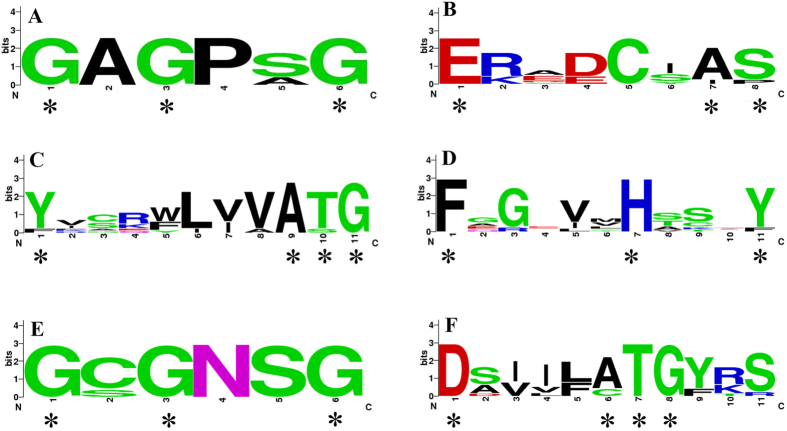Abstract
The phytohormone auxin is essential for plant growth and development, and YUCCA (YUC) proteins catalyze a rate-limiting step for endogenous auxin biosynthesis. Despite YUC family genes have been isolated from several species, systematic expression analyses of YUCs in response to abiotic stress are lacking, and little is known about the function of YUC homologs in agricultural crops. Cucumber (Cucumis sativus L.) is a world cultivated vegetable crop with great economical and nutritional value. In this study, we isolated 10 YUC family genes (CsYUCs) from cucumber and explored their expression pattern under four types of stress treatments. Our data showed that CsYUC8 and CsYUC9 were specifically upregulated to elevate the auxin level under high temperature. CsYUC10b was dramatically increased but CsYUC4 was repressed in response to low temperature. CsYUC10a and CsYUC11 act against the upregulation of CsYUC10b under salinity stress, suggesting that distinct YUC members participate in different stress response, and may even antagonize each other to maintain the proper auxin levels in cucumber. Further, CsYUC11 was specifically expressed in the male flower in cucumber, and enhanced tolerance to salinity stress and regulated pedicel and stamen development through auxin biosynthesis in Arabidopsis.
The phytohormone auxin plays fundamental roles in plant growth and development, such as shoot and root formation1, apical dominance2, plant architecture3, floral development4, gynoecium and fruit development5,6 and various tropic responses7. These biological functions of auxin result from the coordinated actions of auxin biosynthesis, auxin transport and auxin signalling. Auxin biosynthesis was found to take place in tissues such as cotyledons, leaves, flowers, and shoot apex8. Synthesis of auxin can be divided into one tryptophan (Trp)-independent and four Trp-dependent pathways9,10. The Trp-dependent pathways are much well understood than the Trp-independent pathway10,11. The primary Trp-dependent pathway for auxin biosynthesis is a two-step process, in which Trp was first converted into indole-3-pyruvate (IPA) by TRYPTOPHAN AMINOTRANSFERASE OF ARABIDOPSIS1/TRYPTOPHAN AMINOTRANSFERASE RELATED (TAA1/TAR), and then YUCCA (YUC) flavin-containing monooxygenases catalyzed IPA into indole-3-acetic acid (IAA), the major endogenous auxin12,13. The second step catalyzed by YUCs is a rate-limiting step for auxin biosynthesis which plays a significant role in most auxin-mediated developmental processes14,15. This Trp-dependent pathway is highly conserved and widely exists throughout the plant kingdom11.
YUC proteins belong to the flavin-containing monooxygenases (FMOs) which had several conserved domains11,16. The FAD-binding motif and NADPH-binding motif containing the GXGXXG residues were highly conserved across all of the FMOs16,17, with the FAD-binding motif locating near the N-terminus and the NADPH-binding motif locating in the middle of YUC proteins18. The FMO-identifying sequence with FXGXXXHXXXY residues contributes to NADPH binding, and was found in all plant FMOs as well17,18. YUC genes have been identified in many plant species. For example, 11 YUCs were identified in Arabidopsis, in which YUC1-6, YUC10 and YUC11 have been confirmed to be involved in local auxin biosynthesis13,15,19,20. In addition, one YUC gene from petunia21, six YUC genes from tomato22, seven YUC-like genes from rice23 and eight YUC gens from strawberry24 have been isolated.
A wealth of evidence indicated that YUC genes play important roles in multitude of developmental processes. Loss-of-function of YUC family genes resulted in dramatic developmental defects, such as yuc1 yuc4 mutant produced fewer flowers, yuc1 yuc4 yuc10 yuc11 mutant displayed one cotyledon with reduced vascular system, and yuc2 yuc6 plant showed defective stamen development in Arabidopsis15,25,26,27. Similarly, mutation in SPARSE INFLORESCENCE1 (SPI1), the YUC1 ortholog in maize, led to reduced number of branches and spikelets28. Overexpression of YUC genes caused phenotypes mimicking auxin overproduction such as the long hypocotyl phenotype in Arabidopsis14, leaf aberration and abnormal flower architecture in petunia21, and the changes of leaf shape, shoot and root elongation in rice23,29.
In addition to developmental regulation, YUC family genes are also involved in plant response to abiotic stresses. High temperature led to hypocotyl elongation and increased IAA level in Arabidopsis30. PHYTOCHROME-INTERACTING FACTOR 4 (PIF4), which encodes a basic helix-loop-helix (bHLH) transcription factor, regulates high temperature-induced hypocotyl elongation through up-regulation of YUC8 in Arabidopsis31. On the other hand, high temperature resulted in male sterility in the developing anthers of barley and Arabidopsis, which was caused by reduced auxin level and repressed expression of YUC2 and YUC632. The activation-tagged mutant yuc-1D in Arabidopsis showed enhanced resistant to water deficit33. Similarly, overexpression of AtYUC6 increased resistance to drought stress in potato34, whereas inactivation of CONSTITUTIVELY WILTED 1 (rice ortholog to YUCs) impaired the resistant to water deficit in rice35. In Arabidopsis, auxin concentrations were remarkably elevated under shade condition, and the expression of YUC2, YUC5, YUC8 and YUC9 were up-regulated as expected36.
However, systematic expression analyses of YUC family genes in response to abiotic stress are lacking, and little is known about the function of YUC homologs in agricultural crops. Cucumber (Cucumis sativus L.) is an important vegetable crop with great economical and nutritional value. In this study, we isolated 10 YUC family genes in cucumber, and explored the expression of all 10 CsYUCs in response to high temperature, low temperature, salinity stress and shading treatment. Our data showed that distinct CsYUC members mediated different abiotic stresses. Further, CsYUC11 was specifically expressed in male flower in cucumber, and enhanced tolerance to salinity stress and regulated pedicel and stamen development in Arabidopsis.
Results
Identification of CsYUCs in cucumber and gene structure analysis
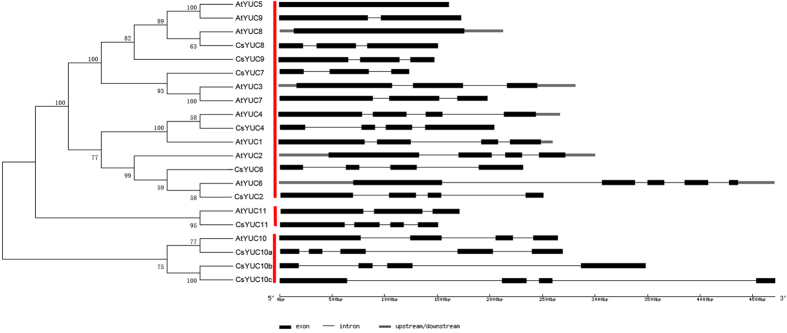
To identify the YUC family members from cucumber, we performed BLAST search in the Cucumber Genome DataBase37 based on the amino acid sequence information of the 11 Arabidopsis YUC genes. 10 CsYUCs were identified and named accordingly based on the best hit in Arabidopsis (Figure S1, Supplemental Table S1). There are no homologs for AtYUC1, AtYUC3 or AtYUC5 in cucumber, but instead, there are three homologs for AtYUC10, which are named as CsYUC10a, CsYUC10b, and CsYUC10c, with the highest similarity in CsYUC10a (56.44%) and least similarity in CsYUC10c (44.16%) (Supplemental Table S1). Protein similarity varies from 43.52% (AtYUC7/CsYUC7) to 78.87% (AtYUC8/CsYUC8) between YUC families in cucumber and Arabidopsis (Supplemental Figure S1). Phylogenetic analysis using the full-length protein sequence of AtYUCs and CsYUCs showed that all the YUCs can be divided into three clades (red lines in Fig. 1). The first clade is the biggest, consisting of 6 CsYUCs and 9 AtYUCs. The second clade is made exclusively of CsYUC11 and AtYUC11, while AtYUC10 and CsYUC10s constitute the third clade.
Figure 1. Phylogenetic analysis and gene structure of YUC family genes in cucumber and Arabidopsis.
The unrooted tree including 11 Arabidopsis YUCs and 10 cucumber YUC genes was constructed by MEGA5 software using the Neighbor-joining (NJ) method. Gene structure was generated in http://gsds.cbi.pku.edu.cn/. Black boxes and lines represent exons and introns, respectively. Upstream and downstream sequences are indicted by gray boxes.
Gene structure analysis was performed to compare the exon-intron organization of CsYUCs and AtYUCs (Fig. 1). CsYUC genes contained three to five exons, in which CsYUC2, CsYUC4, CsYUC7, CsYUC10b, CsYUC10c have the same gene structure as those in Arabidopsis, CsYUC6 has one exon less than AtYUC6, while the remaining four CsYUCs including CsYUC8, CsYUC9, CsYUC10a and CsYUC11 contain one or two more exons than the corresponding AtYUCs (Fig. 1). To further compare the gene structure between CsYUCs and their Arabidopsis homologs, each exon was comparatively analyzed in detail (Supplemental Table S2). For each gene, the first exon is generally the longest, and the third exon is the shortest (Supplemental Table S2). Despite exon numbers are variable in the YUC families, the full length coding sequences are quite similar (around 1200bp) except for CsYUC7 (Supplemental Table S2). The ten CsYUC genes are unevenly distributed on five chromosomes, with CsYUC7, CsYUC9, CsYUC10a located on chromosome 3, while no CsYUC gene is mapped to chromosome 4 or chromosome 5 (Supplemental Figure S2), the accession number and the exact genomic position of each CsYUC gene were provided in Supplemental Table S3.
Given that plant YUC-like flavin monooxygenases (FMOs) have several conserved motifs that play important roles in many aspects of plant development16,18, the full length of the ten CsYUCs protein sequences were aligned and the sequence logo for each FMO domain was produced to explore how conserved of these motifs (Fig. 2, Supplemental Figure S3). From N-terminus to C-terminus, six conserved FMO domains were identified, which are the FAD-binding motif, GC motif, ATG-containing motif 1, FMO-identifying sequence, NADPH-binding motif and ATG-containing motif 2 (Supplemental Figure S3). Previous studies showed that the GC-motif help stabilization of the FAD-binding, ATG-containing motif 1 plays a bridge role between FAD-binding motif and NADPH-binding motif, FMO-identifying sequence contributes to NADPH binding, while the ATG-containing motif 2 provides linkage between NADPH site and the active site24. Among all FMOs in the CsYUCs, the FAD-binding motif at the N-terminus and the NADPH-binding motif in the middle are the most conserved and display consensus core amino acids (asterisks in Fig 2A,E, Supplemental Figure S3). Distinct from other YUC members in cucumber, CsYUC6 and CsYUC7 lack of FAD-binding motif and GC motif (Supplemental Figure S3). CsYUC10b displays variations in the highly conserved sites of GC motif and FMO-identifying sequence, in which the residues of A and S are replaced by I and P in the GC motif, and the residue of Y is replaced by F in the FMO-identifying sequence, implying that CsYUC10b may play different functions among the cucumber CsYUCs. In the ATG-containing motif 1 and 2, CsYUC11 and CsYUC10s display several variants at the conserved amino acid sites, suggesting that YUCs in the clade 2 and clade 3 may play distinct roles than those in the clade 1 (Fig. 1, Supplemental Figure S3).
Figure 2. The conserved flavin monooxygenase (FMO) domains in the CsYUCs.
(A–F) The six relatively conserved FMO domains across all CsYUC proteins. The FAD-binding domain (A), GC-motif (B), ATG-Containing motif 1 (C), FMO-identifying sequence (D), NADPH domain (E), ATG-Containing motif 2 (F). The sequence logos of the 6 motifs are based on full-length alignments of the 10 CsYUC proteins. The overall height of each stack represents the conservation of the sequence at that position, and the height of letters within each stack indicates the relative frequency of the respective amino acid. The asterisk shows the position of the conserved amino acid that is identical cross all FMOs proteins.
Expression patterns of CsYUCs in cucumber
To explore the expression patterns of CsYUCs, semi-quantitative RT-PCR was used to examine the transcript levels in different tissues including stem (S), leaves (L), male floral buds (MB), female floral buds (FB), opened male flowers (MF), opened female flower (FF), tendril (T) and fruit (F) (Fig. 3). In the stem, all CsYUCs show no or very low expression, suggesting that stem is not a place for auxin sysnthesis. For all other tissues, most CsYUCs display ubiquitous expression, but divergence does exist. For example, CsYUC4 and CsYUC10b display somewhat a complementary expression pattern to CsYUC11, in which CsYUC4 and CsYUC10b are highly expressed in all tissues except the opened male flower, whereas CsYUC11 is specifically enriched in the opened male flower but low in other tissues (Fig. 3). Expression of CsYUC2 and CsYUC8 are similarly enriched in leaves, female floral buds, tendrils and fruits, while CsYUC7 expression is exclusively repressed in the fruits (Fig. 3).
Figure 3. Semi-quantitative RT-PCR analysis of CsYUCs in cucumber.
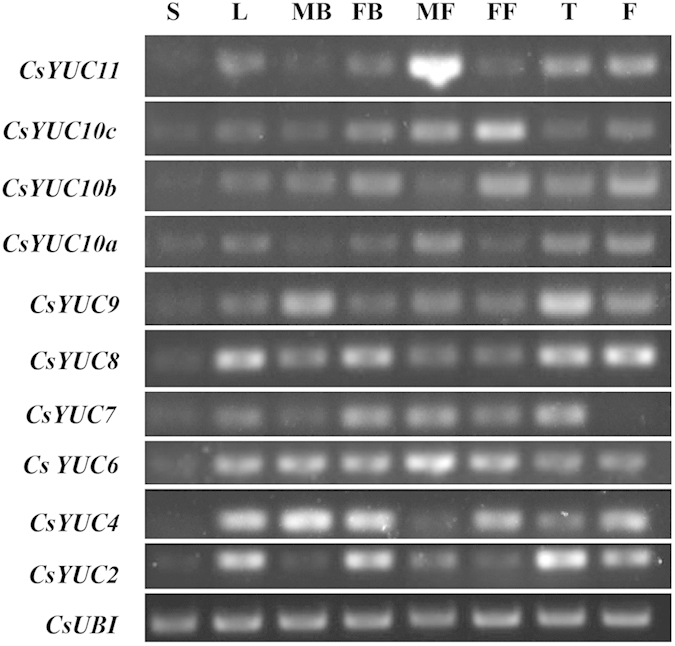
Transcripts of CsYUCs were analyzed in stem (S), leaves (L), male floral buds (MB), female floral buds (FB), opened male flowers (MF), opened female flowers (FF), tendrils (T) and fruits (F). The cucumber UBIQUITIN gene was used as an internal control to normalize the expression data.
Distinct CsYUCs contribute to auxin production under different abiotic stresses
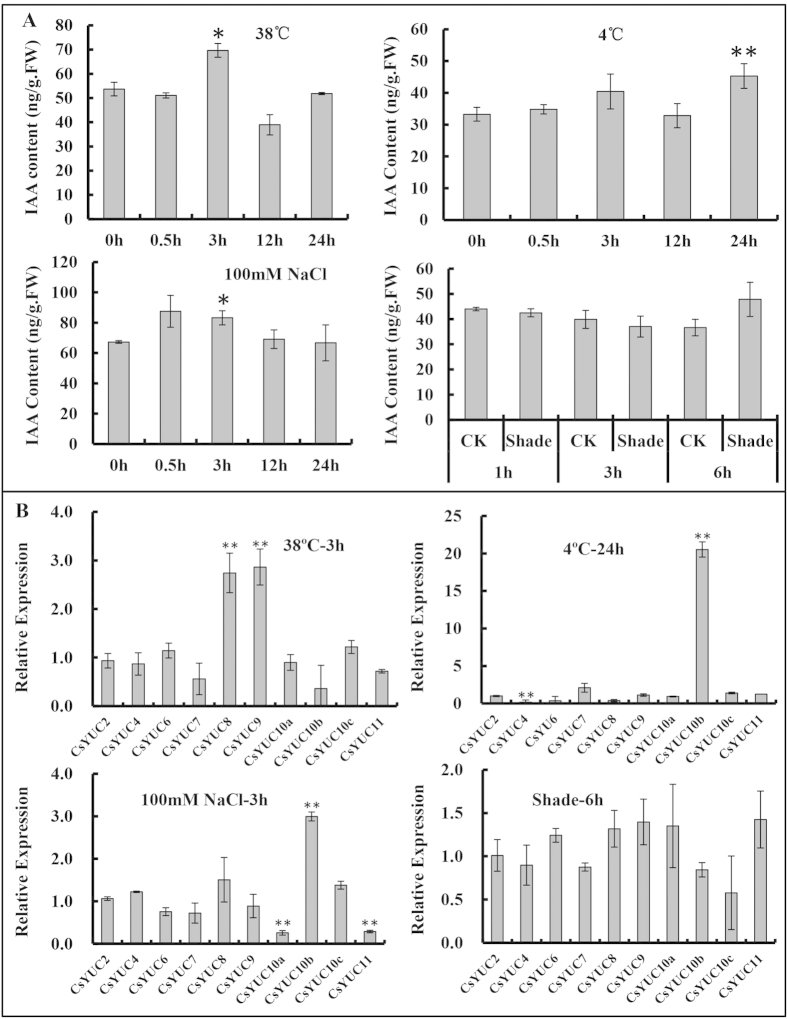
Cucumber is an agricultural crop that is very sensitive to abiotic stress, such as high or low temperature, salinity and shading38,39,40. To investigate whether CsYUCs mediate the stress responses in cucumber, auxin concentration and CsYUCs expression were examined in the cucumber seedlings under high temperature (38 °C), low temperature (4 °C), salinity stress (NaCl 100 mM) and shading treatment. As shown in Fig. 4, IAA abundance accumulated significantly under high temperature for 3 hours, consistent with previous finding that high temperature elevates IAA level in Arabidopsis30 (Fig. 4A). Given that YUC enzymes play essential roles in the auxin biosynthesis11, increased IAA level at high temperature may result from induced CsYUCs expression. Therefore, transcript levels of CsYUCs were examined in cucumber seedlings under high temperature for 3 h. Among all the 10 CsYUCs, CsYUC8 and CsYUC9 were significantly induced by high temperature, indicating that CsYUC8 and CsYUC9 may specifically responsible for the elevated IAA level in response to high temperature in cucumber (Fig. 4A,B), similar to the finding that high temperature induces YUC8 expression and increases endogenous IAA level in Arabidopsis31. Parallel experiments indicated that low temperature and high salinity also elevated free IAA level in cucumber (Fig. 4A), which may mainly due to the dramatically increased CsYUC10b expression (Fig. 4B). Interestingly, the expression of CsYUC10b was induced 20 fold while CsYUC4 was decreased 5.5 fold under low temperature (Fig. 4B). Similarly, transcription of CsYUC10b was increased but CsYUC10a and CsYUC11 were repressed under salinity stress, consistent with the complementary expression patterns as revealed by semiquantitative RT-PCR (Fig. 3), suggesting that different YUC members may antagonize each other to maintain the proper auxin level in cucumber. The relationship between shading and auxin was also explored, but no obvious changes were observed for either auxin content or expression of CsYUC genes in cucumber (Fig. 4A,B).
Figure 4. Endogenous IAA level and expression of CsYUCs in response to different abiotic stresses.
(A) IAA content of 21-days-old seedlings under different abiotic stresses. (B) Expression of CsYUCs in response to abiotic stresses. The bars represent the standard deviation of three biological replicates. Asterisks and double asterisks indicate significant difference between treated and untreated (0 h) control at P < 0.05 and P < 0.01 (unpaired t-test), respectively.
CsYUC11 is specifically enriched in male floral organs
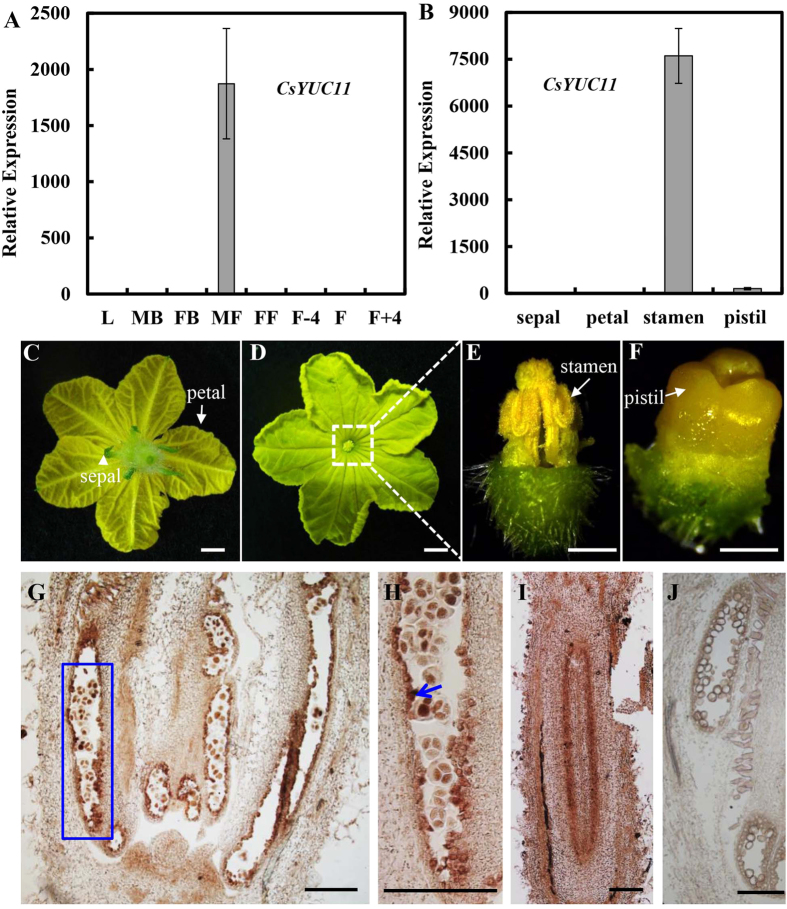
The semiquantitative RT-PCR indicated that CsYUC11 may specifically express in the opened male flowers in cucumber (Fig. 3). To further characterize the expression pattern of CsYUC11, quantitative real time PCR and in situ hybridization analysis were performed in different tissues of cucumber (Fig. 5). qRT-PCR confirmed that CsYUC11 is specifically enriched in the opened male flowers (over 1800 fold higher in male flowers than that in the leaves). To identify CsYUC11 is expressed in which floral organs of male flower, opened male flowers were divided into sepals, petals, stamens and repressed pistils, and qRT-PCR were performed in each type of floral organs (Fig. 5B–F). Our data showed that CsYUC11 is strongly enriched in the stamens, and weakly expressed in the repressed pistils (Fig. 5B). Next, in situ hybridization was used to detect the spatial and temporal distribution of CsYUC11 (Fig. 5G–J). As expected, CsYUC11 signal is mainly detected in the tapetum cell layer and the microspores in anthers (Fig. 5G,H). In the female floral bud, CsYUC11 is weakly expressed in the placenta (Fig. 5I), while no signal was detected upon hybridization with the sense probe of CsYUC11 (Fig. 5J).
Figure 5. Expression analysis of CsYUC11 by qRT-PCR and in situ hybridization in cucumber.
(A–B) Quantitative real time RT-PCR analysis of CsYUC11 in different tissues (A) and different floral organs in opened male flowers (B). L, leaves; MB, male floral buds; FB, female floral buds; MF, opened male flowers; FF, opened female flowers. F − 4, fruits 4 days before anthesis. F, fruits on anthesis. F + 4, fruits 4 days after anthesis. The cucumber UBIQUITIN gene was used as an internal control to normalize the expression. (C–F) Opened male flower showing the four floral organs used for qRT-PCR analysis in (B). (C) Back view of male flower, the arrowhead and arrow indicate sepal and petal, respectively. (D–F) Top view of male flower, the stamen (E) and the repressed pistil (F) are the magnification view of the dotted square in (D). (G–J) in situ hybridization analysis of CsYUC11 in male and female floral buds in cucumber. (G,H) Male floral bud at stage 10 (G,H) is the magnification view of the blue rectangle in (G). Arrow shows the high expression of CsYUC11 in tapetum cell layer. (I) Female floral bud at stage 8. (J) Male floral bud with sense probe. Scale bars represent 1cm in (C,D), 1 mm in (E,F), and 200 μm in (G–J).
Ectopic expression of CsYUC11 in Arabidopsis
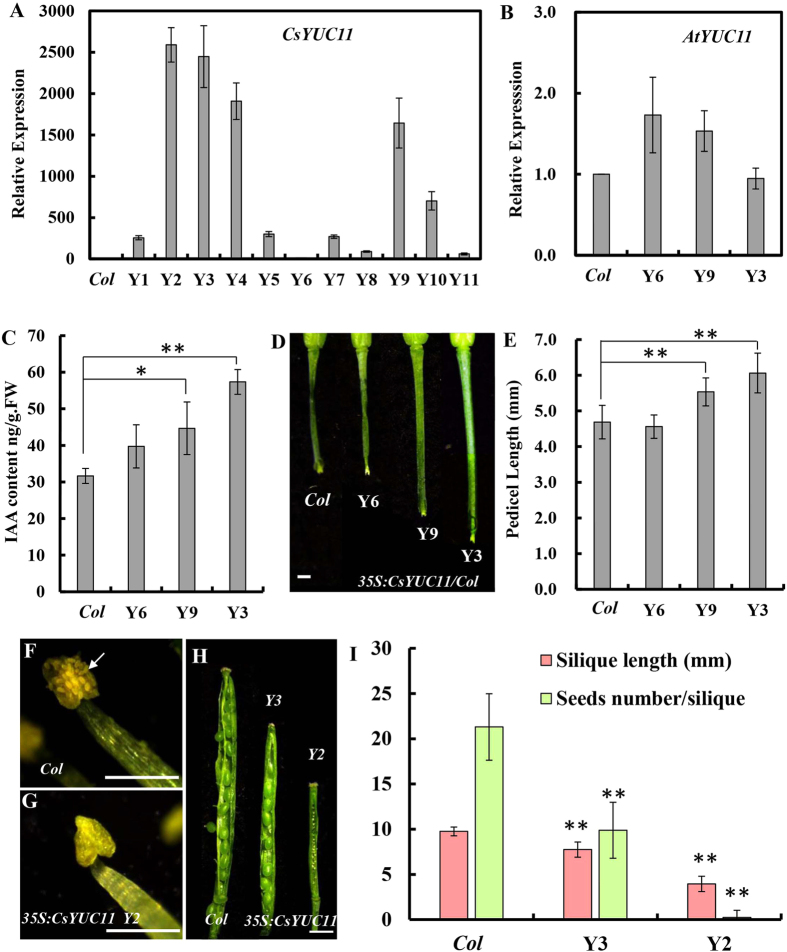
To investigate the biological function of CsYUC11, we ectopically expressed the CsYUC11 coding sequence under the 35S promoter of Cauliflower mosaic virus (CaMV) in Arabidopsis wild-type Columbia (Col), and 11 independent transgenic lines were obtained (Fig. 6A). The phenotypes of the transgenic lines vary from mild to medium to severe, which are in consistent with the respective expression level. Thus we chose three representive lines Y6, Y9 and Y3 for further characterization. To explore whether ectopic expression of CsYUC11 affect the transcription of AtYUC11 in Arabidopsis, we checked the expression of AtYUC11 in the transgenic lines. As shown in Fig. 6B, there is no obvious difference in the expression of AtYUC11, suggesting that the phenotypes of 35S:CsYUC11 lines were due to the function of CsYUC11, not AtYUC11. As the expression level of CsYUC11 increase, the pedicel length as well as the IAA content increase progressively (Fig. 6C,D). The mean pedicel length of Y3 was up to 6.1 ± 0.6 mm while that of Col was only 4.7 ± 0.5 mm (Fig. 6E), implying that CsYUC11 promotes pedicel elongation through auxin biosynthesis in Arabidopsis. Given that CsYUC11 is highly expressed in stamens, we examined the stamen development in the transgenic lines. Despite the number of stamens remain unchanged, the pollens are defective and the anthers fail to open at stage 13 in the severe transgenic lines, a prerequisite for proper pollination and seed set in Arabidopsis (Fig. 6F,G)41. Consequently, the silique length and the seed number per silique were significantly reduced in the transgenic lines (Fig. 6H,I). In the most severe line Y2, only 3 seeds were obtained in the 12 siliques we examined (Fig. 6I, Supplemental Figure S4). Therefore, our data suggested that CsYUC11 regulates silique elongation and seed production through the essential role in stamen development in Arabidopsis.
Figure 6. Ectopic expression of CsYUC11 in Arabidopsis.
(A–B) qRT-PCR analysis of CsYUC11 (A) and AtYUC11 (B) in the inflorescence apex of WT (Col) and 35S:CsYUC11 transgenic lines. (C) Endogenous IAA content of WT (Col) and three transgenic lines. (D) The fruit pedicel of Col and three transgenic lines at stage 17. (E) Quantification of pedicel length in Col and transgenic lines (n = 10). (F–G) Mature stamens in Col (F) and 35S:CsYUC11 transgenic line 2 (G). The arrow indicates the opened pollen sac. (H) The siliques of Col and 35S:CsYUC11 line 3 and 2 at the same developmental stage. (I) Quantification of silique length and seed number per silique in Col and transgenic lines (n = 12). Double asterisks indicate significant difference between Col and transgenic lines at P < 0.01 (unpaired t- test). Scale bars represent 1 mm in D and H, and 250 μm in (F–G).
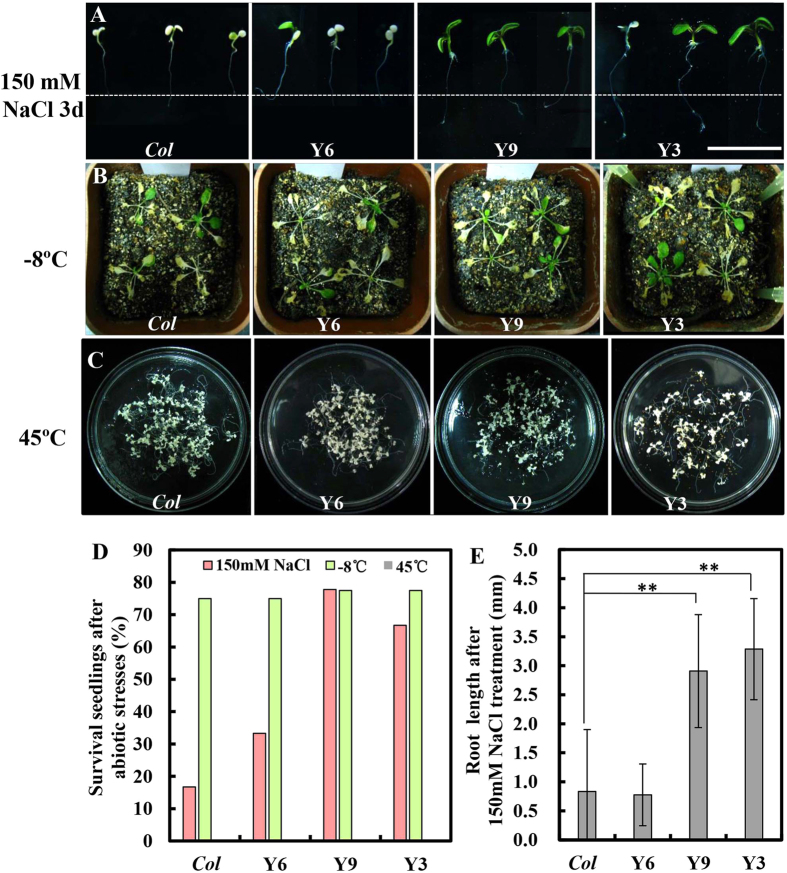
To further characterize the function of CsYUC11 under abiotic stress, 35S:CsYUC11 transgenic Arabidopsis plants were subjected to salinity, freezing and heat stress treatments using the homozygous lines Y6, Y9 and Y3. Compared to the WT, the 35S:CsYUC11 transgenic seedlings displayed reduced suppression of root elongation upon 150 mM NaCl stress for 3 d (Fig. 7A,E), and thus a higher survival rate (Fig. 7D), suggesting that CsYUC11 enhances plant tolerance to salinity stress, which is consistent with the expression data in which salinity stress significantly repressed the transcription of CsYUC11 in cucumber (Fig. 4B). Under freezing (−8 °C) or heat (45 °C) treatments, there is no obvious difference between WT and transgenic plants (Fig. 7B–D), consistent with the fact that expression of CsYUC11 is not respond to freezing or heat stress (Fig. 4B).
Figure 7. CsYUC11 mediates abiotic stress in Arabidopsis.
(A) Root elongation after 150 mM NaCl treatment (E). (B) Tolerance of 3-week-old seedlings at −8 °C for 2 hours. The pictures were taken 7 days after treatments. (C) Tolerance of seedlings after high temperature treatment (45 °C for 2.5 h). (D) Quantification of the survival rate under abiotic stress treatment. Double asterisks indicate significant difference between Col and transgenic lines at P < 0.01 (unpaired t- test). Scale bars represent 1 cm in (A).
Discussion
Structural conservation and divergence of cucumber CsYUCs
The step converting IPA into IAA catalyzed by YUCs is a rate-limiting step for auxin biosynthesis12,13, and thus YUC enzymes were widely distributed and characterized in higher plants including Arabidopsis, rice, tomato, petunia and strawberry21,22,23,24. In this study, we isolated 10 CsYUC family genes in cucumber (Figure S1, Supplemental Table S1). Phylogenetic and gene structure analysis showed that the 10 CsYUCs can be divided into three clades, with most CsYUCs located in the first clade (Fig. 1), similar to those in Arabidopsis, rice and strawberry15,24. CsYUCs were unevenly distributed on five chromosomes (Supplemental Figure S2), and all CsYUCs have similar length of coding sequence (around 1200bp) except for CsYUC7 (Supplemental Table S2). At the protein sequence level, cucumber CsYUCs displayed six conserved motifs (Fig. 2 and Supplemental Table S3). However, CsYUC6 and CsYUC7 lack of the FAD-binding motif and GC motif (Supplemental Figure S3). Given that FAD-binding motif was shown to be essential for YUC monooxygenases activity18,28, CsYUC6 and CsYUC7 may function other than auxin biosynthesis in cucumber. Three cucumber YUC10 genes were identified, and CsYUC10s have several variants in the ATG-containing motif 1 and 2 which are highly conserved in FMO proteins (Fig. 2 and Supplemental Figure S3). In ATG-containing motif 2, CsYUC10b and CsYUC10c share the same divergence at the residue A which substituted by C (Supplemental Figure S3). This substitution has also been identified in the FvYUC10 in strawberry24. In addition, CsYUC10b owns a replacement at the site Y which was the same as strawberry FvYUC10 in the FMO-identifying sequence motif16,24, implying that YUC10 may evolve new function in the horticultural crops including cucumber and strawberry. CsYUC6, −8, −9 and −11 are highly conserved in all the FMO motifs, suggesting that the functions of these genes may be conserved in cucumber.
Cucumber CsYUCs in response to abiotic stress
IAA, as the main endogenous auxin in plants, had two existing forms: free and conjugated42,43. Free IAA can be produced from de novo biosynthesis or released from IAA conjugates11,43. Under high temperature, free IAA level was significantly increased in cucumber seedlings (Fig. 4A). Correspondingly, CsYUC8 and CsYUC9 were selectively upregulated at 38 °C for 3 h (Fig. 4B), suggesting that in response to high temperature, cucumber plants elevate free IAA level to fight against heat injury by activating of CsYUC8 and CsYUC9. In consistent with this finding, previous studies showed that YUC8 and YUC9 expression were upregulated by 29 °C treatment in Arabidopsis seedlings44, and that YUC8 was activated by PIF4 and thus endogenous IAA level was elevated during the high temperature-induced hypocotyl elongation in Arabidopsis31. Therefore, upregulation of YUC8 and YUC9 homologs may be a conserved mechanism to elevate endogenous IAA level in response to high temperature stress.
Despite auxin was shown to play important roles in various aspects of plant development1,5, the specific function of auxin under low temperature and salinity stress remains mysterious. The gravity response was inhibited under cold stress in Arabidopsis inflorescence, in which auxin may be involved45,46. Auxin-related genes including Aux/IAA, GH3, ARF were regulated by salinity stress in sorghum and Arabidopsis47,48. In this study, we found that endogenous IAA level were significantly increased and CsYUC10b transcripts were remarkably upregulated under low temperature and salinity stress (Fig. 4A,B). Further, CsYUC4 was shown to antagonize CsYUC10b under low temperature, while CsYUC10a and CsYUC11 act against CsYUC10b under salinity stress (Fig. 4B), suggesting that different YUC members participate in stress response, and even antagonize each other to create a buffering system for endogenous auxin synthesis in cucumber. Further functional analysis of 35S:CsYUC11 transgenic plants confirmed that CsYUC11 enhanced plant tolerance to salt stress (Fig. 7A,E), but did not mediate response to freezing or heat stress (Fig. 7B–D).
CsYUC11 regulates pedicel elongation and stamen development in Arabidopsis
Several studies have shown the important roles of auxin in stem and pedicel elongation49,50. CORYMBOSA1 (CRM1), a key regulator for pedicel elongation50, belongs to the membrane-associated protein family that functions in polar auxin transport and distribution51. Mutation in CRM1 resulted in shorten pedicel in Arabidopsis50. Similarly, loss-of-function of AUXIN RESISTANT 1 led to reduced auxin signaling and short pedicel in Arabidopsis52. On the other hand, overexpression of wheat TaYUC10 in Arabidopsis produced elongated petiole53. Here we found that ectopic expression of cucumber CsYUC11 displayed elongated pedicel in Arabidopsis (Fig. 6C–E), a typical phenotype for auxin over-accumulation, suggesting that CsYUC11 regulates pedicel elongation through auxin biosynthesis. In addition, we found that CsYUC11 is specifically expressed in the stamen of male flowers, and the pollens are defective and the fertility is reduced in the transgenic lines of 35S:CsYCU11 in Arabidopsis (Fig. 6F–I), supporting the essential function of CsYUC11 in stamen development. However, yuc2 yuc6 double mutants are sterile and YUC6 is specifically expressed in stamens and pollens in Arabidopsis15. Overexpression of wheat TaYUC10 resulted in pollen defects and sterility in Arabidopsis53. Therefore, stamen development is regulated by distinct YUC family genes in different species, and the specific role (positive or negative) of YUCs is species- and member- dependent. Further studies using the overexpression lines and knock-out lines of CsYUCs through genetic transformation in cucumber would shed light on the specific functions of CsYUCs during flower and fruit development.
Materials and Methods
Sequence database, gene structure construction, FMOs-domain identification and phylogenetic analysis
The sequence information of the 11 YUC proteins in Arabidopsis (AtYUCs) were obtained from TAIR (https://www.arabidopsis.org/index.jsp), followed by a BLAST search against the Cucumber Genome Database (http://cucumber.genomics.org.cn/page/cucumber/index.jsp) to obtain the sequence information of the 10 cucumber CsYUCs. The putative protein sequences were aligned using ClustalW and the Neighbor-Joining phylogenetic tree was constructed using the bootstrap analysis with 1000 replications in the MEGA5 software package54. The distribution of the CsYUCs on chromosomes was drawn manually based on the position information from the Cucumber Genome Database (Table S2). The gene structure analysis of CsYUCs was performed using the online tool GSDS (http://gsds.cbi.pku.edu.cn/). The conserved FMOs-domains were identified by the BoxShade web site (http://www.ch.embnet.org/software/BOX_form.html), and the repeats logos were obtained through the online tool (http://weblogo.berkeley.edu/logo.cgi)55.
Plant material and growth conditions
Cucumber inbred line 1461 was used in this study, and grown in the Experimental Field of China Agricultural University at Beijing under standard greenhouse conditions. Water management and pest control were performed according to standard protocols. The Arabidopsis thaliana Columbia (Col) ecotypes and 35S:CsYUC11/Col transgenic plants were grown in soil at 22 °C under 16 h/8 h light/dark condition in a growth chamber.
Abiotic stress treatments and IAA measurement
The 21-day-old cucumber seedlings grown under 16 h/8 h and 25 °C/18 °C day/night condition were used for abiotic stress treatment. Seedlings were incubated at 4 °C or watering with 100 mM NaCl solution. For high temperature treatment, plants were kept in 16 h/8 h and 38 °C/28 °C. For shade treatment, plants were treated with or without simulated shade for 1 h, 3 h and 6 h. Each stress treatment was repeated three times. After stress treatment, cucumber shoot apices were harvested and immediately frozen in liquid nitrogen until further use. About 0.1 ~ 0.2 g samples were used for IAA measurement using enzyme-linked immunosorbent assay as described56. For abiotic treatments in the 35S:CsYUC11 transgenic Arabidopsis, WT and transgenic seeds were growing in Murashige/Skoog (MS) culture medium with 25 mg/L hygromycin for 4d, then the MS plate were replace by new MS plate containing 150 mM NaCl, seedling survival rate and root length were measured after cultured vertically for 3 days. For freezing treatment, the 3-week-old seedlings grown in soil were placed in a temperature chamber at −8 °C for 120 min. Survival rate of WT and transgenic plants were counted after resuming growth at 22 °C under 16 h/8 h light/dark condition for 7 days. For heat treatment, the 10-day-old seedlings grown in Murashige/Skoog (MS) culture medium with 25 mg/L hygromycin were heat treated at 45 °C for 2.5 hours, and survival rate were determined after resuming growth at 22 °C under 16 h/8 h light/dark condition for 5 days.
Semiquantitative RT-PCR and quantitative real-time RT-PCR
Total RNA was extracted using Quick RNA isolation Kit (Waryoung, China). The first strand cDNAs were synthesized using TianScript II RT Kit (Tiangen, China). For semiquantitative RT-PCR, the primers and cycles were empirically optimized. Quantitative real-time RT-PCR was performed as described previously57. The cucumber UBIQUITIN and Arabidopsis ACTIN2 genes were used as an internal control to normalize the expression data. Each qRT-PCR experiment was repeated three times. The primer information is listed in Supplemental Table S4.
In situ hybridization
Cucumber male and female buds were fixed in 3.7% formol-acetic-alcohol (FAA), and in situ hybridization was performed as described58. The primer information is listed in Supplemental Table S4.
Ectopic expression of CsYUC11 in Arabidopsis
The full length coding sequence of CsYUC11 was cloned into pCAMBIA1305.1 vector with 35S promoter through NcoI and SpeI sites. The construct was then introduced into Agrobacterium strain C58 by electroporation and transformed into Arabidopsis wide-type (Col) plants through the floral-dip method59. The transgenic plants were screened on MS medium with 25 mg/L hygromycin. The primer information is listed in Supplemental Table S4.
Additional Information
How to cite this article: Yan, S. et al. Different cucumber CsYUC genes regulate response to abiotic stresses and flower development. Sci. Rep. 6, 20760; doi: 10.1038/srep20760 (2016).
Supplementary Material
Acknowledgments
This study was supported by National Basic Research of China 973 program [2012CB113900], National Natural Science Foundation of China [31171399], Earmarked Fund for Beijing Leaf Vegetables Innovation Team of Modern Agro-industry Technology Research System [BLVT-08], and Chinese Universities Scientific Fund [2012RC012]. The authors are grateful to Dr. Jinsheng Lai for providing qRT-PCR machine, to members of the Zhang lab for technical assistance and stimulating discussions.
Footnotes
Author Contributions S.Y., X.Z. conceived and designed the experiments. S.Y., G.C., L.D., Z.C., X.L., H.W., W.Z., K.N., J.Z. and K.T. performed the experiments. S.Y., G.C., Q.W. and X. Z. analyzed the data. S.Y., Q.W. and X.Z wrote the paper. All authors read and approved the final manuscript.
References
- Vanneste S. & Friml J. Auxin: a trigger for change in plant development. Cell 136, 1005–1016, 10.1016/j.cell.2009.03.001 (2009). [DOI] [PubMed] [Google Scholar]
- Booker J., Chatfield S. & Leyser O. Auxin acts in xylem-associated or medullary cells to mediate apical dominance. The Plant Cell 15, 495–507 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teichmann T. & Muhr M. Shaping plant architecture. Front Plant Sci 6, 233, 10.3389/fpls.2015.00233 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardarelli M. & Cecchetti V. Auxin polar transport in stamen formation and development: how many actors? Front Plant Sci 5, 333, 10.3389/fpls.2014.00333 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandolfini T., Molesini B. & Spena A. Molecular dissection of the role of auxin in fruit initiation. Trends Plant Sci 12, 327–329, 10.1016/j.tplants.2007.06.011 (2007). [DOI] [PubMed] [Google Scholar]
- Sundberg E. & Ostergaard L. Distinct and dynamic auxin activities during reproductive development. Cold Spring Harb Perspect Biol 1, a001628, 10.1101/cshperspect.a001628 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muday G. K. & DeLong A. Polar auxin transport: controlling where and how much. Trends in plant science 6, 535–542 (2001). [DOI] [PubMed] [Google Scholar]
- Ljung K., Bhalerao R. P. & Sandberg G. Sites and homeostatic control of auxin biosynthesis in Arabidopsis during vegetative growth. The Plant Journal 28, 465–474 (2001). [DOI] [PubMed] [Google Scholar]
- Benjamins R. & Scheres B. Auxin: the looping star in plant development. Annu Rev Plant Biol 59, 443–465, 10.1146/annurev.arplant.58.032806.103805 (2008). [DOI] [PubMed] [Google Scholar]
- Zhao Y. Auxin biosynthesis and its role in plant development. Annu Rev Plant Biol 61, 49–64, 10.1146/annurev-arplant-042809-112308 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y. Auxin biosynthesis. Arabidopsis Book 12, e0173, 10.1199/tab.0173 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mano Y. & Nemoto K. The pathway of auxin biosynthesis in plants. J Exp Bot 63, 2853–2872, 10.1093/jxb/ers091 (2012). [DOI] [PubMed] [Google Scholar]
- Zhao Y. Auxin biosynthesis: a simple two-step pathway converts tryptophan to indole-3-acetic acid in plants. Mol Plant 5, 334–338, 10.1093/mp/ssr104 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y. et al. A role for flavin monooxygenase-like enzymes in auxin biosynthesis. Science 291, 306–309, 10.1126/science.291.5502.306 (2001). [DOI] [PubMed] [Google Scholar]
- Cheng Y., Dai X. & Zhao Y. Auxin biosynthesis by the YUCCA flavin monooxygenases controls the formation of floral organs and vascular tissues in Arabidopsis. Genes Dev 20, 1790–1799, 10.1101/gad.1415106 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlaich N. L. Flavin-containing monooxygenases in plants: looking beyond detox. Trends Plant Sci 12, 412–418, 10.1016/j.tplants.2007.08.009 (2007). [DOI] [PubMed] [Google Scholar]
- Krueger S. K. & Williams D. E. Mammalian flavin-containing monooxygenases: structure/function, genetic polymorphisms and role in drug metabolism. Pharmacol Ther 106, 357–387, 10.1016/j.pharmthera.2005.01.001 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou X. et al. Allelic analyses of the Arabidopsis YUC1 locus reveal residues and domains essential for the functions of YUC family of flavin monooxygenases. J Integr Plant Biol 53, 54–62, 10.1111/j.1744-7909.2010.01007.x (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodward C. et al. Interaction of auxin and ERECTA in elaborating Arabidopsis inflorescence architecture revealed by the activation tagging of a new member of the YUCCA family putative flavin monooxygenases. Plant Physiol 139, 192–203, 10.1104/pp.105.063495 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. I. et al. YUCCA6 over-expression demonstrates auxin function in delaying leaf senescence in Arabidopsis thaliana. J Exp Bot 62, 3981–3992, 10.1093/jxb/err094 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobena-Santamaria R. et al. FLOOZY of petunia is a flavin mono-oxygenase-like protein required for the specification of leaf and flower architecture. Genes Dev 16, 753–763, 10.1101/gad.219502 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Exposito-Rodriguez M., Borges A. A., Borges-Perez A. & Perez J. A. Gene structure and spatiotemporal expression profile of tomato genes encoding YUCCA-like flavin monooxygenases: the ToFZY gene family. Plant Physiol Biochem 49, 782–791, 10.1016/j.plaphy.2011.02.022%/ Copyright (c) 2011 Elsevier Masson SAS. All rights reserved. (2011). [DOI] [PubMed] [Google Scholar]
- Yamamoto Y., Kamiya N., Morinaka Y., Matsuoka M. & Sazuka T. Auxin biosynthesis by the YUCCA genes in rice. Plant Physiol 143, 1362–1371, 10.1104/pp.106.091561 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H. et al. Auxin biosynthesis by the YUCCA6 flavin monooxygenase gene in woodland strawberry. J Integr Plant Biol 56, 350–363, 10.1111/jipb.12150%/ (c) 2013 Institute of Botany, Chinese Academy of Sciences. (2014). [DOI] [PubMed] [Google Scholar]
- Mashiguchi K. et al. The main auxin biosynthesis pathway in Arabidopsis. Proc Natl Acad Sci USA 108, 18512–18517, 10.1073/pnas.1108434108 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y., Dai X. & Zhao Y. Auxin synthesized by the YUCCA flavin monooxygenases is essential for embryogenesis and leaf formation in Arabidopsis. Plant Cell 19, 2430–2439, 10.1105/tpc.107.053009 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriechbaumer V., Wang P., Hawes C. & Abell B. M. Alternative splicing of the auxin biosynthesis gene YUCCA4 determines its subcellular compartmentation. Plant J 70, 292–302, 10.1111/j.1365-313X.2011.04866.x %/ (c) 2011 The Authors. The Plant Journal (c) 2011 Blackwell Publishing Ltd. (2012). [DOI] [PubMed] [Google Scholar]
- Gallavotti A. et al. sparse inflorescence1 encodes a monocot-specific YUCCA-like gene required for vegetative and reproductive development in maize. Proc Natl Acad Sci USA 105, 15196–15201, 10.1073/pnas.0805596105 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujino K. et al. NARROW LEAF 7 controls leaf shape mediated by auxin in rice. Mol Genet Genomics 279, 499–507, 10.1007/s00438-008-0328-3 (2008). [DOI] [PubMed] [Google Scholar]
- Gray W. M., Ostin A., Sandberg G., Romano C. P. & Estelle M. High temperature promotes auxin-mediated hypocotyl elongation in Arabidopsis. Proc Natl Acad Sci USA 95, 7197–7202 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J., Qi L., Li Y., Chu J. & Li C. PIF4-mediated activation of YUCCA8 expression integrates temperature into the auxin pathway in regulating arabidopsis hypocotyl growth. PLoS Genet 8, e1002594, 10.1371/journal.pgen.1002594 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakata T. et al. Auxins reverse plant male sterility caused by high temperatures. Proc Natl Acad Sci USA 107, 8569–8574, 10.1073/pnas.1000869107 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M. et al. Activation of a flavin monooxygenase gene YUCCA7 enhances drought resistance in Arabidopsis. Planta 235, 923–938, 10.1007/s00425-011-1552-3 (2012). [DOI] [PubMed] [Google Scholar]
- Kim J. I. et al. Overexpression of Arabidopsis YUCCA6 in potato results in high-auxin developmental phenotypes and enhanced resistance to water deficit. Mol Plant 6, 337–349, 10.1093/mp/sss100 (2013). [DOI] [PubMed] [Google Scholar]
- Woo Y. M. et al. Constitutively wilted 1, a member of the rice YUCCA gene family, is required for maintaining water homeostasis and an appropriate root to shoot ratio. Plant Mol Biol 65, 125–136, 10.1007/s11103-007-9203-6 (2007). [DOI] [PubMed] [Google Scholar]
- Li L. et al. Linking photoreceptor excitation to changes in plant architecture. Genes Dev 26, 785–790, 10.1101/gad.187849.112 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S. et al. The genome of the cucumber, Cucumis sativus L. Nature genetics 41, 1275–1281 (2009). [DOI] [PubMed] [Google Scholar]
- Dong C.-J., Li L., Shang Q.-M., Liu X.-Y. & Zhang Z.-G. Endogenous salicylic acid accumulation is required for chilling tolerance in cucumber (Cucumis sativus L.) seedlings. Planta 240, 687–700 (2014). [DOI] [PubMed] [Google Scholar]
- Zhu Y.-X. et al. Silicon improves salt tolerance by increasing root water uptake in Cucumis sativus L. Plant Cell Reports 34, 1629–46, 10.1007/s00299-015-1814-9 (2015). [DOI] [PubMed] [Google Scholar]
- Naraghi M. & Lotfi M. in IV International Symposium on Cucurbits. ISHS Acta Horticulturae 871, 385–388 (2010). [Google Scholar]
- Alvarez-Buylla E. R. et al. Flower development. Arabidopsis Book 8, e0127, 10.1199/tab.0127 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodward A. W. & Bartel B. Auxin: regulation, action, and interaction. Ann Bot 95, 707–735, 10.1093/aob/mci083 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korasick D. A., Enders T. A. & Strader L. C. Auxin biosynthesis and storage forms. J Exp Bot 64, 2541–2555, 10.1093/jxb/ert080 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavang J. A. et al. Hormonal regulation of temperature-induced growth in Arabidopsis. Plant J 60, 589–601, 10.1111/j.1365-313X.2009.03983.x (2009). [DOI] [PubMed] [Google Scholar]
- Fukaki H., Fujisawa H. & Tasaka M. Gravitropic response of inflorescence stems in Arabidopsis thaliana. Plant Physiol 110, 933–943 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyatt S. E., Rashotte A. M., Shipp M. J., Robertson D. & Muday G. K. Mutations in the gravity persistence signal loci in Arabidopsis disrupt the perception and/or signal transduction of gravitropic stimuli. Plant Physiol 130, 1426–1435, 10.1104/pp.102.010579 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S. et al. Auxin-related gene families in abiotic stress response in Sorghum bicolor. Funct Integr Genomics 10, 533–546, 10.1007/s10142-010-0174-3 (2010). [DOI] [PubMed] [Google Scholar]
- Matsui A. et al. Arabidopsis transcriptome analysis under drought, cold, high-salinity and ABA treatment conditions using a tiling array. Plant Cell Physiol 49, 1135–1149, 10.1093/pcp/pcn101 (2008). [DOI] [PubMed] [Google Scholar]
- Rayle D. L. & Cleland R. E. The Acid Growth Theory of auxin-induced cell elongation is alive and well. Plant Physiol 99, 1271–1274 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi N. & Komeda Y. The role of CORYMBOSA1/BIG and auxin in the growth of Arabidopsis pedicel and internode. Plant Sci 209, 64–74, 10.1016/j.plantsci.2013.04.009%/ Copyright (c) 2013 Elsevier Ireland Ltd. All rights reserved. (2013). [DOI] [PubMed] [Google Scholar]
- Gil P. et al. BIG: a calossin-like protein required for polar auxin transport in Arabidopsis. Genes Dev 15, 1985–1997, 10.1101/gad.905201 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lincoln C., Britton J. H. & Estelle M. Growth and development of the axr1 mutants of Arabidopsis. Plant Cell 2, 1071–1080, 10.1105/tpc.2.11.1071 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N. et al. Isolation and characterization of three TaYUC10genes from wheat. Gene 546, 187–194, 10.1016/j.gene.2014.06.020%/ Copyright (c) 2014 Elsevier B.V. All rights reserved. (2014). [DOI] [PubMed] [Google Scholar]
- Tamura K. et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28, 2731–2739, 10.1093/molbev/msr121 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooks G. E., Hon G., Chandonia J. M. & Brenner S. E. WebLogo: a sequence logo generator. Genome Res 14, 1188–1190, 10.1101/gr.849004%/ Copyright 2004 Cold Spring Harbor Laboratory Press (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maldiney R. et al. A biotin-avidin-based enzyme immunoassay to quantify three phytohormones: auxin, abscisic acid and zeatin-riboside. Journal of Immunological Methods 90, 151–158 (1986). [Google Scholar]
- Jiang L. et al. Transcriptomic analysis reveals the roles of microtubule-related genes and transcription factors in fruit length regulation in cucumber (Cucumis sativus L.). Sci Rep 5, 8031, 10.1038/srep08031 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X. et al. Transcription repressor HANABA TARANU controls flower development by integrating the actions of multiple hormones, floral organ specification genes, and GATA3 family genes in Arabidopsis. Plant Cell 25, 83–101, 10.1105/tpc.112.107854 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S. J. & Bent A. F. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16, 735–743 (1998). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.