Egebjerg J, Bräuner-Osborne H, et al. (2008) N-Hydroxypyrazolyl glycine derivatives as selective N-methyl-D-aspartic acid receptor ligands. J Med Chem 51:4179–4187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels BA, Andrews ED, Aurousseau MR, Accardi MV, Bowie D. (2013) Crosslinking the ligand-binding domain dimer interface locks kainate receptors out of the main open state. J Physiol 591:3873–3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, Murray LW, Arendall WB, 3rd, Snoeyink J, Richardson JS, Richardson DC. (2007) MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res 35:W375–W383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dürr KL, Chen L, Stein RA, De Zorzi R, Folea IM, Walz T, Mchaourab HS, Gouaux E. (2014) Structure and dynamics of AMPA receptor GluA2 in resting, pre-open, and desensitized states. Cell 158:778–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60:2126–2132. [DOI] [PubMed] [Google Scholar]
- Erreger K, Geballe MT, Dravid SM, Snyder JP, Wyllie DJ, Traynelis SF. (2005) Mechanism of partial agonism at NMDA receptors for a conformationally restricted glutamate analog. J Neurosci 25:7858–7866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen KB, Naur P, Kurtkaya NL, Kristensen AS, Gajhede M, Kastrup JS, Traynelis SF. (2009) Modulation of the dimer interface at ionotropic glutamate-like receptor delta2 by D-serine and extracellular calcium. J Neurosci 29:907–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen KB, Tajima N, Risgaard R, Perszyk RE, Jørgensen L, Vance KM, Ogden KK, Clausen RP, Furukawa H, Traynelis SF. (2013) Structural determinants of agonist efficacy at the glutamate binding site of N-methyl-D-aspartate receptors. Mol Pharmacol 84:114–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayward S, Berendsen HJ. (1998) Systematic analysis of domain motions in proteins from conformational change: new results on citrate synthase and T4 lysozyme. Proteins 30:144–154. [PubMed] [Google Scholar]
- Hirai H, Launey T, Mikawa S, Torashima T, Yanagihara D, Kasaura T, Miyamoto A, Yuzaki M. (2003) New role of delta2-glutamate receptors in AMPA receptor trafficking and cerebellar function. Nat Neurosci 6:869–876. [DOI] [PubMed] [Google Scholar]
- Ito-Ishida A, Okabe S, Yuzaki M. (2014) The role of Cbln1 on Purkinje cell synapse formation. Neurosci Res 83:64–68. [DOI] [PubMed] [Google Scholar]
- Jin R, Gouaux E. (2003) Probing the function, conformational plasticity, and dimer-dimer contacts of the GluR2 ligand-binding core: studies of 5-substituted willardiines and GluR2 S1S2 in the crystal. Biochemistry 42:5201–5213. [DOI] [PubMed] [Google Scholar]
- Juknaitė L, Venskutonytė R, Assaf Z, Faure S, Gefflaut T, Aitken DJ, Nielsen B, Gajhede M, Kastrup JS, Bunch L, et al. (2012) Pharmacological and structural characterization of conformationally restricted (S)-glutamate analogues at ionotropic glutamate receptors. J Struct Biol 180:39–46. [DOI] [PubMed] [Google Scholar]
- Kakegawa W, Kohda K, Yuzaki M. (2007) The delta2 ‘ionotropic’ glutamate receptor functions as a non-ionotropic receptor to control cerebellar synaptic plasticity. J Physiol 584:89–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakegawa W, Miyoshi Y, Hamase K, Matsuda S, Matsuda K, Kohda K, Emi K, Motohashi J, Konno R, Zaitsu K, et al. (2011) D-serine regulates cerebellar LTD and motor coordination through the δ2 glutamate receptor. Nat Neurosci 14:603–611. [DOI] [PubMed] [Google Scholar]
- Kasper C, Pickering DS, Mirza O, Olsen L, Kristensen AS, Greenwood JR, Liljefors T, Schousboe A, Wätjen F, Gajhede M, et al. (2006) The structure of a mixed GluR2 ligand-binding core dimer in complex with (S)-glutamate and the antagonist (S)-NS1209. J Mol Biol 357:1184–1201. [DOI] [PubMed] [Google Scholar]
- Klamt A, Schuurmann G. (1993) COSMO: a new approach to dielectric screening in solvents with explicit expressions for the screening energy and its gradient. J Chem Soc, Perkin Trans 2 (5):799–805. [Google Scholar]
- Kumar J, Mayer ML. (2013) Functional insights from glutamate receptor ion channel structures. Annu Rev Physiol 75:313–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C, Yang W, Parr RG. (1988) Development of the Colle-Salvetti correlation-energy formula into a functional of the electron density. Phys Rev B Condens Matter 37:785–789. [DOI] [PubMed] [Google Scholar]
- Lomeli H, Sprengel R, Laurie DJ, Köhr G, Herb A, Seeburg PH, Wisden W. (1993) The rat delta-1 and delta-2 subunits extend the excitatory amino acid receptor family. FEBS Lett 315:318–322. [DOI] [PubMed] [Google Scholar]
- Madden DR, Abele R, Andersson A, Keinänen K. (2000) Large-scale expression and thermodynamic characterization of a glutamate receptor agonist-binding domain. Eur J Biochem 267:4281–4289. [DOI] [PubMed] [Google Scholar]
- Martinez M, Ahmed AH, Loh AP, Oswald RE. (2014) Thermodynamics and mechanism of the interaction of willardiine partial agonists with a glutamate receptor: implications for drug development. Biochemistry 53:3790–3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda K, Miura E, Miyazaki T, Kakegawa W, Emi K, Narumi S, Fukazawa Y, Ito-Ishida A, Kondo T, Shigemoto R, et al. (2010) Cbln1 is a ligand for an orphan glutamate receptor delta2, a bidirectional synapse organizer. Science 328:363–368. [DOI] [PubMed] [Google Scholar]
- Matsuda K, Yuzaki M. (2011) Cbln family proteins promote synapse formation by regulating distinct neurexin signaling pathways in various brain regions. Eur J Neurosci 33:1447–1461. [DOI] [PubMed] [Google Scholar]
- Meyerson JR, Kumar J, Chittori S, Rao P, Pierson J, Bartesaghi A, Mayer ML, Subramaniam S. (2014) Structural mechanism of glutamate receptor activation and desensitization. Nature 514:328–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura E, Matsuda K, Morgan JI, Yuzaki M, Watanabe M. (2009) Cbln1 accumulates and colocalizes with Cbln3 and GluRdelta2 at parallel fiber-Purkinje cell synapses in the mouse cerebellum. Eur J Neurosci 29:693–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabavi S, Kessels HW, Alfonso S, Aow J, Fox R, Malinow R. (2013) Metabotropic NMDA receptor function is required for NMDA receptor-dependent long-term depression. Proc Natl Acad Sci USA 110:4027–4032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naur P, Hansen KB, Kristensen AS, Dravid SM, Pickering DS, Olsen L, Vestergaard B, Egebjerg J, Gajhede M, Traynelis SF, et al. (2007) Ionotropic glutamate-like receptor delta2 binds D-serine and glycine. Proc Natl Acad Sci USA 104:14116–14121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orth A, Tapken D, Hollmann M. (2013) The delta subfamily of glutamate receptors: characterization of receptor chimeras and mutants. Eur J Neurosci 37:1620–1630. [DOI] [PubMed] [Google Scholar]
- Plested AJ, Mayer ML. (2009) AMPA receptor ligand binding domain mobility revealed by functional cross linking. J Neurosci 29:11912–11923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pøhlsgaard J, Frydenvang K, Madsen U, Kastrup JS. (2011) Lessons from more than 80 structures of the GluA2 ligand-binding domain in complex with agonists, antagonists and allosteric modulators. Neuropharmacology 60:135–150. [DOI] [PubMed] [Google Scholar]
- Schäfer A, Horn H, Ahlrichs R. (1992) Fully optimized contracted Gaussian basis sets for atoms Li to Kr. J Chem Phys 97:2571–2577. [Google Scholar]
- Schauder DM, Kuybeda O, Zhang J, Klymko K, Bartesaghi A, Borgnia MJ, Mayer ML, Subramaniam S. (2013) Glutamate receptor desensitization is mediated by changes in quaternary structure of the ligand binding domain. Proc Natl Acad Sci USA 110:5921–5926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogensen SB, Greenwood JR, Bunch L, Clausen RP. (2011) Glutamate receptor agonists: stereochemical aspects. Curr Top Med Chem 11:887–906. [DOI] [PubMed] [Google Scholar]
- Weigend F, Häser M, Patzelt H, Ahlrichs R. (1998) RI-MP2: optimized auxiliary basis sets and demonstration of efficiency. Chem Phys Lett 294:143–152. [Google Scholar]
- Wallace AC, Laskowski RA, Thornton JM. (1996) LIGPLOT: a program to generate schematic diagrams of protein-ligand interactions. Protein Eng 8:127–134. [DOI] [PubMed] [Google Scholar]
- Williams K, Dattilo M, Sabado TN, Kashiwagi K, Igarashi K. (2003) Pharmacology of delta2 glutamate receptors: effects of pentamidine and protons. J Pharmacol Exp Ther 305:740–748. [DOI] [PubMed] [Google Scholar]
- Winn MD. (2003) An overview of the CCP4 project in protein crystallography: an example of a collaborative project. J Synchrotron Radiat 10:23–25. [DOI] [PubMed] [Google Scholar]
- Wollmuth LP, Kuner T, Jatzke C, Seeburg PH, Heintz N, Zuo J. (2000) The Lurcher mutation identifies delta 2 as an AMPA/kainate receptor-like channel that is potentiated by Ca(2+). J Neurosci 20:5973–5980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan H, Low CM, Moody OA, Jenkins A, Traynelis SF. (2015) Ionotropic GABA and Glutamate Receptor Mutations and Human Neurologic Diseases. Mol Pharmacol 88:203–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuzaki M. (2003) The delta2 glutamate receptor: 10 years later. Neurosci Res 46:11–22. [DOI] [PubMed] [Google Scholar]
- Yuzaki M. (2004) The delta2 glutamate receptor: a key molecule controlling synaptic plasticity and structure in Purkinje cells. Cerebellum 3:89–93. [DOI] [PubMed] [Google Scholar]
- Yuzaki M. (2009) New (but old) molecules regulating synapse integrity and plasticity: Cbln1 and the delta2 glutamate receptor. Neuroscience 162:633–643. [DOI] [PubMed] [Google Scholar]
- Yuzaki M. (2010) Synapse formation and maintenance by C1q family proteins: a new class of secreted synapse organizers. Eur J Neurosci 32:191–197. [DOI] [PubMed] [Google Scholar]
- Yuzaki M. (2011) Cbln1 and its family proteins in synapse formation and maintenance. Curr Opin Neurobiol 21:215–220. [DOI] [PubMed] [Google Scholar]
- Zhou LM, Gu ZQ, Costa AM, Yamada KA, Mansson PE, Giordano T, Skolnick P, Jones KA. (1997) (2S,4R)-4-methylglutamic acid (SYM 2081): a selective, high-affinity ligand for kainate receptors. J Pharmacol Exp Ther 280:422–427. [PubMed] [Google Scholar]
- Zuo J, De Jager PL, Takahashi KA, Jiang W, Linden DJ, Heintz N. (1997) Neurodegeneration in Lurcher mice caused by mutation in delta2 glutamate receptor gene. Nature 388:769–773. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.