Abstract
Long-chain n-6 and n-3 PUFA (LC-PUFA), arachidonic acid (AA) (20:4n-6) and DHA (22:6n-3), are critical for optimal brain development. These fatty acids can be consumed directly from the diet, or synthesized endogenously from precursor PUFA by Δ-5 (encoded by FADS1) and Δ-6 desaturases (encoded by FADS2). The aim of this study was to determine the potential importance of maternal genetic variability in FADS1 and FADS2 genes to maternal LC-PUFA status and infant neurodevelopment in populations with high fish intakes. The Nutrition Cohorts 1 (NC1) and 2 (NC2) are longitudinal observational mother-child cohorts in the Republic of Seychelles. Maternal serum LC-PUFA was measured at 28 weeks gestation and genotyping for rs174537 (FADS1), rs174561 (FADS1), rs3834458 (FADS1-FADS2) and rs174575 (FADS2) was performed in both cohorts. The children completed the Bayley Scales of Infant Development II (BSID-II) at 30 months in NC1 and at 20 months in NC2. Complete data were available for 221 and 1310 mothers from NC1 and NC2 respectively. With increasing number of rs3834458 minor alleles, maternal concentrations of AA were significantly decreased (NC1 p=0.004; NC2 p<0.001) and precursor:product ratios for linoleic acid (LA) (18:2n-6)-to-AA (NC1 p<0.001; NC2 p<0.001) and α-linolenic acid (ALA) (18:3n-3)-to-DHA were increased (NC2 p=0.028). There were no significant associations between maternal FADS genotype and BSID-II scores in either cohort. A trend for improved PDI was found among infants born to mothers with the minor rs3834458 allele. In these high fish-eating cohorts, genetic variability in FADS genes was associated with maternal AA status measured in serum and a subtle association of the FADS genotype was found with neurodevelopment.
Keywords: Δ-5 Desaturase (Δ5D), Δ-6 Desaturase (Δ6D), Maternal fish consumption, Neurodevelopment, Seychelles Child Development Study (SCDS)
1. Introduction
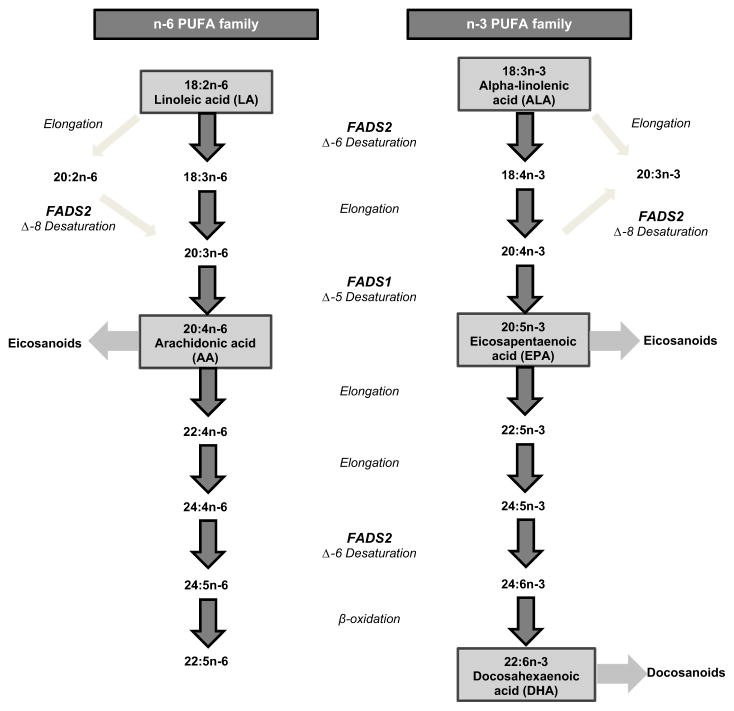
An adequate supply of long-chain PUFA (LC-PUFA) arachidonic acid (AA) (20:4n-6) and DHA (22:6n-3) is critical for optimal brain development [11]. The fetus relies entirely on the maternal supply of these fatty acids in utero. These LC-PUFA can be obtained directly from the diet, but can also be endogenously synthesized in all mammalian systems from their essential n-6 and n-3 PUFA precursors, linoleic acid (LA) (18:2n-6) and α-linolenic acid (ALA) (18:3n-3) respectively. This synthesis involves a series of elongation and desaturation steps, catalyzed by the fatty acid desaturase enzymes [39], where Δ-5 desaturase (Δ5D) and Δ-6 desaturase (Δ6D) are recognized as rate-limiting enzymes [28] (Fig. 1). However, the endogenous synthesis of LC-PUFA, particularly that of DHA from ALA, is recognized to be extremely inefficient [35]. For this reason consumption of preformed DHA from fish sources is recommended.
Fig. 1.
Pathways for endogenous n-6 and n-3 PUFA elongation and desaturation showing the activity of FADS1 and FADS2 and corresponding desaturation steps.
The FADS1 (encoding Δ5D) and FADS2 (encoding Δ6D) genes are located head-to-head in a cluster on chromosome 11 (11q12-q13) [10,24,36]. Carriers of certain genotypes in the FADS gene cluster have consistently been shown to have higher biological status of PUFA precursors, LA and ALA, and lower status of LC-PUFA products, AA and EPA (20:5n-3), probably as a result of having lower expression of the functional enzymes [17,19,23,30,33,47,9]. The highest and the lowest proportion of variability, with respect to the influence of FADS genes on PUFA composition, have been found for AA and for DHA respectively [44].
Among cohorts of pregnant women, maternal genetic variation in FADS has frequently been associated with lower concentrations of AA and EPA in maternal blood and breast milk, as well as infant blood [19,22,26,47]. Importantly, variation of the maternal FADS genotype resulting in lower FADS1 and FADS2 activity has also been associated with lower cognitive development of infants, possibly owing to lower LC-PUFA status during critical stages of fetal development [27,40,6].
These findings suggest that the FADS genotype may be an important determinant not only of LC-PUFA status, but also of the biological effects exerted by LC-PUFA. With respect to dietary variation, it is not fully understood how dietary availability of LC-PUFA impacts on the association between FADS genotype and LC-PUFA status [14]. It has been suggested that FADS genotype may be a greater determinant of LC-PUFA status in populations where fish consumption is low and there is expected to be greater dependence on endogenous synthesis of LC-PUFA [37]. Yet there remain no data on the influence of FADS genotype, on either LC-PUFA status or neurodevelopment, in populations with high fish consumption. These data would be important for risk assessment by providing further evidence on the risks and benefits of fish consumption during pregnancy.
The objective of the current study was to characterize mothers in two high fish-eating cohorts of the Seychelles Child Development Study (SCDS) for FADS1 and FADS2 and to investigate associations among maternal genotype, LC-PUFA status and developmental outcomes of their infants. It was hypothesized that mothers with minor alleles of SNPs in either FADS1 or FADS2 would have higher blood concentrations of precursor n-6 and n-3 PUFA, LA and ALA, and lower concentrations of LC-PUFA products, AA and DHA. We further hypothesized that such variation in the maternal status of these PUFA during pregnancy could impact the cognitive development of their children.
2. Methods
2.1. Study population
The SCDS is a longitudinal observational study conducted in the Republic of Seychelles, an archipelago in the Indian Ocean. The population resides mainly on the island of Mahé and is of mixed African, European and East Asian origin. The overall aim of the SCDS is to investigate the effects of methyl mercury (MeHg) and nutrient exposure, from maternal fish consumption during pregnancy, on child developmental outcomes. The study has found no consistent pattern of adverse associations of prenatal MeHg exposure and neurodevelopment in children of mothers consuming an average of 12 fish meals per week [38,43,45].
Apparently healthy mothers were recruited to Nutrition Cohort 1 (NC1) and Nutrition Cohort 2 (NC2) during their first antenatal visit (from 14 weeks of gestation) at eight health centers across Mahé. NC1 mothers were recruited in 2001 until 300 volunteers had consented [7], and NC2 mothers were enrolled from 2008 until 2011 when the target number of 1500 mothers had consented [43]. Further information on inclusion criteria and power calculations for NC1 [41] and NC2 have previously been described [43]. In NC1, mothers completed a 4-day food diary at 28 weeks gestation to estimate their average daily consumption of fish [4]. In NC2, mothers completed a retrospective Fish Use Questionnaire, also at 28 weeks gestation, to estimate their weekly consumption of fish during pregnancy. This study was conducted according to guidelines laid down in the Declaration of Helsinki and all study procedures involving participants were reviewed and approved by the Seychelles Ethics Board, the Research Subjects Review Board at the University of Rochester, and the Regional Ethics Committee at Lund University, Sweden.
2.2. Blood sampling and analyses
At 28 weeks gestation non-fasting blood samples were collected in both cohorts and serum and whole blood were obtained after processing at the Public Health Laboratory of the Ministry of Health. Maternal serum samples were shipped at −80 °C to the University of Ulster where they were analyzed for fatty acid concentrations. Total lipids were extracted and fatty acid methyl esters (FAME) were prepared by boron trifluroide methanol (BF3) according to an adaptation of the Folch method [8]. FAME were detected and quantified using the gold-standard technique of Gas Chromatography–Mass Spectrometry (GC–MS) (Agilent 7890A-5975C, UK) using heptadecaenoic acid (C17:0) as the internal standard, as previously described [41]. All analytical standards were of >99% purity and purchased from Sigma-Aldrich, UK. Total serum PUFA composition is presented as mg/ml to indicate physiological quantities. In NC1, there were 11 women missing 28 week PUFA status, but values for these women were imputed from PUFA concentrations which were also measured at delivery in the NC1 cohort [7]. As described previously, the efficiency of blood processing in the Seychelles was improved in NC2 compared to NC1 [42].
2.3. Genotyping
Maternal whole blood samples were shipped at −80 °C to the University of Lund, Sweden for genotyping. Four candidate SNPs: FADS1 rs174537 and rs174561, FADS1-FADS2 rs3834458 and FADS2 rs174575; were selected based on evidence of impacting on LC-PUFA in epidemiological studies (Online Resource 1). The rs3834458 is located in the promoter of FADS2 and 5′ of FADS1 and is often referred as intergenic [26]. DNA was extracted from maternal blood using the Qiagen DNA Blood Mini kit (Qiagen, Hilden, Germany). SNPs were genotyped by using the iPLEX® Gold assay on the MassARRAY platform (Sequenom™, San Diego, USA) and by TaqMan allelic discrimination assay on an ABI 7900 instrument (Applied Biosystems, Foster City, CA, USA), according to the manufacturer’s instructions. A random selection of the samples were re-analyzed for quality control purposes with perfect agreement between original and repeat genotyping runs for all SNPs. Mother’s DNA samples missing more than two of the five SNPs recorded in each genotyping batch were considered unreliable and not included in the database, whilst those missing less than two of all SNPs were included. These differences in genotyping efficiency account for different sample sizes for each SNP measured.
2.4. Developmental assessment
When infants were aged approximately 30 months in NC1 and 20 months in NC2, they completed developmental testing with the Bayley Scales of Infant Development (BSID-II). Testing was conducted by specially trained nurses at the Child Development Centre, Mahé. All study forms were shipped to the University of Rochester, where data were double-entered and the Mental Development Index (MDI) and Psychomotor Development Index (PDI) endpoints were scaled according to the child’s age at testing. Test reliabilities for the BSID-II were determined as previously described [41].
2.5. Statistical analyses
Deviations from Hardy–Weinberg equilibrium were tested using chi-square analysis. One SNP (rs174561) was in Hardy Weinberg disequilibrium in both populations (Online Resource 2). As this SNP was genotyped with two different methods (NC1 with Sequenom and NC2 with Taqman) and there was a perfect match between re-runs, we considered the genotyping results correct. Linkage disequilibrium was evaluated using Haploview [3]. Tests for associations between outcomes and SNPs were carried out from a priori analysis plans and all effects were tested using two-sided tests of significance at the α =0.05 level. Unadjusted linear regression was used to estimate the effect of each of the four FADS SNPs on each of the eight individual LC-PUFA measurements and on the precursor:product ratios, LA:AA, ALA:EPA and ALA:DHA. The ratios of LA:AA and ALA:EPA are commonly used as crude indicators of desaturase activity [46]. The minor allele frequencies were sufficiently large in each of these SNPs that heterozygote and variant homozygote effects were estimated separately. In order to make the LC-PUFA results more comparable, we scaled maternal serum PUFA composition in each cohort using homozygote carriers of the common SNP as a reference and the relative differences are presented [31,32].
Multiple linear regression was used to estimate the effect of FADS SNPs on the child’s neurodevelopment, measured as BSID-II scores. Adjustments were made for covariates previously chosen to cover the most important determinants of neurocognitive development in children [41]. These include child sex, maternal age at delivery, presence of two parents in the household, socioeconomic score, and birth weight (NC1) or child age at testing (NC2).
The children in the NC2 cohort were tested approximately 10 months before the age at which testing of NC1 children took place. Over the course of the SCDS, we have observed that the mean scaled score within each cohort decreased with age when followed longitudinally. To make the results more comparable between cohorts, we scaled the BSID-II scores in each cohort by using estimated mean scores in homozygote carriers of the common SNP as a reference and the relative differences are presented.
3. Results
A total of n=222 NC1 mothers and n=1400 NC2 mothers were included in the current study after exclusions owing to missing data in developmental outcomes, LC-PUFA, SNPs and covariates. Descriptive characteristics of the two cohorts, including covariate data, are presented in Table 1. The LC-PUFA values differed between the cohorts and were, apart from LA, generally higher in NC2. This finding may be as a result of potential oxidation of samples during blood processing in NC1, as previously described [42]. NC1 mothers consumed on average 9 fish meals per week [41], whereas NC2 mothers consumed an average of 8.5 estimated fish servings per week [43].
Table 1.
| Maternal Serum PUFA composition (mg/ml) | NC1
|
NC2
|
||||
|---|---|---|---|---|---|---|
| n | Mean | SD | n | Mean | SD | |
| LA | 222 | 1.170 | 0.219 | 1399 | 0.898 | 0.252 |
| AA | 222 | 0.101 | 0.022 | 1399 | 0.202 | 0.081 |
| ALA | 222 | 0.002 | 0.001 | 1399 | 0.037 | 0.006 |
| EPA | 222 | 0.031 | 0.009 | 1399 | 0.183 | 0.084 |
| DHA | 222 | 0.003 | 0.002 | 1399 | 0.052 | 0.008 |
| BSID-II | ||||||
| MDI | 221 | 84.97 | 9.59 | 1358 | 87.71 | 10.69 |
| PDI | 218 | 89.80 | 13.78 | 1356 | 96.78 | 10.58 |
| Demographics | ||||||
| Maternal age (yr) | 222 | 27.7 | 6.0 | 1400 | 27.0 | 6.3 |
| Maternal BMI (kg/m2) | 221 | 25.8 | 6.4 | 1291 | 26.9 | 6.5 |
| Hollingshead SES | 222 | 33.9 | 11.1 | 1353 | 32.0 | 10.3 |
| Girls (% in cohort) | 222 | 51.4 | – | 1401 | 48.0 | – |
| Parents at home (% in cohort) | 221 | 42.1 | – | 1353 | 72.5 | – |
| Child birth weight (g) | 222 | 3232.2 | 470.4 | 1393 | 3178.8 | 498.3 |
| Maternal fish consumption (meals/week) | 221 | 9.1 | 4.1 | 1321 | 8.5 | 4.6 |
NC1, Nutrition Cohort 1; NC2, Nutrition Cohort 2; LA, linoleic acid; AA, arachidonic acid; ALA, α-linolenic acid; BSID-II, Bayley Scales of Infant Development II; MDI, Mental Developmental Index; PDI, Psychomotor Developmental Index; Hollingshead SES, socioeconomic status.
Data shown are for all models.
BSID-II scores are from 30 mo assessment in NC1 and 20 mo assessment in NC2.
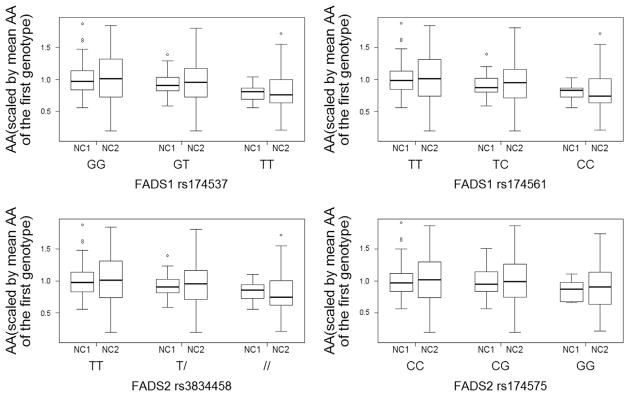
The genotype distributions in NC1 and NC2 are presented in Table 2. The allele frequencies for the FADS SNPs were similar between the Seychellois cohorts (Online Resource 2). For all SNPs, the minor allele frequencies were lower in the Seychellois populations compared to European and African populations [12]. FADS2 rs174575 was found not to be in linkage disequilibrium (LD) with any of the other SNPs, whereas the other three SNPs (FADS1 rs174561 and rs174537, FADS1-FADS2 intergenic rs3834458) had pairwise LD values ranging from 0.83 to 0.93 (data not shown). Therefore, the following results are presented for rs174575 (not in strong LD with other SNPs) and rs3834458 (in strong LD with rs174561 and rs174537). Results for the other SNPs in LD with rs3834458 are presented in Online Resources 3 and 4.
Table 2.
| NC1 | N genotyped | Reference homozygote | Heterozygote | Minor allele homozygote |
|---|---|---|---|---|
|
| ||||
| (%) | ||||
| FADS1 rs174537 | 180 | 63.3 GG | 31.1 GT | 5.6 TT |
| FADS1 rs174561 | 193 | 77.2 TT | 16.1 TC | 6.7 CC |
| FADS1-FADS2 rs3834458 | 220 | 65.9 TT | 28.2 Tdel | 5.9 deldel |
| FADS2 rs174575 | 214 | 59.8 CC | 35.0 CG | 5.1 GG |
| NC2 | ||||
| FADS1 rs174537 | 1413 | 67.9 GG | 28.5 GT | 3.6 TT |
| FADS1 rs174561 | 1413 | 72.1 TT | 24.8 TC | 3.1 CC |
| FADS1-FADS2 rs3834458 | 1414 | 70.4 TT | 26.4 Tdel | 3.2 deldel |
| FADS2 rs174575 | 1413 | 61.2 CC | 33.3 CG | 5.5 GG |
NC1, Nutrition Cohort 1; NC2, Nutrition Cohort 2; del=one base-pair deletion.
n=180–220 in NC1 and n=1413–1414 in NC2 depending on genotyping efficiency for the different SNPs.
rs3834458 is FADS1-FADS2 intergenic (NCBI).
Genetic associations with maternal PUFA composition were evaluated as shown in Table 3. There was a significant association between rs3834458 and serum composition of AA and the ratio of LA:AA in both cohorts. With increasing number of variant alleles, the relative concentrations of AA were reduced in both cohorts (NC2 p<0.001; NC1 p=0.004). The magnitude of association was somewhat larger in NC2 where rs3834458 deldel carriers showed 20% lower AA than TT carriers, compared to 16.5% lower AA shown in NC1 deldel carriers (Fig. 2). The association with the LA:AA ratio was as expected, with ratios being significantly higher with increasing number of variant alleles (NC2 p<0.001; NC1 p<0.001). In the larger NC2 cohort further associations between rs3834458 and PUFA status were found with variant carriers having significantly higher LA (p=0.002) and ALA (p=0.017) concentrations and significantly different ratios of ALA:DHA (p=0.028, all 2 df tests). For ALA:DHA, deldel carriers had a 24.4% higher ratio compared to TT carriers. The results were very similar for the two other SNPs in LD with rs3834458 (Fig. 2).
Table 3.
Relative differences in PUFA concentrations (mg/ml) by FADS genotypes compared to the reference genotype in NC1 and NC2a,b,c.
| SNP | Cohort | Genotype | LA | AA | LA:AA | ALA | DHA | EPA | ALA:DHA | ALA:EPA |
|---|---|---|---|---|---|---|---|---|---|---|
| FADS1-FADS2 rs3834458 | NC1 | TTd | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 |
| Tdel | 1.054 | 0.922 | 1.142 | 1.243 | 1.019 | 1.004 | 1.259 | 1.122 | ||
| Deldel | 1.041 | 0.835 | 1.253 | 1.213 | 0.934 | 0.982 | 1.282 | 1.359 | ||
| P-value | 0.154 | 0.004 | <0.001 | 0.133 | 0.915 | 0.970 | 0.165 | 0.256 | ||
| NC2 | TTd | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | |
| Tdel | 1.044 | 0.933 | 1.106 | 1.011 | 1.019 | 1.014 | 0.987 | 1.000 | ||
| Deldel | 1.120 | 0.799 | 1.426 | 1.064 | 0.940 | 1.023 | 1.244 | 1.036 | ||
| P-value | 0.002 | <0.001 | <0.001 | 0.017 | 0.517 | 0.206 | 0.028 | 0.358 | ||
| FADS2 rs174575 | NC1 | CCd | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 |
| CG | 1.019 | 0.982 | 1.055 | 1.153 | 1.081 | 1.046 | 1.106 | 0.914 | ||
| GG | 1.075 | 0.858 | 1.260 | 0.931 | 0.949 | 0.979 | 0.941 | 0.961 | ||
| P-value | 0.399 | 0.118 | 0.002 | 0.393 | 0.664 | 0.538 | 0.694 | 0.743 | ||
| NC2 | CCd | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | |
| CG | 1.024 | 0.988 | 1.036 | 1.002 | 1.000 | 1.001 | 1.002 | 1.001 | ||
| GG | 0.991 | 0.872 | 1.122 | 0.991 | 0.924 | 0.985 | 1.088 | 1.005 | ||
| P-value | 0.288 | 0.025 | 0.342 | 0.842 | 0.371 | 0.680 | 0.483 | 0.966 |
NC1, Nutrition Cohort 1; NC2, Nutrition Cohort 2; LA, linoleic acid; AA, arachidonic acid; ALA, α-linolenic acid; del=one base-pair deletion.
We present relative difference in PUFA concentrations by genotype compared to the mean concentration in homozygote carriers of the reference genotype. Therefore values greater than one show an average increase compared to the reference SNP and values less than one show an average decrease compared to the reference SNP. Bolded P-values, derived from unadjusted ANOVA, identify groups that differ significantly by genotype.
rs3834458 is FADS1-FADS2 intergenic (NCBI).
Reference genotype.
Fig. 2.
Prenatal arachidonic acid (AA; to compare the two cohorts we report the relative difference compared to the mean AA in carriers of the reference genotype). Boxplots are shown for each genotype in both the NC1 and NC2 cohorts. Differences between the genotypes are significant for all four SNPs in both cohorts except rs174575 in the NC1 cohort (see Table 3 and S3).
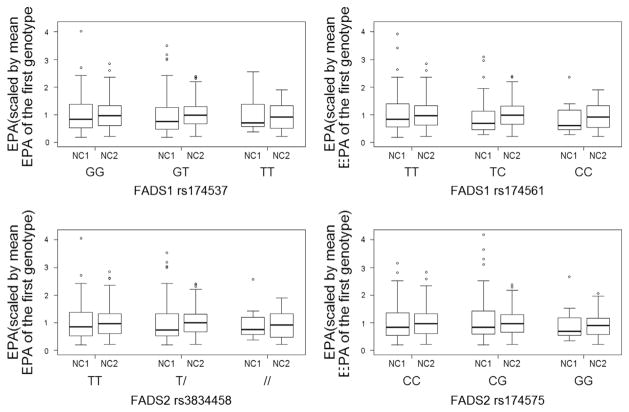
In NC1, rs174575 appeared to modify LA:AA in that G allele carriers showed higher ratios compared to CC carriers (p=0.002, 2 df test). In NC2, rs174575 was associated with lower AA concentrations with increasing number of variant alleles (p=0.025). There were no significant associations between FADS genotype and concentrations of EPA (Fig. 3), DHA or the ratio of ALA:EPA in either cohort.
Fig. 3.
Prenatal Eicosapentaenoic acid (EPA; to compare the two cohorts we report the relative difference compared to the mean EPA in carriers of the reference genotype). Boxplots are shown for each genotype in both the NC1 and NC2 cohorts. Differences between the genotypes are not significant for either cohort or any SNP (see Table 3 and S3).
The genotype of the mothers was evaluated in relation to neurodevelopmental outcomes in the children as shown in Table 4. There were no significant genetic associations between FADS genotype and child developmental scores in either NC1 or NC2. A non-significant trend for infants of rs3834458 del carriers to score higher on the Psychomotor Developmental Index (PDI) was found in both NC1 and NC2 (p=0.07 and 0.07 respectively).
Table 4.
Relative differences in child developmental scores by mother’s genotype compared to the reference genotype from linear regression modelsa,b,c.
| BSID-II Outcome/SNP | Genotype | NC1 BSID-II scores
|
NC2 BSID-II scores
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| n | Relative mean | 95% CI | P | n | Relative mean | 95% CI | P | ||
| MDI | TTd | 145 | 1.00 | (0.982,1.018) | 0.06 | 924 | 1.00 | (0.992,1.008) | 0.18 |
| FADS1-FADS2 rs3834458 | Tdel | 61 | 1.02 | (0.996,1.051) | 343 | 1.01 | (1.000,1.025) | ||
| Deldel | 13 | 1.01 | (0.951,1.069) | 39 | 1.01 | (0.972,1.046) | |||
| MDI | CCd | 127 | 1.00 | (0.981,1.019) | 0.88 | 800 | 1.00 | (0.992,1.008) | 0.50 |
| FADS2 rs174575 | CG | 75 | 1.01 | (0.982,1.030) | 437 | 1.01 | (0.996,1.018) | ||
| GG | 11 | 1.01 | (0.944,1.071) | 68 | 1.01 | (0.984,1.040) | |||
| PDI | TTd | 143 | 1.00 | (0.975,1.025) | 0.07 | 923 | 1.00 | (0.993,1.007) | 0.06 |
| FADS1-FADS2 rs3834458 | Tdel | 60 | 1.04 | (1.001,1.078) | 342 | 1.02 | (1.004,1.027) | ||
| Deldel | 13 | 1.04 | (0.961,1.124) | 39 | 1.02 | (0.985,1.053) | |||
| PDI | CCd | 126 | 1.00 | (0.974,1.026) | 0.91 | 798 | 1.00 | (0.992,1.008) | 0.69 |
| FADS2 rs174575 | CG | 74 | 1.01 | (0.972,1.039) | 435 | 1.01 | (0.996,1.017) | ||
| GG | 11 | 1.00 | (0.916,1.091) | 70 | 1.00 | (0.973,1.024) | |||
NC1, Nutrition Cohort 1; NC2, Nutrition Cohort 2; del=one base-pair deletion; BSID-II, Bayley Scales of Infant Development II; MDI, Mental Developmental Index; PDI, psychomotor developmental index.
β Coefficients, 95% CI and P values presented are from linear regression models adjusted for child sex, maternal age at delivery, presence of two parents in the household, socioeconomic status, and birth weight (for NC1) or child age at testing (NC2). P-values are from the 2 df tests comparing the 3 levels of each genotype.
rs3834458 is FADS1-FADS2 intergenic (NCBI).
Reference genotype.
4. Discussion
We found in two high fish-eating observational mother-child cohorts from the Republic of Seychelles that maternal FADS genotype rs3834458 was significantly associated with LC-PUFA status. The strongest associations were observed for AA and the LA:AA ratio, with variant homozygotes in the much larger cohort (NC2) showing 20% lower AA and 42% higher LA:AA than those with the homozygous reference genotype. These findings support prior studies reporting that carriers of the minor alleles of FADS SNPs, including rs3834458, tend to have a lower blood composition of LC-PUFA, particularly of AA but also of EPA [19,33,47]. However, in our study there was no association of rs3834458, or any other FADS genotype, with either serum EPA or DHA composition. This finding agrees with the majority of studies which have shown DHA status to be less influenced by genetic variation in FADS genes [13,19]. One plausible reason for this finding is that whilst the LC-PUFA desaturation pathway mainly takes place within the endoplasmic reticulum, the final conversion step from docosapentaenoic acid (DPA; C22:5n-3) to DHA (partial β-oxidation) requires a translocation to the peroxisomes [39]. This peroxisomal conversion accounts for DHA being the least efficiently synthesized n-3 LC-PUFA in the body and as a result, the impact of genetic variation may be ‘diluted’ and less likely to influence DHA status [21]. It could also be hypothesized that the reason for a lack of genetic influence on DHA status shown in the current study may be related to high fish consumption and subsequently higher levels of preformed DHA in the population, which may mean lower dependence on the endogenous synthesis pathway. This hypothesis is supported by the recent study by Scholtz et al where the influence of FADS genotype on DHA status became non-significant following fish oil supplementation among a group who had lower DHA status at baseline [37]. However, despite high fish intake in our cohorts, some endogenous activity for production of n-3 LC-PUFA might exist and thereby account for the higher LA:AA found in both cohorts and higher ALA:DHA found in NC2. A recent randomized controlled trial of fish oil supplementation reported that increased dietary intakes of EPA and DHA were associated with higher activity of Δ5D and lower activity of Δ6D [2]. These findings may suggest that the endogenous LC-PUFA synthesis pathway remains active, and perhaps enhanced, despite abundant preformed LC-PUFA in the diet. However we must acknowledge that the ALA:DHA ratio is limited as an indicator of FADS enzyme activity given the notably low conversion of DHA which occurs from ALA endogenously. We also cannot clearly differentiate which of the SNPs is related to the LC-PUFA phenotype, as there is strong linkage for the FADS1 SNPs and the FADS1-FADS2 intergenic rs3834458. However, a less pronounced association was found for FADS2 rs174575 compared to the FADS1 SNPs, which could suggest that the functional association is related to differences in Δ5D rather than Δ6D activities, but we cannot exclude the potential influence of other genes in the LC-PUFA pathway.
The maternal FADS genotype was not significantly associated with infant development in either cohort. However, we did find a trend for improved psychomotor development among infants of mothers with the variant allele for rs3834458 in both NC1 (p=0.07) and NC2 (p=0.07). This suggests a subtle association of the FADS genotype with neurodevelopment, even at high fish intake. We suspect, from the lack of associations between rs3834458 and EPA or DHA, that this trend might be related to lower production of AA, which is a precursor for pro-inflammatory eicosanoids [18]. Data from the English mother–child ALSPAC cohort, also found children of mothers with the rs3834458 minor allele performed better in tests of intelligence quotient at 8 years [40]. In our recent analysis of the associations between maternal PUFA status and neurodevelopment in NC2, we found no direct associations of maternal AA status with cognitive outcomes. However, we did find significant adverse associations between higher maternal n-6/n-3 ratios and both infant psychomotor development and communicative ability at 20 months of age. This suggests that the balance between AA and DHA may be important for neurodevelopment and a possible mechanism would be through influencing the inflammatory milieu [43].
Recent studies suggest that the interaction between PUFA and genetic variability is complex. One recent study suggests that n-3 LC-PUFA supplementation can induce epigenetic regulation of FADS genes and genes encoding the elongase enzymes, ELOVL5 and ELOVL2, in a sex-specific manner [16]. If true, this regulation may explain why different populations, with similar allelic frequencies of FADS SNPs but with distinct diets, may present health disparities [25]. Future studies may need to consider genetic variation in genes controlling eicosanoid synthesis from LC-PUFA, such as 5-lipoxygenase (ALOX5) [25]. Taken together, these data provide a firm basis for considering each population group separately in studies of FADS and PUFA status.
There are some methodological issues to consider with the current study. The processing time of the blood collected was different between the two cohorts, and may partially explain the different serum concentrations of LC-PUFA reported [42]. However, to compensate for this and make the cohorts more comparable, we scaled PUFA values in each cohort to the estimated mean concentration in homozygote carriers of the reference genotype, as described [31,32]. We included developmental testing points in the two cohorts that were reasonably close in age (30 months in NC1 and 20 months in NC2) to enhance comparability. Average MDI scores across the cohorts were similar whereas PDI scores were greater in the NC2 cohort, possibly owing to the younger age at examination. We further included similar a priori covariates in the regression analysis to increase comparability of associations across cohorts. We observed relatively consistent genetic associations in the two cohorts, but fewer were statistically significant in the NC1 cohort, perhaps owing to the smaller sample size. We acknowledge that the precursor:product PUFA ratios are limited as markers of enzyme activity. However, these ratios have been used frequently for this purpose and we believe that our use of serum absolute PUFA concentrations in denoting these ratios is a strength of the current study.
In conclusion, we found that the maternal FADS genotype is an important predictor of maternal AA, but not of EPA and DHA status. To our knowledge this is the first time that such associations have been described within a high fish-eating cohort. These results highlight the importance of considering FADS genotype, even at high fish intake, to aid understanding associations between maternal LC-PUFA status, fish consumption and child development.
Supplementary Material
Acknowledgments
We gratefully acknowledge the participation of all women and children who took part in the study and the nursing staff from the Child Development Centre, Seychelles for their assistance with data collection.
This work was supported by the US National Institute of Health (NIH) (Grants R01-ES010219, P30-ES01247 and T32-ES007271), The Swedish Research Council for Health, Working Life and Welfare, and in-kind support from the Government of Seychelles. The study sponsors had no role in the design, collection, analysis, or interpretation of data, in the writing of this article, or in the decision to submit the article for publication.
The authors’ responsibilities were as follows: KB conceived, designed and conducted the research; GJM, PWD, EvW, GEW, CFS and JJS conceived and designed the SCDS and conducted the research; AJY, MSM, EMM, AA, KW and KB conducted the research; TML, SWT, KG and KE performed the statistical analyses; AJY and KB interpreted the data, drafted the manuscript and KB has primary responsibility for final content. All authors have read and approved the final version to be published.
Abbreviations
- AA
Arachidonic acid
- ALA
Alpha-linolenic acid
- BSID-II
Bayley’s Scales of Infant Development II
- Δ5D
Δ5-desaturase
- Δ6D
Δ6-desaturase
- LA
Linoleic acid
- LC-PUFA
Long-chain polyunsaturated fatty acids
- MDI
Mental Developmental Index
- NC1
Nutrition Cohort 1
- NC2
Nutrition Cohort 2
- PDI
Psychomotor Developmental Index
- SCDS
Seychelles Child Development Study
Appendix A. Supplementary material
Supplementary data associated with this article can be found in the online version at http://dx.doi.org/10.1016/j.plefa.2015.08.004.
Footnotes
Compliance with Ethics Guidelines
This study was conducted in accordance with the ethical standards of the Helsinki Declaration of 1975 and all study procedures involving participants were reviewed and approved by the Seychelles Ethics Board, the Research Subjects Review Board at the University of Rochester, and the Regional Ethics Committee at Lund University, Sweden. Informed consent was obtained from all participants included in the study.
References
- 1.Agostoni C, Trojan S, Bellu R, Riva E, Bruzzese MG, Giovannini M. Developmental quotient at 24 months and fatty acid composition of diet in early infancy: a follow up study. Arch Dis Child. 1997;76:421–424. doi: 10.1136/adc.76.5.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Al-Hilal M, Alsaleh A, Maniou Z, Lewis FJ, Hall WL, Sanders TA, O’Dell SD. Genetic variation at the FADS1-FADS2 gene locus influences delta-5 desaturase activity and LC-PUFA proportions after fish oil supplement. J Lipid Res. 2013;54:542–551. doi: 10.1194/jlr.P032276. http://dx.doi.org/10.1194/jlr.P032276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. http://dx.doi.org/10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 4.Bonham MP, et al. Contribution of fish to intakes of micronutrients important for fetal development: a dietary survey of pregnant women in the Republic of Seychelles. Public Health Nutr. 2009;12:1312–1320. doi: 10.1017/S136898000800387X. http://dx.doi.org/10.1017/S136898000800387X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Calder PC. Polyunsaturated fatty acids, inflammation, and immunity. Lipids. 2001;36:1007–1024. doi: 10.1007/s11745-001-0812-7. [DOI] [PubMed] [Google Scholar]
- 6.Caspi A, et al. Moderation of breastfeeding effects on the IQ by genetic variation in fatty acid metabolism. Proc Natl Acad Sci USA. 2007;104:18860–18865. doi: 10.1073/pnas.0704292104. http://dx.doi.org/10.1073/pnas.0704292104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davidson PW, et al. Neurodevelopmental effects of maternal nutritional status and exposure to methylmercury from eating fish during pregnancy. Neurotoxicology. 2008;29:767–775. doi: 10.1016/j.neuro.2008.06.001. http://dx.doi.org/10.1016/j.neuro.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Folch J, Lees M, Sloane Stanley GH. A simple method for the isolation and purification of total lipides from animal tissues. J Biol Chem. 1957;226:497–509. [PubMed] [Google Scholar]
- 9.Glaser C, Heinrich J, Koletzko B. Role of FADS1 and FADS2 polymorphisms in polyunsaturated fatty acid metabolism. Metabolism. 2010;59:993–999. doi: 10.1016/j.metabol.2009.10.022. http://dx.doi.org/10.1016/j.metabol.2009.10.022. [DOI] [PubMed] [Google Scholar]
- 10.Glaser C, Lattka E, Rzehak P, Steer C, Koletzko B. Genetic variation in poly-unsaturated fatty acid metabolism and its potential relevance for human development and health. Matern Child Nutr. 2011;7(Suppl 2):S27–S40. doi: 10.1111/j.1740-8709.2011.00319.x. http://dx.doi.org/10.1111/j.1740-8709.2011.00319.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haggarty P. Fatty acid supply to the human fetus. Ann Rev Nutr. 2010;30:237–255. doi: 10.1146/annurev.nutr.012809.104742. http://dx.doi.org/10.1146/annurev.nutr.012809.104742. [DOI] [PubMed] [Google Scholar]
- 12. [accessed Jan 2015];HapMap Population Genotype Data from Yoruba in Ibadan, Nigeria (YRI) and Utah Residents with Northern and Western European Ancestry (CEU) 2015 Available at: < http://hapmap.ncbi.nlm.nih.gov/downloads/index.html.en>.
- 13.Harslof LB, Larsen LH, Ritz C, Hellgren LI, Michaelsen KF, Vogel U, Lauritzen L. FADS genotype and diet are important determinants of DHA status: a cross-sectional study in Danish infants. Am J Clin Nutr. 2013;97:1403–1410. doi: 10.3945/ajcn.113.058685. http://dx.doi.org/10.3945/ajcn.113.058685. [DOI] [PubMed] [Google Scholar]
- 14.Hellstrand S, et al. Intake levels of dietary long-chain PUFAs modify the association between genetic variation in FADS and LDL-C. J Lipid Res. 2012;53:1183–1189. doi: 10.1194/jlr.P023721. http://dx.doi.org/10.1194/jlr.P023721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hester AG, et al. Relationship between a common variants in the fatty acid desaturase (FADS) cluster and eicosanoid generation in humans. J Biol Chem. 2014 doi: 10.1074/jbc.M114.579557. http://dx.doi.org/10.1074/jbc.M114.579557. [DOI] [PMC free article] [PubMed]
- 16.Hoile SP, et al. Supplementation with N-3 long-chain polyunsaturated fatty acids or olive oil in men and women with renal disease induces differential changes in the DNA methylation of FADS2 and ELOVL5 in peripheral blood mononuclear cells. PLoS One. 2014 doi: 10.1371/journal.pone.0109896. http://dx.doi.org/10.1371/journal.pone.0109896. [DOI] [PMC free article] [PubMed]
- 17.Howard TD, et al. DNA methylation in an enhancer region of the FADS cluster is associated with FADS activity in human liver. PLoS One. 2014;9:e97510. doi: 10.1371/journal.pone.0097510. http://dx.doi.org/10.1371/journal.pone.0097510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Janssen CI, Kiliaan AJ. Long-chain polyunsaturated fatty acids (LCPUFA) from genesis to senescence: the influence of LCPUFA on neural development, aging, and neurodegeneration. Prog Lipid Res. 2014;53:1–17. doi: 10.1016/j.plipres.2013.10.002. http://dx.doi.org/10.1016/j.plipres.2013.10.002. [DOI] [PubMed] [Google Scholar]
- 19.Koletzko B, Lattka E, Zeilinger S, Illig T, Steer C. Genetic variants of the fatty acid desaturase gene cluster predict amounts of red blood cell docosahexaenoic and other polyunsaturated fatty acids in pregnant women: findings from the Avon Longitudinal Study of Parents and Children. Am J Clin Nutr. 2011;93:211–219. doi: 10.3945/ajcn.110.006189. http://dx.doi.org/10.3945/ajcn.110.006189 ajcn.110.006189 [pii] [DOI] [PubMed] [Google Scholar]
- 20.Lattka E, Illig T, Heinrich J, Koletzko B. FADS gene cluster polymorphisms: important modulators of fatty acid levels and their impact on atopic diseases. J Nutr Nutr. 2009;2:119–128. doi: 10.1159/000235559. http://dx.doi.org/10.1159/000235559. [DOI] [PubMed] [Google Scholar]
- 21.Lattka E, Illig T, Heinrich J, Koletzko B. Do FADS genotypes enhance our knowledge about fatty acid related phenotypes? Clin Nutr. 2010;29:277–287. doi: 10.1016/j.clnu.2009.11.005. http://dx.doi.org/10.1016/j.clnu.2009.11.005. [DOI] [PubMed] [Google Scholar]
- 22.Lattka E, Koletzko B, Zeilinger S, Hibbeln JR, Klopp N, Ring SM, Steer CD. Umbilical cord PUFA are determined by maternal and child fatty acid desaturase (FADS) genetic variants in the Avon Longitudinal Study of Parents and Children (ALSPAC) Br J Nutr. 2013;109:1196–1210. doi: 10.1017/S0007114512003108. http://dx.doi.org/10.1017/S0007114512003108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lattka E, et al. Genetic variants in the FADS gene cluster are associated with arachidonic acid concentrations of human breast milk at 1.5 and 6 mo post-partum and influence the course of milk dodecanoic, tetracosenoic, and trans-9-octadecenoic acid concentrations over the duration of lactation. Am J Clin Nutr. 2011;93:382–391. doi: 10.3945/ajcn.110.004515. http://dx.doi.org/10.3945/ajcn.110.004515. [DOI] [PubMed] [Google Scholar]
- 24.Marquardt A, Stohr H, White K, Weber BH. cDNA cloning, genomic structure, and chromosomal localization of three members of the human fatty acid desaturase family. Genomics. 2000;66:175–183. doi: 10.1006/geno.2000.6196. http://dx.doi.org/10.1006/geno.2000.6196. [DOI] [PubMed] [Google Scholar]
- 25.Mathias RA, Pani V, Chilton FH. Genetic variants in the FADS gene: implications for dietary recommendations for fatty acid intake. Curr Nutr Rep. 2014;3:139–148. doi: 10.1007/s13668-014-0079-1. http://dx.doi.org/10.1007/s13668-014-0079-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Molto-Puigmarti C, Plat J, Mensink RP, Muller A, Jansen E, Zeegers MP, Thijs C. FADS1 FADS2 gene variants modify the association between fish intake and the docosahexaenoic acid proportions in human milk. Am J Clin Nutr. 2010;91:1368–1376. doi: 10.3945/ajcn.2009.28789. http://dx.doi.org/10.3945/ajcn.2009.28789. [DOI] [PubMed] [Google Scholar]
- 27.Morales E, et al. Genetic variants of the FADS gene cluster and ELOVL gene family, colostrums LC-PUFA levels, breastfeeding, and child cognition. PLoS One. 2011;6:e17181. doi: 10.1371/journal.pone.0017181. http://dx.doi.org/10.1371/journal.pone.0017181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakamura MT, Nara TY. Essential fatty acid synthesis and its regulation in mammals. Prostaglandins Leukot Essent Fatty Acids. 2003;68:145–150. doi: 10.1016/s0952-3278(02)00264-8. [DOI] [PubMed] [Google Scholar]
- 29. [accessed 31.01.15];NCBI Reference SNP Cluster Report: rs3834458. 2015 < http://www.ncbi.nlm.nih.gov/projects/SNP/snp_ref.cgi?rs=3834458>.
- 30.Reardon HT, Zhang J, Kothapalli KS, Kim AJ, Park WJ, Brenna JT. Insertion-deletions in a FADS2 intron 1 conserved regulatory locus control expression of fatty acid desaturases 1 and 2 and modulate response to simvastatin. Prostaglandins Leukot Essent Fatty Acids. 2012;87:25–33. doi: 10.1016/j.plefa.2012.04.011. http://dx.doi.org/10.1016/j.plefa.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rentschler G, et al. Polymorphisms in iron homeostasis genes and urinary cadmium concentrations among nonsmoking women in Argentina and Bangladesh. Environ Health Perspect. 2013;121:467–472. 472e461–467. doi: 10.1289/ehp.1205672. http://dx.doi.org/10.1289/ehp.1205672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rentschler G, Kippler M, Axmon A, Raqib R, Skerfving S, Vahter M, Broberg K. Cadmium concentrations in human blood and urine are associated with polymorphisms in zinc transporter genes. Metallomics: Integr Biometal Sci. 2014;6:885–891. doi: 10.1039/c3mt00365e. http://dx.doi.org/10.1039/c3mt00365e. [DOI] [PubMed] [Google Scholar]
- 33.Rzehak P, et al. Evidence for an association between genetic variants of the fatty acid desaturase 1 fatty acid desaturase 2 (FADS1 FADS2) gene cluster and the fatty acid composition of erythrocyte membranes. Br J Nutr. 2009;101:20–26. doi: 10.1017/S0007114508992564. http://dx.doi.org/10.1017/S0007114508992564. [DOI] [PubMed] [Google Scholar]
- 34.Rzehak P, et al. Variants of the FADS1 FADS2 gene cluster, blood levels of polyunsaturated fatty acids and eczema in children within the first 2 years of life. PLoS One. 2010;5:e13261. doi: 10.1371/journal.pone.0013261. http://dx.doi.org/10.1371/journal.pone.0013261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pawlosky R, et al. Physiological compartmental analysis of alpha-linolenic acid metabolism in adult humans. J Lipid Res. 2001;42:1257–1265. [PubMed] [Google Scholar]
- 36.Schaeffer L, et al. Common genetic variants of the FADS1 FADS2 gene cluster and their reconstructed haplotypes are associated with the fatty acid composition in phospholipids. Hum Mol Genet. 2006;15:1745–1756. doi: 10.1093/hmg/ddl117. http://dx.doi.org/10.1093/hmg/ddl117. [DOI] [PubMed] [Google Scholar]
- 37.Scholtz SA, Kerling EH, Shaddy DJ, Li S, Thodosoff JM, Colombo J, Carlson SE. Docosahexaenoic acid (DHA) supplementation in pregnancy differentially modulates arachidonic acid and DHA status across FADS genotypes in pregnancy. Prostaglandins Leukot Essent Fatty Acids. 2014 doi: 10.1016/j.plefa.2014.10.008. http://dx.doi.org/10.1016/j.plefa.2014.10.008. [DOI] [PMC free article] [PubMed]
- 38.Shamlaye CF, et al. The Seychelles child development study on neurodevelopmental outcomes in children following in utero exposure to methylmercury from a maternal fish diet: background and demographics. Neurotoxicology. 1995;16:597–612. [PubMed] [Google Scholar]
- 39.Sprecher H, Luthria DL, Mohammed BS, Baykousheva SP. Reevaluation of the pathways for the biosynthesis of polyunsaturated fatty acids. J Lipid Res. 1995;36:2471–2477. [PubMed] [Google Scholar]
- 40.Steer CD, Lattka E, Koletzko B, Golding J, Hibbeln JR. Maternal fatty acids in pregnancy, FADS polymorphisms, and child intelligence quotient at 8 y of age. Am J Clin Nutr. 2013;98:1575–1582. doi: 10.3945/ajcn.112.051524. http://dx.doi.org/10.3945/ajcn.112.051524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Strain JJ, et al. Associations of maternal long-chain polyunsaturated fatty acids, methyl mercury, and infant development in the Seychelles child development nutrition study. Neurotoxicology. 2008;29:776–782. doi: 10.1016/j.neuro.2008.06.002. http://dx.doi.org/10.1016/j.neuro.2008.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Strain JJ, et al. Maternal PUFA status but not prenatal methylmercury exposure is associated with children’s language functions at age five years in the Seychelles. J Nutr. 2012;142:1943–1949. doi: 10.3945/jn.112.163493. http://dx.doi.org/10.3945/jn.112.163493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Strain JJ, et al. Prenatal exposure to methyl mercury and polyunsaturated fatty acids from fish consumption: Associations with children’s development at 20 months of age in the Republic of Seychelles. Am J Clin Nutr. 2015 doi: 10.3945/ajcn.114.100503. http://dx.doi.org/10.3945/ajcn.114.100503. [DOI] [PMC free article] [PubMed]
- 44.Tanaka T, et al. Genome-wide association study of plasma polyunsaturated fatty acids in the InCHIANTI Study. PLoS Genet. 2009;5:e1000338. doi: 10.1371/journal.pgen.1000338. http://dx.doi.org/10.1371/journal.pgen.1000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van Wijngaarden E, et al. Prenatal methyl mercury exposure in relation to neurodevelopment and behavior at 19 years of age in the Seychelles Child Development Study. Neurotoxicol Teratol. 2013;39:19–25. doi: 10.1016/j.ntt.2013.06.003. http://dx.doi.org/10.1016/j.ntt.2013.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vessby B, Gustafsson IB, Tengblad S, Berglund L. Indices of fatty acid desaturase activity in healthy human subjects: effects of different types of dietary fat. Br J Nutr. 2013;110:871–879. doi: 10.1017/S0007114512005934. http://dx.doi.org/10.1017/S0007114512005934. [DOI] [PubMed] [Google Scholar]
- 47.Xie L, Innis SM. Genetic variants of the FADS1 FADS2 gene cluster are associated with altered (n-6) and (n-3) essential fatty acids in plasma and erythrocyte phospholipids in women during pregnancy and in breast milk during lactation. J Nutr. 2008;138:2222–2228. doi: 10.3945/jn.108.096156. http://dx.doi.org/10.3945/jn.108.096156. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.