BAG6 processing confers plant immunity via autophagy.
Abstract
The Bcl-2-associated athanogene (BAG) family is an evolutionarily conserved group of cochaperones that modulate numerous cellular processes. Previously we found that Arabidopsis thaliana BAG6 is required for basal immunity against the fungal phytopathogen Botrytis cinerea. However, the mechanisms by which BAG6 controls immunity are obscure. Here, we address this important question by determining the molecular mechanisms responsible for BAG6-mediated basal resistance. We show that Arabidopsis BAG6 is cleaved in vivo in a caspase-1-like-dependent manner and via a combination of pull-downs, mass spectrometry, yeast two-hybrid assays, and chemical genomics, we demonstrate that BAG6 interacts with a C2 GRAM domain protein (BAGP1) and an aspartyl protease (APCB1), both of which are required for BAG6 processing. Furthermore, fluorescence and transmission electron microscopy established that BAG6 cleavage triggers autophagy in the host that coincides with disease resistance. Targeted inactivation of BAGP1 or APCB1 results in the blocking of BAG6 processing and loss of resistance. Mutation of the cleavage site blocks cleavage and inhibits autophagy in plants; disease resistance is also compromised. Taken together, these results identify a mechanism that couples an aspartyl protease with a molecular cochaperone to trigger autophagy and plant defense, providing a key link between fungal recognition and the induction of cell death and resistance.
INTRODUCTION
As sessile organisms devoid of an adaptive immune system, plants must continually and rapidly adjust to ever-changing environmental conditions. To cope with both biotic and abiotic environmental pressures, plants have developed a number of defense strategies (Jones and Dangl, 2006). Recent studies have established that control of programmed cell death (PCD) pathways can be an important component that controls the outcome of a given stress response in plants (Dickman et al., 2001; Doukhanina et al., 2006; Lord and Gunawardena, 2012; Kabbage et al., 2013). Apoptosis and autophagy are two principal forms of PCD broadly studied in eukaryotes. It has been suggested that such pathways are conserved, which is evident when comparing worms and flies all the way to humans. However, while conserved core regulators of apoptotic cell death (e.g., Bcl-2 family and caspases) have been identified and extensively characterized in metazoans, such core regulators (or functional equivalents) in general await identification in plants. Conversely, there is clear evidence that key genes necessary for autophagy in mammals are conserved in plants.
As an approach to address whether PCD-related genes have structural and, thus, possibly functional similarities to their mammalian counterparts, we used computational approaches searching for plant genes that structurally resembled apoptotic PCD genes, and as a result we uncovered the Bcl-2-associated athanogene (BAG) protein family in Arabidopsis thaliana (Doukhanina et al., 2006; Kabbage and Dickman, 2008). While plant and human BAG proteins have relatively low sequence identity that initially obscured detection by BLAST searches, these proteins were found to be remarkably similar in several key structural regions. Importantly, these observations extend to functional similarity as well (Doukhanina et al., 2006). In particular, human and plant BAG proteins show high degrees of structural conservation in the functional portions of the protein (e.g., helical structure and hydrophobicity; Doukhanina et al., 2006) that when taken together account for functional similarity. In accordance, our studies have indicated that plant BAG family members are also multifunctional and similar to their animal counterparts, as they block several biotic and abiotic cell death-mediated processes while conferring cytoprotection in situations ranging from pathogen attack to drought stress to plant development (Doukhanina et al., 2006; Williams et al., 2010).
The first BAG gene was discovered in a screen of a mouse embryo cDNA library using recombinant human Bcl-2 protein as bait to identify Bcl-2 interactors and was named BAG1 (Takayama et al., 1995). BAG1 synergistically enhanced cell survival with Bcl-2, suggesting involvement in apoptotic PCD pathways. Subsequent studies revealed a family of BAG proteins and more accurately indicated that BAGs function as molecular cochaperones (Takayama and Reed, 2001). BAG proteins are distinguished by the presence of a common conserved C-terminal BAG domain that mediates their interaction with Hsp70/Hsc70 molecular chaperones (Brive et al., 2001; Takayama and Reed, 2001; Kabbage and Dickman, 2008). Six BAG family members have been identified in humans and regulate, both positively and negatively, the function of Hsp70/Hsc70 chaperones, forming complexes with a range of transcription factors and signaling proteins to modulate diverse physiological processes, including apoptosis, tumorigenesis, neuronal differentiation, stress responses, and cell cycle progression (Kabbage and Dickman, 2008). In contrast to mammals, far less is known about the structure and function of BAG proteins in plants. Among the seven predicted BAG homologs identified in Arabidopsis, four (BAG1-4) have domain organizations similar to animal members. The remaining three members contain a predicted calmodulin binding motif near the BAG domain, a feature unique to plant BAG proteins indicating possible divergent mechanisms associated with plant-specific functions. As noted for mammalian BAGs, plant BAGs also regulate several stress and developmental processes (Kabbage and Dickman, 2008; Williams et al., 2010). Thus structural approaches can be used for prediction of protein function independent of sequence similarity.
Based on our initial results characterizing this gene family, we were particularly interested in Arabidopsis BAG6, owing to its potential involvement with basal immunity. T-DNA knockout lines (bag6) showed enhanced susceptibility to the necrotrophic fungus Botrytis cinerea (Doukhanina et al., 2006). B. cinerea challenge to wild-type Arabidopsis (Col-0) typically resulted in an initial short growth phase emanating from the point of inoculation, transitioning to a rapid delimited cell death phenotype. Following inoculation with B. cinerea, bag6 knockout mutants exhibited a disease phenotype including rapidly spreading cell death and unrestricted fungal growth (Doukhanina et al., 2006). This response was specific to bag6 mutants, as these observations were not noted in knockout lines of other BAG family members. Together, these observations suggest that a specific function of BAG6 is associated with basal resistance.
In this study, we functionally characterize Arabidopsis BAG6 and identify relevant protein interactors and associated pathways. We show that BAG6 is cleaved at a specific caspase-1-like cleavage site downstream of its BAG domain. The cleavage of BAG6 was demonstrated in vitro using recombinant human caspase-1 and in vivo following pathogen-associated molecular pattern (PAMP; chitin) treatment or B. cinerea challenge, indicating that processing of BAG6 is required for basal resistance against this pathogen. Using a combination of pull-downs, mass spectrometry, and yeast two-hybrid approaches, we also show that BAG6 forms a complex with two additional partners, a C2 GRAM domain-containing protein that we designated BAGP1 (for BAG-Associated GRAM Protein) and an aspartyl protease APCB1 (for Aspartyl Protease Cleaving BAG). Both of these partner proteins are required for BAG6 processing. Furthermore, the cleavage of BAG6 leads to autophagy that coincides with and is necessary for disease resistance. Collectively, we demonstrate that autophagy triggered by the cleavage of BAG6 confers resistance to the necrotrophic fungal pathogen B. cinerea. Here, we discuss and provide evidence for the molecular underpinning of basal resistance of Arabidopsis to B. cinerea.
RESULTS
Arabidopsis bag6 Mutants Show Enhanced Susceptibility to B. cinerea
Previously we described the identification and preliminary characterization of the Arabidopsis BAG gene family (Doukhanina et al., 2006; Kabbage and Dickman, 2008). T-DNA knockout lines were obtained for all seven members of the BAG gene family. We determined phenotypes for these lines with respect to a range of abiotic and biotic stresses. Of particular note, bag6 knockout lines, when inoculated with the necrotrophic fungal pathogen B. cinerea, were markedly more susceptible compared with Col-0 plants (Doukhanina et al., 2006). We did not observe this enhanced disease phenotype in any other T-DNA lines or on any lines inoculated with the related fungal necrotroph Sclerotinia sclerotiorum.
When inoculated onto Arabidopsis, B. cinerea exhibited a slow, restricted growth pattern with a distinct constrained ring of dead cells surrounding the inoculation point (Supplemental Figure 1). This observation is further supported by the presence of several defense-associated markers, including a pronounced oxidative burst and callose deposition at the site of inoculation (Supplemental Figure 2). In contrast, bag6 knockout lines (bag6-1, SALK_009534C; bag6-2, SALK_073331C) were permissive to fungal growth and a rapidly spreading cell death was observed displaying the hallmarks of a compatible/susceptible interaction (Supplemental Figure 1) that was accompanied by a much weaker oxidative burst and absence of callose accumulation (Supplemental Figure 2). Together, these data suggest that BAG6 may play a role in plant basal resistance.
Overexpression of BAG6 in Arabidopsis and Tobacco Results in a Lesion Mimic Phenotype
We expressed Arabidopsis BAG6, with vectors containing HA and YFP fused to full-length BAG6 under the control of its native promoter (NP:HA- or YFP-BAG6) or the constitutive cauliflower mosaic virus 35S promoter (35S:HA or YFP-BAG6) and transferred these constructs into bag6 knockout Arabidopsis plants. Transgene expression was confirmed by RT-PCR (data not shown). No morphological abnormalities were observed in bag6 mutant plants or Arabidopsis that was functionally complemented with NP:HA/YFP-BAG6 (Figure 1). However, transgenic plants overexpressing BAG6 (35S:HA or YFP-BAG6) were stunted, had slightly deformed leaves, and displayed a distinct lesion mimic phenotype (Figure 1; Supplemental Figure 3). Lesion mimic mutant phenotypes are observed when plants exhibit disease (lesions) symptoms in the absence of a pathogen (Lorrain et al., 2003). Numerous phenotypes have been described in association with lesion mimics in many plant species (Dangl et al., 1996; Buckner et al., 2000; Zeng et al., 2004). In the BAG6-overexpressing lines, these lesions appeared as small translucent flecks in younger leaves but coalesced to cover entire leaves as they matured (Figure 1). A similar phenotype was observed following the overexpression of Arabidopsis BAG6 in tobacco (Nicotiana tabacum) in a light-dependent manner (Supplemental Figure 4); light sensitivity is known as an important environmental factor affecting lesion mimic development (Johal et al., 1995; Hu et al., 1998; Buckner et al., 2000). The position of the tag did not affect lesion development, as similar phenotypes were observed with C-terminal fusions (Supplemental Figure 3). Overall, the overexpression of BAG6 induces typical lesion mimic symptoms in plants.
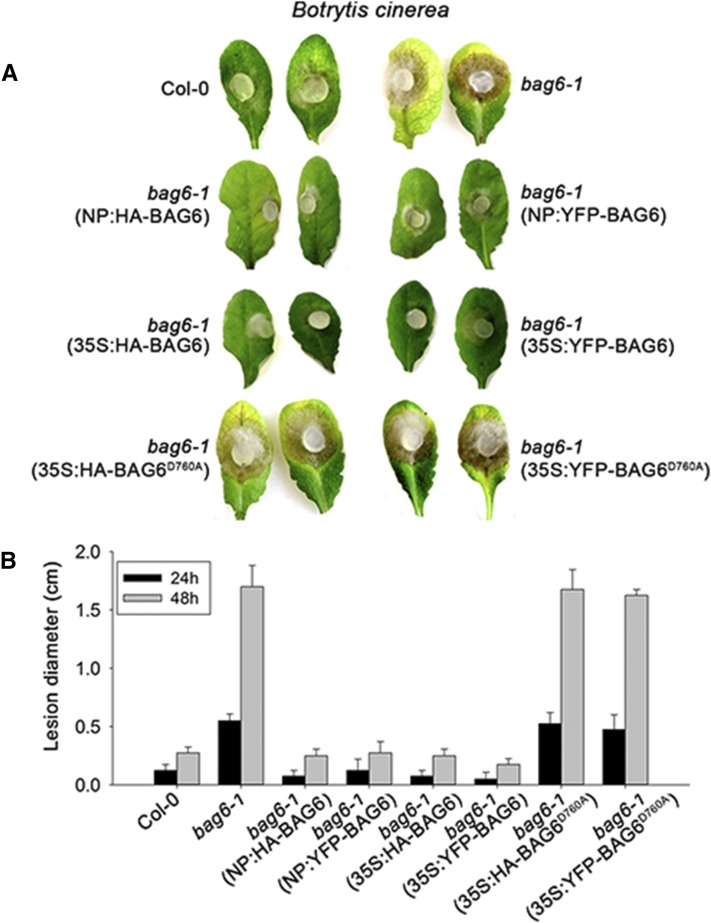
Figure 1.
Overexpression of BAG6 in Arabidopsis Induces Lesion Mimic Development.
Arabidopsis bag6-1 T-DNA insertion mutant was transformed with N-terminal HA- or YFP-tagged BAG6 under either 35S promoter or native promoter (NP) by the floral dip method. Stable transgenic plants harboring BAG6 driven by the 35S promoter (35S:HA-BAG6 and 35S:YFP-BAG6) exhibited apparent lesion mimics on leaves ([D] and [F]), while plants expressing BAG6 driven by the native promoter (NP:HA-BAG6 and NP:YFP-BAG6) ([C] and [H]) were similar to wild-type Col-0 and bag6-1 mutant ([A] and [B]). Overexpression of a cleavage-deficient form of BAG6, BAG6D760A, driven by 35S promoter (35S:HA-BAG6D760A and 35S:YFP-BAG6 D760A), abolished the lesion mimic phenotype in the bag6 mutant background ([E] and [G]). Representative plants were photographed. White circles show the typical lesion mimic phenotype.
The Lesion Mimic Phenotype Is Associated with BAG6 Processing in a Caspase-1-Like-Dependent Manner
We extracted total proteins from transgenic plants expressing BAG6 (35S:HA-BAG6) and performed immunoblot analysis with anti-HA antibody. We detected the BAG6 protein at the predicted molecular mass of the full-length HA-BAG6 (∼100 kD) in transgenic Arabidopsis (Figure 2) and tobacco plants (Supplemental Figure 5). However, an additional lower molecular mass protein (∼75 kD) was also consistently observed in lesion mimic leaves but not in leaf tissue without lesions (Figure 2; Supplemental Figure 5). These observations suggest that BAG6 might be processed in planta and such cleavage might be important for the observed lesion mimic phenotype.
Figure 2.
BAG6 Is Cleaved by Human Caspase-1 in Vitro and by Pant Caspase-1-Like Protease Activity in Vivo.
(A) Structure of BAG6, showing the conserved BAG domain, nuclear localization signal (NLS) and calmodulin binding motif (CaM). The arrow indicates the amino acid position (amino acid 760) cleaved by the caspase-1 protease. aa, amino acid.
(B) Arabidopsis BAG6 is cleaved by human caspase-1 in vitro. HA-tagged full-length BAG6 and BAG6D760A under 35S promoter were transiently expressed in N. benthamiana for 3 d. Equal amounts of purified HA-BAG6 (10 μg) were treated with recombinant human caspase-1 (2 units) at 37°C for 2 h and analyzed by immunoblotting with anti-HA antibody. CBS, Coomassie blue staining
(C) BAG6 is cleaved by plant caspase-1-like protease activity in vivo. Proteins were extracted from Arabidopsis plants expressing HA-BAG6 and HA-BAG6D760A under the 35S promoter or native promoter in response to B. cinerea 24 h after inoculation and subjected to immunoblotting using anti-HA antibody (left panel). Proteins from leaves with or without lesion mimics in Arabidopsis plants expressing HA-BAG6 under 35S promoter were monitored for BAG6 cleavage using anti-HA antibody (right panel). Equal loading was confirmed by SDS-PAGE and Coomassie blue staining.
To test this, we first used the ExPASy Peptide Cutter tool (Wilkins et al., 1999) to search for potential protease cleavage sites in BAG6. This search revealed a single predicted, caspase-1 cleavage site (LATD) with expected cleavage occurring after the aspartate (D) residue at position 760 downstream of its BAG domain (Figure 2A). A caspase-1-like activity at this cleavage site would generate two fragments similar in size to the truncated peptide noted in the lesion mimic transgenic plants (Figure 2; Supplemental Figure 5). To determine whether or not caspase-1 cleaves BAG6, an N-terminal HA-tagged BAG6 (HA-BAG6) was expressed in Nicotiana benthamiana leaves via Agrobacterium tumefaciens-mediated transient expression and purified by anti-HA-agarose affinity chromatography. Treatment of the purified BAG6 protein with human caspase-1 resulted in the formation of an additional lower molecular mass band in accordance with our observation in the lesion mimic immunoblots (Figure 2; Supplemental Figure 5). Importantly, we generated a site-directed mutation in BAG6 (BAG6D760A) where the aspartate (D) residue was replaced by alanine (A); this mutation abolished cleavage by human caspase-1 (Figure 2B) and the development of lesion mimics (Figure 1). These results demonstrate that BAG6 processing likely occurs via a caspase-1-like activity during lesion mimic development.
To further validate these observations and place them in more biologically relevant context, the effect of B. cinerea challenge on BAG6 processing in Arabidopsis was evaluated. Transgenic Arabidopsis leaves expressing HA-tagged BAG6 (NP: or 35S:HA-BAG6) or BAG6D760A (35S:HA-BAG6 D760A) were inoculated with an agar plug containing an actively growing culture of B. cinerea. Total proteins were extracted and subjected to an immunoblot assay with anti-HA antibody. As shown in Figure 2C, the cleaved BAG6 protein accumulated following B. cinerea treatment, whereas BAG6D760A was resistant to B. cinerea-induced cleavage. Similar results were obtained in tobacco treated with chitin, a well established fungal PAMP, or B. cinerea (Supplemental Figure 6). These data are also consistent with the responsible protease being of plant origin and further suggest that BAG6 cleavage may be important for the plant resistant response to B. cinerea.
Processed BAG6 Restricts B. cinerea Growth in Arabidopsis
Given that BAG6 is cleaved in response to PAMP treatment and fungal challenge, we next asked whether BAG6 cleavage is causally involved with host resistance. To address this question, Col-0, bag6 mutants, and transgenic Arabidopsis plants expressing BAG6 and BAG6D760A were challenged by B. cinerea. As previously discussed, bag6 mutants exhibited a runaway cell death when inoculated with this pathogen (Figure 3; Supplemental Figure 1). Complementation of these mutants with BAG6 under the control of both native and constitutive promoters resulted in a delimited lesion phenotype similar to Col-0 plants (Figure 3). Importantly, BAG6D760A, in which BAG6 cleavage is blocked, was unable to complement the bag6 mutant phenotypes in Arabidopsis (Figure 3). Overexpression of BAG6 but not BAG6D760A was also able to provide enhanced resistance to B. cinerea in tobacco, which is normally a compatible host for this pathogen (Supplemental Figure 7). These data further suggest that BAG6 cleavage correlates with basal resistance.
Figure 3.
The Processing of BAG6 Is Required for Resistance to B. cinerea.
(A) Agar plugs containing actively growing cultures of B. cinerea were inoculated onto leaves of Arabidopsis plants. Representative images were taken 48 h after inoculation.
(B) Lesion diameters were measured at 24 and 48 h after inoculation. Data represent means ± sd from three replicates.
Identification of a Plant Caspase-1-Like Protease Required for BAG6 Processing
To identify the responsible protease for BAG6 cleavage, we treated plants with a suite of known target protease inhibitors and evaluated subsequent cleavage profiles following PAMP treatment. Comparison of aspartyl protease (pepstatin), serine protease (aprotitin and leupeptin), leucine protease (bestatin), and cysteine protease (leupeptin, E-64, and N-ethylmaleimide) inhibitors, with respect to BAG6 processing, was conducted. Pepstatin was the only inhibitor that effectively blocked cleavage of BAG6 (Figure 4). These results show that pepstatin treatment inhibits BAG6 cleavage and further suggest that an aspartyl protease activity is required for cleavage and necessary for host resistance.
Figure 4.
The Cleavage of BAG6 by Caspase-1-Like Activity Is Abolished by Aspartyl Protease Activity.
N. benthamiana leaves transiently expressing HA-fused Arabidopsis BAG6 under the 35S promoter were infiltrated with 200 μg/mL chitin (to induce BAG6 cleavage), in combination with individual protease inhibitor (1 μM pepstatin, 1 mM aprotitin, 0.5 mM bestatin, 1 mM leupeptin, 10 μM E-64, and 5 mM N-ethylmaleimide) 30 min before harvesting. BAG6 cleavage was detected by anti-HA immunoblotting. Equal loading was confirmed by SDS-PAGE and Coomassie blue staining (CBS).
Next, we performed a systematic biochemical analysis to identify potential BAG6 binding partners. To do so, we conducted pull-down assays followed by mass spectrometry. HA-BAG6 was transiently expressed in N. benthamiana; total proteins were extracted 3 d after infiltration and HA-BAG6 was purified using immobilized anti-HA antibodies. Equal amounts of protein samples were separated by 10% SDS-PAGE and silver stained. A specific protein band was pulled down with BAG6 and identified by mass spectrometry (Supplemental Figure 8). This analysis identified a sequence homologous to a C2 and GRAM domain-containing protein in the Arabidopsis protein database (http://www.arabidopsis.org), which we designated BAGP1.
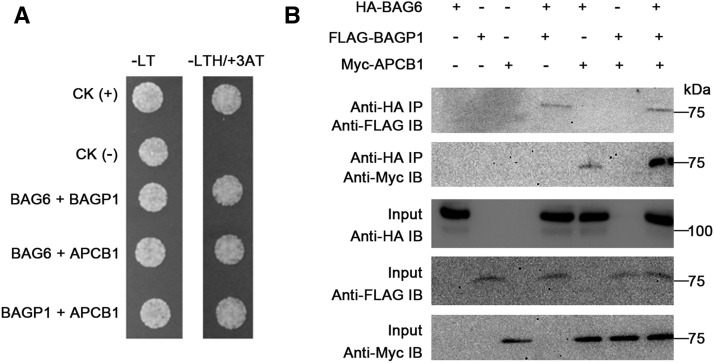
Interestingly, further computational analysis using the database STRING (STRING 9.1; http://string-db.org), which detects known and predicted protein interactions, identified an aspartyl protease as a potential BAGP1 interactor with a score of 0.7 (high confidence). Considering the protease inhibitor results (Figure 4), this aspartyl protease, designated APCB1, was further examined. First, we tested the possibility that this protein physically associates with BAG6. Yeast two-hybrid assays showed that BAG6, PCB1, and BAGP1 all interacted with each other (Figure 5A), possibly as a complex. We also performed coimmunoprecipitation (co-IP) assays by coexpressing HA-BAG6, FLAG-BAGP1, and Myc-PCB1 in Arabidopsis protoplasts. Each protein could immunoprecipitate the other two in vivo, supporting the idea that these three proteins can associate to form a complex (Figure 5B; Supplemental Figure 9).
Figure 5.
BAG6 Interacts with a GRAM Domain-Containing Protein (BAGP1) and an Aspartyl Protease (APCB1) in Yeast and in Planta.
(A) Yeast two-hybrid assay was used to determine the interactions of BAG6 with BAGP1 and APCB1. pGADT7-T and pGBKT7-53, and pGADT7-T and pGBKT-Lam were used as positive CK (+) and negative CK (–), respectively.
(B) BAG6, BAGP1, and APCB1 interact in planta. 35S promoter-driven-HA-BAG6 was coexpressed with FLAG-BAGP1 and Myc-APCB1 in Arabidopsis Col-0 protoplasts. Co-IP was performed with an anti-HA antibody (IP) and proteins analyzed using immunoblotting with either anti-FLAG or anti-Myc antibody (IB).
Role of BAG6 Interacting Partners in Cell Death Regulation
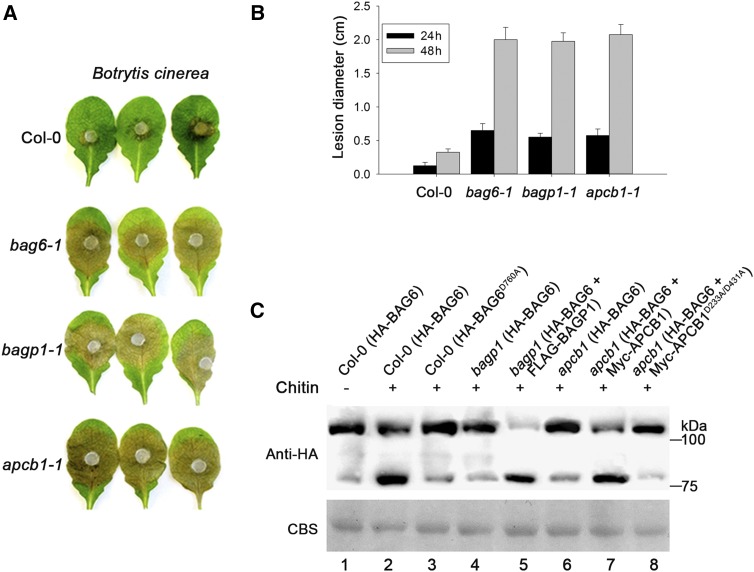
We have shown that cleavage of BAG6 is important for plant immunity against B. cinerea (Figure 3). To investigate whether BAGP1 and APCB1 contribute to plant defense, respective T-DNA insertion lines, bagp1-1 (SALK_072823C) and apcb1-1 (SALK_011244C), were obtained and challenged with B. cinerea. Two days after challenge, Col-0 showed a characteristic restricted cell death phenotype, while bagp1 and apcb1 mutants exhibited enhanced susceptibility in a manner similar to the bag6 mutant (Figure 6A). These results strongly correlate with the possibility that both BAGP1 and APCB1 are also involved in the proteolytic processing of BAG6.
Figure 6.
BAGP1 and APCB1 Are Required for Disease Resistance and BAG6 Cleavage.
(A) Detached leaves of wild-type Col-0, bag6-1, bagp1-1, and apcb1-1 mutants were agar plug inoculated with B. cinerea. Lesion development was monitored and representative photographs were taken 48 h after inoculation.
(B) Lesion diameters were measured at 24 and 48 h after inoculation. Data represent means ± sd from three replicates.
(C) Arabidopsis Col-0, bagp1, and apcb1 protoplasts were transfected with either HA-AtBAG6 or HA-AtBAG6D760A under 35S promoter in the presence or absence of chitin (lanes 1 to 4 and 6). BAG6 cleavage was monitored using immunoblotting. BAG6 cleavage was restored when the bagp1 and apcb1 mutants were complemented (lanes 5 and 7). A catalytically inactive form of APCB1 (Myc-APCB1D223A/D431A) was unable to restore BAG6 cleavage (lane 8). Equal protein samples were separated by 10% SDS-PAGE and stained with Coomassie blue (CBS); the presence of HA-BAG6 was detected by an anti-HA antibody.
To address this possibility, protoplasts of Col-0, bagp1, and apcb1 mutant plants were transfected with HA-BAG6 and treated with chitin (200 μg/mL) for 30 min. Extracts were examined for cleavage of BAG6 using anti-HA antibody. In Col-0 protoplasts, processed BAG6 was clearly observed (Figure 6C, lane 2), whereas both bagp1and apcb1 mutant lines showed no evidence for BAG6 cleavage (Figure 6C, lanes 4 and 6). Individual complementation of each mutant bagp1 and apcb1 line restored cleavage of BAG6 (Figure 6C, lanes 5 and 7), linking their gene products to BAG6 cleavage and plant immunity. Furthermore, two additional and distinct Arabidopsis aspartyl proteases (At2g17760 and At1g01300) were unable to complement the apcb1 mutant line with respect to BAG6 cleavage (data not shown), indicating specificity of the APCB1 protease for BAG6 processing.
Eukaryotic aspartyl proteases generally have a two-domain structure, arising from ancestral duplication. Each domain contributes a catalytic Asp (D) residue, with an extended active site cleft localized between the two lobes of the molecule (Szecsi, 1992). Two aspartic residues (D223 and D431) in APCB1 were identified within the highly conserved aspartyl protease sites. To determine whether aspartyl protease (APCB1) enzyme activity was necessary for processing, both D223 and D431 in APCB1 were replaced with alanine (APCB1D223A/D431A). Subsequently, Myc-tagged APCB1D223A/D431A and HA-BAG6 were coexpressed in protoplasts and cleavage of BAG6 was evaluated following chitin treatment. As shown in Figure 6C (lane 8), mutations within these residues blocked cleavage, in accordance with the requirement for aspartyl protease activity for BAG6 processing. Together, these data connect aspartyl protease activity to the processing of BAG6 and plant basal immunity.
BAG6 Processing Triggers Autophagy in Arabidopsis
We have shown that BAG6 processing requires both APCB1 and BAGP1. Thus, the question remained: How does BAG6 cleavage contribute to immunity? Given that recent reports have suggested a role for autophagy in partial resistance to B. cinerea (Lai et al., 2011; Lenz et al., 2011), we examined whether BAG6 processing is linked to autophagy. Autophagy is a major catabolic process in which proteins and damaged organelles are engulfed and sequestered in characteristic double membrane vesicles termed autophagosomes. This cellular cargo is delivered to lysosomes (mammals) or vacuoles (plants) for degradation and recycling (Levine and Yuan, 2005), maintaining homeostasis and in some cases survival under less than optimum conditions. A growing body of evidence suggests that autophagy plays a regulatory role in resistance during plant-microbe interactions.
Thus, to more directly evaluate autophagy in the context of the B. cinerea/Arabidopsis interaction, we stained B. cinerea challenged Arabidopsis with monodansylcadaverine (MDC), a fluorescent stain for autophagosomes, a double membrane vesicular structure diagnostic for the autophagy process (Biederbick et al., 1995) (Figure 7). Twenty-four hours after leaves were inoculated with B. cinerea, there was a considerable increase in the MDC signal in the areas surrounding the lesions on Col-0, whereas MDC-stained vesicular structures were absent in the B. cinerea-inoculated bag6 mutant (Figure 7C). The phenotype observed in the bag6 mutant was similar to what we observed in the autophagy defective Arabidopsis line atg18 (SALK_081770C). MDC-labeled structures were absent, which was coupled with increased fungal growth (Figures 7A to 7C). In accordance, previous reports (Lai et al., 2011; Lenz et al., 2011) have also linked autophagy mutants (i.e., atg18) to enhanced susceptibility to B. cinerea. Taken together, these results indicate that B. cinerea challenge triggers the formation of MDC-labeled autophagosomal structures in Arabidopsis plants during the resistant response, and when this response is blocked, susceptibility ensues.
Figure 7.
BAG6 Induces Autophagic Structures in Plants.
(A) Wild-type Col-0, bag6 mutant, and the autophagy-deficient mutant atg18 (SALK_081770C) were inoculated with B. cinerea using colonized agar plugs, and lesions monitored over time. Photographs of representative leaves were taken 48 h after inoculation.
(B) Lesion diameters were measured 24 and 48 h after inoculation with B. cinerea. Data represent means ± sd from three replicates.
(C) Autophagic activity was detected using the fluorescent stain MDC. Leaves were vacuum-infiltrated with a 100 μM final concentration of MDC for 30 min at 37°C, 24 h after B. cinerea inoculation. Fluorescence was visualized using an Olympus IX81 inverted fluorescence confocal microscope with excitation and emission wavelengths of 335 and 508 nm, respectively. All images were collected using Olympus DP controller and processed using Olympus Fluoview software. Bar = 20 μm.
(D) Representative TEM images of tobacco tissues expressing the full-length and cleaved form of AtBAG6. Tissues were processed 48 h after infiltration. Arrows indicate autophagosomal structures. Bar = 2 μm.
Cleaved BAG6 Induces Autophagy in Tobacco
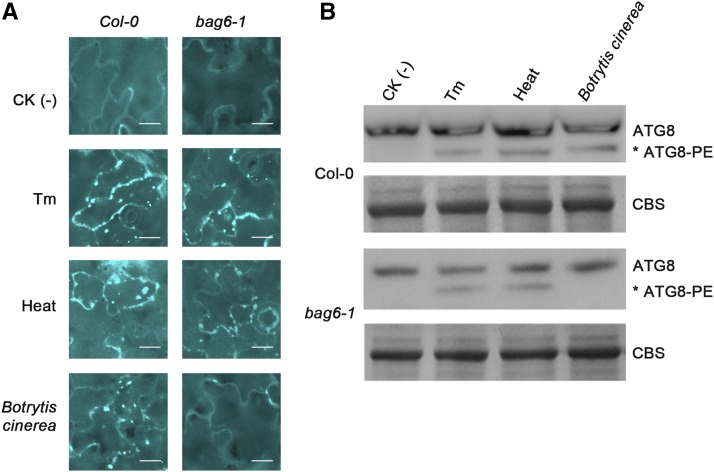
To investigate the contribution of processed BAG6 with respect to autophagy, transmission electron microscopy (TEM) was employed to directly assess the occurrence of autophagic activity. A construct was made that expressed the cleaved 760-amino acid N-terminal portion of BAG6 and was transiently expressed via Agrobacterium in N. benthamiana leaves. Two days after infiltration, the TEM analysis provided unambiguous evidence for the presence of autophagy structures following expression of the cleaved BAG6; these structures were not observed when the full-length BAG6 was transiently expressed (Figure 7D). Clear, diagnostic features of autophagy were observed, including the presence of double membrane autophagosomes with visibly sequestered cargo and older, empty autophagosomes presumably having delivered such cargo. None of these structures were observed in tissue expressing full-length BAG6 (Figure 7D). These results indicated that for autophagy to occur, full-length BAG6 needs to be activated by protease processing. As the BAG domain is the essential hallmark for BAG proteins, these results also suggest that the central BAG domain of BAG6, once exposed after cleavage, might be essential for activating autophagic cell death that leads to plant resistance. Following these observations, we then asked whether the lack of autophagosome formation in the bag6 mutant plants is a result of defective autophagic machinery. Using both tunicamycin and heat treatment (known inducers of endoplasmic reticulum [ER] stress and autophagy), we found that atbag6 plants retained the ability to induce autophagic structures and that the autophagy machinery was not affected in these mutants (Figure 8). Indeed, MDC labeled structures and ATG8 lipidation (required for autophagosome maturation) are clearly evident following these treatments, but absent following B. cinerea inoculation in bag6 mutants. These results suggest that BAG6 is not required for the process of autophagy, but the observed effects are specifically linked to infection by B. cinerea. Following these results, we further assessed whether chemical induction of autophagy is able to rescue the bag6 mutant phenotype. Leaves of bag6 mutant plants were preinfiltrated with known inducers of autophagy, including trehalose (Williams et al., 2015), rapamycin (Sarkar et al., 2009; Xiong and Sheen, 2014), DTT (Pérez-Martín et al., 2014; Zhou et al., 2014), tunicamycin (Liu et al., 2012; Munch et al., 2014), and tamoxifen (Lavieu et al., 2006; Brabec-Zaruba et al., 2007), prior to fungal inoculations. Tunicamycin treatment fully rescued the mutant phenotype to wild-type levels, while trehalose and tamoxifen treatments partially restored this phenotype (>50% of the wild type) (Supplemental Figure 10). DTT and rapamycin did not alter the response of bag6 mutants to B. cinerea challenge. This is not entirely unexpected possibly due to the differing specificities of these compounds. Overall, these results further show that the induction of autophagy is required for basal defense to B. cinerea and link BAG6 to pathogen recognition and the initiation and implementation of autophagy.
Figure 8.
BAG6-Induced Autophagy Is Specific to B. cinerea.
(A) Autophagic activity was detected using MDC staining. Col-0 and bag6 Arabidopsis plants were treated with B. cinerea (24 h), tunicamycin (0.5 μg/mL, 8 h), or heat stress (50°C, 30 min). MDC-stained structures were detected in Col-0 plants in response to all three treatments, but only in response to tunicamycin and heat stress in bag6 plants. Fluorescence was visualized using an Olympus IX81 inverted fluorescence confocal microscope with excitation and emission wavelengths of 335 and 508 nm, respectively. All images were collected using Olympus DP controller and processed using Olympus Fluoview software. Bar = 20 μm.
(B) Similarly, the lipidation pattern of ATG8 was analyzed in Col-0 and bag6 plants following all three treatments. Protein extracts were subjected to SDS-PAGE followed by immunoblotting with anti-ATG8 antibody. The positions of ATG8 and ATG8-PE are indicated. Equal loading was confirmed by SDS-PAGE and Coomassie blue staining (CBS).
DISCUSSION
Regulated proteolysis is a major means by which eukaryotes mediate appropriate cellular decisions in response to environmental cues. In this article, we investigated the underlying mechanisms responsible for basal resistance of Arabidopsis to B. cinerea, a necrotrophic fungal phytopathogen. Our previous studies indicated that inactivation of the Arabidopsis cochaperone BAG6, a member of the plant BAG gene family, compromises basal resistance to this fungus resulting in dramatically enhanced host cell death and susceptibility (Doukhanina et al., 2006). Here, we demonstrate that this process is regulated by at least three interacting partners: (1) BAG6, (2) a GRAM domain-containing protein (BAGP1), and (3) an aspartyl protease (PCB1) where BAGP1 and PCB1 are specifically required for BAG6 processing. Our studies show that PAMP (chitin) treatment, caspase-1 activity, and B. cinerea challenge are necessary for the cleavage of BAG6 leading to autophagic cell death and resistance. These results link fungal PAMP perception and signaling to a specific cell death regime.
The central mediator of this program is a member of the Arabidopsis BAG family of cytoprotective proteins (BAG6). The BAG genes constitute a broadly conserved gene family and function as adapter proteins/cochaperones, forming complexes with signaling molecules and molecular chaperones including Hsp70 (Kabbage and Dickman, 2008; Williams et al., 2010). In mammals, BAG proteins are associated with several key cellular processes, including apoptosis, autophagy, proliferation, differentiation, and stress signaling (Takayama et al., 1999; Song et al., 2001). The functional versatility of this family appears to be maintained in Arabidopsis and quite possibly other plants. Seven BAG proteins have been identified in Arabidopsis (Doukhanina et al., 2006); however, our mechanistic understanding of individual members is limited. Based on our preliminary work with the Arabidopsis BAG family, it appears that these family members have adapted to particular niches/locations for selected functions (Doukhanina et al., 2006; Kabbage and Dickman, 2008; Williams et al., 2010). It is these functions and how they are regulated that are of major interest. For example, we have shown that BAG4 expression in plants confers tolerance to a wide range of abiotic stresses, including cold, UV light, oxidants, drought, and salt, which parallels mammalian studies involving BAG1 (Townsend et al., 2003; Doukhanina et al., 2006; Hoang et al., 2015). Arabidopsis BAG6 was previously shown to be associated with calmodulin and calmodulin-dependent signaling (Kang et al., 2006). BAG7, an ER-localized BAG member, is involved with the unfolded protein response as a result of ER stress, and the deletion of BAG7 compromises heat and cold tolerance (Williams et al., 2010). Interestingly, we recently determined that BAG7 is also processed and, as a result, translocates from the ER to the nucleus where it interacts with the WRKY29 transcription factor resulting in an abiotic stress tolerant phenotype (Y. Li and M.B. Dickman, unpublished data).
BAG6 is the largest Arabidopsis BAG member (1043 amino acids), and in addition to the signature BAG domain, it has a calmodulin binding motif, a feature associated with plant BAGs. Two nuclear localization signals are also predicted. The biological significance of the nuclear localization signal is currently unknown. Localization studies of BAG6 under various stress conditions are in progress. We propose that BAG6 confers resistance via the induction of autophagy-mediated cell death in response to fungal challenge. In this report, we have identified and functionally analyzed the key components along with BAG6 responsible for its function. Our initial observation that inactivation of BAG6 resulted in loss of basal resistance in Arabidopsis to the necrotrophic fungal plant pathogen B. cinerea prompted this study. To explore this observation in more detail, we characterized Arabidopsis bag6 T-DNA knockout lines and plants that heterologously overexpressed BAG6. Yeast two-hybrid approaches, notorious for false positives, proved problematic as very few colonies were observed, particularly when using truncated forms of BAG6 as bait. This is likely due to the pro-death activity of BAG6 as shown in this study and suggests that the BAG6 cell death machinery is conserved in yeast. Clues to explain these observations arose from the finding that lesion mimic phenotypes were commonly detected in transgenic Arabidopsis and tobacco plants that overexpressed BAG6. Immunoblot analysis of these lesion mimics showed a BAG6 protein of predicted size, but we consistently also observed a lower molecular mass protein, suggesting specific cleavage (Figure 2). An ExPASy search for protease cleavage sites revealed a single caspase-1-like site that when cleaved would yield the size of the truncated protein observed in immunoblots of lesion mimic plants. BAG6 treated with recombinant human caspase-1, a PAMP, or fungal inoculation produced a protein also similar in size to the truncated protein observed in the lesion mimics. Caspase-1-like activities have been reported in several plant-microbe interactions. For example, vacuolar processing enzyme (VPE), a cysteine protease localized to the vacuole, is required for TMV-mediated HR-PCD and resistance (Hatsugai et al., 2004). Inoculation of tobacco with an incompatible strain of Pseudomonas syringae or B. cinerea triggered caspase-1-like activity that was suppressed in the VPE mutant background. Furthermore, VPE mutants were also markedly more susceptible to these pathogens. When VPE was overexpressed, cell death markers (e.g., ion leakage) were observed (Rojo et al., 2004). Thus, although strictly speaking, bona fide caspases are not present in plant genomes, caspase-1-like activities are found in plants and in several cases contribute to the outcome of several plant microbe interactions. We suggest that such activity is required for BAG6-mediated basal immunity against B. cinerea.
We also treated BAG6-expressing plants with chitin, a component of fungal cell walls and a well described PAMP. As in B. cinerea inoculation, PAMP/chitin treatment also triggered BAG6 cleavage. To strengthen our assertion that the caspase-1-like site is necessary for cleavage, we generated a mutation (BAG6D760A) at this site and found that cleavage was blocked. Importantly, when BAG6 cleavage was impeded, enhanced susceptibility was observed. Taken together, caspase-1 activity/chitin/B. cinerea can mediate BAG6 cleavage, and as we have shown, this cleavage is necessary for activating basal resistance.
To identify the potential protease responsible for cleavage, we evaluated a suite of protease inhibitors and found that only pepstatin, an aspartyl protease inhibitor, blocked cleavage. In accordance, using the computational tool STRING (STRING 9.1; http://string-db.org), which detects known and predicted protein interactions, we uncovered APCB1, an aspartyl protease, predicted to interact with BAGP1. Common features of aspartyl proteases include an active site cleft that contains two catalytic aspartic acid residues, acidic pH optima for enzymatic activity, inhibition by pepstatin A, a conserved overall fold, and preferential cleavage specificity for peptide bonds between amino acid residues with bulky hydrophobic side chains (Glathe et al., 1998). APCB1 shares significant sequence similarity to typical aspartyl proteases. As with other eukaryotic aspartyl proteases, APCB1 contains two active sites with the conserved, identical motifs Asp-Thr-Gly and Asp-Thr-Gly, respectively. Mutation of the catalytic site of APCB1 specifically abolished cleavage, consistent with the importance of aspartyl protease activity.
Aspartyl proteases have been implicated in regulating a variety of biological processes in mammalian cells; however, examples of their involvement in plant immunity, development, and PCD are limited. Fifty-one aspartyl proteases are predicted in the Arabidopsis genome and are grouped into three categories (typical, atypical, and nucellin-like aspartyl proteases) depending on their domain structure and active-site sequences (Faro et Gal, 2005).
Hyperactivation of the aspartyl protease CONSTITUTIVE DISEASE RESISTANCE1 in Arabidopsis causes spontaneous (lesion mimic) cell death (Xia et al., 2004). In contrast, the aspartyl protease encoded by the PROMOTION OF CELL SURVIVAL1 gene in Arabidopsis functions to promote cell survival during embryogenesis and gametogenesis, and its ectopic overexpression blocks normal PCD processes associated with anther dehiscence (Ge et al., 2005). APCB1 has been suggested to encode a nucellin-like aspartyl protease (Faro and Gal, 2005). Interestingly, this nucellin-like aspartyl protease appears to be BAG6 specific, as two unrelated Arabidopsis aspartyl proteases were unable to complement the apcb1 mutant line with respect to BAG6 cleavage. Nucellins have been associated with plant developmental cell death as they are expressed during barley (Hordeum vulgare) ovule degeneration. Further linking aspartyl protease activity and PCD, in rice (Oryza sativa), ETERNAL TAPETUM1 regulates two aspartyl proteases that are responsible for triggering PCD during development (Niu et al., 2013). Recently a report from Breitenbach et al. (2014) during a screen for systemic acquired resistance (SAR) regulators in Arabidopsis identified an apoplastic aspartyl protease (AED1) that suppresses SAR and suggested an association with feedback regulation during plant innate immunity. Thus, aspartyl protease activities may provide new insights into the modulation of biotic stress in plants.
A C2-GRAM domain-containing protein designated BAGP1 (At3g59660) was identified via pull-downs and mass spectrometry. We demonstrated that this protein is also required for BAG6 cleavage and disease resistance. The C2 domain is a modulatory sequence of ∼40 amino acids consisting of two highly conserved domains separated by a basic region (Clark et al., 1991). The GRAM (for glucosyltransferases, Rab-like GTPase activators, and myotubularins) domain is ∼70 amino acids in length and consists of a seven-stranded β-sandwich and a C-terminal α-helix, found in glucosyltransferases, myotubularins, and other putative membrane-associated proteins (Doerks et al., 2000; Begley et al., 2003). GRAM domain-containing proteins are ubiquitous in eukaryotes and exhibit various protein domain architectures in addition to the GRAM domain. However, C2 domain-containing GRAM proteins are only found in plants and are typically endomembrane localized. The Arabidopsis genome contains three such proteins. The functions of most GRAM proteins are largely unknown. Nevertheless, expression studies in plants indicate a potential role for these proteins in response to stress. Of relevance to this work, locus At3g59660, which we have designated BAGP1, was specifically induced in response to fungal challenge that included B. cinerea (Jiang et al., 2008).
Together, our evidence indicates the requirement for APCB1, BAGP1, and BAG6 for basal resistance of Arabidopsis to B. cinerea. Using yeast two-hybrid and co-IP assays, we demonstrated binding in all permutations: BAGP1, APCB1, and BAG6 all interacted with each other, suggesting a complex. A key question is how is complex formation linked to protease cleavage and ultimately to resistance? Previous reports have linked autophagy to basal resistance to necrotrophs (Lai et al., 2011; Lenz et al., 2011; Kabbage et al., 2013). Blocking autophagy inhibits such resistance and results in greatly enhanced susceptibility. Importantly, B. cinerea can induce autophagy in plants (Lai et al., 2011). In accordance, we show that BAG6 processing triggers autophagy and inhibits B. cinerea pathogenic development. When autophagy is inhibited chemically or genetically, a marked increase in susceptibility to B. cinerea is observed (Lai et al., 2011; Lenz et al., 2011). Finally, using transient assays with the cleaved BAG6 followed by TEM, we provide clear evidence for autophagic structures during this immune response. Thus, the processed BAG6 generated from aspartyl protease cleavage correlates with plant resistance via autophagy. Several genes have been shown to modulate cell death and potentially alter immune responses in Arabidopsis, including LOL1 (Epple et al., 2003), LSD1 (Torres et al., 2005), and metacaspases (Coll et al., 2010). Premature senescence and spreading lesions also occur in autophagy-deficient plants as a result of accumulation of ubiquitinated proteins and with ER stress (Coll et al., 2014; Munch et al., 2014). Therefore, these plants can be primed for pathogen compatibility (Hackenberg et al., 2013). This is of particular note since bag6 mutants have a similar phenotype to those described in other autophagy-deficient systems. Thus, we considered whether bag6 mutants are generally compromised in the execution of autophagy resulting in the observed spreading lesion phenotype. We subjected bag6 mutant plants to ER stresses that are known to induce autophagy (tunicamycin and heat stress) and monitored the development of autophagosome formation and ATG8 lipidation, a process that is crucial for autophagosome maturation. Our results showed that bag6 mutant plants have a functional autophagic machinery as two distinct ER stressors (but not B. cinerea) activate autophagy in the bag6 mutant background. This suggests that BAG6 is not generally required for the process of autophagy (Figure 8) and that the observed phenotype is a direct effect of B. cinerea challenge and not caused by premature cell death and ER stress. Furthermore, we were able to circumvent the bag6 mutation by chemically inducing autophagy in the bag6 background (Supplemental Figure 10), thus recovering the ability of these mutants to respond to fungal challenge. Taken together, these data suggest that pathogen-induced autophagy is lacking in bag6 mutants leading to the diseased state. Therefore, this protein likely provides a link between pathogen perception/detection and the execution of this cell death program leading to resistance. However, many questions remain: (1) How is B. cinerea or its signal recognized by Arabidopsis? (2) How is the presumable complex and associated aspartyl protease become activated? (3) How does BAG domain expression lead to autophagy? (4) What is the function of the BAGP1? (5) How does autophagic cell death inhibit B. cinerea? This latter point is of particular interest and importance. It has been sensibly argued that dead cells provide a hospitable, nutrient-rich environment for necrotrophs like B. cinerea; after all, necrotrophs by definition require dead cells (Govrin and Levine, 2000; Lai et al., 2011). Thus, PCD would limit biotrophic infection but facilitate necrotroph-induced disease as is often observed. However, from these data and our previous studies, it is evident that this interaction is more complex than previously viewed. Here, we show that autophagic PCD inhibits B. cinerea infection. Thus, in the presence of dead cells, B. cinerea is not pathogenic and dead cells are not necessarily sufficient for triggering compatibility. We observed similar results with S. sclerotiorum where the fungal secreted oxalic acid induces cell death with apoptotic-like features in the host to achieve pathogenic success (Kim et al., 2008). Conversely, mutants that are deficient in oxalic acid production trigger a defense response within the host that includes autophagic PCD (Kabbage et al., 2013). However, we cannot rule out that autophagy as a pro-survival mechanism keeps cells alive in both cases, thus restricting fungal induced cell death and pathogenic development (Munch et al., 2014). This potential dual function of autophagy (pro-death versus pro-survival) is intriguing but not entirely understood. There is evidence for both outcomes (Hofius et al., 2011; Kabbage et al., 2013), and further work is required to explain the underpinning mechanisms that contrast pro-survival and pro-death regimes. As we discussed previously and substantiated here, we suggest that it is not cell death as such that is key, but rather the mechanism by which cell death occurs that is critical to the outcome of a given plant-pathogen interaction. While cell death is one possible outcome for autophagy, apoptosis, and necrosis, there are distinctly different (and important) ways by which cells can die.
METHODS
Plant Materials and Growth Conditions
Arabidopsis thaliana Col-0 and relevant T-DNA knockout mutants, bag6-1 (SALK_009534C), bag6-2 (SALK_073331C), bagp1-1 (SALK_072823C), and apcb1-1 (SALK_011244C), were acquired from the Arabidopsis Stock Center (www.arabidopsis.org). Homozygous mutant plants were confirmed by PCR. Arabidopsis and Nicotiana benthamiana plants were grown in soil in a growth chamber under the following conditions: 16/8-h light/dark cycles at 100 μE m−2 s−1 for normal growth or 200 μE m−2 s−1 for lesion mimic phenotype induction, 23°C, and 60% relative humidity. N. benthamiana plants were grown on a solid medium containing basic Murashige and Skoog salts (PhytoTechnology), 2% sucrose, and 0.8% Phytagel (Sigma-Aldrich) in a tissue culture room under 16/8-h light/dark cycles at 100 μE m−2 s−1 for normal growth or 200 μE m−2 s−1 for lesion mimic phenotype induction and 23°C.
BAG6 Constructs and Plant Transformation
Arabidopsis BAG6 full-length cDNA was amplified using a gene-specific primers, cloned into pENTR/D-TOPO (Life Technologies), and recombined via Gateway LR reaction (Life Technologies) into the destination vectors pEarleyGate101, pEarleyGate104, and pEarleyGate201 (Earley et al., 2006) to generate the 35S:BAG6-YFP, 35S:YFP-BAG6, and 35S:HA-BAG6 constructs, respectively. Overlapping PCR was used to construct BAG6 vectors driven by its predicted native promoter. Fragments were ligated into pENTR/D-TOPO and subsequently integrated into the promoterless vector pEarleygate 302 to generate the NP:BAG6-YFP, NP:YFP-BAG6, and NP:HA-BAG6 constructs. The mutated form of BAG6 (BAG6D760A) was constructed by site-directed mutagenesis in the appropriate vectors using the QuikChange II XL site-directed mutagenesis kit (Agilent Technologies) according to the manufacturer’s recommendation. For TEM analysis, the cleaved N-terminal portion of BAG6 upstream of the predicted caspase-1 cleavage site (2280 bp) was amplified and transferred to pEarleygate104 as described previously. This construct was introduced into Agrobacterium tumefaciens GV3101 and expressed transiently into N. benthamiana leaves 2 d prior to TEM analysis. Infiltrated leaf segments were fixed for 5 h at 20°C in 3% glutaraldehyde and 2% formaldehyde in 0.1 M Na cacodylate buffer then were processed for TEM according to standard protocols. Sections were examined with a Phillips Morgagni 268 transmission electron microscope (FEI) at an accelerating voltage of 80 kV. Digital images were recorded with a MegaView III digital camera operated with iTEM software (Olympus Soft Imaging Systems). All constructs were verified by sequencing, introduced into Agrobacterium GV3101 by electroporation, and used for stable transformation or transient expression in tobacco plants (Fisher and Guiltinan, 1995) and Arabidopsis plants. Arabidopsis leaf protoplasts were prepared and transformed as described (Asai et al., 2000).
Immunoblot Analysis
For in vitro human caspase-1 treatment, 2 d after transient expression of 35S:HA-BAG6 and 35S:HA-BAG6D760A under the control of the CaMV 35S promoter in N. benthamiana leaves, total proteins were isolated using standard protein extraction buffer (50 mM HEPES, pH 5.0, 50 mM NaCl, 0.1% CHAPS, 10 mM EDTA, 5% glycerol, and 10 mM DTT), and tagged proteins were purified with monoclonal anti HA-agarose antibody and eluted with specific HA peptide. Equal amounts of protein (10 μg) were treated with recombinant human caspase-1 (2 units) at 37°C for 2 h using standard protocol. BAG6 in vivo cleavage was assayed following chitin treatment (200 μg/mL for 30 min) or B. cinerea inoculation in both N. benthamiana and Arabidopsis. Similarly, total proteins were extracted, purified, and eluted as described above. The presence of HA-BAG6 was detected by anti-HA immunoblotting. For the detection of ATG8 lipidation, protein samples were prepared from leaves treated with B. cinerea (24 h), tunicamycin (0.5 μg/mL, 8 h), or heat stress (50°C, 30 min). Total proteins were separated by 10% SDS-PAGE in the presence of 6 M urea. The blots were probed with the ATG8 antibody according to standard protocols.
Fungal Culture and Disease Assays
B. cinerea strain T-4 was grown on potato dextrose agar at room temperature. Agar plugs containing actively growing mycelia were used for plant inoculation on detached Arabidopsis or tobacco leaves as described previously (Doukhanina et al., 2006).
Protease Inhibitor Assay
35S:HA-BAG6 was introduced into Agrobacterium GV3101 by electroporation and subsequently expressed into N. benthamiana leaves (Fisher and Guiltinan, 1995). Three days after infiltration, leaves were treated with 200 μg/mL chitin to induce BAG6 cleavage, in combination with individual protease inhibitors (1 μM pepstatin, 1 mM aprotinin, 0.5 mM bestatin, 1 mM leupeptin, 10 μM E-64, and 5 mM N-ethylmaleimide) for 30 min. Protein extraction and immunoblots were conducted as described above.
Oxidative Burst Assay
In situ H2O2 was detected by 3,3′-diaminobenzidine (Sigma-Aldrich) staining. Two days after inoculation, fully expanded Arabidopsis leaves were treated with a 1 mg/mL 3,3′-diaminobenzidine solution (0.2 M sodium phosphate buffer, pH 7.0) overnight in the dark at room temperature, and chlorophyll was removed by incubation in 95% ethanol. All samples were visualized using an Olympus SZx10 stereoscope.
Callose Deposition
Two days after inoculation, Arabidopsis leaves were fixed under a vacuum in a 3:1 ratio of ethanol and acetic acid. The clearing solution was changed until the leaves were colorless. Tissues were washed in 70% ethanol and then 50% ethanol for at least 2 h each time and rehydrated in several brief water washes followed by an overnight water wash. Samples were then washed with 0.07 M sodium phosphate buffer, pH 9.0, for 1 h and were incubated in the dark at room temperature for at least 4 h with 0.01% aniline blue solution. After mounting on slides in 50% glycerol, samples were examined with an Olympus IX81 inverted fluorescence confocal microscope using UV illumination and a broadband 4′,6-diamidino-2-phenylindole filter set with excitation and emission wavelengths of 420 and 460 nm, respectively.
Pull-Down Assay
Fifty grams of control and 35S:HA-BAG6 transiently expressing N. benthamiana leaves was ground in liquid nitrogen and suspended in extraction buffer. Proteins were purified with monoclonal anti HA-agarose antibody and eluted with specific HA peptide. Equal protein samples were separated by 10% SDS-PAGE and stained with the SilverQuest Silver staining kit (Invitrogen). The specific protein bands were subjected to mass spectrometry analysis at the UC-Davis Genome Center (http://cores.genomecenter.ucdavis.edu).
Yeast Two-Hybrid Assay
Yeast two-hybrid assays were conducted according to the manufacturer’s recommendations (Clontech). Prey (pGADT7) and bait (pGBKT7) constructs were transformed into AH109 yeast strain to examine the interactions of BAG6 with BAGP1, BAG6 with APCB1, and BAGP1 with APCB1.
Co-IP Assay
Full-length BAGP1 and APCB1 were amplified and cloned downstream of FLAG tag and c-Myc tag under the control of the CaMV 35S promoter in pEarleyGate202 and pEarleyGate203 (Earley et al., 2006), respectively, generating FLAG-BAGP1 and Myc-APCB1. To determine whether aspartyl protease (APCB1) enzyme activity was necessary for processing, aspartic residues 223 and 431 in APCB1 were replaced with alanine (APCB1 D223A/D431A) by site-directed mutagenesis as described previously. Arabidopsis Col-0 protoplasts were transfected with the indicated constructs, incubated for 12 h, and total proteins extracted. For anti-HA immunoprecipitation, total protein was incubated with a monoclonal anti-HA-agarose antibody for 4 h, washed six times with a buffer containing 50 mM HEPES-KOH, 150 mM KCl, 1 mM EDTA, 0.2% Triton X-100, and 1 mM DTT, and bound protein eluted with HA peptide. For anti-FLAG immunoprecipitation, total protein was incubated with an anti-FLAG M2 affinity gel for 4 h and eluted with 3×FLAG peptide. For anti-Myc immunoprecipitation, total protein was incubated with an anti-c-Myc agarose affinity gel for 4 h and eluted with c-Myc peptide. Immunoprecipitates were separated by a 10% SDS-PAGE and detected by anti-HA, anti-Flag, or anti-Myc antibodies (Sigma-Aldrich).
Microscopy Observations
For MDC staining, 6-week-old Arabidopsis plants were treated with B. cinerea (24 h), tunicamycin (0.5 μg/mL, 8 h), or heat stress (50°C, 30 min). Leaves were vacuum-infiltrated with a 100 μM final concentration of MDC (Abcam) for 30 min at 37°C. Fluorescence was visualized using an Olympus IX81 inverted fluorescence confocal microscope with excitation and emission wavelengths of 335 and 508 nm, respectively. For TEM, leaf segments were fixed for 5 h at 37°C in 3% glutaraldehyde and 2% formaldehyde in 0.1 M Na cacodylate buffer and then processed according to standard protocols as described previously (Kabbage et al., 2013). For fluorescent protein detection, samples were directly observed by confocal microscopy under the following conditions: RFP, excitation at 561 nm and detection at 580 to 630 nm; YFP, excitation at 495 nm and detection at 510 to 550 nm. All images were collected using Olympus DP controller and processed using Olympus Fluoview software.
Chemical Induction of Autophagy
Leaves of 6-week-old bag6 mutant plants were infiltrated with trehalose (150 mM), rapamycin (100 nM), tunicamycin (5 μM), DTT (10 mM), or tamoxifen (10 μM) for 12 h, and autophagic structures were detected by MDC staining. After treatment, leaves were inoculated with B. cinerea strain T4; wild-type Col-0 and untreated bag6 mutants served as controls.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: BAG6, TAIR AT2G46240, gene ID 819232; APCB1, TAIR AT1G49050, gene ID 841328; BAGP1, TAIR AT3G59660, gene ID 825135; and ATG18g, TAIR AT1G03380, gene ID 838702.
Supplemental Data
Supplemental Figure 1. Enhanced susceptibility of bag6 mutants to Botrytis cinerea.
Supplemental Figure 2. H2O2 and callose accumulation in Arabidopsis plants expressing BAG6 in response to Botrytis cinerea.
Supplemental Figure 3. Overexpression of BAG6 in Arabidopsis induces a lesion mimic phenotype.
Supplemental Figure 4. Arabidopsis BAG6 expression in Nicotiana tabacum causes lesion mimics in a light-dependent manner.
Supplemental Figure 5. Lesion mimic development coincides with BAG6 cleavage.
Supplemental Figure 6. Arabidopsis BAG6 is cleaved by plant caspase-1-like protease activity in vivo.
Supplemental Figure 7. The processing of BAG6 is required for resistance to Botrytis cinerea in Nicotiana tabacum.
Supplemental Figure 8. Pull-down assays indicate that BAG6 interacts with a C2 GRAM protein.
Supplemental Figure 9. Arabidopsis BAG6, BAGP1, and APCB1 form a trimeric complex in planta.
Supplemental Figure 10. Chemical-induced autophagy partially rescues resistance to Botrytis cinerea in the bag6 mutant.
Supplementary Material
Acknowledgments
We thank Brett Williams and Paul de Figueiredo for stimulating discussions and editorial comments on this manuscript. Funding for this project was provided by the National Science Foundation (MCB-092391) and BARD (US-4041-07C) to M.B.D.
AUTHOR CONTRIBUTIONS
M.K. and M.B.D. designed the research. Y.L., M.K., and W.L. performed the experiments. Y.L., M.K., and M.B.D. analyzed the data. Y.L., M.K., and M.B.D. wrote the article.
Glossary
- PCD
programmed cell death
- PAMP
pathogen-associated molecular pattern
- co-IP
coimmunoprecipitation
- MDC
monodansylcadaverine
- ER
endoplasmic reticulum
- VPE
vacuolar processing enzyme
- SAR
systemic acquired resistance
- TEM
transmission electron microscopy
Footnotes
Articles can be viewed online without a subscription.
References
- Asai T., Stone J.M., Heard J.E., Kovtun Y., Yorgey P., Sheen J., Ausubel F.M. (2000). Fumonisin B1-induced cell death in arabidopsis protoplasts requires jasmonate-, ethylene-, and salicylate-dependent signaling pathways. Plant Cell 12: 1823–1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begley M.J., Taylor G.S., Kim S.A., Veine D.M., Dixon J.E., Stuckey J.A. (2003). Crystal structure of a phosphoinositide phosphatase, MTMR2: insights into myotubular myopathy and Charcot-Marie-Tooth syndrome. Mol. Cell 12: 1391–1402. [DOI] [PubMed] [Google Scholar]
- Biederbick A., Kern H.F., Elsässer H.P. (1995). Monodansylcadaverine (MDC) is a specific in vivo marker for autophagic vacuoles. Eur. J. Cell Biol. 66: 3–14. [PubMed] [Google Scholar]
- Brabec-Zaruba M., Berka U., Blaas D., Fuchs R. (2007). Induction of autophagy does not affect human rhinovirus type 2 production. J. Virol. 81: 10815–10817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitenbach H.H., et al. (2014). Contrasting roles of the apoplastic aspartyl protease APOPLASTIC, ENHANCED DISEASE SUSCEPTIBILITY1-DEPENDENT1 and LEGUME LECTIN-LIKE PROTEIN1 in Arabidopsis systemic acquired resistance. Plant Physiol. 165: 791–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brive L., Takayama S., Briknarová K., Homma S., Ishida S.K., Reed J.C., Ely K.R. (2001). The carboxyl-terminal lobe of Hsc70 ATPase domain is sufficient for binding to BAG1. Biochem. Biophys. Res. Commun. 289: 1099–1105. [DOI] [PubMed] [Google Scholar]
- Buckner B., Johal G.S., Janick-Buckner D. (2000). Cell death in maize. Physiol. Plant. 108: 231–239. [Google Scholar]
- Clark J.D., Lin L.L., Kriz R.W., Ramesha C.S., Sultzman L.A., Lin A.Y., Milona N., Knopf J.L. (1991). A novel arachidonic acid-selective cytosolic PLA2 contains a Ca2+-dependent translocation domain with homology to PKC and GAP. Cell 65: 1043–1051. [DOI] [PubMed] [Google Scholar]
- Coll N.S., Smidler A., Puigvert M., Popa C., Valls M., Dangl J.L. (2014). The plant metacaspase AtMC1 in pathogen-triggered programmed cell death and aging: functional linkage with autophagy. Cell Death Differ. 21: 1399–1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coll N.S., Vercammen D., Smidler A., Clover C., Van Breusegem F., Dangl J.L., Epple P. (2010). Arabidopsis type I metacaspases control cell death. Science 330: 1393–1397. [DOI] [PubMed] [Google Scholar]
- Dangl J.L., Dietrich R.A., Richberg M.H. (1996). Death don’t have no mercy: Cell death programs in plant-microbe interactions. Plant Cell 8: 1793–1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickman M.B., Park Y.K., Oltersdorf T., Li W., Clemente T., French R. (2001). Abrogation of disease development in plants expressing animal antiapoptotic genes. Proc. Natl. Acad. Sci. USA 98: 6957–6962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doerks T., Strauss M., Brendel M., Bork P. (2000). GRAM, a novel domain in glucosyltransferases, myotubularins and other putative membrane-associated proteins. Trends Biochem. Sci. 25: 483–485. [DOI] [PubMed] [Google Scholar]
- Doukhanina E.V., Chen S., van der Zalm E., Godzik A., Reed J., Dickman M.B. (2006). Identification and functional characterization of the BAG protein family in Arabidopsis thaliana. J. Biol. Chem. 281: 18793–18801. [DOI] [PubMed] [Google Scholar]
- Earley K.W., Haag J.R., Pontes O., Opper K., Juehne T., Song K., Pikaard C.S. (2006). Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 45: 616–629. [DOI] [PubMed] [Google Scholar]
- Epple P., Mack A.A., Morris V.R.F., Dangl J.L. (2003). Antagonistic control of oxidative stress-induced cell death in Arabidopsis by two related, plant-specific zinc finger proteins. Proc. Natl. Acad. Sci. USA 100: 6831–6836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faro C., Gal S. (2005). Aspartic proteinase content of the Arabidopsis genome. Curr. Protein Pept. Sci. 6: 493–500. [DOI] [PubMed] [Google Scholar]
- Fisher D.K., Guiltinan M.J. (1995). Rapid, efficient production of homozygous transgenic tobacco plants with Agrobacterium tumefaciens - a seed-to-seed protocol. Plant Mol. Biol. Rep. 13: 278–289. [Google Scholar]
- Ge X., Dietrich C., Matsuno M., Li G., Berg H., Xia Y. (2005). An Arabidopsis aspartic protease functions as an anti-cell-death component in reproduction and embryogenesis. EMBO Rep. 6: 282–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glathe S., Kervinen J., Nimtz M., Li G.H., Tobin G.J., Copeland T.D., Ashford D.A., Wlodawer A., Costa J. (1998). Transport and activation of the vacuolar aspartic proteinase phytepsin in barley (Hordeum vulgare L.). J. Biol. Chem. 273: 31230–31236. [DOI] [PubMed] [Google Scholar]
- Govrin E.M., Levine A. (2000). The hypersensitive response facilitates plant infection by the necrotrophic pathogen Botrytis cinerea. Curr. Biol. 10: 751–757. [DOI] [PubMed] [Google Scholar]
- Hackenberg T., et al. (2013). Catalase and NO CATALASE ACTIVITY1 promote autophagy-dependent cell death in Arabidopsis. Plant Cell 25: 4616–4626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatsugai N., Kuroyanagi M., Yamada K., Meshi T., Tsuda S., Kondo M., Nishimura M., Hara-Nishimura I. (2004). A plant vacuolar protease, VPE, mediates virus-induced hypersensitive cell death. Science 305: 855–858. [DOI] [PubMed] [Google Scholar]
- Hoang T.M.L., Moghaddam L., Williams B., Khanna H., Dale J., Mundree S.G. (2015). Development of salinity tolerance in rice by constitutive-overexpression of genes involved in the regulation of programmed cell death. Front. Plant Sci. 6: 175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofius D., Munch D., Bressendorff S., Mundy J., Petersen M. (2011). Role of autophagy in disease resistance and hypersensitive response-associated cell death. Cell Death Differ. 18: 1257–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu G., Yalpani N., Briggs S.P., Johal G.S. (1998). A porphyrin pathway impairment is responsible for the phenotype of a dominant disease lesion mimic mutant of maize. Plant Cell 10: 1095–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang S.Y., Ramamoorthy R., Ramachandran S. (2008). Comparative transcriptional profiling and evolutionary analysis of the GRAM domain family in eukaryotes. Dev. Biol. 314: 418–432. [DOI] [PubMed] [Google Scholar]
- Johal G.S., Hulbert S.H., Briggs S.P. (1995). Disease lesion mimics of maize - a model for cell-death in plants. BioEssays 17: 685–692. [Google Scholar]
- Jones J.D., Dangl J.L. (2006). The plant immune system. Nature 444: 323–329. [DOI] [PubMed] [Google Scholar]
- Kabbage M., Dickman M.B. (2008). The BAG proteins: a ubiquitous family of chaperone regulators. Cell. Mol. Life Sci. 65: 1390–1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabbage M., Williams B., Dickman M.B. (2013). Cell death control: the interplay of apoptosis and autophagy in the pathogenicity of Sclerotinia sclerotiorum. PLoS Pathog. 9: e1003287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang C.H., et al. (2006). AtBAG6, a novel calmodulin-binding protein, induces programmed cell death in yeast and plants. Cell Death Differ. 13: 84–95. [DOI] [PubMed] [Google Scholar]
- Kim K.S., Min J.Y., Dickman M.B. (2008). Oxalic acid is an elicitor of plant programmed cell death during Sclerotinia sclerotiorum disease development. Mol. Plant Microbe Interact. 21: 605–612. [DOI] [PubMed] [Google Scholar]
- Lai Z., Wang F., Zheng Z., Fan B., Chen Z. (2011). A critical role of autophagy in plant resistance to necrotrophic fungal pathogens. Plant J. 66: 953–968. [DOI] [PubMed] [Google Scholar]
- Lavieu G., Scarlatti F., Sala G., Carpentier S., Levade T., Ghidoni R., Botti J., Codogno P. (2006). Regulation of autophagy by sphingosine kinase 1 and its role in cell survival during nutrient starvation. J. Biol. Chem. 281: 8518–8527. [DOI] [PubMed] [Google Scholar]
- Lenz H.D., et al. (2011). Autophagy differentially controls plant basal immunity to biotrophic and necrotrophic pathogens. Plant J. 66: 818–830. [DOI] [PubMed] [Google Scholar]
- Levine B., Yuan J. (2005). Autophagy in cell death: an innocent convict? J. Clin. Invest. 115: 2679–2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Burgos J.S., Deng Y., Srivastava R., Howell S.H., Bassham D.C. (2012). Degradation of the endoplasmic reticulum by autophagy during endoplasmic reticulum stress in Arabidopsis. Plant Cell 24: 4635–4651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord C.E., Gunawardena A.H. (2012). Programmed cell death in C. elegans, mammals and plants. Eur. J. Cell Biol. 91: 603–613. [DOI] [PubMed] [Google Scholar]
- Lorrain S., Vailleau F., Balagué C., Roby D. (2003). Lesion mimic mutants: keys for deciphering cell death and defense pathways in plants? Trends Plant Sci. 8: 263–271. [DOI] [PubMed] [Google Scholar]
- Munch D., Rodriguez E., Bressendorff S., Park O.K., Hofius D., Petersen M. (2014). Autophagy deficiency leads to accumulation of ubiquitinated proteins, ER stress, and cell death in Arabidopsis. Autophagy 10: 1579–1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu N., Liang W., Yang X., Jin W., Wilson Z.A., Hu J., Zhang D. (2013). EAT1 promotes tapetal cell death by regulating aspartic proteases during male reproductive development in rice. Nat. Commun. 4: 1445. [DOI] [PubMed] [Google Scholar]
- Pérez-Martín M., Pérez-Pérez M.E., Lemaire S.D., Crespo J.L. (2014). Oxidative stress contributes to autophagy induction in response to endoplasmic reticulum stress in Chlamydomonas reinhardtii. Plant Physiol. 166: 997–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojo E., Martín R., Carter C., Zouhar J., Pan S., Plotnikova J., Jin H., Paneque M., Sánchez-Serrano J.J., Baker B., Ausubel F.M., Raikhel N.V. (2004). VPEgamma exhibits a caspase-like activity that contributes to defense against pathogens. Curr. Biol. 14: 1897–1906. [DOI] [PubMed] [Google Scholar]
- Sarkar S., Ravikumar B., Floto R.A., Rubinsztein D.C. (2009). Rapamycin and mTOR-independent autophagy inducers ameliorate toxicity of polyglutamine-expanded huntingtin and related proteinopathies. Cell Death Differ. 16: 46–56. [DOI] [PubMed] [Google Scholar]
- Song J., Takeda M., Morimoto R.I. (2001). Bag1-Hsp70 mediates a physiological stress signalling pathway that regulates Raf-1/ERK and cell growth. Nat. Cell Biol. 3: 276–282. [DOI] [PubMed] [Google Scholar]
- Szecsi P.B. (1992). The aspartic proteases. Scand. J. Clin. Lab. Invest. Suppl. 210: 5–22. [PubMed] [Google Scholar]
- Takayama S., Reed J.C. (2001). Molecular chaperone targeting and regulation by BAG family proteins. Nat. Cell Biol. 3: E237–E241. [DOI] [PubMed] [Google Scholar]
- Takayama S., Sato T., Krajewski S., Kochel K., Irie S., Millan J.A., Reed J.C. (1995). Cloning and functional analysis of BAG-1: a novel Bcl-2-binding protein with anti-cell death activity. Cell 80: 279–284. [DOI] [PubMed] [Google Scholar]
- Takayama S., Xie Z., Reed J.C. (1999). An evolutionarily conserved family of Hsp70/Hsc70 molecular chaperone regulators. J. Biol. Chem. 274: 781–786. [DOI] [PubMed] [Google Scholar]
- Torres M.A., Jones J.D., Dangl J.L. (2005). Pathogen-induced, NADPH oxidase-derived reactive oxygen intermediates suppress spread of cell death in Arabidopsis thaliana. Nat. Genet. 37: 1130–1134. [DOI] [PubMed] [Google Scholar]
- Townsend P.A., Cutress R.I., Sharp A., Brimmell M., Packham G. (2003). BAG-1: a multifunctional regulator of cell growth and survival. Biochim. Biophys. Acta 1603: 83–98. [DOI] [PubMed] [Google Scholar]
- Wilkins M.R., Gasteiger E., Bairoch A., Sanchez J.C., Williams K.L., Appel R.D., Hochstrasser D.F. (1999). Protein identification and analysis tools in the ExPASy server. Methods Mol. Biol. 112: 531–552. [DOI] [PubMed] [Google Scholar]
- Williams B., Kabbage M., Britt R., Dickman M.B. (2010). AtBAG7, an Arabidopsis Bcl-2-associated athanogene, resides in the endoplasmic reticulum and is involved in the unfolded protein response. Proc. Natl. Acad. Sci. USA 107: 6088–6093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams B., Njaci J., Moghaddam L., Long H., Dickman M.B., Zhang X., Mundree S. (2015). Trehalose accumulation triggers autophagy during plant desiccation. PLoS Genet. 11: e1005705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Y., Suzuki H., Borevitz J., Blount J., Guo Z., Patel K., Dixon R.A., Lamb C. (2004). An extracellular aspartic protease functions in Arabidopsis disease resistance signaling. EMBO J. 23: 980–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y., Sheen J. (2014). The role of target of rapamycin signaling networks in plant growth and metabolism. Plant Physiol. 164: 499–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng L.R., Qu S., Bordeos A., Yang C., Baraoidan M., Yan H., Xie Q., Nahm B.H., Leung H., Wang G.L. (2004). Spotted leaf11, a negative regulator of plant cell death and defense, encodes a U-box/armadillo repeat protein endowed with E3 ubiquitin ligase activity. Plant Cell 16: 2795–2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Wang J., Yu J.Q., Chen Z. (2014). Role and regulation of autophagy in heat stress responses of tomato plants. Front. Plant Sci. 5: 174. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.