Abstract
Differentiation of inflammatory macrophages from monocytes is characterized by an orderly integration of epigenetic and transcriptional regulatory mechanisms guided by lineage-determining transcription factors such as PU.1. Further activation of macrophages leads to a stimulus- or microenvironment-specific signal integration with subsequent transcriptional control established by the action of tissue- or signal-associated transcription factors. Here, we assess four histone modifications during human macrophage activation and integrate this information with the gene expression data from 28 different macrophage activation conditions in combination with GM-CSF. Bioinformatically, for inflammatory macrophages we define a unique network of transcriptional and epigenetic regulators (TRs), which was characterized by accessible promoters independent of the activation signal. In contrast to the general accessibility of promoters of TRs, mRNA expression of central TRs belonging to the TR network displayed stimulus-specific expression patterns, indicating a second level of transcriptional regulation beyond epigenetic chromatin changes. In contrast, stringent integration of epigenetic and transcriptional regulation was observed in networks of TRs established from somatic tissues and tissue macrophages. In these networks, clusters of TRs with permissive histone marks were associated with high gene expression whereas clusters with repressive chromatin marks were associated with absent gene expression. Collectively, these results support that macrophage activation during inflammation in contrast to lineage determination is mainly regulated transcriptionally by a pre-defined TR network.
Keywords: epigenetics, inflammatory macrophages, activation, transcription, promoter, enhancer, super enhancer
Introduction
Macrophages are cells of the mononuclear phagocyte system1,2 that respond to environmental signals, are specialized in phagocytosis, and play an important role in tissue homeostasis3,4,5,6,7,8,9. Indeed, macrophages fulfill highly specialized functions depending on their tissue localization10 and their defense mechanisms are context-dependent and established upon signal integration from their microenvironment. Such a functionally diverse reactivity requires significant changes in gene expression achieved mainly by transcriptional regulation in a gene-specific and transcriptional module-specific fashion11,12. To date, neither the module structure nor the master transcriptional and epigenetic regulators (in short 'TRs') of all the potential modules are completely understood in macrophage activation. Transcriptional programing requires the coordinated action of three classes of TRs; first, constitutively expressed transcription factors (TFs), like NF-κB or IRFs, that initiate the transcriptional response thereby inducing numerous secondary TRs that regulate a second wave of gene expression resulting in the expression of a third group of TRs which act to maintain the response. While it is the second class of TRs that mainly induces gene module- and functional specificity of the response, the third class is necessary for the maintenance and prolonged specificity of the response as recently demonstrated8. These TR classes act in concert with pioneer or lineage-determining TFs defining cell type-specific functionality13,14. A similar three step model of hierarchical organization of TR networks was introduced by Ostuni and Natoli in which reversible stimulus-specific programs are shaped by effector TFs and this process was mirrored by extensive modifications of the epigenome15,16.
The pioneering or lineage-determining factor PU.1 is a major regulator of the epigenetic landscape of macrophages17,18,19 binding to the vast majority of accessible sites20. Furthermore, PU.1 is instrumental in recruiting co-factors and epigenetic modifiers like C/EBPβ, RUNX1 and IRF8 to open the chromatin structures under steady-state condition, while chromatin remodeling and transcription factor binding following macrophage activation is independent of PU.118,21. Indeed, regulation of cell-type-specific gene expression has been shown to closely correlate with dynamic changes in histone modifications (HMs)22. For example, trimethylation of H3K27 (H3K27me3) at promoter sites was associated with gene silencing, whereas trimethylation of H3K4 (H3K4me3) correlates with open chromatin states23 and in combination with acetylation of H3K27 (H3K27Ac) marks accessible promoters with ongoing transcription24. Monomethylation of H3K4 (H3K4me1) defines enhancers in the genome25 and concomitant H3K27 acetylation discriminates strong from weak enhancers, while H3K4me1 combined with H3K27 trimethylation defines 'poised' enhancers26.
Assessment of histone modifications combined with transcriptome profiling has been used to map the transcriptional and epigenetic regulation of murine tissue macrophages under homeostatic conditions27,28. Signals from the microenvironment drive tissue-specific gene expression programs, which are controlled by tissue-specific TFs such as Gata6 or Spic and distinct enhancer landscapes.
So far, in humans, histone modifications as a hallmark of epigenetic regulation and their impact on transcription have been mainly studied in monocytes and during monocyte-to-macrophage differentiation, but not macrophage activation24,29,30. While H3K4me1 sites were differentially regulated during differentiation29, H3K27 acetylation at enhancer sites was by far the most dynamic mark and changes in acetylation were mirrored by changes in gene expression during differentiation24. Similar to previous findings in the murine system17,18 PU.1 in combination with C/EBPβ was found to mark enhancer sites in human macrophages but not monocytes29, suggesting conservation of central epigenetic regulatory mechanisms of macrophage differentiation. While in murine macrophages, cell activation has been linked to further transcriptional and epigenetic regulation15,25, similar regulatory mechanisms in human macrophages have not been formally investigated.
We have previously provided a global description of human macrophage activation8, which supported a multi-dimensional model31, suggesting that inflammatory macrophages react with the induction of specific combinations of TFs in response to distinct input signals from their environment. Indeed, every input signal was characterized by a very specific expression pattern of TFs strongly suggesting this to be an important hallmark of macrophage plasticity. Whether both transcriptional and epigenetic mechanisms are required for the regulation of those TFs that drive the observed functional plasticity is still unknown.
Here, we profile the gene expression and chromatin landscape of four populations of human inflammatory macrophages activated with distinct stimuli to determine the contribution of TRs and histone modifications to macrophage activation. We identify common and stimulus-specific regulatory elements including accessible and poised promoters and enhancers. Here, we define a TR network of human inflammation-associated macrophage activation, which is characterized by globally permissive histone modifications in comparison with TRs derived from networks generated for human somatic tissues and murine homeostatic tissue macrophages. Collectively, our results indicate that in contrast to differentiated and functionally restricted somatic cells, inflammatory macrophages possess constitutively accessible loci for nearly all central TRs. We conclude that accessibility of chromatin loci for central TRs is characteristic for activated macrophages and suggest combinatorial transcription factor-mediated regulation of cell activation with newly identified master regulators as a prerequisite for macrophage plasticity.
Results
The common transcriptional program of inflammation-associated macrophages is characterized by permissive histone marks
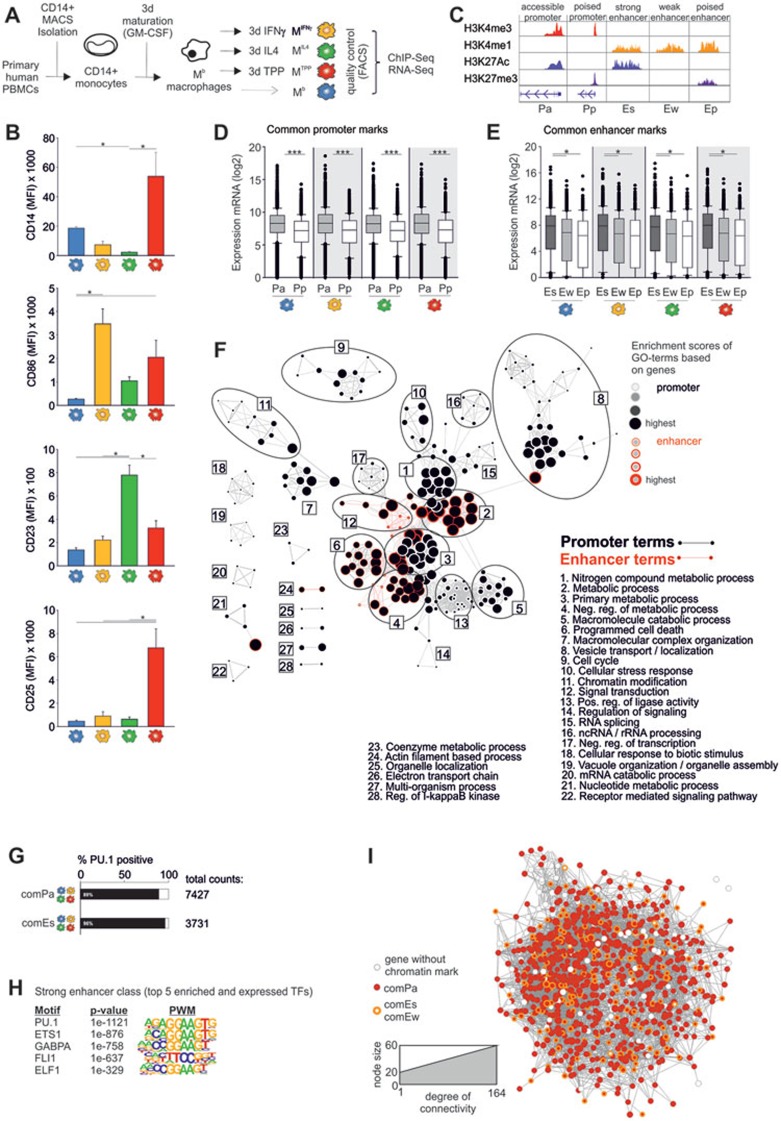
To determine how prolonged exposure to typical macrophage stimuli would alter transcription through long-term changes in histone modifications in human macrophages, we performed transcriptome analysis (RNA-seq, microarray) in combination with chromatin immunoprecipitation (ChIP-seq) for four histone modifications H3K4me3, H3K27Ac, H3K4me1 and H3K27me3. As we wanted to mimic an inflammatory milieu often characterized by elevated systemic levels of GM-CSF, we used GM-CSF as primary differentiation stimulus in our in vitro model32. Therefore, we purified monocytes from peripheral blood, differentiated them with GM-CSF into baseline macrophages (Mb) and further activated these Mb with either IFNγ (MIFNγ, acute inflammation-associated model), IL4 (MIL4, alternative activation model) or a combination of TNF, prostaglandin E2 (PGE2) and Pam3Cys reflecting macrophage activation under chronic inflammatory conditions (MTPP, chronic inflammation-associated model) for 72 h (Figure 1A). This time-point was chosen as we wanted to compare the long-term changes introduced by a specific stimulus on permissive and repressive histone modifications reflective of macrophage activation with our previous transcriptome analysis8. Differentiation and activation of macrophages was validated by analysis of activation state-related surface markers: CD14 for Mb, CD86 for MIFNγ, CD23 for MIL4 and CD25 for MTPP (Figure 1B). Samples were assessed for histone modifications including H3K4me1 (enhancers), H3K4me3 (promoters), H3K27ac (active chromatin states), and H3K27me3 (poised enhancers and promoters) by ChIP-seq defining five distinct chromatin states33,34,35,36 (Figure 1C). First, promoters were identified by their proximity (± 2.5 kb) to transcriptional start sites. Second, active promoters (Pa) were defined by the co-occurrence of the two histone marks H3K4me3 and H3K27Ac, whereas poised promoters (Pp) were identified by simultaneous marks for H3K4me3 and H3K27me3. Enhancers were identified at distal (>2.5 kb up- and downstream) regions from transcriptional start sites (TSS) and categorized into strong enhancers (Es) marked by H3K27Ac and H3K4me1 signals, weak enhancers (Ew) possessing marks for H3K4me1 but lacking H3K27Ac and finally poised enhancer (Ep) sites, which include marks for both H3K4me1 and H3K27me3.
Figure 1.
Defining the epigenetic core signature of human inflammatory macrophages. (A) Schema describing the process of in vitro generation of unstimulated (Mb) or activated macrophages in presence of IL4 (MIL4), IFNγ (MIFNγ) or a combination of TNF, PGE2 and Pam3Cys (MTPP). (B) Macrophage activation was validated by cell surface marker expression (CD14, CD86, CD23, CD25) by flow cytometry and plotted as mean fluorescence intensity (MFI); Statistics for CD14, CD86 and CD25 were performed with Kruskal-Wallis one-way ANOVA on ranks with pairwise multiple comparison according to Dunn's Method, for CD23 P-values were calculated by one-way ANOVA with pairwise multiple comparison according to Bonferroni *P < 0.05; n = 6. (C) Categorization of permissive and repressed histone marks (HM) by the combination of four histone modifications H3K4me3, H3K4me1, H3K27Ac and H3K27me3. (D, E) Visualization (Box-and-Whisker plots) of RNA-seq expression values for genes with HMs common to all four conditions separated into (D) accessible (n = 7 120) and poised promoters (n = 1 204), respectively, (E) strong (n = 2 312), weak (n = 2 055) or poised (n = 376) enhancers. Statistics calculated by (D) Mann-Whitney rank sum test or (E) Kruskal-Wallis one-way ANOVA on ranks with pairwise multiple comparison according to Dunn's Method, ***P < 0.001, *P < 0.05. (F) Visualization of gene ontology enrichment analysis (GOEA) of the common macrophage gene loci with active promoters (black) and strong enhancers (red) using BiNGO and the EnrichmentMap plugin. (G) Determination of PU.1 binding (black bars) by ChIP-seq in common accessible promoters (comPa) and strong enhancers (comEs). (H) Motif enrichment analysis for transcription factor (TF) binding sites in genes with strong enhancer marks shared by all four macrophage subtypes. Displayed are the positional weight matrices (PWM) and the binomial P-values for enrichment of the top five similarly expressed TFs. (I) Visualization of promoter and enhancer information for the 10% most connected genes of the human macrophage activation network8. RNA-seq and ChIP-seq data were derived from three independent experiments.
We focused first on promoters and enhancers that were similarly accessible in all four activation conditions. A total of 7 427 common loci with histone marks matching the criteria of Pa (corresponding to 7 280 genes) were observed while 1 247 loci (corresponding to 1 239 genes) matched the HM characteristics for Pp (Supplementary information, Figure S1A, Table S1A and S1B). The high number of accessible and relatively low number of poised promoters indicated that macrophages share a common and epigenetically active cellular program. Next, we assessed enhancer regions and could demonstrate that the largest number of common enhancers were strong enhancers (3 731 Es), followed by weak (3 110 Ew) and poised enhancers (476 Ep) (Supplementary information, Figure S1B and Table S1C-S1E). These common accessible enhancers and promoters define the 'common histone signature' of human macrophages. The larger number of common accessible promoters in comparison to common strong enhancers suggests that transcriptional regulation of these common gene loci are dependent on transcriptional activators binding to the promoter regions and that enhancers are mainly used to support the transcriptional activity of a gene locus marked by an active promoter, an observation that also has been made in the murine myeloid cell compartment very recently27.
To determine whether the different chromatin states are reflected by differential gene expression we plotted gene expression in relation to chromatin states for each macrophage subtype individually (Figure 1D and 1E). In all four activation states the average gene expression was significantly higher in genes with accessible versus poised promoters (Figure 1D). Similarly, genes with strong enhancers showed in average higher expression than genes with weak or poised enhancers (Figure 1E). These observations support that the chromatin state of promoters and enhancers is indicative for gene expression according to the established understanding of H3K4me3, H3K4me1 and H3K27Ac as permissive histone marks37. Analysis of the combination of enhancers and promoters further supported this observation, as genes marked by an active promoter and a strong enhancer show higher expression than genes with a weak, poised or no enhancer as exemplified for Mb (Supplementary information, Figure S1C).
To decipher cellular functions of those genes marked by permissive histone modifications in activated macrophages, we separately performed gene ontology enrichment analysis (GOEA) for genes with common Pa and Es. Significantly enriched GO-terms from both analysis were visualized in a combined network. Genes marked by Pa were enriched in GO-terms that are shown as nodes, while enrichment of permissive enhancers corresponds to the border of the nodes (Figure 1F). Single GO-terms were finally grouped according to their relationship of biological functions. This analysis resulted in a network consisting of 28 sub-clusters. Genes marked by common Pa- and/or Es mainly participate in metabolic processes, maintenance, or 'housekeeping' functions. Furthermore, this analysis revealed that common enhancers are mainly associated with gene functions related to metabolic processes.
As PU.1 has been described as a pioneering lineage-factor for macrophages, we performed ChIP-seq analysis to determine whether PU.1 is involved in these common macrophage functions. Gene loci belonging to the 'common histone signature' of human macrophages showed a striking enrichment for PU.1 binding of 89% for common active promoter sites and 96% for common strong enhancer sites (Figure 1G). Furthermore, PU.1 was the top expressed TF predicted to bind at common strong enhancers (Figure 1H) followed by other ETS family members with very similar binding motifs like ETS1 and GABPA. At common active promoters the PU.1 motif was again highly enriched by bioinformatic prediction (P-value 1E−832), but TFs of the ETS family, among them ELK4 and FLI1 displayed even higher enrichment P-values (Supplementary information, Figure S1D).
If gene loci commonly expressed during human macrophage activation are best characterized by accessible promoters, then we hypothesized that the previously defined major hub genes of the multidimensional model of human macrophage activation8, which participate in metabolic processes and housekeeping functions should also be characterized by permissive histone modifications at their promoters (Figure 1I and Supplementary information, Figure S1E). Indeed, among the 869 most highly connected genes 94% were marked by a Pa but only 27% by an Es or Ew, further supporting the hypothesis that permissive histone modifications at the promoter were highly important for the common activation-associated class of genes (Figure 1I). Together, these data support that the common human macrophage program, reflecting largely core cellular functions, is defined by active promoters which can be associated with common enhancers and binding of the lineage TF PU.1 to allow for transcriptional regulation of these genes by additional TRs.
Inflammatory macrophage activation results in distinct patterns of histone modifications
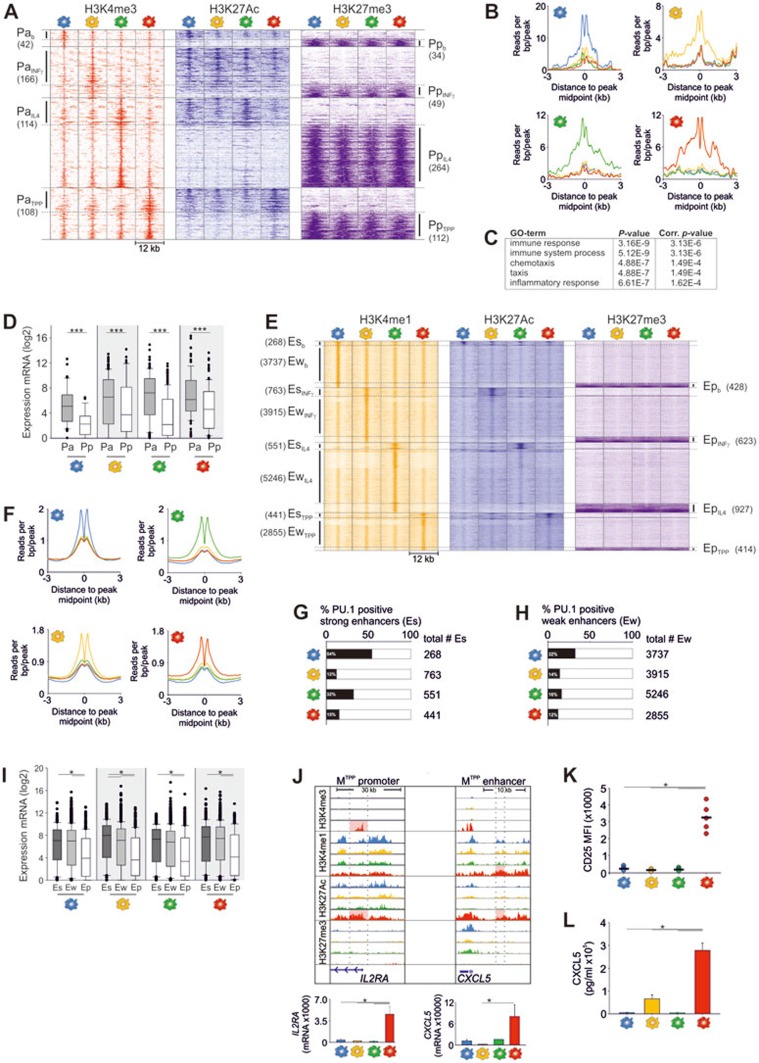
Exogenous signals can regulate gene expression by modifying histone marks at promoter sites23 but also enhancer regions as shown for the usage of latent enhancers to fortify stimulus-specific gene expression in murine macrophages20. Along these lines, we determined the magnitude of activation signal-specific changes of the epigenetic landscape in human inflammatory macrophages. Identification of activation-specific promoters and enhancers followed two criteria: (1) candidate positions showed two times higher tag counts for H3K4me3 or H3K4me1 than the input control at this position and (2) tag signals for H3K4me3 or H3K4me1 were increased in the respective activation state in comparison to the other macrophage activation states (FC ≥ 2). Following these criteria, we identified a relatively small number of signal-specific Pa ranging from only 42 in Mb to 166 in MIFNγ (Supplementary information, Figure S2A and Table S2A-S2C). The number of signal-specific poised promoters was similarly low with a different distribution showing the highest number (n = 264) in MIL4 (Supplementary information, Figure S2A and Table S2E-S2H). Visualization of the activation-specific Pa positions over a range of 6 kb surrounding the peak midpoints (Figure 2A) and histograms of normalized reads per bp/peak (Figure 2B), verified elevated H3K4me3 peak signals at the respective positions in comparison with other activation states.
Figure 2.
Macrophage activation-specific promoter and enhancer landscapes. (A) Pile-up heatmaps and histograms of genomic positions with accessible (Pa) and poised (Pp) promoter marks specific for each macrophage activation condition. Heatmaps and histograms of read densities for H3K4me3 marked genomic positions were centered to H3K4me3 peak midpoints and signals were determined with 500 bp windows 6 kb up- and downstream of the peak midpoints. Numbers of activation specific Pa and Pp displayed in brackets. (B) Histograms of read densities for H3K4me3 signals at activation specific accessible promoter regions ± 3 kb from peak midpoint. (C) Top five gene ontology terms of a gene ontology enrichment analysis (GOEA) for genes with an accessible promoter specifically found in MTPP. (D) Box-and-Whisker plots showing expression (RNA-seq data) of genes with condition-specific accessible (Pa) or poised promoters (Pp) (Mann-Whitney rank sum test, ***P < 0.001). (E) Pile-up heat maps and histograms of genomic positions with strong, weak and poised enhancer marks specific for each macrophage condition (same visualization values as in A). (F) Histograms of read densities for H3K4me1 signals at conditions specific strong and weak enhancer regions ± 3 kb from peak midpoint. (G, H) Percentages (black bars) of (G) strong and (H) weak enhancer positions with PU.1 binding determined by ChIP-seq. (I) Box-and-Whisker plots for expression values of macrophage condition-specific genes with strong, weak or poised enhancer marks (Kruskal-Wallis one-way ANOVA on ranks with pairwise multiple comparison according to Dunn's Method, *P < 0.05). (J) Visualization of HM peaks at two representative gene loci for MTPP-specific genes with an accessible promoter (IL2RA, left panel) and strong enhancer (CXCL5, right panel) including corresponding RNA-seq expression data (One Way Repeated Measures ANOVA on ranks (IL2RA) or Kruskal-Wallis one-way ANOVA (CXCL5) with pairwise multiple comparison according to Tukey, *P < 0.05). (K) Scatter plot of the mean fluorescence intensities (MFI) of CD25 expression in inflammatory macrophages evaluated by flow cytometry. Group means are indicated as horizontal bars (one-way ANOVA with pairwise multiple comparison according to Bonferroni *P < 0.05; n = 5). (L) Quantification of secreted CXCL5 into supernatants by inflammatory macrophages (one-way ANOVA with pairwise multiple comparison according to Bonferroni *P < 0.05; n = 7; SEM). Mb (blue), MIFNγ (yellow), MIL4 (green) and MTPP (red); RNA-seq and ChIP-seq were derived from three independent experiments.
We further investigated if the signal-specific promoters would also show high PU.1 binding. Approximately half of the signal-specific accessible promoters contained the PU.1 motif as determined by ChIP-seq, except for Mb-specific Pa which displayed up to 71% PU.1 positive sites (Supplementary information, Figure S2B), suggesting that PU.1 binding is not a prerequisite for the activity of the stimulus-specific promoters. Genes with signal-specific Pa were mainly associated with immune functions as exemplified for MTPP (Figure 2C). Similar to common promoters, the average expression levels of genes associated with signal-specific Pa were significantly higher than the expression of genes with signal-specific Pp (Figure 2D). Specificity of histone modifications at Pa sites was further illustrated by exploring the expression levels of e.g., MTPP-associated genes in other macrophage subtypes (Supplementary information, Figure S2C). This was even true for genes associated with Pp in MTPP (Supplementary information, Figure S2D). Similar results were obtained when analyzing genes associated with signal-specific accessible or poised promoters in Mb, MIl4 and MIFNγ (data not shown).
Recently, the concept of latent enhancers was introduced for murine macrophages suggesting that exogenous signals can shape the epigenetic landscape15. We therefore defined activation-specific enhancer sites (Figure 2E, Supplementary information, Figure S2E and Table S2I-S2T). Overall, we identified more activation-specific strong (up to 763 Es in MIFNγ), weak (from 2 855 Ew in MTPP to 5 246 Ew in MIL4) and poised enhancers (highest occurrence in MIL4 with 927 Ep sites) compared with promoters. Genes marked by strong and weak enhancers participated in effector functions, as GO-terms like 'signaling', 'regulation of leukocyte activation' and 'regulation of immune system process' were enriched (Supplementary information, Table S2U). As already observed for H3K4me3 at activation-specific promoters, read densities for activation-specific H3K4me1 marks were elevated in comparison with H3K4me1 signals of other macrophage activation states at the same position (Figure 2F). In conclusion, the relatively high number of activation-specific enhancers strongly supported the hypothesis that epigenetic modifications at enhancers shape the activation-associated transcriptome to induce stimulus-specific functions, while epigenetic modifications at promoters are required to shape the common macrophage transcriptome allowing for the exertion of homeostatic functions.
Next, we assessed whether stimulus-specific enhancers would also require PU.1 binding for their activity. While we observed high PU.1 binding at common enhancers sites (Figure 1), the percentage of all enhancers containing PU.1 binding sites by ChIP-seq was lower (from 54% to 59%, Supplementary information, Figure S2F). Analysis of activation-specific enhancers for PU.1 binding showed highest percentages of PU.1 bound to strong and weak enhancers in Mb (Es: 54%, Ew: 32%) while signal-specific enhancers in all other activation states showed a reduction in PU.1 positive sites (from 12% to 32%, Figure 2G and 2H). In line with the finding by Mancino et al.21, this observation led us to the assumption that PU.1 binding is not required for stimulus-specific enhancers and that additional regulators account for transcriptional regulation of these gene loci. Indeed, using TF-binding prediction we found a significant enrichment of binding motifs for PU.1 in Mb (enrichment p-value 1E-116), while TFs like IRF1 in MIFNγ, STAT6 in MIL4 and FOSL2 in MTPP were predicted to bind to activation-specific accessible enhancers (Supplementary information, Table S2V-S2Y). Exemplarily, we visualized the top five enriched positional weight matrices including their enrichment P-values for expressed TFs, which are predicted to bind at MTPP-specific enhancer regions (Supplementary information, Figure S2G).
To investigate the impact of enhancer state on transcriptional activity of genes, we linked stimulus-specific enhancer sites to gene expression. The highest average expression levels were observed for genes marked by strong enhancers followed by weak and poised enhancers for all macrophage activation states (Figure 2I). Next, we visualized the histone marks for the two prototypic gene loci (IL2RA and CXCL5) which are differentially expressed at transcriptional as well as protein level in MTPP supporting that expression can be dependent on a combination of specific accessible promoters and enhancers (Figure 2J, 2K and 2L). Overall, our findings clearly extend earlier observations in murine macrophages15 and human monocytes24 demonstrating that the accessibility of promoters and enhancers in human macrophages is shaped in an input-signal specific manner.
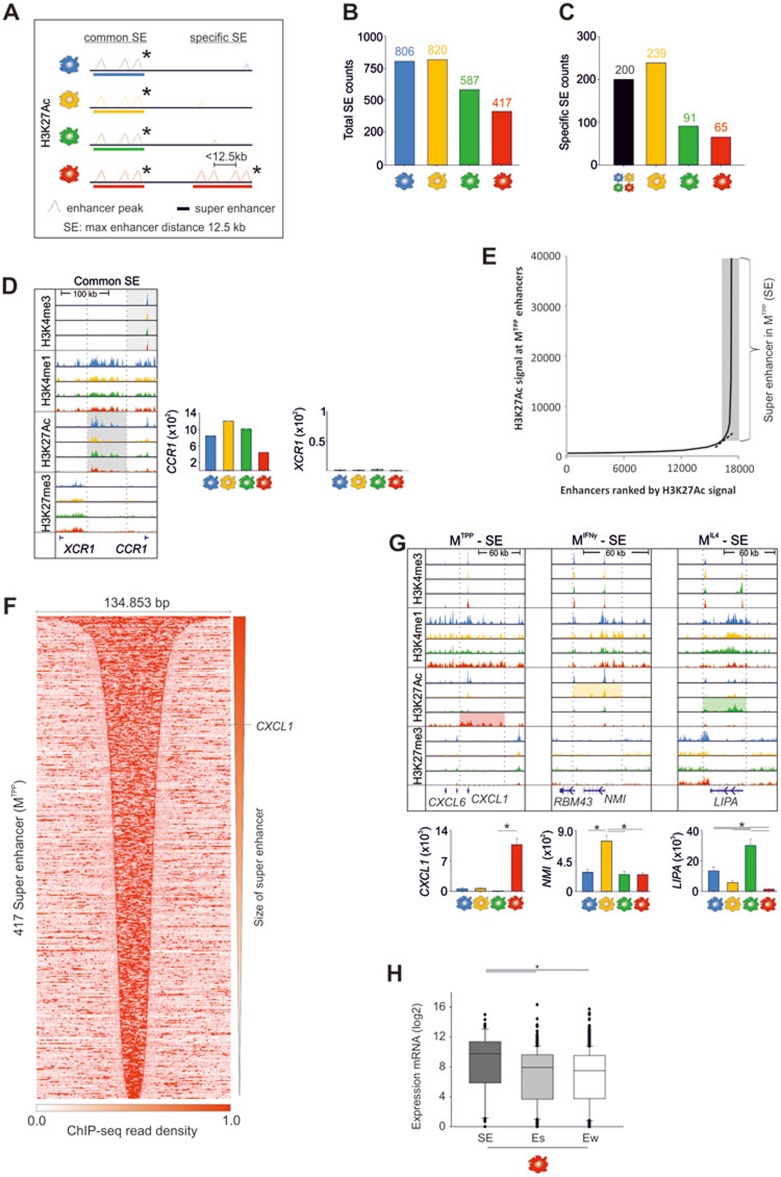
Super enhancers act in an activation-specific manner
So-called 'super enhancers' (SEs) have been a recent extension of the cooperative concept of accessible promoters and enhancers driving gene expression in a cell-type-specific fashion38,39. Defined as regions with disproportionately high TF-binding sites and activation-associated histone marks, SEs can be identified by multiple H3K27Ac peaks located within regions of <12.5kb (Figure 3A). Up to 820 SEs were identified in each of the four different macrophage conditions (Figure 3B and Supplementary information, Table S3A-S3D) with 200 SEs common to all 4 macrophage conditions (Figure 3C and Supplementary information, Table S3E). To illustrate the role of a common SE on expression, we visualized the genomic region for CCR1 and XCR1 (Figure 3D). Based on our own and published expression data40, this common SE can be linked to CCR1 gene regulation. The majority of genes in close proximity to common SEs displayed high transcriptional activity with highest average expression in MIFNγ and MTPP (Supplementary information, Figure S3A).
Figure 3.
Prevalence of super enhancers during macrophage activation. (A) Schematic overview of super enhancer (SE) characteristics. (B) Total numbers of identified SE in each macrophage condition. (C) Number of SEs common to all conditions and condition-specific SEs. (D) Representative example of a common SE site located in proximity to the CCR1 gene locus on chromosome 3. Gene expression of CCR1 and XCR1 is shown as a bar graph. (E) Dotplot identifying SEs due to their excessive enrichment of H3K27Ac HM signals (shown here MTPP macrophages). (F) Pile-up heat map of H3K27Ac read densities for 417 specific MTPP SEs. Midpoints reflect centers of each SE. (G) Visualization of genomic loci with subtype specific SE structures in MTPP (CXCL1), MIFNγ (NMI), and MIL4 (LIPA) macrophages. Mean RNA expression values were plotted as bar graphs (n = 3, one-way ANOVA with pairwise multiple comparison according to Tukey, *P < 0.05). (H) Box-and-Whisker plots of mRNA expression values for MTPP-specific genes with SE marks, strong, or weak enhancers (one-way ANOVA with pairwise multiple comparison according to Dunn's Method, *P < 0.05). Mb (blue), MIFNγ (yellow), MIL4 (green) and MTPP (red); RNA-seq and ChIP-seq were derived from three independent experiments
The number of subtype-specific SEs ranged from 65 (MTPP) to 239 (MIFNγ) (Figure 3C and Supplementary information, Table S3F-S3H). As an example SEs were visualized for MTPP as a dot plot (Figure 3E) and pile-up heat map (Figure 3F) revealing their exceptionally high H3K27Ac signals distributed over broad areas of genomic DNA loci. For each of the macrophage subtypes, we identified genes with significantly higher expression in the subtype characterized by a SE in close proximity (Figure 3G), suggesting that there is a functional relationship between expression and SEs. Moreover, comparing gene expression in relation to SEs, strong and weak enhancers, genes in close proximity to SEs showed significantly higher mean expression (Figure 3H and Supplementary information, Figure S3B) suggesting that input-signal specific induction of highly elevated gene expression is associated with specific changes in histone modifications at SE loci.
Permissive histone modifications mark loci of central TRs within the TR network of inflammatory macrophages
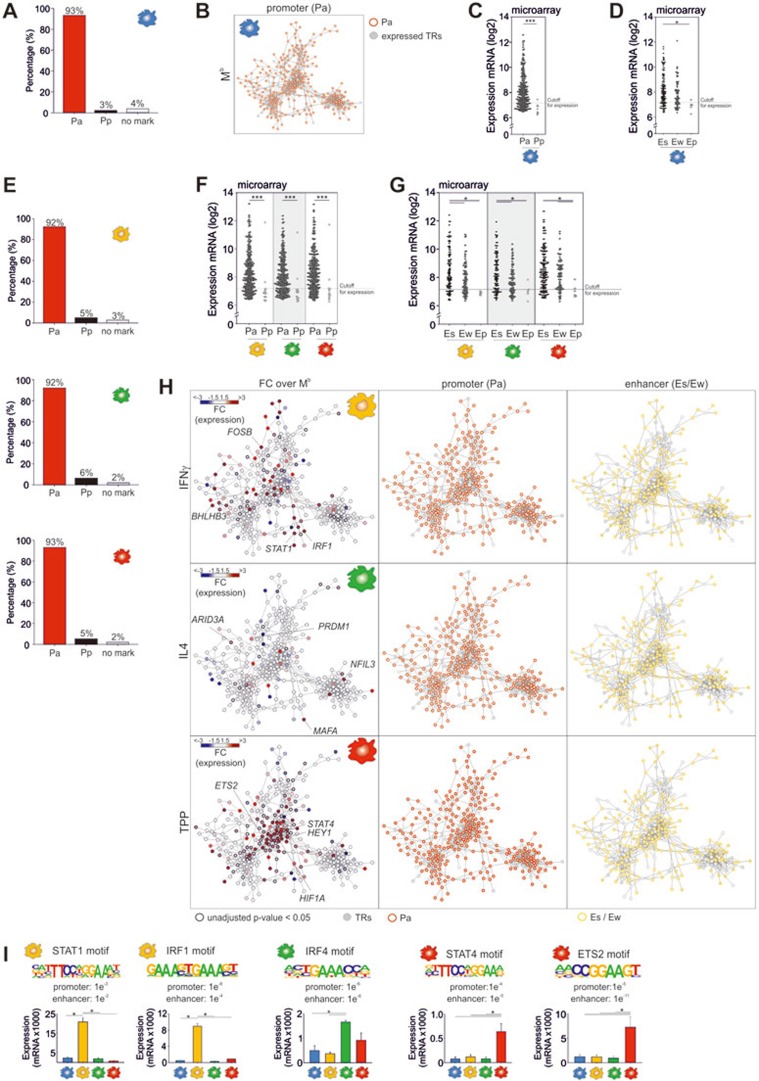
Based on our observation that genes expressed in inflammatory macrophages can be grouped according to their epigenetic modification patterns into genes with either common permissive or activation-specific histone marks, we asked, whether gene loci of TRs expressed during activation can be classified into one of the two categories. To define this distinct gene set of TRs (Supplementary information, Table S4A) we made use of our previously published multi-dimensional model of macrophage activation8. Briefly, GM-CSF-derived macrophages (Mb) were further activated by 28 different stimuli, amongst them IL-10, IL-4, IFNβ, IFNγ, free fatty acids, glucocorticoids, and bacterial compounds to mimic various inflammatory conditions (Supplementary information, Figure S4A). We identified 485 TRs which were expressed in at least one of the 29 conditions. Co-regulation network analysis revealed that 297 TRs were highly interconnected. We termed this network the 'macrophage activation TR network', in which each node represents one specific TR with co-regulated TRs connected in the network. All of the herein presented networks are accessible online at VisuTranscript (http://www.s-khb.de/visutrans/content/index_eng.html). Strikingly, nearly all promoters (93%, Figure 4A) for central TRs participating in macrophage activation were already accessible in untreated Mb (Figure 4B) and over 69% of TRs further marked by strong or weak enhancers (Supplementary information, Figure S4B, left panel). While the majority of TRs marked by an active promoter or strong/weak enhancers in Mb showed expression levels above background levels, a subset of TRs with active promoters or strong/weak enhancers only showed relatively low expression levels (Figure 4C and 4D) supporting that TRs within the network are marked by constitutively permissive histone modifications independent of active transcription.
Figure 4.
Epigenetic landscape of central transcriptional regulators. (A) Bar charts visualizing the distribution of accessible (Pa) or poised (Pp) promoter marks of TRs being part of the macrophage activation TR network in baseline macrophages (Mb). (B) Macrophage TR network with integrated information of accessible promoter (Pa) in baseline macrophages (Mb). (C, D) Scatter plots for microarray expression data of 297 TRs in baseline macrophages (Mb) (defined as described in Supplementary information, Figure S4A) grouped by their (C) promoter state as accessible (Pa) or poised (Pp) or (D) enhancer state as strong (Es), weak (Ew) or poised (Ep) (statistics calculated by (C) Mann-Whitney rank sum test or (D) Kruskal-Wallis One Way ANOVA on ranks with pairwise multiple comparison according to Dunn's Method, *P < 0.05, ***P < 0.001, cutoff for expression: 7.2). (E) Bar charts visualizing the distribution of accessible (Pa) or poised (Pp) promoter marks of TRs being part of the macrophage activation TR network in IFNγ (yellow), IL4 (green) or TPP (red) activated macrophages. (F, G) Scatter plots for microarray expression data of 297 TRs in IFNγ (yellow), IL4 (green) or TPP (red) activated macrophages with statistics as described in C and D. (H) Visualization of the macrophage activation TR network. Left panel: overlay of the corresponding RNA-seq expression data depicted as fold-change (FC) compared with the expression in Mb (FC with an unadjusted P < 0.05 are indicated by wider black borders). Mid panel: information on accessible promoter marks (red border). Right panel: strong or weak enhancer marks (orange border). (I) Positional weight matrices and binomial P-values of enrichments for predicted master regulators in Mb (blue), MIFNγ (yellow), MIL4 (green) or MTPP (red) and their respective gene expression values in all macrophage conditions (one-way ANOVA with pairwise multiple comparison according to Tukey test, *P < 0.05, n = 3). RNA-seq and ChIP-seq were derived from three independent experiments.
Next, we asked, if regulation of TR expression is reflected by stimulus-specific changes of histone modifications at TR loci. As observed for Mb the large majority of the specific TRs showed accessible promoters (>92%, Figure 4E, Supplementary information, Table S4B) in all investigated activation states and up to 70% showed strong or weak enhancers (Supplementary information, Figure S4B and Table S4B). This was reflected in the expression levels as genes marked by Pa showed higher expression, similarly to genes with Es which also were expressed at higher levels than genes with Ew or Ep (Figure 4F and 4G).
Next, expression changes in comparison to Mb, as well as epigenetic information for promoters and enhancers were mapped onto the network (Figure 4H). MIFNγ, MIL4, and MTPP were characterized by a distinct and specific pattern of differentially regulated TRs (Figure 4H, left panel), illustrating that there is an enormous transcriptional regulation of TRs in an input-signal specific fashion in human macrophages. In contrast, the patterns for accessible promoter and enhancer marks within the network were uniform with the majority of the TRs within the network showing accessible promoters and weak/strong enhancers (Figure 4H, middle and right panel), strongly suggesting that gene expression of TRs within the network is transcriptionally and not epigenetically regulated. Similar findings were found, when looking at all 485 TRs (Supplementary information, Table S4C). When analyzing TRs not expressed in any of the 29 stimulation conditions only a minority showed accessible promoters (12%-15%) or strong enhancers (12%-17%, Supplementary information, Table S4D). These data support that the permissive histone landscape is a unique feature for macrophage-activation associated TRs. To confirm that an open chromatin landmark within the network is a hallmark of monocyte-derived macrophages we assessed genes related to the central nervous system or randomly chosen genes, which both showed fewer accessible promoters (Supplementary information, Figure S4C), further supporting the hypothesis that the accessibility of TR loci is a special feature of macrophages.
To identify potential master regulators for each stimulation condition, we determined TFs that showed significantly enriched expression in MIFNγ, MIL4 or MTPP and were predicted to bind to accessible promoters and strong or weak enhancers of gene loci within the network. Prominent examples of such TFs were STAT1 and IRF1 for MIFNγ, IRF4 for MIL4 and STAT4 and ETS2 for MTPP (Figure 4I and Supplementary information, Table S4E-S4G).
Overall, we determined the network of active TRs during human macrophage activation and showed that this network is characterized by an open chromatin with significant differences in TR expression between stimulation conditions. Our results support a model where the existence of open TR loci is a pre-requisite for quick adaptions of macrophages to environmental signals, which are governed by a layer of transcriptional control mechanisms, e.g., binding of transcription factors, co-repressors or -activators or non-coding RNAs which regulate TR mRNA transcription at open loci.
The tissue-defined TR network is epigenetically and transcriptionally regulated
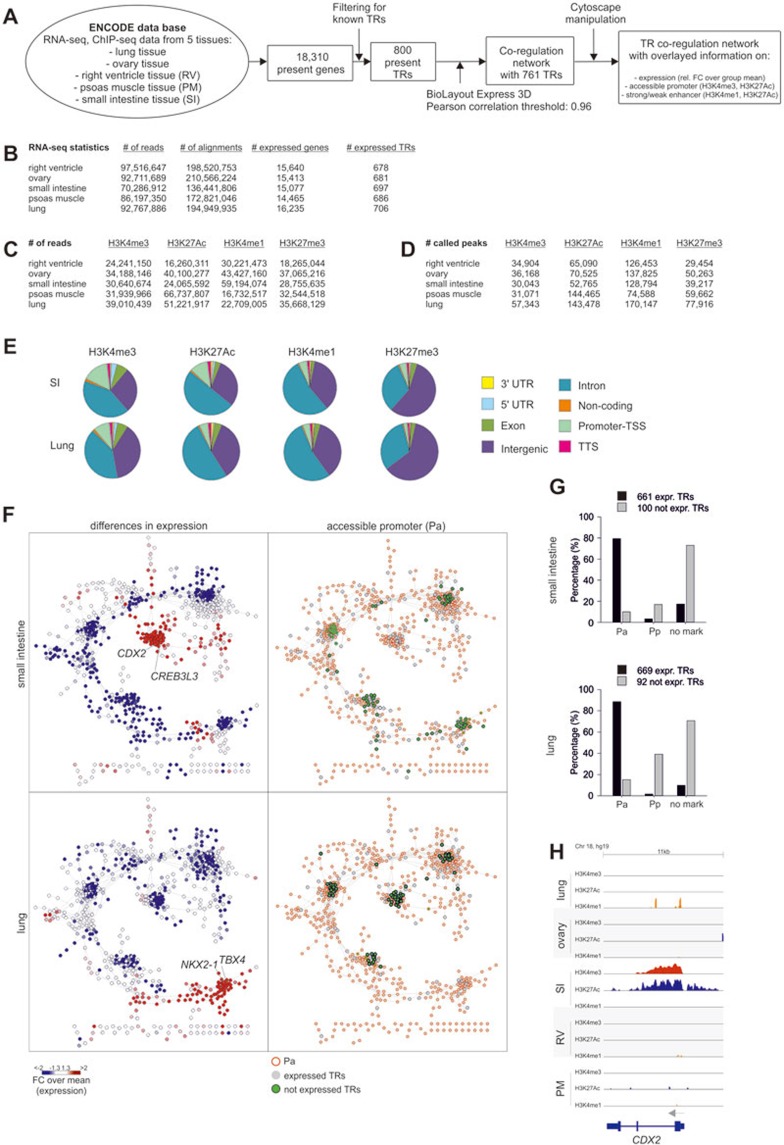
To understand whether the open chromatin state of the human macrophage activation TR network is unique to this cell type, we defined changes in histone modifications of TRs within five human tissues (intestine, lung, ovary, heart, and muscle)41 (Figure 5A). The chosen tissue samples showed comparable RNA-seq statistics (Figure 5B), but up to threefold differences in the number of peaks for histone modifications within the genome (Figure 5C and 5D). Nevertheless, the overall distribution of individual histone modification across the genome was comparable between the five different tissues (Figure 5E and Supplementary information, Figure S5A). We built a TR network for these five tissues (Figure 5A and 5F, Supplementary information, Figure S5B) following the approach introduced for the human macrophage activation TR network. Plotting differences in gene expression onto the network revealed that each of the five subclusters within the network was associated with a particular tissue (Figure 5F, left panel and Supplementary information, Figure S5B, left panel). TRs being not expressed in a particular tissue were located within the other clusters in the network that marked the remaining tissues (Figure 5F, right panel and Supplementary information, Figure S5B, right panel). More importantly, these genes were characterized by the lack of accessible promoters (Figure 5F, right panel and Supplementary information, Figure S5B, right panel) strongly supporting that differences in gene expression of TRs between tissues are epigenetically regulated following previously defined models22. In fact, relating promoter states with gene expression of TRs for each of the tissues clearly revealed a dichotomous distribution with expressed TRs being defined by accessible promoters, while TR genes not expressed were characterized by absence of accessible promoters (Figure 5G, Supplementary information, Figure S5C). On a gene locus level, we illustrate this for TFs that are specifically expressed in one tissue but not any other including CDX2 being expressed only in small intestine (Figure 5H) or NKX2-1 expressed only in lung tissue (Supplementary information, Figure S5D). Overall, these data suggest that tissue-associated TRs seem to strictly follow a coordinated epigenetic and transcriptional regulation.
Figure 5.
Epigenetic control of transcriptional regulators in human tissues. (A) Overview on the bioinformatics workflow for the generation of a human tissue-related TR network. (B) Statistics for consolidated RNA-seq data (n = 1; Road map consortium41) for each human tissue with information on number of reads, alignments, expressed genes and TRs. (C, D) Statistics for consolidated ChIP-seq data (n = 1) of the Road map consortium41 for histone modifications with information on (C) read counts and (D) called peaks. (E) Distribution of histone modifications to genomic regions calculated as percentages for small intestine (SI) and lung tissue. (F) Left Panel: Human tissue-related TR network for small intestine and lung tissues overlaid with expression values in form of fold-changes (FC) over the mean expression value of all five tissues included in the analysis. Right panel: Expressed TRs are marked in grey (RNA-seq expression values <10), not expressed TRs are marked in green with black borders. Red borderlines indicate an accessible promoter. (G) Bar charts for TRs belonging to the human tissue-related TR network grouped into expressed (black) and not expressed (grey) TRs with different promoter states: accessible promoter (Pa), poised promoter (Pp), or lacking an H3K4me3 mark (no mark). (H) Representative example (CDX2) of a TR with tissue specific expression and promoter marks.
The TR network of tissue macrophages is more similar to the tissue TR network
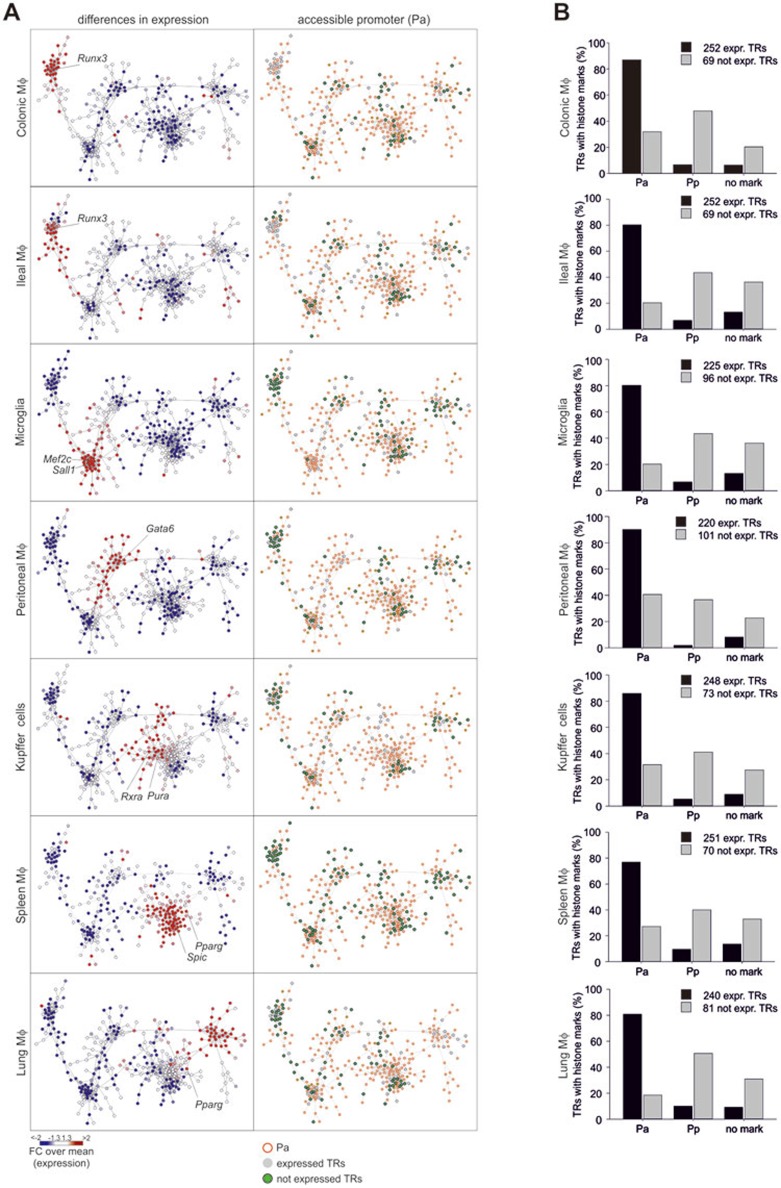
We finally addressed the question whether the open chromatin state in the human macrophage TR network is already a feature of macrophages during tissue homeostasis, or whether this is restricted to TRs associated with monocyte-derived macrophages undergoing activation. Since sufficient global transcriptome and epigenome data for human tissue macrophages have not been compiled yet, we analyzed the data previously described by Amit et al.27 and developed the TR network of seven murine tissue macrophage populations (Figure 6A and Supplementary information, Figure S6A, Table S5A). We identified 554 TRs to be expressed in at least 1 of the 7 populations of which 321 TRs were highly interconnected. Plotting differential gene expression onto the network revealed that each of the six subclusters within the network was associated with a particular tissue macrophage population (Figure 6A, left panel). All TFs previously described to be expressed in a tissue-specific fashion27 were part of the tissue-specific subclusters within the TR network, e.g., Runx3, previously described as being ileal and colonic macrophage-associated was located within the gut subcluster. This was similarly true for Mef2c (microglia), Gata6 (peritoneal macrophages), Rxra (Kupffer cells) or Spic (spleen macrophages). However, our TR network approach revealed additional information. Tissue macrophages are not defined by single TRs but rather specific clusters of co-regulated TRs. At the same time, two-thirds of all TRs within the network are expressed in any given macrophage population suggesting that changes in the expression of only a small number of TRs within the network is responsible for tissue-specific transcriptional programs (Figure 6A). When mapping promoter information onto the network, 77% to 90% of the expressed TRs but only 19% to 41% of the TRs being not expressed within the respective macrophage population showed an accessible promoter (Figure 6A, right panel and Figure 6B). A similar result was obtained when using all 554 TRs being expressed at least in one of the seven macrophage populations (data not shown). For TRs not included in the TR network (n = 358), the percentage of gene loci with accessible promoters dropped to 5% - 8% (Supplementary information, Figure S6B). We also determined the landscape of strong enhancers and identified the highest percentage of strong enhancers within the group of expressed TRs within the network (Supplementary information, Figure S6C), followed by TRs within the network being not expressed (Supplementary information, Figure S6C) and TRs not being part of the network (Supplementary information, Figure S6B). To visualize the tissue-associated modification of histones in murine tissue macrophages we chose two specific tissue TRs (Gata6 and Sall1) and plotted the respective H3K4me3, H3K27Ac and H3K4me1 signals in comparison with other tissues as histograms (Supplementary information, Figure S6D). Overall, whereas the human macrophage activation TR network was characterized by a completely open promoter landscape, tissue macrophages and tissues showed a more tissue-restricted and integrated pattern of epigenetic and transcriptional regulation.
Figure 6.
Epigenetic regulation of TRs in murine tissue macrophages. Epigenetic regulation of TRs in murine tissue macrophages. (A) TR network for murine tissue macrophages build from 321 TRs (defined as described in Supplementary information, Figure S6A). Differences in expression are plotted onto the network as fold-changes (FC) over the mean expression (left panel) of all seven tissue macrophages. Right panel shows accessible promoter (Pa) states as red borderlines. Expressed TRs (RNA-seq expression values <10) in grey, not expressed: green with black borderlines. (B) Bar charts for the distribution of expressed (black) and not expressed (grey) TRs belonging to the murine tissue macrophage TR network with accessible (Pa), poised (Pp) or no promoter marks. Mφ: macrophages. RNA-seq and ChIP-seq were derived from two independent experiments27.
Discussion
A general model in regulation of gene expression has emerged indicating that transcriptional regulation is tidily integrated with epigenetic regulation22. Highly expressed genes are defined by an accessible chromatin landscape, while non-expressed genes are defined by inaccessible chromatin. Developing a network approach to define networks of transcriptional and epigenetic regulators (TRs) we demonstrate that genes being part of a tissue-associated TR network strictly follow the general model (Figure 5), while TRs that can be expressed in an input signal-specific manner during inflammation-associated macrophage activation (Figure 4) show a different regulation pattern. In contrast to the tissue-related TR network, the complete TR network of inflammatory macrophages is pre-defined by an open chromatin suggesting that gene expression within this network is guided solely by transcriptional regulation ruled by input-specific master TRs. In fact, TRs within this network are expressed in different input signal-specific combinations during macrophage activation (Figure 4). We postulate that the enormous plasticity of gene expression as observed in our previously introduced multi-dimensional model of human macrophage activation8 is a direct consequence of expression-dependent regulation within this specialized TR network. Consequently, we predict that other cell types showing functional plasticity may contain similar cell-type-associated activation TR networks, while TRs in cells that are not characterized by functional plasticity are precisely regulated by epigenetic mechanisms, e.g., TRs being not expressed are epigenetically silenced, while expressed TRs are defined by accessible promoters and strong enhancers.
Overall gene expression in inflammatory macrophages follows the general model proposed for macrophages and their activation (Figures 1 and 2). As expected, we identified a large set of common gene loci for four human macrophage subtypes that showed highest gene expression for those loci with accessible promoters and strong enhancers (Figure 1). Functionally, these common gene loci constitute genes associated with housekeeping functions or cellular maintenance. As suggested by previous findings in murine macrophages15,25, we also identified significant remodeling of the enhancer and promoter landscapes in human macrophages during activation (Figure 2). Expression levels of genes associated with input signal-specific epigenetic marks were strictly related to the state of the epigenetic landscape at these sites with highest expression observed in genes with accessible promoters and strong enhancers. We further extended this rule to so-called super enhancers38 (Figure 3).
Gene loci within the macrophage activation TR network seem to possess a non-classical type of regulation, as expression of these TRs is stimulus dependent, yet their chromatin state corresponds to the generally permissive state observed for common genes. While the large majority of common promoters and enhancers was characterized by binding of the lineage determining factor PU.1, only a fraction of activation-specific sites bound PU.1. We presume that these sites are the human equivalent of what has been recently defined as latent enhancers in murine macrophages15 and are not dependent on PU-1 binding. Taken together, our data indicate that while inflammatory macrophages seem to follow the general model of gene expression regulation, for an important class of genes, which we define as the macrophage activation TR network, an exception to the rule is apparent. This exception to the rule could be cell-type intrinsic for macrophages or might be associated with cell activation itself. Future work will also address, whether activation by M-CSF will lead to a comparable TR network in monocyte-derived macrophages generated. Since sufficient epigenetic and transcriptional data of human tissue macrophages are not yet available to us, we utilized murine tissue macrophage data to construct the global tissue macrophage TR network27 (Figure 6). Similar to the tissue-related network, we find a more strictly epigenetic regulation within this network; TRs not being expressed in macrophages derived from a particular tissue lack the respective histone marks for permissive chromatin, while the same TRs being expressed in macrophages from another tissue are characterized by accessible promoters and strong or at least weak enhancers. Interestingly, the number of TRs not being expressed but characterized by an accessible promoter was the highest for peritoneal and colonic macrophages, for which epigenetic regulation was shown to be most closely related to monocytes27. Furthermore, it is now well accepted that tissue macrophages in these two organs are continuously replaced by monocyte-derived cells in adulthood5 and therefore it is very well possible that the epigenetic and transcriptional regulation of these cells is more closely related to human monocyte-derived macrophages during activation. Overall, these findings suggest that the observed uncoupling of epigenetic and transcriptional regulation of TRs within the human macrophage activation TR network is a hallmark of activation of monocyte-derived macrophages rather than a property of tissue macrophages. We have evidence from re-analyzing a smaller dataset of murine bone-marrow derived macrophages activated with seven stimuli previously published by Natoli and colleagues20 that the epigenetic landscape in this TR network is similar to what we have observed during human macrophage activation (Schmidt, unpublished results). To definitely address whether there is a difference between monocyte-derived and tissue macrophages in epigenetic and transcriptional regulation of TRs during activation, global epigenetic and transcriptional landscapes of cells during homeostasis and different activation states (including inflammation) need to be evaluated for all tissues, a task that will require large international consortium efforts. These efforts will require the application of novel technologies that allow the assessment of histone modifications in very small cell numbers derived from inflamed tissues. However, since macrophages play a critical role during many inflammatory conditions including obesity, diabetes, atherosclerosis, cancer and neurodegenerative diseases, such knowledge might be critical for the development of more specific, molecularly defined therapeutic intervention strategies.
In recent years, a multitude of epigenetic and transcriptional mechanisms regulating gene expression in macrophages has been described27,42,43,44,45. The uncoupling of epigenetic and transcriptional regulation within the human macrophage activation TR network is reminiscent of regulation observed for primary response genes induced by Toll-like receptor (TLR) ligands. They are enriched for CpG island and H3K4me3-marked promoters, paused RNA polymerase II near the TSS in unstimulated cells and do not require SWI/SNF-dependent nucleosome remodeling during activation44,46,47,48. Most of the TRs within the human macrophage activation TR network displayed RNA pol II binding irrespective of gene expression levels further supporting transcriptional regulation being the major mechanism of gene expression regulation in macrophage activation (Schmidt, unpublished results). A very likely mechanism establishing an open chromatin landscape within the macrophage activation TR network was the binding of the lineage-determining factor PU.1 to promoter and enhancer sites, since it has been previously demonstrated that PU.1 is required for the macrophage-specific open chromatin landscape at promoters and enhancers in general17,18,19,49. Indeed, >91% of TRs within the macrophage activation TR network showed binding of PU.1 to the promoter region or to cis enhancers (data not shown), strongly suggesting that PU.1 is involved in establishing and maintaining the open chromatin landscape at these gene loci irrespective of their actual transcription.
Among normal somatic cells macrophages are one of the most plastic cells42. Plasticity of macrophages has been associated with their responsiveness to a wide spectrum of environmental signals that lead to a signal input-specific cascade of signal transduction resulting in a signal-specific transcriptional response. A prerequisite for regulation of plastic cell responses by macrophages is the combinatorial and subsequent action of lineage-determining factors such as PU.1 and C/EBPs followed by signal-dependent TFs (e.g., STAT transcription factors, NF-κB family members, CREB and others). In many recent studies mainly using TLR4 stimulation as a model, cellular plasticity of macrophages has been defined as the capacity to dynamically respond to an environmental signal in a coordinated fashion by combined transcriptional and epigenetic regulatory mechanisms. However, much less attention has been ascribed to the fact that macrophages can react rather quickly to a myriad of different signals and it is this capacity that represents their overall plasticity. By introducing the macrophage activation TR network we provide a general framework that can serve as a platform for fast transcriptional responses without the need for protracted epigenetic mechanisms to many different stimuli macrophages can encounter in their particular microenvironments. In general, cellular plasticity is not restricted to macrophages as it has been linked to epithelial-mesenchymal transition (EMT) in cancer50. However, regulatory mechanisms suggested to be involved in EMT such as polycomb repressor-, histone deacetylase-, or histone demethylase-mediated gene silencing still follow the general model of orderly integration of transcriptional and epigenetic regulation without uncoupling of those two mechanisms. Very recently, however, cell plasticity was associated with broadly permissive chromatin in stem cells and their progenies within intestinal crypts51. In fact, the expression of a single TF despite a similarly open chromatin landscape defined whether cells appeared phenotypically and functionally as enterocytes or secretory cells. In addition to our findings in macrophages, this is currently the only other example of uncoupling of epigenetic and transcriptional control being associated with cellular plasticity in response to microenvironmental signals. However, we propose transcriptional regulation in an open chromatin landscape to be a major hallmark of cellular plasticity and predict that this mechanism will also play a role in other settings.
Taken together, using transcriptional profiling of a large number of different activation states of human monocyte-derived inflammatory macrophages, assessing important histone marks during activation, and utilizing these data for network analysis and data visualization, we have identified those transcriptional and epigenetic regulators that — in different combinations — represent the macrophage activation TR network. Hallmark of this network is its open chromatin landscape associated with strong PU.1 binding to promoters and enhancers within the network. We propose a model of human macrophage plasticity during inflammatory immune responses that is mainly defined by signal integration-dependent transcriptional regulation of network TRs. Elucidating further specific and combinatorial transcriptional regulatory mechanisms within this network in response to input-signals will be an important endeavor for finding novel drug targets in chronic inflammatory diseases.
Materials and Methods
Ethics statement, reagents, antibodies and cell culture procedures
For information on buffy coats, reagents for monocyte isolation, fluorescence-labeled antibodies and cell culture procedures please refer to the Supplementary information, Data S1.
RNA isolation, RNAseq library preparation, sequencing and data processing
RNA isolation, quality tests and RNA sequencing experiments were performed as described earlier52. About 5-10 μg of total RNA were used for cDNA synthesis (Illumina TruSeq RNA Sample Preparation Kit). For detailed information and bioinformatics data processing please see Supplementary information, Data S1.
Chromatin immunoprecipitation followed by deep sequencing (ChIP-seq) and data processing
H3K4me3 histone modification (HM) was carried out as previously described8. ChIP for HMs (H3K4me1, H3K4me2, H3K27ac, H3K27me3) and PU.1 were carried out according to a cross-linking chromatin immunoprecipitation (X-ChIP) protocol53. Briefly, either 0.5 × 106 cells or 6 × 106 cells were cross-linked with 1% formaldehyde for HM or PU.1 ChIP, respectively. Nuclei were lysed and chromatin was sheared by sonication. Shared chromatin was incubated overnight at 4 °C with protein G Dynabeads (Invitrogen) that were previously coupled with 0.1-3 μg of antibody for HM (H3K4me1 (Abcam), H3K27Ac (Abcam), H3K27me3 (Millipore)) and 5 μg for PU.1 ChIP (SantaCruz, sc-352). After immunoprecipitation and washing steps, DNA was eluted and decrosslinked overnight at 65 °C. Purification of DNA fragments was performed with solid-phase reversible immobilization beads (Agencourt, AMPure XP, Beckman Coulter) and quantified with the Bioanalyzer 2100/Tapestation system (Agilent). Between 0.5 and 1 ng of purified DNA was used for amplification with the Illumina sequencing compatible NEXTflex adapter oligonucleotide kit (Bioo Scientific). Primers used for the ChIP quality controls are listed in the Extended Experimental Procedures. ChIP-seq libraries were sequenced on HiScanSQ and HiSeq 1000 sequencer (Illumina) in a single read run for 57 cycles. Short sequence reads were aligned to the human reference genome (hg18) with Bowtie (v0.85) (Supplementary information, Table S5B). Detailed information on the computational analysis is provided in the Extended Experimental Procedures. In brief, annotated SAM files are converted to tag directories using HOMER using the makeTagDirectory module (makeTagDirectory /HistonDestinationFolder/ SAM-file.sam -genome). These directories are further used for peak calling (findPeaks /HistonDestinationFolder/ -i /Input_HistonDestinationFolder/ -style histone > peak_file.txt) or converted to the bedGraph format normalized to 106 total tag counts with HOMER (pos2bed.pl peak_file.txt > bed_file.bed) using the makeUCSCfile module. For the analysis of histone ChIP-seq data input samples were utilized as control files during peak detection, whereas IgG control files were used during peak correction of the PU.1 ChIP-seq data.
Identification and classification of promoter and enhancer sites
The identification of HM signal enriched sites with aligned ChIP-seq reads was performed with the findPeaks function available in HOMER (v4.6) utilizing the histone style option and input experiments as controls18. The exact criteria for the categorization into two promoter and three enhancer states as well as common promoter and enhancer sites present in all macrophage conditions are described in Supplementary information, Data S1.
In brief, H3K4me3 signals near TSS (± 2.5 kb) and H3K4me1 HM signals up- or downstream the TSS (>2.5 kb / <100 kb) were used to define promoter and enhancer sites in activated macrophage data sets, while H3K27Ac (active) and H3K27me3 (repressive) HMs were used to classify sites into accessible (H3K4me3/H3K27Ac) and poised (H3K4me3/H3K27me3) promoters and strong (H3K4me1/H3K27Ac), weak (H3K4me1) and poised (H3K4me1/H3K27me3) enhancers. For the detection of multiple HM signals at a genomic locus the mergePeaks function of HOMER with the prefix option was used to find direct overlaps (at least 1 bp) for significant HM peak sites. Common macrophage sites were identified by a direct overlap of each peak position with H3K4me3 (promoter) and H3K4me1 (enhancer) HM signals, whereas remaining positions are filtered for sites (I) only present in one macrophage activation state without direct overlap in other macrophages or (II) show at least two times more normalized HM tag counts (normalized to 10 million total tag counts) in one activation state in comparison to the others and are thus defined as specific. Human RefSeq annotations allowed the association of peak sites to gene symbols with the annotatePeaks (annotatePeaks.pl peak_file.txt genome > peaks_file_annotated.txt) function in HOMER.
For the visualization of genomic peak positions, tag signals were normalized prior to analysis to 107 total tag numbers and displayed as pile-up heatmaps with Java-Treeview (v1.1.6.r4)54. Read densities for H3K4me3 marked genomic positions were calculated with the annotatePeaks.pl (annotatePeaks.pl hg18 -size 6000 -hist 25 -d) function of HOMER and displayed as histograms. Peaks were centered to the peak midpoint.
Identification of super enhancers
Super enhancers28 were identified according to their characteristics by massive H3K27Ac marks using HOMER with the following settings: findPeaks.pl -style super, -L 0. In addition, these settings stitch super enhancer peaks together if they are located within 12.5 kb of each other.
TR co-regulation network construction and meta-information visualization
A collective list of human and murine transcriptional regulators encompassing TFs, co-factors, RNA-binding proteins and chromatin remodelers originating from TFCat data base55 was used to filter expressed genes for transcriptional and epigenetic regulators (TRs) (Supplementary information, Table S4A). The expression matrices of these TRs were loaded into BioLayout and co-regulation networks were generated with variable Pearson correlations depending on the size of the TR network. The predicted TF-TF pairs together with their correlation coefficients were visualized by Cytoscape using Force-directed layout. Further information at transcriptional level such as FC values calculated against group mean value and the corresponding epigenetic information for the respective samples were mapped onto the TR network one by one. The observed results remained comparable in TR networks which were built based on different Pearson correlation co-efficients as cutoff (Supplementary information, Table S6).
Gene ontology enrichment analysis
Gene names were assigned to HM-marked promoter regions ± 2 500 bp around TSS or to TSS for enhancer marks within 100-kb distance. GOEA was performed with Cytoscape v2.8.356 and the BiNGO plugin (v2.44) with a FDR-corrected hypergeometric p-value of 0.001. The Cytoscape plugin enrichment map (v1.2) displayed the GOEA results as a network of GO-terms derived from enriched sets of genes with accessible promoters and strong or weak enhancer. Statistical options were set to 0.001 for the FDR corrected q-value and subnetworks with at least 2 GO-terms were shown.
VisuTranscript
To visualize and provide the networks described here, we developed a HTML5 based, modular web application with responsive design and a cloud based backend in form of a WebPortal called VisuTranscript. For detailed information we refer to the Supplementary information, Data S1.
Accession numbers
The accession numbers for the sequencing data reported in this paper are GEO: GSE36952, GSE66593, GSE16256, GSE63341, GSE47188, GSE66594, GSE16256, GSE63339.
Author Contributions
SVS, WK and TU designed and performed experiments, carried out bioinformatics analysis, assembled and wrote the manuscript; WK, A-LH and HT performed cultures of human primary cells, flow cytometry, quality control steps and ChIP; WK, A-LH and MK performed library production, RNA-seq and ChIP-seq; JX, KB, PG, JS, KK performed alignments, TR network generations, bioinformatics analysis and statistics; HS designed the Web portal for display of network data; MB and JLS designed research, supervised experiments and reviewed the paper.
Competing Financial Interests
The authors declare no competing financial interests.
Acknowledgments
We thank Andrew Pospisilik (MPI of Immunobiology and Epigenetics) for critically reading the manuscript. We thank Johannes Oldenburg for providing us with buffy coats from healthy individuals. This work was supported by the German Research Foundation (SFB 832, SFB 704, INST 217/577-1). JLS and MB are members of the Excellence Cluster ImmunoSensation. JLS is an associated member of the German epigenome program DEEP. The research leading to these results has received funding from the People Program (Marie Curie Actions) of the European Union's Seventh Framework Program FP7/2077-2013 under REA grant agreement no. 317445.
Footnotes
(Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary Information
Histone marks in common macrophage core signature genes
Activation-specific specific promoter and enhancer marks
Expression analysis of genes encompassing super enhancer
Epigenetic landscape of transcriptional regulators in human inflammatory macrophages
Human tissue-related TR networks show a differential epigenetic regulation of TR loci
Epigenetic landscape of TR of murine tissue macrophages
(A–B) Chromosomal positions of (A) common accessible promoters or (B) common poised promoters. (C–E) Chromosomal positions of common (C) strong, (D) weak or (E) poised enhancer promoters.
(A–D) Activation specific accessible promoters in (A) Mb, (B) IFNγ, (C) IL4 or (D) TPP stimulated human inflammatory macrophages. (E–H) Activation specific poised promoters in (E) Mb, (F) IFNγ, (G) IL4 or (H) TPP stimulated human inflammatory macrophages. (I–T) Activation specific (I–L) strong, (M–P) weak and (Q–T) poised enhancer positions in baseline macrophages and three human macrophage activation states. (U) Gene ontology enrichment analysis for activation specific strong and weak enhancer in MTPP. (V–Y) Positional weight matrices (PWMs) of the top 5 enriched motifs for TFs at activation specific accessible promoters and enhancers in (V) Mb, (W) IFNγ, (X) IL4 or (Y) TPP stimulated human inflammatory macrophages.
(A–D) Super enhancer in (A) baseline macrophages, (B) IFNγ, (C) IL4 or (D) TPP stimulated human macrophages. (E) Super enhancer found in all macrophage conditions. (F–H) Activation specific super enhancer in (F) IFNγ, (G) IL4 or (H) TPP stimulated human macrophages.
(A) List of human and murine transcription factors and transcriptional regulators. (B–D) Statistics on promoter and enhancer states of TRs (B) belonging to the human macrophage activation TR network, (C) being expressed in human inflammatory macrophages, (D) being not expressed in human inflammatory macrophages. (E–G) Positional weight matrices and expression levels for TRs in (E) IFNγ, (F) IL4 or (G) TPP stimulated human inflammatory macrophages.
Statistics of RNA-seq and ChIP-seq data of (A) murine tissue macrophages and (B) human inflammatory macrophages.
Statistics for TR networks of human activated macroophages generated by different Pearson correlation co-efficients
Extended experimental procedures
References
- 1van Furth R, Cohn ZA, Hirsch JG, Humphrey JH. Spector WG, Langevoort HL. The mononuclear phagocyte system: a new classification of macrophages, monocytes, and their precursor cells. Bull World Health Organ 1972; 46:845–852. [PMC free article] [PubMed] [Google Scholar]
- 2Hume DA. The mononuclear phagocyte system. Curr Opin Immunol 2006; 18:49–53. [DOI] [PubMed] [Google Scholar]
- 3Jenkins SJ, Hume DA. Homeostasis in the mononuclear phagocyte system. Trends in immunology 2014; 35:358–367. [DOI] [PubMed] [Google Scholar]
- 4Haldar M, Murphy KM. Origin, development, and homeostasis of tissue-resident macrophages. Immunol Rev 2014; 262:25–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5Ginhoux F, Jung S. Monocytes and macrophages: developmental pathways and tissue homeostasis. Nat Rev Immunol 2014; 14:392–404. [DOI] [PubMed] [Google Scholar]
- 6Wynn TA, Chawla A, Pollard JW. Macrophage biology in development, homeostasis and disease. Nature 2013; 496:445–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7Van Dyken SJ, Locksley RM. Interleukin-4- and interleukin-13-mediated alternatively activated macrophages: roles in homeostasis and disease. Annu Rev Immunol 2013; 31:317–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8Xue J, Schmidt SV, Sander J, et al. Transcriptome-based network analysis reveals a spectrum model of human macrophage activation. Immunity 2014; 40:274–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9Underhill DM, Goodridge HS. Information processing during phagocytosis. Nat Rev Immunol 2012; 12:492–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10Davies LC, Jenkins SJ, Allen JE, Taylor PR. Tissue-resident macrophages. Nat Immunol 2013; 14:986–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11Medzhitov R, Horng T. Transcriptional control of the inflammatory response. Nat Rev Immunol 2009; 9:692–703. [DOI] [PubMed] [Google Scholar]
- 12Smale ST. Transcriptional regulation in the innate immune system. Curr Opin Immunol 2012; 24:51–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13Friedman AD. Transcriptional control of granulocyte and monocyte development. Oncogene 2007; 26:6816–6828. [DOI] [PubMed] [Google Scholar]
- 14Carotta S, Wu L, Nutt SL. Surprising new roles for PU.1 in the adaptive immune response. Immunol Rev 2010; 238:63–75. [DOI] [PubMed] [Google Scholar]
- 15Ostuni R, Natoli G. Lineages, cell types and functional states: a genomic view. Curr Opin Cell Biol 2013; 25:759–764. [DOI] [PubMed] [Google Scholar]
- 16Garber M, Yosef N, Goren A, et al. A high-throughput chromatin immunoprecipitation approach reveals principles of dynamic gene regulation in mammals. Mol Cell 2012; 47:810–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17Ghisletti S, Barozzi I, Mietton F, et al. Identification and characterization of enhancers controlling the inflammatory gene expression program in macrophages. Immunity 2010; 32:317–328. [DOI] [PubMed] [Google Scholar]
- 18Heinz S, Benner C, Spann N, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 2010; 38:576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19Barozzi I, Simonatto M, Bonifacio S, et al. Coregulation of transcription factor binding and nucleosome occupancy through DNA features of mammalian enhancers. Mol Cell 2014; 54:844–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20Ostuni R, Piccolo V, Barozzi I, et al. Latent enhancers activated by stimulation in differentiated cells. Cell 2013; 152:157–171. [DOI] [PubMed] [Google Scholar]
- 21Mancino A, Termanini A, Barozzi I, et al. A dual cis-regulatory code links IRF8 to constitutive and inducible gene expression in macrophages. Genes Dev 2015; 29:394–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22Kouzarides T. Chromatin modifications and their function. Cell 2007; 128:693–705. [DOI] [PubMed] [Google Scholar]
- 23Alvarez-Errico D, Vento-Tormo R, Sieweke M, Ballestar E. Epigenetic control of myeloid cell differentiation, identity and function. Nat Rev Immunol 2015; 15:7–17. [DOI] [PubMed] [Google Scholar]
- 24Saeed S, Quintin J, Kerstens HH, et al. Epigenetic programming of monocyte-to-macrophage differentiation and trained innate immunity. Science 2014; 345:1251086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25Kaikkonen MU, Spann NJ, Heinz S, et al. Remodeling of the enhancer landscape during macrophage activation is coupled to enhancer transcription. Mol Cell 2013; 51:310–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26Creyghton MP, Cheng AW, Welstead GG, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci USA 2010; 107:21931–21936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27Lavin Y, Winter D, Blecher-Gonen R, et al. Tissue-resident macrophage enhancer landscapes are shaped by the local microenvironment. Cell 2014; 159:1312–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28Gosselin D, Link VM, Romanoski Casey E, et al. Environment drives selection and function of enhancers controlling tissue-specific macrophage identities. Cell 2014; 159:1327–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29Pham TH, Benner C, Lichtinger M, et al. Dynamic epigenetic enhancer signatures reveal key transcription factors associated with monocytic differentiation states. Blood 2012; 119:e161–e171. [DOI] [PubMed] [Google Scholar]
- 30Schmidl C, Renner K, Peter K, et al. Transcription and enhancer profiling in human monocyte subsets. Blood 2014; 123:e90–e99. [DOI] [PubMed] [Google Scholar]
- 31Schultze JL, Freeman T, Hume DA, Latz E. A transcriptional perspective on human macrophage biology. Semin Immunol 2015; 27:44–50. [DOI] [PubMed] [Google Scholar]
- 32Metcalf D, Nicola N.A.. The Hemopoietic Colony-Stimulating Factors: from Biology to Clinical Applications. Cambridge, UK: Cambridge University Press, 1995.
- 33Shimbo T, Du Y, Grimm SA, et al. MBD3 localizes at promoters, gene bodies and enhancers of active genes. PLoS Genet 2013; 9:e1004028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34Ernst J, Kheradpour P, Mikkelsen TS, et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature 2011; 473:43–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35Zentner GE, Tesar PJ, Scacheri PC. Epigenetic signatures distinguish multiple classes of enhancers with distinct cellular functions. Genome Res 2011; 21:1273–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36Suzuki A, Makinoshima H, Wakaguri H, et al. Aberrant transcriptional regulations in cancers: genome, transcriptome and epigenome analysis of lung adenocarcinoma cell lines. Nucleic Acids Res 2014; 42:13557–13572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37Shlyueva D, Stampfel G, Stark A. Transcriptional enhancers: from properties to genome-wide predictions. Nat Rev Genet 2014; 15:272–286. [DOI] [PubMed] [Google Scholar]
- 38Hnisz D, Abraham BJ, Lee TI, et al. Super-enhancers in the control of cell identity and disease. Cell 2013; 155:934–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39Whyte WA, Orlando DA, Hnisz D, et al. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell 2013; 153:307–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40Kaufmann A, Salentin R, Gemsa D, Sprenger H. Increase of CCR1 and CCR5 expression and enhanced functional response to MIP-1 alpha during differentiation of human monocytes to macrophages. J Leukoc Biol 2001; 69:248–252. [PubMed] [Google Scholar]
- 41Roadmap Epigenomics Consortium, Kundaje A, Meuleman W, et al. Integrative analysis of 111 reference human epigenomes. Nature 2015; 518:317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42Zhang DX, Glass CK. Towards an understanding of cell-specific functions of signal-dependent transcription factors. J Mol Endocrinol 2013; 51:T37–T50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43Lawrence T, Natoli G. Transcriptional regulation of macrophage polarization: enabling diversity with identity. Nat Rev Immunol 2011; 11:750–761. [DOI] [PubMed] [Google Scholar]
- 44Adelman K, Kennedy MA, Nechaev S, et al. Immediate mediators of the inflammatory response are poised for gene activation through RNA polymerase II stalling. Proc Natl Acad Sci USA 2009; 106:18207–18212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45Saccani S, Trabucchi M. Regulation of stimulus-inducible gene expression in myeloid cells. Semin Immunol 2015; 27:33–43. [DOI] [PubMed] [Google Scholar]
- 46Bhatt DM, Pandya-Jones A, Tong AJ, et al. Transcript dynamics of proinflammatory genes revealed by sequence analysis of subcellular RNA fractions. Cell 2012; 150:279–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47Hargreaves DC, Horng T, Medzhitov R. Control of inducible gene expression by signal-dependent transcriptional elongation. Cell 2009; 138:129–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48Ramirez-Carrozzi VR, Braas D, Bhatt DM, et al. A unifying model for the selective regulation of inducible transcription by CpG islands and nucleosome remodeling. Cell 2009; 138:114–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49Heinz S, Romanoski CE, Benner C, et al. Effect of natural genetic variation on enhancer selection and function. Nature 2013; 503:487–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50Tam WL, Weinberg RA. The epigenetics of epithelial-mesenchymal plasticity in cancer. Nat Med 2013; 19:1438–1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51Kim TH, Li F, Ferreiro-Neira I, et al. Broadly permissive intestinal chromatin underlies lateral inhibition and cell plasticity. Nature 2014; 506:511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52Beyer M, Mallmann MR, Xue J, et al. High-resolution transcriptome of human macrophages. PLoS One 2012; 7:e 45466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53Blecher-Gonen R, Barnett-Itzhaki Z, Jaitin D, Amann-Zalcenstein D, Lara-Astiaso D, Amit I. High-throughput chromatin immunoprecipitation for genome-wide mapping of in vivo protein-DNA interactions and epigenomic states. Nat Protoc 2013; 8:539–554. [DOI] [PubMed] [Google Scholar]
- 54Saldanha AJ. Java Treeview--extensible visualization of microarray data. Bioinformatics 2004; 20:3246–3248. [DOI] [PubMed] [Google Scholar]
- 55Fulton DL, Sundararajan S, Badis G, et al. TFCat: the curated catalog of mouse and human transcription factors. Genome Biol 2009; 10:R29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 2003; 13:2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Histone marks in common macrophage core signature genes
Activation-specific specific promoter and enhancer marks
Expression analysis of genes encompassing super enhancer
Epigenetic landscape of transcriptional regulators in human inflammatory macrophages
Human tissue-related TR networks show a differential epigenetic regulation of TR loci
Epigenetic landscape of TR of murine tissue macrophages
(A–B) Chromosomal positions of (A) common accessible promoters or (B) common poised promoters. (C–E) Chromosomal positions of common (C) strong, (D) weak or (E) poised enhancer promoters.
(A–D) Activation specific accessible promoters in (A) Mb, (B) IFNγ, (C) IL4 or (D) TPP stimulated human inflammatory macrophages. (E–H) Activation specific poised promoters in (E) Mb, (F) IFNγ, (G) IL4 or (H) TPP stimulated human inflammatory macrophages. (I–T) Activation specific (I–L) strong, (M–P) weak and (Q–T) poised enhancer positions in baseline macrophages and three human macrophage activation states. (U) Gene ontology enrichment analysis for activation specific strong and weak enhancer in MTPP. (V–Y) Positional weight matrices (PWMs) of the top 5 enriched motifs for TFs at activation specific accessible promoters and enhancers in (V) Mb, (W) IFNγ, (X) IL4 or (Y) TPP stimulated human inflammatory macrophages.
(A–D) Super enhancer in (A) baseline macrophages, (B) IFNγ, (C) IL4 or (D) TPP stimulated human macrophages. (E) Super enhancer found in all macrophage conditions. (F–H) Activation specific super enhancer in (F) IFNγ, (G) IL4 or (H) TPP stimulated human macrophages.
(A) List of human and murine transcription factors and transcriptional regulators. (B–D) Statistics on promoter and enhancer states of TRs (B) belonging to the human macrophage activation TR network, (C) being expressed in human inflammatory macrophages, (D) being not expressed in human inflammatory macrophages. (E–G) Positional weight matrices and expression levels for TRs in (E) IFNγ, (F) IL4 or (G) TPP stimulated human inflammatory macrophages.
Statistics of RNA-seq and ChIP-seq data of (A) murine tissue macrophages and (B) human inflammatory macrophages.
Statistics for TR networks of human activated macroophages generated by different Pearson correlation co-efficients
Extended experimental procedures