Abstract
This study reports on a comprehensive comparison of the effects of soy and meat proteins given at the recommended level on physiological markers of metabolic syndrome and the hepatic transcriptome. Male rats were fed semi-synthetic diets for 1 wk that differed only regarding protein source, with casein serving as reference. Body weight gain and adipose tissue mass were significantly reduced by soy but not meat proteins. The insulin resistance index was improved by soy, and to a lesser extent by meat proteins. Liver triacylglycerol contents were reduced by both protein sources, which coincided with increased plasma triacylglycerol concentrations. Both soy and meat proteins changed plasma amino acid patterns. The expression of 1571 and 1369 genes were altered by soy and meat proteins respectively. Functional classification revealed that lipid, energy and amino acid metabolic pathways, as well as insulin signaling pathways were regulated differently by soy and meat proteins. Several transcriptional regulators, including NFE2L2, ATF4, Srebf1 and Rictor were identified as potential key upstream regulators. These results suggest that soy and meat proteins induce distinct physiological and gene expression responses in rats and provide novel evidence and suggestions for the health effects of different protein sources in human diets.
Metabolic syndrome is becoming a global epidemic. It is a cluster of metabolic abnormalities characterized by central obesity, dyslipoproteinemia, hypertension, and glucose intolerance, leading to increased risks of cardiovascular diseases and type 2 diabetes1. Dietary intervention has been regarded as an important therapy for metabolic syndrome2. For example, dietary restriction3 and incorporation of bioactive nutrients (fiber4, ω-3 polyunsaturated fatty acids5) have been widely studied. Protein is an essential nutrient for the whole life cycle of the human. It has become increasingly recognized that, apart for providing a source of amino acids (AA) for protein synthesis, dietary protein also exerts a wide range of biological functions, such as the regulation of food intake6, lipid metabolism7, glucose homeostasis and insulin secretion8. An example is the soy plant protein which has been recognized as having a hypocholesterolemic effect9,10. Meat protein is an important dietary source of animal protein for human nutritional requirements. Compared to plant protein, meat protein distinguishes itself for its richness in all the essential amino acids11. However, except for their differences in amino acid composition, our understanding regarding the effects of different protein sources on modulating metabolic health is still limited. Therefore, this study was designed to investigate and compare the impacts of different dietary protein sources (soy and meat) in rats on markers of metabolic syndrome (body weight, adipose weight, triacylglycerol (TAG), cholesterol, glucose and insulin levels and the insulin resistance index HOMA-IR). To provide clues for the possible molecular mechanisms that may underlay the effects of the different protein sources, liver gene expression profiles were studied by using RNA-sequence methodology.
Results
Growth performance and body composition
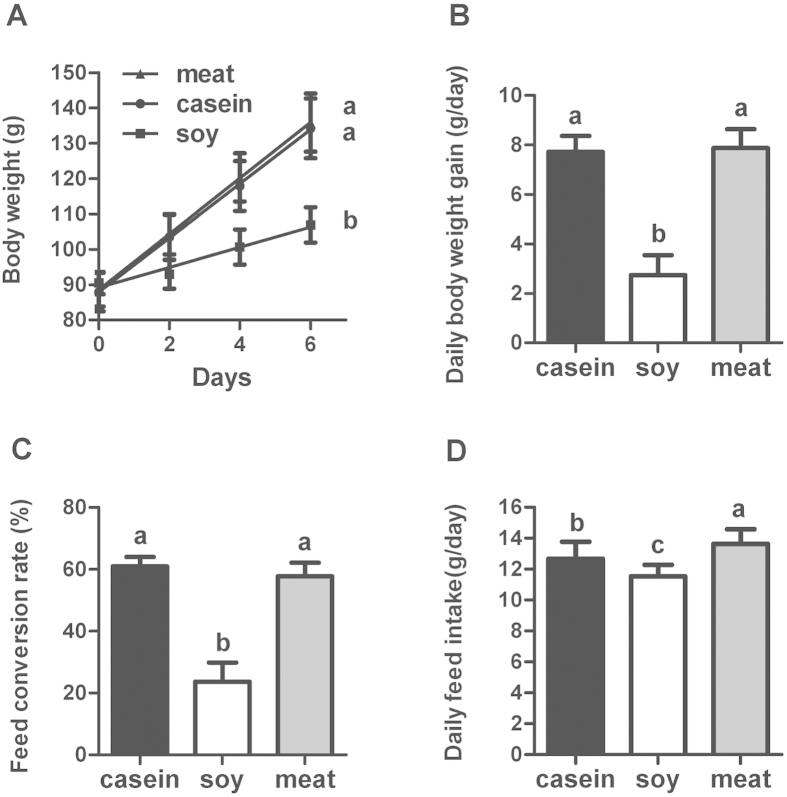
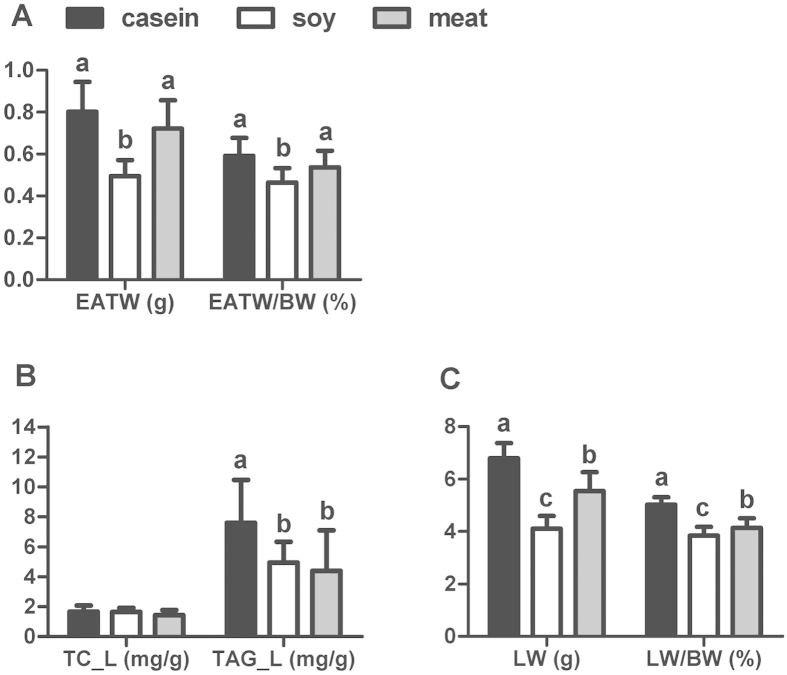
Rats in each group had similar initial body weights (Fig. 1). Compared to the casein group, that served as control in our experiment, the group fed dietary soy protein had significantly reduced daily feed intake (decreased by 9%, P < 0.05), body weight gain (decreased by 65%, P < 0.05) and feed conversion rate (decreased by 61%, P < 0.05). However, feeding rats meat protein only increased the daily feed intake (increased by 7.6%, P < 0.05), but did not affect the body weight gain or the feed conversion rate (P > 0.05). To evaluate the effect of dietary soy and meat proteins on body adiposity, epididymal adipose tissue weight and hepatic lipid content were measured (Fig. 2). Only soy protein reduced the epididymal adipose tissue weight (P < 0.05) compared with casein and meat protein. Both soy and meat proteins significantly reduced the liver weight and TAG content (P < 0.05). The total cholesterol (TC) content of liver did not differ between the groups.
Figure 1. Growth performance of rats fed casein, soy and meat protein diets.
(A) Body weight; (B) Daily body weight gain; (C) Feed conversion rate (BWG/FI). (D) Daily feed intake. Values are shown as means ± SD. The numbers of biological repetitions of casein, soy and meat protein groups were 10, 10 and 40, respectively. For panel (A), linear regression was performed for body weight of rats. The intercept representing initial body weight of rats was not different for three groups. The slope represents growth rate of rats, and its statistical significance (P < 0.05) is illustrated by different letters on the right side of lines. For panels (B–D), different letters above bars indicate significant difference at P < 0.05 analyzed by one-way ANOVA and LSD multiple comparisons.
Figure 2. Adipose tissue weight, liver weight, liver TC and TAG content of rats fed casein, soy and meat protein diets.
(A) EATW: absolute weight of epididymal adipose tissue; EATW/BW: relative weight of epididymal adipose tissue to body weight. (B) TC-L: total cholesterol in the liver; TAG-L: triacylglycerol in the liver. (C) LW: absolute weight of liver; LW/BW: relative weight of liver to body weight. Values are shown as means ± SD. The numbers of biological repetitions of casein, soybean and meat protein groups were 10, 10 and 40, respectively. Different letters above bars indicate significant difference at P < 0.05 tested by one-way ANOVA and LSD multiple comparisons.
Plasma profiling
Plasma TAG concentrations were increased by both soy and meat proteins (P < 0.05, Table 1), but only meat protein reduced plasma TC concentrations (P < 0.05). Both soy and meat proteins reduced plasma total protein concentrations (P < 0.05), but only soy protein increased plasma urea concentrations (increased by 53.7%, P < 0.05). Plasma glucose concentrations and insulin levels were significantly reduced by both soy and meat proteins (P < 0.05). As a result, the insulin resistance index (HOMA-IR) was improved by both soy and meat proteins (P < 0.05). Total plasma AA concentrations were significantly increased only by meat protein (P < 0.05, Table 2), and compared to casein this was due to increased concentrations of both nutritionally essential AAs (EAAs) and nonessential AAs (NEAAs) (P < 0.05). The only exception was that methionine was reduced by meat protein (reduced by 17.1%, P < 0.05). Similarly, feeding soy protein resulted higher plasma concentrations of NEAAs and arginine (P < 0.05) compared to casein. However, compared to the meat proteins, the soy protein had much lower concentrations of methionine (decreased by 53.3%, P < 0.05) and also valine concentrations were lower (decreased by 21.2%, P = 0.054).
Table 1. Plasma triacylglycerol, cholesterol, glucose, insulin, total protein and urea concentrations of rats fed casein, soy or meat protein diets.
| Casein (n = 10) | Soy (n = 7)1 | Meat(n = 39)2 | |
|---|---|---|---|
| TAG, mmol/L | 0.35 ± 0.06b | 0.45 ± 0.08a | 0.43 ± 0.06a |
| TC, mmol/L | 2.06 ± 0.41a | 1.99 ± 0.21ab | 1.84 ± 0.25b |
| glucose, mmol/L | 8.51 ± 0.63a | 6.89 ± 1.69b | 7.44 ± 1.56b |
| insulin, mIU/L | 32.39 ± 23.27a | 10.77 ± 6.43b | 18.31 ± 12.46b |
| HOMA-IR | 12.14 ± 8.15a | 3.66 ± 2.71b | 6.53 ± 5.31b |
| total protein, g/L | 51.51 ± 3.86a | 45.76 ± 3.12b | 48.65 ± 3.93b |
| urea, mmol/L | 3.91 ± 0.66b | 6.01 ± 1.43a | 3.88 ± 1.31b |
Values are shown as means ± SD. (n = 7)1, blood of three rats in soy group was collected but yielded insufficient plasma to allow analyses; (n = 39)2, blood of one rat in meat group was collected but yielded insufficient plasma to allow analyses. Different superscripts indicate statistical significance at P < 0.05 analyzed by one-way ANOVA and LSD multiple test. TAG, triacylglycerol; TC, total cholesterol; HOMA-IR was calculated using the formula: HOMA-IR = [glucose (mmol/L) × insulin (mIU/L)/22.5], using fasting values.
Table 2. Plasma amino acid concentrations of the rats fed casein, soy or meat protein diets.
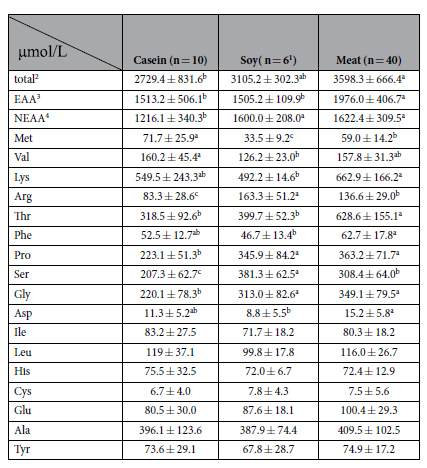
Values are shown as means ± SD. (n = 6)1, blood of four rats in soy group was collected but yielded insufficient plasma to allow analyses. Different superscripts indicate the statistical significance at P < 0.05 analyzed by one-way ANOVA and LSD multiple test. Total2: the sum of 17 kinds of amino acids in plasma including Arg, Pro, Met, Val, Ser, Gly, Lys, Thr, Phe, Asp, Ile, Leu, Cys, Glu, Ala, Tyr, His. EAA3: the sum of 9 kinds of essential amino acids in plasma including Arg, Met, Val, Lys, Thr, Phe, Ile, Leu, His. NEAA2: the sum of 8 kinds of non-essential amino acids in plasma including Pro, Ser, Gly, Asp, Cys, Glu, Ala, Tyr.
Differentially expressed genes in the liver
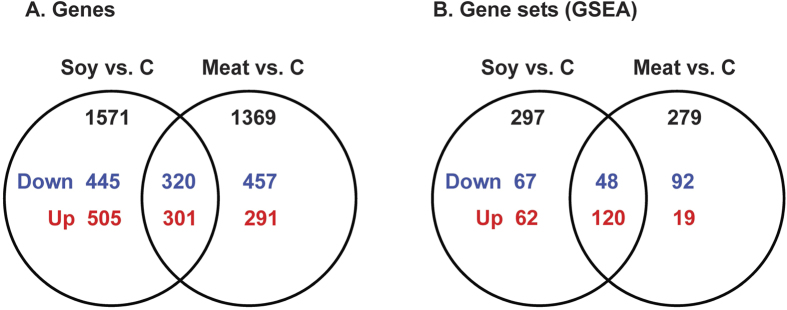
RNA-sequencing was performed to identify differentially expressed genes in rat liver. Soy and meat protein groups were pairwise compared to casein, and the expression of 1571 and 1369 genes were significantly changed by soy and meat proteins, respectively (P < 0.05, Fig. 3A). When presented in a Venn plot, it became clear that about half of these changed genes were specific for each protein group. In total, 320 down-regulated genes and 301 up-regulated genes were commonly altered by soy and meat proteins. See Supplementary Table S1 online for all genes.
Figure 3. Venn plot of differentially expressed genes and gene sets in the liver of rats fed soy and meat proteins when compared to casein.
(A) Genes: significant level was set at P < 0.05; (B) Gene sets (GSEA): significant level was P < 0.05 & FDR < 0.25. Soy vs. C: Soy vs. Casein; Meat vs. C: Meat vs. Casein; down: down-regulated; up: up-regulated.
Functional implications of differential gene expression
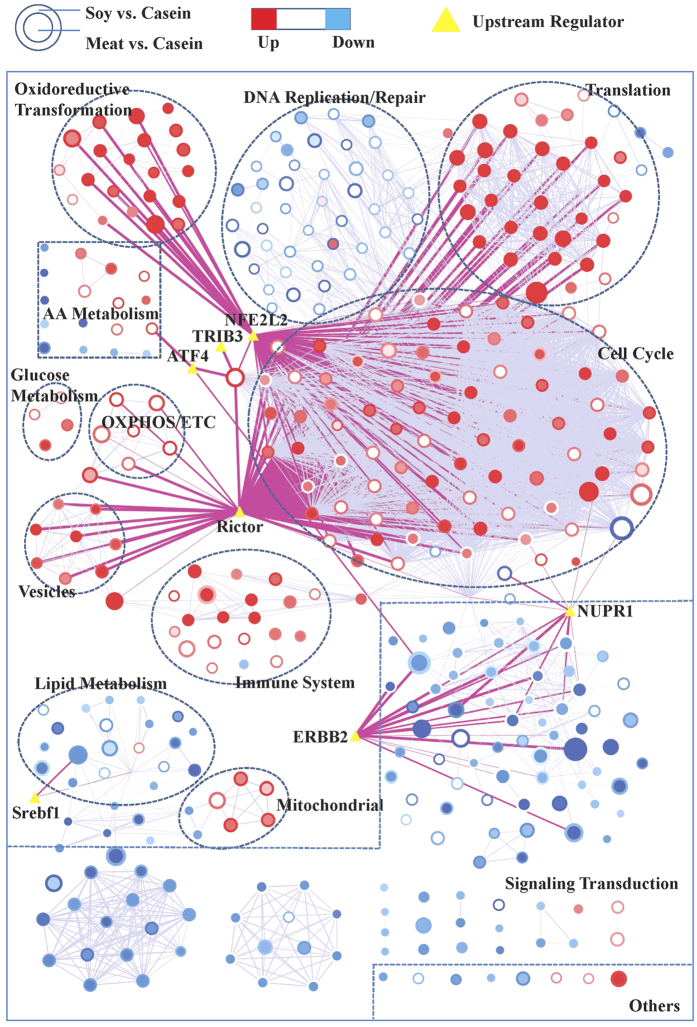
To gain better insight into the underlying biologic phenomena these changes in gene expression brought about, GSEA was conducted. Compared to the casein group, 297 and 279 gene sets were significantly changed by the dietary soy and meat proteins, respectively, when compared with casein (P < 0.05 & FDR < 0.25, Fig. 3B). See Supplementary Table S2 online for all significant gene sets. To enhance the interpretation, all significant gene sets were summarized in an enrichment map, and functionally related gene sets were semi-automatically annotated and manually labeled to highlight the prevalent biologic functions among the related gene sets (Fig. 4). A high-resolution map that includes names of all gene-sets is shown in Supplementary Fig. S1 online. Five clusters of gene sets describing (control of) the cell cycle, mRNA translation, oxidoreductive transformation, immune system and vesicles were increased by both soy and meat proteins compared with the casein diet. Other processes relating to oxidative phosphorylation/electron transport chain and gluconeogenesis were increased by soy protein only. Gene sets relating to AA metabolic pathways were increased by soy protein but were decreased by meat protein. More specifically, gene sets describing the metabolism of methionine, cysteine, phenylalanine, threonine, glycine, serine, alanine, aspartate and glutamine were increased by soy protein only, while gene sets describing the metabolism of tryptophan, histidine and branched-chain amino acids (BCAAs) were repressed by meat protein only.
Figure 4. Gene set network of liver transcriptome of the rats fed soy and meat proteins compared to the rats fed casein.
The network was produced by using Cytoscape v3.2 and Enrichment Map plugin v2.1. Nodes represent enriched gene sets in GSEA analysis of liver transcriptome. The node size is proportional to the total number of genes within each gene-set (from 15 to 500). The border area of the round node represents the comparison of soy protein group to the casein group (Soy vs. Casein), whereas the inner area of the round node represents the comparison of meat protein group to the casein group (Meat vs. Casein). The colors of nodes indicate the directions of changes of gene-sets with red for up-regulation and blue for down-regulation. The enrichment P value is mapped to the node color as a color gradient. The color changes from light to bright with the P value decreasing from 0.05 to 0. The weighted grey lines between the round nodes represent the “overlap” score (Jaccard and overlap coefficients >0.375) depending on the number of genes two gene-sets share. The more genes two gene-sets share, the wider the line. The yellow triangles in the map represent the upstream regulators predicted by using Ingenuity Pathway Analysis, and they were overlapped with putative target genes in the genes sets using the Post Analysis tool in the Enrichment Map v2.1 plugin. Pink lines represent the overlap P value < 0.05 (Fisher’s Exact Test) between gene sets and upstream regulators. Nodes of high similarity were automatically arranged close together, and circles were semi-automatically annotated and manually labelled.
Gene sets describing lipid metabolism were repressed by both soy and meat proteins (Fig. 4), but to different extents. More specifically, the gene set describing regulation of cholesterol biosynthesis by Srebf (sterol regulatory element-binding transcription factor) was reduced by both soy and meat proteins. Only soy protein reduced expression of genes involved in TAG and cholesterol biosynthesis. Gene sets relating to bile acid biosynthesis were increased by soy protein but reduced by meat protein. In addition, gene sets describing fatty acid degradation (beta oxidation) and biosynthesis were reduced by both soy and meat proteins. However, the PPARα signaling pathway was reduced by meat protein only, which was also confirmed by real-time PCR array analysis. This showed that mRNA expression of 68 PPARα target genes were significantly reduced by meat protein (P value < 0.05 & fold change >2.0) but not by soy protein (see Supplementary Table S3 online). In addition, various clusters of signal transduction pathways were suppressed; the TGFβ and focal adhesion pathways were reduced by both soy and meat proteins. Notably, the NOTCH, ERBB, mTOR and insulin signaling pathways were reduced by meat protein only.
Upstream regulators
To explain the shifts in gene expression profiles, upstream regulator analysis was performed. Results of this analysis identified several transcriptional regulators that were predicted to be activated or inhibited (Table 3). The transcription factor NFE2L2 (nuclear factor erythroid 2-like 2) stood out because it was predicted to be the top activated upstream regulator for both soy and meat proteins. In line with results obtained by GSEA, its target genes overlapped with the cluster of gene sets describing oxidoreductive transformation reactions, but also with some genes involved in (control of) cell cycle. In addition, Rictor (rapamycin-insensitive companion of TOR, complex 2) was predicted to be the top inhibited upstream regulator for both soy and meat proteins and its target genes overlapped with various gene sets relating to cell cycle, translation, as well as oxidative phosphorylation and the electron transport chain. Transcription factor Srebf1 was predicted to be inhibited by soy, and to a lesser extent by meat proteins, and its target genes overlapped with lipid metabolism. Several other regulators were predicted to play a role in the transcriptional responses caused by only one specific dietary protein. Notably, two regulators, activating transcription factor 4 (ATF4) and tribbles pseudokinase 3 (TRIB3), were predicted as being specific for soy protein, and their target genes overlapped with AA metabolism, which was increased by soy protein. On the other hand, PPARG and RXRA were predicted to be upstream regulators for meat proteins only.
Table 3. Upstream regulators predicted for the hepatic transcriptome changes in the rats fed soy or meat protein diets compared to rats fed casein.
| UpstreamRegulator | Molecule Type | Z Score1 |
|
|---|---|---|---|
| Soy vs.newline/>Casein | Meat vs.newline/>Casein | ||
| NFE2L2 | transcription regulator | 4.368 | 3.273 |
| NUPR1 | transcription regulator | 2.814 | 2.699 |
| ATF4 | transcription regulator | 3.498 | |
| SPDEF | transcription regulator | 2.646 | |
| EIF4E | translation regulator | 2.311 | |
| PTTG1 | transcription regulator | 2.213 | |
| PPARG | ligand-dependent nuclear receptor | 2.377 | |
| RXRA | ligand-dependent nuclear receptor | −2.458 | |
| CTNNB1 | transcription regulator | −2.055 | |
| MLXIPL | transcription regulator | −2.407 | |
| CCND1 | transcription regulator | −2.414 | |
| FOXM1 | transcription regulator | −2.533 | |
| TRIB3 | kinase | −2.954 | |
| ERBB2 | kinase | −2.310 | −3.075 |
| Srebf1 | transcription regulator | −2.956 | −2.179 |
| Rictor | transcription regulator | −6.090 | −4.308 |
All upstream regulators were significant at P < 0.01 as calculated by the Fisher’s Exact Test. Z score1: determine whether an upstream regulator is significantly activated (>2.0) or inhibited (<−2.0).
NFE2L2: nuclear factor, erythroid 2-like 2; NUPR1: nuclear protein 1; ATF4: activating transcription factor 4; SPDEF: SAM pointed domain-containing Ets transcription factor; EIF4E: Eukaryotic translation initiation factor 4E; PTTG1: Securin; PPARG: Peroxisome proliferator-activated receptor gamma; RXRA: Retinoid X receptor alpha (RXR-alpha); CTNNB1: Catenin beta-1; MLXIPL: Carbohydrate-responsive element-binding protein/MLX-interacting protein-like; CCND1: Cyclin-D1; FOXM1: forkhead box protein M1; TRIB3: tribbles pseudokinase 3; ERBB2: v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 2;Srebf1: sterol regulatory element-binding transcription factor 1; Rictor: rapamycin-insensitive companion of TOR, complex 2.
Discussion
We are the first to report on a comprehensive comparison of the effects of dietary soy and meat proteins given at the recommended level on markers of the metabolic syndrome and hepatic mRNA expression. Our work relates to some previously published papers that also reported on the effects of dietary proteins, but most of these studies only focused on lipid metabolism and applied high fat diets in their studies12,13,14,15, or focused on only one kind of dietary protein, mostly soy protein15 or fish protein13,14. In addition, other studies investigated the effects of different nutritional levels of dietary protein, such as dietary protein restriction16 or a high protein diet17, on metabolism in animals. However, unlike these studies, by measuring and comparing the widespread responses on plasma, hepatic gene expression and physiological markers, our study provides a comprehensive comparison between two different protein sources (soy and meat) versus casein, all provided at the recommended level. There are thus several key points which makes our study unique when compared to other studies: 1) In our study the rats were fed nutritionally balanced, semi-synthetic diets (AIN-93G18) that differed only regarding protein source; the casein (reference) was fully replaced by purified proteins from soy or meat. 2) We compared the effects of the different dietary protein sources on the physiological and metabolic responses in rats in a comprehensive way, instead of only focusing on specific metabolic or physiological processes such as lipid metabolism. 3) In order to find potential molecular mechanism that may mediate the differential effects, liver gene expression profiles were determined by using RNA-sequence methodology.
Compared to casein, soy and meat proteins induced distinct physiological and molecular effects in rats that suggested an improvement of metabolic health, which is likely caused by the different composition of soy and meat proteins. Soy protein is a mixture of β-conglycinin and glycinin that account for 90% of the total protein19. Meat proteins are much more structurally diverse being composed of myofibrillar (50–55%), sarcoplasmic (30–34%) and connective tissue proteins (10–15%)20. Compared with casein, these structurally different proteins in soy and meat proteins are likely to digested differently resulting in a diverse range of peptides being absorbed from the gastro-intestinal tract. Our previous study indeed showed that even dietary meat proteins from different animal types were specifically digested, resulting in the production of different peptides in the stomach and jejunum of rats21. Except for protein composition, the AA compositions, and particularly the fraction of EAAs in dietary proteins, are the most important criteria and markers for protein quality assessment22.
According to the AA compositions found in the protein powders (see Supplementary Table S4 online), diets (see Supplementary Table S5 online) and plasma of rats (Table 2), methionine was found to be the first limiting AA in soy protein. In order to evaluate the AA composition of protein powders, the AA ratios of the EAAs were calculated by using casein +1.5% cysteine (AIN-93G) as the reference protein (see Supplementary Table S6 online)23. We found that the AA ratio of methionine in the soy protein was the lowest (52%) among all the EAAs, but in the meat protein it was 111%. Considering the AA composition in the various diets (see Supplementary Table S5 online), methionine was also the lowest EAA (53.7% lower than casein) in the soy protein diet compared to casein group. However, in meat protein diets, the content of methionine was similar to that of the casein group. It has been demonstrated that the plasma AA pattern, in particular the most limiting AA, would cause a direct response when rats are fed an AA imbalanced diet24. Further analysis of plasma AA concentrations showed that methionine was significantly reduced by 53.3% by the dietary soy protein, which coincided with the lower methionine content in the soy protein powder and diet. The plasma methionine concentration in the meat protein group was also reduced, compared with casein, but to a lesser extent (reduced by 17.1%). Therefore, we conclude that soy protein is an imbalanced protein source with methionine being the most limiting AA, which is not the case for the meat protein. This is also in accordance with previous observations11,25.
According to our results, dietary soy protein triggered significant phenotype changes in the rats, including the significantly reduced feed intake and body weight gain. This indicates soy protein may inhibit the appetite and growth of rats, which did not happen to the rats fed meat protein. Correlation analysis found a highly positive correlation between the body weight gain and feed intake (correlation coefficient = 0.8, P < 0.05), which indicates the reduced body weight probably resulted from the reduced feed intake induced by the soy protein. At the same time, the soy protein was found to be methionine deficient. It is known that EAA deficiency can lead to feed intake repression, and its mechanism has been thoroughly reviewed26. Therefore, we reasoned that the growth inhibitory effect of soy protein is due to the repressed feed intake which may be induced by methionine deficiency. Based on the above, it seems that diet restriction and methionine restriction occurred concomitantly in the soy protein group. However, the rats fed soy protein weighed 20% less than the casein group but consumed only 9% less feed. As a result, the feed intake per gram of body mass (0.65 g/g body weight) was actually higher for the soy protein group than for the casein group (0.57 g/g body weight). Therefore, there was actually no diet restriction on a body weight basis but only the methionine restriction in the soy protein group.
In response to the different protein sources, protein and AA metabolism was regulated differently. Only dietary soy protein increased the metabolism of NEAAs and sulfur-containing AAs in the liver and plasma urea level, indicating increased AA degradation in rats27. We believe that this can be related to the methionine limitation in soy protein. In this condition the NEAAs are abundantly available, but since these cannot all be used for body protein synthesis, due to the lack of methionine, they are deaminated to produce urea24,28. These changes were predicted to be regulated by upstream regulators ATF4 (Z score = 3.498) and TRIB3 (Z score = −2.954). ATF4 is usually up-regulated when AA deprivation occurs29 and it regulates a wide array of genes involved in AA transport, metabolism and energy management30. TRIB3 acts as a negative feedback regulator of ATF4 and is also involved in the regulation of gene expression in instances of AA limitation31. In contrast to soy protein, dietary meat proteins did not change the plasma urea concentrations. Expression level of genes involved in EAA metabolism in the liver were significantly reduced by meat protein only. However, these genes did not overlap with the ATF4 and TRIB3 target genes, indicating that additional molecular mechanisms for AA metabolism are initiated in response to dietary soy and meat proteins.
Our results indicated that both the dietary soy and meat proteins have beneficial effects on lipid metabolism in rats. For the rats fed soy protein, although their feed intakes per gram of body mass were higher than the casein group, their body fat mass including adipose mass and liver TAG content was significantly reduced, which we believe can be related to the significantly repressed lipid and fatty acid biosynthesis in the liver. Unlike soy protein, the dietary meat protein did not reduce adipose mass in rats. As observed for soy protein, the dietary meat protein also reduced the hepatic TAG content and expression of genes involved in lipid synthesis. The transcription factor Srebf1, which is a master regulator of lipogenesis32, was predicted to be an inhibited upstream regulator for soy, and to a lesser extent meat proteins. However, expression of genes of the PPARα signaling pathway was only reduced by dietary meat protein according to both the RNA-sequencing and qPCR results. With regards to cholesterol metabolism, dietary soy protein significantly inhibited cholesterol biosynthesis in the liver indicating its potential cholesterol-reducing effect, which has also been reported in previous studies33,34. However, no significant changes were found in plasma TC levels in rats fed soy protein. This could be because the intervention period in the present study was relatively short compared to previous studies which usually had more than 2-weeks of dietary intervention33,34. Dietary meat proteins reduced plasma TC levels in rats, indicating a hypocholesterolemic effect of meat protein. Taken together, dietary soy and meat protein may have the similar effects on lipogenesis process via the transcription factor Srebf1 but have different effects on the PPARα signaling pathway in the liver. Both soy and meat protein have beneficial effects on cholesterol homeostasis.
It has been acknowledged that a reduction of body fat occurs when the energy requirements for growth and maintenance exceeds energy intake35. Our transcriptomic results revealed that the energy metabolism (oxidative phosphorylation and electron transport chain) in the liver was not affected by dietary meat proteins, and this coincided with the unchanged adipose mass in rats fed meat protein. However, unlike the meat protein group, the hepatic expression of genes involved in oxidative phosphorylation and electron transport chain were significantly increased in the rats fed soy protein. At the same time, these genes significantly overlapped with the target genes of Rictor, which was predicted to be the top inhibited upstream regulator for the soy protein (Z score = −6.090). Rictor is a key regulatory/structural subunit of mTORC2 and plays an important role in regulating hepatic glycolysis and lipogenesis via glucokinase and Srebf136. Therefore, we speculate that the body fat reducing effect of the soy protein may be related to the increased energy metabolism and decreased lipogenesis mediated through the transcription regulators Rictor and Srebf1.
Insulin resistance is a major component of metabolic syndrome37. A previous study suggested that soy protein may reduce insulin secretion by the pancreas which in turn was related to the plasma amino acid pattern after consumption of soy protein38. However, some epidemiological studies have suggested that animal protein may increase risk of diabetes39. Uhe et al. (1992) observed that ingestion of beef, chicken or fish proteins did not change insulin level in human subjects40. In the present study we found that plasma insulin levels were significantly reduced by both soy and meat proteins compared to the casein group, indicating that both soy and meat proteins have potential effects on improving insulin sensitivity.
Taken together, our results showed that dietary soy and meat proteins induce widespread but distinct physiological and gene expression responses in rats. These responses may be attributed to the different AA contents in soy and meat protein sources, especially of the EAA. Soy protein was shown to be deficient in methionine. According to various methionine restriction studies, the biological responses to methionine restriction are variable41,42,43. Despite this heterogeneity, many of the biological processes that were changed by the dietary soy protein in our study were similar to the results reported in previous methionine restriction studies41,42,43. These commonly changed biological processes included the increased feed intake per gram of body weight, the reduced body weight gain, adiposity and lipid metabolism, and the increased energy metabolism and insulin sensitivity41,42,43. At the same time, our transcriptomic results indicated that the mTOR (mammalian target of rapamycin) signaling pathway was significantly changed by the dietary meat protein (Supplementary Fig. S1 online). It has been recognized that mTOR is a master sensor of amino acid levels in cells and regulates widespread biological processes including protein synthesis and autophagy44. Furthermore, other research suggests that many pathways, such as mTOR, insulin, PPAR and Srebf signaling, have close crosstalk in regulating the cellular responses to cope with changes in nutrient status or stress45,46. Based on the above, these widespread physiological and gene expression responses to different dietary protein sources found in our study are biologically relevant. Our study thus provides important novel scientific evidence and suggestions for the health effects of different protein sources in human diets.
Materials and Methods
Ethics statement
Male Sprague Dawley (SD) rats were purchased from the Shanghai Laboratory Animal Research Center, Chinese Academy of Science at 3 weeks of age. The animals were housed in pairs in a controlled environment with a 12h light-dark cycle (12:12 h reversed light/dark cycle, 23 °C). The animal experiment was performed according to the guide for care and use of laboratory animals of Nanjing Agriculture University (Nanjing, China) and the Jiangsu Provincial Academy of Agricultural Sciences (The license number was SCXK (Su) 2002–0029). All efforts were made to minimize the number of animals used and to minimize their suffering. The rats were acclimated for 1 week before the study and they had free access to water and standard rat chow throughout the experiment.
Diets
The standard, nutritionally balanced semi-synthetic AIN-93G18 was used as reference diet in this study. The protein source in this diet is casein (milk protein). The other five experimental diets (soy, beef, pork, chicken and fish protein) were also prepared according to the AIN-93G formula, except that casein was fully replaced by one of the five proteins (Table 4). To ensure consistency between the diets, large batches of ingredients of diets were purchased from Dyets Inc. (Bethlehem, PA). Soy protein isolates (food grade) were purchased from Linyi Shansong biological products company (Linyi, China) and treated with 80% methanol to remove isoflavones. Meat protein sources were prepared as follows: raw meat materials, i.e., pork and beef Longissimus dorsi muscle, chicken pectoralis major muscle and fish (carp) dorsal muscle were cooked in a 72 °C water bath to an internal temperature of 70 °C. The cooked meat were chilled to 4 °C and at 16h, the cooked meat was freeze-dried and twice defatted with methylene chloride/methanol (2:1, v:v). After the fat extraction, the residual solvent was removed by evaporation and the resulting protein powder was passed through a 30 Mesh (0.595 mm) sieve. The final protein powders consisted of more than 90% of protein and 6–9% of water. AA composition of protein powders (see Supplementary Table S4 online) and diets (see Supplementary Table S5 online) was determined using a Hitachi L-8900 AA analyzer (Tokyo, Japan). The presence of iron in meat results from it being a component of the heme in myoglobin and thus, as there are large differences in contents of different meats, iron was not balanced during diet preparation. Other minerals in the diets were balanced based on the actual mineral composition in protein powders (see Supplementary Table S7 online). The mineral formulations for the six diets are listed in the Supplemental Table S8 online. All diets were prepared by Jiangsu-Xietong, Inc. (Nanjing, China).
Table 4. Ingredient composition and nutritional content of diets.
| g/Kg diet | Casein | Soy | Pork | Beef | Chicken | Fish |
|---|---|---|---|---|---|---|
| diet composition, g/Kg diet | ||||||
| Protein1 | 200.0 | 203.0 | 190.0 | 195.0 | 192.0 | 191.0 |
| Cornstarch | 397.5 | 397.5 | 397.5 | 397.5 | 397.5 | 397.5 |
| Dyetros | 132 | 132 | 132 | 132 | 132 | 132 |
| Sucrose | 100 | 100 | 100 | 100 | 100 | 100 |
| Soybean oil | 70 | 70 | 70 | 70 | 70 | 70 |
| Cellulose | 50 | 50 | 50 | 50 | 50 | 50 |
| Mineral mix2 | 35.0 | 31.9 | 30.3 | 33.4 | 31.4 | 29.2 |
| Vitamin mix3 | 10 | 10 | 10 | 10 | 10 | 10 |
| L-Cystine4 | 3.0 | 0 | 0 | 0 | 0 | 0 |
| Choline Bitartrate | 2.5 | 2.5 | 2.5 | 2.5 | 2.5 | 2.5 |
| nutritional level, U/Kg | ||||||
| Energy, Kcal | 4056.0 | 4056.0 | 4056.0 | 4056.0 | 4056.0 | 4056.0 |
| Protein, g | 177 | 177 | 177 | 177 | 177 | 177 |
| Fat, g | 70 | 70 | 70 | 70 | 70 | 70 |
| Carbohydrate, g | 679.5 | 679.5 | 679.5 | 679.5 | 679.5 | 679.5 |
Protein1, the amount of protein powder was adjusted and balanced according to the protein content in soy and meat protein powder. Mineral mix2, the formulation of mineral mixes for the six diets is listed in Supplementary Table S8 online. Vitamin mix3: the formulation of vitamin mix as described 18. L-Cystine4: the amino acid composition of soy and meat protein diets were not modified.
Experimental protocol
When the rats arrived at the laboratory, all rats were fed the standard AIN-93G diet for 7 days to adapt to the purified diets and new environment. Subsequently, the rats were randomly allocated to one of the six diet groups (n = 10 per group), i.e. casein, soy, pork, beef, chicken or fish protein groups. Rats were fed these diets for 7 days. Body weights and dietary intakes were measured every 2 days. On the day of sacrifice, rats were deprived of feed for 4 h prior to sacrifice but were given free access to water. Rats were anaesthetized with ether inhalation. Blood was taken by orbital puncture and plasma was isolated. Liver and epididymal adipose tissues were obtained, weighed and snap frozen in liquid nitrogen. All samples were stored at −80 °C until analysis. To simplify the comparison between the soy and meat proteins versus casein, all data from the four meat protein groups, including the physiological, plasma and gene expression data, were combined and analyzed as single meat protein group.
Liver lipid contents and plasma parameters detection
TAG and TC contents in the liver were determined using commercial kits purchased from Nanjing Jiancheng Bioengineering Institute (Nanjing, China). Plasma TAG, TC, glucose, urea, and total protein concentrations were analyzed using a Hitachi 7180 auto analyzer (Tokyo, Japan). Plasma insulin concentrations were determined using a radioimmunoassay kit purchased from Beijing North Institute of Biological Technology (Beijing, China). The homeostasis model assessment for insulin resistance (HOMA-IR)47 was calculated according to the equation IR = (fasting insulin in mU/L × fasting glucose in mM)/22.5. Plasma free AA concentrations were determined using a Hitachi L-8900 AA analyzer (Tokyo, Japan).
Gene expression profiling in liver by RNA-sequencing
RNA isolation, library construction and sequencing
Total liver RNA was isolated by using the RNAiso Reagent Kit (Takara, Dalian, China). RNA-sequencing of three biological replicates from each group was commissioned to BGI Tech (Shenzhen, China). In brief, the isolated RNA samples were treated with DNase I to remove DNA, and enriched by using oligo (dT) magnetic beads. The mRNA was fragmented into short fragments (about 200 bp), and converted to double strand cDNA. The cDNA was purified with magnetic beads, and sequencing adaptors were ligated to the fragments. The fragments were enriched by PCR amplification. The library products were sequenced on an Illumina HiSeq 2000 machine (BGI, Shenzhen, China).
Identification of differentially expressed genes
After cleaning up the raw sequence data by removing adapter sequences and low quality reads, reads were quantified and annotated using the workflow implemented in the Bioconductor package QuasR48. Reads were mapped against the Rat Genome Sequencing Consortium Rnor_5.0 rat genome assembly, and annotated based on the University of California Santa Cruz known Genes track49,50. Nonspecific filtering of the count table was carried out to increase detection power51, based on the requirement that a gene should have an expression level greater than 1 count per million reads mapped for at least 3 libraries across all samples. Differences in library size were adjusted for by the trimmed mean of M-values normalization method52, implemented in the Bioconductor package edgeR53. Counts were then log-transformed and the observed mean-variance trend was converted into precision weights by the voom function54 in the Bioconductor package limma55. Differentially expressed genes were identified by using linear models and a moderated t-statistic55. Genes that satisfied the criterion of P < 0.05 were considered to be significantly regulated.
Biological interpretation of gene expression data
Changes in gene expression were related to biologically meaningful changes using gene set enrichment analysis (GSEA)56. It is well accepted that GSEA has multiple advantages over analyses performed on the level of individual genes56,57. Gene-sets were retrieved from the expert-curated Kyoto Encyclopedia of Genes and Genomes, Biocarta, Reactome and WikiPathways pathway databases. Only gene-sets consisting of more than 15 and fewer than 500 genes were taken into account, which resulted in the inclusion of 2,272 gene sets. For each comparison, genes were ranked on their t-value that was calculated by the empirical Bayes method. Statistical significance of GSEA results was determined using 1,000 permutations. The Enrichment Map v2.1.0 plugin for Cytoscape v3.2.0 was used for visualization and interpretation of the GSEA results58. Enrichment maps were generated with gene-sets that passed conservative significance thresholds (P < 0.05, False Discovery Rate (FDR) < 0.25). Upstream regulator analysis in Ingenuity Pathway Analysis (content version 21249400 released 23 September 2014; Ingenuity Systems) was used to identify the cascade of potential upstream regulators that may explain the observed gene expression changes in the data set. For this study, focus was on transcriptional regulators. Target genes of several top regulators were overlapped to the genes in the gene set network using Post Analysis tool in Enrichment Map v2.1.0.
Quantitative PCR array analyzing PPAR signaling pathway
The PPAR (peroxisome proliferator-activated receptor) signaling pathway was profiled using RT2 profiler PCR array (330231 PARN-149ZA, Qiagen, Hilden, Germany), according to the manufacturer’s instructions. The RT2 Profiler PCR array profiles the expression of 84 target genes besides housekeeping and control genes. Gene amplification was performed on an Applied Biosystems 7500 Real-Time PCR System (Foster City, CA).
Statistical methods
Except for the RNA-seq data, all statistical analyses were performed using the SPSS version 16.0 (Chicago, IL). The diet effect on measured variables were analyzed by one-way analysis of variance (ANOVA) and means were compared by least-significant difference (LSD) multiple comparison. Statistical significance was set at P < 0.05. Values are shown as means ± SD.
Additional Information
How to cite this article: Song, S. et al. Dietary soy and meat proteins induce distinct physiological and gene expression changes in rats. Sci. Rep. 6, 20036; doi: 10.1038/srep20036 (2016).
Supplementary Material
Acknowledgments
We would like to thank professors Ron Tume, Feng Gao and Weihua Chen (Nanjing Agricultural University) for helpful suggestions during the preparation of animal experiment. This work was funded by grants 31471600 (National Natural Science Foundation of China), NCET-11-0668 (Ministry of Education of the P. R. China), and CXZZ13_0286 (Jiangsu Provincial Department of Education, China).
Footnotes
Author Contributions G.Z., C.L., M.M., X.X. and S.S. designed research; S.S., M.L., F.Z. and W.Z. conducted research; S.S., C.L. and G.H. analyzed data; S.S., C.L. and G.H. wrote the paper; G.Z. and C.L. had primary responsibility for final content. All authors read and approved the final manuscript.
References
- Gade W., Schmit J., Collins M. & Gade J. Beyond obesity: the diagnosis and pathophysiology of metabolic syndrome. Clin Lab Sci 23, 51–61; quiz 62–55 (2010). [PubMed] [Google Scholar]
- Andersen C. J. & Fernandez M. L. Dietary strategies to reduce metabolic syndrome. Rev Endocr Metab Disord 14, 241–254, 10.1007/s11154-013-9251-y (2013). [DOI] [PubMed] [Google Scholar]
- Matarese L. E. & Pories W. J. Adult weight loss diets: metabolic effects and outcomes. Nutr Clin Pract 29, 759–767, 10.1177/0884533614550251 (2014). [DOI] [PubMed] [Google Scholar]
- He B. et al. Transmissible microbial and metabolomic remodeling by soluble dietary fiber improves metabolic homeostasis. Sci Rep 5, 10604, 10.1038/srep10604 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert B. B. et al. Higher omega-3 index is associated with increased insulin sensitivity and more favourable metabolic profile in middle-aged overweight men. Sci Rep 4, 6697, 10.1038/srep06697 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromentin G. et al. Peripheral and central mechanisms involved in the control of food intake by dietary amino acids and proteins. Nutr Res Rev 25, 29–39, 10.1017/S0954422411000175 (2012). [DOI] [PubMed] [Google Scholar]
- El Khoury D. & Anderson G. H. Recent advances in dietary proteins and lipid metabolism. Curr Opin Lipidol 24, 207–213, 10.1097/MOL.0b013e3283613bb7 (2013). [DOI] [PubMed] [Google Scholar]
- Ranawana V. & Kaur B. Role of proteins in insulin secretion and glycemic control. Adv Food Nutr Res 70, 1–47, 10.1016/B978-0-12-416555-7.00001-1 (2013). [DOI] [PubMed] [Google Scholar]
- Carroll K. K. & Kurowska E. M. Soy consumption and cholesterol reduction: review of animal and human studies. J Nutr 125, 594S–597S (1995). [DOI] [PubMed] [Google Scholar]
- Potter S. M. Overview of proposed mechanisms for the hypocholesterolemic effect of soy. J Nutr 125, 606S–611S (1995). [DOI] [PubMed] [Google Scholar]
- Pereira P. M. & Vicente A. F. Meat nutritional composition and nutritive role in the human diet. Meat Sci 93, 586–592, 10.1016/j.meatsci.2012.09.018 (2013). [DOI] [PubMed] [Google Scholar]
- Brandsch C., Shukla A., Hirche F., Stangl G. I. & Eder K. Effect of proteins from beef, pork, and turkey meat on plasma and liver lipids of rats compared with casein and soy protein. Nutrition 22, 1162–1170, 10.1016/j.nut.2006.06.009 (2006). [DOI] [PubMed] [Google Scholar]
- Shukla A. et al. Dietary fish protein alters blood lipid concentrations and hepatic genes involved in cholesterol homeostasis in the rat model. Br J Nutr 96, 674–682 (2006). [PubMed] [Google Scholar]
- Hosomi R., Fukunaga K., Arai H., Nishiyama T. & Yoshida M. Effects of dietary fish protein on serum and liver lipid concentrations in rats and the expression of hepatic genes involved in lipid metabolism. J Agric Food Chem 57, 9256–9262, 10.1021/jf901954r (2009). [DOI] [PubMed] [Google Scholar]
- Madani S., Lopez S., Blond J. P., Prost J. & Belleville J. Highly purified soybean protein is not hypocholesterolemic in rats but stimulates cholesterol synthesis and excretion and reduces polyunsaturated fatty acid biosynthesis. J Nutr 128, 1084–1091 (1998). [DOI] [PubMed] [Google Scholar]
- Kang W., Lee M. & Baik M. Dietary Protein Restriction Alters Lipid Metabolism and Insulin Sensitivity in Rats. Asian Australas. J. Anim. Sci. 24, 1274–1281 (2011). [Google Scholar]
- Schwarz J., Tome D., Baars A., Hooiveld G. J. & Muller M. Dietary protein affects gene expression and prevents lipid accumulation in the liver in mice. PLoS One 7, e47303, 10.1371/journal.pone.0047303 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves P. G., Nielsen F. H. & Fahey G. C. Jr. AIN-93 purified diets for laboratory rodents: final report of the American Institute of Nutrition ad hoc writing committee on the reformulation of the AIN-76A rodent diet. J Nutr 123, 1939–1951 (1993). [DOI] [PubMed] [Google Scholar]
- Friedman M. & Brandon D. L. Nutritional and health benefits of soy proteins. J Agric Food Chem 49, 1069–1086 (2001). [DOI] [PubMed] [Google Scholar]
- Tornberg E. Effects of heat on meat proteins-Implications on structure and quality of meat products. Meat Sci 70, 493–508, 10.1016/j.meatsci.2004.11.021 (2005). [DOI] [PubMed] [Google Scholar]
- Wen S. et al. Discrimination of in vitro and in vivo digestion products of meat proteins from pork, beef, chicken and fish. Proteomics 15, 3688–3698, 10.1002/pmic.201500179 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tome D. Criteria and markers for protein quality assessment-a review. Br J Nutr 108 Suppl 2, S222–229, 10.1017/S0007114512002565 (2012). [DOI] [PubMed] [Google Scholar]
- Sarwar G. The protein digestibility-corrected amino acid score method overestimates quality of proteins containing antinutritional factors and of poorly digestible proteins supplemented with limiting amino acids in rats. J Nutr 127, 758–764 (1997). [DOI] [PubMed] [Google Scholar]
- Leung P. M., Rogers Q. R. & Harper A. E. Effect of amino acid imbalance on plasma and tissue free amino acids in the rat. J Nutr 96, 303–318 (1968). [DOI] [PubMed] [Google Scholar]
- Berry T., Becker D., Rasmussen O., Jensen A. & Norton H. The limiting amino acids in soybean protein. J Anim Sci 21, 558–561 (1962). [Google Scholar]
- Gietzen D. W., Hao S. & Anthony T. G. Mechanisms of food intake repression in indispensable amino acid deficiency. Annu Rev Nutr 27, 63–78, 10.1146/annurev.nutr.27.061406.093726 (2007). [DOI] [PubMed] [Google Scholar]
- Hoffenberg R. Measurement of the synthesis of liver-produced plasma proteins with special reference to their regulation by dietary protein and amino acid supply. Proc Nutr Soc 31, 265–272 (1972). [DOI] [PubMed] [Google Scholar]
- Sidransky H. Regulatory effect of amino acids on polyribosomes and protein synthesis of liver. Prog Liver Dis 4, 31–43 (1972). [PubMed] [Google Scholar]
- Kilberg M. S., Shan J. & Su N. ATF4-dependent transcription mediates signaling of amino acid limitation. Trends Endocrinol Metab 20, 436–443, 10.1016/j.tem.2009.05.008 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. I. et al. HepG2/C3A cells respond to cysteine deprivation by induction of the amino acid deprivation/integrated stress response pathway. Physiol Genomics 33, 218–229, 10.1152/physiolgenomics.00263.2007 (2008). [DOI] [PubMed] [Google Scholar]
- Ord D., Meerits K. & Ord T. TRB3 protects cells against the growth inhibitory and cytotoxic effect of ATF4. Exp Cell Res 313, 3556–3567, 10.1016/j.yexcr.2007.07.017 (2007). [DOI] [PubMed] [Google Scholar]
- Eberle D., Hegarty B., Bossard P., Ferre P. & Foufelle F. SREBP transcription factors: master regulators of lipid homeostasis. Biochimie 86, 839–848, 10.1016/j.biochi.2004.09.018 (2004). [DOI] [PubMed] [Google Scholar]
- Tachibana N. et al. Intake of soy protein isolate alters hepatic gene expression in rats. J Agric Food Chem 53, 4253–4257, 10.1021/jf0481103 (2005). [DOI] [PubMed] [Google Scholar]
- Takahashi Y. & Ide T. Effects of soy protein and isoflavone on hepatic fatty acid synthesis and oxidation and mRNA expression of uncoupling proteins and peroxisome proliferator-activated receptor gamma in adipose tissues of rats. J Nutr Biochem 19, 682–693, 10.1016/j.jnutbio.2007.09.003 (2008). [DOI] [PubMed] [Google Scholar]
- Lass A., Zimmermann R., Oberer M. & Zechner R. Lipolysis-a highly regulated multi-enzyme complex mediates the catabolism of cellular fat stores. Prog Lipid Res 50, 14–27, 10.1016/j.plipres.2010.10.004 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagiwara A. et al. Hepatic mTORC2 activates glycolysis and lipogenesis through Akt, glucokinase, and SREBP1c. Cell Metab 15, 725–738, 10.1016/j.cmet.2012.03.015 (2012). [DOI] [PubMed] [Google Scholar]
- Guo S. Insulin signaling, resistance, and the metabolic syndrome: insights from mouse models into disease mechanisms. J Endocrinol 220, T1–T23, 10.1530/JOE-13-0327 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noriega-Lopez L. et al. Pancreatic insulin secretion in rats fed a soy protein high fat diet depends on the interaction between the amino acid pattern and isoflavones. J Biol Chem 282, 20657–20666, 10.1074/jbc.M701045200 (2007). [DOI] [PubMed] [Google Scholar]
- Beasley J. M. & Wylie-Rosett J. The role of dietary proteins among persons with diabetes. Curr Atheroscler Rep 15, 348, 10.1007/s11883-013-0348-2 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhe A. M., Collier G. R. & O’Dea K. A comparison of the effects of beef, chicken and fish protein on satiety and amino acid profiles in lean male subjects. J Nutr 122, 467–472 (1992). [DOI] [PubMed] [Google Scholar]
- Anthony T. G., Morrison C. D. & Gettys T. W. Remodeling of lipid metabolism by dietary restriction of essential amino acids. Diabetes 62, 2635–2644, 10.2337/db12-1613 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaveroux C. et al. Molecular mechanisms involved in the adaptation to amino acid limitation in mammals. Biochimie 92, 736–745, 10.1016/j.biochi.2010.02.020 (2010). [DOI] [PubMed] [Google Scholar]
- Orgeron M. L. et al. The impact of dietary methionine restriction on biomarkers of metabolic health. Prog Mol Biol Transl Sci 121, 351–376, 10.1016/B978-0-12-800101-1.00011-9 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jewell J. L., Russell R. C. & Guan K. L. Amino acid signalling upstream of mTOR. Nat Rev Mol Cell Biol 14, 133–139, 10.1038/nrm3522 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimobayashi M. & Hall M. N. Making new contacts: the mTOR network in metabolism and signalling crosstalk. Nat Rev Mol Cell Biol 15, 155–162, 10.1038/nrm3757 (2014). [DOI] [PubMed] [Google Scholar]
- Yuan H. X., Xiong Y. & Guan K. L. Nutrient sensing, metabolism, and cell growth control. Mol Cell 49, 379–387, 10.1016/j.molcel.2013.01.019 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cacho J., Sevillano J., de Castro J., Herrera E. & Ramos M. P. Validation of simple indexes to assess insulin sensitivity during pregnancy in Wistar and Sprague-Dawley rats. Am J Physiol Endocrinol Metab 295, E1269–1276, 10.1152/ajpendo.90207.2008 (2008). [DOI] [PubMed] [Google Scholar]
- Gaidatzis D., Lerch A., Hahne F. & Stadler M. B. QuasR: quantification and annotation of short reads in R. Bioinformatics 31, 1130–1132, 10.1093/bioinformatics/btu781 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karolchik D. et al. The UCSC Genome Browser database: 2014 update. Nucleic Acids Res 42, D764–770, 10.1093/nar/gkt1168 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twigger S. N. et al. What everybody should know about the rat genome and its online resources. Nat Genet 40, 523–527, 10.1038/ng0508-523 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourgon R., Gentleman R. & Huber W. Independent filtering increases detection power for high-throughput experiments. Proc Natl Acad Sci USA 107, 9546–9551, 10.1073/pnas.0914005107 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M. D. & Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol 11, R25, 10.1186/gb-2010-11-3-r25 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M. D., McCarthy D. J. & Smyth G. K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140, 10.1093/bioinformatics/btp616 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law C. W., Chen Y., Shi W. & Smyth G. K. voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol 15, R29, 10.1186/gb-2014-15-2-r29 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie M. E. et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43, e47, 10.1093/nar/gkv007 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 102, 15545–15550, 10.1073/pnas.0506580102 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abatangelo L. et al. Comparative study of gene set enrichment methods. BMC Bioinformatics 10, 275, 10.1186/1471-2105-10-275 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merico D., Isserlin R., Stueker O., Emili A. & Bader G. D. Enrichment map: a network-based method for gene-set enrichment visualization and interpretation. PLoS One 5, e13984, 10.1371/journal.pone.0013984 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.