Abstract
Little is known regarding the effect of intestinal microbiota modifiers, such as probiotics and conventionalization with exogenous bacteria, on host hepatic drug metabolism. Therefore, the goal of this study was to determine the effect of these modifiers on the expression of various drug-metabolizing enzymes of the host liver. VSL3 is a probiotic that contains eight live strains of bacteria. Five groups of mice were used: 1) conventional mice (CV), 2) conventional mice treated with VSL3 in drinking water, 3) germ-free (GF) mice, 4) GF mice treated with VSL3, and 5) GF mice exposed to the conventional environment for 2 months. All mice were 3 months old at tissue collection. GF conditions markedly downregulated the cytochrome P450 (P450) 3a gene cluster, but upregulated the Cyp4a cluster, whereas conventionalization normalized their expression to conventional levels [reverse-transcription quantitative polymerase chain reaction (qPCR) and western blot]. Changes in the Cyp3a and 4a gene expression correlated with alterations in the pregnane X receptor and peroxisome proliferator–activated receptor α–DNA binding, respectively (chromatin immunoprecipitation–qPCR). VSL3 increased each bacterial component in the large intestinal content of the CV mice, and increased these bacteria even more in GF mice, likely due to less competition for growth in the GF environment. VSL3 given to conventional mice increased the mRNAs of Cyp4v3, alcohol dehydrogenase 1, and carboxyesterase 2a, but decreased the mRNAs of multiple phase II glutathione-S-transferases. VSL3 given to germ-free mice decreased the mRNAs of UDP-glucuronosyltransferases 1a9 and 2a3. In conclusion, conventionalization and VSL3 alter the expression of many drug-metabolizing enzyme s in the liver, suggesting the importance of considering “bacteria-drug” interactions for various adverse drug reactions in patients.
Introduction
Intestinal microbiota modifiers, such as probiotics, antibiotics, and conventionalization to the environment, may both positively and negatively impact human health (Boyle et al., 2006; Carvalho et al., 2012; Kim, 2015; Vandenplas et al., 2015). Although the excessive use of antibiotics has raised concerns regarding their potential adverse health effects, leading to more stringent usage in medical practice, less is known regarding the safety of probiotics. Probiotics are defined as live microorganisms that confer a health benefit to the host when administered in adequate amounts (Sanders, 2008). VSL#3 (also called VSL3) is a combinatorial probiotic that is used for human intestinal disorders, such as inflammatory bowel disease and ulcerative colitis (Bibiloni et al., 2005; Penner and Fedorak, 2005; Lee et al., 2012; Mardini and Grigorian, 2014). VSL3 contains eight live bacterial strains that are considered beneficial for the host, including Bifidobacterium breve, Bifidobacterium longum, Bifidobacterium infantis, Lactobacillus acidophilus, Lactobacillus plantarum, Lactobacillus paracasei, Lactobacillus bulgaricus, and Streptococcus thermophilus. Even though there is evidence of beneficial effects and safety of VSL3 in animal models and clinical trials, little is known regarding the effect of VSL3 on the expression of various drug-metabolizing enzymes in the liver, which is the major organ for drug detoxification. It is important to obtain this critical information, because altered expression of hepatic drug-metabolizing enzymes by VSL3 may lead to altered drug effects when VSL3 is coadministered with drugs.
Conventionalization, which is the exposure of the host to a microbial background in the environment, has attracted interest regarding its potential health benefits to humans. The “hygiene hypothesis” suggests that newborns delivered by cesarean section and raised in an overly clean environment may lack sufficient stimulation of the immune system, and may be prone to develop chronic inflammatory conditions and obesity (Collado et al., 2015). In mice, colonization of germ-free (GF) mice during the neonatal period is helpful to establish immune signaling, whereas colonization of GF mice at 5 weeks of age leads to specific changes in chemokine signaling (Yamamoto et al., 2012). Conventionalization of the intestines of 8-week-old GF mice to a typical environmental microbial background impacts host intermediary metabolism, including increased body weight, stimulated hepatic glycogenesis and triglyceride synthesis, as well as altered bile acid composition (Claus et al., 2011). However, relatively less is known regarding the effect of colonization on xenobiotic metabolism in the liver of the host.
A major class of the phase I hepatic drug-metabolizing enzymes are the cytochrome P450s (P450s), of which the Cyp1-3 family members are considered to be mainly responsible for xenobiotic metabolism, whereas the Cyp4 family members are responsible for lipid metabolism (Hardwick, 2008). It is well known that the aryl hydrocarbon receptor (AhR) transcriptionally upregulates Cyp1 gene expression, the constitutive androstane receptor upregulates Cyp2b gene expression, and the pregnane X receptor (PXR) upregulates Cyp3a gene expression in the liver. Therefore, AhR, constitutive androstane receptor, and PXR are often referred to as xenobiotic-sensing transcription factors (Xu et al., 2005). The peroxisome proliferator–activated receptor α (PPARα) is a lipid sensor that upregulates Cyp4a gene expression (Kroetz et al., 1998). Other important phase I enzymes involved in xenobiotic bioactivation and detoxification include alcohol dehydrogenases (Adhs), aldehyde dehydrogenases (Aldhs), carboxyesterases (Cess), flavin-containing monooxygenases (Fmos), aldo-keto reductases (Akrs), epoxide hydrolases (Ephxs), and NAD(P)H dehydrogenase quinone 1 (Nqo1). Phase II metabolism or conjugation reactions consist of the glutathione S-transferases (Gsts), UDP glucuronosyltransferases (Ugts), and sulfotransferases (Sults) that usually function as detoxification enzymes in the liver (Jancova et al., 2010). The previously noted xenobiotic- and lipid-sensing transcription factors also regulate the expression of other non-P450 phase I enzymes as well as phase II enzymes in the liver (Alnouti and Klaassen, 2008; Knight et al., 2008; Buckley and Klaassen, 2009; Pratt-Hyatt et al., 2013). The GF mouse is an important model that allows mechanistic investigations of the role of intestinal microbiota in host drug metabolism. We have reported that certain Cyp3a genes are downregulated, whereas certain Cyp4a genes are upregulated in livers of GF mice (Selwyn et al., 2015a,b).
The goal of this study was to systematically characterize the gene expression profiles of 94 drug-metabolizing enzymes (including Cyp3a and Cyp4a) in livers of five groups of mice: 1) conventional mice (CV), 2) CV treated with VSL3 in drinking water, 3) GF mice, 4) GF mice treated with VSL3, and 5) GF mice exposed to the conventional environment (GF+CV) for 2 months. We hypothesize that introducing exogenous bacteria, such as probiotics or conventionalization with bacteria from the environment, would partially restore mucosal mRNA expression of Cyp3a and 4a genes as well as other important drug-metabolizing enzymes.
Materials and Methods
Animals and Procedures.
Male C57BL/6J CV mice were purchased from Jackson Laboratories (Bar Harbor, ME). The initial breeding colony of GF C57BL/6J/UNC mice was established with mice purchased from the National Gnotobiotic Rodent Resource Center (University of North Carolina, Chapel Hill, NC). All mice used in the present study were housed in an Association for Assessment and Accreditation of Laboratory Animal Care International–accredited facility at the University of Kansas Medical Center (Kansas City, KS), with a 14-hour light/10-hour dark cycle, in a temperature and humidity-controlled environment, and all mice had ad libitum access to autoclaved rodent chow and water. Starting at 2 months of age, CV and GF mice (male, n = 6–8/group) were given the VSL3 probiotics in drinking water for 28 days, which includes a total of 4.5 × 106 CFU/ml of the following strains: L. acidophilus, L. bulgaricus, L. paracasei, L. plantarum, B. breve, B. infantis, B. longum, and S. thermophilus (Sigma-Tau Pharmaceuticals, Inc., Gaithersburg, MD). In a separate study, 1-month-old GF mice (male, n = 4/group) were taken out of the germ-free isolator, and were housed in the same environment as the CV mice for two months.
All mice were euthanized with an overdose of pentobarbital at approximately 3 months of age. Livers were immediately frozen in liquid nitrogen. Small and large intestinal contents were flushed using phosphate-buffered saline containing 10 mM dithiothreitol, and centrifuged at 20,000g for 30 minutes at 4°C, as described previously (Zhang et al., 2012). All samples were stored in a −80°C freezer until further analyses. All animal procedures were approved by the Institutional Animal Care and Use Committee at the University of Kansas Medical Center.
Bacterial DNA Extraction and Quantification.
Total genomic bacterial DNA was extracted from the small and large intestinal contents of CV and GF mice (treated with or without VSL3), as well as conventionalized GF (GF+CV) mice using the QIAmp DNA Stool Kit (Qiagen, Valencia, CA) according to the manufacturer’s instructions. The concentration of total DNA was determined using a Qubit 2.0 Fluorometer (Life Technologies, Grand Island, NY). The 16S ribosomal RNA (rRNA) primers for the detection of B. infantis and S. thermophilus were described previously (Vitali et al., 2003; Furet et al., 2004). The 16S rRNA primers for the detection of L. acidophilus, L. bulgaricus, L. plantarum, L. paracasei, B. breve, and B. longum were designed based on the 16S rRNA bacterial amplicon sequences of these bacteria, and specificity was determined using the National Center for Biotechnology Information (NCBI) Basic Local Alignment Search Tool against the 16S ribosomal RNA sequences (bacteria and archaea) database, as shown in Supplemental Table 1. The sequences for a pair of primers that recognize the universal bacterial 16S rRNA sequences were provided by the University of North Carolina gnotobiotic core facilities. All primers were synthesized by Integrated DNA Technologies (Coralville, IA). The abundance of the genomic DNA encoding the bacterial 16S rRNAs in the intestinal content of mice was determined by quantitative polymerase chain reaction (qPCR) using a CFX384 Real-Time PCR Detection System (Bio-Rad, Hercules, CA). Results are expressed as the mean delta-delta cycle value (calculated as 2^[−(Cq − average reference Cq)]) of the quantitative PCR as compared with the universal bacteria, per nanogram of DNA from the intestinal content. To validate the primers that target the VSL3 bacterial components of VSL3, the bacterial DNA from the pure VSL3 powder was extracted using the E.Z.N.A. Stool DNA Kit (Omega Bio-Tek Inc., Norcross, GA), and qPCR assays were performed using 1, 3, 10, and 30 ng of total VSL3 DNA. Results are expressed as the mean delta-delta cycle value of the quantitative PCR compared with the average quantitation cycle (Cq) of the universal bacteria.
RNA Isolation and mRNA Quantification of DPGs.
Total RNA was isolated from livers using RNA-Bee reagent (Tel-Test Inc., Friendswood, TX) per the manufacturer’s instructions. (Table 1) The concentration of total RNA was determined at 260 nm using a NanoDrop 1000 Spectrophotometer (Thermo Scientific, Wilmington, DE). Reverse transcription was performed using an iScript cDNA Synthesis Kit (Bio-Rad). The resulting cDNA products were amplified by qPCR using SsoAdvanced Universal SYBR Green Supermix in a Bio-Rad CFX384 Real-Time PCR Detection System. The primers for all qPCR reactions were synthesized by Integrated DNA Technologies, and the primer sequences are shown in Supplemental Table 2. The results are expressed as the percentage of the expression of 18S rRNA.
TABLE 1.
Summary of DPGs that are differentially regulated in response to gut microbiota modifiers
| Gut Microbiota Modifiers | Upregulated Drug-Metabolizing Enzymes | Downregulated Drug-Metabolizing Enzymes |
|---|---|---|
| VSL3 (as compared with control groups of the same mouse model) | Phase I: Cyp4v3 (4.26-fold), Adh1 (2.23-fold), Ces2a (2.07-fold) | Phase I: Cyp3a44 (67.42%), Cyp3a11 (59.88%) |
| Phase II: none observed | Phase II: Gstm1 (65.50%), Gstm2 (49.86%), Gstm3 (61.58%), Gsto1 (63.62%), Ugt1a9a (54.41%), Ugt2a3a (52.24%) | |
| Germ free (as compared with control CV mice) | Phase I: Cyp4a cluster (Cyp4a14 [9.84-fold], 4a10 [53.34-fold], 4a31 [5.86-fold], 4a32 [6.06-fold], 4a12a/b [2.20-fold]), Cyp4f17 (1.86-fold), Cyp4f14 (5.30-fold), Cyp4f18 (19.83-fold), Cyp4v3 (2.88-fold), Cyp1a2 (1.64-fold), Aldh1a1 (2.03-fold), Aldh3a1 (3.08-fold), Aldh3a2 (2.03-fold) | Phase I: Cyp3a cluster (Cyp3a41a/b [16.18%], 3a44 [11.33%], 3a11 [10.69%], 3a25/59 [32.44%]), Aldh1b1 (41.30%), Fmo5 (54.43%) |
| Phase II: Ugt1a9 (1.68-fold), Ugt2b1 (1.83-fold) | Phase II: Gstpi (57.71%), Gstm1 (53.15%), Gstm2 (29.70%), Gstm3 (34.28%), Gsto1 (49.36%), Sult5a1 (36.39%) | |
| Conventionalization (as compared with control GF mice) | Phase I: Cyp3a cluster (Cyp3a41a/b [6.38-fold], 3a44 [1.60-fold], 3a11 [7.56-fold], 3a25/59, 3a16 [7.12-fold]), Cyp4f17 (1.47-fold), Ces2a (1.86-fold) | Phase I: Cyp4a cluster (Cyp4a14 [0.98%], 4a10 [6.51%], 4a31 [11.42%], 4a32 [7.43%], 4a12a/b [59.22%]), Aldh3a2 [20.17%], Ces1e/1g [42.60%], Akr1d1 (39.02%) |
| Phase II: Gstpi (2.01-fold), Ugt2a36/37/38 (6.01-fold), Sult5a1 (2.88-fold) | Phase II: Ugt1a9 (53.19%) |
*Differentially regulated in GF livers.
Western Blotting.
Hepatic microsomes were isolated from CV, GF, and GF+CV control mice, as well as VSL3-treated CV and GF mice, and protein concentrations were determined using the Qubit Protein Assay Kit (Life Technologies) according to the manufacturer’s instructions. The samples (50 μg of protein) were subjected to polyacrylamide gel electrophoresis and transferred onto a polyvinylidene difluoride membrane. Primary antibody against mouse Cyp3a11 (anti-rat CYP3A1/2 monoclonal antibody, clone 2-13-1, 1:500) was a generous gift from Dr. Frank Gonzalez at the National Cancer Institute (Bethesda, MD). Primary antibody against Cyp4a14 (goat polyclonal IgG, 1:500) was purchased from Santa Cruz Biotechnology (Dallas, TX; SC-46087). Primary antibody against mouse β-actin was purchased from Abcam (Cambridge, MA; ab8227, 1:500). Horseradish peroxidase–linked secondary antibodies were purchased from Sigma-Aldrich (St. Louis, MO) and used at 1:2000 (namely, rabbit anti-mouse A9044 for Cyp3a11, rabbit anti-goat A5420 for Cyp4a14, and goat anti-rabbit A6154 for β-actin). Proteins were detected using chemiluminescence (Thermo Fisher Scientific, Life Technologies). Intensities of the protein bands were quantified using ImageJ software (National Institutes of Health, Bethesda, MD).
Enzyme Activities of Cyp3a and Cyp4a.
Livers of mice were homogenized using a 2-ml glass homogenizer (Wheaton Co., (Milville, NJ)) in ice-cold Sucrose Tris buffer of 250 mM sucrose in 10 mM Tris-HCl (pH 7.5). The homogenate was centrifuged (100,000g, 60 min) at 4°C. The crude membrane pellet was resuspended in Sucrose Tris buffer and stored at −80 °C. P450 enzyme activity determination was carried out following instructions from Promega (Madison, WI). Twenty micrograms of protein was diluted to 12.5 µl and added to each well of a 96-well plate. Then 12.5 µl of the 4× P450 reaction mixture (12 µM luciferin–indole 3-propionic acid for CYP3A; 320 µM luciferin-4A for CYP4A) was added into each well and mixed gently. The plate was incubated at room temperature for 10 minutes. Twenty-five microliters of the 2× NADPH regeneration system was added into the P450 assays, mixed, and incubated at the same temperature for 20 minutes. Fifty microliters of the reconstituted luciferin detection reagent was added to the P450 assays and incubated at room temperature for another 20 minutes to stabilize the luminescent signal. Luminescence was recorded using a luminometer (PlateLumino system; MidSci Co., Valley Park, MO).
Chromatin Immunoprecipitation.
Chromatin immunoprecipitation (ChIP) was performed using approximately 200 mg of frozen livers from CV, GF, and GF+CV mice, using the MAGnify Chromatin Immunoprecipitation System (Life Technologies), with modifications. In brief, livers were finely minced into less than 1-mm cubes using razor blades in cold 1× Dulbecco’s phosphate-buffered saline in a sterile 10-cm culture dish on ice, and transferred into an ice-cold Dounce homogenizer (VWR International, Radnor, PA) to further grind the liver into a homogenous solution with a glass pestle. Samples were subjected to crosslinking using freshly prepared formaldehyde (final concentration: 1%), and were rotated for 20 minutes at room temperature using an ELMI Intelli Mixer (ELMI Company, Riga, Latvia). The crosslinking was reversed by glycine (final concentration: 0.125 M) with rotation for 5 minutes at room temperature, followed by centrifugation to collect the pellets. The pellets were washed with cold Dulbecco’s phosphate-buffered saline and resuspended using cold ChIP lysis buffer with a protease inhibitor cocktail (Sigma-Aldrich, St. Louis), rotated at 4°C for 15 minutes, and centrifuged to obtain the pellets. The pellets were resuspended in ChIP nuclear lysis buffer with protease inhibitors and incubated on ice for 15 minutes. Chromatins were then fragmented into a 300–500-bp average size range using a Bioruptor UCD200 connected to a water-cooling system (Diagenode, Denville, NJ). The sonication condition was 10× (30 seconds on + 30 seconds off) at 4°C, and was repeated after 10 minutes at the highest intensity. The fragment size was confirmed by electrophoresis. ChIP-grade antibodies—namely, SC-25381 (Santa Cruz) for PXR, NB600-636 (Novus Biologicals, Littleton, CO) for PPARα, and MMS-126R for RNA polymerase II (Covance, Emeryville, CA)—were used for immunoprecipitation. An IgG antibody (ab18413; Abcam) was used as a negative control. The immunoprecipitation procedures are described in detail per the manufacturer’s protocol (MAGnify Chromatin Immunoprecipitation System; Life Technologies).
ChIP-qPCR Primer Design and qPCR Reactions.
PXR and PPARα genomic DNA binding sites were obtained by reanalyzing the ChIP sequencing data in control C57BL/6 male mouse livers from previous publications (Cui et al., 2010; Lee et al., 2014). Nuclear receptor enrichment peaks were visualized by the Integrated Genome Viewer (Robinson et al., 2011), and the known DNA-binding motifs of PXR and PPARα—namely, direct-repeat-3 (DR-3), DR-4, everted-repeat-6 (ER-6), and ER-8 for PXR, as well as DR-1 and DR-2 for PPARα (Cui et al., 2010; Lee et al., 2014)—were determined using NUBIScan version 2.0 (http://www.nubiscan.unibas.ch/). The qPCR primers were designed around the targeted motifs using the NCBI Primer Design Tool (http://www.ncbi.nlm.nih.gov/tools/primer-blast/), and their specificities were confirmed using University of California, Santa Cruz BLAT (https://genome.ucsc.edu/cgi-bin/hgBlat?command=start). The promoter sequences of Cyp3a and Cyp4a clusters were retrieved using the Mammalian Promoter Database (http://mpromdb.wistar.upenn.edu/). The qPCR primers for RNA polymerase-II (RNA-Pol-II) were designed using the queried promoter sequences. Real-time qPCR reactions of the ChIP DNA were performed using SsoAdvanced Universal SYBR Green Supermix in a Bio-Rad CFX384 Real-Time PCR Detection System. The qPCR primer sequences, targeted genomic regions, and putative motifs are listed in Supplemental Table 3.
Statistical Analysis.
Data are presented as the mean ± S.E.M. Differences among multiple groups were determined by analysis of variance followed by Duncan’s post-hoc test (P < 0.05).
Results
Bacterial Quantification in the Large Intestinal Content of VSL3-Treated CV and GF mice, as well as in GF+CV Mice.
To confirm the colonization of VSL3 bacterial components in the large intestinal contents of CV and GF mice treated with VSL3, qPCR analysis of bacterial 16S rRNA was performed using DNA isolated from the large intestinal content as described in Materials and Methods. The bacteria found in VSL3 were also quantified in large intestinal content DNA samples of the GF+CV mice (conventionalized with the regular environment) to determine whether these bacteria are present in the conventional animal-housing environment. Primers that recognize the universal bacterial 16S rRNA sequences were used to quantify changes in the total amount of bacteria following VSL3 treatment of CV and GF mice, as well as after conventionalization of GF+CV mice. To validate the robustness of all the 16S rRNA qPCR primers for bacterial detection, the bacterial DNA from the pure VSL3 powder at various concentrations was subjected to qPCR assays, as shown in Supplemental Fig. 1. Addition of 1–10 ng of VSL3 bacterial DNA input resulted in an increase in total bacteria detected (average Cq = 12.24, 10.83, and 10.02), whereas a decrease in the signal was observed at the highest concentration (30 ng), which is likely due to inhibition by an overwhelming amount of DNA templates (average Cq = 18.26). Similarly, the eight individual bacteria in VSL3 were detectable with high robustness at low and middle concentrations, whereas the middle and highest concentrations of VSL3 DNA input result in inhibition (Supplemental Fig. 1). In summary, the robustness of these 16S rRNA qPCR primers was confirmed for the follow-up analyses.
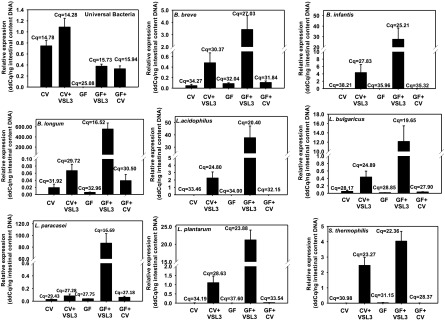
As shown in Fig. 1, VSL3 treatment results in a 41% increase in the total bacteria in the large intestinal content of CV mice (as assessed by quantifying the universal bacteria). As expected, only background signals were detected in vehicle-treated GF mice (0.08% of vehicle-treated CV signals). As a result of the VSL3 treatment, there was a 652-fold increase in the signal detected in the large intestinal content of the GF mice. Conventionalization of GF mice also led to a marked increase in the signal detected in the GF+CV large intestinal content (564-fold) as compared with the GF mice, and this is a result of the bacterial colonization from the conventional housing environment. The conventionalization of GF mice restored approximately 50% of total bacteria in the large intestine compared with the CV mice.
Fig. 1.
The 16S rRNA abundance of universal bacteria, as well as the eight bacterial components in VSL3 (B. breve, B. infantis, B. longum, L. acidophilus, L. bulgaricus, L. plantarum, L. paracasei, and S. thermophilus) in the large intestinal content. DNA samples from CV, CV+VSL3, GF, GF+VSL3, and GF+CV groups. Large intestinal content DNA was extracted as described in Materials and Methods, and 5.6 ng of total DNA was loaded in each well of the qPCR reactions. Results are expressed as delta-delta cycle value (calculated as 2^[−(Cq − average reference Cq)]) of the quantitative PCR (ddCq) as compared with the universal bacteria.
The Bifidobacterium bacteria in VSL3 are B. breve, B. infantis, and B. longum, and they were detected only at background levels in the large intestinal contents of CV, GF, and GF+CV mice. VSL3 given to CV mice in the drinking water for 2 months increased the colonization of each of the Bifidobacterium components in the large intestinal content. Among these bacteria, B. infantis appeared to colonize the most in CV mice. Interestingly, in GF mice, VSL3 resulted in an even greater increase in the Bifidobacterium genus. B. longum, which colonized only to a minor extent in the large intestine of CV mice, became the dominant colonizer in the large intestinal content of GF mice. Within the Lactobacillus genus, L. acidophilus, L. bulgaricus, L. paracasei, and L. plantarum are VSL3 components, and these bacteria were also minimally present in the large intestinal content of CV, GF, and GF+CV mice. In the large intestinal content of CV mice, VSL3 increased each of the Lactobacillus components—namely, 405-fold for L. acidophilus, 9.7-fold for L. bulgaricus, 47-fold for L. plantarum, and to a lesser extent (4.4-fold) for L. paracasei. In the large intestinal content of GF mice, VSL3 resulted in an even greater increase in all of these Lactobacilli. The VSL3 component Streptococcus thermophilus increased 209-fold in CV and 442-fold in GF large intestinal content, and it was not present in the large intestinal content of the conventionalized GF mice. In summary, VSL3 treatment resulted in increased colonization of each of the eight VSL3 bacteria components in the large intestinal content of CV mice, and an even higher colonization of these bacteria in the GF mice, likely due to the lack of competition with the endogenous residential bacteria in the large intestine. Conventionalization increased the total bacteria in the large intestinal content of the GF+CV mice, but generally did not markedly increase the VSL3 bacteria components in these mice, likely because these VSL3 bacteria are not abundant in the conventional housing environment.
In summary, VSL3 increased all the bacteria in the probiotic mixture in the large intestinal content of CV mice, and increased these bacteria even more in GF mice, likely due to less competition for bacterial growth in a GF environment.
Expression of the Cyp3a Gene Cluster in Livers of CV and GF Mice following VSL3 Treatment or Conventionalization.
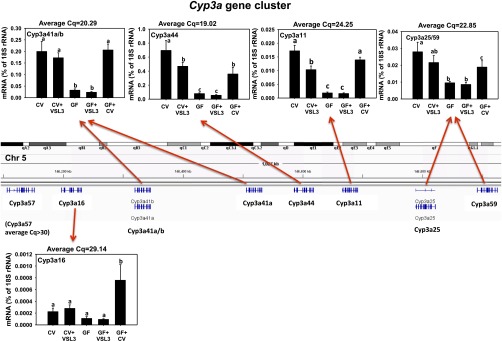
The Cyp3a gene family is well known to be responsible for oxidation of many drugs and other xenobiotics (Wilkinson, 1996). Similar to the human CYP3A gene cluster, the majority of the Cyp3a genes in mice also form a cluster (on chromosome 5), and these genes are Cyp3a57, 3a16, 3a41a and 3a41b, 3a44, 3a11, 3a25, and 3a59 (Fig. 2). Due to high sequence homology, a common pair of qPCR primers was designed for Cyp3a41a and 3a41b, and a common pair of qPCR primers was designed for Cyp3a25 and 3a59. Interestingly, the expression of the Cyp3a genes is “region-specific,” in that the mRNAs of Cyp3a41a/b, 3a44, 3a11, and 3a25/59 genes, which are located in the right portion of the Cyp3a gene cluster, all displayed a similar pattern following VSL3 treatment or conventionalization; more specifically, 1) VSL3 moderately decreased the mRNAs of Cyp3a44 and Cyp3a11, and tended to decrease Cyp3a41a/b and 3a25/59 (although a statistical significance was not achieved); 2) GF conditions markedly decreased the mRNAs of all these P450s, and VSL3 was not able to normalize their expression in livers of GF mice; and 3) conventionalization of GF mice partially restored the mRNAs of all of these P450s to CV levels. In GF mice that were conventionalized, Cyp3a41a/b and Cyp3a11 mRNAs were completely normalized to CV levels, whereas Cyp3a44 and 3a25/29 mRNAs in livers of GF+CV mice were higher than in livers of GF mice, but were only partially normalized when compared with expression in CV mice. Interestingly, for Cyp3a57 and 3a16, which are located on the left of the Cyp3a cluster, Cyp3a57 mRNA was minimally expressed in livers of all groups (Cq > 30, data not shown), whereas Cyp3a16, which is a perinatal-specific Cyp3a isoform, was lowly expressed in CV, CV+VSL3, GF, and GF+VSL3, and was only increased in the GF mice that were conventionalized. In summary, the chromatin region in the Cyp3a gene cluster that is codownregulated by GF conditions but coupregulated by conventionalization is located in the right portion of the cluster. The region-specific expression of Cyp3a mRNAs in the Cyp3a cluster suggests that certain epigenetic factors and/or transcription factors may contribute to the responsiveness to GF or conventionalization conditions as well as the basal expression of the Cyp3a genes.
Fig. 2.
The mRNA expression of the Cyp3a gene cluster (Cyp3a41a/b, 3a44, 3a11, 3a25/59, 3a57, and 3a16) in liver samples from CV, CV+VSL3, GF, GF+VSL3, and GF+CV groups. The genomic locations of the Cyp3a genes are displayed using the Integrated Genome Viewer. Reverse-transcription qPCR for each gene was performed as described in Materials and Methods. Data are expressed as the percentage of the housekeeping gene 18S rRNA. Statistical analysis was performed using analysis of variance followed by Duncan’s post-hoc test with P < 0.05 considered statistically significant. Treatment groups that are not statistically different are labeled with the same letter.
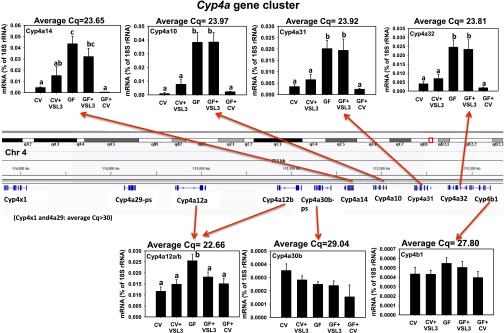
Expression of the Cyp4a/b/x Gene Cluster in Livers of CV and GF Mice following VSL3 Treatment or Conventionalization.
The Cyp4a gene family members encode important fatty acid and prostaglandin ω-hydroxylases, and are highly inducible by PPARα ligands, such as the hypolipidemic drugs (Kroetz et al., 1998). Similar to the human CYP4A cluster, the mouse Cyp4a genes form a cluster that are located on chromosome 4, together with Cyp4x1 and Cyp4b1 (Fig. 3). Interestingly, the four Cyp4a genes in the middle of this cluster—namely, Cyp4a14, 4a10, 4a31, and 4a32—all displayed a similar expression pattern: 1) VSL3 tended to increase the mRNAs of these P450s in CV mouse livers, although statistical significance was not achieved; 2) GF conditions markedly increased the mRNAs of these P450s to the level of both GF and GF-VSL3 mice; 3) VSL3 did not alter the mRNAs of these P450s in GF mice; and 4) conventionalization markedly reduced the mRNAs of these P450s to conventional levels. In fact, there appeared to be an even further decrease in Cyp4a14 mRNA in livers of conventionalized GF mice, although statistical significance was not achieved. In contrast, the Cyp4a genes located on the left (Cyp4x1, 4a29, 4a12a/b, and 30b), as well as Cyp4b1 located on the right boundary of the Cyp4a cluster, did not display the coregulatory pattern. Cyp4x1 and 4a29 mRNAs were minimally expressed (Cq > 30, data not shown), whereas Cyp4a30b and 4b1 mRNAs were not readily altered in any of the treatment groups; Cyp4a12a/b mRNA was higher in livers of GF mice compared with CV mice, and was not altered by VSL3. In summary, similar to the Cyp3a gene cluster, the expression of the Cyp4a/b/x gene cluster was also region-specific, in that there appeared to be an active transcription region in the middle of the cluster that was coregulated by VSL3, GF, and conventionalization conditions, whereas genes at the left and right boundaries of the cluster were either not expressed or did not display a similar expression pattern as the four Cyp4a genes in the middle of the cluster.
Fig. 3.
The mRNA expression of the Cyp4a gene cluster (Cyp4a14, 4a10, 4a31, 4a32, 4x1, 4a29, 4a12a/b, 4a30b, and 4b1) in liver samples from CV, CV+VSL3, GF, GF+VSL3, and GF+CV groups. The genomic locations of the Cyp4 genes are displayed using the Integrated Genome Viewer. Reverse-transcription qPCR for each gene was performed as described in Materials and Methods. Data are expressed as the percentage of the housekeeping gene 18S rRNA. Statistical analysis was performed using analysis of variance followed by Duncan’s post-hoc test with P < 0.05 considered statistically significant. Treatment groups that are not statistically different are labeled with the same letter.
In summary, the mRNA expression patterns for the Cyp3a and Cyp4a gene clusters indicate that VSL3 had much less of an effect on their gene expression (a moderate decrease in the mRNAs of a couple of Cyp3as); GF conditions markedly decreased some Cyp3a mRNAs, but increased some Cyp4a mRNAs in a region-specific manner on the chromosomes, suggesting the presence of coregulatory transcriptional mechanisms; and conventionalization of GF mice at least partially normalizes the Cyp3a and 4a gene expression to CV levels.
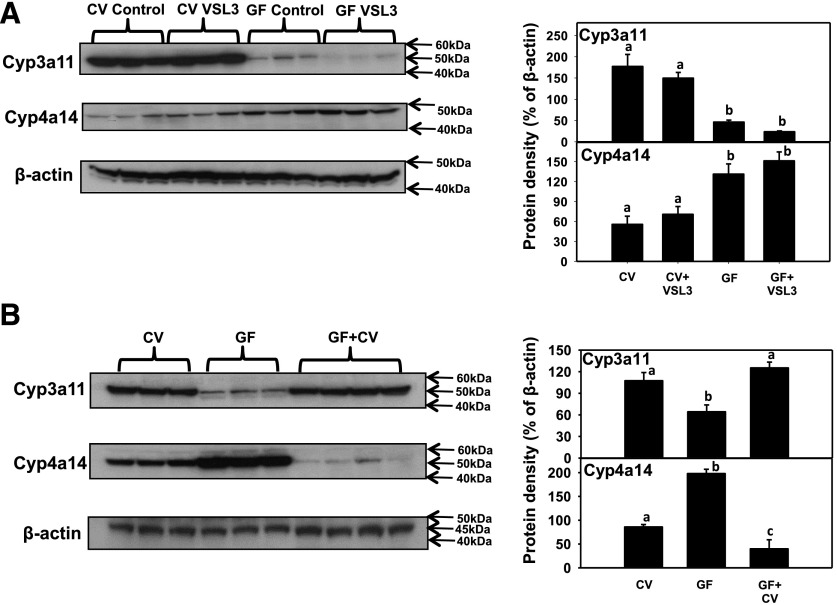
Western Blotting Analysis of Cyp3a11 and 4a14 Protein Expression in Livers of CV and GF Mice following VSL3 Treatment or Conventionalization.
Because the mRNAs of Cyp3a and 4a genes were altered by the intestinal microbiome, the protein expression of the representative P450—namely, Cyp3a11 and 4a14—was determined by western blotting analysis (Fig. 4). VSL3 did not alter the protein expression of Cyp3a11 or 4a14 in either CV or GF mice. As expected, livers from GF- and VSL3-treated GF mice had a marked decrease in Cyp3a11 protein, but a marked increase in Cyp4a14 protein (Fig. 4A). Conventionalization of GF mice restored the protein expression of Cyp3a11 to CV control levels, and decreased Cyp4a14 protein to CV control levels (Fig. 4B). Cyp4a14 mRNA also tended to be lower in livers of conventionalized GF mice as compared with livers of CV mice, although a statistical significance was not achieved (Fig. 3). In summary, protein expression of Cyp3a11 and 4a11 was consistent with the mRNA data of these genes in livers of CV and GF mice following VSL3 treatment and conventionalization.
Fig. 4.
(A) Protein expression in livers of CV and GF mice treated with vehicle or VSL3 (n = 3 per group). (B) Protein expression by western blots in livers of control CV, GF, and GF+CV mice (n = 3 per group). Western blotting of Cyp3a11, Cyp4a14, and β-actin proteins in hepatic microsomes was quantified as described in Materials and Methods. Quantification of protein band intensities after normalization to the loading control β-actin was performed using ImageJ software. Statistical analysis was performed using analysis of variance followed by Duncan’s post-hoc test with P < 0.05 considered statistically significant. Treatment groups that are not statistically different are labeled with the same letter.
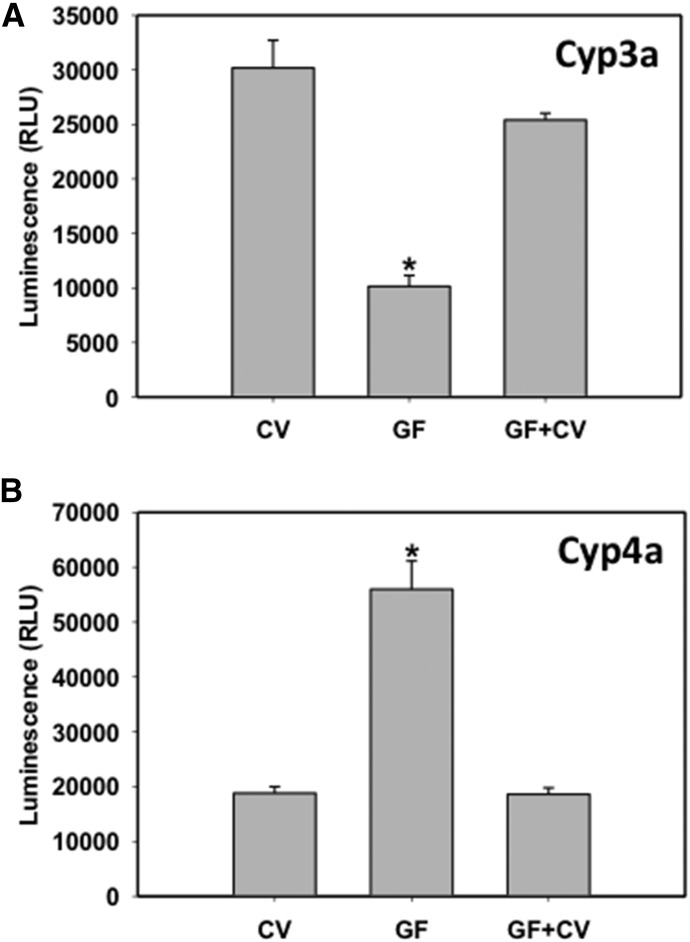
The effects of conventionalization on Cyp3a11 and Cyp4a14 activity in GF mice are shown in Fig. 5. The results demonstrate that Cyp3a11 activity was markedly decreased in GF mice and increased to conventional levels after exposure to the conventional environment. However, Cyp4a14 activity was upregulated in GF mice but was normalized to conventional levels by exposure to the conventional environment. The results are consistent with mRNA and protein expression levels in GF mice under conventionalization.
Fig. 5.
Enzyme activities of Cyp3a (A) and Cyp4a (B) in crude membranes of livers from CV, GF, and GF+CV mice. P450 enzyme activity determination was carried out according to the manufacturer’s protocol, as described in Materials and Methods. *Statistically significant differences as compared with CV mice.
Expression of the Cyp4f Gene Cluster in Livers of CV and GF Mice following VSL3 Treatment or Conventionalization.
The CYP4F family in human liver microsomes is known to catalyze the omega oxidation of 3-hydroxy fatty acid and the initial oxidative O-demethylation of pafuramidine, which is an experimental drug for the treatment of pneumocystic pneumonia (Wang et al., 2006; Dhar et al., 2008). Regarding the mouse orthologs, the Cyp4f gene family members that form a cluster on chromosome 17 include Cyp4f39, 4f17, 4f37, 4f40, 4f15, 4f14, 4f13, and 4f41-ps (Supplemental Fig. 2). Cyp4f38, 4f41-ps, and 4f40 were minimally expressed in livers of all mouse groups (average Cq > 30). Cyp4f13, 4f15, 4f16, and 4f37 mRNAs were not readily altered by any treatment. Cyp4f17 mRNA was not altered by VSL3 in CV or GF mice; however, its mRNA was upregulated in GF conditions (with or without VSL3 treatment), as well as in livers of conventionalized GF mice. Cyp4f14 mRNA tended to be increased by VSL3 in CV mouse livers (although a statistical significance was not achieved), and was upregulated in GF mouse livers. In summary, genes within the Cyp4f cluster do not appear to be coregulated; Cyp4f17 and 4f14 mRNAs were the only Cyp4f genes that were differentially regulated by at least one of the three bacterial modification treatments. Both Cyp4f14 and 4f17 were upregulated in GF conditions, whereas Cyp4f17 was further upregulated by conventionalization.
Expression of Other P450s in Livers of CV and GF Mice following VSL3 Treatment or Conventionalization.
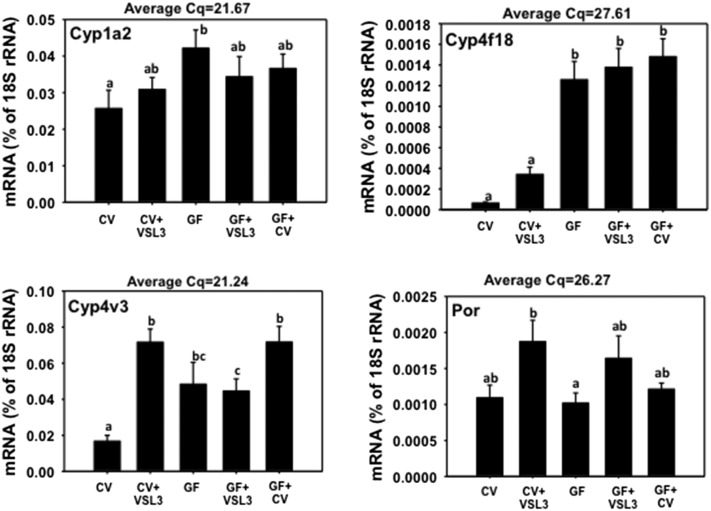
As shown in Fig. 6, the mRNA of Cyp1a2, which is a prototypical target gene of the AhR in the liver, was not readily altered by VSL3 or conventionalization; however, it was upregulated in livers of GF mice as compared with CV mice. The mRNAs of Cyp2b10 and 3a13 were not altered in any group of mice (Supplemental Fig. 3). Note that the Cyp3a13 gene is not part of the Cyp3a gene cluster described in Fig. 2. Cyp4f18 mRNA was markedly increased in livers of GF mice (both control and VSL3-treated groups) as well as in livers of conventionalized GF mice. VSL3 also tended to increase Cyp4f18 mRNA in CV livers, although a statistical significance was not achieved. Cyp4v3 mRNA was increased markedly in livers from the VSL3-treated CV mice, GF mice, VSL3-treated GF mice, and conventionalized GF mice. The mRNA of cytochrome P450 oxidoreductase, which is required for the electron transfer from NADPH to the P450s in the endoplasmic reticulum, was not markedly different in any of the groups.
Fig. 6.
The mRNA expression of Cyp1a2, 4f18, 4v3, and P450 oxidoreductase (Por) in liver samples from CV, CV+VSL3, GF, GF+VSL3, and GF+CV groups. Reverse-transcription qPCR for each gene was performed as described in Materials and Methods. Data are expressed as the percentage of the housekeeping gene 18S rRNA. Statistical analysis was performed using analysis of variance followed by Duncan’s post-hoc test with P < 0.05 considered statistically significant. Treatment groups that are not statistically different are labeled with the same letter.
Expression of Adh1 and Aldhs in Livers of CV and GF Mice following VSL3 Treatment or Conventionalization.
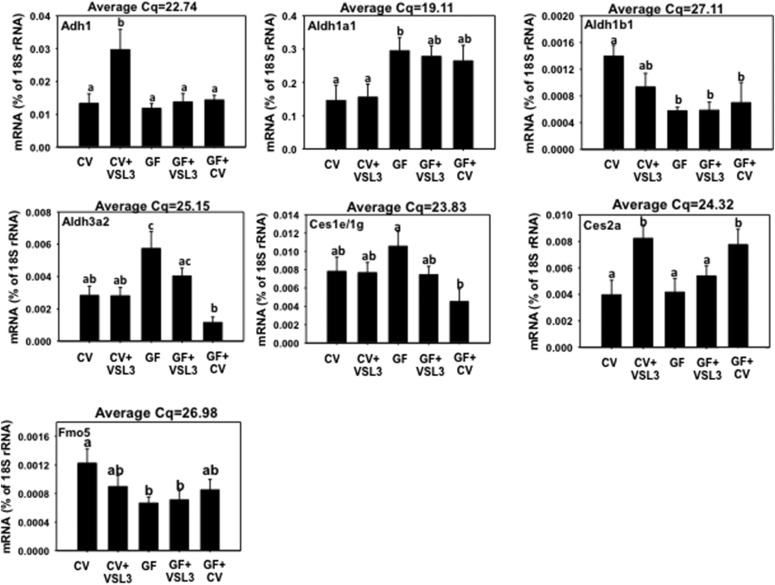
Adhs and Aldhs are critical phase I enzymes in the liver that metabolize alcohols and aldehydes. Adhs convert alcohols into aldehydes, whereas Aldhs further metabolize aldehydes into acids. VSL3 markedly increased the mRNA of Adh1 in livers of CV mice (Fig. 7). However, the expression of Aldh1a7, 4a1, and 7a1 mRNAs was similar in all groups (Supplemental Fig. 3). GF conditions resulted in an increase in the mRNAs of Aldh1a1 and Aldh3a2, but a decrease in the mRNA of Aldh1b1 (Fig. 7). Conventionalization of GF mice reduced Aldh3a2 mRNA back to conventional levels, but did not return the mRNAs of Aldh1a1 or Aldh1b1 back to conventional levels.
Fig. 7.
The mRNA expression of Adh1, Aldh1a1, Aldh1b1, Aldh3a2, Ces1e/1g, Ces2a, and Fmo5 in liver samples from CV, CV+VSL3, GF, GF+VSL3, and GF+CV groups. Reverse-transcription qPCR for each gene was performed as described in Materials and Methods. Data are expressed as the percentage of the housekeeping gene 18S rRNA. Statistical analysis was performed using analysis of variance followed by Duncan’s post-hoc test with P < 0.05 considered statistically significant. Treatment groups that are not statistically different are labeled with the same letter.
Expression of Cess, Fmos, Akrs, Ephx1, and Nqo1 in Livers of CV and GF Mice following VSL3 Treatment or Conventionalization.
Cess are a group of important phase I enzymes that hydrolyze carboxylic esters to form alcohols and carboxylates. The mRNAs of Ces1e/1g, Ces2c, and Ces3a were not readily altered by VSL3, GF, or conventionalized conditions; however, Ces2a mRNA was upregulated by VSL3 in livers of CV mice but not in GF mice, whereas conventionalization of GF mice also increased Ces2a mRNA (Fig. 7; Supplemental Fig. 3). Fmos are monooxygenases that oxygenate drugs and other xenobiotics. Fmo1, 2, and 4 mRNAs were similar in livers of all five groups of mice (Supplemental Fig. 4); however, Fmo5 mRNA was downregulated in livers of GF mice (Fig. 7). Akrs, Ephx1, and Nqo1 are phase I enzymes that are involved in oxidation/reduction reactions. In general, the mRNAs of these genes were not readily altered by VSL3, GF, or conventionalized conditions, except that Akr1d1 mRNA was lower in livers of conventionalized GF mice (Supplemental Fig. 4).
Expression of Gsts in Livers of CV and GF Mice following VSL3 Treatment or Conventionalization.
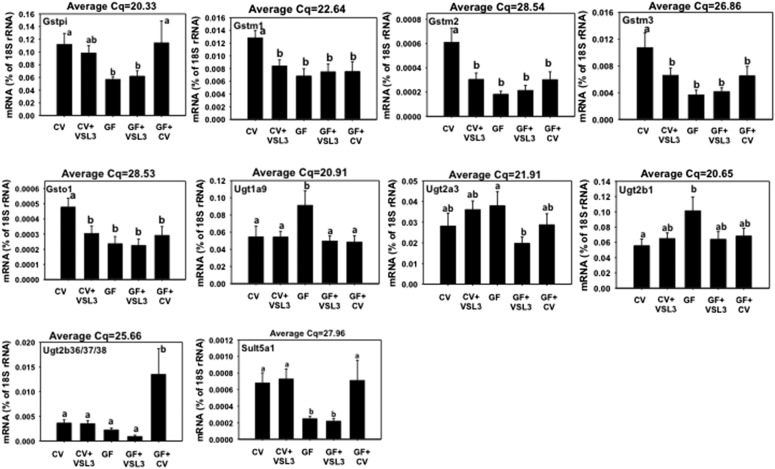
Gsts are an important family of phase II enzymes that detoxify electrophiles by conjugating them with glutathione. The mRNAs of Gstm1, m2, m3, and o1 were all downregulated by VSL3 in livers of CV mice (Fig. 8); Gstm4 mRNA also tended to be decreased by VSL3 in livers of CV mice, although a statistically significant difference was not achieved (Supplemental Fig. 5). Other Gst isoforms were not readily altered by VSL3 in livers of either CV or GF mice, except Gsta1, a3, and a4 mRNAs, which tended to be increased in livers of VSL3-treated CV mice (as compared with control CV mice), although a statistically significant difference was not achieved; this tendency disappeared in livers of VSL3-treated GF mice (Supplemental Fig. 6). GF conditions resulted in decreased mRNAs of Gstpi, m1, m2, m3, and o1 (Fig. 8). The mRNAs of Gstm4 and t2 also tended to be lower in livers of GF mice, although a statistically significant difference was not achieved (Supplemental Fig. 6). Conventionalization of GF mice restored the Gstpi mRNA, but did not normalize the mRNAs of Gstm1, m2, m3, or o1 (Fig. 8). In summary, multiple Gstm isoforms as well as Gsto1 were downregulated by VSL3; these genes as well as Gstpi were also downregulated by GF conditions.
Fig. 8.
The mRNA expression of Gstpi, Gstm1-m3, Gsto1, Ugt1a9, Ugt2a3, Ugt2b1, Ugt2b36/37/38, and Sult5a1 in liver samples from CV, CV+VSL3, GF, GF+VSL3, and GF+CV groups. Reverse-transcription qPCR for each gene was performed as described in Materials and Methods. Data are expressed as the percentage of the housekeeping gene 18S rRNA. Statistical analysis was performed using analysis of variance followed by Duncan’s post-hoc test with P < 0.05 considered statistically significant. Treatment groups that are not statistically different are labeled with the same letter.
Expression of Ugts in Livers of CV and GF Mice following VSL3 Treatment or Conventionalization.
Ugts are a group of phase II enzymes that catalyze the glucuronidation of substrates. As shown in Fig. 8 and Supplemental Fig. 6, in general, most of the 12 Ugt mRNAs examined were not altered by any of the treatments, and these genes include Ugt1a1, 1a5, 1a6, 1a7, 2b5, 2b34, 2b35, and 2b36. VSL3 had no effect on the Ugt mRNA expression in livers of CV mice; however, it decreased the mRNAs of Ugt1a9 and 2a3 in livers of GF mice. GF conditions upregulated the mRNAs of Ugt1a9 and 2b1. Conventionalization of GF mice reduced Ugt1a9 mRNA back to CV levels, and tended to reduce Ugt2a3 and 2b1 mRNAs to CV levels (although a statistical significance was not achieved), but markedly increased the mRNA of Ugt2b36/37/38.
Expression of Sults in Livers of CV and GF Mice following VSL3 Treatment or Conventionalization.
Sults are a group of phase II enzymes that catalyze the transfer of the sulfate group from the cosubstrate 3′-physphoadenosine-5′-phosphosulfate to alcohols or amines. 3′-Phosphoadenosine 5′-phosphosulfate synthase 2 is involved in the synthesis of the cosubstrate of the sulfation reactions. The mRNAs of Sult2b1, 2a1, and 3a1 were minimally expressed in male mouse livers (average Cq > 30, data not shown). The mRNAs of Sult1b1, 1d1, and 3′-phosphoadenosine 5′-phosphosulfate synthase 2 were not altered by any of the treatments (Supplemental Fig. 6). VSL3 had minimal effects on the expression of Sults; however, GF conditions markedly decreased Sult5a1 mRNA (Fig. 8; Supplemental Fig. 6). GF mice colonized with bacteria from the CV environment had lower expression of Sult1a1 compared with GF mice colonized with VSL3 bacteria, indicating that different bacteria components have different effects on Sult1a1 gene expression.
ChIP-qPCR of DNA-Binding Fold Enrichment of PXR and RNA-Pol-II to the Cyp3a Gene Loci, as well as DNA-Binding Fold Enrichment of PPARα and RNA-Poll-II to the Cyp4a Gene Loci.
To determine the mechanistic involvement of PXR and PPARα in modulating the transcriptional regulation of the Cyp3a and Cyp4a clusters, ChIP was performed in livers of CV, GF, and conventionalized GF mice (two independent pull-downs per receptor). Because VSL3 had minimal effects on the Cyp3a and Cyp4a gene expression, ChIP of VSL3 samples was not performed.
The constitutive PXR-DNA binding sites to the Cyp3a cluster in mouse liver were selected based on our previous publication (Cui et al., 2010), and the constitutive PPARα-DNA binding sites to the Cyp4a cluster in mouse liver were selected based on NCBI Gene Expression Omnibus Database query data set GSE61817 (Lee et al., 2014) (Supplemental Fig. 7). The PXR- and PPARα-DNA binding sites that are proximal to the transcription start sites of target genes were analyzed for putative DNA-binding motifs (namely, DR-3, DR-4, ER-6, and ER-8 for PXR, as well as DR-1 and DR-2 for PPARα), and qPCR primers were designed centering the key motifs as noted in Supplemental Table 3.
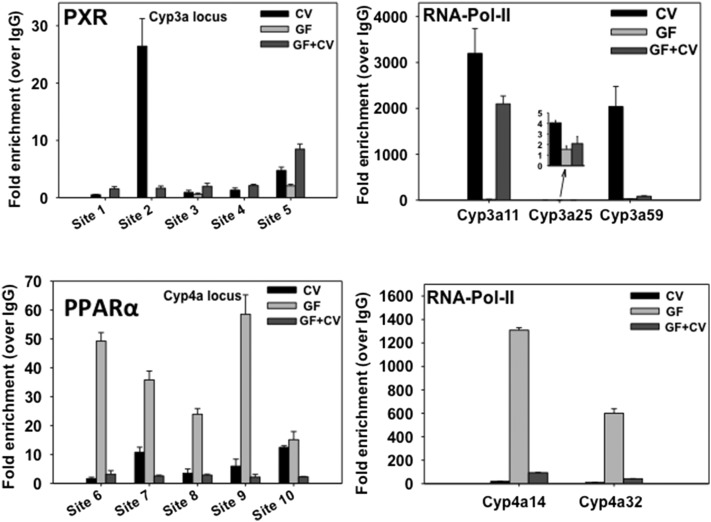
As shown in Fig. 9, among the five selected PXR-DNA binding sites, site 2 (−1.6 kb upstream of Cyp3a11) displayed the highest PXR-DNA binding in livers of CV mice (26-fold), and GF conditions markedly decreased the PXR-DNA binding, whereas conventionalization moderately restored the PXR-DNA binding (1.66-fold). Site 5 (−8.9 kb upstream of Cyp3a59) displayed the second-highest PXR-DNA binding fold enrichment in livers of CV mice (4.77-fold), and GF conditions reduced the PXR-DNA binding to 2.08-fold, whereas conventionalization increased the PXR-DNA binding (8.47-fold). Site 1 (−90 bp upstream of Cyp3a11) as well as site 3 and site 4 (−144 and −1.9 kb upstream of Cyp3a25, respectively) had low PXR-DNA binding in livers of CV mice, and GF conditions further reduced PXR-DNA binding to background levels, whereas conventionalization increased PXR-DNA binding in these regions. To confirm the functional significance of PXR-DNA binding to the Cyp3a cluster on gene transcription, quantification of RNA-Pol-II to the promoters of Cyp3a11, 3a25, and 3a59 were analyzed by ChIP (due to high sequence similarity, the primers targeting specific promoters of other P450 genes were not designed). Consistent with the PXR-DNA binding data, there was a marked decrease in RNA-Pol-II binding to the Cyp3a11 promoter (from 3200-fold in CV mice to 15-fold in GF conditions), whereas conventionalization restored RNA-Pol-II binding approximately 2100-fold (Fig. 9). RNA-Pol-II binding to Cyp3a59 promoter decreased from 2039-fold (CV) to 30-fold (GF), whereas conventionalization moderately increased RNA-Pol-II binding (86-fold). Constitutive RNA-Pol-II binding to the Cyp3a25 promoter was low (4.06-fold), whereas GF conditions further decreased the fold enrichment to 1.55-fold, and RNA-Pol-II binding to conventionalized conditions was approximately 2.09-fold.
Fig. 9.
ChIP-qPCR of the DNA binding for PXR and RNA-Pol-II to the Cyp3a gene loci, as well as PPARα and RNA-Pol-II to the Cyp4a gene loci. For PXR, sites 1–5 were selected based on the reanalysis of a published PXR-ChIP sequencing experiment in control adult conventional male mouse livers (Cui et al., 2010), and qPCR primers were designed centering the known PXR-DNA binding motifs (DR-3, DR-4, ER-6, and ER-8) in these regions. For PPARα, sites 6–10 were selected based on the reanalysis of a published PPARα ChIP sequencing experiment in control adult conventional male mouse livers (NCBI Gene Expression Omnibus Database query data set GSE61817; Lee et al., 2014), and qPCR primers were designed centering the known PPARα-DNA binding motif DR-2 in these regions. For RNA-Pol-II, qPCR primers were designed centering the TATA box within the promoters of the target genes. ChIP assays were performed using specific antibodies against PXR, PPARα, RNA-Pol-II, and IgG as described inMaterials and Methods. Data were first normalized to genomic DNA input, and then expressed as fold enrichment over IgG control.
In regard to the Cyp4a cluster, PPARα binding to site 6 (approximately 4 kb upstream of Cyp4a10) increased from 1.67-fold (CV) to 49-fold (GF), whereas conventionalization reduced PPARα-binding to 3.21-fold (Fig. 9). Similarly, PPARα binding to site 7 (approximately 1 kb downstream of the transcription start site and within the first intron of Cyp4a10) increased from 10.75-fold (CV) to 36-fold (GF), whereas conventionalization reduced the PPARα binding to 2.59-fold. PPARα binding to the other regions—namely, site 8 and site 9 (approximately −8.1 kb upstream and 930 bp downstream [within the first intron] of Cyp4a31, respectively) and site 10 (approximately 4 kb upstream of Cyp4a32)—followed a similar pattern, which was low PPARα binding in livers of CV mice, increased PPARα binding in livers of GF mice, and reduced PPARα binding in livers of conventionalized GF mice. The RNA-Pol-II binding to the promoters of Cyp4a14 and 4a32 was consistent with the PPARα-binding profiles. Due to high sequence similarity in the promoter regions, RNA-Pol-II binding to Cyp4a10 and 4a31 was not performed.
In conclusion, the present study has shown that VSL3 in the drinking water of CV and GF mice resulted in successful colonization of the VSL3 bacterial components in the large intestine, but in general, VSL3 has a relatively minor effect on hepatic drug-metabolizing enzyme expression in mice. Germ-free conditions resulted in the most prominent changes in hepatic drug-metabolizing enzyme expression, most notably a consistent downregulation of many genes in the Cyp3a cluster, but a consistent upregulation of many genes in the Cyp4a cluster. Conventionalization of GF mice at least partially restores the expression of these genes to CV levels. The GF- and conventionalization-mediated changes in Cyp3a and 4a genes are associated with altered PXR and PPARα binding to the targeted DNA sequences within these genes.
Discussion
One of the interesting observations of the present study is the coregulation of the Cyp3a and Cyp4a genes in specific genomic regions of polycistronic clusters. It is possible that distinct genomic regions within polycistron clusters are “hot zones” for transactivation mediated by nuclear receptors, such as PXR and PPARα, and this may be due to distinct histone epigenetic mechanisms (Barrera and Ren, 2006; Wang et al., 2009) that allow a permissive chromatin environment for PXR and PPARα to transcribe certain regions of a gene cluster. Indeed, previous studies using ChIP sequencing have also identified region-specific localization of PXR and PPARα to the Cyp3a and Cyp4a clusters in mouse liver, respectively (Cui et al., 2010 for PXR; Lee et al., 2014 for PPARα) (Supplemental Fig. 2). The interaction between intestinal microbiota and the hepatic histone epigenetic marks, as well as the subsequent effects on nuclear receptor recruitment of target genes, should be addressed in future studies.
The altered PXR and PPARα signaling in livers of GF and conventionalized GF mice is likely due to altered levels of bacterial metabolites in GF and conventionalization conditions. For PXR, secondary bile acids, such as lithocholic acid, as well as indole 3-propionic acid, are known endogenous PXR activators (Staudinger et al., 2001; Venkatesh et al., 2014). For PPARα, it has been shown that the circadian rhythm gene Clock transactivates the expression of PPARα (Oishi et al., 2005), and the Clock:Bmal1 target genes (such as Per1, 2, and 3) are markedly increased in livers of GF mice (data not shown), suggesting that the germ-free conditions may upregulate the PPARα signaling by enhancing the Bmal1:Clock signaling. Future studies should include colonized bacteria in conventionalized GF mice to determine which bacteria are likely responsible for increasing the PXR signaling but suppressing the PPARα signaling in the liver.
Previously, the bona fide PXR and PPARα target genes that encode drug-metabolizing enzymes in mice have been determined using pharmacological and genetic approaches (Aleksunes and Klaassen, 2012). Cyp3a11 as well as Gstm1-3 have been shown to be bona fide PXR target genes (Aleksunes and Klaassen, 2012). We have observed a downregulation of these genes in GF livers, correlating with decreased PXR binding, and confirmed the critical involvement of PXR in the hepatic regulation of these genes following modifications in the intestinal microbiota (Fig. 2 and 9). Similarly, Cyp4a14, Aldh1a1, and Aldh3a2 are bona fide PPARα targets in the liver (Aleksunes and Klaassen, 2012), and we have demonstrated an upregulation of all of these genes in livers of GF mice, and this increase in Cyp4a14 and Aldh3a2 is completely reversed by conventionalization (Figs. 3 and 7). The basal expression of Aldh1b1 and Sult5a1 has been shown to be suppressed by PPARα, as noted by increased Aldh1b1 and Sult5a1 mRNA in livers of PPARα-null mice (Aleksunes and Klaassen, 2012); the present study has also demonstrated a decrease in Aldh1b1 and Sult5a1 mRNAs in livers of GF mice, which was reversed by conventionalization of the GF mice (Figs. 7 and 8). Therefore, the alteration of Sult5a1 mRNAs in GF and conventionalized conditions is likely mediated through PPARα. PPARα-ChIP data on the Cyp4a cluster further demonstrated the role of PPARα in the hepatic regulation of target genes following modifications in the intestinal microbiota. Interestingly, Gstm1, m3, and m4 are also common target genes of both PXR and PPARα, evidenced by PXR-dependent upregulation following pregnenolone-16-alpha-carbonitrile treatment and PPARα-dependent upregulation following clofibrate treatment (Aleksunes and Klaassen, 2012). In the present study, the downregulation of Gstm1-3 mRNAs (and a tendency to decrease Gstm4 mRNA) by VSL3 treatment and GF conditions suggests that the PXR effect is dominant over PPARα in regulating the expression of these genes. Conversely, Ugt1a9 mRNA has been shown to be increased by both PXR and PPARα ligands (PCN and clofibrate, respectively) (Aleksunes and Klaassen, 2012), whereas in the present study, Ugt1a9 mRNA is increased in livers of GF mice but reduced to CV levels in livers of conventionalized GF mice. Thus, Ugt1a9 mRNA regulation may involve more PPARα than PXR following changes in intestinal microbiota. Certain drug-metabolizing enzymes, such as Ugt2b1, are downregulated by PPARα, evidenced by decreased gene expression following PPARα-ligand treatment but increased gene expression in PPARα-null mice (Aleksunes and Klaassen, 2012), but its mRNA is actually increased in livers of GF mice, where PPARα signaling appears to be enhanced (Figs. 7 and 8). Ugt1a1, 1a5, 2b35, and 2b36 mRNAs have been shown to be increased by PXR and PPARα ligands (PCN and clofibrate, respectively) (Aleksunes and Klaassen, 2012), but they are not changed in GF or conventionalized conditions (Fig. 8). The inconsistency in these observations suggests that additional regulatory factors are present in the expression of these Ugts.
The present findings of a decrease in expression of the Cyp3a genes and increase in expression of Cyp4a genes in livers of GF mice are consistent with our previous studies (Selwyn et al., 2015a,b). The present results are also consistent with our previous studies in GF mice regarding the regulation of many other drug-metabolizing enzymes, such as Cyp1a2, Aldh1b1, Aldh3a2, Gstpi, Gstm3, and Sult5a1 (Selwyn et al., 2015a,b). The present observation of normalized Cyp3a11 gene expression in livers of GF mice after conventionalization is also consistent with previous studies using conventionalization or secondary bile acid replacement approaches (Toda et al., 2009; Claus et al., 2011). The contribution of the present study is that, in addition to the previous knowledge on the effect of GF conditions on drug-processing gene expression, results of this study have systematically addressed the effects of the probiotic VSL3 and conventionalization on the expression of major drug-metabolizing enzymes in the liver, and determined the putative PXR and PPARα binding to Cyp3a and Cyp4a at the cluster level, which provides mechanistic explanations of the gene expression profiles following changes in intestinal microbiota. There are certain genes for which the mRNAs are moderately altered in livers of GF mice in a previous RNA sequencing (RNA-Seq) (Selwyn et al., 2015b), but were not altered in livers of GF mice the present study (such as Ces2a, Akr1c19, Cyp2b10, Cyp3a16, etc.). This inconsistency is likely due to different techniques used (RNA-Seq versus reverse-transcription qPCR), vehicle effects, and/or statistical methods. Only moderate changes were reported between some genes in CV and GF mice in the previous study, whereas the trend is still present in the current study with many genes, but is not statistically significant.
The starting ages of the mice on VSL3 or conventionalization are different (2 months old for VSL3 treatment versus 1 month old for conventionalization). It is possible that the difference in the starting age will influence the regulation of drug-metabolizing genes. It has been shown that early-age conventionalization has more impact on the immune response signaling than late-age exposure to the conventional microbial environment (Yamamoto et al., 2012). Thus, it is also possible that early-age exposure to VSL3 may have a different effect on drug-metabolizing enzyme expression. Regarding the duration of the VSL3 treatment, it has been shown that VSL3 supplementation for just 3 days profoundly alters the ileal microbiota composition in conventional mice, and improves the disease scores of dextran sodium sulfate–induced colitis (Mar et al., 2014). Therefore, the 2-month treatment with VSL3 should be sufficient to alter the intestinal microbiota composition. However, it is likely that the duration of VSL3 is not long enough to markedly alter the expression of drug-metabolizing enzymes, and chronic treatment with VSL3 may produce different results.
The PXR binding to site 2 as well as RNA-Pol-II binding to Cyp3a59 in livers of GF+CV mice did not completely restore the CV conditions. It is possible that a moderate increase in the PXR/RNA-Pol-II binding is sufficient to transactivate the target gene expression; it is also possible that additional regulatory factors, such as permissive chromatin epigenetic marks and other transcription factors, facilitate the complete restoration of the Cyp3a11 gene expression in livers of GF+CV mice. This will need to be tested using an unbiased detection method, such as RNA-Seq, in future studies.
One potential concern regarding conventionalization procedures is that the types of bacteria introduced to GF+CV mice may be facility-specific. It is likely that the exogenous bacteria gained in the GF+CV mice do not necessarily recapitulate the exogenous bacteria configuration in CV mice in terms of both quantity and composition, evidenced by a further increase in Cyp4f18, Cyp4v3, and Ugt2b36/37/38 mRNAs in livers of GF+CV mice as compared with CV mice. Specific bacterial strains in the intestines of conventionalized GF mice that are responsible for the changes in the PXR/PPARα signaling and the expression of drug-metabolizing enzymes of the host liver are not known, but a previous study using conventionalized C3H mice showed that Enterococcaceae, Enterobacteriaceae, Lactobacillaceae, Erysipelotrichaceae, and Peptostreptococcaceae are the first bacterial families to settle in the intestine after exposure to the local environment, whereas Coriobacteriaceae appears to link the liver and the intestine in host energy metabolism pathways (Claus et al., 2011). Future studies using 16S rRNA and metatranscriptome sequencing approaches will be helpful to determine the specific bacterial strains that modulate the changes in hepatic drug-metabolizing enzyme expression, and subsequently administering these bacterial strains to GF mice will validate its contribution.
Supplementary Material
Acknowledgments
The authors thank Dr. Jerry Cangelosi as well as his laboratory members Connie Tzou and Kris Weigel, and Dr. Scott Meschke in the Department of Environmental and Occupational Health Sciences, University of Washington (Seattle, WA), for their discussion and advice on bacterial quantification, as well as previous members in Dr. Klaassen’s laboratory for help in tissue collection.
Abbreviations
- Adh
alcohol dehydrogenase
- AhR
aryl hydrocarbon receptor
- Akr
aldo-keto reductase
- Aldh
aldehyde dehydrogenase
- Ces
carboxylesterase
- ChIP
chromatin immunoprecipitation
- CV
conventional mice
- P450
cytochrome P450
- DR
direct-repeat
- Ephx
epoxide hydrolase
- ER
everted-repeat
- Fmo
flavin-containing monooxygenase
- GF
germ free
- Gst
glutathione S-transferase
- NCBI
National Center for Biotechnology Information
- Nqo1
NAD(P)H dehydrogenase quinone 1
- PPARα
peroxisome proliferator–activated receptor α
- PXR
pregnane X receptor
- qPCR
quantitative polymerase chain reaction
- RNA-Pol-II
RNA polymerase-II
- RNA-Seq
RNA sequencing
- rRNA
ribosomal RNA
- Sult
sulfotransferase
- Ugt
UDP glucuronosyltransferase
Authorship Contributions
Participated in research design: Klaassen, Selwyn, Cui.
Conducted experiments: Selwyn, Cui, Cheng.
Wrote or contributed to the writing of the manuscript: Cui, Klaassen, Selwyn, Cheng.
Footnotes
This work was supported by the National Institutes of Health [Grants GM111381 and ES019487] as well as start-up funds from University of Washington Center of Ecogenetics and Environmental Health [Grant P30 ES0007033].
 This article has supplemental material available at dmd.aspetjournals.org.
This article has supplemental material available at dmd.aspetjournals.org.
References
- Aleksunes LM, Klaassen CD. (2012) Coordinated regulation of hepatic phase I and II drug-metabolizing genes and transporters using AhR-, CAR-, PXR-, PPARα-, and Nrf2-null mice. Drug Metab Dispos 40:1366–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alnouti Y, Klaassen CD. (2008) Regulation of sulfotransferase enzymes by prototypical microsomal enzyme inducers in mice. J Pharmacol Exp Ther 324:612–621. [DOI] [PubMed] [Google Scholar]
- Barrera LO, Ren B. (2006) The transcriptional regulatory code of eukaryotic cells--insights from genome-wide analysis of chromatin organization and transcription factor binding. Curr Opin Cell Biol 18:291–298. [DOI] [PubMed] [Google Scholar]
- Bibiloni R, Fedorak RN, Tannock GW, Madsen KL, Gionchetti P, Campieri M, De Simone C, Sartor RB. (2005) VSL#3 probiotic-mixture induces remission in patients with active ulcerative colitis. Am J Gastroenterol 100:1539–1546. [DOI] [PubMed] [Google Scholar]
- Boyle RJ, Robins-Browne RM, and Tang ML (2006) Probiotic use in clinical practice: what are the risks? Am J Clin Nutr 83:1256-1264; quiz 1446-1257. [DOI] [PubMed] [Google Scholar]
- Buckley DB, Klaassen CD. (2009) Induction of mouse UDP-glucuronosyltransferase mRNA expression in liver and intestine by activators of aryl-hydrocarbon receptor, constitutive androstane receptor, pregnane X receptor, peroxisome proliferator-activated receptor alpha, and nuclear factor erythroid 2-related factor 2. Drug Metab Dispos 37:847–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho BM, Guadagnini D, Tsukumo DM, Schenka AA, Latuf-Filho P, Vassallo J, Dias JC, Kubota LT, Carvalheira JB, Saad MJ. (2012) Modulation of gut microbiota by antibiotics improves insulin signalling in high-fat fed mice. Diabetologia 55:2823–2834. [DOI] [PubMed] [Google Scholar]
- Claus SP, Ellero SL, Berger B, Krause L, Bruttin A, Molina J, Paris A, Want EJ, de Waziers I, Cloarec O, et al. (2011) Colonization-induced host-gut microbial metabolic interaction. MBio 2:e00271–e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collado MC, Rautava S, Isolauri E, Salminen S. (2015) Gut microbiota: a source of novel tools to reduce the risk of human disease? Pediatr Res 77:182–188. [DOI] [PubMed] [Google Scholar]
- Cui JY, Gunewardena SS, Rockwell CE, Klaassen CD. (2010) ChIPing the cistrome of PXR in mouse liver. Nucleic Acids Res 38:7943–7963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhar M, Sepkovic DW, Hirani V, Magnusson RP, Lasker JM. (2008) Omega oxidation of 3-hydroxy fatty acids by the human CYP4F gene subfamily enzyme CYP4F11. J Lipid Res 49:612–624. [DOI] [PubMed] [Google Scholar]
- Furet JP, Quénée P, Tailliez P. (2004) Molecular quantification of lactic acid bacteria in fermented milk products using real-time quantitative PCR. Int J Food Microbiol 97:197–207. [DOI] [PubMed] [Google Scholar]
- Hardwick JP. (2008) Cytochrome P450 omega hydroxylase (CYP4) function in fatty acid metabolism and metabolic diseases. Biochem Pharmacol 75:2263–2275. [DOI] [PubMed] [Google Scholar]
- Jancova P, Anzenbacher P, Anzenbacherova E. (2010) Phase II drug metabolizing enzymes. Biomed Pap Med Fac Univ Palacky Olomouc Czech Repub 154:103–116. [DOI] [PubMed] [Google Scholar]
- Kim DH. (2015) Gut Microbiota-Mediated Drug-Antibiotic Interactions. Drug Metab Dispos 43:1581–1589. [DOI] [PubMed] [Google Scholar]
- Knight TR, Choudhuri S, Klaassen CD. (2008) Induction of hepatic glutathione S-transferases in male mice by prototypes of various classes of microsomal enzyme inducers. Toxicol Sci 106:329–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroetz DL, Yook P, Costet P, Bianchi P, Pineau T. (1998) Peroxisome proliferator-activated receptor alpha controls the hepatic CYP4A induction adaptive response to starvation and diabetes. J Biol Chem 273:31581–31589. [DOI] [PubMed] [Google Scholar]
- Lee JH, Moon G, Kwon HJ, Jung WJ, Seo PJ, Baec TY, Lee JH, Kim HS. (2012) [Effect of a probiotic preparation (VSL#3) in patients with mild to moderate ulcerative colitis]. Korean J Gastroenterol 60:94–101. [DOI] [PubMed] [Google Scholar]
- Lee JM, Wagner M, Xiao R, Kim KH, Feng D, Lazar MA, Moore DD. (2014) Nutrient-sensing nuclear receptors coordinate autophagy. Nature 516:112–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mar JS, Nagalingam NA, Song Y, Onizawa M, Lee JW, Lynch SV. (2014) Amelioration of DSS-induced murine colitis by VSL#3 supplementation is primarily associated with changes in ileal microbiota composition. Gut Microbes 5:494–503. [DOI] [PubMed] [Google Scholar]
- Mardini HE, Grigorian AY. (2014) Probiotic mix VSL#3 is effective adjunctive therapy for mild to moderately active ulcerative colitis: a meta-analysis. Inflamm Bowel Dis 20:1562–1567. [DOI] [PubMed] [Google Scholar]
- Oishi K, Shirai H, Ishida N. (2005) CLOCK is involved in the circadian transactivation of peroxisome-proliferator-activated receptor alpha (PPARalpha) in mice. Biochem J 386:575–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penner RM, Fedorak RN. (2005) Probiotics in the management of inflammatory bowel disease. MedGenMed 7:19. [PMC free article] [PubMed] [Google Scholar]
- Pratt-Hyatt M, Lickteig AJ, Klaassen CD. (2013) Tissue distribution, ontogeny, and chemical induction of aldo-keto reductases in mice. Drug Metab Dispos 41:1480–1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. (2011) Integrated genomics viewer. Nat Biotechnol 29:24–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders ME (2008) Probiotics: definition, sources, selection, and uses. Clin Infect Dis 46 Suppl 2:S58-61; discussion S144-151. [DOI] [PubMed] [Google Scholar]
- Selwyn FP, Cheng SL, Bammler TK, Prasad B, Vrana M, Klaassen C, Cui JY. (2015a) Developmental regulation of drug-processing genes in livers of germ-free mice. Toxicol Sci 147:84–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selwyn FP, Cui JY, Klaassen CD. (2015b) RNA-Seq quantification of hepatic drug processing genes in germ-free mice. Drug Metab Dispos 43:1572–1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staudinger JL, Goodwin B, Jones SA, Hawkins-Brown D, MacKenzie KI, LaTour A, Liu Y, Klaassen CD, Brown KK, Reinhard J, et al. (2001) The nuclear receptor PXR is a lithocholic acid sensor that protects against liver toxicity. Proc Natl Acad Sci USA 98:3369–3374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toda T, Saito N, Ikarashi N, Ito K, Yamamoto M, Ishige A, Watanabe K, Sugiyama K. (2009) Intestinal flora induces the expression of Cyp3a in the mouse liver. Xenobiotica 39:323–334. [DOI] [PubMed] [Google Scholar]
- Vandenplas Y, Huys G, Daube G. (2015) Probiotics: an update. J Pediatr (Rio J) 91:6–21. [DOI] [PubMed] [Google Scholar]
- Venkatesh M, Mukherjee S, Wang H, Li H, Sun K, Benechet AP, Qiu Z, Maher L, Redinbo MR, Phillips RS, et al. (2014) Symbiotic bacterial metabolites regulate gastrointestinal barrier function via the xenobiotic sensor PXR and Toll-like receptor 4. Immunity 41:296–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitali B, Candela M, Matteuzzi D, Brigidi P. (2003) Quantitative detection of probiotic Bifidobacterium strains in bacterial mixtures by using real-time PCR. Syst Appl Microbiol 26:269–276. [DOI] [PubMed] [Google Scholar]
- Wang MZ, Saulter JY, Usuki E, Cheung YL, Hall M, Bridges AS, Loewen G, Parkinson OT, Stephens CE, Allen JL, et al. (2006) CYP4F enzymes are the major enzymes in human liver microsomes that catalyze the O-demethylation of the antiparasitic prodrug DB289 [2,5-bis(4-amidinophenyl)furan-bis-O-methylamidoxime]. 2,5-bis 4-amidinophenyl furan-bis-O-methylamidoxime Drug Metab Dispos 34:1985–1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Schones DE, Zhao K. (2009) Characterization of human epigenomes. Curr Opin Genet Dev 19:127–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson GR. (1996) Cytochrome P4503A (CYP3A) metabolism: prediction of in vivo activity in humans. J Pharmacokinet Biopharm 24:475–490. [DOI] [PubMed] [Google Scholar]
- Xu C, Li CY, Kong AN. (2005) Induction of phase I, II and III drug metabolism/transport by xenobiotics. Arch Pharm Res 28:249–268. [DOI] [PubMed] [Google Scholar]
- Yamamoto M, Yamaguchi R, Munakata K, Takashima K, Nishiyama M, Hioki K, Ohnishi Y, Nagasaki M, Imoto S, Miyano S, et al. (2012) A microarray analysis of gnotobiotic mice indicating that microbial exposure during the neonatal period plays an essential role in immune system development. BMC Genomics 13:335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Limaye PB, Lehman-McKeeman LD, Klaassen CD. (2012) Dysfunction of organic anion transporting polypeptide 1a1 alters intestinal bacteria and bile acid metabolism in mice. PLoS One 7:e34522. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.