Abstract
The RAS association domain family protein 1a (RASSF1A), a tumor suppressor gene at 3p21.3, plays a very important role in various cancers, including the head and neck squamous cell carcinoma (HNSCC). Hypermethylation of CpG islands in the RASSF1A promoter region contribute to epigenetic inactivation. However, the association between RASSF1A promoter methylation and HNSCC remains unclear and controversial. Therefore, a meta-analysis was performed in the study to identify the association. We identified the eligible studies through searching PubMed, EMBASE, Web of Science, and China National Knowledge Infrastructure (CNKI) databases with a systematic searching strategy. The information on characteristics of each study and prevalence of RASSF1A methylation were collected. Pooled odds ratios (ORs) with corresponding confidence intervals (CIs) were calculated. Meta-regression was performed to analyze heterogeneity and funnel plots were applied to evaluate publication bias. A total of 550 HNSCC patients and 404 controls from twelve eligible studies were included in the meta-analysis. Overall, a significant association was observed between RASSF1A methylation status and HNSCC risk under a random-effects model (OR = 2.93, 95% CI: 1.58–5.46). There was no significant publication bias observed. The meta-analysis suggested that there was a significant association between aberrant RASSF1A methylation and HNSCC.
Head and neck cancer is the sixth most common cancer worldwide accounting for approximately 6% of all newly diagnosed malignancies. HNSCC makes up over 90% of head and neck cancer, and commonly arises from the mucosal lining in this region1. Epidemiological data demonstrates that heavy smoking and alcohol consumption contribute to HNSCC tumorigenesis2. Human papillomavirus (HPV) can also implicate the increased incidence of HNSCC in the United States3. Despite the advances in therapy, the overall survival rates of HNSCC have not improved significantly over the past several decades and more than 50% of patients have experienced local relapse and distant metastasis4. Early diagnosis of HNSCC might improve its prognosis, but it is usually not detected in the early stages of HNSCC. Therefore, the efforts to identify novel molecular predictors for HNSCC are instrumental for early diagnosis in the early stage of cancer development.
DNA methylation of cytosine-guanosine dinucleotides (CpG) islands within the promoter region of genes is an alternative mechanism of gene inactivation to gene deletion or mutation. Teschendorff5 observed that invasive cancers displayed increased DNA methylation at the risk CpG sites in contrast to normal tissue, but lower levels in contrast to pre-cancerous lesions. This revealed that aberrant DNA methylation of risk CpG loci was prior to the onset of cancer, indicating that epigenetic diversity in normal cells increased the risk of cancer. Aberrant DNA methylation is frequently considered to be critical in the early stage of cancer development, including HNSCC6. Previous studies had investigated the association between hypermethylation of tumor suppressor genes and HNSCC and evaluated the value of them as potential biomarkers of HNSCC7,8,9,10,11,12. RASSF1A, a kind of tumor suppressor gene, is one of eight isoforms of RASSF1 which is involved in cell cycle control, microtubule stabilization, cellular adhesion and motility as well as apoptosis13. Epigenetic inactivation of RASSF1A by hypermethylation is originally described in lung and breast cancer14. Since then, it has emerged that RASSF1A is one of the most frequently hypermethylated genes so far described and was reported as a prognostic indicator in renal cell carcinoma, non-small cell lung cancer, neuroblastoma, endometrial cancer and breast cancer15,16,17,18,19,20,21. Furthermore, hypermethylation of RASSF1A within promoter CpG islands is frequently observed in the HNSCC cell lines22. All of these findings indicate that RASSF1A might play an important role in the development of HNSCC.
To date, a number of studies have investigated the association between aberrant methylation of RASSF1A and HNSCC through a comparison of the methylation prevalence of RASSF1A between cancerous tissues and controls. However, the obtained results of these studies are inconclusive and inconsistent23,24. Therefore, we conducted a meta-analysis of 12 published studies to conclude the association.
Results
Study characteristics
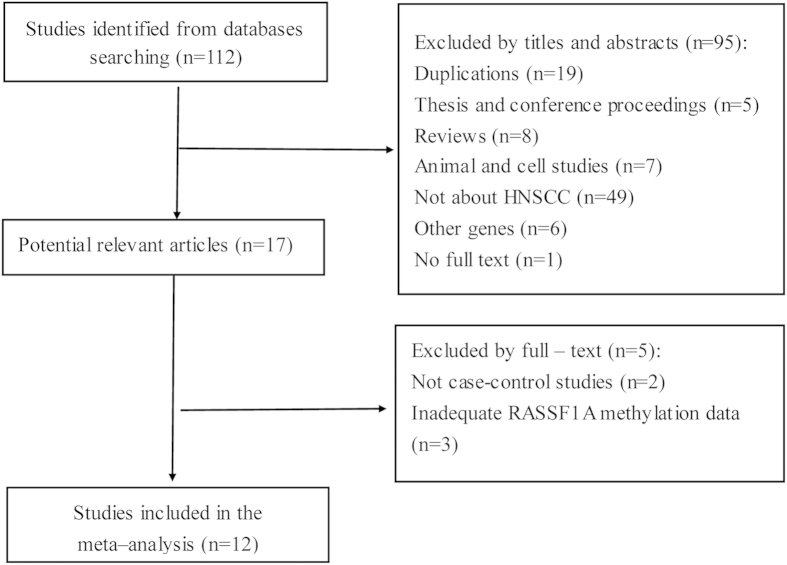
In total, the electronic search strategy initially identified 112 potentially relevant studies. Firstly, these potentially relevant studies were screened for inclusion based on their titles and abstracts. As a result, 19 duplications and 76 studies (four thesis, one conference proceeding, eight reviews, two animal studies, five cell lines, 49 not about HNSCC, six without RASSF1A and one without full text) were excluded. The remaining 17 citations were retrieved for full-text assessment. Upon the assessment, two articles which were not case-control studies and three articles with inadequate RASSF1A methylation data were excluded. Figure 1 showed the whole process of study selection and exclusion, with specification of reasons. Lastly, 12 studies, published between 2002 and 2012 with 18 to 111 cases, met the inclusion criteria and were included in our meta-analysis. The individual characteristics of the 12 included studies are summarized in Table 1.
Figure 1. Selection of studies in the meta-analysis.
Table 1. General Characteristics of the Included Studies.
| First author | Year | Location | Race | Mean/median age (range) (y) | Gender (M/F) | Case (n) |
Control (n) |
Method | Control style | Control source | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| M+ | U | M+ | U | |||||||||
| Hogg34 | 2002 | UK | Caucasians | NA | NA | 4 | 24 | 0 | 24 | BSP | A | NMT |
| Dong26 | 2003 | USA | Caucasians | NA | NA | 7 | 46 | 0 | 46 | MSP | A | ANT |
| Maruya22 | 2004 | USA | Caucasians | 58 (31–81) | 26/6 | 0 | 32 | 1 | 32 | MSP | A | NMT |
| Xu23 | 2006 | China | Asians | 60 (41–76) | 35/13 | 34 | 48 | 11 | 48 | MSP | A | ANT |
| Righini27 | 2007 | French | Caucasians | 57 (33–74) | NA | 14 | 90 | 0 | 30 | MSP | A | NMT |
| Wan31 | 2007 | China | Asians | NA | 17/15 | 13 | 32 | 4 | 28 | MSP | H | OCT |
| 0 | 10 | MSP | H | OCT | ||||||||
| Ghosh33 | 2008 | India | Asians | NA | NA | 23 | 111 | 9 | 52 | MSRA | H | DLT |
| Steinmann28 | 2009 | Germany | Caucasians | 57 (41–77) | NA | 10 | 54 | 0 | 23 | MSP | A | CMT |
| Su24 | 2010 | Taiwan | Asians | 55 (37–82) | 47/5 | 9 | 31 | 12 | 31 | Q-MSP | A | ANT |
| 0 | 12 | Q-MSP | H | NMT | ||||||||
| Laytragoon-Lewin29 | 2010 | Sweden | Caucasians | 62 (42–101) | 30/11 | 8 | 18 | 4 | 18 | MSP | A | NMT |
| Paluszczak30 | 2011 | Poland | Caucasians | 58 (41–75) | 35/6 | 13 | 41 | 9 | 41 | MSP | A | NMT |
| Koutsimpelas31 | 2012 | Germany | Caucasians | 62 (45–83) | 19/4 | 3 | 23 | 0 | 3 | MSP | H | GT |
Abbreviation: NA, not available; M, male; F, female; M+, methylated; U, unmethylated; A, Autologous (the control from the HNSCC patients themselves); H, Heterogeneous (the control from other individuals); NMT, normal mucosa tissue; ANT, adjacent non-tumor tissue; OCT, oral cavity tissue; DLT, dysplastic lesions tissue; CMT, cheek mucosa tissue; GT, gingiva tissue.
The meta-analysis consisted of 550 cases of HNSCC tissues and 404 controls, with a total sample size of 954. Among the 12 included studies, the study populations were Caucasians in eight articles22,25,26,27,28,29,30,31 and Asians in four articles23,24,32,33. A total of nine studies conducted methylation-specific polymerase chain reaction (MSP) to assess the gene methylation status. Three articles used quantitative methylation-specific polymerase chain reaction (Q-MSP), bisulfite sequencing PCR (BSP) and methylation sensitive restriction analysis (MSRA) respectively to evaluate the RASSF1A methylation in cases and controls. The genomic location of the analyzed regions of eight studies included was the promoter. The genomic location of the analyzed regions of the remaining four articles was the CpG islands of the promoter. Hogg34 analyzed the methylation status of CpG islands in the promoter region of RASSF1A from LCTSGR1 at 3p21.3 in the HNSCC patients. 11 of the articles were published in English, and 1 was published in Chinese. The specimens were cancerous tissues of HNSCC cases and non-cancerous tissues of controls. The control group was comprised of HNSCC patients, benign disease patients and healthy volunteers.
Meta-analysis results
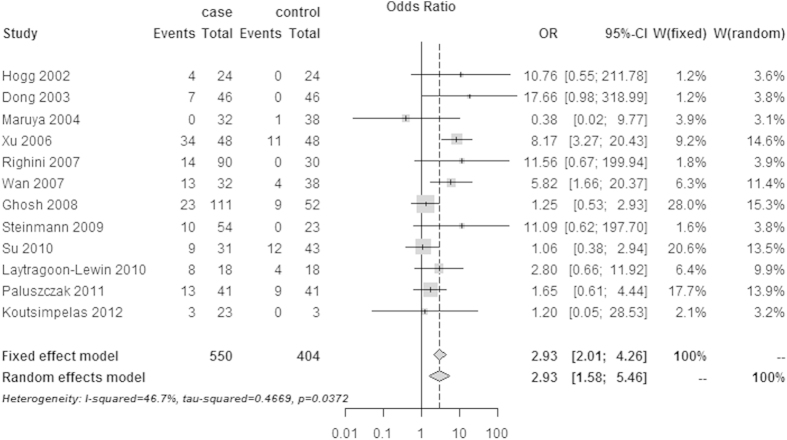
The pooled ORs and corresponding ORs 95% CIs for the association between RASSF1A promoter methylation and HNSCC were shown in Fig. 2. A random-effects model was employed because a significant heterogeneity was observed among 12 included studies by the χ2-based Cochran Q statistic test and I2 statistics (I2 = 46.7%, Q = 20.65, P = 0.0372). In the overall meta-analysis, the RASSF1A promoter methylation was significantly associated with HNSCC, with a combined OR of 2.93 (95% CI: 1.58–5.46) under the random-effects model.
Figure 2. The estimates for RASSF1A methylation frequency associated with HNSCC in the meta-analysis.
Meta-regression analysis and subgroup analysis
A significant heterogeneity was found among the studies. Therefore we conducted a meta-regression to explore the source of heterogeneity with restricted maximum likelihood method (REML method35). Based on previous studies, we assumed that the heterogeneity might arise from the ethnicity, control types, age of patients, RASSF1A methylation detection methods, case sample size, HPV infection status, gender proportion smoking status and histology types. However, only the data about ethnicity, control types, methods of RASSF1A methylation detection and case sample size were collected completely. Then, we conducted a multiple regression model with following four variables: races (Asians and Caucasians), control types (autologous control heterogeneous control), methods used to RASSF1A methylation detection (MSP, Q-MSP and BSP) and case sample size (≥40 and <40). According to the result of the meta-regression analysis result, all 95% confidence intervals included 0 for the coefficients which indicated that none of the variables can explain the heterogeneity between-studies in Table 2. Furthermore, we performed a subgroup analysis of those variables in Table 3. The ORs were 10.76 (95% CI: 0.55–211.78) in the BSP group, 1.06 (95% CI: 0.38–2.94) in the Q-MSP group, and 3.30 (95%CI: 2.17–5.01) in the MSP group (MSRA was classified as MSP group) under the fixed-effects model, respectively. The heterogeneity did not change significantly in the subgroup analysis of detection methods. Similar results on the change of heterogeneity were found in other subgroup analysis.
Table 2. Meta-regression analysis.
| Heterogeneity sources | Coefficient | 95%CI | P | |
|---|---|---|---|---|
| Lower | Upper | |||
| Races | −1.22 | −2.58 | 0.14 | 0.08 |
| Control types | −0.19 | −1.46 | 1.07 | 0.76 |
| Methods | ||||
| MSP | −1.78 | −5.29 | 1.73 | 0.32 |
| Q-MSP | −3.42 | −7.19 | 0.35 | 0.08 |
| Case sample size | 0.38 | −1.02 | 1.78 | 0.59 |
Races: Asians and Caucasians; Control types: autologous control and heterogeneous control; Methods: MSP (methylation-specific polymerase chain reaction), Q-MSP (quantitative methylation-specific polymerase chain reaction) and BSP (bisulfite sequencing polymerase chain reaction); Case sample size: <40 and ≥40.
Table 3. Subgroup analysis of the association between RASSF1A and HNSCC.
| Group | Case | Control | M-H pooled OR* | D+L pooled OR† | Heterogeneity | ||||
|---|---|---|---|---|---|---|---|---|---|
| M+ | U | M+ | U | OR (95% CI) | OR (95% CI) | I2(%) | P | τ2 | |
| Total | 138 | 550 | 50 | 404 | 2.93 (2.01–4.26) | 2.93 (1.58–5.46) | 46.7 | 0.0372 | 0.4669 |
| Races | |||||||||
| Asians | 79 | 222 | 36 | 181 | 2.63 (1.64–4.22) | 2.75 (0.96–7.89) | 77.6 | 0.0039 | 0.8887 |
| Caucasians | 59 | 328 | 14 | 223 | 3.45 (1.85–6.44) | 2.83 (1.35–5.93) | 6.9 | 0.0869 | 0.3772 |
| Control types | |||||||||
| Autologous | 99 | 384 | 37 | 293 | 3.13 (2.00–4.90) | 3.19 (1.30–7.83) | 59.6 | 0.0112 | 0.9318 |
| Heterogeneous | 95 | 309 | 13 | 111 | 2.52 (1.35–4.73) | 2.69 (1.06–6.87) | 29.7 | 0.2236 | 0.3298 |
| Methods | |||||||||
| BSP | 4 | 24 | 0 | 24 | 10.76 (0.55–211.78) | 10.76 (0.55–211.78) | — | — | — |
| Q—MSP | 9 | 31 | 12 | 43 | 1.06 (0.38–2.94) | 1.06 (0.38–2.94)) | — | — | — |
| MSP | 125 | 495 | 28 | 337 | 3.30 (2.17–5.01) | 3.27 (1.67–6.40) | 44.7 | 0.0615 | 0.442 |
| Case sample size | |||||||||
| <40 | 37 | 160 | 21 | 164 | 2.30 (1.25–4.21) | 2.28 (1.00–5.20) | 26.1 | 0.2389 | 0.2678 |
| ≥40 | 101 | 390 | 29 | 240 | 3.36 (2.07–5.43) | 3.87 (1.46–10.25) | 62.7 | 0.0199 | 0.7714 |
Abbreviation: RASSF1A, RAS association domain family protein 1a; HNSCC, head and neck squamous cell carcinoma; BSP, bisulfite sequencing polymerase chain reaction; Q-MSP, quantitative methylation-specific polymerase chain reaction; MSP, methylation-specific polymerase chain reaction; M+, methylated; U, unmethylated.
*The fixed-effects model.
†The random-effects model.
The numbers with bold font were the results under the model applied to calculate the pooled ORs. When I2 > 50% and P < 0.1 for the Q statistic, the pooled ORs was calculated using a random-effects model. Otherwise, a fixed-effects model was applied.
Sensitivity Analysis
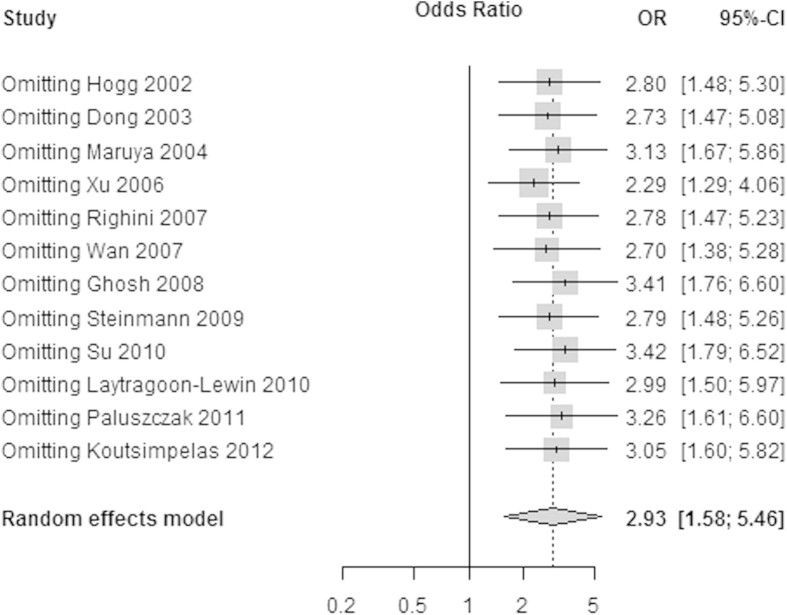
Sensitivity analysis was performed by omitting a single study under the random-effects model. The results of sensitivity analysis showed that the pooled ORs ranged from 2.29 (95% CI: 1.29–4.06) to 3.42 (95% CI: 1.79–6.52). This demonstrated that none of the studies dramatically influenced the pooled ORs in Fig. 3. The REML method was used to estimate the variance between studies.
Figure 3. The sensitivity analysis by omitting a single study under the random-effects method.
Publication Bias
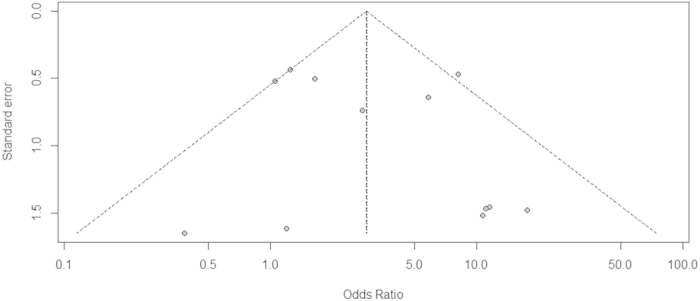
We performed a Begg’s funnel plot36 and Peter test37 to assess the publication bias of the included studies. The shape of the Begg’s funnel plot showed no obvious asymmetry and absence of symmetry indicated publication bias (Fig. 4). No publication bias was detected by Peter test (P = 0.73) and Begg’s rank correlation test36 (P = 0.87), respectively. Furthermore, the fail-safe number38 was applied to evaluate the publication bias. If this number was relatively large to the number of observed studies, we could feel fairly confident in the summary conclusions. The fail-safe number (Z = 21.60, Nfs0.05 = 161.47, Nfs0.01 = 73.94) indicated that the pooled ORs were stable in our meta-analysis.
Figure 4. The Begg’s funnel plot for assessment of publication bias in the meta-analysis (each study is represented by a point).
Discussion
Previous studies have demonstrated that epigenetic alteration is an important event in the carcinogenic progression. Particularly, increased methylation in the promoter region of tumor suppressor gene can account for a progressive reduction of its expression, silencing and selective proliferative advantage in certain cells, which plays a vital role in the development of human cancer39. The aberrant methylation has been observed in the promoter region of RASSF1A in various cancers, including HNSCC6.
Our meta-analysis included 550 HNSCC tissues and 404 controls from 12 published studies. Overall, the pooled OR of RASSF1A methylation in cancer tissues and controls under the random-effects model was 2.93 (95% CI: 1.58–5.46), which suggested a significant association of the methylation of RASSF1A promoter with HNSCC. The overall heterogeneity between included studies was interpreted by the χ2-based Cochran Q statistic test and I2 statistics and meta-regression was used to explore the sources of the heterogeneity. When I2 > 50% and P < 0.1 for the Q statistic, the between-study heterogeneity was considered significant and the pooled ORs was calculated using a random-effects model (the DerSimonian-Laird estimate)40. Otherwise, a fixed-effects model (the Mantel-Haenszel test) was applied40. In the subgroups of races, the pooled ORs were 3.45 (95% CI: 1.85–6.44) in Caucasians subgroup under the fixed-effects model and 2.75 (95% CI: 0.96–7.89) in Asians subgroup under the random-effects model, respectively. This indicated that hypermethylation of RASSF1A had a stronger association with increased risk of HNSCC in Caucasians. Similarly, the methylation rates of the MGMT gene and GSTP1 gene in non-small cell lung cancer were also significantly higher in Caucasians than in Asians41, and this divergence might be due in large part to a combination of differences in allele frequencies and complex epistasis or interactions between the gene and environment42. The summary OR was 3.19 (95% CI: 1.30–7.83) in the autologous control subgroup under the random-effects model, and was 2.52 (95% CI: 1.35–4.73) in the heterogeneous control subgroup under the fixed-effects model. Interestingly, this was consistent with a previous study of non-small cell lung cancer (NSCLC)43, which indicated an increased likelihood of RASSF1A methylation in heterogeneous controls compared to autologous controls. The reason for this might be because the benign lesions had a higher probability of RASSF1A methylation as an early stage of carcinoma. In the method subgroup, the ORs were 10.76 (95% CI: 0.56–211.78) in the BSP group, 1.06 (95% CI: 0.38–2.94) in the Q-MSP group, and 3.30 (95%CI: 2.17–5.01) in the MSP group under the fixed-effects model. The differences of ORs in these subgroups were potentially caused by the different sensitivities and specificities of the method used to the detection of gene methylation. Q-MSP is a sensitive quantitative assay with normalization of the amplifiable DNA content of samples. The cut-off point of Q-MSP was derived from the best distinguish point. However, the cut-off point of MSP is defined by visual detection of the presence or absence of PCR product compared to the intensity of controls44. Therefore, MSP (a nonquantitative and nonfluorometric method) would be hard to detect low levels of promoter methylation, while Q-MSP can detect up to 1/1000 methylated alleles45 and this would have an impact on the results. BSP, a method of genomic sequencing, can provide a more direct and quantitative analysis for most CPG sites within a defined region than MSP and Q-MSP46. The 95% confidence intervals of ORs of BSP method subgroup and Q-MSP method subgroup included 1. This was potentially attributed to the relatively small sample size (less than 60) in these subgroups. The pooled OR also differed according to different sample size. In the <40 cases subgroup, the OR was 2.30 (95% CI: 1.25–4.21) under the fixed-effects model. In the ≥40 cases subgroup, the OR 3.87 (95% CI: 1.46–10.25) under the random-effects model. However, there was no significant difference between different sample size subgroups.
Some potential limitations of the study should be taken into consideration when interpreting the results of meta-analysis. Firstly, due to the 12 included studies were retrospective, there might be a potential unidentified confounding bias, information bias and selection bias. Secondly, although we explored and evaluated the source of heterogeneity in four variables, we could not explore heterogeneity from other aspects because of the insufficient demographic and clinical data. Thirdly, previous studies demonstrated that time of sampling47 and fixation techniques48 potentially influenced methylation status in paraffin-embedded tumors. The 12 included studies varied in time of sampling and fixation techniques and these could result in heterogeneity. Additionally, since the number of included studies and samples were relatively small, further investigations with a large number of samples were required.
Conclusions
Our meta-analysis identified an association between aberrant rmethylation of RASSF1A promoter with HNSCC, which indicated that hypermethylation of RASSF1A promoter might be a potential biomarker in the process of HNSCC. Prospective studies with larger sample size are needed to confirm these results in the future.
Methods
The meta-analysis was performed according to the latest meta-analysis guidelines (PRISMA).
Studies identification
Studies were identified via an electronic search of a range of computerized databases, including PubMed, Embase, Web of Science and CNKI using the following key words: ‘squamous cell carcinoma or cancer’, ‘oropharyngeal or oropharynx or head and neck or tonsil’, ‘RAS association domain family protein 1A’, ‘RASSF1A’, ‘methylation’ and ‘hypermethylation’. Articles were searched in the databases form Jan 1, 2000 to May 8, 2015 without language limitation. Two independent reviewers screened the titles and abstracts identified by the electronic search to identify relevant studies. The inclusion criteria of the meta-analysis were as follows: (1) case-control study design; (2) presentation of data necessary for calculating odds ratios (ORs); (3) studies primarily evaluating the incidence of RASSF1A methylation in HNSCCs and corresponding autologous/heterogeneous control, including non-tumor tissue, plasma and sputum of HNSCC patients. The excluded studies were as follows: duplication, review, animal study, experimental study and adequate specific data.
Data extraction
Data retrieved from the eligible studies including first author’s name, year of publication, published journal and country, patient ethnicity, population size, methods used to determine methylation status, histology, control type, and methylation status of RASSF1A promoter in extracted cancer tissues and controls. Data extraction was conducted by two reviewers independently using a standard data extraction form. If there were disagreement between them, a third reviewer was used to reach a consensus.
Statistical analysis
All statistical analyses were conducted by using the Meta package (version 2.2-1) in R (version 3.0.2; http://www.r-project.org/). The pooled odds ratios (ORs) of different studies and corresponding 95% confidence intervals (CIs) were calculated to evaluate the strength of the association between RASSF1A methylation and HNSCC risk. In order to assess the percentage of variability across studies attributable to heterogeneity beyond by sampling error, the χ2-based Cochran Q statistic test and I2 statistics were employed. When I2 > 50% and P < 0.1 for the Q statistic, the between-study heterogeneity was considered significant and the pooled ORs was calculated using a random-effects model (the DerSimonian-Laird estimate)40. Otherwise, a fixed-effects model (the Mantel-Haenszel test) was applied40. If the heterogeneity was significant, to explore and assess the source of heterogeneity, a meta-regression (restricted maximum-likelihood estimator method, REML35) was initially performed and a subgroup analysis was followed according to the results of meta-regression. Sensitivity analysis was employed to assess the effects of single study on pooled ORs after omitting one study. Publication bias was assessed by a funnel plot for Egger’s test. When the individual studies had cells with zero counts, the default was to add 0.5 to all zero counts in the Meta package. Statistical significance was defined as a two-tailed P value of 0.05 in our study.
Additional Information
How to cite this article: Meng, R.-W. et al. Aberrant Methylation of RASSF1A Closely Associated with HNSCC, a Meta-Analysis. Sci. Rep. 6, 20756; doi: 10.1038/srep20756 (2016).
Acknowledgments
This work is supported by grants from The National Natural Science Foundation of China (81373092).
Footnotes
Author Contributions All authors contributed significantly to this work. M.X.L. designed the research study; R.W.M. and D.D.D. performed the research study and extracted the data; R.W.M., Y.C.L. and X.C. analyzed the data; R.W.M., H.S. and X.N. wrote and revised the manuscript, and Y.X.H. and C.L. prepared the Figures 1,2,3–4 and supplemental Tables 1–3. All authors reviewed the manuscript. In addition, all authors approved the final draft.
References
- Alibek K., Kakpenova A. & Baiken Y. Role of infectious agents in the carcinogenesis of brain and head and neck cancers. Infect Agent Cancer 8, 7, 10.1186/1750-9378-8-7 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata M., Takayama K., Choi B. C. & Pak A. W. A nested case-control study on alcohol drinking, tobacco smoking, and cancer. Cancer Detect Prev 20, 557–565 (1996). [PubMed] [Google Scholar]
- Chaturvedi A. K. et al. Human papillomavirus and rising oropharyngeal cancer incidence in the United States. J Clin Oncol 29, 4294–4301, 10.1200/jco.2011.36.4596 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forastiere A., Koch W., Trotti A. & Sidransky D. Head and neck cancer. N Engl J Med 345, 1890–1900, 10.1056/NEJMra001375 (2001). [DOI] [PubMed] [Google Scholar]
- Teschendorff A. E. et al. The dynamics of DNA methylation covariation patterns in carcinogenesis. PLoS Comput Biol 10, e1003709, 10.1371/journal.pcbi.1003709 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laytragoon-Lewin N., Rutqvist L. E. & Lewin F. DNA methylation in tumour and normal mucosal tissue of head and neck squamous cell carcinoma (HNSCC) patients: new diagnostic approaches and treatment. Med Oncol 30, 654, 10.1007/s12032-013-0654-0 (2013). [DOI] [PubMed] [Google Scholar]
- Smiraglia D. J. et al. Differential targets of CpG island hypermethylation in primary and metastatic head and neck squamous cell carcinoma (HNSCC). J Med Genet 40, 25–33 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiung D. T. et al. Global DNA methylation level in whole blood as a biomarker in head and neck squamous cell carcinoma. Cancer Epidemiol Biomarkers Prev 16, 108–114, 10.1158/1055-9965.epi-06-0636 (2007). [DOI] [PubMed] [Google Scholar]
- De Schutter H., Geeraerts H., Verbeken E. & Nuyts S. Promoter methylation of TIMP3 and CDH1 predicts better outcome in head and neck squamous cell carcinoma treated by radiotherapy only. Oncol Rep 21, 507–513 (2009). [PubMed] [Google Scholar]
- Carvalho A. L. et al. Detection of promoter hypermethylation in salivary rinses as a biomarker for head and neck squamous cell carcinoma surveillance. Clin Cancer Res 17, 4782–4789, 10.1158/1078-0432.ccr-11-0324 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss D., Stockmann C., Schrodter K. & Rudack C. Protein expression and promoter methylation of the candidate biomarker TCF21 in head and neck squamous cell carcinoma. Cell Oncol (Dordr) 36, 213–224, 10.1007/s13402-013-0129-5 (2013). [DOI] [PubMed] [Google Scholar]
- Ovchinnikov D. A. et al. DNA Methylation at the Novel CpG Sites in the Promoter of MED15/PCQAP Gene as a Biomarker for Head and Neck Cancers. Biomark Insights 9, 53–60, 10.4137/bmi.s16199 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donninger H., Vos M. D. & Clark G. J. The RASSF1A tumor suppressor. J Cell Sci 120, 3163–3172, 10.1242/jcs.010389 (2007). [DOI] [PubMed] [Google Scholar]
- Dammann R. et al. Epigenetic inactivation of a RAS association domain family protein from the lung tumour suppressor locus 3p21.3. Nat Genet 25, 315–319, 10.1038/77083 (2000). [DOI] [PubMed] [Google Scholar]
- Jo H. et al. Association of promoter hypermethylation of the RASSF1A gene with prognostic parameters in endometrial cancer. Oncol Res 16, 205–209 (2006). [DOI] [PubMed] [Google Scholar]
- Kioulafa M., Kaklamanis L., Mavroudis D., Georgoulias V. & Lianidou E. S. Prognostic significance of RASSF1A promoter methylation in operable breast cancer. Clin Biochem 42, 970–975, 10.1016/j.clinbiochem.2009.04.003 (2009). [DOI] [PubMed] [Google Scholar]
- Misawa A. et al. RASSF1A hypermethylation in pretreatment serum DNA of neuroblastoma patients: a prognostic marker. Br J Cancer 100, 399–404, 10.1038/sj.bjc.6604887 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai Y. et al. Methylation level of the RASSF1A promoter is an independent prognostic factor for clear-cell renal cell carcinoma. Ann Oncol 21, 1612–1617, 10.1093/annonc/mdp577 (2010). [DOI] [PubMed] [Google Scholar]
- Martins A. T. et al. High RASSF1A promoter methylation levels are predictive of poor prognosis in fine-needle aspirate washings of breast cancer lesions. Breast Cancer Res Treat 129, 1–9, 10.1007/s10549-010-1160-0 (2011). [DOI] [PubMed] [Google Scholar]
- Wang J., Wang B., Chen X. & Bi J. The prognostic value of RASSF1A promoter hypermethylation in non-small cell lung carcinoma: a systematic review and meta-analysis. Carcinogenesis 32, 411–416, 10.1093/carcin/bgq266 (2011). [DOI] [PubMed] [Google Scholar]
- Cho Y. H. et al. Prognostic significance of gene-specific promoter hypermethylation in breast cancer patients. Breast Cancer Res Treat 131, 197–205, 10.1007/s10549-011-1712-y (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruya S. I. et al. Differential methylation status of tumor-associated genes in head and neck squamous carcinoma: Incidence and potential implications. Clinical Cancer Research 10, 3825–3830, 10.1158/1078-0432.ccr-03-0370 (2004). [DOI] [PubMed] [Google Scholar]
- Xu C. B. & Ten B., LI, C.Q. & Jin, C. S. Promoter methylation of RASSF1A gene in laryngeal squamous cell carcinoma and protein expression. Journal of Jilin University (Medicine Edition) 32, 326–329 (2006). [Google Scholar]
- Su P. F. et al. p16(INK4A) promoter hypermethylation is associated with invasiveness and prognosis of oral squamous cell carcinoma in an age-dependent manner. Oral Oncol 46, 734–739, 10.1016/j.oraloncology.2010.07.002 (2010). [DOI] [PubMed] [Google Scholar]
- Hogg R. P. et al. Frequent 3p allele loss and epigenetic inactivation of the RASSF1A tumour suppressor gene from region 3p21.3 in head and neck squamous cell carcinoma. European Journal of Cancer 38, 1585–1592, http://dx.doi.org/ 10.1016/S0959-8049%2801%2900422-1 (2002). [DOI] [PubMed] [Google Scholar]
- Dong S. M. et al. Epigenetic inactivation of RASSF1A in head and neck cancer. Clin Cancer Res 9, 3635–3640 (2003). [PubMed] [Google Scholar]
- Righini C. A. et al. Tumor-specific methylation in saliva: a promising biomarker for early detection of head and neck cancer recurrence. Clin Cancer Res 13, 1179–1185, 10.1158/1078-0432.ccr-06-2027 (2007). [DOI] [PubMed] [Google Scholar]
- Steinmann K., Sandner A., Schagdarsurengin U. & Dammann R. H. Frequent promoter hypermethylation of tumor-related genes in head and neck squamous cell carcinoma. Oncol Rep 22, 1519–1526 (2009). [DOI] [PubMed] [Google Scholar]
- Laytragoon-Lewin N. et al. DNA Content and Methylation of p16, DAPK and RASSF1A Gene in Tumour and Distant, Normal Mucosal Tissue of Head and Neck Squamous Cell Carcinoma Patients. Anticancer Res 30, 4643–4648 (2010). [PubMed] [Google Scholar]
- Paluszczak J., Misiak P., Wierzbicka M., Wozniak A. & Baer-Dubowska W. Frequent hypermethylation of DAPK, RARbeta, MGMT, RASSF1A and FHIT in laryngeal squamous cell carcinomas and adjacent normal mucosa. Oral Oncol 47, 104–107, 10.1016/j.oraloncology.2010.11.006 (2011). [DOI] [PubMed] [Google Scholar]
- Koutsimpelas D. et al. Promoter methylation of MGMT, MLH1 and RASSF1A tumor suppressor genes in head and neck squamous cell carcinoma: pharmacological genome demethylation reduces proliferation of head and neck squamous carcinoma cells. Oncol Rep 27, 1135–1141, 10.3892/or.2012.1624 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Y., Gao W. X., Chen Y. X., Cui Y. B. & Liu Y. M. Methylation and expression of RASSF1A in oralpre malignant lesions and squamous cel l carcinomas. J Modern Stomatol 21, 30–33 (2007). [Google Scholar]
- Ghosh S. et al. Alterations of 3p21.31 tumor suppressor genes in head and neck squamous cell carcinoma: Correlation with progression and prognosis. International Journal of Cancer 123, 2594–2604, 10.1002/ijc.23834 (2008). [DOI] [PubMed] [Google Scholar]
- Hogg R. P. et al. Frequent 3p allele loss and epigenetic inactivation of the RASSF1A tumour suppressor gene from region 3p21.3 in head and neck squamous cell carcinoma. Eur J Cancer 38, 1585–1592 (2002). [DOI] [PubMed] [Google Scholar]
- Veroniki A. A. et al. Methods to estimate the between-study variance and its uncertainty in meta-analysis. Res Synth Methods, 10.1002/jrsm.1164 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begg C. B. & Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics 50, 1088–1101, http://dx.doi.org/ 10.2307/2533446 (1994). [DOI] [PubMed] [Google Scholar]
- Peters J. L., Sutton A. J., Jones D. R., Abrams K. R. & Rushton L. Comparison of two methods to detect publication bias in meta-analysis. JAMA 295, 676–680, 10.1001/jama.295.6.676 (2006). [DOI] [PubMed] [Google Scholar]
- Rosenthal R. The file drawer problem and tolerance for null results. Psychological Bulletin May 86, 638–641 (1979). [Google Scholar]
- Arantes L. M., de Carvalho A. C., Melendez M. E., Carvalho A. L. & Goloni-Bertollo E. M. Methylation as a biomarker for head and neck cancer. Oral Oncol 50, 587–592, 10.1016/j.oraloncology.2014.02.015 (2014). [DOI] [PubMed] [Google Scholar]
- Stuck A. E., Rubenstein L. Z. & Wieland D. Bias in meta-analysis detected by a simple, graphical test. Asymmetry detected in funnel plot was probably due to true heterogeneity. BMJ 316, 469; author reply 470–461 (1998). [PMC free article] [PubMed] [Google Scholar]
- Toyooka S. et al. Smoke exposure, histologic type and geography-related differences in the methylation profiles of non-small cell lung cancer. Int J Cancer 103, 153–160, 10.1002/ijc.10787 (2003). [DOI] [PubMed] [Google Scholar]
- Fraser H. B., Lam L. L., Neumann S. M. & Kobor M. S. Population-specificity of human DNA methylation. Genome Biol 13, R8, 10.1186/gb-2012-13-2-r8 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W. J. et al. Associations between RASSF1A promoter methylation and NSCLC: a meta-analysis of published data. Asian Pac J Cancer Prev 14, 3719–3724 (2013). [DOI] [PubMed] [Google Scholar]
- Hsu C. Y. et al. Prognosis of glioblastoma with faint MGMT methylation-specific PCR product. J Neurooncol 122, 179–188, 10.1007/s11060-014-1701-1 (2015). [DOI] [PubMed] [Google Scholar]
- Yates D. R. et al. Methylational urinalysis: a prospective study of bladder cancer patients and age stratified benign controls. Oncogene 25, 1984–1988, 10.1038/sj.onc.1209209 (2006). [DOI] [PubMed] [Google Scholar]
- Herman J. G., Graff J. R., Myohanen S., Nelkin B. D. & Baylin S. B. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci U S A 93, 9821–9826 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ushijima T., Nakajima T. & Maekita T. DNA methylation as a marker for the past and future. J Gastroenterol 41, 401–407, 10.1007/s00535-006-1846-6 (2006). [DOI] [PubMed] [Google Scholar]
- Hamilton M. G. et al. Determination of the methylation status of MGMT in different regions within glioblastoma multiforme. J Neurooncol 102, 255–260, 10.1007/s11060-010-0307-5 (2011). [DOI] [PubMed] [Google Scholar]