Abstract
A genome-wide association study was performed in 1,130 premenopausal women to detect common variants associated with three serum iron-related phenotypes. Total iron binding capacity was strongly associated (p=10−14) with variants in and near the TF gene (transferrin), the serum iron transporting protein, and with variants in HFE (p= 4×10−7), which encodes the human hemochromatosis gene. Association was also detected between percent iron saturation (p=10−8) and variants in the chromosome 6 region containing both HFE and SLC17A2, which encodes a phosphate transport protein. No significant associations were detected with serum iron, but variants in HFE were suggestive (p=10−6). Our results corroborate prior studies in older subjects and demonstrate that the association of these genetic variants with iron phenotypes can be detected in premenopausal women.
Keywords: iron, premenopausal, GWAS, transferrin, hemochromatosis
Introduction
Iron deficiency anemia is a common disease, particularly among women of reproductive age due to menstrual blood loss[1, 2]. Iron overload is relatively uncommon, but causes toxicity in a variety of cell types and tissues[3]. Iron balance is regulated at the level of the enterocyte and absorbed iron is bound to serum transferrin for transport to the tissues. Serum iron concentration, along with the unsaturated iron binding capacity of transferrin, reflect to a large extent the state of the iron cycle in an individual. Consequently, clinical measurement of serum iron, total iron binding capacity (TIBC) and percent iron saturation may be used in assessing critical parameters indicating iron status[4]. Substantial portions of the variation in iron phenotypes among individuals have been attributed to genetic factors in several populations[5, 6]. Importantly, Benyamin[7], Tanaka[8],McLaren[9] and Li[10] have conducted genome-wide association studies (GWAS) of serum iron and related measures to detect common genetic polymorphisms contributing to the observed variation in these measures. They found significant evidence of association between these iron-related phenotypes and single nucleotide polymorphisms (SNPs) in genes known to be involved in iron metabolism (hemochromatosis, HFE; transferrin, TF; transmembrane protease serine 6, TRMPSS6), as well as at other loci.
These GWAS have included few or no premenopausal women, either because of concerns regarding the influence of menstrual blood loss on serum iron levels, or because younger women were absent from the cohorts. Thus, it is unclear at which age the genetic polymorphisms contributing to variation in serum iron-related phenotypes have their effect[11]. The aim of this study was to provide data regarding genes that influence iron metabolism in healthy premenopausal women; and further, by comparing these results with those from published studies, to provide information on the period of life in which genetic variants may have their primary effect. To address these aims, we performed GWAS studies of serum iron, total iron binding capacity (TIBC) and percent iron saturation in a cohort of 1,130 healthy, premenopausal European-American women.
Material and Methods
Subjects
Full sibships consisting of healthy European-American premenopausal women were recruited for a genetic study of bone mineral density in Indiana, along with additional unrelated women or half-sibships meeting the same criteria[12]. DNA was isolated from whole blood, and an additional blood sample was collected to obtain serum at the baseline visit (age 20–45) or during a second visit 4–5 years later (for subjects yet to reach menopause by that time)[13]. Studies were performed at the General Clinical Research Center of the Indiana University School of Medicine. Informed, written consent (Indiana University IRB: 8502-23) was obtained from all participants.
Iron measures
Iron and unsaturated iron binding capacity (UIBC) were measured on stored serum samples collected in subjects after an overnight fast using a spectrophotometric assay (Randox Laboratories; Antrim, Northern Ireland, United Kingdom). TIBC was calculated as the sum of serum iron and UIBC. Iron saturation was calculated as serum iron / TIBC and expressed as a percentage.
Genotyping and Imputation
Genotyping was performed on the Illumina Human610Quadv1_B BeadChips (Illumina, San Diego, CA, USA) by the Center for Inherited Disease Research (CIDR) using the Illumina Infinium II assay protocol. This array contains 592,532 markers. Genotype calls and quality control filters were applied as described previously[12]. Briefly, DNA samples with a 98% SNP call rate and genotypes consistent with reported family structures were included. A principal component analysis was performed in Eigenstrat[14] to cluster these samples with HapMap reference samples (CEU, YRI, CHB, and JPT). Only samples consistent with European ancestry were retained. Subsequently, individual SNP metrics were computed. SNPs having a call rate less than 95%, estimated minor allele frequency (MAF) less than 0.03 or significant deviation (p<0.0001) from Hardy Weinberg equilibrium were removed from further analysis. The genotype dataset taken forward for imputation contained 539,566 SNPs.
The IMPUTE2 software[15, 16] was used to generate genotype data for the 1000 Genomes Phase I SNP map in chromosomal regions with genome-wide significant evidence of association (p<5×10−8) to one or more of the iron phenotypes. The integrated variant set for samples of European descent was used as the source of reference haplotypes. The original genotype call was not overwritten for the set of SNPs genotyped on the Illumina array. A large subset of rare and low-frequency variants (MAF < 0.03) or those of lower imputation certainty (IMPUTE2 info score < 0.3) were excluded from association analyses.
Statistical analysis
For each of the phenotypic measures, distributions were evaluated and transformations applied as necessary to remove skewness or kurtosis. Pearson correlation coefficients (r) between the measures were computed using SASv9.1 (SAS Institute, Cary, NC, USA), and a subset of phenotypes selected for GWAS analyses to minimize the number of tests performed.
For each genotyped SNP in the filtered dataset, association tests were performed separately for each of the selected phenotypes. These models were fitted with R package GWAFv2.1, using a mixed model to account for relatedness within our sample[17]. Genome-wide results for each phenotype were evaluated for evidence of inflation of the association statistics. Genomic control as implemented in METAL[18] was applied as needed to correct for cryptic admixture or phenotypic non-normality. Imputed SNPs in chromosomal regions with evidence of association to one or more phenotypes were tested for association in the same manner.
Results
Evaluation of the iron phenotype distributions showed that only iron concentration was not normally distributed (skewness=0.71); this was corrected by a natural-logarithm transformation (skewness=0.11). Summary statistics for the iron phenotypes and subject demographics are provided in Table 1, and Pearson correlation coefficients (r) between the four iron phenotypes are shown in Table 2. Serum samples from study visits 4–5 years after baseline were included for 7 premenopausal subjects among the subset of our GWAS cohort included in the current report, when serum from the baseline visit was unavailable. Table 1 shows that demographic parameters are unchanged from the previous publication. Pairwise values of |r| greater than 0.5 with all other phenotypes were observed for UIBC; therefore, TIBC, iron, and iron saturation were selected as outcome measures for quantitative GWAS analyses.
Table 1.
Iron phenotypes and sample demographics
| Variable | n | Mean | SD | Min | Max |
|---|---|---|---|---|---|
| Age, years | 1130 | 34.3 | 7.7 | 20 | 50 |
| Weight, kg | 1130 | 71.6 | 16.9 | 39.6 | 133.9 |
| TIBC, mcg/dl | 1122 | 371.7 | 56.0 | 220.5 | 577.3 |
| Iron, mcg/dl | 1130 | 97.52 | 42.6 | 14.9 | 292.6 |
| ln(iron)1 | 1130 | 4.5 | 0.5 | 2.7 | 5.7 |
| Iron saturation, % | 1122 | 26.6 | 11.7 | 3.5 | 75.3 |
| UIBC, mcg/dl | 1122 | 274.1 | 66.1 | 85.0 | 483.7 |
Summary statistics for iron levels after natural-log transformation, as used in the statistical analyses presented. Untransformed values (Iron, mcg/dl) in the previous row of the Table are included for reference.
102 (9.0%) of the study subjects were iron deficient, as defined by serum iron levels below the normal range (<50 mcg/dl).
Table 2.
Pearson correlation coefficients (r) for the assayed or calculated iron-related phenotypes (n=1130)
| Variable | TIBC | Iron | Iron saturation |
|---|---|---|---|
| Iron | 0.08 | ||
| Iron saturation | −0.22 | 0.90 | |
| UIBC | 0.77 | −0.55 | −0.79 |
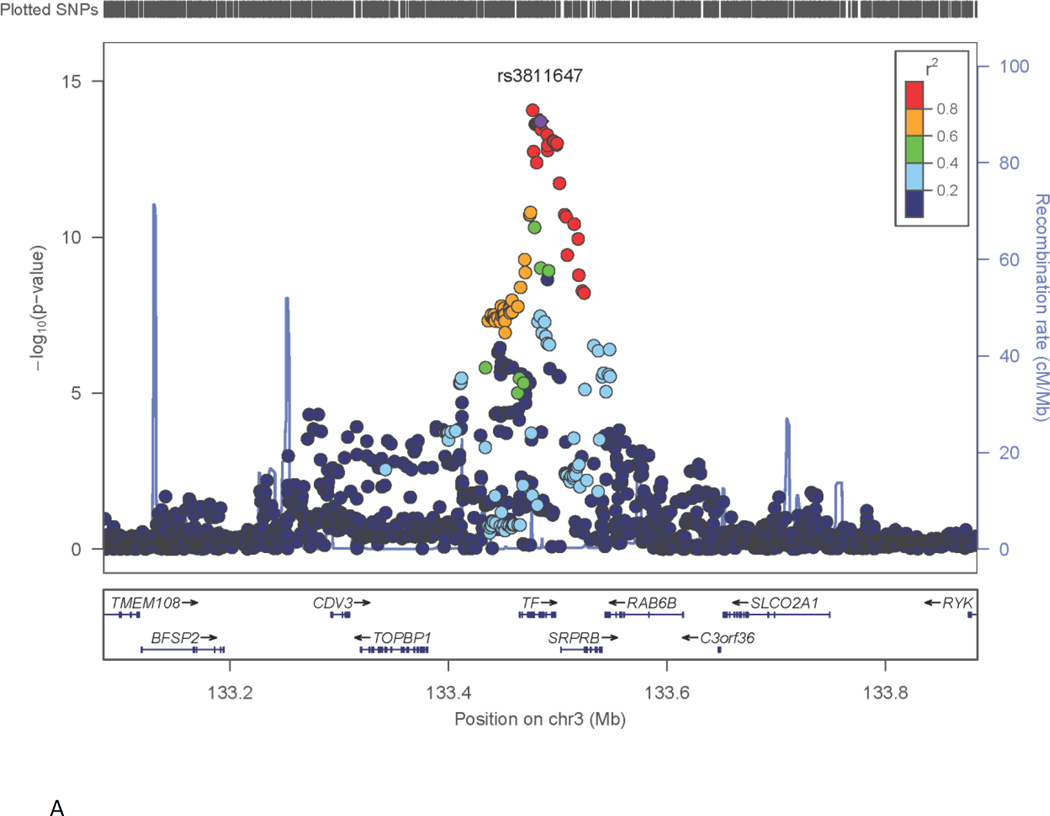
Results for the genome screen for TIBC are shown in Fig. S1A (Supplementary material). There was evidence of inflation of the genome-wide association statistics (λ=1.18) and genomic control was applied. Evidence of association for TIBC far surpassing the typical threshold for genome-wide significance (5×10−8) was found on chromosome 3 (133.48 Mb, build 37 coordinates), with the most significant results found for a group of relatively common SNPs (MAF=0.33) in the region of the transferrin (TF) gene (Fig. 1A). SNPs at or above the genome-wide significance level spanned the full length of the TF gene, extending from the region 5’ of TF, to the 5’ region of the neighboring gene, SRPRB8.
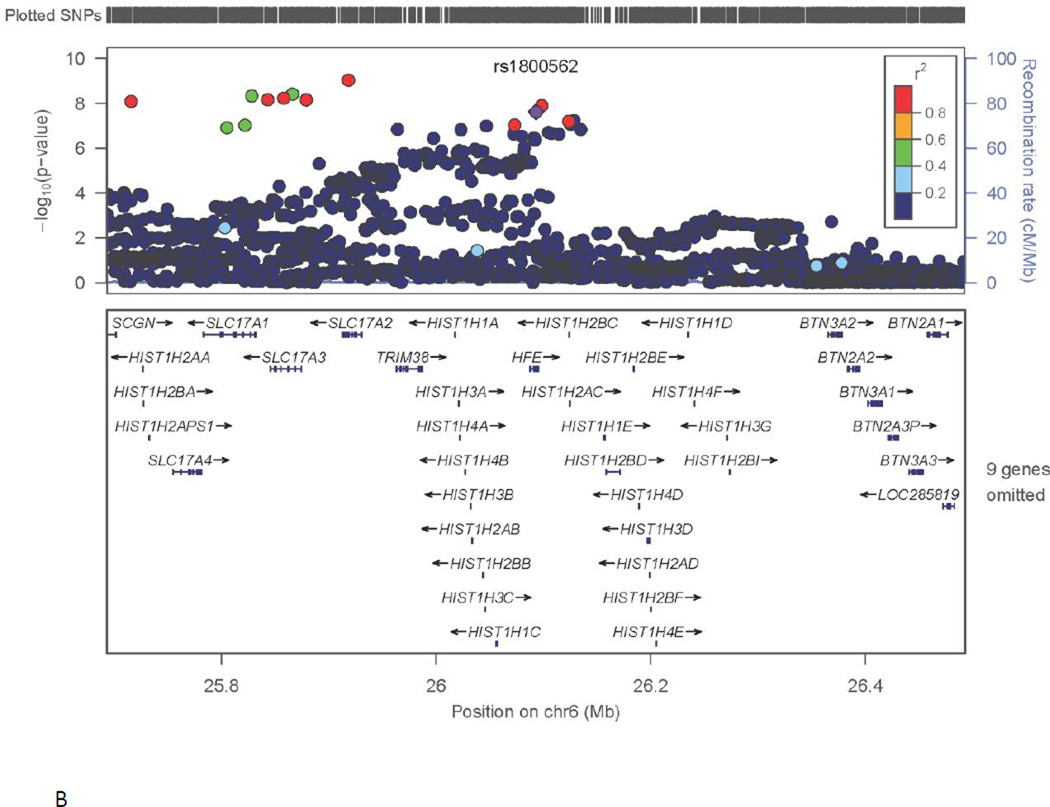
Fig. 1. Genome-wide significant results for key phenotypes: A) TIBC and TF gene on chromosome 3; B) Iron Saturation and HFE gene on chromosome 6.
X-axis is the physical position on the chromosome (Mb). Y-axis denotes the –log10(p-value) for association. The most significantly associated SNP is shown with purple diamond and its dbSNP identifier is indicated. The extent of LD (as measured by r2) between each SNP and the most significantly associated SNP is indicated by the color scale at top right. Larger values of r2 indicate greater LD.
Results of the association analysis of the iron saturation phenotype are shown in Fig. S1B. Inflation of the full set of test statistics was minimal (λ=1.05) and corresponding corrections were not applied. Genome-wide significant evidence for association (p=2×10−8) was observed for iron saturation near the major histocompatibility complex (MHC) region on chromosome 6, with SNPs of modest allele frequency (MAF=0.06–0.08). These included the nonsynonymous variant rs1800562 (C282Y) in the hereditary hemochromatosis (HFE) locus (Fig. 1B). SNPs in this region also approached the significance threshold with the other iron phenotypes as well (p≤1×10−6; Tables S1A-S1C, Supplementary material).
Results of the GWAS for iron level are shown in Fig. S1C. As for iron saturation, genomic control correction was not necessary (λ=1.04). None of the genotyped or imputed SNPs demonstrated genome-wide evidence of association.
Discussion and Conclusions
Our GWAS, in a sample of healthy premenopausal women for three iron phenotypes, detected highly significant associations with several loci previously reported in samples of older subjects. The strongest evidence for association was observed with TIBC in the region of the gene for transferrin (TF) on chromosome 3 with p<10−13 after genomic control correction. This is consistent with the finding observed in older individuals [9, 19], using a case-control approach for iron deficiency. This association clearly reflects the importance of genetic variants in transferrin, the main protein responsible for transporting iron in blood, to variation among individuals in the iron cycle.
The iron saturation phenotype demonstrated significant evidence of association on chromosome 6, with the hemochromatosis gene (HFE) variant C282Y [20] and nearby SNPs. Evidence of association to this region has been observed in studies of iron-related measures in older individuals [8, 9]. Recessive defects in this gene are responsible for hemochromatosis, a hereditary iron storage disease. The wild-type HFE protein modulates signaling of iron-bound transferrin through receptors co-localized to the hepatocyte membrane, stimulating hepcidin production and down-regulating enteric iron transporter ferroportin, resulting in decreased iron absorption [3]. The reason for the association of iron saturation and HFE variants among individuals without hemochromatosis is unclear, but may reflect the dependence of TIBC values on both iron-saturated and unsaturated binding sites; TIBC also demonstrates suggestive evidence of association to the HFE region in our sample (p=4×10−7).
A highly significant association of serum iron concentration with SNPs in TRMPSS6 on chromosome 22, a gene involved in regulation of iron absorption in which mutations cause iron-deficiency anemia, has been reported in studies of iron phenotypes in older subjects of both sexes [8, 19]. Only minimal evidence of association at this locus was present in our study (p=0.005-0.01 for iron phenotypes). It is possible that this lack of association could be due to limited power, but it is consistent with a smaller effect of variation at this locus in premenopausal women.
There was large degree of overlap between our most significant GWAS findings and those of studies conducted by others for a variety of iron-related phenotypes. However, large GWAS studies of hemoglobin levels have typically detected effects at only a subset of these loci[21, 22]. Anemia only occurs at more severe iron deficiency, but we were unable to make a direct comparison of genetic association results for hemoglobin due to lack of hematologic measures on our subjects. We also were unable to assess other measures of total body iron stores such as ferritin or the soluble transferrin receptor; however, our strongest association findings with serum iron coincide with the loci detected by GWAS studies of these measures[7, 9]. Assessments of iron or ferritin may be complicated by acute changes in levels during infection or inflammatory responses[23]; however, our subjects were healthy young adult volunteers recruited for a genetic study of bone density, unlikely to have infection or inflammatory responses that might cause acute changes in iron and ferritin levels at time of sampling. Within this population, FGF23 and other factors related to mineral homeostasis have also been linked to serum iron concentration [24]. FGF23 has also been associated with ferritin concentration and hemoglobin, but not with inflammation[25, 26].
Our GWAS for iron phenotypes in a sample of healthy premenopausal women provides strong evidence that the genetic variants which have been shown to influence iron phenotypes in older people also influence these traits in during the premenopausal period. Continued studies of these effects across the lifespan will help elucidate the complex genetic architecture of both normal variation and disease states involving iron metabolism.
Supplementary Material
Acknowledgements
This work was supported by National Institutes of Health grants R21 AR061078 (MJE), K23AR057096 (EAI), R01 AG041517 (MJE), P01 AG-18397 and M01 RR-00750. Genotyping services were provided by The Center for Inherited Disease Research (CIDR), which is fully funded through a contract from the National Institutes of Health to The Johns Hopkins University (contract number HHSN268200782096C). These funding sources played no role in the design or conduct of the study or in the decision to submit the article for publication.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Cogswell ME, Looker AC, Pfeiffer CM, Cook JD, Lacher DA, et al. Assessment of iron deficiency in US preschool children and nonpregnant females of childbearing age: National Health and Nutrition Examination Survey 2003–2006. Am J Clin Nutr. 2009;89:1334–1342. doi: 10.3945/ajcn.2008.27151. [DOI] [PubMed] [Google Scholar]
- 2.Heath AL, Skeaff CM, Williams S, Gibson RS. The role of blood loss and diet in the aetiology of mild iron deficiency in premenopausal adult New Zealand women. Public Health Nutr. 2001;4:197–206. doi: 10.1079/phn200054. [DOI] [PubMed] [Google Scholar]
- 3.Fleming RE, Ponka P. Iron overload in human disease. N Engl J Med. 2012;366:348–359. doi: 10.1056/NEJMra1004967. [DOI] [PubMed] [Google Scholar]
- 4.Moron C, Viteri FE. Update on common indicators of nutritional status: food access, food consumption, and biochemical measures of iron and anemia. Nutr Rev. 2009;67(Suppl 1):S31–S35. doi: 10.1111/j.1753-4887.2009.00156.x. [DOI] [PubMed] [Google Scholar]
- 5.Fairweather-Tait SJ, Guile GR, Valdes AM, Wawer AA, Hurst R, et al. The contribution of diet and genotype to iron status in women: a classical twin study. PLoS One. 2013;8:e83047. doi: 10.1371/journal.pone.0083047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gichohi-Wainaina WN, Melse-Boonstra A, Swinkels DW, Zimmermann MB, Feskens EJ, et al. Common Variants and Haplotypes in the TF, TNF-alpha, and TMPRSS6 Genes Are Associated with Iron Status in a Female Black South African Population. J Nutr. 2015;145:945–953. doi: 10.3945/jn.114.209148. [DOI] [PubMed] [Google Scholar]
- 7.Benyamin B, McRae AF, Zhu G, Gordon S, Henders AK, et al. Variants in TF and HFE explain approximately 40% of genetic variation in serum-transferrin levels. Am J Hum Genet. 2009;84:60–65. doi: 10.1016/j.ajhg.2008.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tanaka T, Roy CN, Yao W, Matteini A, Semba RD, et al. A genome-wide association analysis of serum iron concentrations. Blood. 2010;115:94–96. doi: 10.1182/blood-2009-07-232496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McLaren CE, Garner CP, Constantine CC, McLachlan S, Vulpe CD, et al. Genome-wide association study identifies genetic loci associated with iron deficiency. PLoS One. 2011;6:e17390. doi: 10.1371/journal.pone.0017390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li J, Lange LA, Duan Q, Lu Y, Singleton AB, et al. Genome-wide admixture and association study of serum iron, ferritin, transferrin saturation and total iron binding capacity in African Americans. Hum Mol Genet. 2015;24:572–581. doi: 10.1093/hmg/ddu454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Medina-Gomez C, Kemp JP, Estrada K, Eriksson J, Liu J, et al. Meta-analysis of Genome-Wide Scans for Total Body BMD in Children and Adults Reveals Allelic Heterogeneity and Age-Specific Effects at the WNT16 Locus. PLoS Genetics. 2012;8:e1002718. doi: 10.1371/journal.pgen.1002718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koller DL, Ichikawa S, Lai D, Padgett LR, Doheny KF, et al. Genome-wide association study of bone mineral density in premenopausal European-American women and replication in African-American women. J Clin Endocrinol Metab. 2010;95:1802–1809. doi: 10.1210/jc.2009-1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hui SL, Koller DL, Foroud TM, Econs MJ, Johnston CC, et al. Heritability of changes in bone size and bone mass with age in premenopausal white sisters. J Bone Miner Res. 2006;21:1121–1125. doi: 10.1359/jbmr.060412. [DOI] [PubMed] [Google Scholar]
- 14.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 15.Howie B, Fuchsberger C, Stephens M, Marchini J, Abecasis GR. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat Genet. 2012;44:955–959. doi: 10.1038/ng.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen MH, Yang Q. GWAF: an R package for genome-wide association analyses with family data. Bioinformatics. 2010;26:580–581. doi: 10.1093/bioinformatics/btp710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McLaren CE, McLachlan S, Garner CP, Vulpe CD, Gordeuk VR, et al. Associations between single nucleotide polymorphisms in iron-related genes and iron status in multiethnic populations. PLoS One. 2012;7:e38339. doi: 10.1371/journal.pone.0038339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Beutler E, Felitti V, Gelbart T, Waalen J. Haematological effects of the C282Y HFE mutation in homozygous and heterozygous states among subjects of northern and southern European ancestry. Br J Haematol. 2003;120:887–893. doi: 10.1046/j.1365-2141.2003.04215.x. [DOI] [PubMed] [Google Scholar]
- 21.Chambers JC, Zhang W, Li Y, Sehmi J, Wass MN, et al. Genome-wide association study identifies variants in TMPRSS6 associated with hemoglobin levels. Nat Genet. 2009;41:1170–1172. doi: 10.1038/ng.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kullo IJ, Ding K, Jouni H, Smith CY, Chute CG. A genome-wide association study of red blood cell traits using the electronic medical record. PLoS One. 2010;5 doi: 10.1371/journal.pone.0013011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Suominen P, Punnonen K, Rajamaki A, Irjala K. Serum transferrin receptor and transferrin receptor-ferritin index identify healthy subjects with subclinical iron deficits. Blood. 1998;92:2934–2939. [PubMed] [Google Scholar]
- 24.Imel EA, Peacock M, Gray AK, Padgett LR, Hui SL, et al. Iron modifies plasma FGF23 differently in autosomal dominant hypophosphatemic rickets and healthy humans. J Clin Endocrinol Metab. 2011;96:3541–3549. doi: 10.1210/jc.2011-1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Braithwaite V, Jarjou LM, Goldberg GR, Prentice A. Iron status and fibroblast growth factor-23 in Gambian children. Bone. 2012;50:1351–1356. doi: 10.1016/j.bone.2012.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Braithwaite V, Prentice AM, Doherty C, Prentice A. FGF23 is correlated with iron status but not with inflammation and decreases after iron supplementation: a supplementation study. Int J Pediatr Endocrinol. 2012;2012:27. doi: 10.1186/1687-9856-2012-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.