Abstract
TCP proteins are plant-specific transcription factors implicated to perform a variety of physiological functions during plant growth and development. In the current study, we performed for the first time the comprehensive analysis of TCP gene family in a diploid cotton species, Gossypium arboreum, including phylogenetic analysis, chromosome location, gene duplication status, gene structure and conserved motif analysis, as well as expression profiles in fiber at different developmental stages. Our results showed that G. arboreum contains 36 TCP genes, distributing across all of the thirteen chromosomes. GaTCPs within the same subclade of the phylogenetic tree shared similar exon/intron organization and motif composition. In addition, both segmental duplication and whole-genome duplication contributed significantly to the expansion of GaTCPs. Many these TCP transcription factor genes are specifically expressed in cotton fiber during different developmental stages, including cotton fiber initiation and early development. This suggests that TCP genes may play important roles in cotton fiber development.
TCP proteins constitute a family of plant-specific transcription factors widely distributed in angiosperms1,2. The TCP gene family was termed after its founding members: TEOSINTE BRANCHED 1 in Zea mays, CYCLOIDEA in Antirrhinum majus and PCF in Oryza sativa1. Since its initial identification and characterization in 1999, the TCP family has become one of the focuses of plant studies due to its importance in the evolution and developmental control of plant form2. TCP proteins are defined by a 59-residue-long basic helix-loop-helix (bHLH) structure called TCP domain, which provides this family with the ability to bind GC-rich DNA sequence motifs2. According to the secondary structure prediction, the basic region of TCP domain is followed by two helices separated by a loop2. In addition, phylogenetic analysis showed that TCP proteins can be classified into two subfamilies based on their DNA binding domain structure2. To date, more than 20 TCP family members have been identified in a number of monocot and eudicot plants, such as Arabidopsis3, Oryza sativa4, Vitis vinifera and Populus trichocarpa5.
In plants, the TCP transcription factor family has been implicated to perform a variety of physiological functions during plant growth and development, such as branching, regulation of the circadian clock, seed germination, gametophyte development, hormone pathways, leaf development, mitochondrial biogenesis, flower development and cell cycle regulation2,6,7,8,9,10,11,12. Recently, evidences indicated that TCP proteins also play a significant role in fiber development13,14, which makes it necessary to identify and characterize TCP family members in cotton, one of the most important economic crops and natural fiber sources all over the world15.
Cotton comprises both diploid and tetraploid species, belonging to the Gossypium genus. The most commonly cultivated cotton species for fiber and oil production is upland cotton (Gossypium hirsutum), an AD tetraploid evolved from A-genome diploids such as G. arboreum and D-genome diploids like G. raimondii at around 1–2 million years ago16. Up to now, only two TCP family members have been functionally characterized in cotton, suggesting that TCP genes may play key roles in fiber development13,14. Therefore, there is an urgent need to perform a genome wide analysis of this family in cotton. The recent completion of the sequencing of G. arboreum genome allowed us to characterize all cotton TCP genes.
In the current study, we performed for the first time the comprehensive analysis of TCP gene family in G. arboreum, including phylogenetic analysis, chromosome location, gene duplication status, gene structure and conserved motif analysis, as well as tissue specific expression profiles. Our findings will lay solid foundation to better understand the function and evolutionary history of GaTCPs, and will help further investigation of the detailed molecular and biological functions of TCP members in cotton.
Results
Identification of the TCP gene family in G. arboretum
The TCP transcription factor family is featured by a highly conserved TCP domain at the N-terminus. To identify this family in G. arboretum, the profile hidden Markov Models of TCP domain (PF03634) downloaded from Pfam was used as query to run Hmmsearch against the G. arboretum genome. As a result, 58 putative TCP genes were identified. After the removal of 22 redundant sequences based on multiple sequence alignment, 36 candidate TCP genes sequences were manually inspected with ScanProsite to confirm the existence of the conserved TCP domain. As expected, they all contained the TCP domain, suggesting that they were members of the TCP gene family. The 36 TCP genes were further named as GaTCP1 to GaTCP25 according to Arabidopsis TCP nomenclature suggestions. Table 1 summarized their gene symbols, corresponding gene names, sequence length, molecular weights, isoelectric points and chromosome location.
Table 1. TCP gene family in G. arboretum.
| Gene Name | Gene symbol | Length (aa) | MW (Da) | pI | Chr. Location |
|---|---|---|---|---|---|
| GaTCP1 | Cotton_A_09911 | 397 | 43545.24 | 9.21 | chr3:16012758:16014321 |
| GaTCP2 | Cotton_A_26168 | 410 | 44917.18 | 6.77 | chr10:15878745:15879977 |
| GaTCP3 | Cotton_A_23161 | 409 | 44273.72 | 6.78 | chr13:94068971:94070200 |
| GaTCP4 | Cotton_A_22289 | 443 | 48760.48 | 6.06 | chr10:78860246:78861667 |
| GaTCP5 | Cotton_A_31971 | 325 | 36033.79 | 5.8 | chr1:70185179:70186156 |
| GaTCP6 | Cotton_A_23025 | 250 | 26502.48 | 9.58 | chr6:20222965:20223732 |
| GaTCP7a | Cotton_A_08973 | 258 | 26739.99 | 9.72 | chr5:10164781:10165557 |
| GaTCP7b | Cotton_A_14593 | 243 | 25308.51 | 9.99 | chr6:109156974:109157705 |
| GaTCP8 | Cotton_A_24144 | 486 | 50969.89 | 7.36 | chr5:48534877:48536337 |
| GaTCP9a | Cotton_A_10947 | 338 | 35365.29 | 9.08 | chr4:104148560:104149576 |
| GaTCP9b | Cotton_A_14431 | 385 | 41254.45 | 8.95 | chr7:47526308:47527465 |
| GaTCP10 | Cotton_A_20110 | 448 | 48746.25 | 6.6 | chr7:116121419:116122765 |
| GaTCP11 | Cotton_A_24059 | 200 | 21687.42 | 8.32 | chr10:109535667:109536269 |
| GaTCP12 | Cotton_A_37122 | 501 | 55859.83 | 6.82 | chr7:89657212:89658717 |
| GaTCP13a | Cotton_A_27227 | 309 | 34245.51 | 8.71 | chr12:56165133:56166062 |
| GaTCP13b | Cotton_A_14726 | 285 | 31976.83 | 7.94 | chr11:102879533:102880390 |
| GaTCP14a | Cotton_A_09220 | 395 | 42218.14 | 6.91 | chr13:44785981:44787168 |
| GaTCP14b | Cotton_A_02703 | 418 | 44468.80 | 8.60 | chr8:96754543:96755799 |
| GaTCP14c | Cotton_A_27685 | 406 | 43980.41 | 6.88 | chr6:58440407:58441627 |
| GaTCP15a | Cotton_A_06142 | 342 | 37377.38 | 8.53 | chr6:68046188:68047216 |
| GaTCP15b | Cotton_A_33342 | 365 | 39684.16 | 9.54 | chr2:18664013:18665110 |
| GaTCP16 | Cotton_A_10509 | 196 | 21078.67 | 8.56 | chr8:103306077:103306667 |
| GaTCP17 | Cotton_A_19125 | 266 | 30312.08 | 7.78 | chr1:83153095:83153967 |
| GaTCP18a | Cotton_A_07573 | 329 | 37748.89 | 9.02 | chr11:53845238:53846327 |
| GaTCP18b | Cotton_A_01394 | 367 | 41500.33 | 9.08 | chr6:120385396:120386499 |
| GaTCP19a | Cotton_A_21588 | 341 | 36882.22 | 6.26 | chr13:4529921:4530946 |
| GaTCP19b | Cotton_A_09964 | 335 | 35753.32 | 9.42 | chr3:14665842:14666849 |
| GaTCP20a | Cotton_A_40823 | 300 | 32008.71 | 7.93 | chr3:18568141:18569043 |
| GaTCP20b | Cotton_A_07501 | 298 | 31710.22 | 7.28 | chr9:49546327:49547223 |
| GaTCP20c | Cotton_A_39272 | 298 | 31420.10 | 9.64 | chr11:43972643:43973539 |
| GaTCP20d | Cotton_A_22689 | 306 | 32794.32 | 9.27 | chr1:99325941:99326861 |
| GaTCP21 | Cotton_A_26482 | 255 | 26370.49 | 9.66 | chr13:2155968:2156735 |
| GaTCP22 | Cotton_A_27060 | 553 | 58300.67 | 6.73 | chr2:54926878:54928539 |
| GaTCP23 | Cotton_A_03998 | 418 | 44401.83 | 6.72 | chr10:80131534:80132790 |
| GaTCP24 | Cotton_A_02913 | 463 | 50231.92 | 7.01 | chr9:73463153:73464544 |
| GaTCP25 | Cotton_A_37650 | 431 | 47737.38 | 6.47 | chr12:125507830:125509978 |
Evolutionary analysis of the TCP transcription factor family
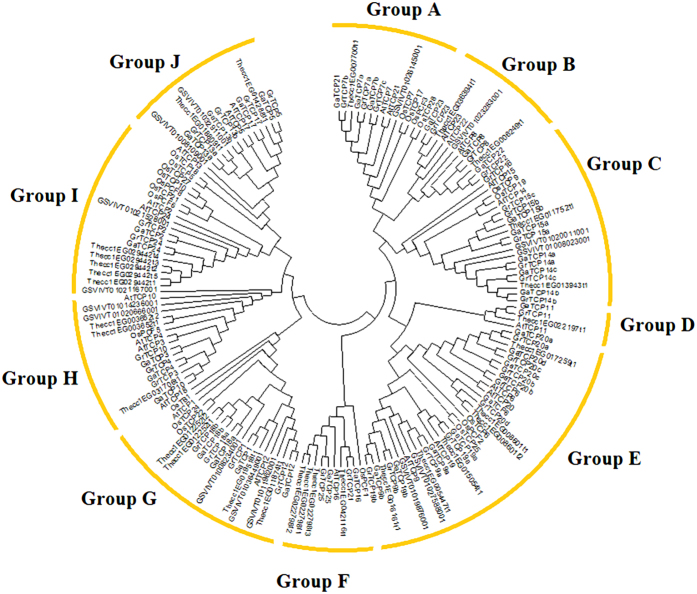
In order to explore the evolutionary relationships of the TCP transcription factor family, an unrooted phylogenetic tree was generated using the full length TCP protein sequences from G. arboreum, G. raimondii, Theobroma cacao, Vitis vinifera, Arabidopsis thaliana and Oryza sativa. As illustrated in the Neiboring-Joining phylogenetic tree (Fig. 1), the TCP transcription factor family was classified into ten distinct subgroups designated as Group A to Group J. Group E is composed of 33 members, constituting the largest clade among all subgroups, while Group G is the second largest clade, consisting of 21 TCP protein sequences. The smallest clade is Group D, which contains only four TCP proteins. Generally speaking, all subgroups contain at least five plant species except Group D, exhibiting an interspersed distribution. This may imply that the divergence of these species took place after the TCP transcription factor family expanded. Noticeable, the TCPs from G. arboretum and G. raimondii formed quite a few clusters of homologs in all subfamilies due to their high sequence similarity and close evolutionary relationship that the divergence of the two cotton species occurred only 2–13 million years ago17. In addition, the cotton TCPs (GrTCPs and GaTCPs) were more closely allied to TCP proteins from T. cacao than from other plant species, consistent with the fact that G. arboreum, G. raimondii and T. cacao originated from a common ancestor 18–58 million years ago17. Based on multiple sequence alignments, orthologous TCP gene pairs between T. cacao and G. arboreum were identified: GaTCP14b/Thecc1EG013943t1, GaTCP15b/Thecc1EG011752t1, GaTCP11/Thecc1EG022197t1, GaTCP23/Thecc1EG036394t1, GaTCP22/Thecc1EG006249t1, GaTCP19a/Thecc1EG015054t1, GaTCP20a/Thecc1EG017259t1, GaTCP16/Thecc1EG042116t1, GaTCP12/Thecc1EG011874t1, and GaTCP1/Thecc1EG019518t1. Moreover, Group B and Group D did not contain O. sativa TCP proteins, suggesting that the TCP family members in the two subgroups were either lost in O. sativa or obtained after the divergence of monocots and eudicots. Additionally, the phylogenetic analysis also showed that the TCP members from different plant species were not evenly distributed in some subgroups. GaTCPs, for example, were overrepresented than AtTCPs in Group C and Group E, in which GaTCPs were over two times the number of AtTCPs. This may indicate that the TCPs went through differential expansion in G. arboreum and in Arabidopsis.
Figure 1. Phylogenetic tree of TCP proteins from Gossypium arboreum, G. raimondii, Theobroma cacao, Vitis vinifera, Arabidopsis thaliana and Oryza sativa.
The phylogenetic tree was generated using the Neighbor-Joining (NJ) method implemented in the MEGA 6.0 software with JTT model and pairwise gap deletion option. The bootstrap analysis was conducted with 1000 iterations.
Chromosomal distribution and gene duplication
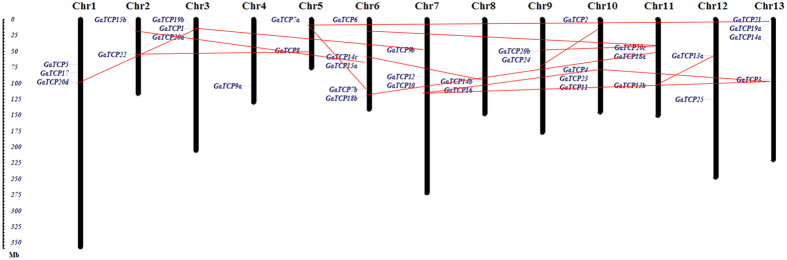
The 36 G. arboretum genes were mapped onto chromosomes in order to elucidate their chromosomal distribution and gene duplication status. As shown in Fig. 2, the 36 GaTCPs were scattered throughout all 13 chromosomes of G. arboretum. Chromosome 6 had the highest number of five TCP genes, followed by Chromosome 10 and Chromosome 13 with four TCP genes. The lowest number of GaTCPs were observed in Chromosome 4 with one TCP gene. In general, the TCP genes were more evenly distribute across G. arboretum chromosomes than that in G. raimondii18.
Figure 2. Chromosomal location and gene duplication status of TCP genes from Gossypium arboreum on 13 chromosomes.
The scale represents megabases (Mb). The chromosome numbers are indicated above each vertical bar. The red lines connect the duplicated gene pairs.
In addition, the gene duplication events were further investigated to reveal the expansion mechanism of the TCP gene family in G. arboretum. The criteria described in previous studies were employed to identify paralogous genes pairs. As a result, 15 pairs of putative paralogous TCP genes were found in G. arboretum with high gene and protein sequence identity and similarity, accounting for about 70% of the whole GaTCP gene family. As illustrated in Fig. 2, all of the gene pairs were distributed on different chromosomes, while no tandem duplication events could be observed, suggesting that segmental duplications contributed a lot to the amplification of the GaTCP gene family. Additionally, in order to gain more insights of the evolutionary history of the GaTCP gene family, the DnaSP program was used to calculate the approximate dates of duplication events, dating the duplication events of GaTCPs between 11.28 Mya (million years ago) to 36.51 Mya, with an average of 19.7 Mya. The detailed analysis for the duplicated gene pairs was listed in Table 2.
Table 2. Dates of duplication for the duplicated gene pairs.
| Gene 1 | Gene 2 | ks | T = Ks/2λ |
|---|---|---|---|
| GaTCP2 | GaTCP24 | 0.3869 | 12.89667 |
| GaTCP3 | GaTCP10 | 0.3868 | 12.89333 |
| GaTCP10 | GaTCP4 | 0.4783 | 15.94333 |
| GaTCP7a | GaTCP7b | 0.5899 | 19.66333 |
| GaTCP9b | GaTCP19b | 0.4529 | 15.09667 |
| GaTCP13b | GaTCP13a | 0.6527 | 21.75667 |
| GaTCP14b | GaTCP14c | 0.5637 | 18.79 |
| GaTCP15a | GaTCP15b | 0.6751 | 22.50333 |
| GaTCP17 | GaTCP5 | 1.0293 | 34.31 |
| GaTCP18a | GaTCP18b | 1.0954 | 36.51333 |
| GaTCP20a | GaTCP20d | 0.4212 | 14.04 |
| GaTCP20c | GaTCP6 | 0.3384 | 11.28 |
| GaTCP20b | GaTCP20a | 0.4097 | 13.65667 |
| GaTCP20d | GaTCP20b | 0.4599 | 15.33 |
| GaTCP22 | GaTCP8 | 0.9276 | 30.92 |
Gene structure and conserved motifs
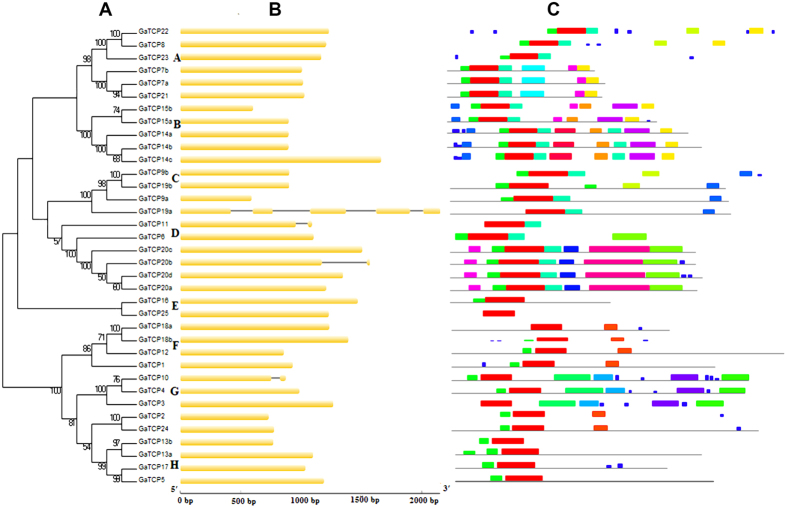
To get a better understanding of the diversification of the TCP genes in G. arboretum, the exon/intron organization and conserved motifs of GaTCPs were analyzed. A new Neiboring-Joining phylogenetic tree was constructed using the protein sequences of GaTCPs, dividing the TCP family into eight subclades. As shown in Fig. 3, over 80% of GaTCP genes were intronless, which was quite similar to the structure of G. raimondii TCP genes18. Generally speaking, most GaTCPs within the same subclades exhibited similar gene structure in terms of numbers and lengths of introns and exons. Subclade A and H, for instance, contained intronless genes with similar exon lengths. In contrast, great structure variants were observed in Subclade C. In addition, the MEME programs was used to predict motif composition, identifying twenty conserved motifs in GaTCPs. Subsequently, the program InterProScan was employed to annotate these motifs. The results showed that the only motif that hit for the database was the conserved TCP domain (the red motif), which was found in all GaTCPs. Moreover, similar to the exon/intron organization, members belonging to the same subclades also showed similar motif composition, indicating their functional similarities. Additionally, some motifs were only presented at specific subclades, suggesting that they may perform subclade specific functions.
Figure 3. Phylogenetic analysis, exon/intron organization and motif composition s of Gossypium arboreum TCP genes.
(A) The phylogenetic tree was generated using the Neighbor-Joining (NJ) method implemented in the MEGA 6.0 software with JTT model and pairwise gap deletion option. The bootstrap analysis was conducted with 1000 iterations. (B) The exon/intron distribution of G. arboreum TCP genes. Exons and introns are represented by green boxes and black lines, respectively. (C) The motif compositions of G. arboreum TCP genes. Each color represents a specific motif.
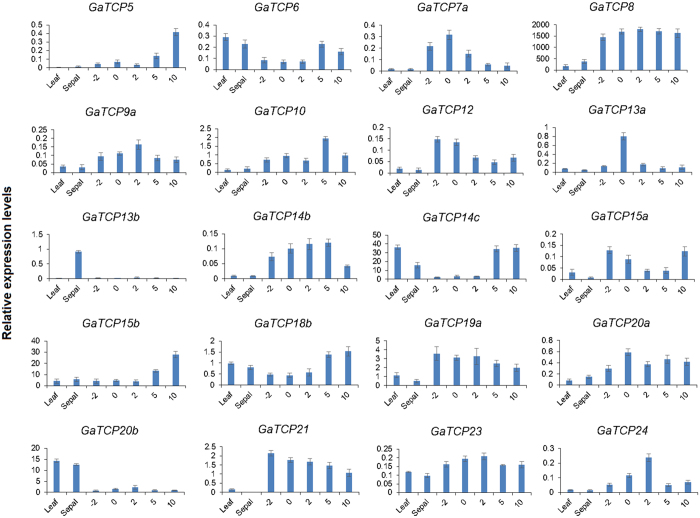
Expression profiles of cotton TCP genes at different developmental stages
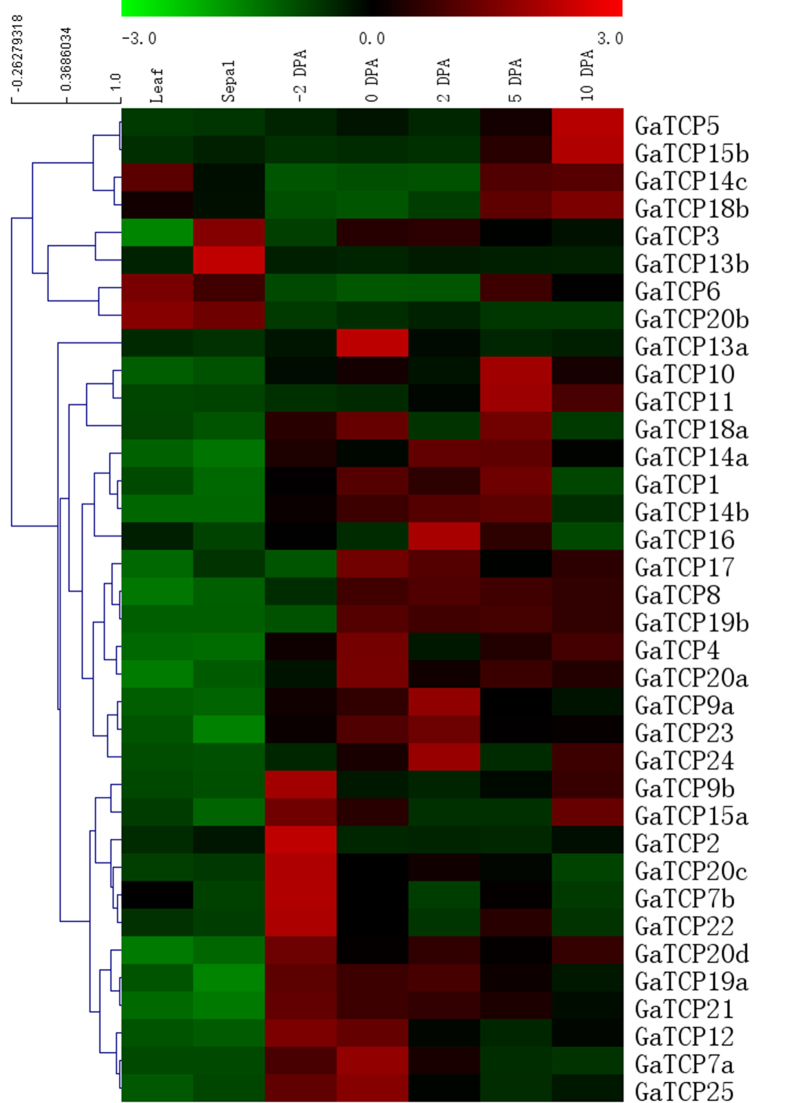
In order to shed light on the potential physiological functions of the TCP gene family in G. arboretum at different developmental stages as well as in cotton fiber development, their expression profiles were investigated using Real-Time Quantitative Reverse Transcription PCR on several different organs, including leaves, sepals and fibers at −2, 0, 2, 5 and 10 DPA. As shown in Figs 4 and 5, the majority of the TCP genes exhibited diverse expression profiles, while a few of them showed similar expression patterns. For example, a number of genes, including GaTCP5, GaTCP8, GaTCP12, GaTCP13a, GaTCP14b, GaTCP15a, GaTCP15b, GaTCP19a, GaTCP21, were exclusively highly expressed in fiber, while GaTCP13b and GaTCP20b were preferentially expressed in leaves or sepals at the high levels. Additionally, several TCP genes had high expression levels in all the tissues examined, such as GaTCP6, GaTCP18b and GaTCP23. Among those genes that were highly expressed in fibers, GaTCP7a, GaTCP12, GaTCP13a and GaTCP24 were highly expressed at the fiber initiation stage (from −2 to 2 DPA), whereas the expression levels of GaTCP5, GaTCP10, GaTCP14c and GaTCP15b were significantly high at the fiber elongation stage (from 5 to 10 DPA). Remarkably, GaTCP8 had extremely high expression levels at all the fiber developmental stages tested.
Figure 4. Expression profiles of G. arboreum TCP genes in different tissues and at different fiber developmental stage.
The expression levels are represented by the color bar.
Figure 5. Expression profiles of 20 G. arboreum TCP genes in different tissues and at different fiber developmental stage.
The X-axis represents different tissues or developmental stages while the Y-axis represents relative expression levels against the reference gene SAD1. Error bars are drawn based on standard deviation for three replicates.
Discussion
The TCP transcription factor family plays important roles in many biological processes during plant growth and development, such as fiber development13,14, seed germination19,20, leaf development21, hormone signal transduction22 and flower development21,22,23. The recent availability of G. arboreum genome sequences17 allowed us to perform a comprehensive analysis of this family in cotton, including phylogenetic analysis, chromosome location, gene duplication status, gene structure and conserved motif analysis, as well as tissue specific expression profiles.
Evolutionary conservation and divergence of the TCP gene family in cotton
The G. arboreum genome contains almost the same number of TCP genes as in G. raimondii18 and have more than 50% more than in Arabidopsis2, which is in consistency with the number of protein coding genes in each species17,24,25. It has been reported that Arabidopsis and G. arboreum evolved from a common ancestor at around 93 Mya and subsequently underwent paleopolyploidy events24,26. Our phylogenetic analysis showed that TCP genes in cotton and Arabidopsis might go through differential expansion caused by gene duplication, an important source of raw genetic materials to the evolution of complex plant systems27,28. This is supported by the fact that the number of paralogous TCP gene pairs accounted for over 70% of the entire TCP gene family in G. arboreum and that segmental duplication is a predominant duplication event for GaTCPs. Previous studies indicated that gene duplication contributed to the amplification of gene family members on various scales, such as tandem duplication, segmental duplication and whole-genome duplication, and that the expansion of regulatory genes can hardly ever be achieved simply through single gene duplication alone27,28,29, implying that genome duplication may also contribute to the amplification of the GaTCP gene family. According to a recent study, a recent and an ancient whole genome duplication event have occurred in G. arboreum at approximately 13–20 and 115–146 Mya, respectively17. Our results indicated that the average duplication date of GaTCPs was around 19.7 Mya, which is consistent with the recent whole genome duplication event. This suggests that genome duplication may also play a significant role in the expansion of the GaTCP gene family.
In addition to gene duplication status, differences in exon/intron organizations can also shed light on the evolutionary history of gene families. In this study, we compared the gene structure of GaTCPs with their homologous counterparts in Arabidopsis18. The results showed that eight of ten pairs of TCP genes shared conserved exon/intron distribution in terms of exon length and intron numbers, whereas two pairs displayed some extent of divergence. AtTCP12, for instance, contained one more intron than its counterpart GaTCP12. Such intron loss of gain may result from insertion/deletion events in the process of evolution30.
According to previous reports, upland cotton (G. hirsutum), an AD tetraploid, evolved from A-genome diploids G. arboreum and D-genome diploids G. raimondii at around 1–2 Mya16. Due to high similarities between the A and D genomes in terms of gene sequence and genome organization, the TCP family members in G. arboreum and G. raimondii formed a lot of clusters of homologs in the phylogenetic tree and shared almost identical exon/intron structures as well as motif compositions18. Our phylogenetic analysis also showed that GaTCPs and GrTCPs were more closely allied to TCP proteins from T. cacao, a close relative of cotton in the Malvaceae family, than from other plant species, consistent with the fact that G. arboreum, G. raimondii and T. cacao originated from a common ancestor 33 Mya17. Since GaTCPs and GrTCPs duplicated at around 19.7 million years ago, these duplications happened after their divergence from T. cacao and Arabidopsis but before the reunion of the A and D genome diploids that gave rise to upload cotton.
Functional divergence of the TCP gene family in cotton
It has been widely recognized that duplicated genes undergo one of the following evolutionary fates: pseudogenization (in which one copy becomes unexpressed or functionless), conservation of gene function (in which both copies maintain the same function), subfunctionalization (in which the ancestral function is subdivided between copies), and neofunctionalization (in which one copy acquires a new function)27. In the present study, the expression profiles of GaTCPs at different developmental stages were investigated to reveal their functional divergence during plant growth and development.
Our results showed that the majority of the paralogous GaTCP gene pairs exhibited differential expression profiles. GaTCP7a, for example, was preferentially expressed in fiber at the initiation stage, while its paralogous counterpart GaTCP23 was highly expressed in fiber at both the initiation stage and the elongation stage. GaTCP14c had extremely high expression level in leaf and fiber at the elongation stage, whereas GaTCP14b was relatively highly expressed in fiber at the initiation stage. In general, the expression patterns of GaTCP genes imply that the TCP family may perform multiple physiological functions in G. arboreum, especially in fiber initiation and elongation. Remarkably, some of these findings have already been experimentally confirmed through analysis of mutant cotton species with reduced and/or overexpressed TCP activities. For instance, the expression level of GaTCP15b was significantly high in fiber at the elongation stage. Hao et al. demonstrated that GbTCP, the homologous counterpart of GaTCP15b in G. barbadense, confers cotton fiber elongation by regulating JA biosynthesis and response and other pathways using RNAi silencing technique14. In addition, Wang et al. demonstrated that the GhTCP14 from upland cotton functions as a crucial regulator in auxin-mediated elongation of cotton fiber cells13, which is in agreement with our result that GaTCP14c was highly expressed in fiber at the elongation stage. However, further functional analysis of GaTCPs are still needed in order to determine which evolutionary fates the duplicated GaTCP genes undergo during the process of sequence and functional evolution.
Comparison of the TCP gene family between G. arboreum and G. raimondii
Molecular systematics studies show that the most cultivated cotton species G. hirsutum is produced by interspecific hybridization of A-genome (like G. arboreum) and D-genome (like G. raimondii) diploid progenitor species, which makes it necessary to make a comparison analysis on gene structure, chromosomal distribution and gene duplication of the TCP gene family between G. arboreum and G. raimondii. Based on genome scans of the two cotton species, a total of 38 and 36 TCP genes were identified in G. raimondii and G. arboreum, respectively. Comparison of gene structures indicated that the orthologous TCP gene pairs exhibited a highly conserved distribution of exons and introns either in terms of intron numbers or gene length. In addition, these TCP genes also shared similar gene duplication status that both segmental duplication and whole-genome duplication contributed significantly to the expansion of TCP genes. In contrast, great variety was observed in their chromosomal distribution. For instance, The GrTCPs were unevenly distributed across 11 out of the 13 G. raimondii chromosomes, ranging widely from 0 to 8 genes per chromosome, whereas the GaTCPs were more evenly scattered throughout all 13 chromosomes of G. arboreum. The cause of this great variety is still unclear. It might be related to the high Long Terminal Repat (LTR) activities that contributed to the twofold increase in the size of the G. arboreum genome.
Materials and Methods
Identification of TCP genes and proteins
The genome sequence of G. arboreum was downloaded from the Cotton Genome Project (CGP) (http://cgp.genomics.org.cn/). To identify the TCP family in G. arboretum, the profile hidden Markov Models of TCP domain (PF03634) downloaded from Pfam was used as query to run Hmmsearch against the G. arboretum genome (P-value = 0.0011). The candidate TCP genes were further aligned to remove redundant sequences31. Subsequently, the TCP sequences were manually inspected with ScanProsite to confirm the presence of the conserved TCP domain32. The TCP gene and protein sequences from Theobroma cacao, Vitis vinifera, Arabidopsis thaliana and Oryza sativa were retrieved from PlantTFDB plant transcription factor database, while the GrTCP sequences were obtained from previous studies18.
Phylogenetic analysis
Cluster X program was employed to perform multiple sequence alignments with default parameters33. Unrooted phylogenetic trees were subsequently constructed using the Neighbor-Joining (NJ) method implemented in the MEGA 6.0 software with JTT model and pairwise gap deletion option34. The bootstrap analysis was conducted with 1000 iterations.
Chromosomal location and gene duplication
The physical location data of GaTCP genes were retrieved from G. arboreum genome. Mapping of these GaTCP genes was then performed using MapInspect software. Gene duplication was defined according to the criteria descripted in previous studies: the aligned region of two sequences covers over 70% of the longer sequence and the similarity of the aligned region is over 70%35,36. In addition, the DnaSp software37 was employed to calculate Ka (nonsynonymous substitution rate) and Ks (synonymous substitution rate), which was further used to estimate the date of duplication events with the formula T = Ks/2λ, assuming clock-like rate (λ) of 1.5 synonymous substitutions per 108 years for cotton24.
Gene structure and conserved motif
The exon/intron organizations of GaTCPs were inferred through comparison of genomic sequences and CDS sequences in the gene structure display server38. The program MEME39 was employed to identify conserved motifs in GaTCPs with the following parameters: the optimum width of motif, 6–250; the maximum number of motif, 20; the number of repetitions, any. In addition, motif annotation was performed using the program InterProScan40.
RNA isolation and Real-time quantitative RT-PCR analysis
Total RNA was isolated from G. arboreum leaves, sepals and fibers at −2, 0, 2, 5 and 10 days post anthesis (DPA) using the mirVanaTM miRNA Isolation Kit (Ambion, USA). Subsequently, the NanoDrop ND-1000 Spectrophotometer was employed to determine RNA concentration and quality. cDNA was then synthesized from 1 μg of total RNA with poly-T primers using the TaqMan® MicroRNA Reverse Transcription Kit (Applied Biosystems, USA). RT-qPCR was later conducted on 7300 Real-Time PCR System (Applied Biosystems, USA) according to the manufacture’s protocol. The amplification parameters were as follows: enzyme activation at 95 °C for 10 min, 45 cycles of denaturation at 95 °C for 15 s and annealing/elongation at 60 °C for 60 s. The relative expression levels was calculated according to previous studies18. A reference genes SAD1 was used to normalize the expression values. There were three biological replicates and each biological replicate was run three times. Finally, the software MultiExperiment Viewer was used to construct heatmap representation for expression patterns.
Additional Information
How to cite this article: Ma, J. et al. Comprehensive analysis of TCP transcription factors and their expression during cotton (Gossypium arboreum) fiber early development. Sci. Rep. 6, 21535; doi: 10.1038/srep21535 (2016).
Acknowledgments
This work is partially support by the Cotton Incorporated.
Footnotes
Author Contributions J.M., F.L., Q.W., K.W., D.C.J. and B.Z. designed the experiments and analyzed the data. J.M. and B.Z. wrote the manuscript text. J.M. performed the experiments. All authors reviewed the manuscript.
References
- Cubas P., Lauter N., Doebley J. & Coen E. The TCP domain: a motif found in proteins regulating plant growth and development. Plant J. 18, 215–222, doi: 10.1046/j.1365-313X.1999.00444.x (1999). [DOI] [PubMed] [Google Scholar]
- Martin-Trillo M. & Cubas P. TCP genes: a family snapshot ten years later. Trends Plant Sci. 15, 31–39, doi: 10.1016/j.tplants.2009.11.003 (2010). [DOI] [PubMed] [Google Scholar]
- Riechmann J. L. et al. Arabidopsis Transcription Factors: Genome-Wide Comparative Analysis among Eukaryotes. Science 290, 2105–2110, doi: 10.2307/3081600 (2000). [DOI] [PubMed] [Google Scholar]
- Yao X., Ma H., Wang J. & Zhang D. Genome-Wide Comparative Analysis and Expression Pattern of TCP Gene Families in Arabidopsis thaliana and Oryza sativa. J. Integr. Plant Biol. 49, 885–897, doi: 10.1111/j.1744-7909.2007.00509.x (2007). [DOI] [Google Scholar]
- Navaud O., Dabos P., Carnus E., Tremousaygue D. & Herve C. TCP transcription factors predate the emergence of land plants. J. Mol. Evol. 65, 23–33, doi: 10.1007/s00239-006-0174-z (2007). [DOI] [PubMed] [Google Scholar]
- Pagnussat G. C. et al. Genetic and molecular identification of genes required for female gametophyte development and function in Arabidopsis. Development 132, 603–614, doi: 10.1242/dev.01595 (2005). [DOI] [PubMed] [Google Scholar]
- Sarvepalli K. & Nath U. Hyper-activation of the TCP4 transcription factor in Arabidopsis thaliana accelerates multiple aspects of plant maturation. Plant J. 67, 595–607, doi: 10.1111/j.1365-313X.2011.04616.x (2011). [DOI] [PubMed] [Google Scholar]
- Takeda T. et al. RNA interference of the Arabidopsis putative transcription factor TCP16 gene results in abortion of early pollen development. Plant Mol. Biol. 61, 165–177, doi: 10.1007/s11103-006-6265-9 (2006). [DOI] [PubMed] [Google Scholar]
- Giraud E. et al. TCP transcription factors link the regulation of genes encoding mitochondrial proteins with the circadian clock in Arabidopsis thaliana. Plant cell 22, 3921–3934, doi: 10.1105/tpc.110.074518 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammani K. et al. An Arabidopsis dual-localized pentatricopeptide repeat protein interacts with nuclear proteins involved in gene expression regulation. Plant cell 23, 730–740, doi: 10.1105/tpc.110.081638 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda T. et al. The OsTB1 gene negatively regulates lateral branching in rice. Plant J. 33, 513–520 (2003). [DOI] [PubMed] [Google Scholar]
- Aguilar-Martinez J. A. & Sinha N. Analysis of the role of Arabidopsis class I TCP genes AtTCP7, AtTCP8, AtTCP22, and AtTCP23 in leaf development. Front. Plant Sci. 4, 406, doi: 10.3389/fpls.2013.00406 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M. Y. et al. The cotton transcription factor TCP14 functions in auxin-mediated epidermal cell differentiation and elongation. Plant Physiol. 162, 1669–1680, doi: 10.1104/pp.113.215673 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao J. et al. GbTCP, a cotton TCP transcription factor, confers fibre elongation and root hair development by a complex regulating system. J. Exp. Bot. 63, 6267–6281, doi: 10.1093/jxb/ers278 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J. et al. Expression profiles of miRNAs in Gossypium raimondii. J. Zhejiang Univ. Sci. B 16, 296–303, doi: 10.1631/jzus.B1400277 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson A. H. et al. Repeated polyploidization of Gossypium genomes and the evolution of spinnable cotton fibres. Nature 492, 423–427, doi: 10.1038/nature11798 (2012). [DOI] [PubMed] [Google Scholar]
- Li F. et al. Genome sequence of the cultivated cotton Gossypium arboreum. Nat. Genet. 46, 567–572, doi: 10.1038/ng.2987 (2014). [DOI] [PubMed] [Google Scholar]
- Ma J. et al. Genome-wide identification and expression analysis of TCP transcription factors in Gossypium raimondii. Sci. Rep. 4, 6645, doi: 10.1038/srep06645 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatematsu K., Nakabayashi K., Kamiya Y. & Nambara E. Transcription factor AtTCP14 regulates embryonic growth potential during seed germination in Arabidopsis thaliana. Plant J. 53, 42–52, doi: 10.1111/j.1365-313X.2007.03308.x (2008). [DOI] [PubMed] [Google Scholar]
- Rueda-Romero P., Barrero-Sicilia C., Gomez-Cadenas A., Carbonero P. & Onate-Sanchez L. Arabidopsis thaliana DOF6 negatively affects germination in non-after-ripened seeds and interacts with TCP14. J. Exp. Bot. 63, 1937–1949, doi: 10.1093/jxb/err388 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieffer M., Master V., Waites R. & Davies B. TCP14 and TCP15 affect internode length and leaf shape in Arabidopsis. Plant J. 68, 147–158, doi: 10.1111/j.1365-313X.2011.04674.x (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Z. et al. TCP1 modulates brassinosteroid biosynthesis by regulating the expression of the key biosynthetic gene DWARF4 in Arabidopsis thaliana. Plant cell 22, 1161–1173, doi: 10.1105/tpc.109.069203 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyama T., Ohme-Takagi M. & Sato F. Generation of serrated and wavy petals by inhibition of the activity of TCP transcription factors in Arabidopsis thaliana. Plant Signal. Behav. 6, 697–699 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K. et al. The draft genome of a diploid cotton Gossypium raimondii. Nat. Genet. 44, 1098–1103, doi: 10.1038/ng.2371 (2012). [DOI] [PubMed] [Google Scholar]
- Arabidopsis Genome I. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408, 796–815, doi: 10.1038/35048692 (2000). [DOI] [PubMed] [Google Scholar]
- Blanc G. & Wolfe K. H. Widespread paleopolyploidy in model plant species inferred from age distributions of duplicate genes. Plant cell 16, 1667–1678, doi: 10.1105/tpc.021345 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J. Evolution by gene duplication: an update. Trends Ecol. Evol. 18, 292–298, doi: 10.1016/S0169-5347(03)00033-8 (2003). [DOI] [Google Scholar]
- Flagel L. E. & Wendel J. F. Gene duplication and evolutionary novelty in plants. New Phytol. 183, 557–564, doi: 10.1111/j.1469-8137.2009.02923.x (2009). [DOI] [PubMed] [Google Scholar]
- Van de Peer Y., Maere S. & Meyer A. The evolutionary significance of ancient genome duplications. Nat. Rev. Genet. 10, 725–732, doi: 10.1038/nrg2600 (2009). [DOI] [PubMed] [Google Scholar]
- Lecharny A., Boudet N., Gy I., Aubourg S. & Kreis M. Introns in, introns out in plant gene families: a genomic approach of the dynamics of gene structure. J. Struct. Funct. Genomics 3, 111–116 (2003). [PubMed] [Google Scholar]
- Thompson J. D., Higgins D. G. & Gibson T. J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Castro E. et al. ScanProsite: detection of PROSITE signature matches and ProRule-associated functional and structural residues in proteins. Nucleic Acids Res. 34, W362–365, doi: 10.1093/nar/gkl124 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J. D., Gibson T. J., Plewniak F., Jeanmougin F. & Higgins D. G. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25, 4876–4882 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A. & Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729, doi: 10.1093/molbev/mst197 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T. et al. Genome-wide identification of NBS genes in japonica rice reveals significant expansion of divergent non-TIR NBS-LRR genes. Mol. Genet. Genomics 271, 402–415, doi: 10.1007/s00438-004-0990-z (2004). [DOI] [PubMed] [Google Scholar]
- Yang S., Zhang X., Yue J. X., Tian D. & Chen J. Q. Recent duplications dominate NBS-encoding gene expansion in two woody species. Mol. Genet. Genomics 280, 187–198, doi: 10.1007/s00438-008-0355-0 (2008). [DOI] [PubMed] [Google Scholar]
- Librado P. & Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25, 1451–1452, doi: 10.1093/bioinformatics/btp187 (2009). [DOI] [PubMed] [Google Scholar]
- Guo A. Y., Zhu Q. H., Chen X. & Luo J. C. [GSDS: a gene structure display server]. Yi Chuan 29, 1023–1026 (2007). [PubMed] [Google Scholar]
- Bailey T. L., Williams N., Misleh C. & Li W. W. MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 34, W369–373, doi: 10.1093/nar/gkl198 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quevillon E. et al. InterProScan: protein domains identifier. Nucleic Acids Res. 33, W116–120, doi: 10.1093/nar/gki442 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]