The deposition of H3K27me3 at MIR156A and MIR156C mediates the vegetative phase change in Arabidopsis.
Abstract
Vegetative phase change in flowering plants is regulated by a decrease in the level of miR156. The molecular mechanism of this temporally regulated decrease in miR156 expression is still unknown. Most of the miR156 in Arabidopsis thaliana shoots is produced by MIR156A and MIR156C. We found that the downregulation of these genes during vegetative phase change is associated with an increase in their level of histone H3 lysine 27 trimethylation (H3K27me3) and requires this chromatin modification. The increase in H3K27me3 at MIR156A/MIR156C is associated with an increase in the binding of PRC2 to these genes and is mediated redundantly by the E(z) homologs SWINGER and CURLY LEAF. The CHD3 chromatin remodeler PICKLE (PKL) promotes the addition of H3K27me3 to MIR156A/MIR156C but is not responsible for the temporal increase in this chromatin mark. PKL is bound to the promoters of MIR156A/MIR156C, where it promotes low levels of H3K27ac early in shoot development and stabilizes the nucleosome at the +1 position. These results suggest a molecular mechanism for the initiation and maintenance of vegetative phase change in plants.
INTRODUCTION
Plants undergo two major developmental transitions after germination. The first occurs when the shoot transitions from a juvenile to an adult phase of vegetative growth (vegetative phase change); this is followed by the transition to a reproductive phase of development, which is marked by the production of specialized reproductive structures, such as flowers or cones (Poethig, 2013). Many perennial plants alternate between the adult vegetative phase and the reproductive phase, but vegetative phase change is typically unidirectional: Once plants have transitioned to the adult vegetative phase, they usually remain in this phase for the rest of their life. The stability of the juvenile and adult vegetative phases has fascinated plant biologists for a long time (Wareing, 1959; Brink, 1962), but the molecular mechanism of this stability is still unknown.
The repressive chromatin modification, histone H3 lysine 27 trimethylation (H3K27me3), plays a major role in controlling the timing of developmental transitions in plants (reviewed in Köhler et al., 2012; Derkacheva and Hennig, 2014; Kim and Sung, 2014). After germination, it contributes to the downregulation of embryonic genes in both the root and shoot and to the repression of SHOOTMERISTEMLESS (STM) in leaves. H3K27me3 also plays an important role in the regulation of flowering time in Arabidopsis thaliana, where it prevents the premature expression of genes involved in floral morphogenesis (Goodrich et al., 1997) and represses the expression of the floral repressor FLOWERING LOCUS C (FLC) during the process of vernalization (Bastow et al., 2004; Sung and Amasino, 2004).
H3K27me3 is a product of Polycomb Repressive Complex 2 (PRC2) activity (Simon and Kingston, 2013). The four subunits of this complex (Polycomb Group proteins [PcG]) were originally identified in Drosophila melanogaster and consist of the histone methyl transferase, Enhancer of Zeste [E(z)], Suppressor of Zeste12 [Su(z)12], Extra sex combs (ESC), and p55. Arabidopsis has three genes encoding E(z) homologs (MEDEA [MEA], CURLY LEAF [CLF], and SWINGER [SWN]), three genes encoding homologs of Su(z)12 (EMBRYONIC FLOWER2 [EMF2], FERTILIZATION INDEPENDENT SEED2, and VERNALIZATION2 [VRN2]), five p55-related genes (MSI1-MSI5), and one ESC homolog (FERTILIZATION INDEPENDENT ENDOSPERM [FIE]). The mutant phenotypes of the genes encoding these proteins, and the composition of the protein complexes identified in planta (Köhler et al., 2003; Wood et al., 2006; De Lucia et al., 2008; Derkacheva et al., 2013), reveal that Arabidopsis possesses several functionally distinct PRC2 complexes. The complex with the largest role in postembryonic development is referred to as the EMF2 complex and consists of FIE, EMF2, MSI1, and either SWN or CLF (Derkacheva et al., 2013).
The molecular mechanism of H3K27me3 deposition in plants has been particularly well studied in the case of FLC in Arabidopsis. After a prolonged exposure to cold, the transcription of FLC is repressed by the presence of H3K27me3 across the entire locus (Bastow et al., 2004; Sung and Amasino, 2004). The deposition of H3K27me3 is mediated by several PHD finger proteins (Bastow et al., 2004; Sung and Amasino, 2004; Sung et al., 2006; Kim and Sung, 2013) and a long noncoding RNA (Heo and Sung, 2011), which become associated with PRC2 during the cold and direct it to FLC. At other loci, the deposition of H3K27me3 is regulated by chromatin remodeling factors that facilitate or repress the association of PcG proteins with chromatin. For example, the SWI2/SNF2 family chromatin remodeler BRAHMA inhibits the deposition of H3K27me3 by blocking the accessibility of chromatin to CLF and SWN (Li et al., 2015). The CHD3 chromatin remodeler PICKLE (PKL) is present at many genes that are enriched for H3K27me3 and promotes H3K27me3 and the transcriptional repression of several of these genes (Perruc et al., 2007; Zhang et al., 2008). However, PKL is also present at highly expressed genes, as well as at H3K27me3-enriched genes whose expression is either upregulated or unchanged in pkl mutants (Aichinger et al., 2009, 2011; Zhang et al., 2012; Jing et al., 2013). How PKL promotes H3K27me3 and why it sometimes activates and sometimes represses the expression of genes with which it is associated is unknown.
In Arabidopsis and in many other plants, vegetative phase change is regulated by a decrease in the level of the microRNA, miR156. miR156 is present at high levels after germination and declines during shoot development, leading to an increase in its direct targets, transcripts encoding SQUAMOSA PROMOTER BINDING (SBP/SPL) transcription factors (Wu and Poethig, 2006; Wang et al., 2009; Wu et al., 2009). Genome-wide analysis of the distribution of H3K27me3 indicates that several MIR156 genes possess this mark (Lafos et al., 2011). It has also been reported that miR156 expression is upregulated in mutants with reduced levels of H3K27me3 (Picó et al., 2015). However, whether H3K27me3 is temporally deposited at MIR156 genes and, if so, whether this contributes to the decrease in their expression during vegetative phase change remains unclear. Here, we show that the downregulation of miR156 during vegetative phase change is mediated by a decrease in the level of H3K27ac and an increase in the level of H3K27me3 at MIR156A and MIR156C. We also show that PKL promotes vegetative phase change by stabilizing the +1 nucleosome and by promoting low levels of H3K27ac and high levels of H3K27me3 at MIR156A and MIR156C. Our results suggest a mechanism for vegetative phase change and support the hypothesis that the balance between H3K27ac and H327me3 plays an important role in gene expression.
RESULTS
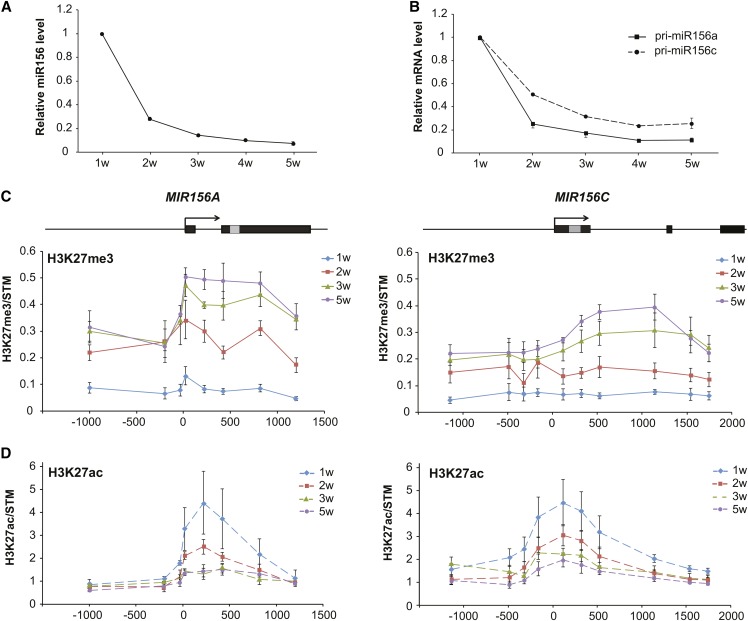
Under short-day conditions, the abundance of mature miR156 in shoot apices of wild-type plants declined to ∼10% of the starting level during the first three weeks after germination and changed very little after this time (Figure 1 A). This result is consistent with previous studies (Wang et al., 2009; Wu et al., 2009; Bergonzi et al., 2013; Wahl et al., 2013), although our results revealed that the decline in miR156 levels early in development is much larger than has previously been reported. Most of the miR156 in vegetative tissue is produced by MIR156A and MIR156C, which generate approximately equal amounts of this transcript (Yang et al., 2013). RT-qPCR revealed that the abundance of the primary transcripts (pri-miRNAs) of these genes decreased in a pattern similar to that of mature miR156 (Figure 1B). This observation indicates that the abundance of miR156 depends primarily on the transcription of MIR156A/MIR156C, rather than on the processing of pri-miR156a and pri-miR156c into miR156. pri-miR156a declined more rapidly and to a lower level than pri-miR156c, suggesting that the transcription of MIR156A and MIR156C is regulated by similar, but not completely identical mechanisms.
Figure 1.
Transcriptional Repression of MIR156A/MIR156C Is Associated with an Increase in H3K27me3 and a Decrease in H3K27ac.
(A) and (B) RT-qPCR analysis of temporal variation in mature miR156 (A), and pri-miR156a and pri-miR156c (B) in Col shoot apices. Values are mean ± se from three independent biological replicates. w, weeks.
(C) and (D) ChIP analysis of temporal variation in H3K27me3 (C) and H3K27ac (D) at MIR156A and MIR156C in Col shoot apices. The positions of the sites analyzed by qPCR are indicated by the diagrams above the graphs. Black boxes represent exons, the gray box is the position of the miR156 hairpin, and arrows indicate the direction of transcription, based on the data presented by Yang et al. (2013). Data are presented as the ratio of (MIR156A/H3) to (STM/H3) or (MIR156C/H3) to (STM/H3).
To identify genes involved in the temporal regulation of miR156 expression, we screened for ethyl methanesulfonate-induced mutations that enhance or suppress the early phase change phenotype of sqn-1. SQUINT (SQN) is the Arabidopsis ortholog of Cyclophilin 40 (Berardini et al., 2001) and acts in association with HEAT SHOCK PROTEIN90 to promote the activity of ARGONAUTE1 (AGO1) (Earley and Poethig, 2011; Iki et al., 2012). The phenotype of sqn-1 reflects a decrease in the activity of AGO1-dependent miRNAs, with its effect on vegetative phase change being largely explained by a reduction in the activity of miR156 (Smith et al., 2009). Among the suppressors identified in this screen was a splice-site mutation in PKL, pkl-11 (Supplemental Figure 1A). Plants doubly mutant for sqn-1 and pkl-11 closely resembled pkl-11 in both leaf morphology and in the timing of abaxial trichome production (Supplemental Figure 1B). This result indicates that PKL is required for the effect of sqn-1 on vegetative phase change.
The Decrease in MIR156A/MIR156C Transcription Is Associated with an Increase in H3K27me3 and a Decrease in H3K27ac
Loss-of-function mutations of PKL affect the expression of genes associated with H3K27me3 and reduce the level of H3K27me3 at some of the affected genes (Perruc et al., 2007; Zhang et al., 2008; Aichinger et al., 2009; Zhang et al., 2012). This observation suggested that pkl might suppress the precocious vegetative phase phenotype of sqn-1 by reducing the level of H3K27me3 at MIR156A/MIR156C, thereby increasing the expression of these genes. We tested this hypothesis by measuring the abundance of H3K27me3 at MIR156A/MIR156C in plants of different ages using chromatin immunoprecipitation (ChIP). Chromatin from shoot apices of wild-type plants grown in short days (SDs) was immunoprecipitated with antibodies to H3K27me3 (see Methods for the sampling strategy). Sites in the promoter and transcribed regions of MIR156A/MIR156C (Yang et al., 2013) were amplified by qPCR, and these results were normalized to the results from H3 ChIP and then to the results obtain for a site in located just after the transcription start site of the STM gene to correct for experimental variation in the recovery of H3K27me3 and H3K27ac. This site has been used as a normalization control in previous studies of epigenetic factors controlling FLC expression (Coustham et al., 2012; Crevillén et al., 2014; Yang et al., 2014). It has substantial amounts of both H3K27me3 and H3K27ac, allowing it to be used for the normalization of both of these epigenetic marks (Supplemental Figure 2). The results obtained without normalization to STM are shown in Supplemental Figure 3.
H3K27me3 was low in the promoters and transcribed regions of MIR156A/MIR156C at 1 week and increased across these loci in successively older shoots (Figure 1C). It increased most quickly in the region immediately downstream of the transcription start site (TSS) and was relatively high in this region even at 5 weeks. Consistent with the expression pattern of their primary transcripts (Figure 1B), H3K27me3 increased very rapidly from 1 to 2 weeks at MIR156A and increased more slowly at MIR156C. H3K27ac is a mark of active chromatin and is negatively correlated with H3K27me3 in Arabidopsis (Charron et al., 2009), possibly because it inhibits the binding of PRC2 (Tie et al., 2009; Reynolds et al., 2012; Kimura, 2013). A parallel analysis of H3K27ac levels at MIR156A/MIR156C revealed that this mark is present in a region immediately after the TSS, in a pattern complementary to, but more restricted than, the distribution of H3K27me3 (Figure 1D). H3K27ac levels decreased from 1 to 3 weeks and remained constant after this time. These results support the hypothesis that a decrease in H3K27ac and a concomitant increase in H3K27me3 contribute to the downregulation of MIR156A/MIR156C during vegetative phase change.
PKL and SWN Promote Vegetative Phase Change by Repressing the Transcription of MIR156A/MIR156C
To test the hypothesis that vegetative phase change is regulated by an increase in H3K27me3, we characterized the phenotype of mutations that reduce this chromatin mark. For this purpose, we examined strong alleles of PKL, pkl-1 and pkl-10 (GK_273E06), two null alleles of CLF, clf-28 and clf-29, and a null allele (swn-3) and a hypomorphic allele (swn-7) of SWN (Supplemental Figure 1C). CLF and SWN encode the histone methyl transferase subunit of PRC2. Plants deficient for both of these genes have no detectable H3K27me3 in vegetative tissue (Bouyer et al., 2011; Lafos et al., 2011), suggesting that all of the H3K27me3 at MIR156A/MIR156C is a product of one or both of these enzymes.
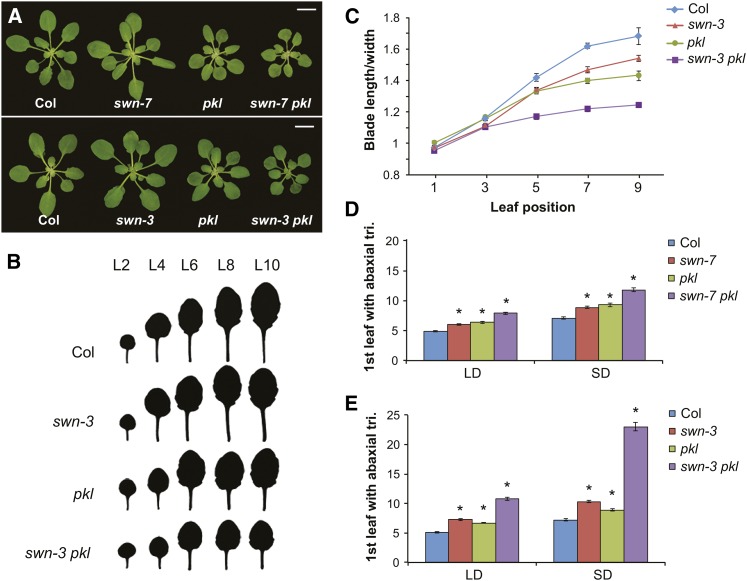
In Arabidopsis, juvenile leaves are small and round and lack trichomes on their abaxial surface, whereas adult leaves are elongated, have serrated margins, and produce abaxial trichomes (Telfer et al., 1997). swn-3 and swn-7 had slightly rounder leaves (Figures 2A to 2C) and produced abaxial trichomes approximately two plastochrons later than Col in both long days (LDs) and SDs (Figures 2D and 2E). The phenotype of swn-7 was slightly weaker than that of swn-3, which is consistent with the observation that swn-7 produces a small amount of the SWN transcript (Supplemental Figure 1C). pkl-1 and pkl-10 were morphologically identical and produced an identical delay in abaxial trichome production (Supplemental Figure 4A). We used pkl-10 (hereafter, pkl) for all experiments because it is a null allele (Supplemental Figure 1C). pkl had nearly the same effect on leaf shape and abaxial trichome production as swn-3 and swn-7 but produced smaller leaves than these mutations (Figure 2). clf-28 and clf-29 had elongated, up-curled leaves, which is opposite to the phenotype of swn and pkl (Supplemental Figure 4B). In LDs, clf-29 produced a small, but significant delay in abaxial trichome production (Supplemental Figure 4C). However, in SDs, clf-29 produced abaxial trichomes slightly earlier than Col because abaxial trichome production was delayed in Col but not in clf-29 under these conditions (Supplemental Figure 4C).
Figure 2.
PKL and SWN Act in Parallel to Promote Vegetative Phase Change.
(A) Three-week-old Col and mutant plants growing in SDs. Bars = 1 cm.
(B) Shape of successive rosette leaves of Col and mutant plants (SD). swn-3 pkl has smaller, smoother, and rounder leaves than Col or the single mutants.
(C) The length:width ratio of the lamina of successive rosette leaves of Col and mutant plants (SD) demonstrates that the leaves of swn-3 pkl resemble juvenile leaves. n = 24 for each genotype.
(D) and (E) swn-7, swn-3, and pkl delay the production of leaves with abaxial trichomes under both LD and SD conditions; (E) swn-3 and pkl interact synergistically in double mutants. All mutants were significantly different from Col in both LDs and SDs. *P < 0.01, Student’s t test, n = 23 or 24 for each genotype.
To determine the functional relationships between SWN, CLF, and PKL, we produced clf-28 swn-7, clf-29 pkl, swn-7 pkl, and swn-3 pkl double mutants. As has previously been reported (Chanvivattana et al., 2004; Aichinger et al., 2009), clf-28 swn-7 never progressed past the early seedling stage and eventually dedifferentiated into callus, making it impossible to determine the effect of this double mutant combination on vegetative phase change. clf pkl mutants had a largely additive phenotype: Although these plants were smaller than either single mutant, they displayed a combination of traits from each single mutant (Supplemental Figures 4B and 4C). Specifically, the basal rosette leaves of clf-29 pkl curled downwards, like pkl, whereas the apical leaves curled upwards, like clf (Supplemental Figure 4B); abaxial trichome production in clf-29 pkl was not significantly different than in pkl (Supplemental Figure 4C). In swn-7 pkl double mutants, abaxial trichome production was delayed by three plastochrons in LDs and five plastochrons in SDs relative to Col (Figure 2D). swn-3 pkl double mutants had a more severe phenotype. In this genotype, abaxial trichome production was delayed by six plastochrons in LDs and 16 plastochrons in SDs (Figure 2E), which is significantly greater than the sum of the effects of the single mutations. The leaves of swn-3 pkl mutants were also less serrated and rounder than the leaves of the single mutant lines (Figures 2B and 2C). The synergistic effect of pkl swn on these phase-specific traits suggests that PKL and SWN operate in different, functionally related processes.
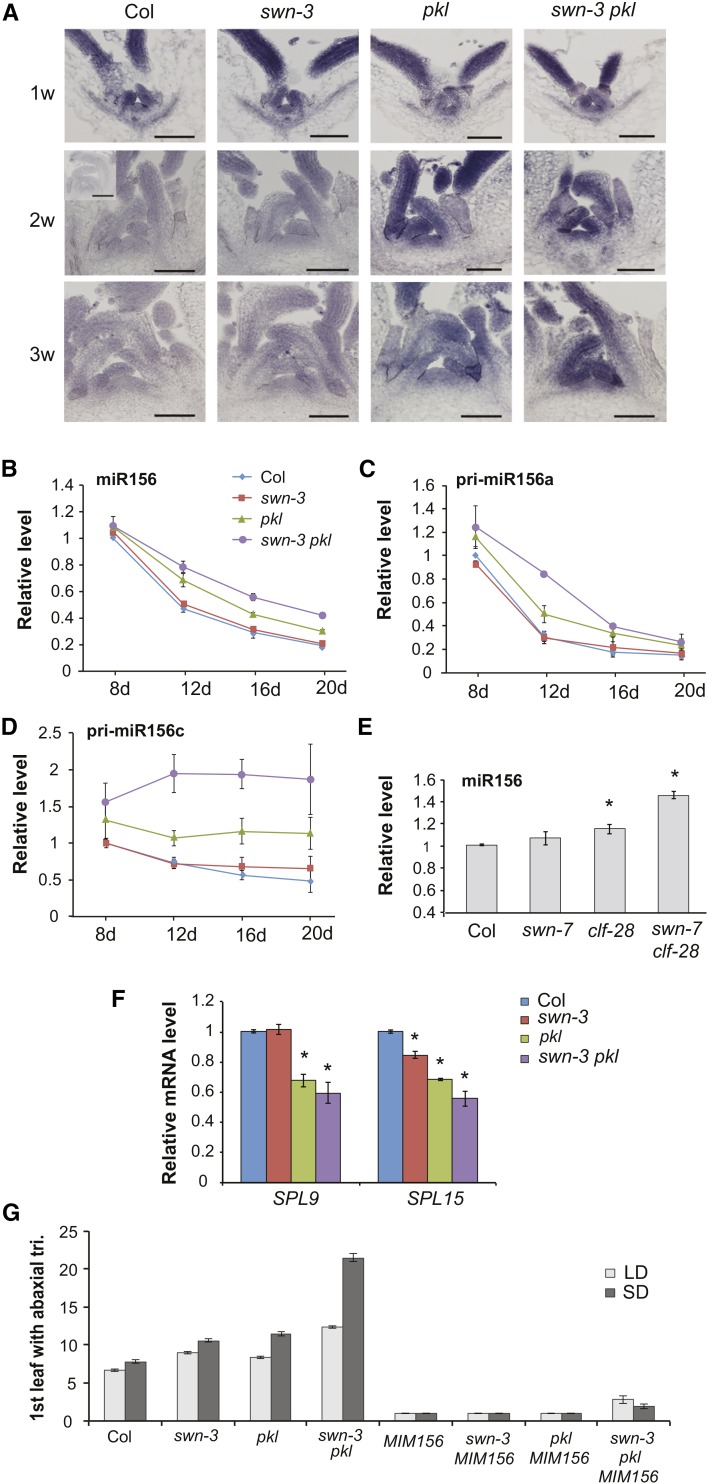
To determine if PKL and SWN regulate vegetative phase change by affecting the expression of miR156, we examined the effect of pkl, swn-3, and swn-3 pkl on the expression pattern of miR156 by in situ hybridization and RT-qPCR. In situ hybridization revealed that miR156 was expressed uniformly and at high levels in the shoot apical meristem and leaf primordia of Col at 1 week and was expressed at much lower levels at 2 and 3 weeks (Figure 3A). The expression pattern of miR156 in swn-3 was similar to Col at all three times (Figure 3A). pkl and swn-3 pkl were indistinguishable from Col at 1 week, but both genotypes had significantly elevated levels of miR156 at 2 and 3 weeks (Figure 3A). This analysis also revealed that there was no change in the spatial pattern of miR156 expression over time and no difference in spatial pattern of miR156 expression in swn-3, pkl, and swn-3 pkl relative to Col. RT-qPCR analysis of miR156 levels in shoot apices of 8-, 12-, 16-, and 20-d-old plants showed that miR156 levels were slightly, but consistently, elevated in swn-3, although this effect was not statistically significant. However, miR156 declined more slowly than normal in pkl and declined even more slowly in swn-3 pkl. The effect of pkl and swn on the expression of pri-miR156a (Figure 3C) and pri-miR156c (Figure 3D) was largely consistent with their effect on miR156. swn-3 had no significant effect on the expression of these transcripts early in development, although they were sometimes elevated in this mutant after 12 d. In pkl, pri-miR156a and pri-miR156c were elevated, but not significantly, at 8 d but were elevated at later time points. In swn-3 pkl, pri-miR156a and pri-miR156c were also slightly elevated at 8 d and declined much more slowly than in Col (Figures 3C and 3D). Indeed, pri-miR156c did not undergo any decrease in expression in swn-3 pkl from 8 to 20 d (Figure 3D). These results suggest that SWN and PKL operate in parallel to promote the transcriptional repression of MIR156A/MIR156C during vegetative phase change.
Figure 3.
PKL and SWN Promote Vegetative Phase Change by Repressing the Transcription of MIR156A and MIR156C.
(A) In situ localization of mature miR156 in the shoot apices of Col and mutant plants at 1, 2, and 3 weeks of development. The insert in the 2w Col panel shows hybridization to a mir156a miR156c double mutant (Yang et al., 2013), as a control for the background signal. Bars = 50 µm.
(B) to (D) RT-qPCR analysis of temporal variation in miR156 (B), pri-miR156a (C), and pri-miR156c (D) in the shoot apices of Col and mutant plants. miR156 decreases more slowly than normal in swn-3 pkl. Values are relative to Col at 8 d and represent the mean ± se from three biological replicates.
(E) RT-qPCR analysis of miR156 in 2-week-old Col and mutant plants. miR156 is significantly elevated in clf-28 (*P < 0.05) and swn-7 clf-28 (*P < 0.01, Student’s t test). Values are relative to Col and represent the mean ± se from three biological replicates.
(F) RT-qPCR analysis of SPL9 and SPL15 transcripts in 20-d-old Col and mutant plants. SPL9 is significantly reduced in pkl and swn-3 pkl, and SPL15 is significantly reduced in all three genotypes (*P < 0.05, Student’s t test). Values are relative to Col and represent the mean ± se from two biological replicates.
(G) First leaf with abaxial trichomes in Col, mutant, and 35S:MIM156 transgenic plants. 35S:MIM156 suppresses the phenotype of swn-3, pkl, and swn-3 pkl. n = 19 to 24 for each genotype.
One possible explanation for the synergistic interaction between pkl and swn-3 is that SWN and CLF redundantly repress MIR156A/MIR156C and PKL promotes the activity of both proteins. According to this hypothesis, the relatively weak phenotype of pkl is attributable to a slight reduction in the activity of both SWN and CLF, whereas the synergistic phenotype of swn-3 pkl is attributable to the complete loss of SWN coupled with a reduction in CLF activity. If this hypothesis is correct, the expression of miR156 should be elevated in swn clf double mutants. swn clf seedlings develop slowly and eventually form a callus; however, they produce leaf primordia and remain differentiated for ∼2 weeks after germination, making it possible to compare miR156 expression in mutant and wild-type plants just prior to their dedifferentiation. miR156 expression was not significantly affected by swn-7, was slightly elevated in clf-28, and was significantly higher in swn-7 clf-28 than in either clf-28 or Col (Figure 3E). This result is consistent with the results of other investigators (Picó et al., 2015) and supports the conclusion that SWN and CLF redundantly repress the expression of miR156.
The functional significance of the effect of swn-3, pkl, and swn-3 pkl on miR156 expression was investigated by measuring the expression of SPL9 and SPL15, which are direct targets of miR156. SPL genes are both cleaved and translationally repressed by miR156, so their transcript levels do not necessarily reflect the full effect of miR156 on their expression (Gandikota et al., 2007; Yang et al., 2012). Nevertheless, the abundance of these transcripts was consistent with the level of miR156 in these genotypes: SPL9 and SPL15 were unaffected (SPL9) or only slightly lower (SPL15) in swn-3 but were reduced to ∼70% of Col in pkl and to ∼50 to 60% of Col in swn-3 pkl (Figure 3F). We also examined the effect of a 35S:MIM156 transgene on the phenotype of swn-3, pkl, and swn-3 pkl. This transgene constitutively expresses a transcript with a noncleavable miR156 target site and acts as a sponge for miR156 (Franco-Zorrilla et al., 2007). 35S:MIM156 completely suppressed the abaxial trichome phenotype of swn-3 and pkl and nearly completely suppressed the abaxial trichome phenotype of swn-3 pkl (Figure 3G). These results suggest that the effect of pkl and swn-3 pkl on vegetative phase change is largely attributable to their effect on the expression of miR156. However, the observation that swn-3 has a slight effect on vegetative phase change and the expression of SPL15 without having a significant effect on miR156 expression suggests that it may also affect the expression of SPL genes independently of miR156.
PKL Increases H3K27me3 and Reduces H3K27ac at MIR156A/MIR156C
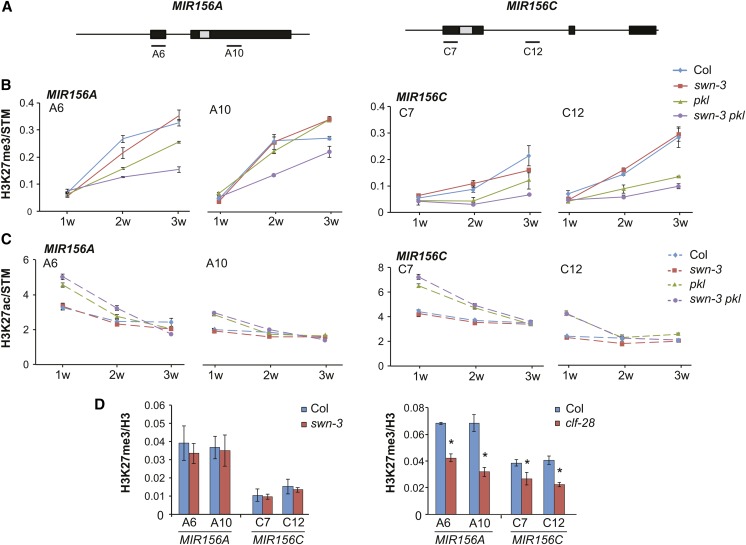
To determine if the increase in miR156 expression in pkl and swn-3 pkl is correlated with changes in H3K27me3 and H3K27ac, we used ChIP to measure the levels of these marks at positions near the TSS and the middle of MIR156A/MIR156C in mutant and wild-type plants of different ages (Figure 4). The results were normalized to the level of H3K27me3 or H3K27ac at STM, which was not significantly affected by these mutations (Supplemental Figure 2). In Col, H3K27me3 increased nearly 3-fold at MIR156A between 1 and 2 weeks and then increased more slowly between 2 and 3 weeks. At MIR156C, H3K27me3 increased gradually from 1 to 3 weeks. The swn-3 mutants sometimes displayed reduced amounts of H3K27me3 at both genes at 2 or 3 weeks, but this effect was not reproducible. pkl had no effect on H3K27me3 at 1 week but had significantly lower levels of H3K27me3 near the 5′ ends of both genes at 2 and 3 weeks. swn-3 pkl had normal levels of H3K27me3 at 1 week and significantly reduced levels of H3K27me3 at both the 5′ and 3′ ends of these genes at 2 and 3 weeks. Indeed, swn-3 pkl nearly completely blocked the increase in H3K27me3 at MIR156C and had a similarly strong effect on the 5′ site at MIR156A. These results are consistent with the expression patterns of pri-miR156a and pri-miR156c in these mutants (Figures 3C and 3D) and suggest that H3K27me3 is required for the transcriptional repression of MIR156A/MIR156C during vegetative phase change.
Figure 4.
PKL, SWN, and CLF Promote the Deacetylation and/or Trimethylation of H3K27.
(A) Genomic location of the sites analyzed by ChIP.
(B) and (C) ChIP analysis of H3K27me3 (B) and H3K27ac (C) in the shoot apices of Col and mutant plants at 1, 2, and 3 weeks. pkl and swn-3 pkl have reduced levels of H3K27me3 at 2 and 3 weeks and elevated levels of H3K27ac at 1 and 2 weeks. Data are presented as the ratio of (MIR156A/H3) to (STM/H3) or (MIR156C/H3) to (STM/H3). Values are mean ± se for the technical replicates from one representative experiment.
(D) ChIP analysis of H3K27me3 in the shoot apices of 2-week-old Col and mutant plants. swn-3 had no detectable effect on H3K27me3, while clf-28 reduced H3K27me3 by ∼40% at MIR156A and by ∼25% at MIR156C. Values are mean ± se from three biological replicates for swn-3 and from one experiment for clf-28; asterisk indicates significant difference from Col, P < 0.01, Student’s t test.
We compared the effect of swn-3 and clf-28 on H3K27me3 levels at MIR156A/MIR156C at 2 weeks in order to determine the relative importance of SWN and CLF at these loci (Figure 4D). We did not normalize these results to STM because clf is known to reduce H3K27me3 levels at this gene (Schubert et al., 2006). swn-3 had no effect on H3K27me3 at MIR156A/MIR156C, but clf-28 reduced H3K27me3 at MIR156A by ∼40% and produced a smaller, but still significant, reduction in H3K27me3 at MIR156C. Thus, CLF contributes more significantly to H3K27me3 at MIR156A/MIR156C than SWN. However, both CLF and SWN repress miR156 expression, as miR156 expression was greater in swn clf double mutants than in either single mutant (Figure 3E).
In animals, PKL-related proteins are components of NuRD, a multiprotein complex that contains a histone deacetylase (McDonel et al., 2009; Allen et al., 2013). To determine if PKL regulates the level of H3K27ac, we measured the abundance of this mark in parallel to H3K27me3 (Figure 4C). swn-3 had no effect on H3K27ac at MIR156A/MIR156C at 1, 2, or 3 weeks, but both pkl and swn-3 pkl seedlings had significantly elevated levels of H3K27ac at MIR156A/MIR156C at 1 week and slightly elevated levels of H3K27ac near the 5′ ends of these genes at 2 weeks. There was no significant difference in the level of H3K27ac in pkl and swn-3 pkl, implying that this effect is entirely attributable to pkl. This result demonstrates that PKL promotes low levels of H3K27ac at MIR156A/MIR156C early in shoot development. It is important to emphasize that there is little, if any, difference in the level of miR156, pri-miR156a, or pri-miR156c in pkl and wild-type plants at 1 week (Figures 3B to 3D), despite the significant difference in the level of H3K27ac in these plants (Figure 4C). This implies that the increased level of H3K27ac in pkl at 1 week is not an indirect result of its effect on transcription.
PKL and PRC2 Bind to MIR156A/MIR156C
It has been suggested (Aichinger et al., 2009, 2011) that PKL functions redundantly with PKR2 to promote H3K27me3 by promoting the transcription of PcG genes. To test this hypothesis, we used RT-qPCR to measure the transcripts of CLF, FIE, EMF2, VRN2, and MSI1 in 12- and 16-d-old shoot apices of wild-type, swn-3, pkl, and swn-3 pkl mutants. Consistent with the results of a previous study (Zhang et al., 2012), we did not observe a consistent difference in the expression of these genes in mutant and wild-type plants (Supplemental Figures 5A and 5B). pkl also had no consistent effect on SWN expression (Supplemental Figure 5C). These results demonstrate that the effect of pkl on H3K27me3 is not an indirect result of reduced PcG gene expression. To determine if the temporal increase H3K27me3 is attributable to a increase in the expression of PKL or PcG genes, we measured the transcripts of PKL, PKR1, SWN, CLF, VRN2, FIE, VRN2, MSI1, EMF2, and MEA by RT-qPCR over a 3-week period. None of these transcripts varied significantly during this time period (Supplemental Figure 5D). This result suggests that the temporal increase in H3K27me3 at MIR156A/MIR156C is either mediated by a posttranscriptionally regulated increase in PRC2 proteins or by a temporally regulated factor that increases the affinity of PcG proteins for MIR156A and MIR156C.
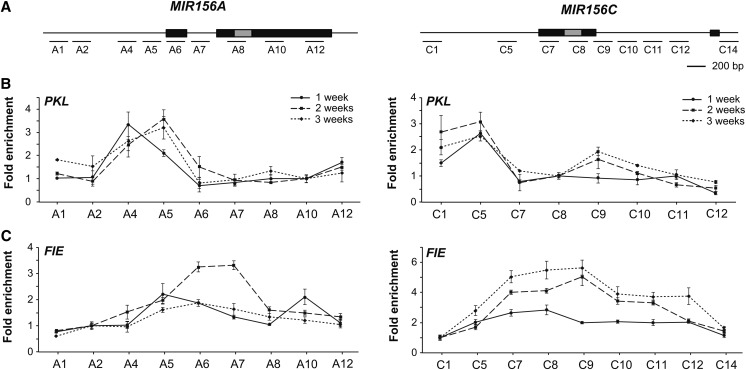
To determine if PKL could potentially promote the association of PcG proteins with MIR156A/MIR156C, we took advantage of pkl-1 plants expressing a pPKL:PKL-FLAG transgene that rescues the pkl-1 mutant phenotype (Zhang et al., 2012). Chromatin from shoot apices of 1-, 2-, and 3-week-old plants was immunoprecipitated with an antibody to FLAG, and sites from across MIR156A/MIR156C were assayed by qPCR (Figure 5A) These results were normalized to sites near these genes (Figure 5A) that were not bound by PKL, as shown by a comparison of the qPCR results for these sites to the results obtained for TA2 (Supplemental Figure 6), a retrotransposon whose chromatin state is not affected by pkl (Perruc et al., 2007; Zhang et al., 2012). PKL was bound to the promoters of MIR156A/MIR156C adjacent to the TSS and was present at approximately the same level at this site in 1-, 2-, and 3-week-old seedlings (Figure 5B). We then used a fie-11 pFIE:FIE-HA transgenic line (Wood et al., 2006) to characterize the association of PRC2 with MIR156A/MIR156C. FIE is the only ESC-like protein in Arabidopsis and is therefore present in all PRC2 complexes. At MIR156A, FIE was present at very low levels at 1 week, increased significantly at 2 weeks, and then returned to its original level at 3 weeks (Figure 5C). At MIR156C, FIE levels increased gradually across the entire transcribed region from 1 to 3 weeks (Figure 5C). These results suggest that the increase in H3K27me3 at MIR156A/MIR156C is the result of an increase in the affinity of PRC2 for these genes. Our data also suggest that PKL could potentially facilitate the interaction between PRC2 and MIR156A/MIR156C.
Figure 5.
PKL and PRC2 Bind to MIR156A and MIR156C.
(A) Location of the genomic sites analyzed by ChIP.
(B) Sites associated with PKL-FLAG in MIR156A and MIR156C chromatin in shoot apices of 1-, 2-, and 3-week-old PKL-FLAG plants, relative to non-transgenic plants. PKL binds in the promoter, near the TSS of MIR156A and MIR156C.
(C) Sites associated with FIE-HA in MIR156A and MIR156C chromatin in shoot apices of 1-, 2-, and 3-week-old FIE-HA plants, relative to nontransgenic plants. FIE binds near the TSS of MIR156A at 2 weeks and binds in a broader region of MIR156C at 2 and 3 weeks. Fold enrichment of each fragment was first calculated as the ratio of PKL-Flag to Col or FIE-HA to C24 and then normalized against a nonbinding site within MIR156A/MIR156C (A10 and C8 for PKL-Flag; A2 and C1 for FIE-HA). Values are presented as mean ± se for the technical replicates from one representative experiment.
PKL Promotes Nucleosome Occupancy
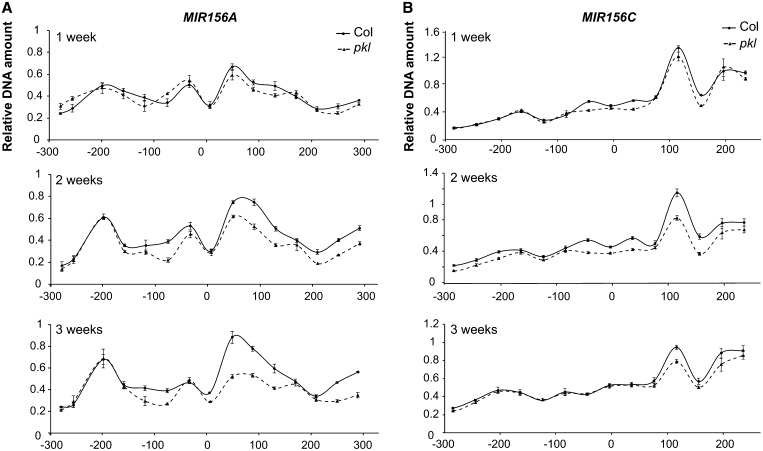
PKL is capable of promoting nucleosome sliding in vitro (Ho et al., 2013), but it is unknown if it promotes nucleosome remodeling in planta. To address this question, we used a micrococcal nuclease (MNase) assay to measure nucleosome occupancy near the MIR156A/MIR156C TSS in Col and pkl. We were particularly interested in nucleosome occupancy near the MIR156A/MIR156C TSS because the +1 nucleosome affects transcription efficiency (Henikoff, 2008; Petesch and Lis, 2012), and our ChIP results indicate that PKL is located near this site (Figure 5B). Chromatin was isolated from Col and pkl rosettes at 1, 2, and 3 weeks after germination and treated with MNase, and the abundance of DNA fragments from a 600-bp region surrounding the TSS of MIR156A and MIR156C was then assayed by qPCR. We used rosettes rather than shoot apices because of the large amount of tissue required for this assay. These data were normalized using a site from At4g07700 (Kumar and Wigge, 2010), whose MNase sensitivity did not change over time and which was not affected by pkl (Supplemental Figure 7).
MIR156A had two protected sites in the 300-nucleotide region upstream of the TSS and a broad protected region extending 200 nucleotides downstream of the TSS (Figure 6A). MIR156C had a narrow, highly protected site 100 nucleotides downstream of the TSS (Figure 6B) and two very weakly protected sites upstream of the TSS. pkl had no significant effect on the MNase sensitivity of MIR156A/MIR156C in 1-week-old seedlings and also had a minor, if any, effect on the MNase sensitivity of the promoters of these genes at later time points. However, there was a significant increase in MNase sensitivity of the protected regions downstream of the TSS of MIR156A/MIR156C in pkl at both 2 and 3 weeks (Figure 6). This result suggests that PKL promotes nucleosome occupancy at the +1 position during the period when MIR156A/MIR156C transcription is repressed but not when these genes are highly expressed. These data indicate that PKL acts as a nucleosome remodeler in planta, but also suggest that this function is modulated by other factors that contribute to the transcriptional activity of the loci with which it is associated.
Figure 6.
PKL Stabilizes the +1 Nucleosome.
MNase sensitivity of MIR156A (A) and MIR156C (B) chromatin isolated from 1-, 2-, and 3-week-old Col and pkl shoot apices. There is no significant difference between Col and pkl at 1 week (P > 0.05), but there is a significant increase in the MNase sensitivity of sites in the position of the +1 nucleosome (100 to 200 nucleotides) in pkl at 2 and 3 weeks (P < 0.05). Values were normalized to the −73 fragment of At4g07700 and are the results of one representative experiment.
DISCUSSION
The Mechanism of Vegetative Phase Change
A central question in developmental biology is how cells and tissues stably maintain their identity over time and how they switch from one stable state to the next. One of the most striking examples of this phenomenon in plants is vegetative phase change, which involves the transition between juvenile and adult phases of vegetative growth (Wareing, 1959). The first person to suggest a molecular mechanism for this phenomenon was R.A. Brink (Brink, 1962). Brink proposed that vegetative phase change is mediated by “self-perpetuating accessory materials” that undergo “orderly changes in state.” Brink predicted that these materials would include components of chromatin and coined the term “parachromatin” to distinguish this dynamic form of chromatin from the constitutive heterochromatin found near centromeres and at other chromosomal locations. This hypothesis was based on his discovery of paramutation in maize (Zea mays), which he showed involved an interaction between alleles that led to a heritable change in the expression of one of the alleles. Brink was struck by the similarity between the stable, but reversible, changes in gene expression that resulted from paramutation, and the stable, but reversible, changes in vegetative identity that accompany the juvenile-to-adult transition in plants and was prescient in recognizing that these phenomena might have mechanistic similarities. It is now generally recognized that changes in chromatin structure are the basis for the stability of many developmental states. However, the mechanism of vegetative phase change, and the basis for the stability of the juvenile and adult phases, is still unknown. The results presented here suggest that, as Brink predicted, changes in chromatin structure play key roles in these phenomena.
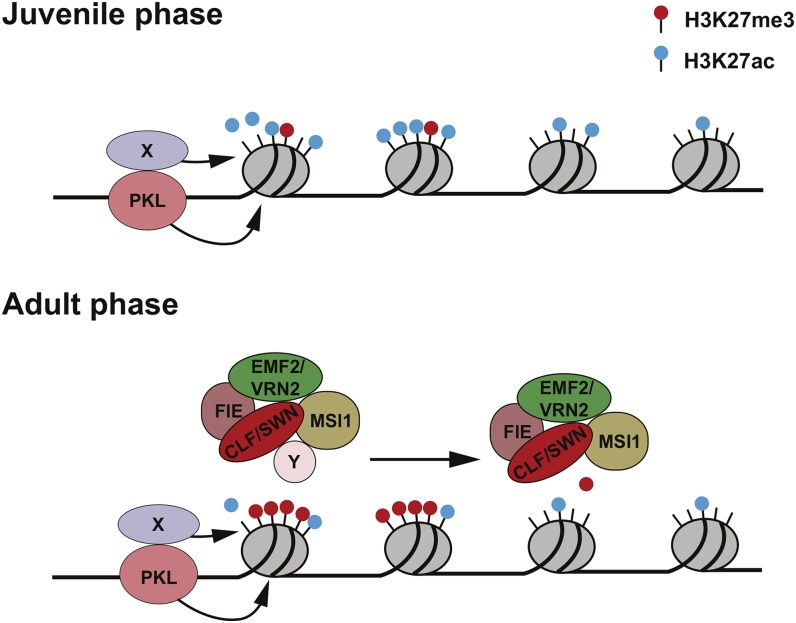
A model for the mechanism of vegetative phase change is shown in Figure 7. Vegetative phase change in Arabidopsis is mediated by a decline in the transcription of MIR156A/MIR156C. We found that this decrease is temporally correlated with an increase in the amount of H3K27me3 at these genes and that mutations that interfere with the deposition of this epigenetic mark slow the decrease in MIR156A expression and almost completely block the decrease in MIR156C expression. This observation is significant because it suggests that H3K27me3 is required for the downregulation of these genes. H3K27me3 may therefore play a different role in vegetative phase change than it does in vernalization, where it is required for the continued repression of the master regulator of this process, FLC, but is not required for the initial decline in FLC expression (Gendall et al., 2001).
Figure 7.
Model for the Regulation of MI156A/C during Vegetative Phase Change.
PKL is bound constitutively to the promoters of MIR156A/MIR156C and reduces H3K27ac by virtue of its association with a histone deacetylase (X). The transition to the adult phase occurs either when the level of H3K7ac drops to level that is no longer inhibitory to PRC2 binding or when a temporally regulated factor (Y) increases the affinity of SWN-PRC2 or CLF-PRC2 for these genes. PRC2 then methylates H3K27, moving from the 5′ to the 3′ end of the gene.
The increase in H3K27me3 at MIR156A/MIR156C is associated with an increase in the amount of PRC2 bound to these genes. One factor that might contribute to this is a decrease in the amount of H3K27ac at these loci. In both Drosophila (Tie et al., 2009) and mammals (Reynolds et al., 2012; Kim et al., 2015), H3K27ac antagonizes the binding of PRC2. A temporally regulated decrease in the rate of H3K27 acetylation or an increase in the rate of H3K27 deacetylation could therefore play an important role in the increased affinity of PRC2 for MIR156A/MIR156C. Another way in which PRC2 could be recruited to MIR156A/MIR156C is by association with a temporally regulated factor that binds to these genes. For example, PRC2 is directed to the FLC locus by the cold-induced PHD protein VIN3 (Sung and Amasino, 2004), which becomes physically associated with PRC2 during the cold (De Lucia et al., 2008). Among the candidates for such interacting factors are the B3 domain transcription factors VAL1 and VAL2. Like pkl, plants lacking these factors have elevated levels of pri-miR156a/pri-miR156c and reduced levels of H3K27me3 at MIR156A/MIR156C (Picó et al., 2015). However, the effect of val1 val2 on MIR156A/MIR156C expression and H3K27me3 was observed during a period when there was no major change in the expression of MIR156A/MIR156C in wild-type plants (Picó et al., 2015), so the relevance of this effect for vegetative phase change is unclear. It has been suggested (Picó et al., 2015) that the effect of val1 val2 on miR156 expression might be attributable to the inappropriate expression of the seed-specific transcription factor FUS3, which promotes the transcription of MIR156A in developing seeds (Wang and Perry, 2013) and is expressed postembryonically in these mutants (Yang et al., 2013a). FUS3 is also expressed postembryonically in pkl roots (Dean Rider et al., 2003), which raised the possibility that it might be responsible for the effect of these mutations on miR156 expression. We examined this possibility by comparing the expression patterns of FUS3, pri-miR156a, and pri-miR156c in Col, pkl, swn, and swn pkl. We observed no significant difference in FUS3 expression in Col, pkl, and swn, but did observe a variable, transient increase in FUS3 transcripts at 12 d in swn pkl (Supplemental Figure 7). We do not think this explains the phenotype of swn pkl because the increase in FUS3 transcripts is highly variable and transient, whereas miR156 expression is elevated in swn pkl for a prolonged period. Furthermore, we found that fus3-3 swn pkl triple mutants produced the same number of juvenile leaves as swn pkl, demonstrating that FUS3 is not required for the delayed phase change phenotype of swn pkl.
The Function of PKL
PKL is a member of subfamily II of CHD proteins, which includes the animal protein Mi2β/CHD4 (Ho et al., 2013). This ATP-dependent nucleosome remodeling protein is associated with the nucleosome remodeling and deacetylation complex (NuRD), one of the most abundant histone-modifying protein complexes in mammals (McDonel et al., 2009). In mammalian embryonic stem cells, NuRD promotes transcriptional repression by deacetylating H3K27, which facilitates the binding of PcG proteins and the addition of H3K27me3 to target genes (Reynolds et al., 2012). The basis for its role in gene activation is less clear, but the evidence suggests that NuRD controls gene expression by modulating the balance between H3K27ac and H3K27me3 (Reynolds et al., 2012). Although there is no evidence that PKL functions as part of a large protein complex (Ho et al., 2013), our results demonstrate that PKL regulates the level of H3K27ac, suggesting that it might be associated with, or promote the activity of, a histone deacetylase. Interestingly, pkl produced an increase in H3K27ac before it had a noticeable effect on H3K27me3. This observation suggests that PKL may regulate the balance between H3K27ac and HeK27me3 and provides an explanation for why PKL can both repress and activate gene expression.
Alternatively, PKL could promote the binding and/or activity of PRC2 through its role in nucleosome remodeling. Previous work has shown that PKL promotes nucleosome sliding in vitro in an ATP-dependent fashion (Ho et al., 2013). Along with our observation that PKL promotes nucleosome occupancy at the +1 position, this result suggests that PKL could operate either to stabilize the +1 nucleosome or to promote the addition of nucleosomes at this position. Interestingly, the binding pattern of PKL at MIR156A/MIR156C did not change from 1 to 3 weeks after germination, but loss of PKL only affected nucleosome occupancy in 2- and 3-week-old seedlings and only affected H3K27me3 at MIR156A/MIR156C at these time points. This observation suggests that other nucleosome remodelers play a more important role in regulating MIR156A/MIR156C transcription early in development, when these genes are most active. An excellent candidate is the chromatin remodeler BRAHMA, which has recently been shown to promote gene expression by antagonizing the activity of PRC2 (Li et al., 2015). It may be that PKL constitutively promotes PRC2 activity by promoting nucleosome occupancy at the +1 position, but this function is overridden early in development by the activity of other nucleosome remodelers. Deciphering how PKL contributes to H3K27ac and nucleosome occupancy, and whether these functions are responsible for the temporal pattern of PRC2 binding at MIR156A/MIR156C, is an important subject for future research.
The hypothesis that PKL promotes H3K27me3 through its effect on H3K27ac and/or nucleosome occupancy is consistent with the synergistic interaction of pkl with swn. This genetic interaction implies that PKL and SWN operate in different, functionally related processes and most likely reflects the combined effect of a reduction in the amount of functional PRC2 (due to swn) and a reduction in the ability of the remaining PRC2 to act on MIR156A/MIR156C (due to pkl). All of the H3K27me3 in shoot tissue is produced by SWN and CLF (Bouyer et al., 2011; Lafos et al., 2011), so this hypothesis implies that the synergistic phenotype of swn pkl is attributable to the effect of pkl on CLF activity. Although we did not test the effect of pkl on CLF activity, the results presented here, as well as other research (Picó et al., 2015), demonstrate that CLF promotes H3K27me3 of MIR156A/MIR156C, so this is a reasonable hypothesis.
One argument against the conclusion that H3K27me3 is important for vegetative phase change is the observation that clf has no effect on vegetative phase change. However, clf has a pleiotropic phenotype and elevates the expression of many genes, some of which could obscure the direct effect of clf on miR156 expression and vegetative phase. For example, FT is highly overexpressed in clf mutants (Jiang et al., 2008; Shen et al., 2014) and promotes the transcription of SPL3, SPL4, and SPL5 (Jung et al., 2012), which are direct targets of miR156 (Wu and Poethig, 2006). Overexpression of SPL3/4/5 promotes vegetative phase change (Wu and Poethig, 2006) and could counteract any delay in this process brought an effect of clf on miR156 expression.
Although the timing of vegetative phase change is quite predictable under any given set of environmental conditions, this transition can be delayed by a variety of factors (Poethig, 2013). Rejuvenation of adult shoots can also occur (Brink, 1962). Determining whether these phenomena are mediated by changes in H3K27me3 or other types of chromatin modifications is an important question for future research.
METHODS
Plant Material and Growth Conditions
All of the stocks used in this study were in the Col genetic background, except for the FIE-HA line, which was in C24. pkl (GK_273E06), swn-3 (SALK_050195), swn-7 (SALK_109121), clf-28 (SALK_139371), and clf-29 (SALK_021003) were obtained from the ABRC. pkr1-1 was obtained from Claudia Köhler, PKL-FLAG was provided by Joe Ogas, and FIE-HA was a gift from Chris Helliwell and Doris Wagner. Seeds were sown on Farfard #2 potting soil, stratified at 4°C for 2 to 4 d, and then transferred to Conviron growth chambers maintained at a constant 22°C and either LDs (16 h light:10 h dark) or SDs (10 h light:14 h dark). Illumination was provided by a 5:3 combination of white (USHIO F32T8/741) and red-enriched (Interlectric F32T8/WS Gro-Lite) fluorescent lights, at a photosynthetically active fluence rate of 150 µmoles m−2 s−1. Unless otherwise specified, all of the data presented in this article were obtained from plants growing in SDs. Plant age was measured from the date pots were transferred to growth chambers.
Unless otherwise specified, all gene expression or ChIP analyses were performed on shoot apices bearing leaf primordia 5 mm or less in length, in order to ensure that samples from different time points contained leaves of the same maturation state. For samples collected up to 8 d after planting, cotyledons were removed and the entire shoot apex was used; at this stage, most of the tissue in the shoot is from juvenile leaves. At later times, four or more leaves and leaf primordia were removed, leaving only transition and/or adult leaf primordia.
qPCR
Total RNA was isolated using TRIzol (Invitrogen) followed by Turbo DNase (Ambion) treatment, according to the manufacturer's instructions. One microgram of RNA was reverse transcribed with SuperScript III reverse transcriptase (Invitrogen), and qPCR was performed using a Bio-Rad CFX96 real-time System. Quantitative analysis of the mature miR156 transcript was performed according to Varkonyi-Gasic et al. (2007), using snoR101 as an internal control (Bergonzi et al., 2013). Primers used for qPCR are listed in Supplemental Table 1.
In Situ Hybridization
In situ hybridization was performed as previously described (Xu et al., 2010). Shoot apices of 1-, 2-, and 3-week-old plants were fixed in 3.7% formaldehyde, embedded in Paraplast, and then sectioned longitudinally with a microtome. miR156 was detected using an antisense miR156 Locked Nucleic Acid probe purchased from Exiqon.
Chromatin Immunoprecipitation
ChIP was performed according to Saleh et al. (2008), with some modifications. Shoot apices (0.5 to 1.0 g) were cross-linked in 1% formaldehyde. Nuclei were isolated using extraction buffer 1 (0.4 M sucrose, 10 mM Tris-HCl, pH 8.0, 10 mM MgCl2, 5 mM β-mercaptoethanol, 1 mM PMSF, and 0.1% Triton X-100), followed by extraction buffer 2 (0.25 M sucrose, 10 mM Tris-HCl, pH 8.0, 10 mM MgCl2, 5 mM β-mercaptoethanol, 1 mM PMSF, and 1% Triton X-100), and resuspended in nuclei lysis buffer (50 mM Tris-HCl, pH 8.0, 10 mM EDTA, and 1% SDS). DNA was then diluted in buffer (1.2 mM EDTA, 16.7 mM Tris-HCl, pH 8.0, 167 mM NaCl, and 0.01% SDS) and sonicated. Immunoprecipitation and reverse cross-linking was performed according to Saleh et al. (2008). DNA was finally isolated using a QIA Quick PCR purification kit (Qiagen), and qPCR was performed on a Bio-Rad CFX96 real-time system. Antibodies against H3, H3K27me3, and H3K27ac were purchased from Abcam (ab1791), Millipore (07-449), and Abcam (ab4729), respectively. Enrichment of H3K27me3 and H3K27ac was calculated as the ratio of H3K27me3/H3 or H3K27ac/H3 normalized to the value obtained from a site at STM. Binding of PKL-Flag and FIE-HA to MIR156A and MIR156C chromatin was determined using an anti-Flag antibody (Sigma-Aldrich; F3165) and an anti-HA antibody (Roche; 11583816001), respectively. Relative enrichment was calculated as 2−ΔΔCt= 2−(Ct (PKL-Flag ChIP)−Ct (PKL−Flag input))/2−(Ct (Col ChIP)−Ct (Col input)). For PKL-FLAG, the data were further normalized against a region at the 3′-end of MIR156A and MIR156C that displayed no significant binding; for FIE-HA, the data were further normalized against a region at the 5′-end of MIR156A and MIR156C that showed no significant binding, as demonstrated by a comparison of these sites to the −71 region of the TA2 locus, which is not affected by pkl (Perruc et al., 2007; Zhang et al., 2012). All assays were replicated two or more times using independently isolated samples. The number of replicates represented in each figure is provided in the figure legend.
MNase Assay
One to two grams of tissue from 1-, 2-, and 3-week-old rosettes was harvested for MNase assays. Nuclei were isolated according to Han et al. (2012) and were then subjected to MNase digestion for 3 to 5 min at 37°C. Mononucleosomes were gel-purified using a GenJET gel extraction kit (Fisher). The extracted DNA was then amplified by qPCR using a series of primers that amplify 90- to 110-bp fragments spaced 35 to 45 bp apart (Supplemental Table 1). Relative nucleosome occupancy was calculated as the fraction of MNase-digested DNA relative to mock undigested DNA (2−(Ct (MNase)−Ct (mock))), followed by normalization to a fragment at the −73 position in the gypsy-like retrotransposon, AT4G07700 (Kumar and Wigge, 2010; Han et al., 2012).
Accession Numbers
Sequence data from this article can be found in Supplemental Table 1 and in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: MIR156A, AL031369, At2g25095; MIR156C, AL049607, At4g31877; PKL, NM_128074, At2g25170; SWN, NM_116433, At4g02020; CLF, NM_127902, At2g23380; STM, NM_104916, At1g62360; TA2, CP002687, At4g06488; FIE, NM_112965, At3g20740; EMF2, NM_124502, At5g51230; VRN2, NM_117787, At4g16845; MEA, NM_100139, At1g02580; MSI1, NM_125208, At5g58230; PKR1, NM_123848, At5g44800; FUS3, NM_113591, At3g26790; SNOR101, NM_101919, At1g20690; ACT2, NM_112764, At3g18780; At4g07700, CP002687.
Supplemental Data
Supplemental Figure 1. The phenotype of pkl-11 and the effect of pkl-10, swn-3, and swn-7 on gene expression.
Supplemental Figure 2. The abundance of H3K27me3 and H3K27ac at the STM locus in shoot apices of Col and mutant plants at various times after germination.
Supplemental Figure 3. H3K27me3 increases and H3K27ac decreases at MIR156A and MIR156C during shoot development.
Supplemental Figure 4. The phenotype of pkl, clf, and pkl clf.
Supplemental Figure 5. PcG gene expression in Col and mutant plants.
Supplemental Figure 6. Abundance of the sites used for normalization in the PKL-FLAG and HA-FIE ChIP experiments, relative to the retrotransposon TA2.
Supplemental Figure 7. pkl has no effect on the MNase sensitivity of At4g07700.
Supplemental Figure 8. RT-qPCR analysis of the temporal pattern of FUS3 expression in the shoot apices of Col and mutant plants.
Supplemental Table 1. PCR primers.
Supplementary Material
Acknowledgments
We thank Justin Goodrich and Frazer Thorpe for information about the phenotype of swn mutants. We also thank Li Yang and members of the Poethig lab for useful discussions and helpful comments on this manuscript. This work was funded by a grant from the National Institutes of Health (GM51893).
AUTHOR CONTRIBUTIONS
M.X. conceived and performed experiments, analyzed data, and wrote the manuscript. T.H. performed experiments and analyzed data. M.R.S. identified and characterized the pkl-11 mutation. R.S.P. wrote the grant that funded this work and conceived experiments, analyzed data, and wrote the manuscript.
Glossary
- ChIP
chromatin immunoprecipitation
- SD
short day
- TSS
transcription start site
- MNase
micrococcal nuclease
- LD
long day
References
- Aichinger E., Villar C.B., Di Mambro R., Sabatini S., Köhler C. (2011). The CHD3 chromatin remodeler PICKLE and polycomb group proteins antagonistically regulate meristem activity in the Arabidopsis root. Plant Cell 23: 1047–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aichinger E., Villar C.B., Farrona S., Reyes J.C., Hennig L., Köhler C. (2009). CHD3 proteins and polycomb group proteins antagonistically determine cell identity in Arabidopsis. PLoS Genet. 5: e1000605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen H.F., Wade P.A., Kutateladze T.G. (2013). The NuRD architecture. Cell. Mol. Life Sci. 70: 3513–3524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastow R., Mylne J.S., Lister C., Lippman Z., Martienssen R.A., Dean C. (2004). Vernalization requires epigenetic silencing of FLC by histone methylation. Nature 427: 164–167. [DOI] [PubMed] [Google Scholar]
- Berardini T.Z., Bollman K., Sun H., Poethig R.S. (2001). Regulation of vegetative phase change in Arabidopsis thaliana by cyclophilin 40. Science 291: 2405–2407. [DOI] [PubMed] [Google Scholar]
- Bergonzi S., Albani M.C., Ver Loren van Themaat E., Nordström K.J., Wang R., Schneeberger K., Moerland P.D., Coupland G. (2013). Mechanisms of age-dependent response to winter temperature in perennial flowering of Arabis alpina. Science 340: 1094–1097. [DOI] [PubMed] [Google Scholar]
- Bouyer D., Roudier F., Heese M., Andersen E.D., Gey D., Nowack M.K., Goodrich J., Renou J.P., Grini P.E., Colot V., Schnittger A. (2011). Polycomb repressive complex 2 controls the embryo-to-seedling phase transition. PLoS Genet. 7: e1002014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brink R.A. (1962). Phase change in higher plants and somatic cell heredity. Q. Rev. Biol. 37: 1–22. [Google Scholar]
- Chanvivattana Y., Bishopp A., Schubert D., Stock C., Moon Y.H., Sung Z.R., Goodrich J. (2004). Interaction of Polycomb-group proteins controlling flowering in Arabidopsis. Development 131: 5263–5276. [DOI] [PubMed] [Google Scholar]
- Charron J.B., He H., Elling A.A., Deng X.W. (2009). Dynamic landscapes of four histone modifications during deetiolation in Arabidopsis. Plant Cell 21: 3732–3748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coustham V., Li P., Strange A., Lister C., Song J., Dean C. (2012). Quantitative modulation of polycomb silencing underlies natural variation in vernalization. Science 337: 584–587. [DOI] [PubMed] [Google Scholar]
- Crevillén P., Yang H., Cui X., Greeff C., Trick M., Qiu Q., Cao X., Dean C. (2014). Epigenetic reprogramming that prevents transgenerational inheritance of the vernalized state. Nature 515: 587–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean Rider S. Jr., Henderson J.T., Jerome R.E., Edenberg H.J., Romero-Severson J., Ogas J. (2003). Coordinate repression of regulators of embryonic identity by PICKLE during germination in Arabidopsis. Plant J. 35: 33–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Lucia F., Crevillen P., Jones A.M., Greb T., Dean C. (2008). A PHD-polycomb repressive complex 2 triggers the epigenetic silencing of FLC during vernalization. Proc. Natl. Acad. Sci. USA 105: 16831–16836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derkacheva M., Hennig L. (2014). Variations on a theme: Polycomb group proteins in plants. J. Exp. Bot. 65: 2769–2784. [DOI] [PubMed] [Google Scholar]
- Derkacheva M., Steinbach Y., Wildhaber T., Mozgová I., Mahrez W., Nanni P., Bischof S., Gruissem W., Hennig L. (2013). Arabidopsis MSI1 connects LHP1 to PRC2 complexes. EMBO J. 32: 2073–2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earley K.W., Poethig R.S. (2011). Binding of the cyclophilin 40 ortholog SQUINT to Hsp90 protein is required for SQUINT function in Arabidopsis. J. Biol. Chem. 286: 38184–38189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco-Zorrilla J.M., Valli A., Todesco M., Mateos I., Puga M.I., Rubio-Somoza I., Leyva A., Weigel D., García J.A., Paz-Ares J. (2007). Target mimicry provides a new mechanism for regulation of microRNA activity. Nat. Genet. 39: 1033–1037. [DOI] [PubMed] [Google Scholar]
- Gandikota M., Birkenbihl R.P., Höhmann S., Cardon G.H., Saedler H., Huijser P. (2007). The miRNA156/157 recognition element in the 3′ UTR of the Arabidopsis SBP box gene SPL3 prevents early flowering by translational inhibition in seedlings. Plant J. 49: 683–693. [DOI] [PubMed] [Google Scholar]
- Gendall A.R., Levy Y.Y., Wilson A., Dean C. (2001). The VERNALIZATION 2 gene mediates the epigenetic regulation of vernalization in Arabidopsis. Cell 107: 525–535. [DOI] [PubMed] [Google Scholar]
- Goodrich J., Puangsomlee P., Martin M., Long D., Meyerowitz E.M., Coupland G. (1997). A Polycomb-group gene regulates homeotic gene expression in Arabidopsis. Nature 386: 44–51. [DOI] [PubMed] [Google Scholar]
- Han S.K., Sang Y., Rodrigues A., Wu M.F., Rodriguez P.L., Wagner D.; BIOL425 F2010 (2012). The SWI2/SNF2 chromatin remodeling ATPase BRAHMA represses abscisic acid responses in the absence of the stress stimulus in Arabidopsis. Plant Cell 24: 4892–4906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S. (2008). Nucleosome destabilization in the epigenetic regulation of gene expression. Nat. Rev. Genet. 9: 15–26. [DOI] [PubMed] [Google Scholar]
- Heo J.B., Sung S. (2011). Vernalization-mediated epigenetic silencing by a long intronic noncoding RNA. Science 331: 76–79. [DOI] [PubMed] [Google Scholar]
- Ho K.K., Zhang H., Golden B.L., Ogas J. (2013). PICKLE is a CHD subfamily II ATP-dependent chromatin remodeling factor. Biochim. Biophys. Acta 1829: 199–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iki T., Yoshikawa M., Meshi T., Ishikawa M. (2012). Cyclophilin 40 facilitates HSP90-mediated RISC assembly in plants. EMBO J. 31: 267–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang D., Wang Y., Wang Y., He Y. (2008). Repression of FLOWERING LOCUS C and FLOWERING LOCUS T by the Arabidopsis Polycomb repressive complex 2 components. PLoS One 3: e3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing Y., Zhang D., Wang X., Tang W., Wang W., Huai J., Xu G., Chen D., Li Y., Lin R. (2013). Arabidopsis chromatin remodeling factor PICKLE interacts with transcription factor HY5 to regulate hypocotyl cell elongation. Plant Cell 25: 242–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung J.H., Ju Y., Seo P.J., Lee J.H., Park C.M. (2012). The SOC1-SPL module integrates photoperiod and gibberellic acid signals to control flowering time in Arabidopsis. Plant J. 69: 577–588. [DOI] [PubMed] [Google Scholar]
- Kim D.H., Sung S. (2013). Coordination of the vernalization response through a VIN3 and FLC gene family regulatory network in Arabidopsis. Plant Cell 25: 454–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D.H., Sung S. (2014). Genetic and epigenetic mechanisms underlying vernalization. Arabidopsis Book 12: e0171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T.W., et al. (2015). Ctbp2 modulates NuRD-mediated deacetylation of H3K27 and facilitates PRC2-Mediated H3K27me3 in active embryonic stem cell genes during exit from pluripotency. Stem Cells 33: 2442–2455. [DOI] [PubMed] [Google Scholar]
- Kimura H. (2013). Histone modifications for human epigenome analysis. J. Hum. Genet. 58: 439–445. [DOI] [PubMed] [Google Scholar]
- Köhler C., Hennig L., Bouveret R., Gheyselinck J., Grossniklaus U., Gruissem W. (2003). Arabidopsis MSI1 is a component of the MEA/FIE Polycomb group complex and required for seed development. EMBO J. 22: 4804–4814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler C., Wolff P., Spillane C. (2012). Epigenetic mechanisms underlying genomic imprinting in plants. Annu. Rev. Plant Biol. 63: 331–352. [DOI] [PubMed] [Google Scholar]
- Kumar S.V., Wigge P.A. (2010). H2A.Z-containing nucleosomes mediate the thermosensory response in Arabidopsis. Cell 140: 136–147. [DOI] [PubMed] [Google Scholar]
- Lafos M., Kroll P., Hohenstatt M.L., Thorpe F.L., Clarenz O., Schubert D. (2011). Dynamic regulation of H3K27 trimethylation during Arabidopsis differentiation. PLoS Genet. 7: e1002040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Chen C., Gao L., Yang S., Nguyen V., Shi X., Siminovitch K., Kohalmi S.E., Huang S., Wu K., Chen X., Cui Y. (2015). The Arabidopsis SWI2/SNF2 chromatin Remodeler BRAHMA regulates polycomb function during vegetative development and directly activates the flowering repressor gene SVP. PLoS Genet. 11: e1004944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonel P., Costello I., Hendrich B. (2009). Keeping things quiet: roles of NuRD and Sin3 co-repressor complexes during mammalian development. Int. J. Biochem. Cell Biol. 41: 108–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perruc E., Kinoshita N., Lopez-Molina L. (2007). The role of chromatin-remodeling factor PKL in balancing osmotic stress responses during Arabidopsis seed germination. Plant J. 52: 927–936. [DOI] [PubMed] [Google Scholar]
- Petesch S.J., Lis J.T. (2012). Overcoming the nucleosome barrier during transcript elongation. Trends Genet. 28: 285–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picó S., Ortiz-Marchena M.I., Merini W., Calonje M. (2015). Deciphering the role of POLYCOMB REPRESSIVE COMPLEX1 variants in regulating the acquisition of flowering competence in Arabidopsis. Plant Physiol. 168: 1286–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poethig R.S. (2013). Vegetative phase change and shoot maturation in plants. Curr. Top. Dev. Biol. 105: 125–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds N., Salmon-Divon M., Dvinge H., Hynes-Allen A., Balasooriya G., Leaford D., Behrens A., Bertone P., Hendrich B. (2012). NuRD-mediated deacetylation of H3K27 facilitates recruitment of Polycomb Repressive Complex 2 to direct gene repression. EMBO J. 31: 593–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleh A., Alvarez-Venegas R., Avramova Z. (2008). An efficient chromatin immunoprecipitation (ChIP) protocol for studying histone modifications in Arabidopsis plants. Nat. Protoc. 3: 1018–1025. [DOI] [PubMed] [Google Scholar]
- Schubert D., Primavesi L., Bishopp A., Roberts G., Doonan J., Jenuwein T., Goodrich J. (2006). Silencing by plant Polycomb-group genes requires dispersed trimethylation of histone H3 at lysine 27. EMBO J. 25: 4638–4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L., Thong Z., Gong X., Shen Q., Gan Y., Yu H. (2014). The putative PRC1 RING-finger protein AtRING1A regulates flowering through repressing MADS AFFECTING FLOWERING genes in Arabidopsis. Development 141: 1303–1312. [DOI] [PubMed] [Google Scholar]
- Simon J.A., Kingston R.E. (2013). Occupying chromatin: Polycomb mechanisms for getting to genomic targets, stopping transcriptional traffic, and staying put. Mol. Cell 49: 808–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith M.R., Willmann M.R., Wu G., Berardini T.Z., Möller B., Weijers D., Poethig R.S. (2009). Cyclophilin 40 is required for microRNA activity in Arabidopsis. Proc. Natl. Acad. Sci. USA 106: 5424–5429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung S., Amasino R.M. (2004). Vernalization in Arabidopsis thaliana is mediated by the PHD finger protein VIN3. Nature 427: 159–164. [DOI] [PubMed] [Google Scholar]
- Sung S., Schmitz R.J., Amasino R.M. (2006). A PHD finger protein involved in both the vernalization and photoperiod pathways in Arabidopsis. Genes Dev. 20: 3244–3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telfer A., Bollman K.M., Poethig R.S. (1997). Phase change and the regulation of trichome distribution in Arabidopsis thaliana. Development 124: 645–654. [DOI] [PubMed] [Google Scholar]
- Tie F., Banerjee R., Stratton C.A., Prasad-Sinha J., Stepanik V., Zlobin A., Diaz M.O., Scacheri P.C., Harte P.J. (2009). CBP-mediated acetylation of histone H3 lysine 27 antagonizes Drosophila Polycomb silencing. Development 136: 3131–3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varkonyi-Gasic E., Wu R., Wood M., Walton E.F., Hellens R.P. (2007). Protocol: a highly sensitive RT-PCR method for detection and quantification of microRNAs. Plant Methods 3: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl V., Ponnu J., Schlereth A., Arrivault S., Langenecker T., Franke A., Feil R., Lunn J.E., Stitt M., Schmid M. (2013). Regulation of flowering by trehalose-6-phosphate signaling in Arabidopsis thaliana. Science 339: 704–707. [DOI] [PubMed] [Google Scholar]
- Wang F., Perry S.E. (2013). Identification of direct targets of FUSCA3, a key regulator of Arabidopsis seed development. Plant Physiol. 161: 1251–1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J.-W., Czech B., Weigel D. (2009). miR156-regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana. Cell 138: 738–749. [DOI] [PubMed] [Google Scholar]
- Wareing P. (1959). Problems of juvenility and flowering in trees. Biol. J. Linn. Soc. Lond. 56: 282–289. [Google Scholar]
- Wood C.C., Robertson M., Tanner G., Peacock W.J., Dennis E.S., Helliwell C.A. (2006). The Arabidopsis thaliana vernalization response requires a polycomb-like protein complex that also includes VERNALIZATION INSENSITIVE 3. Proc. Natl. Acad. Sci. USA 103: 14631–14636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G., Park M.Y., Conway S.R., Wang J.W., Weigel D., Poethig R.S. (2009). The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell 138: 750–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G., Poethig R.S. (2006). Temporal regulation of shoot development in Arabidopsis thaliana by miR156 and its target SPL3. Development 133: 3539–3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M., Hu T., McKim S.M., Murmu J., Haughn G.W., Hepworth S.R. (2010). Arabidopsis BLADE-ON-PETIOLE1 and 2 promote floral meristem fate and determinacy in a previously undefined pathway targeting APETALA1 and AGAMOUS-LIKE24. Plant J. 63: 974–989. [DOI] [PubMed] [Google Scholar]
- Yang C., Bratzel F., Hohmann N., Koch M., Turck F., Calonje M. (2013a). VAL- and AtBMI1-mediated H2Aub initiate the switch from embryonic to postgerminative growth in Arabidopsis. Curr. Biol. 23: 1324–1329. [DOI] [PubMed] [Google Scholar]
- Yang H., Howard M., Dean C. (2014). Antagonistic roles for H3K36me3 and H3K27me3 in the cold-induced epigenetic switch at Arabidopsis FLC. Curr. Biol. 24: 1793–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Wu G., Poethig R.S. (2012). Mutations in the GW-repeat protein SUO reveal a developmental function for microRNA-mediated translational repression in Arabidopsis. Proc. Natl. Acad. Sci. USA 109: 315–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Xu M., Koo Y., He J., Poethig R.S. (2013). Sugar promotes vegetative phase change in Arabidopsis thaliana by repressing the expression of MIR156A and MIR156C. eLife 2: e00260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Bishop B., Ringenberg W., Muir W.M., Ogas J. (2012). The CHD3 remodeler PICKLE associates with genes enriched for trimethylation of histone H3 lysine 27. Plant Physiol. 159: 418–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Rider S.D. Jr., Henderson J.T., Fountain M., Chuang K., Kandachar V., Simons A., Edenberg H.J., Romero-Severson J., Muir W.M., Ogas J. (2008). The CHD3 remodeler PICKLE promotes trimethylation of histone H3 lysine 27. J. Biol. Chem. 283: 22637–22648. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.