Abstract
Host stress is well known to result in flare-ups of many bacterial, viral and parasitic infections. The mechanism by which host stress is exploited to increase pathogen loads, is poorly understood. Here we show that Salmonella enterica subspecies enterica serovar Typhimurium employs a dedicated mechanism, driven by the scsA gene, to respond to the host stress hormone cortisol. Through this mechanism, cortisol increases Salmonella proliferation inside macrophages, resulting in increased intestinal infection loads in DBA/2J mice. ScsA directs overall Salmonella virulence gene expression under conditions that mimic the intramacrophagic environment of Salmonella, and stimulates the host cytoskeletal alterations that are required for increased Salmonella proliferation inside cortisol exposed macrophages. We thus provide evidence that in a stressed host, the complex interplay between a pathogen and its host endocrine and innate immune system increases intestinal pathogen loads to facilitate pathogen dispersal.
Apt responses to stressors promote an organism’s chance of survival as they prepare the body for the innate fight-or-flight response1. During periods of stress, hormones are released into the circulation or tissues in order to mediate the stress response. The resulting increase in circulating stress hormones such as epinephrine, norepinephrine and cortisol, however, coincides with increased susceptibility to infectious diseases, resulting in recrudescence of viral2, parasitic3 or bacterial diseases4. These flare-ups are commonly ascribed to general effects on the overall host immune status disrupting the host-pathogen equilibrium. Evidence is however growing that pathogens, such as Salmonella, can exploit the neuroendocrine alteration due to a stress reaction as a signal for growth and pathogenic processes4,5 and by doing so, promote pathogen survival through increased dispersal in the host or to novel hosts5,6.
Depending on the Salmonella strain and host, salmonellosis, one of the globally most important zoonotic bacterial diseases, can result in (self-limited) gastroenteritis, septicemia and even death. Worldwide, typhoidal7 and nontyphoidal8 Salmonella infections result in an estimated 20 million and 98.3 million human cases each year, of which 200, 000 and 155, 000 result in death. As such, Salmonella infections still pose major public health concerns worldwide. The pathogen is notorious for its ability to cause persistent infections both in humans, with the most notable example being typhoid Mary9, and a number of animal species, such as poultry and pigs4. During periods of host stress, Salmonella is able to sense stress hormones and to exploit their presence to its advantage, increasing the pathogen’s likeliness of dispersal and thus its chances of successful maintenance in the host population5. Both catecholamines10,11 and glucocorticoids5 have been suggested to contribute to stress-induced recrudescence of a Salmonella infection. Recently, we showed that glucocorticoids such as cortisol and dexamethasone, promote Salmonella proliferation inside porcine macrophages and Salmonella infections in pigs, representing a prime example for stress-induced flare-up of infections5.
Until now, the underlying mechanism of these stress-induced flare-ups is poorly understood. Therefore, the aim of the present study was to unravel the mechanism of how the infected macrophages respond to cortisol exposure and to identify Salmonella Typhimurium genes responsible for the cortisol-induced increased survival of the bacterium. Our study reveals that the scsA gene drives glucocorticoid-induced recrudescence of Salmonella inside host macrophages by altering bacterial virulence gene expression, which eventually results in recrudescence of infection.
Results and Discussion
Cortisol induces the macrophage cytoskeletal rearrangements that facilitate intracellular bacterial replication
We demonstrated that the cortisol-induced increased proliferation of Salmonella is due to glucocorticoid-induced host cytoskeletal changes that result in the formation of newly formed bacterial replicative niches12,13,14, the Salmonella-containing vacuoles (SCVs). During increased replication of Salmonella Typhimurium in cortisol exposed macrophages, the SCV was shown to divide together with the bacterium, resulting in a single bacterium per SCV (Fig. 1c). This process of increased SCV production required F-actin polymerization and microtubule formation (Fig. 1a) and did not coincide with obvious cortical actin redistribution (Supplementary Fig. S1). Proteomic and transcriptomic analyses confirmed the cortisol-induced increased expression of several cytoskeleton-associated proteins in Salmonella-infected macrophages, including tubulin beta chain (TUBB2A), F-actin capping protein subunit 2 (CAPZB), thymosin beta-4 (TMSB4X), actin-related protein 3B (ACT3), and tropomyosin 3 (TPM3) (Supplementary Table S1 and Supplementary Fig. S2). We thus conclude that cortisol induces cytoskeletal rearrangement activities in Salmonella-infected macrophages that underpin the formation of novel SCVs and thus allow increased bacterial replication.
Figure 1. Cytoskeletal rearrangements in porcine macrophages are required for increased intracellular proliferation of Salmonella.
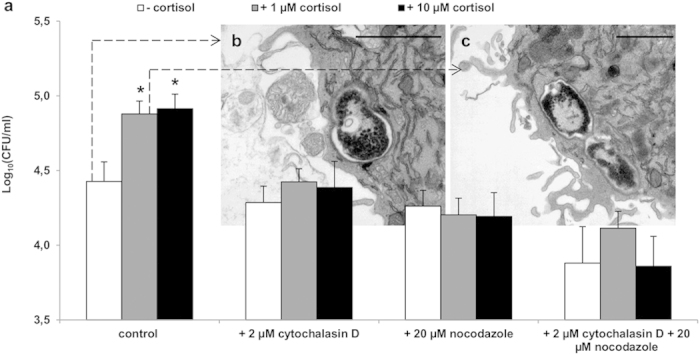
(a) Number of intracellular Salmonella Typhimurium bacteria in porcine macrophages that were treated with unsupplemented medium, 2 μM cytochalasin D (inhibitor of F-actin polymerization), 20 μM nocodazole (inhibitor for microtubule formation) or the combination of both, for 24 hours after invasion. White bars illustrate medium without cortisol, grey bars represent medium with 1 μM cortisol and black bars indicate medium with 10 μM cortisol. An asterisk (*) refers to a significant difference compared to the respective controls without cortisol (P ≤ 0.05). Panels (b,c) represent TEM images, 6 hours after control (b) or 1 μM cortisol (c) treatment of Salmonella Typhimurium WT-infected porcine macrophages, showing SCV formation (scale bar, 1 μm).
Cortisol increased intracellular replication is driven by scsA
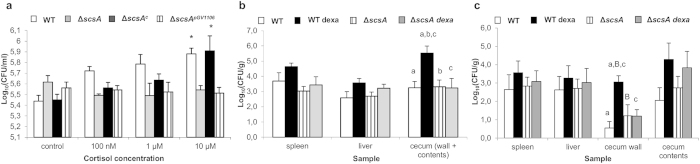
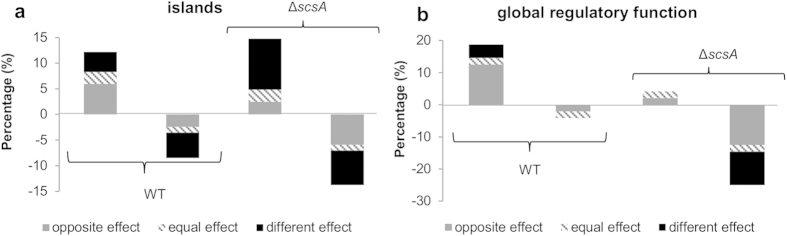
Using in vivo expression technology15 (IVET; Supplementary Table S2) and intracellular proliferation tests (Fig. 2a), we identified the Salmonella gene scsA as the key driver for cortisol-induced intracellular replication of Salmonella in porcine and murine macrophages (Fig. 2a and Supplementary Fig. S3a). Deletion of scsA abolished this increase in proliferation, an effect that was restored by the complementation of scsA (ΔscsAc) (Fig. 2a). We then explained the crucial contribution of the scsA gene by its key role in the cortisol-induced dramatic alteration of Salmonella gene expression under phagosome mimicking conditions (Supplementary Table S3 and Fig. 3). Indeed, under these conditions, cortisol altered the expression of more than 20% of Salmonella genes involved in global regulation and pathogenicity islands, both key groups for the bacterium’s virulence (KEGG database). Deletion of the scsA gene largely disturbed this cortisol-induced expression pattern (Fig. 3). The scsA gene was shown to directly or indirectly mediate the cortisol-induced increased expression of several major systems that are indispensable for intramacrophage survival: SPI-2 associated proteins (sscA chaperone, sseI effector and ssaG structural protein); phoP, belonging to the PhoQ/PhoP two-component regulatory system16; and the sigma factors σS (rpoS) and σE (rpoE)17,18. The involvement of scsA in the cytoskeletal rearrangements, required for intracellular replication, was shown by its impact on the expression of Salmonella genes involved in host cytoskeletal reformations (sipC)19 and SCV/Salmonella-induced filaments formation (pipB)20 (Supplementary Table S3) and by its impact on the expression of the host actin-related protein ACT3 (Supplementary Fig. S2).
Figure 2. ScsA mediates cortisol-induced increase in proliferation of Salmonella, both in vitro and in vivo.
(a) Shown is the effect of cortisol on the log10 values + standard deviation of intracellular Salmonella Typhimurium WT, ΔscsA, ΔscsAc and ΔscsApGV1106 bacteria in porcine macrophages. An asterisk (*) refers to a significant difference compared to the condition without cortisol (P ≤ 0.05). Panels (b,c) illustrate the effect of dexamethasone exposure on the recovery of Salmonella WT and ΔscsA from organs of DBA/2J mice. The log10 value of the CFU/gram sample is given as the mean + standard error of the mean. Significant differences are signed with a, b, c (P ≤ 0.017) and a tendency with B (P ≤ 0.033).
Figure 3. Cortisol effect on gene expression during intracellular environment mimicking conditions.
For both WT and ΔscsA strain, the percentage of genes up or down regulated by cortisol is shown quantitatively compared to the total number of genes belonging to the (a) islands (894 genes) or (b) regulatory function (48 genes) group. Grey bars represent the percentage of genes of which the cortisol effect in WT is the opposite in ΔscsA strain. Striped bars reflect the genes that act similar after cortisol treatment. Black bars depict the genes that are up or down regulated after cortisol treatment in WT but not in ΔscsA, or vice versa. Microarray data have been deposited in the Gene Expression Omnibus at NCBI with series accession numbers GSE55430.
ScsA mediates the glucocorticoid-induced increase of macrophage-associated Salmonella loads in the cecal wall of DBA/2J mice
We then demonstrated the crucial role of both scsA and macrophages in glucocorticoid-driven Salmonella Typhimurium recrudescence in a mouse model of infection. Previously, it was shown that a single dexamethasone stimulus can be used to mimic stress-related flare-up of a Salmonella infection in pigs5. We now first optimized a model in DBA/2J mice to reproduce dexamethasone-induced Salmonella proliferation. As shown in Supplementary Fig. S4, organs of Salmonella-infected DBA/2J mice that were subsequently injected with dexamethasone showed significantly higher numbers of Salmonella Typhimurium bacteria, compared to DBA/2J mice that were injected with a saline solution. In BALB/c mice, no significant differences were observed in the Salmonella load between dexamethasone and HBSS-injected animals.
Using this DBA/2J model, we then demonstrated that scsA is required for the glucocorticoid-induced increase in Salmonella infection load in the murine cecum (wall + contents) in vivo (Fig. 2b). In a second experiment, we showed that the scsA-dependent increase of intestinal Salmonella Typhimurium loads coincides with higher numbers of Salmonella Typhimurium positive macrophages in the cecal wall, but not with intraluminal proliferation of Salmonella Typhimurium bacteria (Figs 2c, 4, 5 and 6), which is in agreement with macrophages being the main host cells for Salmonella in the murine cecum21. No such phenomenon was observed in spleen and liver, strengthening the hypothesis that the scsA-dependent increase of Salmonella is tissue specific and limited to the intestinal tract. Previous published work by Van Parys et al. (2011) indeed showed that scsA gene expression is relevant in the gut wall, but not in the tonsils and ileocaecal lymph nodes of pigs15. Moreover, our findings are in agreement with previously published work5 in which glucocorticoids significantly increased the number of Salmonella bacteria in the intestinal wall (ileum, cecum and colon) of pigs. Using DBA/2J mice, which are considered intermediately susceptible to Salmonella Typhimurium infections, we were able to mimic stress-related increased replication of Salmonella Typhimurium in a persistently infected host. The infection dynamics in this model are similar to the course of a Salmonella infection in pigs, which is characterized by an acute phase around day 3–5 post infection (p.i.) and the development of a chronic infection once the acute phase is controlled22. As such, our results demonstrate a novel bacterial mechanism that senses host stress, resulting in a flare up of bacterial loads, being of great importance for food-producing animals like pigs. From an epidemiological point of view, this is highly relevant to promote sustaining the infection at population level.
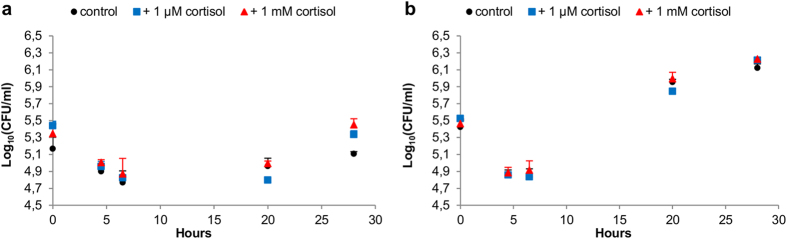
Figure 4. Effect of cortisol on the growth of Salmonella Typhimurium in cecal contents.
The log10 values of the CFU/ml + standard deviation are given at different time points (t = 0, 4.5, 6.5, 20, 28 hours under microaerobic conditions at 37 °C) after inoculation in cecal contents. Salmonella Typhimurium (a) WT and (b) ΔscsA growth was examined in cecal contents with (1 μM or 1 mM) or without cortisol.
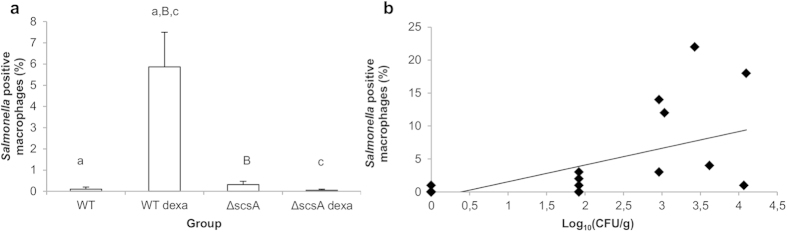
Figure 5. Effect of glucocorticoids on the number of Salmonella positive macrophages.
Panel (a) shows the effect of dexamethasone exposure on the colocalization of Salmonella WT and ΔscsA with F4/80+ cells in the cecal wall of DBA/2J mice. Given is the percentage of the Salmonella positive macrophages + standard error of the mean compared to the total number of counted macrophages. Significant differences are signed with a, c (P ≤ 0.017) and a tendency with B (P ≤ 0.033). Panel (b) indicates a significant (P ≤ 0.05) positive correlation between the percentage of Salmonella positive macrophages per mouse and their respective log10 value of the CFU/gram cecal wall (correlation coefficient = 0.62).
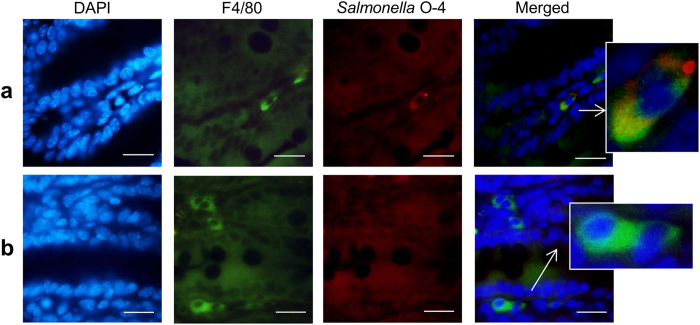
Figure 6. Salmonella colocalization with F4/80+ cells in the cecum.
Cecal sections of Salmonella-infected DBA/2J mice were stained for immunofluorescence using an F4/80 antibody targeting macrophages (green) and a Salmonella O–4 antibody (red). Nuclei were stained with DAPI (blue). Shown are representative images demonstrating (a) colocalization of Salmonella and F4/80+ cells or (b) Salmonella negative F4/80+ cells (scale bar, 20 μm).
ScsA is involved in scsBCD regulation
The scsA gene is part of a scs locus, consisting of two operons, a first one consisting of a single gene (scsA) and a second operon consisting of three genes (scsB-C-D)23. The scs genes encode for proteins with a predicted Cys-X-X-Cys motif and an enriched containment of hydrophobic amino acids between the Cys residues, being characteristic for the thioredoxin superfamily24. The Scs proteins also show homology with Dsb proteins of Escherichia coli, being disulfide bond formation proteins that are involved in the correct folding of target proteins by catalyzing disulfide bond isomerization25. Many virulence factors are secretion proteins that are posttranslationally modified via disulfide bond formation and inactivation of genes involved in the Dsb system leads to reduced virulence in many pathogens25. Recently, the role of ScsB as an electron transporter was confirmed and ScsB proteins were described as a new class of DsbD proteins26. Therefore, we hypothesize that the scsA gene is also involved in the formation of disulfide bonds and correct folding of proteins. Using microarray analysis, we showed that deletion of the scsA gene results in the upregulation of the scsBCD operon (Supplementary Table S4), suggesting that scsA acts as a regulator for the scsBCD operon. Gupta et al. (1997) stated that expression of both operons is required for the correct functioning of the scs locus. As such, deletion of scsA probably results in the incomplete and impaired operation of this locus, which could lead to impaired virulence.
We conclude that the scsA gene-driven glucocorticoid-induced proliferation of Salmonella inside host macrophages results in increased intestinal infection loads that facilitate dispersal to novel hosts. These results oppose the general idea that stress-associated exacerbation of infectious diseases merely is the result of lowered general host immune status. Rather, the acquisition of a dedicated virulence mechanism to increase chances of escape from a stressed and possibly less fit host should be considered an evolutionary advantage, which promotes pathogen maintenance in the host population.
Methods
Bacterial strains
Salmonella Typhimurium strain 112910a was used as the wild type strain (WT), in which the spontaneous nalidixic acid resistant derivative strain (WT nal) was constructed27. Salmonella Typhimurium ΔscsA deletion mutant was constructed according to the one-step inactivation method described by Datsenko and Wanner28 and slightly modified for use in Salmonella Typhimurium as described previously29. Gene complementation of the deletion mutant ΔscsA was constructed using the vector plasmid pGV110630. The resulting complemented deletion mutant is further abbreviated as ΔscsAc. As a control, we electroporated the empty pGV1106 plasmid into the deletion mutant, hereafter named ΔscsApGV1106. Primers used to create the gene-specific linear PCR fragment for gene deletion (scsA forward and reverse) and complementation (scsAc forward and reverse) are given in Supplementary Table S5. Salmonella Typhimurium WT and ΔscsA expressing green fluorescent protein (GFP) were used for confocal laser scanning microscopy (CLSM).
Infection experiments in cell cultures
Porcine alveolar macrophages (PAM) were isolated and cultured as previously described5. PAM were seeded in 175 cm2 cell culture flasks (5 × 107 cells) for the proteome analysis, in 24-well plates (1 × 106 cells) for the invasion and proliferation assays, on a cover slip (1 × 106 cells) for transmission electron microscopy (TEM) and CLSM, and in 6-well plates (3 × 106 cells) for the IVET experiment and RNA extraction. The cells were infected with Salmonella Typhimurium at a multiplicity of infection (MOI) of 10:1, with the exception for TEM and IVET (50:1). To synchronize the infection, the inoculated plates or flasks were centrifuged at 365 × g for 10 min and incubated for 30 min at 37 °C under 5% CO2. After washing the cells, extracellular bacteria were treated with gentamicin (100 μg/ml) for 1 hour. To assess the intracellular proliferation of Salmonella, the medium containing 100 μg/ml gentamicin was replaced after 1 hour incubation with fresh medium containing 20 μg/ml gentamicin, supplemented with cortisol when required.
Proteome analysis
To map out the effects of cortisol-induced increased survival of Salmonella Typhimurium on the protein expression of host cells, iTRAQ analysis was performed. Twenty-four hours after infection, protein extraction, digest, labeling, nano LC-MSMS analysis and data analysis of the samples, were performed as previously described31. The experiment was conducted in duplicate and the labeling of the samples was as follows: run 1 (untreated PAM sample 1: 114 – untreated PAM sample 2: 115 – cortisol treated PAM sample 1: 116 – cortisol treated PAM sample 2: 117) – run 2 (untreated PAM sample 3: 114 – untreated PAM sample 4: 115 – cortisol treated PAM sample 3: 116 – cortisol treated PAM sample 4: 117).
Role of F-actin and microtubuli
To assess the intracellular proliferation of Salmonella, the medium containing 100 μg/ml gentamicin was replaced after 1 hour incubation with fresh medium containing 20 μg/ml gentamicin, with or without 1 μM or 10 μM cortisol, 2 μM cytochalasin D (inhibitor of F-actin polymerization) and/or 20 μM nocodazole (inhibitor for microtubule formation). Twenty-four hours after infection, the number of viable bacteria was determined by plating 10-fold dilutions on brilliant green agar (BGA).
TEM
PAM cells previously infected with Salmonella Typhimurium parental strain (in the absence or presence of 1 μM cortisol) were analyzed at 2, 6 and 16 hours after infection. TEM was performed with glutaraldehyde fixation in 0.05 M sodium cacodylate buffer, 1% osmium tetroxide postfixation, and en bloc staining for 1 h in a 1% solution of uranyl acetate.
CLSM
Sixteen hours after infection and cortisol treatment, PAM were fixed, permeabilized and stained with phalloidin-Texas Red as previously described32. Nuclei were stained with 4′,6′-diamidino-2-phenylindole (DAPI) (Vector).
IVET screening
An IVET transformants pool was used, covering the major part of the Salmonella Typhimurium genome, to identify Salmonella Typhimurium genes that are intracellularly expressed in PAM after exposure to cortisol. The IVET pool was constructed by the use of a pIVET1 plasmid as previously explained15. Shortly, the pIVET1 plasmid is a derivate of the suicide vector pGP704 and contains a promoterless synthetic operon of purA coupled to lacZY, preceded by a BglII site. Salmonella Typhimurium genomic DNA was purified and subsequently digested with Sau3AI, resulting in a library of 1–4 kb overlapping genomic DNA fragments. These fragments were cloned in the BglII site of the pIVET1 plasmid, upstream to promoterless wild type copies of purA and lacZY. After transferring the plasmids to Escherichia coli DH5αpir, approximately 100 000 clones were pooled. Using a Plasmid Midi Kit (Qiagen), the pIVET1 fusion plasmids were isolated and electroporated in the conjugative E. coli SM10 λpir and again, approximately 1,00, 000 different E. coli SM10 λpir clones were pooled. These fusion plasmids were then mobilized into a Salmonella Typhimurium ΔpurA strain by conjugation using the E. coli SM10 λpir as donor strain. PurA encodes for adenylosuccinate synthetase, an enzyme which is essential for the synthesis of adenosine monophosphate. The wild type locus of the gene was not disrupted since the integration of the pIVET1 plasmid in the chromosome resulted in a single cross over. Finally, Salmonella Typhimurium ΔpurA transformants were obtained, in which te native promoter drives the expression of the purA-lacZY fusion, while the cloned promoter drives the expression of the wild type gene. The Salmonella Typhimurium ΔpurA strain is severely attenuated in pigs15 and in macrophages (Supplementary Fig. S3b). This means that if the cloned DNA contains a promoter which is activated intracellularly in macrophages, the purA gene and the lacZY operon will be expressed and the bacterium will survive. If the cloned DNA fragment does not contains a promoter, or a promoter that is not activated intracellularly, no adenosine monophosphate will be synthsized and the bacterium will be severely attenuated.
The Salmonella Typhimurim IVET pool was inoculated into PAM and the infected cells were treated with medium with or without 1 μM cortisol. Sixteen hours after infection, PAM were washed and lysed for 10 min with 500 μl 1% Triton X-100. This was added to 9.5 ml of LB broth enriched with the additives, 50 μg/ml ampicillin, 20 μg/ml nalidixic acid, 1.35% adenine and 0.337% thiamine and grown with aeration at 37 °C. After 3 hours, the bacterial culture was centrifuged at 2300 x g for 10 min at 37 °C and the pellet was resuspended in 3 ml PAM medium without antibiotics. This was considered as one passage and in total three passages were performed. Finally, the cells were lysed and plated on MacConkey agar supplemented with the additives and 1% filter-sterilized lactose to assess the lacZY expression level of the IVET transformants. This allowed detection of IVET transformants containing promoters expressed intracellularly in PAM but not on MacConkey agar. These fusion strains formed white to pink colonies on MacConkey lactose agar (low-level lacZY expression), whereas fusion strains containing promoters that are constitutively expressed showed red colonies (high-level lacZY expression). As we were interested in genes that are intracellularly induced, but not extracellularly, all the colonies showing low-level lacZY expression were picked up, purified and sequenced as previously described15.
Invasion and proliferation assays
Based on literature and IVET screening, the effect of cortisol on the intracellular proliferation of ΔscsA and ΔscsAc was determined in comparison to the WT strain. Since Salmonella Typhimurium ΔscsAc is resistant to kanamycin, polymyxin B was used instead of the commonly used gentamicin to kill the extracellular Salmonella bacteria. The medium containing 5 μg/ml polymyxin B was replaced after 1 hour incubation with fresh medium containing 5 μg/ml polymyxin B with or without cortisol ranging from 0,1–10 μM. After 24 hours, the cells were washed 3 times, lysed and 10-fold dilutions were plated on BGA plates.
Microarray analysis
The effect of cortisol exposure (1 μM) on Salmonella gene expression was analyzed using RNA isolated from parental and ΔscsA strains grown to logarithmic phase in a medium that reflects the intracellular environment. The Salmonella strains were grown to logarithmic phase in a low pH minimal medium (MM 5.8), which was slightly modified to achieve a maximal SPI-2 expression, as previously described33. The analysis was performed as previously described5.
Analysis of eukaryotic mRNA expression
Sixteen hours after cortisol treatment, total RNA was isolated using TRI Reagent® combined with the RNeasy mini kit (Qiagen) according to the manufacturer’s guidelines. RNA concentration was measured by absorbance at 260 nm using Nanodrop spectrophotometer and purity of the RNA samples was checked using an Experion RNA StdSens Analysis kit. Total RNA (500 ng) was reverse transcribed to cDNA with the iScript cDNA synthesis kit34. Primers used for the amplification were designed using Primer3 software. Specificity of the primers was tested by performing a BLAST search against the genomic NCBI database. The list of genes and sequences of the primers used for quantitative PCR analysis can be found in Supplementary Table S5. HIS34 and BLM35 were used as housekeeping genes. All reference genes had a stable expression, in all the samples tested, as calculated using the geNorm software. Real-time quantitative PCR reactions were performed as earlier described34. For each condition, RNA was extracted from three replicates and data were collected from two independent experiments. Real-time quantitative PCR reactions were run in duplicate.
Ethics statement
All animal experiments were carried out in strict accordance with the recommendation in the European Convention for the Protection of Vertebrate Animals used for Experimental and other Scientific Purposes. The experimental protocols and care of the animals were approved by the Ethical Committee of the Faculty of Veterinary Medicine, Ghent University (EC2011/116 and EC2012/160).
Optimization of a stress mimicking mouse model
The scsA-dependent increased intracellular replication of Salmonella Typhimurium was also observed in murine macrophages (RAW cells; Supplementary Fig. S3a). Therefore, a mouse model was developed that mimicks stress-related increased replication of Salmonella Typhimurium. It was evaluated whether dexamethasone increases the number of Salmonella Typhimurium bacteria in the gut of Salmonella-infected mice, in order to create a mouse model allowing screening of bacterial genes that might be involved in dexamethasone-induced enhanced replication of Salmonella. Depending on the bacterial strain and host, a Salmonella infection can vary from a (self-limited) gastrointestinal infection, to septicemia and even death. To mimick these differences in disease outcome, we tested the effects of glucocorticoids in two different mouse models. DBA/2J mice are intermediately sensitive to Salmonella Typhimurium infections, resulting in carrier animals, whereas BALB/c mice are highly susceptible to Salmonella Typhimurium infections36. Eighteen, four-week-old DBA/2J mice and eighteen, four-week-old BALB/c mice, were housed in filter-topped cages at 25 °C under natural day-night rhythm with ad libitum access to feed and water and enriched with mouse houses and play tunnels. Five days after arrival, all mice were infected with a total of 1 × 106 CFU of Salmonella Typhimurium WT nal by the orogastric route. At day 7 p.i. six BALB/c mice were subcutaneously injected once with 100 mg/kg dexamethasone. Simultaneously, six BALB/c mice received a subcutaneous injection of 25 mg/kg dexamethasone, which was repeated after three hours. Fourteen days p.i. six DBA/2J mice were subcutaneously injected once with 100 mg/kg dexamethasone and simultaneously six DBA/2J mice received a subcutaneous injection of 25 mg/kg dexamethasone (repeated after three hours). Six mice of each strain received a subcutaneous injection of 200 μl HBSS- (24 hours before euthanasia) and were used as control group. Twenty-four hours after the last injection of dexamethasone, all animals were humanely euthanized and samples of spleen, liver and cecum were collected. Samples were processed and the number of Salmonella bacteria was determined as previously described5. The detection limit for direct plating was 83 CFU/gram tissue or contents.
Role of scsA during the glucocorticoid response of Salmonella in vivo
The DBA/2J mouse model was used to verify whether scsA plays an essential part in the glucocorticoid-related multiplication in vivo. Three- to four-week-old DBA/2J mice were used and randomly allocated in two groups of sixteen mice. The animals were housed in filter-topped cages at 25 °C under natural day-night rhythm with ad libitum access to feed and water and enriched with mouse houses and play tunnels. After an acclimatization period of one week, mice were inoculated with a total of 1 × 106 CFU of Salmonella Typhimurium WT nal or its isogenic scsA knock-out mutant. At day 14 p.i., eight animals of each group were subcutaneously injected with 100 mg/kg dexamethasone and eight mice were subcutaneously injected with 200 μl HBSS- and served as a control group. Twenty-four hours later, all mice were humanely euthanized. Spleen, liver and cecum (wall + contents) samples were examined for the number of Salmonella Typhimurium bacteria. Samples were processed and the number of Salmonella bacteria was determined as previously described5. The detection limit for direct plating was 83 CFU/gram tissue or contents.
Contribution of scsA to glucocorticoid-dependent intestinal Salmonella loads in vivo
The in vivo experiment described above, showed that Salmonella responds to glucocorticoids in an scsA-dependent way, with increased intestinal loads as a result. In this experiment we investigated whether this results from an increased number of intramural Salmonella containing macrophages, or increased extracellular proliferation in the intestinal contents. Therefore, 35 four-week-old DBA/2J mice were randomly divided in 5 groups of 7 mice. Per group, the animals were distributed over 3 filter-topped cages. One cage contained 3 mice and the two other cages comprised 2 mice. After an acclimatization period, one group was inoculated with HBSS- and served as a negative control group, two groups were inoculated with 1 × 106 CFU of Salmonella Typhimurium WTnal and two groups were infected with 1 × 106 CFU of Salmonella Typhimurium ΔscsA. At day 14 p.i., three mice of the control group, seven Salmonella Typhimurium WTnal- and seven Salmonella Typhimurium ΔscsA-infected mice were subcutaneously injected with 100 mg/kg dexamethasone. The other mice were subcutaneously injected with 200 μl HBSS-. Twenty-four hours later, all mice were humanely euthanized. Spleen, liver, cecum and cecum contents were examined for the number of Salmonella Typhimurium bacteria. Samples were processed and the number of Salmonella bacteria was determined as previously described5. The detection limit for direct plating was 83 CFU/gram tissue or contents. Samples of the cecum were fixed in 4% phosphate buffered formaldehyde for immunofluorescence. They were processed by standard methods, and embedded in paraffin.
Immunofluorescence of cecal sections
Cecal sections were de-paraffinized in xylene and hydrated through a graded series of alcohols. Heat-mediated antigen retrieval using citrate buffer (pH 6.0) was included to achieve optimum staining. After washing three times for 5 minutes with phosphate-buffered saline (PBS), the samples were incubated for 15 minutes with PBS supplemented with 0.3% Triton and 2% goat serum, washed three times with PBS and incubated with PBS supplemented with 10% goat serum, for 30 minutes. To prevent non-specific binding, the sections were subsequently incubated for 2 hours with an unconjugated Fab fragment goat anti-mouse IgG (200 μg/ml; Jackson ImmunoResearch Labs). After washing three times for 20 minutes, the samples were incubated for 90 minutes with a rabbit polyclonal antibody raised against a peptide mapping at the C-terminus of F4/80 of mouse origin (1/400 dilution; Santa Cruz biotechnology) and a mouse monoclonal antibody targeting the O–4 antigen of Salmonella Typhimurium (1/1000, Santa Cruz biotechnology). After washing three times for 20 minutes, the samples were incubated with a goat anti-rabbit Alexa Fluor 488 (1/300; Life Technologies) targeting the F4/80 antibody, together with an Alexa Fluor 568 goat anti-mouse IgG1 (1/500; Life Technologies) targeting the Salmonella antibody. After an incubation of 1 hour, the sections were washed and incubated with DAPI. Finally the sections were mounted using ProLong Gold antifade mountant (Life Technologies). Salmonella containing cells were quantified blinded and visually by counting the number of F4/80 positive cells that co-localized with Salmonella. Per animal, three different sections were analyzed and per section, ten different areas were examined at a magnification of 400 X.
Salmonella growth in cecal contents
The cecal contents of three seven-week-old DBA/2J mice was collected and ten times diluted in aqua dest. To remove larger particles, the suspension was filtered using a cell strainer (70 μm). Salmonella Typhimurium WT or ΔscsA bacteria were added to the cecal suspension (approximately 104 CFU/ml) supplemented with HBSS-, 1 μM or 1 mM cortisol. Growth of Salmonella was examined at different time points (0, 4.5, 6.5, 20, 28 hours under microaerobic conditions at 37 °C) by plating ten-fold dilutions on BGA plates. This experiment was conducted in twofold.
Statistical Analysis
All in vitro experiments were conducted at least in triplicate with 3 repeats per experiment, unless otherwise stated. All statistical analyses were performed using SPSS version 22 (SPSS Inc., Chicago, IL, USA). Normally distributed data were analyzed using unpaired Student’s t-test or one-way ANOVA to address the significance of difference between mean values with significance set at P ≤ 0.05. Bonferroni as post hoc test was used when equal variances were assessed. If equal variances were not assessed or if the data were not normally distributed, they were analyzed using the non parametric Kruskal-Wallis analysis, followed by a Bonferroni-corrected Mann-Whitney U test.
Additional Information
Accession Number: Microarray data have been deposited in the Gene Expression Omnibus at NCBI with series accession numbers GSE30924 and GSE55430
How to cite this article: Verbrugghe, E. et al. Host Stress Drives Salmonella Recrudescence. Sci. Rep. 6, 20849; doi: 10.1038/srep20849 (2016).
Supplementary Material
Acknowledgments
This work was supported by the Institute for the Promotion of Innovation by Science and Technology in Flanders [IWT Landbouw 70574] and the Research Foundation-Flanders [3G008012]. The technical assistance of Nathalie Van Rysselberghe, Anja Van den Bussche, Myriam Claeys, Christian Puttevils, Delphine Ameye and Sarah Loomans is greatly appreciated. The pGV1106 plasmid was a kind gift of Prof. Dr. J.P. Hernalsteens and Prof. Dr. H. Degreve.
Footnotes
Author Contributions E.V., A.T. and N.S. performed the microarray analysis. E.V. and W.B. carried out the TEM analyses. E.V., M.D. and D.D. performed the iTRAQ analysis. E.V., A.V.P., R. H. and B.L. executed the animal and in vitro infection experiments. E.V., F.B., F.H., H. F. and F.P. conceived the study, participated in its design and coordination. E.V. and F.P. co-drafted the manuscript. All authors read and approved the final manuscript.
References
- Cannon W. Wisdom Of The Body. United States: W.W. Norton & Company. ISBN 0393002055 (1932). [Google Scholar]
- Rozlog L. A., Kiecolt-Glaser J. K., Marucha P. T., Sheridan J. F. & Glaser R. Stress and immunity: implications for viral disease and wound healing. J. Periodontol. 70, 786–92 (1999). [DOI] [PubMed] [Google Scholar]
- Oppliger A. et al. Environmental stress increases the prevalence and intensity of blood parasite infection in the common lizard Lacerta vivipara. Ecol. Let. 1, 129–138 (1998). [Google Scholar]
- Verbrugghe E. et al. The complex interplay between stress and bacterial infections in animals. Vet. Microbiol. 155, 115–27 (2012). [DOI] [PubMed] [Google Scholar]
- Verbrugghe E. et al. Stress induced Salmonella Typhimurium recrudescence in pigs coincides with cortisol induced increased intracellular proliferation in macrophages. Vet. Res. 42, 118 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanter B. A., Sauer K. & Davies D. G. Bacteria present in carotid arterial plaques are found as biofilm deposits which may contribute to enhanced risk of plaque rupture. mBio 5, e01206 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crump J. A., Luby S. P. & Mintz E. D. The global burden of typhoid fever. Bull. World Health Organ. 82, 346–53 (2004). [PMC free article] [PubMed] [Google Scholar]
- Majowicz S. E. et al. The global burden of nontyphoidal Salmonella gastroenteritis. Clin. Infect. Dis. 50, 882–889 (2010). [DOI] [PubMed] [Google Scholar]
- Soper G. A. The work of a chronic typhoid germ distributor. J. Am. Med. Assoc. 48, 2019–22 (1907). [Google Scholar]
- Bearson B. L. & Bearson S. M. The role of the QseC quorum-sensing sensor kinase in colonization and norepinephrine-enhanced motility of Salmonella enterica serovar Typhimurium. Microb. Pathog. 44, 271–278 (2008). [DOI] [PubMed] [Google Scholar]
- Bearson B. L., Bearson S. M. D., Uthe J. J. & Dowd S. E. Iron regulated genes of Salmonella enterica serovar Typhimuirum in response to norepinephrine and the requirement of fepDGC for norepinephrine-enhanced growth. Microb. Infect. 10, 807–816 (2008). [DOI] [PubMed] [Google Scholar]
- Guignot J. et al. Microtubule motors control membrane dynamics of Salmonella-containing vacuoles. J. Cell. Sci. 117, 1033–1045 (2004). [DOI] [PubMed] [Google Scholar]
- Guiney D. G. & Lesnick M. Targeting of the actin cytoskeleton during infection by Salmonella strains. Clin. Immunol. 114, 248–255 (2005). [DOI] [PubMed] [Google Scholar]
- Henry T., Gorvel J. P. & Méresse S. Molecular motors hijacking by intracellular pathogens. Cell. Microbiol. 8, 23–32 (2006). [DOI] [PubMed] [Google Scholar]
- Van Parys A. et al. Tissue-specific Salmonella Typhimurium gene expression during persistence in pigs. PLoS One 6, e24120 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groisman E. A. The pleiotropic two-component regulatory system PhoP-PhoQ. J. Bacteriol. 183, 1835 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Testerman T. et al. The alternative sigma factor σE control antioxidant defences required for Salmonella virulence and stationary-phase survival. Mol. Microbiol. 43, 771–782 (2002). [DOI] [PubMed] [Google Scholar]
- Skovierova H. et al. Identification of the σE regulon of Salmonella enterica serovar Typhimurium. Microbiol. 152, 1347–1359 (2006). [DOI] [PubMed] [Google Scholar]
- Hayward R. D. & Koronakis V. Direct nucleation and bundling of actin by the SipC protein of invasive Salmonella. EMBO J. 18, 4926–4934 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knodler L. A. et al. Salmonella effectors within a single pathogenicity island are differentially expressed and translocated by separate type III secretion systems. Mol. Microbiol. 43, 1089–1103 (2002). [DOI] [PubMed] [Google Scholar]
- Lai M. A. et al. Innate immune detection of flagellin positively and negatively regulates Salmonlla infection. PLos One 8, e72047 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyen F. et al. The fibronectin binding protein ShdA is not a prerequisite for long term faecal shedding of Salmonella Typhimurium in pigs. Vet. Mic. 115, 284–290 (2006). [DOI] [PubMed] [Google Scholar]
- Gupta S. D., Wu H. C. & Rick P. D. A Salmonella Typhimurium genetic locus which confers copper tolerance on copper-sensitive mutants of Escherichia coli. J. Bacteriol. 179, 4977–4984 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anwar N., Sem X. H. & Rhen M. Oxidoreductases that act as conditional virulence suppressors in Salmonella enterica serovar Typhimurium. PLoS One 8, e64948 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Łasica A. M. & Jagusztyn-Krynicka E. K. The role of Dsb proteins of Gram-negative bacteria in the process of pathogenesis. FEMS Microbiol. Rev. 31, 626–636 (2007). [DOI] [PubMed] [Google Scholar]
- Cho S. H. et al. A new family of membrane electron transporters and its substrates, including a new cell envelope peroxiredoxin, reveal a broadened reductive capacity of the oxidative bacterial cell envelope. mBio 3, e00291–11 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyen F. et al. Limited role for SsrA/B in persistent Salmonella Typhimurium infections in pigs. Vet. Mic. 128, 364–373 (2008). [DOI] [PubMed] [Google Scholar]
- Datsenko K. A. & Wanner B. L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97, 6640–6645 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyen F. et al. Role of SPI-1 in the interactions of Salmonella Typhimurium with porcine macrophages. Vet. Mic. 113, 35–44 (2006). [DOI] [PubMed] [Google Scholar]
- Leemans J. et al. Broad–host-range cloning vectors derived from the W-plasmid Sa. Gene 19, 361–364 (1982). [DOI] [PubMed] [Google Scholar]
- Verbrugghe E. et al. T-2 toxin induced Salmonella Typhimurium intoxication results in decreased Salmonella numbers in the cecum contents of pigs, despite marked effect on Salmonella-host cell interactions. Vet. Res. 43, 22 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenbroucke V. et al. The mycotoxin deoxynivalenol promotes uptake of Salmonella Typhimurium in porcine macrophages, associated with ERK1/2 induced cytoskeleton reorganization. Vet. Res. 40, 64 (2009). [DOI] [PubMed] [Google Scholar]
- Knox L. F., Wosten M. M. & Groisman E. A. A small protein that mediates the activation of a two-component system by another two-component system. EMBO J. 19, 1861–1872 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenbroucke V. et al. The mycotoxin deoxynivalenol potentiates intestinal inflammation by Salmonella Typhimurium in porcine ileal loops. PLoS One 6, e23871 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uddin M. J. et al. Age-related changes in relative expression stability of commonly used housekeeping genes in selected porcine tissues. BMC Res. Notes 4, 441 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hormaeche C. E. Natural resistance to Salmonella Typhimurium in different inbred mouse strains. Immunology 37, 311–318 (1979). [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.