Abstract
This study investigates the temporal and spatial interchange of the aromatic amino acid phenylalanine (Phe) between human retinal pigment epithelial cell line (ARPE-19) and tachyzoites of the apicomplexan protozoan parasite Toxoplasma gondii (T. gondii). Stable isotope labelling by amino acids in cell culture (SILAC) is combined with Raman micro-spectroscopy to selectively monitor the incorporation of deuterium-labelled Phe into proteins in individual live tachyzoites. Our results show a very rapid uptake of l-Phe(D8) by the intracellular growing parasite. T. gondii tachyzoites are capable of extracting l-Phe(D8) from host cells as soon as it invades the cell. l-Phe(D8) from the host cell completely replaces the l-Phe within T. gondii tachyzoites 7–9 hours after infection. A quantitative model based on Raman spectra allowed an estimation of the exchange rate of Phe as 0.5–1.6 × 104 molecules/s. On the other hand, extracellular tachyzoites were not able to consume l-Phe(D8) after 24 hours of infection. These findings further our understanding of the amino acid trafficking between host cells and this strictly intracellular parasite. In particular, this study highlights new aspects of the metabolism of amino acid Phe operative during the interaction between T. gondii and its host cell.
Toxoplasma gondii is the most prevalent protozoal infection of the Central Nervous System. It causes a spectrum of human and animal diseases ranging from abortion to neurological disorders1. This intracellular apicomplexan protozoan parasite has the capacity to infect all vertebrate species within virtually any phagocytic and nonphagocytic cell type. It survives within its own membrane-bound compartment referred to as the parasitophorous vacuole (PV)2 The broad range of cells amenable to infection by T. gondii reflects the plasticity of the parasite’s metabolism and versatility in scavenging nutrients to support its intracellular growth3,4. Nutrient acquisition from the host is a strategic utilization of essential nutrients, such as amino acids (AA), lipids, and glucose to support the parasite’s metabolic requirements. T. gondii and other apicomplexans are fast-replicating organisms and thus are critically dependent on host-cell nutrients as carbon and energy sources for the extensive membrane biogenesis required during their replication5.
Even though T. gondii can synthesise some AA de novo, this parasite lacks synthetic pathways for some AA, which must be retrieved from the host cells6,7,8,9. Therefore, the pools of amino acids in the host’s cytosol constitute an important reservoir to complement the endogenous production of AAs in the parasite. For example, the replication rate of T. gondii is proportional to the concentration of tryptophan in the host cell10,11.
Phenylalanine (Phe) is one of the most vital AA needed by both mammalian cells and T. gondii. The T. gondii genome encodes hydroxylase enzymes, which can metabolize both Phe and Tyrosine (Tyr) to l-DOPA, with a substrate preference for Tyr12. However, little is known regarding the transport of Phe from mammalian cell hosts to the parasite, the formation of precursor pool, protein synthesis, or protein turnover within the parasite. Unravelling these complex processes requires novel approaches that are capable of monitoring the shuttling of AA from or toward the host during T. gondii infection.
Isotopically labelled AA tracer is frequently used to trace the kinetics of incorporation of a labelled AA into newly synthesized proteins. It can also be used to characterize the acquisition of AA by the growing parasite. Various isotope labelling approaches exist for studying protein dynamics and turnover. These methods include stable isotope labelling by amino acids in cell culture (SILAC), chemical derivatization (GC-MS, ICAT, iTRAQ, TMT), and enzymatically catalyzed incorporation (18O labelling). However, these approaches are invasive, cost- and/or time-prohibitive, and might not be amenable to the biological system of interest. Also, these methods cannot elucidate the kinetic events that occur rapidly and change from cell to cell. Thus, these techniques alone cannot quantitatively describe the complex dynamic interactions between engaging host and parasite systems.
Raman micro-spectroscopy (RMS) is a non-invasive, label-free imaging technique that can enable non-invasive molecular analysis of dynamic events in live cells in-vitro13. Its utility has been confirmed in studies covering a wide spectrum of applications, including cellular differentiation14,15,16, apoptosis17,18,19, drug-cell interactions20. Also, previous studies have demonstrated the ability of RMS to detect molecular changes during parasite development within host cells21. However, this approach could not provide enough specificity to reveal which molecules (e.g., nucleotides, glucose, lipids or amino acids) are altered during host-T. gondii interaction.
Combining stable-isotope labelling with RMS can increase the chemical specificity of Raman bands, and identify the origin of the molecular species and monitor its metabolism22,23,24,25,26,27. In this study, we show for the first time that it is possible to combine time-resolved RMS and stable-isotope labelling approach to monitor the exchange of molecules between individual cells in real-time. We monitor the kinetic of labelled Phe acquisition by T. gondii from the host cell during the first 48 hours of the parasite’s intracellular development. Identification of T. gondii metabolic requirements during infection provides insight into the nutritional cues being sensed by the parasite and could reveal unique targets for novel therapeutics.
Results
Host cell uptake of l-Phe(D8)
First, we investigated the rate at which l-Phe(D8) tracer replaces l-Phe within the host cells. ARPE-19 cells were metabolically labelled by adding l-Phe(D8) to customised Phe-free DMEM medium. The only source of Phe for the cells is the customised culture medium. The uptake of l-Phe(D8) is expected to be driven mostly by new protein biosynthesis during cell division and protein turnover in non-dividing cells. The doubling time of ARPE-19 cells grown in vitro was determined using cell counting at a series of 7 time points collected over a period of 7 days, revealing a doubling time of 24 hours. To achieve complete substitution of l-Phe(D8), ARPE-19 cells were maintained in serum-free DMEM overnight. Then, cells were maintained in complete DMEM with serum, but free of any Phe for 24 hours. The culture medium was then replaced by l-Phe(D8)-supplemented medium.
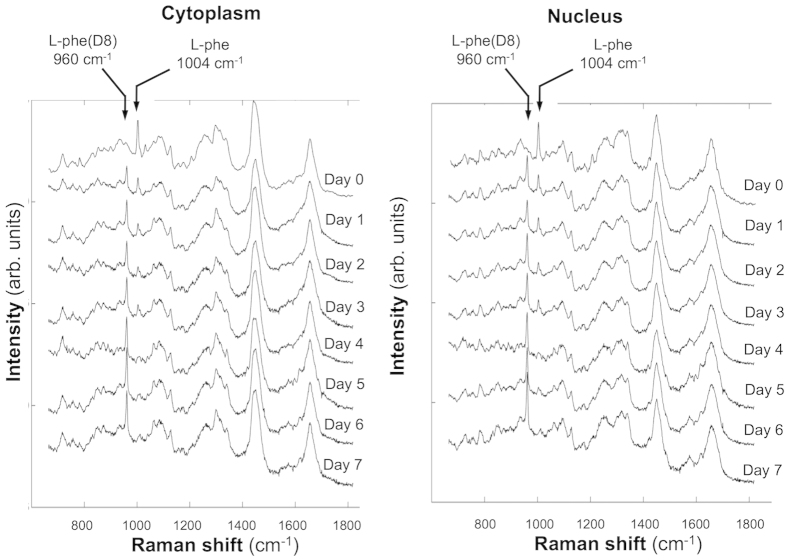
Figure 1 shows the average Raman spectra measured from the cytoplasm and nucleus of ARPE-19 cells growing in l-Phe(D8)-containing medium. The spectra in Fig. 1 were obtained from cells fixed with 4% paraformaldehyde and washed with PBS. This treatment maintained the morphology of the cells and ensured that the Raman spectra were not contaminated by signals from the culture medium,thus allowing discrimination between spectra of cytoplasm and nucleus. Prior to supplementing the culture medium with l-Phe(D8), the sharp Raman band corresponding to the ring-breathing mode of l-Phe could be identified at 1004 cm−1. After using culture medium containing l-Phe(D8) the frequency of this vibrational mode shifted to 960 cm−1. Figure 1 shows the time-dependent increase in the intensity of the 960 cm−1 Raman band corresponding to l-Phe(D8). This coincides with a decrease in the intensity of the 1004 cm−1 band, which corresponds to l-Phe. After one doubling time (1 day), the ratio I960/(I960 + I1004) is ~0.53 for the spectra measured in the cytoplasm and ~0.60 in the nucleus, indicating a similar distribution of newly synthesized proteins in the host cells. The substitution increases with time, as the ratio I960/(I960 + I1004) in both the cytoplasm and nucleus becomes ~0.72 at 4 days, and reaches 0.9–1.0 at days 5. At this point, the Raman band at 1004 cm−1 cannot be detected.
Figure 1. Time-course Raman spectra showing l-Phe(D8) uptake by growing human retinal cells (ARPE-19 cells).
Raman spectra were measured in the cytoplasm and nucleus of ARPE-19 cells cultured in l-Phe(D8)-containing medium, after fixation in 4% paraformaldehyde and washing in PBS, at the indicated time points. Spectra are shifted vertically for clarity.
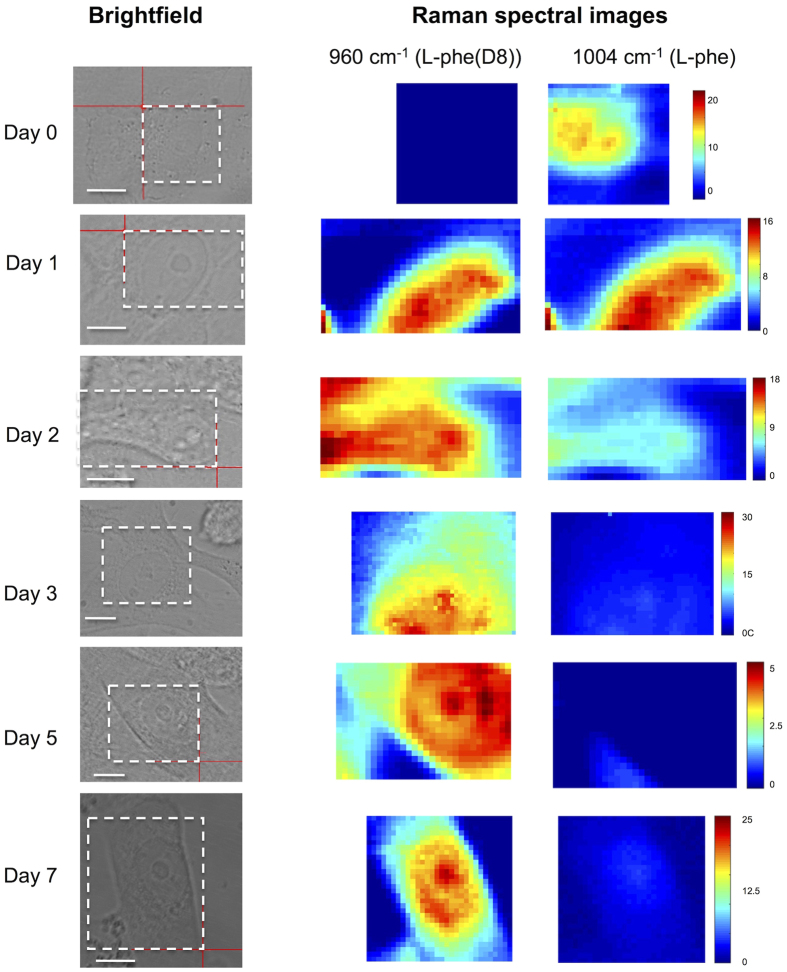
Raman micro-spectroscopy (RMS) was also used to evaluate the intracellular distribution and accumulation of l-Phe(D8) over time (Fig. 2). After short incubation times in l-Phe(D8) medium, the Raman spectral maps corresponding to the 1004 cm−1 and 960 cm−1 bands indicate similar patterns for the distribution of l-Phe and l-Phe(D8), supporting their biological equivalence. Comparison between the Raman spectra recorded from locations in the cell cytoplasm and nucleus indicates that the incorporation of l-Phe(D8) occurs at the similar rates.
Figure 2. Time-course Raman spectral maps of fixed human retinal cells showing cellular incorporation l-Phe(D8).
Raman spectral images were obtained by plotting the height of Raman bands assigned to the ring-breathing mode vibrations of l-Phe (1004 cm−1) and l-Phe(D8) (960 cm−1). Cells were grown in l-Phe(D8)-supplemented medium, fixed in 4% paraformaldehyde and washed in PBS prior to Raman measurements. The areas scanned by Raman spectroscopy are highlighted by white dashed-line rectangles (the red crosses should be ignored). Scale bars: 10 μm.
While the 1004 cm−1 and 960 cm−1 Raman bands are assigned to ring-breathing vibrations of l-Phe and l-Phe(D8), respectively, the intensity of the 1660 cm−1 Raman band corresponds mainly to the Amide I vibrational modes, with some contributions from C = C vibrations in unsaturated lipids. Thus, the intensity of this band can be related to the overall level of protein synthesis. Thus, the ratio between the band intensities of the ring-breathing modes of Phe (I960 + I1004) and the Amide I (I1660) can be linked to the level of incorporation of Phe into proteins. Figure 3a shows the calculated values of the ratio (I960 + I1004)/I1660 for cells grown in-vitro in culture media containing l-Phe and l-Phe(D8) at different time points. Regardless of the incubation time, the uptaken l-Phe-(D8) is incorporated into proteins at similar levels as l-Phe under normal culture conditions.
Figure 3. Potential detection of metabolic products in cells following cellular uptake of l-Phe(D8).
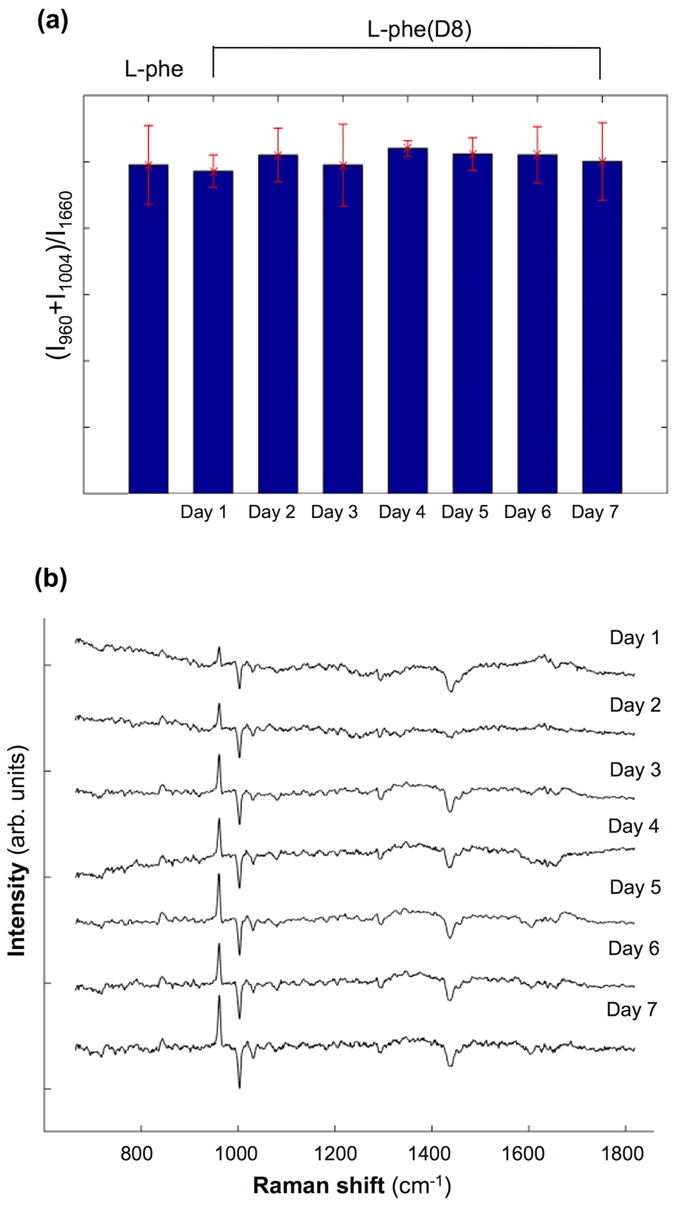
(a) Calculated ratio (I1004 + I960)/I1660 for ARPE-19 cells grown in l-Phe and l-Phe(D8) culture media. (b) Computed difference Raman spectra acquired from cells cultured in l-Phe and l-Phe(D8) daily for 7 days. Spectra are shifted vertically for clarity.
To determine whether the uptaken l-Phe(D8) is also metabolized by the ARPE-19 cells, the difference between the average Raman spectra for cells cultured in medium containing l-Phe and l-Phe(D8) were calculated (see Fig. 3a). The results indicate that apart from the downshift of the ring-breathing mode of Phe from 1004 cm−1 to 960 cm−1, no other major changes can be observed that could be related to the isotope substitution.
Of particular interest is the absence of changes associated with tyrosine (Tyr), a metabolic product of Phe. Typically, in the Raman spectra of cells, Tyr can be identified as a spectral doublet at 828 cm−1 and 854 cm−1, which corresponds to a Fermi resonance between the ring-breathing mode and the overtone of the out-of-plane ring-bending mode28. Upon substitution of H with D in Tyr, the frequency of the out-of-plane ring-bending mode was found to drop to 362 cm−1 (with an overtone at 724 cm−1) while the ring-breathing mode occurs at ~791 cm−1 28. Because of the difference in the frequencies for these two vibrational modes, the Fermi resonance is removed and only a single peak at ~791 cm−1 can be observed in the Raman spectrum of Tyr(D4). The presence of this peak in Raman spectra from cells incubated in l-Phe(D8) medium would indicate conversion of l-Phe(D8) into Tyr(D7). The calculated difference spectra in Fig. 3a show no changes in the Raman bands in the 770–790 cm−1, but rather an increased peak at ~840 cm−1. It is also important to mention that the detection of the 791 cm−1 band of Tyr(D7) may be hindered by the overlap with the Raman bands corresponding to nucleic acids (788 cm−1 O-P-O phosphodiester stretching vibrations). Therefore, no conversion of l-Phe(D8) into Tyr(D7) can be detected. These results indicate that the l-Phe(D8) is directly incorporated into newly synthesized proteins.
Differential uptake of l-Phe(D8) by extracellular versus intracellular T. gondii
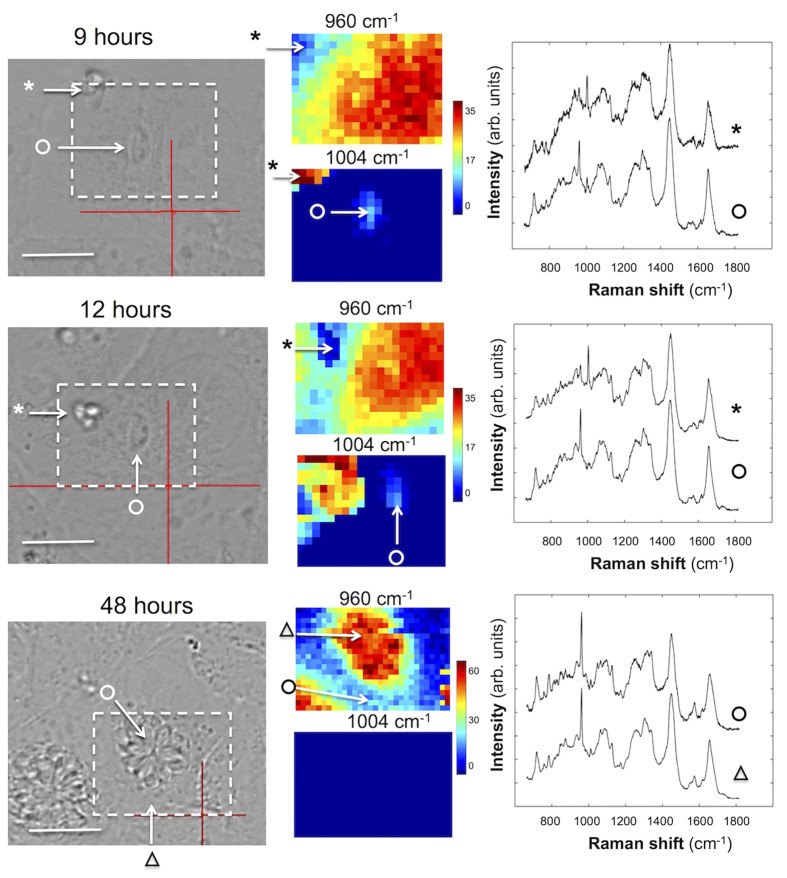
We know that T. gondii tachyzoites are incapable of replication while outside mammalian cells. But, we do not know whether extracellular T gondii tachyzoites are able to acquire Phe directly from the culture media. Hence, it is important to confirm that the information provided by the Raman spectra reflects solely molecular exchange between T. gondii and host cells and that there is no contribution from the culture medium. In these experiments, the infection was carried out while maintaining the cells in culture medium containing l-Phe(D8). Figure 4 compares the Raman spectra for T. gondii infecting ARPE-19 cells and extracellular T. gondii tachyzoites that have not yet invaded the host ARPE-19 cells. At 9 hours post infection (PI), individual T. gondii tachyzoites were observed inside ARPE-19 cells. Raman spectra indicated a high intensity band at 960 cm−1 assigned to l-Phe(D8) while the corresponding band at 1004 cm−1 corresponding to l-Phe had decreased by about 4-fold. At 12 hours the 1004 cm−1 band could be detected in the spectra of T. gondii. These results indicate a very rapid uptake of l-Phe(D8) by T. gondii even before the start of the parasite division (occurs at approximately12 hours PI).
Figure 4. Comparison between Raman spectra of live T. gondii tachyzoites within host ARPE-19 cells (indicated by symbol “o”) and live T. gondii that did not enter host cells and remained extracellular (indicated by “*”).
Here the cells were in culture medium containing l-Phe(D8). The Raman spectra show rapid uptake of l-Phe(D8) by the intracellular T. gondii, while no uptake of l-Phe(D8) is observed for the extracellular T. gondii. At 12 h parasites start to divide (middle) and at 48 h, a large parasitophorous vacuole full of parasites can be observed (bottom) (indicated by symbol “o”). All spectra are shifted vertically for clarity. Scale bars: 10 μm.
In contrast, the Raman spectra of T .gondii that did not enter the ARPE-19 cells (marked by * in Fig. 4), contain intense Raman bands at 1004 cm−1 corresponding to l-Phe and only small peaks (~5-fold lower) at 960 cm−1, both at 9 hours and 12 hours. The lack of change in either the 1004 cm−1 or 960 cm−1 bands from 9 hours PI to 12 hours PI indicates that this peak is likely the contribution of l-Phe(D8) molecules in the underlying ARPE-19 cell. These results indicate that T. gondii cannot uptake Phe directly from the culture medium and that the Raman bands at 960 cm−1 identified in the intracellular T. gondii (marked by o in Fig. 4) originate from l-Phe(D8) acquired only from the host ARPE-19 cells after infection.
The images recorded at 48 hours PI also show that T. gondii developed and replicated successfully as the parasitophorous vacuole (PV) contained many tachyzoites. No differences were observed in the growth of T. gondii infecting ARPE-19 cells cultured in l-Phe or l-Phe(D8). These results show that the substitution of l-Phe with l-Phe(D8) and the exposure to 785 nm laser light during Raman spectral measurements did not adversely affect the viability of the cells.
Time- and spatially-resolved exchange of proteins between host and pathogen
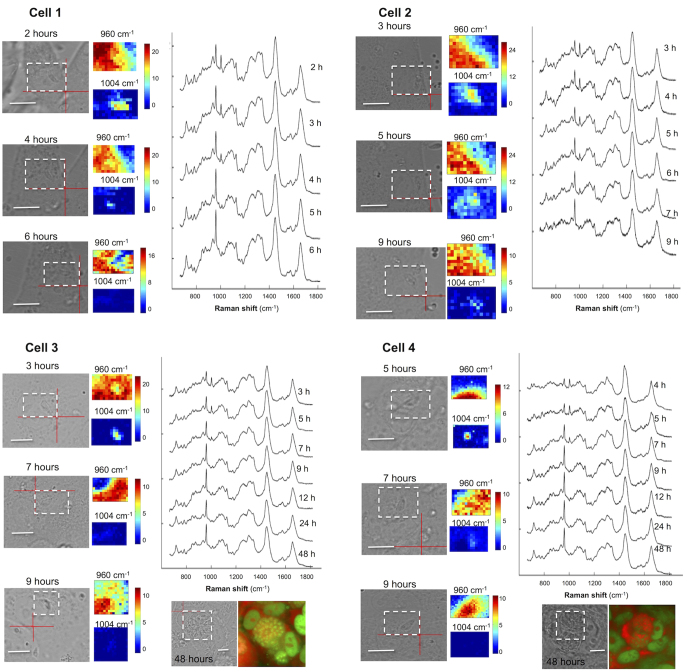
To obtain detailed information regarding metabolism and exchange of Phe between the host ARPE-19 cells and T. gondii, time-resolved Raman spectral mapping of T. gondii was performed from 1 hour to 48 hours PI. For these experiments, the culture medium was replaced with l-Phe(D8)-free media to avoid any false labelling of the parasite while in the extracellular medium before cell invasion.
Figure 5 presents the time resolved Raman spectra and spectral maps corresponding to the 960 cm−1 and 1004 cm−1 of four live T. gondii tachyzoites during the invasion of ARPE-19 cells. Because the intensities of Raman bands are affected by both molecular and morphological changes of the cells, the spectral maps provide only qualitative information. Nevertheless, at positions corresponding to T. gondii, an intense Raman band assigned to l-Phe(D8) at 960 cm−1 can be observed at as early as 2 hours PI. A corresponding decrease in the intensity of the 1004 cm−1 band assigned to l-Phe can also be detected. It is also interesting to note that Raman bands at 1004 cm−1 can be detected in the cytoplasm of the APRE-19 cells, suggesting release of proteins by T. gondii into the host. The intensity of the 960 cm−1 band increases with time, indicating a rapid uptake of l-Phe(D8) by T. gondii. By 9–12 hours PI, the Raman band at 1004 cm−1 is no longer detectable in the Raman spectra of T. gondii, suggesting that the initial l-Phe in T. gondii is completely substituted with l-Phe(D8) acquired from the host cells. Although during this time T. gondii grow in size, they divide at ~12 hours PI. By this point, l-Phe has been fully replaced by l-Phe(D8).
Figure 5. Typical time-course Raman spectra and maps corresponding to the 1004 cm−1 (l-Phe) and 960 cm−1 (l-Phe(D8)) bands of live T. gondii infecting ARPE-1 cells.
The measured areas are indicated by the white dashed rectangles. Acridine orange (AO) staining was carried out at 48 h after infection. All spectra are shifted vertically for clarity. Scale bars: 10 μm.
Dynamics of uptake of l-Phe(D8) at single parasite level
Figure 6 shows the kinetics of Phe exchange between T. gondii and the host ARPE-1 cells. The uptake was expressed as the percentage of l-Phe(D8) from the total Phe calculated as the ratio I960/(I960 + I1004). While other techniques require large populations of cells and provide only average results, Raman microscopy can measure the rates of Phe transport for individual live T. gondii. Figure 6 shows that more than 90% of T. gondii have a rapid uptake of Phe from the host cells, as l-Phe(D8) reached ~50% at 2–3 hours PI. It is interesting to note that after ~3 hours PI, two trends can be observed. For the majority of T. gondii the transport rate seems to decrease slightly and the complete substitution of l-Phe with l-Phe(D8) is typically observed at 6–9 hours. For a smaller group of T. gondii, the transport rates remain constant and the saturation with l-Phe(D8) is reached as early as 6 hours, which is approximately half the time required for T. gondii to divide.
Figure 6. Uptake of l-Phe(D8) by individual T. gondii tachyzoites.
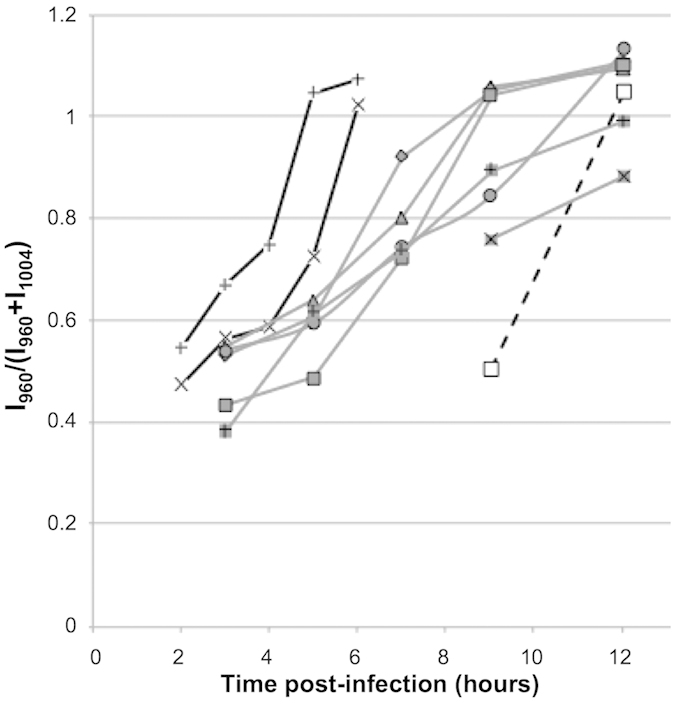
Individual tachyzoites were monitored from the moment they invaded the host cells. l-Phe(D8) uptake by the tachyzoites was measured by Raman microscopy every hour for 12 hr.
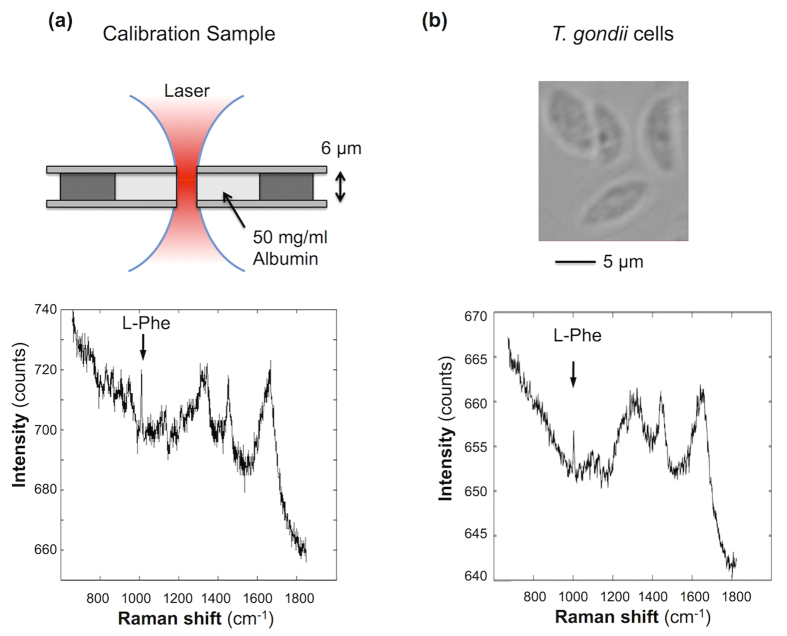
To quantify the transport rates of l-Phe to T. gondii, a calibration model was built for the Raman micro-spectrometer based on human serum albumin solution in a sample with known thickness (Fig. 7a). The intensity of the 1004 cm−1 Raman band I1004 assigned to l-Phe is directly proportional to the number of l-Phe molecules NPhe irradiated within the sample: I1004 = k × NPhe. For a 50 mg/ml of albumin solution within the cylindrical laser volume (height 6 μm and ~2 μm2 area), the number of l-Phe residues is ~1.6 × 108. Based on the intensity of the 1004 cm−1 Raman band for the calibration samples (Fig. 7a), the calibration constant was calculated to be k = 1.2 ± 0.1 × 10−7 counts/molec.
Figure 7. Quantification model for l-Phe(D8) in ARPE-19 cells and T. gondii based on Raman micro-spectroscopy.
(a) Schematic of the calibration sample of 50 mg/ml human serum albumin in water and the corresponding Raman spectrum. (b) Images of isolated T. gondii tachyzoites and typical Raman spectrum.
Although it was not possible to measure the exact height of the cells used in the time-course experiment described in Figs 5 and 6, the size of T. gondii was estimated to be ~2 μm thickness and 8 μm length based on the bright field microscopy images. Using the height of the 1004 cm−1 band in the measured Raman spectra for isolated T. gondii (Fig. 7b) and an estimated height of ~2 μm, the concentration of l-Phe molecules in T. gondii was estimated to be 10 ± 4 × 106 molecules/μm3. For a typical value of ~8 μm for the length of T. gondii, the total number of Phe molecules in a T. gondii was estimated to be 2.6 ± 1 × 108. Based on the results in Fig. 6 indicating that all l-Phe are replaced by l-Phe(D) within 7–9 hours PI, the transport rate of l-Phe(D8) from the host cells to the T. gondii is 0.5–1.6 × 104 molecules/s.
Discussion
Even though phenylalanine (Phe) is a vital component of proteins in all living organisms, the sources of this aromatic amino acid (AA) are organism-dependent. T. gondii can synthesize Phe through the shikimate pathway given an adequate supply of a specific precursor, phenylpyruvate, from infected host cell6. Despite the capacity for de novo synthesis, T. gondii still scavenges Phe and other amino acids, which are essential for the host9.
In this study, we leveraged the capabilities of a combined stable-isotope labelled Phe and Raman micro-spectroscopy (RMS) approach to investigate the transport of Phe from host cell to live T. gondii, both in time- and spatial-domains. Specifically, we examined the relationship among the processes of Phe transport, possible pool build up, and the incorporation of Phe into newly synthesized protein. Our findings revealed that the time taken for the transport of l-Phe(D8) to T. gondii was remarkably quick. T. gondii tachyzoites acquired l-Phe(D8) from host cells at a very rapid rate, ~50% at 2–3 hours post infection (PI), and l-Phe became completely substituted with l-Phe(D8) at ~7–9 hours PI, which even precedes parasite division.
Uptake of l-Phe(D8) might consist of active transport through transporters in the parasite membrane into a free AA internal pool. The Raman spectral results indicated that the uptaken l-Phe(D8) was not metabolized/cleaved into tyrosine (Tyr) by the parasite. These results suggest that l-Phe(D8) uptaken by T. gondii might be incorporated into proteins without conversion to Tyr, similar to previous reports that only low levels of turnover of Phe into Tyr were observed when mammalian cells are grown in Tyr-containing culture media29. Unexpectedly, the extracellular T. gondii tachyzoites which require high energy to empower their gliding motility and invasion into host cells were found incapable of utilizing l-Phe(D8) from culture medium even after 24 hours.
The rapid accumulation of l-Phe(D8) within T. gondii supports the role of T. gondii in behavioural changes in mice or psychiatric behaviours in infected humans30,31,32,33. It has been suggested that T. gondii causes these changes in vivo and in vitro by affecting equilibrium of neurotransmitters, especially dopamine32,34. This hypothesis has been supported by the discovery of two genes, AAH1 and AAH2, in T. gondii genome encoding for two isoforms of aromatic amino acid hydroxylases AAH (tyrosine and phenylalanine hydroxylases), which catalyze Phe to Tyr and Tyr to 3,4 dihydroxyphenylalanine (l-Dopa) (the precursor to dopamine), which may alter dopamine pathway, leading to alteration in the behaviour of T. gondii-infected host12. AAH1 and AAH2 differ in their temporal expression. AAH1 was reported to be constitutively expressed across the parasite life cycle, while the expression of AAH2 was found to be expressed during the dormant bradyzoite stage12. This pattern has led to the assumption that AAH2 is the candidate effector of the parasite’s manipulation of host dopamine. However, a recent study reported that a ΔAAH2 mutant T. gondii strain was not able to increase the production of dopamine in infected mouse brains and that over-expression of the AAH2 gene could not augment dopamine content in vitro35. This, to some extent, agrees with our findings that the uptaken l-Phe(D8) was not metabolized into Tyr by the parasite.
This reduction in Tyr during T. gondii infection might also have biological effects. Patients suffering from chronic immune activation and inflammatory conditions, which are prominent features in toxoplasmosis, were found to have elevated serum levels of Phe together with elevated Phe to Tyr ratios (Phe/Tyr). These changes in AA levels were associated with neuropsychiatric symptoms, such as depression and mood changes36. The rapid uptake of l-Phe(D8) by T. gondii indicate the significant need for this AA by T. gondii even at an early stage of the intracellular infection cycle. Also, the two predicted AAH genes in T. gondii reflect the parasite’s ability to internalize and digest Phe from the host12. In the present study the amount of l-Phe(D8) uptaken from the host cells and incorporated into protein exceeds the expected amount of new protein formed based on the expected increase in size of the growing T. gondii as indicated by the bright field images. It is reasonable to hypothesize that l-Phe(D8) may be transported into T. gondii and enter the AA pool of the host cell. This is then exchanged with the original l-Phe residues in its native form and without preliminary degradation similar to the exchange reaction in bacteria29.
Another possible mechanism that may facilitate the rapid uptake of l-Phe(D8) is the release of proteins by T. gondii into the host cells during the early stage of infection. A range of proteins delivered from rhoptries and dense granules, which are secretory organelles unique to the phylum Apicomplexa, are released by T. gondii into the parasitophorous vacuole (PV) within 10 minutes after invasion to subvert host cell function38,39. Even though the concentration of l-Phe was expected to be very low given the large volume of the mammalian cells, Raman signals corresponding to the 1004 cm−1 Raman band, assigned to l-Phe molecules, were detected inside the host cytoplasm at 2–5 hours PI.
T. gondii and other protozoans can synthesize few AAs because they have lost key enzymes needed to make some AAs de novo. Hence, they rely on host to supply them with AAs, the transport of which is mediated by several plasma membrane carriers (TransportDB). It is very likely that the import of Phe via the plasma membrane and the subsequent entry within the intravacuolar parasites occurs via transporters. Even though AA import is essential for protozoans only a few AA transporters have been functionally characterized, including the AAAP (amino acid/auxin permease) transporter family whose number vary significantly among protozoan species, and may transport one or multiple AAs, reflecting fundamental differences among protozoan species in AA requirements or salvage mechanisms40. However, we calculated the transfer rate of l-Phe(D8) molecules from host to T. gondii to be 0.5–1.6 × 104 molecules/s. Because an average transporter can have a transport rate of ~103–104 molecules/s, our results agree with the prediction that four AAAP transporters are involved in the AA transfer in T. gondii B7 strain (TransportDB). On the other hand, two transport systems for Phe have been identified in the protozoan Trypanosoma brucei brucei, characterized by a high and a very high affinity41. When the Vmax were expressed in terms of molecules/s/single cell, we estimated transport rates of 9.4 × 104 and 56.4 × 104 of Phe molecules/s for the high and the very high transport system, respectively. These transport rates are 10 to 50-fold higher than the rates for T. gondii measured in our study, supporting the idea that differences exist in the trafficking of Phe between different protozoans.
Conclusions
Common imaging techniques often do not allow distinguishing between newly imported AAs and those existing within the cell, or visualizing individual AA. To overcome these challenges, we used a RMS-based imaging approach to directly monitor the uptake of individual deuterium-labelled Phe by individual T. gondii tachyzoites. Using this approach, we showed for the first time that it is possible to follow in real time the uptake and intracellular distribution of l-Phe(D8) within individual live T. gondii-infected cells. T. gondii tachyzoites acquired l-Phe(D8) from pre-labelled host cells at a very rapid rate (~50% at 2–3 hours PI) and l-Phe became fully substituted with l-Phe(D8) at ~7–9 hours PI. Based on calibration model, it was also possible to estimate the transfer rate of l-Phe(D8) molecules from host to the T. gondii to 0.5–1.6 × 104 molecules/s. Although the import of Phe by the host cell plasma membrane has been documented, the transporter(s) responsible of the import of Phe from host cell to the parasite growing within the PV has not yet been identified. Future analyses should address whether disruption of Phe salvage pathway in T. gondii may have therapeutic value. Our approach can facilitate the study of other intracellular microbial systems in which metabolic exchanges between the host cell and the intracellular pathogen are prominent.
Methods
Reagents
l-Phenylalanine (D8, 98%) (Cat no. DLM-372-1) was purchased from Cambridge Isotope Laboratories, Inc. (Andover, MA, USA) whereas l-Phenylalanine (Cat no. P5482), paraformaldehyde, trypan blue, human serum albumin, and acridine orange were purchased from Sigma Aldrich (St. Louis, MO, USA). Dulbecco’s modified Eagle’s medium low glucose, pyruvate (DMEM, Cat no. 11885) was purchased from Life technologies, Grand Island, NY. Fetal bovine serum (FBS) and penicillin/streptomycin were obtained from Invitrogen Corp., Carlsbad, CA.
Cell culture
The human retinal pigment epithelial cell line (ARPE-19) was kindly provided by Dr Kevin Webb from University of Nottingham and used at passage 19. As an immune-privileged and easily accessible organ, the eye constitutes a favourable target for T. gondii. Hence, we selected this cell line as a model to investigate the molecular changes that accompany T. gondii infection to the eye. ARPE-19 cells were maintained in DMEM supplemented with 1% penicillin/streptomycin and 5% heat-inactivated FBS. Cells were incubated in 5% CO2 at 37 °C and were passaged twice weekly. Cell viability was determined by trypan blue exclusion assay and only cell cultures with > 99% viability were used in the experiments.
Growth and preparation of parasites
Tachyzoites of Toxoplasma gondii strain RH were maintained in vitro by serial passage on monolayers of human brain microvascular endothelial cells (hBMECs) at 37 °C in 5% CO2 as described previously42. Infected host cell monolayers were scraped and parasites were isolated from host cells by passage through 25- and 27-gauge needles. Parasites were purified using PD-10 Desalting columns pre-packed with Sephadex G-25 medium as described previously43. Purified parasites were centrifuged at 800 × g, washed twice with sterile 1x Phosphate Buffered Saline (PBS; 8 g/l NaCl, 0.2 g/l KCl, 0.2 g/l KH2PO4, 1.15 g/l Na2HPO4), re-suspended in fresh DMEM medium, and quantified using a hemacytometer. The final volume of parasite suspension was adjusted with DMEM to achieve a ratio of 2:1 parasite/host cell for subsequent infection experiments. Parasites’ viability was checked by using trypan blue exclusion assay and parasite suspensions with more than 98% viability were used.
Stable isotope labelling by amino acids (SILAC)
Amino acid-type selective labelling was achieved by formulating a medium containing all amino acids in unlabelled form except for Phe, which was added with the desired isotope label. To minimize the background signal of the l-Phe, ARPE-19 cells were synchronized overnight in serum-deficient, Phe-free customised DMEM culture medium. The cells were subsequently incubated with SILAC DMEM medium supplemented with various concentrations of Phe(D8). Cells were subjected to Raman imaging analysis daily to determine the time taken by l-Phe(D8) to substitute l-Phe within host cells. The concentration of 66 mg/500 ml of l-Phe(D8) (i.e. twice the concentration of l-Phe present in the normal media) was found to be the lowest concentration that showed a detectable shift of the Phe peak in acquired Raman spectra.
Infection of host cells with T. gondii
ARPE-19 cells (1 × 104 cells/ml) were seeded in titanium cell-chambers, which are fitted in 6-well plastic cell culture plates, with a volume of 2 ml DMEM/chamber. The titanium cell chambers were custom-built with MgF2 coverslips (0.17 mm thick) at the bottom to enable acquisition of Raman spectra. Cells were allowed to grow overnight by incubation at 37 °C in a humidified atmosphere of 5% CO2 −95% air. Then, cells were serum starved for 18 hours, followed by incubation in l-Phe(D8)-supplemented DMEM for a minimum of 5 doubling times to ensure that cells were uniformly labelled and that l-Phe was completely substituted by l-Phe(D8). Before introducing the parasites to cells, the culture medium was replaced with l-Phe-free media to avoid any false labelling of the parasite with l-Phe(D8) while in the extracellular medium before cell invasion. Parasites are grown in a medium containing l-Phe, but they were transferred to l-Phe-free media before adding them to the cells and during host cell invasion. This avoided introducing l-Phe to l-Phe(D8)-labelled cells and ensured that the only source of Phe to the parasites was from the l-Phe(D8)-labelled host cells. Infection was initiated by adding T. gondii tachyzoites at a multiplicity of infection (MOI) of 1 cell to 2 parasites (1C:2P) in 2-ml fresh l-Phe(D8)-free DMEM. The control chambers received only 2-ml DMEM (mock-infected). Culture plates were then incubated to allow infection to progress within ARPE-19 cells. Parasite and control cultures were sampled at 0, 6, 24 and 48 hours post infection (PI). At each sampling time two chambers (one infected and one control) were collected from the 6-well culture plate and subjected to Raman Spectroscopy imaging. At least 10 infected cells were analyzed per chamber. In a separate experiment, we incubated free extracellular tachyzoites that are recovered from l-Phe culture media in l-Phe(D8)-supplemented media. These parasites failed to take l-Phe(D8) from media even after 24 h.
Time course Raman imaging
Raman spectra were recorded using a home-built confocal Raman micro-spectrometer optimized for live-cell studies. An inverted microscope (IX71, Olympus, Essex, UK) was chosen for the setup as it allows live cell measurement without additional disturbance of objective dipping in culture media required by up-right microscopes. The laser is focused through a quartz coverslip at the bottom of specially designed sample holders. An environmental enclosure (Solent, Segensworth, UK) was integrated to the microscope to maintain suitable culture conditions (37 °C temperature and 5% CO2) for living cultured cells. Under these conditions, Raman spectral measurements of individual live cells maintained in their original culture media were obtained over several hours without disturbing the cells while avoiding bacterial contamination. A 785 nm~120 mW (before objective) Ti:sapphire laser (Spectra-Physics, Oxford, UK) was used to excite Raman spectra. The beam diameter was expanded to match the objective pupil. A water-immersion 60x objective, NA 1.20 (Olympus) was used to focus the laser on individual cells as well as for collection of Raman scattered photons. The size of the laser spot was approximately 700 nm on the sample. Light collected by the objective was collimated and focused on a 50 μm diameter optical fibre connected to a spectrometer equipped with a 830 lines/mm grating (spectral resolution of ~1.5 cm−1 in the 600–1800 cm−1 region) and cooled deep-depletion back-illuminated CCD detector (Andor Technologies, Belfast, UK). Raman spectral imaging was performed by scanning the cells through the laser focus in a raster pattern (1 μm step size) using a high-precision step-motor stage (Prior, Cambridge, UK) and acquiring Raman spectra at each position (1s/pixel). The spectrometer wavenumber axis was calibrated prior to each experiment using tylenol at an accuracy of 0.5 cm−1. The raster-scan Raman measurements of live cells at the early stages of invasion (1–12 hours) was limited to areas ranging between 10 μm x 15 μm and 15 μm x 20 μm around the parasites. The imaging time for these measurements was 3–6 min. At the late stages of infection (e.g. 24 and 48 hours), the areas occupied by parasites were larger (up to 30 μm x 30 μm), thus the measurements required 15–20 min.
Fluorescence staining
Acridine orange (AO) staining was used to distinguish the morphological appearance of the intracellular developing T. gondii tachyzoites from host cell organelles. AO is a DNA intercalating dye and is used to differentially stain RNA and DNA inside eukaryotic cells. Cells were fixed in 4% paraformaldehyde for 15 min, followed by washing with 1x PBS twice. Then, cells were incubated with 5% AO staining solution for 10 min. AO fluorescence imaging of T. gondii-infected cells was performed on the same cell after completion of the Raman spectral imaging. Imaging was performed by using wide-field fluorescent microscope integrated on the confocal Raman micro-spectrometer. The retrospective positioning and identification of the cells was based on two thin marks engraved on the cell chambers (retro-positioning accuracy was 5 μm).
Image analysis and data processing
Data pre-processing and analysis was done by using in-house built functions in Matlab (The MathWorks, Natick, MA). Data pre-processing involved cosmic rays removal and background subtraction. Noise reduction was performed using singular value decomposition. The height of each peak of interest was measured after a local linear baseline was subtracted. Human serum albumin (CAS number 70024-90-7, Sigma Aldrich, UK) was used to calibrate the Raman spectrometer (30 Phe residues per albumin molecule).
Additional Information
How to cite this article: Naemat, A. et al. Tracing amino acid exchange during host-pathogen interaction by combined stable-isotope time-resolved Raman spectral imaging. Sci. Rep. 6, 20811; doi: 10.1038/srep20811 (2016).
Acknowledgments
The work was supported by Petplan Charitable Trust grant 11–38 (H. Elsheikha and I. Notingher). A. Naemat acknowledges the support from COM-SATS Institute of Information Technology, Pakistan. The development of the Raman micro-spectrometer was supported by the Biotechnology and Biological Sciences Research Council UK research grant BB/G010285/1.
Footnotes
Author Contributions I.N. and H.M.E. designed the study. A.N. performed the Raman imaging and fluorescent staining experiments with inputs and guidance from I.N. and H.M.E. R.A.B. performed the calibration of the Raman spectrometer with support from I.N., A.N., I.N. and H.M.E. contributed to the analysis and interpretation of data. H.M.E. performed cell culture and parasite growth experiments. I.N. and H.E. wrote the paper. All authors critically revised and approved the final version of the manuscript.
References
- Elsheikha H. M. Congenital toxoplasmosis: priorities for further health promotion action. Public Health 122(4), 335–353 (2008). [DOI] [PubMed] [Google Scholar]
- Sibley L. D. Toxoplasma gondii: perfecting an intracellular life style. Traffic 4, 581–586 (2003). [DOI] [PubMed] [Google Scholar]
- Polonais V. & Soldati-Favre D. Versatility in the acquisition of energy and carbon sources by the Apicomplexa. Biol. Cell 102, 435–445 (2010). [DOI] [PubMed] [Google Scholar]
- Macrae J. I. et al. Mitochondrial metabolism of glucose and glutamine is required for intracellular growth of Toxoplasma gondii. Cell Host Microbe 12(5), 682–692 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta N., Zahn M. M., Coppens I., Joiner K. A. & Voelker D. R. Selective disruption of phosphatidylcholine metabolism of the intracellular parasite Toxoplasma gondii arrests its growth. J. Biol. Chem. 280, 16345–16353 (2005). [DOI] [PubMed] [Google Scholar]
- Keeling P. J. et al. Shikimate pathway in apicomplexan parasites. Nature 397, 219–220 (1999). [DOI] [PubMed] [Google Scholar]
- Fox B. A., Gigley J. P. & Bzik D. J. Toxoplasma gondii lacks the enzymes required for de novo arginine biosynthesis and arginine starvation triggers cyst formation. Int. J. Parasitol. 34, 323–331 (2004). [DOI] [PubMed] [Google Scholar]
- Possenti A. et al. Global proteomic analysis of the oocyst/sporozoite of Toxoplasma gondii reveals commitment to a host-independent lifestyle. BMC Genomics 14, 183 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tymoshenko S. et al. Metabolic needs and capabilities of Toxoplasma gondii through combined computational and experimental analysis. PLoS Comput. Biol. 11(5), e1004261 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster J. P. & McConkey G. A. Toxoplasma gondii-altered host behavior: Clue as to mechanism of action. Folia Parasitol. (Praha) 57, 95–104 (2003). [DOI] [PubMed] [Google Scholar]
- Carruthers V. B. & Suzuki Y. Effects of Toxoplasma gondii infection on the brain. Schizophr. Bull. 33, 745–751 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaskell E. A., Smith J. E., Pinney J. W., Westhead D. R. & McConkey G. A. A unique dual activity amino acid hydroxylase in Toxoplasma gondii. PLoS One 4, e4801 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein K. et al. Label-free live-cell imaging with confocal Raman microscopy. Biophys. J. 102(2), 360–368 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascut F. C. et al. Toward label-free Raman-activated cell sorting of cardiomyocytes derived from human embryonic stem cells. J. Biomed. Opt. 16, 045002 (2011). [DOI] [PubMed] [Google Scholar]
- Pascut F. C. et al. I. Non-invasive label-free monitoring the cardiac differentiation of human embryonic stem cells in-vitro by Raman spectroscopy. Biochim. Biophys. Acta 1830, 3517–3524 (2013). [DOI] [PubMed] [Google Scholar]
- Takanezawa S., Morita S., Ozaki Y. & Sako Y. Raman spectral dynamics of single cells in the early stages of growth factor stimulation. Biophys. J. 108(9), 2148–2157 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada M. et al. Label-free Raman observation of cytochrome c dynamics during apoptosis. Proc. Natl. Acad. Sci. USA. 109(1), 28–32 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y. J. et al. Investigation of biochemical property changes in activation-induced CD8 + T cell apoptosis using Raman spectroscopy. J. Biomed. Opt. 20(7), 75001 (2015). [DOI] [PubMed] [Google Scholar]
- Zhang D. et al. Raman spectrum reveals the cell cycle arrest of Triptolide-induced leukemic T-lymphocytesapoptosis. Spectrochim. Acta A Mol. Biomol. Spectrosc. 141, 216–222 (2015). [DOI] [PubMed] [Google Scholar]
- El-Mashtoly S. F. et al. Label-free imaging of drug distribution and metabolism in colon cancer cells by Raman microscopy. Analyst 139(5), 1155–1161 (2014). [DOI] [PubMed] [Google Scholar]
- Naemat A. et al. Analysis of interaction between the apicomplexan protozoan Toxoplasma gondii and host cells using label-free Raman spectroscopy. Analyst 140(3), 756–64 (2015). [DOI] [PubMed] [Google Scholar]
- Huang W. E. et al. Raman-FISH: combining stable-isotope Raman spectroscopy and fluorescence in situ hybridization for the single cell analysis of identity and function. Environ. Microbiol. 9(8), 1878–1889 (2007). [DOI] [PubMed] [Google Scholar]
- van Manen H. J., Lenferink A. & Otto C. Noninvasive imaging of protein metabolic labeling in single human cells using stable isotopes and Raman microscopy. Anal. Chem. 80(24), 9576–9582 (2008). [DOI] [PubMed] [Google Scholar]
- Noothalapati Venkata H. N. & Shigeto S. Stable isotope-labeled Raman imaging reveals dynamic proteome localization to lipid droplets in single fission yeast cells. Chem. Biol. 19(11), 1373–1380 (2012). [DOI] [PubMed] [Google Scholar]
- Li M., Huang W. E., Gibson C. M., Fowler P. W. & Jousset A. Stable isotope probing and Raman spectroscopy for monitoring carbon flow in a food chain and revealing metabolic pathway. Anal. Chem. 85(3), 1642–1649 (2013). [DOI] [PubMed] [Google Scholar]
- Kubryk P. et al. Exploring the potential of Stable Isotope (Resonance) Raman Microspectroscopy and Surface-Enhanced Raman Scattering for the analysis of microorganisms at single cell level. Anal. Chem. 87(13), 6622–6630 (2015). [DOI] [PubMed] [Google Scholar]
- Matthäus C. et al. Noninvasive imaging of intracellular lipid metabolism in macrophages by Raman microscopy in combination with stable isotopic labeling. Anal. Chem. 84(20), 8549–8556 (2012). [DOI] [PubMed] [Google Scholar]
- Siamwiza M. N. et al. Interpretation of the doublet at 850 and 830 cm−1 in the Raman spectra of tyrosyl residues in proteins and certain model compounds. Biochemistry 14(22), 4870–4876 (1975). [DOI] [PubMed] [Google Scholar]
- Eagle H., Piez K. A. & Fleischman R. The utilization of phenylalanine and tyrosine for protein synthesis by human cells in tissue culture. J. Biol. Chem. 228(2), 847–861 (1957). [PubMed] [Google Scholar]
- Skallova A., Kodym P., Frynta D. & Flegr J. The role of dopamine in Toxoplasma-induced behavioural alterations in mice: an ethological and ethopharmacological study. Parasitology 133, 525–535 (2006). [DOI] [PubMed] [Google Scholar]
- Vyas A., Kim K. S., Giacomini N., Boothroyd J. C. & Sapolsky R. M. Behvioral changes induced by Toxoplasma infection of rodents are highly specific to aversion of cat odors. Proc. Natl. Acad. Sci. USA 104, 6442–6447 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prandovszky E. et al. The neurotropic parasite Toxoplasma gondii increases dopamine metabolism. PLoS One 6(9), e23866 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingram W. M., Goodrich L. M., Robey E. A. & Eisen M. B. Mice infected with low-virulence strains of Toxoplasma gondii lose their innate aversion to cat urine, even after extensive parasite clearance. PLoS One 8, e75246 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stibbs H. H. Changes in brain concentrations of catecholamines and indoleamines in Toxoplasma gondii infected mice. Ann. Trop. Med. Parasitol. 79, 153–157 (1985). [DOI] [PubMed] [Google Scholar]
- Wang Z. T., Harmon S., O’Malley K. L. & Sibley L. D. Reassessment of the role of aromatic amino acid hydroxylases and the effect of infection by Toxoplasma gondii on host dopamine. Infect. Immun. 83(3), 1039–1047 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neurauter G. et al. Chronic immune stimulation correlates with reduced phenylalanine turnover. Curr. Drug Metab. 9, 622–627 (2008). [DOI] [PubMed] [Google Scholar]
- Widner B., Fuchs D., Leblhuber F. & Sperner-Unterweger B. Does disturbed homocysteine and folate metabolism in depression result from enhanced oxidative stress ? J. Neurol. Neurosurg. Psychiatry 70, 419 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey K. L., Jongco A. M., Kim K. & Ward G. E. The Toxoplasma gondii rhoptry protein ROP4 is secreted into the parasitophorous vacuole and becomes phosphorylated in infected cells. Eukaryot. Cell 3(5), 1320–1330 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn J. D., Ravindran S., Kim S. K. & Boothroyd J. C. The Toxoplasma gondii dense granule protein GRA7 is phosphorylated upon invasion and forms an unexpected association with the rhoptry proteins ROP2 and ROP4. Infect. Immun. 76(12), 5853–5861 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean P., Major P., Nakjang S., Hirt R. P. & Embley T. M. Transport proteins of parasitic protists and their role in nutrient salvage. Front Plant Sci 5, 153 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasne M.-P. Amino acid transporters in Trypanosoma brucei brucei. PhD thesis, University of Glasgow, P. 94 (2001).
- Elsheikha H. M., McKinlay C. L., Elsaied N. A. & Smith P. A. Effects of Neospora caninum infection on brain microvascular endothelial cells bioenergetics. Parasit. Vectors 6, 24 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsheikha H. M. et al. Generally applicable methods to purify intracellular coccidia from cell cultures and to quantify purification efficacy using quantitative PCR. Vet. Parasitol. 135(3–4), 223–234 (2006). [DOI] [PubMed] [Google Scholar]