Group VII ERF transcription factors regulate the low-oxygen response by binding to a newly identified promoter element in hypoxia-responsive genes, leading to their transactivation.
Abstract
The response of Arabidopsis thaliana to low-oxygen stress (hypoxia), such as during shoot submergence or root waterlogging, includes increasing the levels of ∼50 hypoxia-responsive gene transcripts, many of which encode enzymes associated with anaerobic metabolism. Upregulation of over half of these mRNAs involves stabilization of five group VII ethylene response factor (ERF-VII) transcription factors, which are routinely degraded via the N-end rule pathway of proteolysis in an oxygen- and nitric oxide-dependent manner. Despite their importance, neither the quantitative contribution of individual ERF-VIIs nor the cis-regulatory elements they govern are well understood. Here, using single- and double-null mutants, the constitutively synthesized ERF-VIIs RELATED TO APETALA2.2 (RAP2.2) and RAP2.12 are shown to act redundantly as principle activators of hypoxia-responsive genes; constitutively expressed RAP2.3 contributes to this redundancy, whereas the hypoxia-induced HYPOXIA RESPONSIVE ERF1 (HRE1) and HRE2 play minor roles. An evolutionarily conserved 12-bp cis-regulatory motif that binds to and is sufficient for activation by RAP2.2 and RAP2.12 is identified through a comparative phylogenetic motif search, promoter dissection, yeast one-hybrid assays, and chromatin immunopurification. This motif, designated the hypoxia-responsive promoter element, is enriched in promoters of hypoxia-responsive genes in multiple species.
INTRODUCTION
Plant viability depends on oxygen. Low ambient oxygen concentrations can become a growth-limiting factor, especially for non-oxygen-evolving organs (like roots) or green tissues under dark conditions (Drew, 1997; Bailey-Serres and Voesenek, 2008; Voesenek and Bailey-Serres, 2015), leading to a decrease in ATP production and a subsequent energy crisis affecting numerous plant processes. To circumvent a negative energy status, plants have developed several strategies, including a switch to anaerobic fermentation to regenerate NAD+ required to sustain glycolytic ATP production (Drew, 1997; Bailey-Serres and Voesenek, 2008).
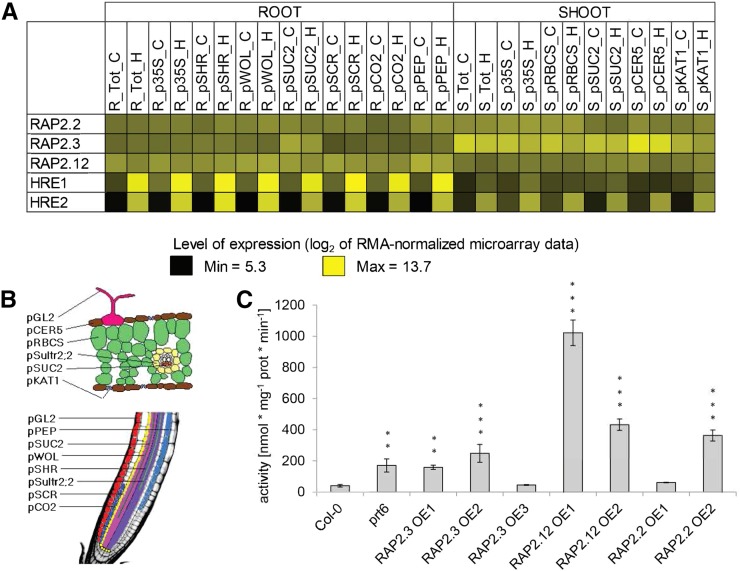
The basis for this cellular acclimation is the transcriptional activation of genes encoding key enzymes of fermentation, for example, alcohol dehydrogenase (ADH) and pyruvate decarboxylase (PDC) (summarized in Drew, 1997). A genome-scale evaluation of cell-type-specific transcriptional and translational status in Arabidopsis thaliana (Col-0) led to the definition of a set of 49 core hypoxia-responsive genes (HRGs), which are ubiquitously induced upon hypoxia (Mustroph et al., 2009), during submergence (Lee et al., 2011), and under one or both conditions in multiple plant species (Mustroph et al., 2010). The HRGs encode enzymes involved in sucrose catabolism, anaerobic fermentation (i.e., ADH and PDC), reactive oxygen species regulation, gene transcription, as well as proteins of unknown function, several of which were subsequently characterized. The transcriptional upregulation of ∼50% of the A. thaliana HRGs is the consequence of low-oxygen-dependent stabilization of group VII ethylene response factor transcription factors (ERF-VII TFs) (Gibbs et al., 2011; Licausi et al., 2011a; Kosmacz et al., 2015). A. thaliana encodes five ERF-VIIs, two of which, HYPOXIA RESPONSIVE ERF1 (HRE1) and HRE2, are HRGs (Figure 1A; Mustroph et al., 2009; Licausi et al., 2010). The ERF-VII transcripts RELATED TO APETALA2.12 (RAP2.12), RAP2.2, and RAP2.3 accumulate constitutively under normoxic conditions (Figure 1A; Büttner and Singh, 1997; Papdi et al., 2008; Hinz et al., 2010) and undergo translation under control and hypoxic conditions in seedlings (Mustroph et al., 2009; Juntawong et al., 2014).
Figure 1.
Expression and Function of ERF-VIIs.
(A) Expression of ERF-VIIs in Arabidopsis organs (roots, R; shoots, S) and cell types under normoxic (C) and hypoxic (H) conditions. Translatome (mRNA associated with polysome) data are from Mustroph et al. (2009)
(B) Overview of promoters used to define cell types assayed in translatome analyses for (A).
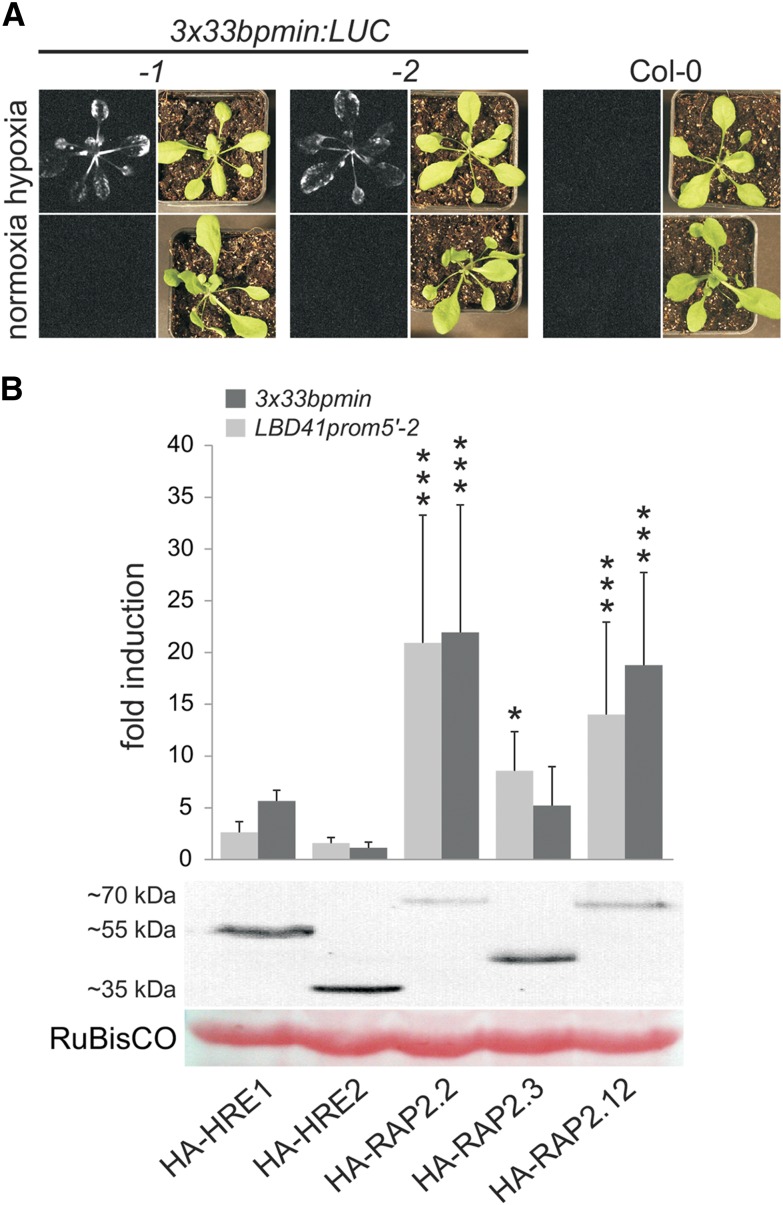
(C) Activity of the fermentation enzyme ADH in 7-d-old seedlings of ERF-VII overexpression lines, in nmol*mg−1 prot*min−1. Values are means ± sd of four independent samples. Significant differences in comparison to Col-0 are marked with asterisks for **P < 0.01 and ***P < 0.001 (one-way ANOVA, Tukey HSD Test).
A. thaliana ERF-VIIs and those of other plant species are characterized by a conserved N-terminal motif (Met-Cys-Gly-Gly-Ala-Ile/Leu, MCGGAI/L, termed the MC motif). In A. thaliana, Cys2 is essential for the oxygen-triggered degradation of the ERF-VIIs via the N-end rule pathway of targeted proteolysis (Gibbs et al., 2011, 2014; Licausi et al., 2011a; Abbas et al., 2015; Kosmacz et al., 2015). This pathway involves several protein-modifying enzymes, including the following: METHIONINE AMINOPEPTIDASE (MAP1/2), which cleaves the N-terminal methionine to expose the N-terminal Cys (Gibbs et al., 2014); oxidation of this Cys by two core HRGs encoding PLANT CYSTEINE OXIDASE (PCO1/2) (Weits et al., 2014); ARGINYL-tRNA PROTEIN TRANSFERASE (ATE1/2), which catalyzes the transfer of the Arg of amino-acylated tRNAArg to the N terminus of the oxidized NH2-Cys-ERF-VII; and the E3 ligase PROTEOLYSIS6, which recognizes the NH2-Arg-ERF-VII as degron for ubiquitination and thus proteasomal degradation (Gibbs et al., 2011, 2014; Licausi et al., 2011a).
In seedlings grown in air, some fraction of cellular RAP2.12 is masked from turnover through an association with the plasma membrane, likely via interaction with Acyl-CoA Binding Proteins (ACBP1/2) (Licausi et al., 2011a). As external oxygen levels fall below 10%, RAP2.12 migrates into the nucleus concomitant with induction of a major portion of the core HRGs, including ADH1 and PDC1 (Licausi et al., 2011a; Kosmacz et al., 2015). Of the 49 core HRGs, only seven might not be regulated by the N-end rule pathway, as their expression is unchanged in the prt6, ate1 ate2, and ged1 mutants (Gibbs et al., 2011; Riber et al., 2015). During reoxygenation, RAP2.12 becomes rapidly destabilized (Licausi et al., 2011a; Kosmacz et al., 2015), presumably aided by PCO1/2 catalysis of NH2-Cys2 oxidation (Weits et al., 2014). Overexpression of native forms of RAP2.2, RAP2.3, and RAP2.12 via the CaMV 35S promoter (prom35S) activates a similar set of genes only under hypoxic conditions, whereas N-terminally mutated versions of these proteins that stabilize their accumulation activate HRGs under normoxia (Hinz et al., 2010; Licausi et al., 2011a; Papdi et al., 2015). There is indirect evidence that the hypoxia-induced ERF-VIIs HRE1 and HRE2 function to maintain HRG transcription under low-oxygen conditions (Licausi et al., 2010), which may be necessary due to RAP2.12 inactivation during the stress by direct interaction with the trihelix-domain TF HYPOXIA RESPONSE ATTENUATOR1 (HRA1), also encoded by a core HRG (Giuntoli et al., 2014). Reduction of RAP2.2 and RAP2.12 transcript accumulation strongly reduces but does not abolish HRG activation relative to wild-type plants (Licausi et al., 2011a; Bui et al., 2015). This could be due to residual activity of RAP2.2 or RAP2.12, or that of RAP2.3, which is also linked to HRG regulation (Papdi et al., 2015). We hypothesize that despite subtle distinctions in their spatiotemporal and regulated expression, these three constitutively expressed ERF-VIIs function redundantly to activate HRGs to provide enzymes necessary for anaerobic metabolism as cellular oxygen levels decline below some threshold.
Redundant regulation by related TFs is often achieved by recognition of the same cis-regulatory elements in one or multiple target genes (Bernard et al., 2012). Based on in vitro assays, A. thaliana RAP2.2 can bind the sequence 5′-ATCTA-3′ present in the promoters of the carotenoid biosynthesis pathway genes PHYTOENE SYNTHASE (PSY) and PHYTOENE DESATURASE (PDS) (Welsch et al., 2007). This five-nucleotide motif is present (Hsu et al., 2011) or overrepresented (Licausi et al., 2010) in promoters of HRGs; moreover, promoters that contain 5′-ATCTA-3′ motifs were transactivated by RAP2.2 and RAP2.12 in protoplasts (Papdi et al., 2008; Licausi et al., 2011a; Giuntoli et al., 2014; Weits et al., 2014). Nonetheless, PSY and PDS transcript levels are not elevated in N-end rule mutants, transgenics that overexpress RAP2.2 or RAP2.12, or under hypoxic conditions (Mustroph et al., 2009; Gibbs et al., 2011; Licausi et al., 2011a). Motivated to solve this paradox, we performed promoter deletion analyses in this study to systematically define a cis-element that is sufficient for A. thaliana ERF-VII-mediated transcriptional activation in planta, and we further validated TF binding by mutational analyses, yeast one-hybrid assay, and chromatin immunopurification (ChIP).
Early work on the Adh1 gene of maize (Zea mays) identified a region with a duplicated and bipartite GC-rich and GT-rich cis-element called the anaerobic-responsive element (ARE) that is necessary and sufficient for hypoxia-responsive activation in protoplasts (Walker et al., 1987). The two interdependent subregions overlap with the promoter region that constitutively binds nuclear factors (Ferl and Nick, 1987; Ferl, 1990; Olive et al., 1991). Radiolabeled concatemers of the ARE showed decreased electrophoretic migration when incubated with nuclear extracts from maize cells. The requirement of the GC-rich region for protein binding led to the postulation of a GC binding protein 1 (GCBP-1) (Olive et al., 1991). Each GC-motif [GC(G/C)CC] of the Zm Adh1 ARE has an adjacent GT-rich motif [(T)GGTTT], which is a functional MYB TF binding site (Walker et al., 1987; Hoeren et al., 1998). A somewhat similar motif discovered in the At ADH1 promoter has a bipartite GT- and GC-motif region that is necessary for promoter activity in hypoxia-stressed plants (Dolferus et al., 1994). A. thaliana MYB2 was found to bind the GT-motif in vitro (Hoeren et al., 1998), but At ADH1 shows a wild-type-like hypoxic upregulation when MYB2 is knocked out (Licausi et al., 2010). MYB binding consensus sequences and GC-motifs are enriched 5′ of HRGs of A. thaliana including ADH1, but in a nonadjacent manner (Klok et al., 2002; Liu et al., 2005; Mohanty et al., 2005; Licausi et al., 2010). Despite recognition of a common core of HRGs in A. thaliana and other species (Mustroph et al., 2009; Christianson et al., 2010; Mustroph et al., 2010; Narsai et al., 2011), a cis-regulatory element that binds specific TFs to promote hypoxia-responsive transcriptional activation has not been unambiguously established.
Here, we demonstrate that redundant action of RAP2.2 and RAP2.12 is sufficient to activate ∼95% of hypoxia-responsive transcription of five core HRGs in A. thaliana. A comparative phylogenetic motif search analysis of HRG promoters in multiple species followed by detailed promoter deletion and mutation of the core HRG LOB DOMAIN-CONTAINING PROTEIN41 (LBD41) guided the discovery of a cis-element activated by RAP2.2 and RAP2.12. The consensus sequence 5′-AAACCA(G/C)(G/C)(G/C)GC-3′, designated the A. thaliana hypoxia-responsive promoter element (HRPE), was shown to be necessary and sufficient for ERF-VII transactivation of LBD41 and PCO1. A yeast-one-hybrid assay and ChIP analyses were used to further validate RAP2.2 and RAP2.12 interaction with the HRPE. These results explain ERF-VII redundancy and establish an evolutionarily conserved hierarchical network of genes transcriptionally activated by stabilization of ERF-VIIs.
RESULTS
RAP2.2 and RAP2.12 Are Redundant Major Hypoxia Gene Regulators
The A. thaliana ERF-VIIs have been proposed to act redundantly in HRG regulation, with RAP2.2 and RAP2.12 playing a predominant role in the rapid response to a decline in oxygen availability (Licausi et al., 2011a; Bui et al. 2015). As a first step to examine the redundancy of the three ERF-VII RAPs, N-terminally tagged versions of these TFs were stably overexpressed in Col-0, and ADH-specific activity was measured in homogenates from seedlings cultivated in ambient air (normoxia) (Figure 1). At least one overexpression line for each ERF-VII RAP displayed significantly enhanced ADH activity in comparison to Col-0, at levels similar to or higher than the prt6 mutant (Figure 1C) in which ERF-VIIs are stable under normoxia (Gibbs et al., 2011; Licausi et al., 2011a). This suggests that stabilization of any of the three ERF-VII RAPs is sufficient to upregulate ADH1 and its gene product.
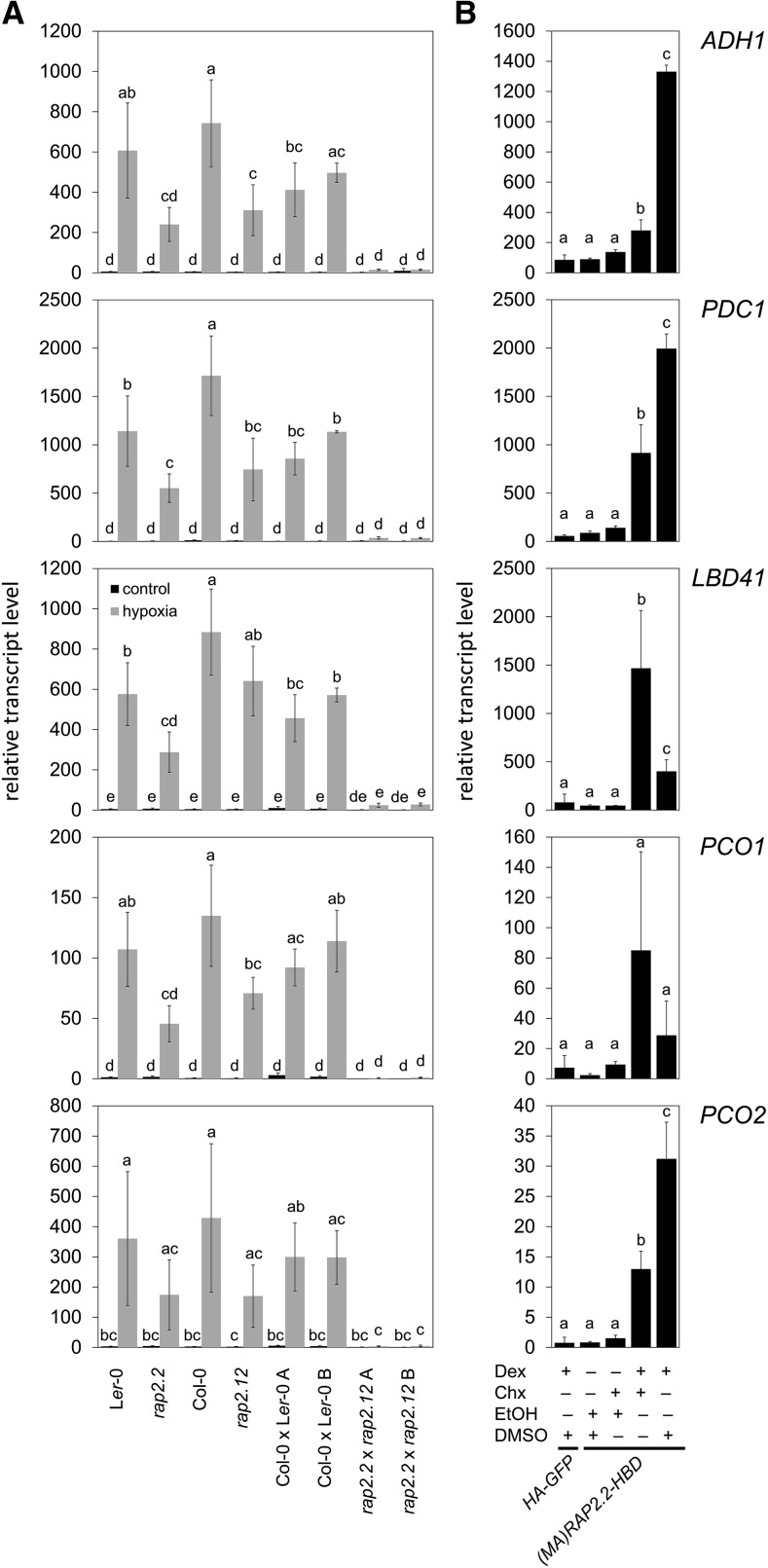
A genetic approach was used to test whether RAP2.2 or RAP2.12 was sufficient for upregulation of five core HRGs (LBD41, PCO1, PCO2, ADH1, and PDC1). These genes were selected because of their regulation by early hypoxia and in N-end rule pathway mutants. Single and double mutant (loss-of-function) homozygotes for alleles of RAP2.12 (rap2.12-2) and RAP2.2 (rap2.2-5) were established (Supplemental Table 1 and Supplemental Figure 1). In seedlings stressed for 2 h with hypoxia, transcript levels of the five HRGs were highly increased in Ler-0 and Col-0 wild-type backgrounds (Figure 2), with some (natural) variation between accessions that was not further studied. The knockout of RAP2.2 in Ler-0 significantly lowered transcript levels of all five HRGs to a minimum of 40% of wild-type induction. Similar results were obtained for RAP2.12 knockout in Col-0, indicating that both TFs contribute to hypoxia-dependent gene activation (Figure 2A). RAP2.2 and RAP2.12 share the highest amino acid sequence similarity among ERF-VIIs and therefore are the most likely to be functionally redundant. This was supported by the observation that two independently bred rap2.2-5 rap2.12-2 double knockout lines exhibited only 1 and 5% of HRG transcript levels under hypoxia, respectively, compared with the controls, which were two independently bred Col-0 × Ler-0 lines with homozygous RAP2.2 and RAP2.12 wild-type alleles from the same cross (Figure 2A). The significant reduction in HRG transcript accumulation in the single and double knockouts corresponded to lower survival of short-term submergence in the dark by rosette stage plants (Supplemental Figure 2). Both double mutants displayed significantly lower survival than rap2.12-1. However, because Ler-0 rosettes were more susceptible to the stress than Col-0, as reported previously (Vashisht et al., 2011), the phenotype of the double mutants could not be unambiguously linked to the loss of RAP2.2 and RAP2.12 (Supplemental Figure 2).
Figure 2.
RAP2.2 and RAP2.12 Act Redundantly as Direct Inducers of Hypoxia-Responsive Genes.
(A) RT-qPCR analyses of five HRGs in 7-d-old ERF-VII double or single knock out seedlings and respective wild types under hypoxia and normoxia.
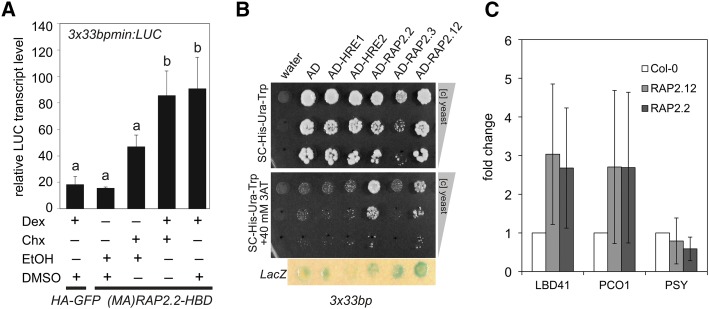
(B) RT-qPCR analyses of the same HRGs in mesophyll protoplasts from A. thaliana Col-0 transiently expressing either p35S:HA-GFP or p35S:(MA)RAP2.2-HBD. Chx or the solvent DMSO was applied 30 min before Dex or the solvent ethanol, and protoplasts were incubated for an additional 4 h under LD conditions. Transcript levels were normalized to ELONGATION FACTOR 1A (EF1a) mRNA. Values are means ± sd from three biological replicates (each with three technical replicates). Different letters indicate values that vary significantly at P < 0.05 (Tukey HSD test).
We hypothesized that the basis for redundancy of RAP2.2 and RAP2.12 in HRG regulation is a cis-element recognized by both ERF-VIIs. To find genes directly activated by ERF-VIIs, a translational fusion of a stabilized RAP2.2 to a glucocorticoid receptor [(MA)RAP2.2-HBD] was expressed in rosette leaf protoplasts (Figure 2B). The cytoplasmic-to-nuclear translocation of RAP2.2-HBD was initiated by application of the synthetic glucocorticoid dexamethasone (Dex) concomitant with inhibition of de novo protein synthesis with cycloheximide (Chx). After 4 h of Dex treatment, transcript levels of four of the five HRGs tested increased significantly in a (MA)RAP2.2-HBD-dependent manner (Figure 2B). ADH1, PDC1, and PCO2 levels showed a significant increase after Dex+Chx treatment, which was further enhanced when Chx was absent (Figure 2B). This points to a regulatory role of RAP2.2 for all of these HRG mRNAs, whose levels are constitutively higher in the prt6 mutant (Gibbs et al., 2011) and reduced under hypoxia in HRA1 overexpression lines (Giuntoli et al., 2014). Interestingly, LBD41 showed a significant induction in the presence of Dex and Chx, but a significantly lower induction in the absence of Chx (Figure 2B). A similar trend was evident for PCO1. These data hint at possible further regulatory steps initiated by RAP2.2 involving de novo protein synthesis-dependent gene activation, such as synthesis of a second protein that further enhances activation or limits the activity of a repressor.
Comparative Phylogenetic Footprinting Uncovers Conserved and Overrepresented Motifs
To predict potential hypoxia- and ERF-VII-responsive motifs, a comparative phylogenetic-based motif discovery strategy was performed on the 1-kb sequence upstream of the ATG start site of the 49 core HRGs (Mustroph et al., 2009). As a control, the strategy was applied to 49 low phosphate (Pi)-responsive genes (Bustos et al., 2010; wild-type shoot −P; wild-type shoot +P) to determine if the well-characterized PHR1 binding site, which has a GNATATNC consensus (Rubio et al., 2001), could be recognized.
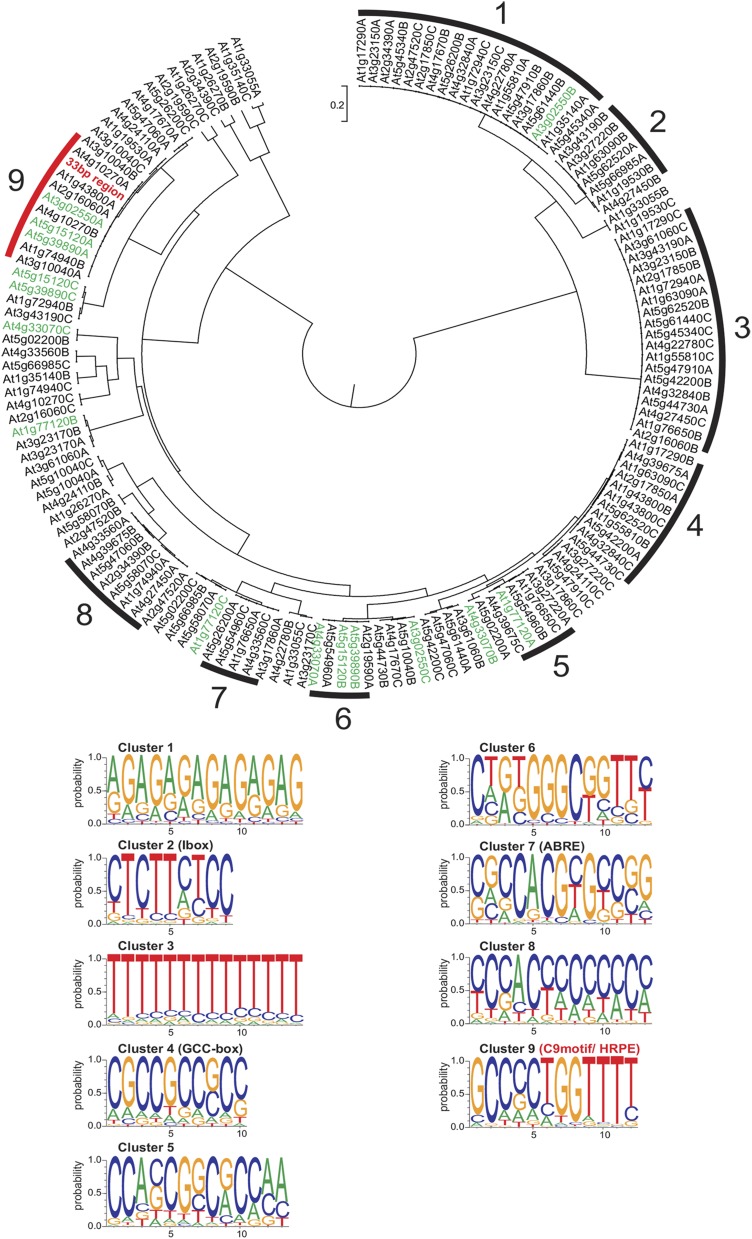
First, promoter alignments with the multiple expectation maximization for motif elicitation tool were used to identify at most three conserved patterns for each homologous gene set from 25 plant species (Supplemental Data Set 1). Each pattern was named by its Arabidopsis Gene Identifier and a letter (Figure 3). The resulting 147 predictions were processed by the motif comparison program STAMP (Mahony and Benos, 2007), enabling production of an unrooted dendrogram with clusters comprising at least five promoter patterns of high similarity with a branch length of <1% (0.01) between two nodes (Figure 3). The consensus sequence of each cluster was regarded as a motif. The core HRGs yielded nine clusters, three of which showed noninformative low sequence complexity (Clusters 1, 3, and 8), with similar sequences found in three of seven motifs from the low-Pi data set (Clusters 2, 5, and 6), indicating that they do not play a specific role in hypoxia-responsive regulation (Supplemental Figure 3). Motifs from each cluster were resubmitted to STAMP to obtain position-specific scoring matrices (PSSMs), which are a more accurate display of consensus sequences (Supplemental Data Set 2A). The PSSMs were used to search for similarities to published cis-motifs in the PLACE, Agris, and Athamap databases (Higo et al., 1999; Davuluri et al., 2003; Steffens et al., 2004) (Supplemental Data Set 2B). Validating this method, we found that Cluster 7 from the low-Pi-response motif set corresponded to the PHR1 binding site (P1BS) (Supplemental Figure 3).
Figure 3.
Shared and Conserved Motifs Uncovered by Comparative Phylogenetic Footprinting.
Tree shows phylogenetic footprints for each of the 49 core HRGs (Mustroph et al., 2009), grouped by similarity into nine distinct clusters (Clusters 1 to 9). Clusters were defined as groups of at least five patterns with a branch length of <0.01 between two nodes, representing one consensus sequence. Sequence logos display consensus sequences, and names of known matching cis-elements are indicated in parentheses. The 33-bp region (red) of the LBD41 promoter clusters with the C9motif. Genes tested in Figure 2 are marked in green.
Among HRGs, the Cluster 2 motif (C2motif) corresponded to the Ibox found upstream of light-regulated genes (Giuliano et al., 1988) and Cluster 7 (C7motif) strongly resembled the abscisic acid-responsive element (i.e., Guiltinan et al., 1990; Stålberg et al., 1996; Choi et al., 2000), pointing to possible involvement of bZIP TFs in hypoxia-dependent gene regulation (Figure 3). Indeed, it was previously shown that At ADH1 can also be induced by ABA through G-BOX BINDING FACTOR3 (Jarillo et al., 1993; de Bruxelles et al., 1996; Lu et al., 1996). A GCC-core, which had the potential to act as an ERF binding site (Ohme-Takagi and Shinshi, 1995), was present in the C4motif. Neither the more ambiguous C5motif [CCANCGNC(G/A)GG] nor the C6motif [(GGGC) flanked by loosely conserved sequences] matched any known cis-element. The C9motif displayed a partially cryptic sequence [AAAACCA(G/C)(G/C)(G/C)GC] consisting of a strongly conserved AC-rich region identified by PLACE as an MYB TF binding site and a GC-rich region consisting of a loosely conserved spacer and a strongly conserved terminal GC pair (Figure 3). This motif was present in either orientation in HRG promoters. Although there was no database match for the C9motif, the bipartite MYB binding site with a neighboring GC-region somewhat resembles that of the ARE of maize Adh1 (Walker et al., 1987).
Cluster-specific consensus sequences were possibly shared by more HRGs than those listed in each cluster. The RSA-tool matrix scan (Medina-Rivera et al., 2015) was used to search for the occurrences of PSSMs from all clusters in the promoters (3 kb upstream of ATG) of all 49 core HRGs (Supplemental Data Set 3). Hypergeometric tests for enrichment were made by comparison with their occurrences in the full genome of A. thaliana (Supplemental Table 2). Among three other motifs (Clusters 3, 6, and 8) that were significantly enriched (P < 0.05), the C9motif scored the lowest P value (P = 7.65*10−06), indicating the highest enrichment. The C9motif was found one to three times in the 3-kb upstream promoter of 22 HRGs with a significance value of >4.5 and additionally in 17 HRGs with a lower significance value. At least three occurrences were detected in LBD41, HRA1, and the uncharacterized HRG At4g10270 (Supplemental Data Set 3). The C9motif was not identified within the 3-kb upstream region of 10 core HRGs, among them HYPOXIA-RESPONSIVE UNKNOWN PROTEIN9 (At5g10040) and putative HALOACID DEHALOGENASE-LIKE HYDROLASE (AT5G44730) (Supplemental Data Set 3).
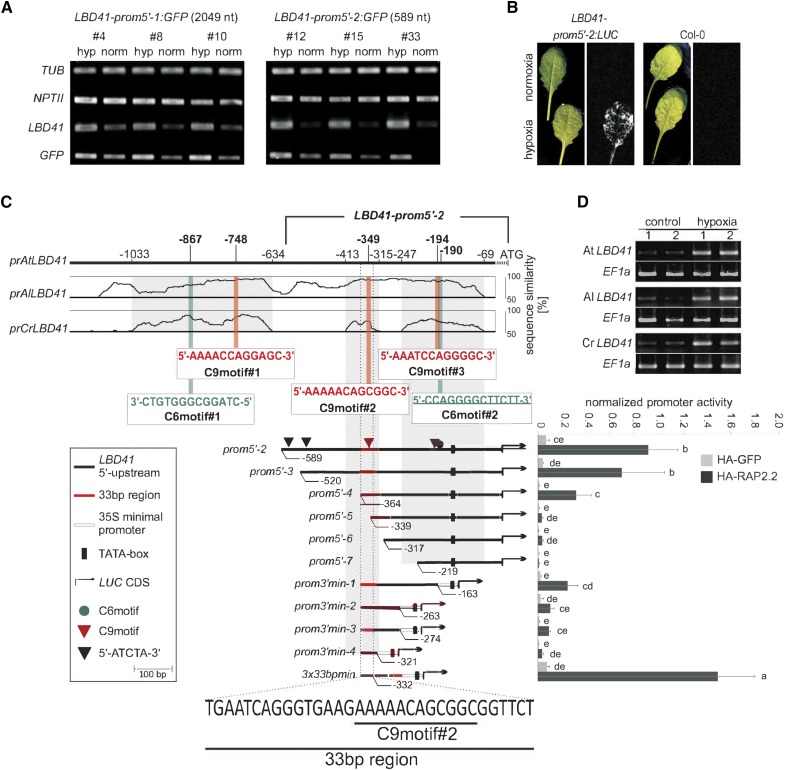
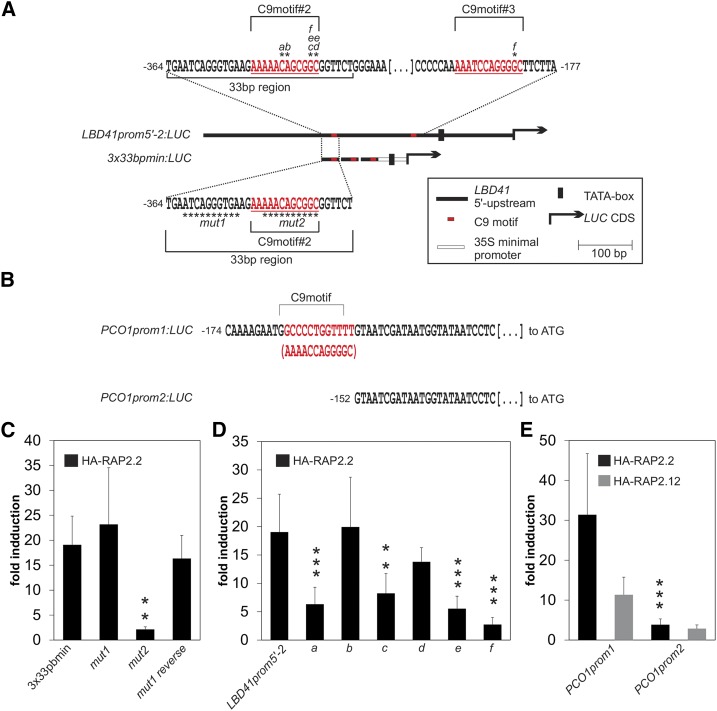
Further support of functional motifs was sought by scanning the upstream region of hypoxia-responsive LBD41 for areas of >50% sequence similarity in putative homologs of Arabidopsis lyrata and Capsella rubella, close relatives of A. thaliana (Figure 4). The A. lyrata and C. rubella LBD41 homolog transcripts were highly upregulated by short-term hypoxia (2 h) of rosette leaves, as clearly evident by standard RT-PCR (Figure 4D). The At LBD41 C9motifs are at positions −748, −349, and −194 and C6motifs at positions −867 and −190, relative to the start codon. The downstream C6 and C9motifs overlap (Figure 4C). Importantly, all C6 and C9motifs in the At LBD41 promoter appear to be conserved in the homologous promoters of A. lyrata and C. rubella (Figure 4C).
Figure 4.
Conserved Hypoxic-Response Promoter Element Narrowed Down to a 33-bp Region of the LBD41 Promoter.
(A) Stable Arabidopsis promoter:GFP transgenics containing two fragments of the LBD41 promoter (LBD41prom5′-1:GFP and LBD41prom5′-2:GFP). Plants were treated with hypoxia for 2 h, and expression of native LBD41 and GFP is shown in comparison to TUBULIN and NPTII.
(B) Stable Arabidopsis promoter:LUC transgenics containing the 589-bp LBD41 promoter region (LBD41prom5′-2:LUC). Bioluminescence in d-luciferin-treated reporter lines after 9 h of hypoxic treatment.
(C) Upper part: Percentage identity plot shows pairwise alignments of upstream sequences from At LBD41 with putative homologous genes of A. lyrata (Al LBD41) and C. rubella (Cr LBD41). Sequence similarities of >50% in all species are in gray regions. Numbers indicate distances from the ATG start codon of LBD41 in base pairs. Positions and sequences of C9motifs and C6motifs are indicated and numbered. Lower part: Deletion series of the 589-bp LBD41 promoter. Map indicates relative length and regions of 5′- and 3′-deleted promoter versions cloned 5′ to a LUC coding sequence. Graph shows basal promoter activities measured in presence of HA-GFP and activities induced by HA-RAP2.2. Data are means ± sd of at least six replicates. Different letters indicate values that vary significantly at P < 0.05 (Tukey HSD test). Normalized promoter activity is LUC activity normalized to p35S:RUC activity.
(D) Standard RT-PCR analysis of homologous LBD41 and EF1a under normoxia (control) and 2 h hypoxia in A. thaliana, A. lyrata, and C. rubella. Panel shows representative results from three biological replicates, each with technical duplicates.
Molecular Promoter Dissection Confirms Function of the C9motif through ERF-VIIs
To test if the C6, C9, or other motifs participate in ERF-VII regulation of transcription under hypoxia, the 5′ upstream of the LBD41 start codon was analyzed in a stable GFP reporter system using two constructs, LBD41prom5′-1:GFP and LBD41prom5′-2:GFP, with 2049 and 589 bp, respectively (Figure 4A). The incubation of two independent homozygous lines for each promoter under low-oxygen conditions resulted in a strong increase in endogenous LBD41 and GFP mRNA levels relative to ambient air conditions (Figure 4A). By contrast, there was no change in transcript levels of the reference gene TUBULIN or the NPTII selection marker linked to the GFP reporter (Figure 4A). To reduce the complexity of our analysis, we focused on the shorter promoter (LBD41prom5′-2), which was also sufficient to promote firefly luciferase (LUC) activity under hypoxia in transgenics expressing an LBD41prom5′-2:LUC reporter construct. Rosette leaf luminescence was evident in these transgenics following 9 h of hypoxia but was absent when grown in ambient air and in the Col-0 control (Figure 4B). These results indicate that a 589-bp region of LBD41 is sufficient for hypoxia-responsive transcriptional activation.
This promoter region was dissected by creating constructs with five deletions from the 5′-end (LBD41prom5′-3 to 7) and four deletions from the 3′-end (LBD41prom3′min-1 to 4) fused to LUC (Figure 4C). To determine if these constructs were transactivated by RAP2.2, they were cotransfected into mesophyll protoplasts with N-terminally HA-tagged (stabilized) RAP2.2 (HA-RAP2.2) driven by the 35S promoter. Promoter activity was quantified by monitoring LUC activity relative to cotransfected Renilla Luciferase gene (RUC) (Wehner et al., 2011). HA-RAP2.2 strongly enhanced the basal activity of LBD41prom5′-2 (Figure 4C). This 589-bp region contains two 5′-ATCTA-3′ sequences (Figure 4C) as well as predicted motifs of Clusters 1, 2, 4, 6, 8, and 9. For clarity, only the overrepresented C6motifs (#1 and #2) and C9motifs (#1 and #2) are highlighted in Figure 4C. Removal of one of two 5′-ATCTA-3′ motifs (LBD41prom5′-3) did not significantly reduce promoter activity. A larger deletion (LBD41prom5′-4) that removed the second 5′-ATCTA-3′ motif and other sequences significantly decreased LUC expression but did not abolish RAP2.2-mediated transactivation. However, targeted deletion of C9motif#2 in LBD41prom5′-5 reduced promoter activity to basal levels (Figure 4C), highlighting the importance of this element.
The 3′ sequences of the 589-bp LBD41 promoter were tested with four deletion constructs. To compensate for removal of the putative TATA-box, the 52-bp minimal prom35S was inserted between the 3′ deletions and the LUC coding sequence. A 3′-deleted LBD41 promoter, containing the single C6 and two C9motifs, fused to an artificial TATA-box (LBD41prom3′min-1), reached a similar induction level to that of LBD41prom5′-4 (Figure 4C). Further 3′ deletions that removed C6motif#2 and C9motif#3 incrementally eroded promoter activity. Together, the 5′ and 3′ deletion series emphasized that the region containing C9motifs#2 and 3 and C6 motif#2 provide sequences sufficient for HA-RAP2.2-dependent transcriptional activation.
The upstream C9motif#2 was necessary for induction. A promoter with only this sequence (LBD41prom3′min-4) showed limited transactivation by HA-RAP2.2, but a triplicated 33-bp sequence (−364 to −332 bp of LBD41) containing three copies of C9motif#2 fused to the minimal promoter exceeded the transactivation of the LBD41prom5′-2 (Figure 4C). Thus, the 3x33bpmin promoter provides sufficient cis-regulatory sequences for HA-RAP2.2-dependent transcriptional activation in protoplasts. This promoter also generated LUC-mediated luminescence in rosette leaves of stable transgenics following 9 h of hypoxia but not under normoxia (Figure 5A). Altogether, these results demonstrate that the evolutionarily conserved C9motif-containing 33-bp region of the LBD41 promoter in Brassicaceae is sufficient for hypoxia-dependent gene activation mediated by RAP2.2.
Figure 5.
A LBD41 Promoter 33-bp Region Is Responsive to Hypoxia and ERF-VII Activation.
(A) Stable A. thaliana lines bearing a triplicated 33-bp region of the LBD41 promoter fused to a 35S minimal promoter and a LUC coding sequence (3x33bpmin:LUC). Hypoxia-dependent bioluminescence upon d-luciferin application is seen only in leaves of 3x33bpmin-containing reporter lines after 9 h of hypoxia, whereas no signal is observed in leaves of Col-0.
(B) Comparison of LUC activity from the 589-bp-long LBD41prom5′-2:LUC with the 3x33bpmin:LUC reporter construct in the presence of transiently expressed HA-ERF-VIIs in mesophyll protoplasts from Col-0. Data are means ± sd of six replicates; asterisks indicate significant differences from basal promoter activity at ***P < 0.001 and *P < 0.05 (Tukey HSD test). Fold induction is the ratio between the transcription factor-induced and basal promoter activity using p35S:HA-GFP as the control. Promoter activity values (LUC activity normalized to p35S:RUC activity) are shown in Supplemental Figure 4.
ERF-VIIs Redundantly Transactivate the LBD41 and 3x33bpmin Promoters
Transient transformation of protoplasts was performed to test if the other four ERF-VIIs transactivate the LBD41prom5′-2:LUC and 3x33bpmin:LUC constructs. To eliminate oxygen-regulated N-end rule-mediated ERF-VII turnover, N-terminally HA-tagged versions were used (Figure 5B; Supplemental Figure 4). The three RAPs effected 5- to 20-fold transactivation of both promoters. This indicates that the 3x33bp minimal promoter is sufficient to explain ERF-VII-regulated LBD41 promoter activation. Interestingly, transactivation by HRE1 and HRE2, the ERF-VIIs encoded by HRGs, was more limited. Whereas HA-HRE1 increased LUC activity 5.7-fold above basal levels, HA-HRE2 did not transactivate either LBD41 construct (Figure 5B). This raises the possibility that HRE1 and HRE2 have functions distinct from the transactivation activity of the RAP ERF-VIIs.
To further evaluate the 33-bp region, substitution mutagenesis was performed on the 12-nucleotide C9motif#2 and flanking sequences. Substitutions within the C9motif#2 (3x33bpmin_mut2) but not the 5′ flanking region (3x33bpmin_mut1) strongly reduced transactivation by HA-RAP2.2 (Figure 6). The reverse complement of 3x33bpmin_mut1 was still transactivated, demonstrating that the C9motif can work in either direction, consistent with the discovery of C9motifs in the reverse orientation in HRGs (Supplemental Data Set 3). To rule out that the triplication of the 33-bp region functioned effectively due to altered spacing or creation of a transcription factor binding site, we tested if C9motif#2 was necessary for HA-RAP2.2 transactivation of the longer LBD41prom5′-2 using four single-nucleotide substitutions and two two-nucleotide substitutions of strongly conserved positions in C9motif#2 at position −349 (Figure 6A). Except for an A-to-T mutation within the AC-rich subregion (LBD41prom5′-2_b), which had no effect, all five other single substitutions significantly lowered transactivation relative to LBD41prom5′-2, to a minimum of 33% activity (Figure 6D). This residual activity is likely due to C9motif#3 because a further decline in promoter transactivation was achieved by adding a disruptive substitution in the C9motif#3 at −194 (LBD41prom5′-2_f). A more distal C9motif at −748 (C9motif#1, Figure 3C) was not tested. These results demonstrate that C9motif#2 and C9motif#3 of LBD41 are essential for transcriptional activation by RAP2.2. Due to our informatics survey and experimental results, we renamed the C9motif the HRPE.
Figure 6.
The C9motif (HRPE) Functions in ERF-VII-Targeted Transactivation.
(A) and (B) Maps of LBD41 and PCO1 promoter fragments cloned 5′ to the LUC coding region with C9motifs and base substitutions or deletions indicated.
(C) Comparison of HA-RAP2.2-induced LUC activity of wild-type and mutated versions of 3x33bpmin:LUC.
(D) Comparison of HA-RAP2.2-induced LUC activity of the 589-bp LBD41prom5′-2:LUC reporter and point mutations in two C9motifs.
(E) Effect of HA-RAP2.2 and HA-RAP2.12 on C9motif-containing and C9motif-less PCO1 promoter versions. All measurements were performed in a transient protoplast transactivation system using p35S:RUC for normalization. Data are means ± sd of six replicates; asterisks indicate significant differences from wild-type-induced promoter activity at ***P < 0.001 and **P < 0.01 (Tukey HSD test). Fold induction, ratio between the transcription factor-induced and basal promoter activity using p35S:HA-GFP. Promoter activity values (LUC activity normalized to p35S:RUC activity) are shown in Supplemental Figure 4.
To demonstrate that the HRPE comprises a cis-element regulated by ERF-VIIs that functions independently of the sequence context of the LBD41 or a minimal prom35S, we analyzed a putative HRPE at −164 bp upstream of the start codon in the promoter of PCO1, a core HRG (Mustroph et al., 2009; Weits et al., 2014). This sequence (5′-GCCCCTGGTTTT-3′) was identified as At5g15120A in Cluster 9 (Figure 3). It is identical to the HRPE consensus sequence, has the highest significance value of all HRPEs detected via the RSA-tool matrix scan (Supplemental Data Set 3), and is a reverse complement of C9motif#2 of LBD41 (Figure 6B; Supplemental Data Set 3). We compared the responses of two PCO1 promoter versions to transactivation by HA-RAP2.2 and HA-RAP2.12 (Figure 6E). The version that contained the HRPE (PCO1prom1) responded to both ERF-VIIs, with transactivation values comparable to those of LBD41prom5′-2 (Figure 6E). However, a 22-nucleotide deletion of the PCO1 HRPE (PCO1prom2) reduced promoter activity (Figure 6E) to that of the most disruptive nucleotide substitutions of LBD41prom5′-2 (Figure 6D, substitution f). This analysis supports the conclusion that the evolutionarily conserved HRPE is an overrepresented sequence in the core HRGs that is transactivated by the ERF-VIIs RAP2.2 and RAP2.12 in protoplasts and in planta.
ERF-VIIs Directly Interact with HRPE-Containing 33-bp Sequence
The rapid transcriptional activation of HRGs by hypoxia could be mediated by N-end rule impairment and ERF-VII stabilization and/or increased nuclear localization along with continued synthesis of these proteins (Mustroph et al., 2009; Gibbs et al., 2011, 2014; Licausi et al., 2011a; Kosmacz et al., 2015). Genes that contain RAP2.2/RAP2.12 binding sites in their promoters should therefore be constitutively transcribed in N-end rule mutants, such as prt6 or ate1 ate2. Such primary targets of rapidly stabilized ERF-VIIs might have their transcript levels elevated prior to the effects of newly synthesized negative regulators (i.e., HRA1; Giuntoli et al., 2014). Indeed, HRPE-containing genes were significantly enriched among early HRGs and prt6 and ate1 ate2 constitutively upregulated genes, compared with all A. thaliana genes (Supplemental Table 2). In addition, all five HRGs tested for activation by (MA)RAP2.2-HBD possessed one or more HRPE and are early-response genes (activated after 30 min of hypoxia; van Dongen et al., 2009) (Figure 2B; Supplemental Data Set 3), supporting the possibility that ERF-VII accumulation directly targets a network of HRGs through direct binding to this cis-element.
To test this hypothesis, p35S:(MA)RAP2.2-HBD plasmid was transiently expressed in mesophyll protoplasts of the 3x33bp:LUC-1 transgenic (Figure 7). After Dex treatment to promote nuclear localization of RAP2.2-HBD, LUC transcript abundance rose significantly. Dex treatment after Chx application resulted in similar transcript induction, indicating that protein synthesis was not required for 3x33bp:LUC activation by (MA)RAP2.2-HBD (Figure 7A). Thus, nuclear localization of RAP2.2 could lead to direct binding of RAP2.2 to the HRPE within the 33-bp region and transcriptional activation.
Figure 7.
Constitutive ERF-VIIs Directly Bind to the 33-bp Region of the LBD41 Promoter.
(A) RT-qPCR analysis of LUC in a stable 3x33bpmin:LUC transgenic line in mesophyll protoplasts transiently expressing either HA-GFP or (MA)RAP2.2-HBD. Chx or DMSO was applied 30 min before Dex or ethanol, and protoplasts were incubated for an additional 4 h under long-day conditions. Transcript levels were normalized to EF1a. Data are means ± sd from three biological replicates (each with three technical replicates). Different letters indicate values that vary significantly at P < 0.05 (Tukey HSD test).
(B) Y1H assay with stable 3x33bp:HIS 3x33bp:LacZ yeast strains transiently expressing AD-ERF-VII fusions. Water and an AD-only expressing vector were used as negative controls. Panels show equally concentrated yeast transformants and 1:10- and 1:100-fold dilutions after 5 d of growth on selective medium in the absence and presence of 40 mM of the HIS gene inhibitor 3-AT. Blue circles indicate β-galactosidase (LacZ) activity as double positive control.
(C) ChIP of RAP2.2- and RAP2.12-associated promoter regions. FLAG-tagged overexpression lines were used (Figure 1), and HRPE-containing promoter regions of LBD41 and PCO1 as well as the 5′-ATCTA-3′-containing region of PSY were tested by subsequent RT-qPCR. Fold enrichment was calculated in comparison to Col-0 wild type. Data are means ± sd from three biological replicates, each with technical duplicates.
The heterologous yeast one-hybrid (Y1H) system was used to provide evidence of direct ERF VII-HRPE interaction. A strain with a stably integrated 3x33bp:HIS reporter was transfected with the activation domain (AD) alone or with AD-ERF-VII baits for all five A. thaliana ERF-VIIs. In the absence of the HIS3 inhibitor 3-amino-1,2,4-triazole (3-AT), cells grew on His-lacking medium due to leaky reporter gene expression. A Trp marker located on the bait vector allowed for growth of transfected cells in the absence of Trp (Figure 7B). The overexpression of the AD alone was not sufficient to restore strong growth through enhanced HIS3 expression. In contrast to the promotion of growth by AD-RAP2.2 and AD-RAP2.12 as well as activity of a LacZ reporter regulated by the 3x33bp concatemer, AD-HRE1 and AD-HRE2 had no visible influence on growth or reporter gene expression (Figure 7B). The same was seen for the AD-RAP2.3 bait, but growth of cells with this construct was reduced under 3-AT-free conditions, despite effective transfection, making it impossible to assess binding between this ERF-VII and 3x33bp-containing promoter (Figure 7B). As a negative control, we confirmed that none of the ERF-VIIs transactivated the Caenorhabditis elegans Snail-type TF 1 cis-element, which bears no resemblance to the LBD41 33-bp region (Supplemental Figure 5).
RAP2.2 and RAP2.12 Bind to LBD41 and PCO1 Promoters in HRPE Regions
To obtain evidence in planta that ERF-VIIs bind promoter regions with HRPEs, 7-d-old seedlings of air-grown transgenics stably expressing N-terminally FLAG-tagged RAP2.2 or RAP2.12 (Figure 1) were used for ChIP followed by evaluation of the LBD41, PCO1, and PSY promoters (Figure 7C). Wild-type Col-0 was used as a negative control. ChIP with p35S:FLAG-RAP2.2 and p35S:FLAG-RAP2.12 yielded a FLAG-tagged protein that migrated at ∼70 kD, although the calculated molecular masses are 42.5 and 39.8 kD, respectively. If these proteins function as homo- or heterodimers, the larger mass could correspond to stable dimers formed by cross-linking. The copurified genomic DNA that had been sheared to 200 to 400 bp was analyzed by qPCR with primers that flank regions of LBD41 or PCO1 promoters that contain an HRPE (Figure 7C). Enrichment of the PCR product was observed for both genes in FLAG-RAP2.2 and FLAG-RAP2.12 but not in control (Col-0) ChIP samples. The promoter region of PSY lacks an HRPE but contains 5′-ATCTA-3′ elements, previously reported to bind in vitro to RAP2.2 (Welsch et al., 2007). This promoter showed no evidence of binding of either ERF-VII relative to the control. These results support the conclusion that RAP2.2 and RAP2.12 can bind the LBD41 and PCO1 promoters in HRPE-containing regions in planta.
The HRPE Shows Similarity to the ARE in Sequence and Function
Both the HRPE and the previously described Zm Adh1 ARE (Walker et al., 1987) have GC- and GT-rich components. The ARE of At ADH1 also has a GC- and a GT-rich region, but this region did not confer specific hypoxia responsiveness in a previous analysis (Dolferus et al., 1994). To investigate the At ADH1 ARE further, we tested its activation by RAP2.2 and RAP2.2 in the Y1H assay and in transfected protoplasts. Three copies of the 32-nucleotide ARE from At ADH1 (Dolferus et al., 1994; Supplemental Figures 6C and 7) were fused in tandem to a minimal prom35S and a LUC reporter (3xAREmin:LUC). As seen for 3x33bp:HIS, yeast containing a 3xARE:HIS3 reporter grew on 3-AT-containing plates in the absence of His when genetically combined with AD-RAP2.2 or AD-RAP2.12 fusions but not AD baits with the other three ERF-VIIs (Figure 8A). Consistent with this, RAP2.2 transactivated the 3xAREmin:LUC in protoplasts as well (Figure 8B). The level of induction of the At ARE construct was 5-fold compared with 41-fold for the 3x33bpmin:LUC due to high basal activity. The RSA-tool matrix scan predicts a C9motif/HRPE in At ADH1 that partially overlaps with the previously defined ARE (Supplemental Figure 6). By contrast, scans of the Zm Adh1 promoter reveal that the characterized ARE does not correspond to a predicted C9motif/HRPE; however, one of these is present in an adjacent 5′ region identified by Ferl and Nick (1987) as a factor binding region (Supplemental Figure 7).
Figure 8.
Functional Similarity between the Arabidopsis HRPE and Arabidopsis ADH1 ARE.
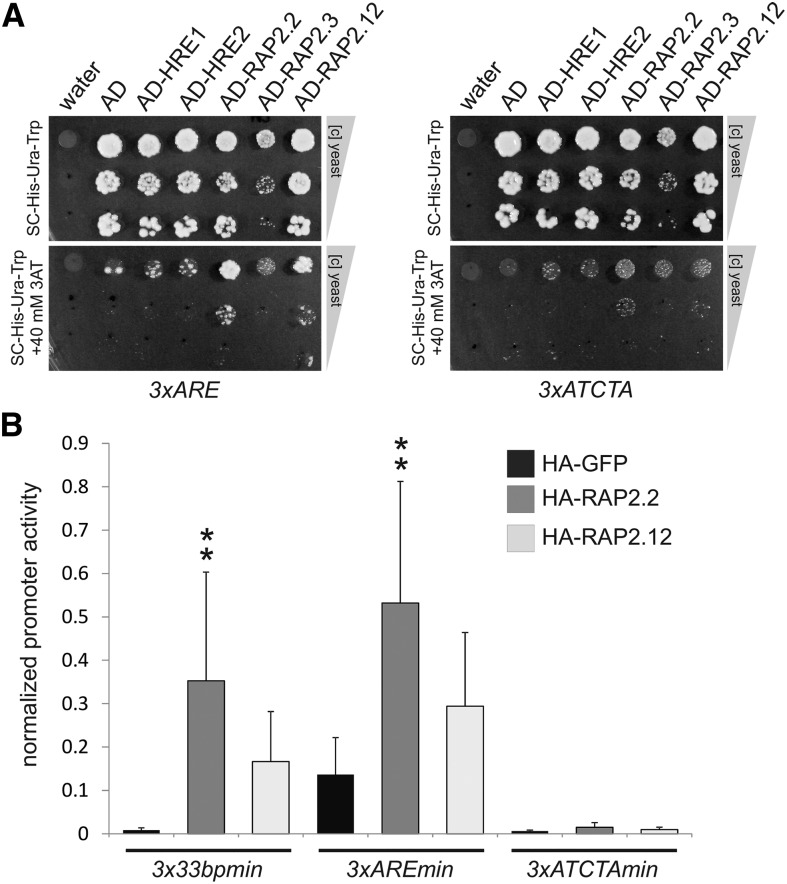
(A) Y1H assay with stable 3xARE:HIS and 3xATCTA:HIS yeast strains transiently expressing AD-ERF-VII fusions. Water and an AD-only expressing vector were used as negative controls. Panels show equally concentrated yeast transformants and 1:10- and 1:100-fold dilutions after 5 d of growth on selective medium in the absence and presence of 40 mM of the HIS gene inhibitor 3-AT.
(B) Effect of HA-RAP2.2 and HA-RAP2.12 on 3x33bpmin:LUC in comparison to 3xAREmin:LUC and 3xATCTAmin:LUC in transiently transformed mesophyll protoplasts from A. thaliana Col-0, cotransfected with p35S:RUC. Data are means ± sd of six replicates; asterisks indicate significant differences from wild-type-induced promoter activity at **P < 0.01 (Tukey HSD test). Normalized promoter activity is LUC activity normalized to p35S:RUC activity.
The previously identified cis-element with a 5′-ATCTA-3′ core (Welsch et al., 2007) has no obvious sequence similarity to the Zm Adh1 or At ADH1 ARE or the HRPE. However, an interaction between RAP2.2 and 5′-ATCTA-3′ was established by gel retardation assay (Welsch et al., 2007). To test if RAP2.2 can bind to multiple targets, we generated a triplicated 5′-ATCTA-3′ promoter from a construct that was used in the reported experiment (5′-CAATCTAAATATCTAAAATATAAA-3′) (Welsch et al., 2007). Although fused to a minimal promoter, the 3xATCTAmin:LUC reporter was not transactivated by HA-RAP2.2 or HA-RAP2.12 in protoplasts (Figure 8A). This negative result was supported by the observation with a HIS3 reporter in the Y1H system (Figure 8B) and by ChIP analysis (Figure 7C).
DISCUSSION
Functional Redundancy of ERF-VIIs
The A. thaliana ERF-VII transcription factor family has been investigated in terms of their roles in gene regulation, low-oxygen sensing, and low-oxygen survival (Hinz et al., 2010; Licausi et al., 2010, 2011a; Gibbs et al., 2011; Weits et al., 2014; Riber et al., 2015), as well as their roles in germination and skotomorphogenesis (development in the dark) (Gibbs et al., 2014; Abbas et al., 2015). Despite this, the quantitative impact of each member and hierarchical mechanisms underlying their redundancy are not satisfyingly understood. We show here that RAP2.2 and RAP2.12 act equally and redundantly as predominant activators of the majority of HRGs (Figures 1C, 2A, and 5B). In comparison to the hre1 hre2 double mutant, which shows impaired maintenance of upregulation of HRGs after 4 h of severe oxygen deprivation (Licausi et al., 2010), the double knockout rap2.2-5 rap2.12-2 genotype shows minimal upregulation of HRG transcripts after as early as 2 h of hypoxia (Figure 2A). This is consistent with the limited early (1.5 h stress) HRG activation in artificial microRNA (amiRNA) lines with reduced RAP2.2 and RAP2.12 transcript levels and implies that these act as main inducers of the hypoxic response (Licausi et al., 2011a), as recently confirmed by an independent rap2.2-4 rap2.12-3 double knockout analysis (Bui et al., 2015). These findings are in accordance with constitutive expression of both genes (Figure 1A; Mustroph et al., 2009; Licausi et al., 2010) and the sequestration of RAP2.12 at the plasma membrane under normoxia, which allows for relocalization to the nucleus as oxygen levels drop below 10% (Licausi et al., 2011a; Kosmacz et al., 2015). Moreover, nuclear accumulation of RAP2.2 is sufficient to activate HRGs under normoxic conditions even when de novo protein synthesis is inhibited (Figure 2B).
The predominance of RAP2.2 and RAP2.12 in HRG activation is reflected by their strong trans-activity on marker and reporter genes (Figures 1C, 5, and 7). It is assumed that homologous binding domains of TFs bind similar sites with similar affinities (Bernard et al., 2012). Therefore, the HRPE-containing 33-bp region, which is directly bound by RAP2.2 and RAP2.12 in the Y1H system and is sufficient for their transactivation, provides an explanation for their redundancy (Figures 5 and 7). We find that the double null rap2.2-5 rap2.12-2 fails to elevate HRG (ADH1, PDC1, PCO1, PCO2, and LBD41) expression under hypoxia. However, our attempt to link the loss of RAP2.2 and RAP2.12 to survival of low-oxygen stress failed due to the greater susceptibility of the Ler-0 accession to submergence than Col-0, as reported by Vashisht et al. (2011), because the rap2.2-5 rap2.12-2 double mutant was in a nonuniform genetic background (Supplemental Figures 1 and 2). Attempts by others using mutants and amiRNA lines to decipher the necessity of RAP2.2 and RAP2.12 for low-oxygen/submergence survival were stymied by residual expression of RAP2.2 and RAP2.12 (Licausi et al., 2011a). Thus, it remains unclear if both are required for acclimation to transient hypoxia or submergence due to factors such as cell-specific or regional variation in production (Mustroph et al., 2009). Nonetheless, the collective studies suggest that reduced HRG transcription, as seen in the rap2.2 rap2.12 double mutant and amiRNA silencing lines, results in a handicap under low-oxygen stress (Licausi et al., 2011a; Bui et al., 2015).
A third ERF-VII RAP, RAP2.3, was also linked to hypoxia signaling by knockout and overexpression analyses (Papdi et al., 2015). The increase in ADH activity in RAP2.3 overexpression lines and significant induction of reporter constructs through RAP2.3 further hint at its involvement in HRG activation (Figures 1C and 5). Like RAP2.2 and RAP2.12, RAP2.3 is constitutively expressed (Figure 1A; Licausi et al., 2010), a target of the N-end rule pathway (Gibbs et al., 2011, 2014), and an interactor with ACBPs (Li and Chye, 2004; Li et al., 2008), as demonstrated for RAP2.12 (Licausi et al., 2011a). We provide evidence for redundancy of RAP2.3 to RAP2.2 and RAP2.12 that includes shared transactivation capacity of the 3x33bpmin promoter derived from LBD41 (Figure 5B), which is likely a result of direct protein-DNA interaction. Furthermore, the double null homozygote rap2.2-5 rap2.12-2 still shows a slight but not significant hypoxic induction of ADH1, PDC1, and LBD41 (Figure 2A), which could be linked to RAP2.3 activity. Although the impact of RAP2.3 could be minor in comparison to RAP2.2 and RAP2.12 in seedlings exposed to hypoxia, this third factor may be of importance in specific organs, developmental ages, or environmental contexts. Intriguingly, the AP2 DNA binding domain of RAP2.3 binds GIBBERELIN INSENSITIVE (GAI) and RELATED TO GAI (RGA1), an interaction that inhibits RAP2.3 activity (Marín-de la Rosa et al., 2014). Analysis of a triple mutant under hypoxic conditions is required to prove the physiological relevance of this ERF-VII.
The hypoxia-induced ERF-VII genes HRE1 and HRE2 have been previously identified as regulators of the low-oxygen response. Overexpression of HRE1 leads to increased specific activity of the fermentative enzymes ADH and PDC, whereas its knockout has the opposite effect (Licausi et al., 2010; Hess et al., 2011). Notably, the ERF-VIIs HRE1 and HRE2 are also N-end rule targets (Gibbs et al., 2011). Overexpression of stabilized versions of HRE1 and HRE2 enhanced seed germination at low oxygen concentrations and provided seedling resilience under hypoxic conditions (Gibbs et al., 2011). Furthermore, expression of HRGs is reduced after long-term (i.e., >4 h) hypoxic treatment in hre1 hre2 knockouts or HRE1-RNAi transgenics (Licausi et al., 2010; Yang et al., 2011). Despite their similarity to the RAP ERF-VIIs, evidence is mounting that HRE1 and HRE2 may not be effective transactivators of HRGs. The weak-to-absent transactivation of the LBD41 and 3x33bpmin promoter by HRE1 and particularly HRE2 (Figure 5B) indicates that these ERFs are not redundant to the RAPs. Consistently, Bui et al. (2015) reported that HRE1 and HRE2 did not transactivate promoters of three HRGs that were upregulated by the three other ERF-VIIs, and Abbas et al. (2015) found that HRE1 and HRE2 did not promote apical hook formation under hypoxic conditions. The hypoxia-inducible ERF-VIIs may possibly regulate later steps of the hypoxic response as a so-called rearguard, whereas the ERF-VII RAPs could be the foreguard (Voesenek and Bailey-Serres, 2013). Further studies are needed to determine if HRE1 and HRE2 are direct targets or downstream targets within the gene network regulated by RAP2.2 and RAP2.12.
Under hypoxic conditions, Chx impaired full ADH1 upregulation (Hoeren et al., 1998), an observation confirmed for ADH1 as well as PDC1 and PCO2 under RAP2.2 transactivation (Figure 2B). This points to possible involvement of a secondary activator downstream of RAP2.2 that must be synthesized during hypoxia. Since higher transactivation is not seen for the 3x33bpmin:LUC reporter when Chx is absent (Figure 7A), it is unlikely that the proposed secondary activator binds to the same cis-element as RAP2.2 or RAP2.12. LBD41 and PCO1 induction was lower in the absence of Chx, pointing to the involvement of a labile transcriptional repressor. Thus, hypoxia-induced HRGs may have different binding affinities to positive regulators produced under hypoxia and may be differentially regulated by proteins synthesized during hypoxia. Added to this is the observation that all of these markers are negatively regulated by HRA1, which limits RAP2.12 activity (Giuntoli et al., 2014).
Advantages of Comparative Phylogenetic Footprinting
The identification of the HRPE was aided by the use of cross-species comparison of HRG promoters. Three in silico promoter motif prediction methods are frequently used to accelerate the identification of cis-elements (Das and Dai, 2007): (1) search for overrepresented patterns in coregulated promoters of one genome (Rombauts et al., 1999), (2) detection of conserved patterns in homologous promoters of multiple genomes (Tagle et al., 1988), and (3) combinations of (1) and (2) (Gelfand et al., 2000). In previous predictions of hypoxically regulated motifs, only pattern recognition methods with coregulated gene sets of single species were used with string-based algorithms (Liu et al., 2005; Hsu et al., 2011; Licausi et al., 2011b) or probabilistic models (Klok et al., 2002; Mohanty et al., 2005; Licausi et al., 2010, 2011b). None of the predicted motifs showed an exact match with the partially degenerate C9motif/HRPE, which was identified through comparative phylogenetic footprinting and was overrepresented among promoters of the 49 core HRGs (Supplemental Table 2).
A challenge of pattern recognition among unrelated promoters is the increase in misdetection due to the varying lengths of nonconserved promoter sequences (Buhler and Tompa, 2002). A phylogenetic footprinting approach can overcome this problem if evolutionarily distance between species is chosen properly (Cliften et al., 2001). However, when using a single homologous gene group for phylogenetic footprinting, potential cis-elements may not be detected in a stimulus-specific context.
The combination of phylogenetic analyses with classical motif recognitions in coregulated genes can be more efficient than the respective approach alone (Wang and Stormo, 2003, 2005; Sinha et al., 2004; MacIsaac et al., 2006) and has been performed in microorganisms (Gelfand et al., 2000; McGuire et al., 2000), animals (Prakash et al., 2004), and plants (Wang et al., 2009). Some strategies use conservation as a measure for true positive motifs that were identified by classical approaches beforehand (Cliften et al., 2003; Wang and Stormo, 2003). Instead, comparative phylogenetic footprinting, as described here, filters conserved elements by their occurrences in coregulated genes and optionally by overrepresentation (Figure 3). Therefore, it does not depend on limitations of algorithms that use motif overrepresentation in coregulated promoters but offers stimulus-specific motif predictions (Figure 3). Moreover, complex TF cascades could involve cis-elements that are not overrepresented but are crucial that could be overlooked by classical approaches. Indeed, besides the C9motif/HRPE, several underrepresented motifs with high complexity are conserved and shared by HRG promoters (Figure 3). Moreover, our approach was validated by the successful identification of the previously identified and validated low-phosphate-responsive P1BS (Supplemental Figure 3; Bustos et al., 2010). Phylogenetic footprinting could therefore be of value in wider cross-genome comparisons of hypoxically inducible promoters or in other groups of stress-regulated or cell-type specifically expressed genes to determine other broadly conserved motifs.
The HRPE Is an Authentic cis-Response Element
Plant genomes contain well characterized cis-elements that regulate genes under biotic and abiotic stresses (Giuliano et al., 1988; Guiltinan et al., 1990; Yamaguchi-Shinozaki and Shinozaki, 1994; Rushton et al., 1996; Stålberg et al., 1996; Choi et al., 2000). The HRPE exhibits crucial characteristics of such authentic cis-elements. First, it is clearly enriched in promoters of HRGs compared with the whole genome of A. thaliana (Supplemental Table 2), a property widely used to predict true positive cis-elements (Das and Dai, 2007). Second, conservation is typical for authentic cis-elements because selective pressure on functional motifs decelerates their evolution (Wasserman et al., 2000; Moses et al., 2003). The HRPE shows conservation among angiosperms in several HRGs, which correlates with its function (Figures 3 and 4). Third, cis-elements often show significant position preferences, as demonstrated in humans (Cooper et al., 2006), yeast (Harbison et al., 2004), and plants (Maruyama et al., 2004; Tran et al., 2004; Zou et al., 2011). The influence of position is demonstrated for the HRPE by the decrease in promoter activity of serial 3′ deletions (Figure 4C). The copy number appears to be of similar importance, as deduced from lower promoter activities after substitution mutations in either one or both HRPE versions (Figure 6). Moreover, triplication of the HRPE-containing 33-bp region led to stronger promoter activation than a single copy version (Figure 4C). Presence, number and position of cis-elements are crucial parameters to predict stress-induced expression patterns and are therefore part of a cis-regulatory code (Zou et al., 2011). In contrast, the direction of the HRPE was not important since a reverse complement could be transactivated by RAP2.2 (Figure 6C), and it is present and functional as the reverse complement in the PCO1 promoter (Figure 6E). We show that both RAP2.2 and RAP2.12 interact directly with the HRPE-containing 33-bp region (Figure 7), and the HRPE is crucial for 33-bp activity (Figure 6). Moreover, ChIP confirmed that RAP2.2 and RAP2.12 bind directly to promoter regions containing a HRPE of two HRGs (LBD41 and PCO1) (Figure 7C). Taken together, RAP2.2 and RAP2.12 act as essential activators under hypoxia through the HRPE, which is recognizable in the promoters of 39 of 49 HRGs.
RAP2.2 and RAP2.12 Apparently Do Not Act through the 5′-ATCTA-3′ Sequence under Hypoxia
The 5′-ATCTA-3′ sequence was suggested to act as a direct binding site for RAP2.2 and RAP2.12 (Welsch et al., 2007; Licausi et al., 2011a). Although this motif is overrepresented in promoters of HRGs (Licausi et al., 2011b), there was no evidence of evolutionary conservation of this sequence in hypoxia-regulated promoters (Figure 3). A 126-bp deletion from the 5′ part of hypoxia-responsive HB1 promoter regions removes two 5′-ATCTA-3′ motifs and reduces transactivation of RAP2.12 in protoplasts (Licausi et al., 2011a). By contrast, deletions of two nonconserved 5′-ATCTA-3′ sequences from LBDprom5′-2 lowered basal promoter activity in stable transgenics, but not RAP2.2-mediated induction (Figure 4C). Moreover, PCO1prom5′-1, which is strongly transactivated by RAP2.2 and RAP2.12, has no 5′-ATCTA-3′ motifs (Figure 6E). Based on our findings, we conclude that LBD41 and PCO1 are direct targets of RAP2.2 via the HRPE (Figures 2B and 7). Despite the finding that DNA fragments containing 5′-ATCTA-3′ bind heterologously expressed RAP2.2 in electrophoretic mobility shift assays (Welsch et al., 2007), we were unable to confirm this interaction via Y1H and protoplast transactivation assays (Figure 8). Instead, we provide independent demonstration of HRPE and At ARE interaction and/or activation mediated by RAP2.2 and RAP2.12, including transactivation in protoplasts (Figures 5B and 8B), activation in stable transgenics (Figure 5A), activation in a Y1H system (Figures 7B and 8A), and chromatin binding of regions with these elements in vivo by ChIP (Figure 7C). Our observation that the PSY gene with a triplicated 5′-ATCTA-3′ motif in its promoter is neither constitutively activated in N-end rule pathway mutants nor through RAP2.12 overexpression or hypoxia (Mustroph et al., 2009; Gibbs et al., 2011; Licausi et al., 2011a) reduces the likelihood for a hypoxic function of 5′-ATCTA-3′. In addition, lowered RAP2.2 expression does not lead to reduced PSY transcript accumulation (Welsch et al., 2007). Finally, replacement of the Zm ARE by 5′-ATCTA-3′ resulted in a loss of nuclear factor binding and hypoxic response in maize protoplasts (Olive et al., 1991). Nonetheless, we cannot rule out that ERF-VII binding to 5′-ATCTA-3′ is condition-specific or developmentally determined. In fact, an ERF from group IXc was shown to alter its binding site affinity in a stress-specific manner (Cheng et al., 2013), and there is evidence from ChIP analyses that RAP2.3 binds genomic regions with GCC-boxes (Marín-de la Rosa et al., 2014). Future ChIP-seq analyses with the ERF-VIIs under hypoxic and normoxic conditions are required to expand our understanding of the direct gene targets of these factors, the sequences they bind, and temporal and conditional parameters of their DNA and protein-protein interactions.
The HRPE Is Similar but Not Identical to the ARE
Our results indicate both commonalities and distinctions between the Zm Adh1 and At ADH1 AREs and the HRPE. The Zm Adh1 ARE is composed of a GC-rich and a GT-rich region that is duplicated (i.e., ARE_MAIZE_sub1 and _sub2; Supplemental Figures 6C and 7) (Walker et al., 1987), both with similarities to the HRPE (Supplemental Figure 6C). Although the consensus sequence of the two Zm Adh1 ARE sequences was described (Walker et al., 1987; Olive et al., 1990, 1991), a PSSM of the ARE was not constructed, hindering matches with partially degenerated motifs such as the HRPE (Supplemental Data Set 2). However, our data suggest that the ARE sequences characterized in Zm Adh1 do not correspond to a motif that can be recognized as an HRPE by the search algorithms used (Supplemental Data Set 4), although it also possesses GC- and GT-rich regions. Moreover, the putative GC pair important for TF binding in the At LBD41 HRPE (Figure 6D) is not in the exact same position in the Zm Adh1 ARE (Supplemental Figure 6C). Instead, our search predictions discovered a Zm Adh1 HRPE that corresponds to factor binding region C (Ferl and Nick, 1987), which might be studied in the future.
For the At ADH1 ARE, the GT-rich region was characterized as a MYB binding element for which a PSSM exists (Hoeren et al., 1998). In this study, the use of the RSA-tool with this PSSM uncovered an overlap of this GT-rich sequence with the HRPE for 15 of the 49 core genes (Supplemental Data Set 4). For example, the LBD41 C9motif#1 overlaps with a GT-rich region. Notably, the At ADH1 ARE GT-rich region and the GC-rich region are switched in comparison to the Zm Adh1 ARE, making the situation difficult to resolve. Interestingly, an HRPE and a GT-rich region also overlap in the Zm Adh1 and Zm LBD41 promoters (Supplemental Data Set 4). In the At ADH1 promoter, the HRPE overlaps with the GC-rich region of the ARE but not with a GT-rich region (Supplemental Figure 7). These observations point to putative differences in the substructure of these motifs.
The HRPE and ARE show similarities within the promoter and appear to function as activating elements. Both function in forward and reverse complement orientation, and their activity multiplies when present in multiple copies (Olive et al., 1990; Figures 4 and 6). An evolutionarily conservation was evident for the HRPE (Figure 3) and is also assumed to be a characteristic of the ARE, since it is found in putatively homologous promoters in A. thaliana and maize (Walker et al., 1987; Dolferus et al., 1994). Strikingly, both elements are sufficient for reporter gene induction under hypoxic conditions (Walker et al., 1987; Olive et al., 1990; Figure 5), and both are bound and activated by RAP2.2 and RAP2.12 (Figures 5, 7, and 8). Activation of the 3xARE from At ADH1 by RAP2.2 was weaker than activation of the 3x33bp (HRPE), after considering the higher basal expression of the 3xARE (Figure 8). The binding affinity of RAP2.2 may be stronger for the HRPE than for the ARE. An alternative explanation is that the At ADH1 HRPE and ARE partially overlap (Supplemental Figure 7); therefore, activation of the ARE could be due to the presence of the partial HRPE.
Despite the similarities between the HRPE and Zm ARE in sequence and function, both possess specific features. The interdependence between two adjacent Zm ARE subregions (Walker et al., 1987) is not present in the similar motif of At ADH1. Two HRPE copies in LBD41prom5′-2 both contribute to RAP2.2 induction (Figure 4), but their relative distance of 141 bp exceeds the tolerable limit for spacing between sub-region I and II of the Zm ARE (Olive et al., 1990). In addition, the A. thaliana ADH1 promoter is hypoxia-responsive when one of two ARE subregion-like sequences is deleted (Dolferus et al., 1994) and PCO1prom5′-1 responded to RAP2.2 and RAP2.12 with its single HRPE (Figure 6), whereas for the Zm ARE, both subregions were important for full activation (Walker et al., 1987). Further comparative sequence analyses and biochemical validation using the native ADH1 promoters are required to better determine if ARE and HRPE are similar or distinct in different species.
The GC-rich and GT-rich motif in the Zm ARE are distinct binding sites for the yet unidentified GCBP-1 and ARF-B(2), respectively (Ferl, 1990; Olive et al., 1991), raising the possibility that the HRPE is bound by other TFs besides ERF-VIIs. Based on single nucleotide substitutions, it is not possible to locate the exact binding site for RAP2.2 and RAP2.12 (Figure 6). The A. thaliana version of the ARE sequence is bound by MYB2 at the GT-rich motif (Hoeren et al., 1998), which displays a bona fide MYB binding site with an AAC core (Lüscher and Eisenman, 1990; Urao et al., 1993). However, nucleotide substitutions at this position lead to a general loss of promoter function, not only specific hypoxic induction (Dolferus et al., 1994; Hoeren et al., 1998). In line with this, MYB2 is not essential for At ADH1 upregulation under hypoxia (Licausi et al., 2010). We therefore predict that RAP2.2 and RAP2.12 likely bind to the GC-rich motif rather than to the GT-rich motif. GC-rich cis-elements can be bound by ERFs (Jaglo-Ottosen et al., 1998; Menke et al., 1999; Xue, 2003), even if these GC-rich elements differ from the classical ERF binding site 5′-TAAGAGCCGCC-3′ (Ohme-Takagi and Shinshi, 1995). A CG core is universal for ERF-DNA interaction, whereas flanking regions serve as specific recognition sites (Yang et al., 2009). Accordingly, a strongly conserved terminal GC pair in the HRPE is crucial for promoter activation in response to RAP2.2 (Figure 6D). For RAP2.3, binding to GC-containing promoter elements was also demonstrated (Franco-Zorrilla et al., 2014; Marín-de la Rosa et al., 2014). An equivalent region in the Zm ARE is important for binding to the proposed GCBP-1 (Olive et al., 1991). It is tempting to speculate that GCBP-1 is one or several of the 14 putative maize ERF-VIIs. If homologies between both binding factors can be demonstrated, this would also strongly hint at a phylogenetic relationship between the HRPE and the ARE.
In conclusion, we identified an evolutionarily conserved promoter element that is necessary and sufficient for hypoxic induction of genes through the ERF-VIIs RAP2.2 and RAP2.12 in A. thaliana. The HRPE shares similarities with the previously identified ARE from maize and A. thaliana but is not identical. These findings identify 39 of the 49 core HRGs as the putative direct targets of the ERF-VIIs that are stabilized in oxygen-deficient cells. A future challenge is to determine how HRGs that lack a detectable HRPE, for example, HYPOXIA-RESPONSIVE UNKNOWN PROTEIN9 (At5g10040) and putative HALOACID DEHALOGENASE-LIKE HYDROLASE (AT5G44730), are regulated in an N-end rule-dependent manner. One possibility is that these transcripts are indirectly targeted by ERF-VIIs. Furthermore, recent observations of transient TF action, the “hit-and-run” transcription model (Para et al., 2014; Varala et al., 2015), indicate that the picture on transcriptional regulation might be even more complex, with group VII ERF RAPs being early binders and other TFs being involved in later steps of transcriptional regulation under oxygen deficiency. The current systematic analysis provides a clear demonstration of a transcription factor–cis-regulatory sequence module that coordinates the response that provides protection from transient low oxygen and submergence stress.
METHODS
Organisms and Growth Conditions
For all reporter-based experiments, Arabidopsis thaliana ecotype Col-0 was used. The rap2.12-2 mutant allele (Sail_1215_H10; Col-0) containing a T-DNA insert in the second exon of AT1G53910 (Sessions et al., 2002) was obtained from the Nottingham Arabidopsis Stock Centre (NASC). For RAP2.2, previously reported alleles still produced gene transcript; therefore, the rap2.2-5 mutant allele (AY201781/ GT5337, Ler-0) containing a maize Ds transposon insertion in the second exon of AT3G14230 was obtained from the Martienssen lab (Cold Spring Harbor Laboratory; Sundaresan et al., 1995). A. thaliana wild-type Col-0 and Ler-0, Arabidopsis lyrata, and Capsella rubella seeds were obtained from the NASC. Y1H strain Y1H-aS2 was kindly provided by A.J.M. Walhout (Reece-Hoyes et al., 2011).
Except for the protoplast experiments, seeds were surface-sterilized and stratified for 3 d at 4°C in darkness after imbibition on Murashige and Skoog (MS) salt-containing plates (1× MS salts, 0.8% [w/v] agar, and 1% [w/v] Suc, pH 5.7). To obtain seedlings for transcript analyses, stratified seeds were germinated under long-day (LD) conditions (23°C; 16 h/8 h light/dark cycle; 100 µmol photons m–2 s–1, bulb type Osram Lumilux 21-W/cool white 840) for 7 d. For other experiments 7-d-old seedlings were planted into soil (soil:vermiculite; 2:1) and transferred to short-day (SD) conditions (23°C; 8 h/16 h light/dark cycle; 100 µmol photons m–2 s–1, bulb type Osram Lumilux 36-W/cool white 840) and grown for additional 13 d for LUC imaging of intact plants or 14 d for hypoxic treatment of single leaves. For determination of the survival after submergence, 7-d-old seedlings were transplanted to pots containing a soil:vermiculite:sand mixture (2:1:1) and grown for an additional 2 weeks under SD conditions. For protoplast experiments, A. thaliana seeds were directly sown on soil (soil:vermiculite; 2:1) and stratified for 3 d at 4°C and grown for 3 to 4 weeks under SD conditions.
Hypoxia Treatment and Submergence
For hypoxic induction of gene expression, 7-d-old seedlings on vertically positioned open plates, cut 3-week-old rosette leaves with petioles in water-filled tubes, or 20-d-old well-watered soil-grown plants in pots were placed in a positive pressure system, allowing for constant 100% nitrogen flushing under LD conditions in the light (treatments started 2 h after onset of light, 100 µmol photons m–2 s–1, 100% humidity). Control plants were placed under ambient LD conditions in air. For transcript analyses, treatments were for 2 h. For LUC reporter analyses, treatment was for 9 h. Following treatments, plant material was immediately frozen in liquid nitrogen and stored at −80°C or further processed for bioluminescence imaging.
To measure survival under hypoxic conditions, 3-week-old plants grown under SD conditions were submerged in water (equilibrated at room temperature) for 3 d in darkness, with treatment commencing 3 h after onset of the light period. Plants were desubmerged and recovered for 15 d under well-watered SD conditions before evaluation. To evaluate survival, the number of visibly dead and viable meristems after the recovery phase was recorded, along with the number of green leaves (>1 mm length) and shoot fresh weight after 15 d of recovery. Means of three independent biological replicate experiments with five plants each were expressed as percent survival rate relative to the control.
Phylogenetic and Motif Enrichment Analysis of Conserved Motifs in LBD41 and Other Core Hypoxia-Responsive Promoters
For phylogenetic analyses of conserved sequence motifs in the LBD41 promoter, homologous genes were identified within sequenced Brassicacea using the Phytozome database (Goodstein et al., 2012). The 1500 bp upstream of the annotated transcription start site plus the 5′-untranslated region of A. thaliana LBD41, A. lyrata putative-LBD41, and the 1500 bp upstream of the start codon of C. rubella putative LBD41 homolog were obtained. Pairwise alignments were performed with mVISTA (Frazer et al., 2004) using the alignment program Shuffle-LAGAN (Brudno et al., 2003). Results for sequence similarities were confirmed by a multiple sequence alignment with CHAOS and DIALIGN (Brudno et al., 2004).
A list of 49 core HRGs of A. thaliana (Mustroph et al., 2009) and a control data set, i.e., 49 low phosphate-responsive genes of A. thaliana with the highest expression values in the shoot, was assembled from publically available microarray data (Bustos et al., 2010), as described by Klecker et al. (2014). For each of these genes, the PLAZA database (Proost et al., 2009) was used to identify putative homologs in 25 plant species detected as BLAST best hits. The PLAZA workbench was used to obtain upstream intergenic sequences with a maximum length of 1 kb, starting from the predicted transcription start sites. Each set of 49 genes of Arabidopsis and the putative homologous promoter sequences were analyzed by the unsupervised multiple expectation maximization for motif elicitation (Bailey and Elkan, 1994) with the following parameter settings: distribution of motif occurrences, 0 or 1 per sequence; number of different motifs, 3; minimum number of sites, 2; maximum number of sites, 200; minimum motif width, 6; maximum motif width, 14. When sequence lists exceeded the maximally allowed number of 60,000 characters, a smaller set of putatively homologous genes was filtered by hand using the integrative orthology viewer in PLAZA. For motif comparison, the resulting 147 sequence alignments were submitted to STAMP with unchanged default parameters (Mahony and Benos, 2007). The resulting tree, given in Newick format, was displayed via MEGA (Tamura et al., 2011). A motif cluster was defined as a group of at least five patterns on branches shorter than 0.01, indicating very high similarity. Motifs from each cluster were resubmitted to STAMP to generate PSSMs. These were used to find matches with known cis-elements in the plant-specific promoter databases AthaMap, AGRIS, and PLACE (Higo et al., 1999; Davuluri et al., 2003; Steffens et al., 2004), a function provided by STAMP. Similar matches with E-values of lower than 1e−05 in two of three databases were considered significant. For sequence logo generation, TRANSFAC motif formatted PSSMs were submitted to WebLogo 3 (Crooks et al., 2004).
PSSMs of each cluster were converted to tab-formatted matrices and used as a query in the RSA-tool matrix scan (Medina-Rivera et al., 2015). Upstream 3000-bp promoter sequences from the 49 core HRGs and from all 27,206 protein-coding genes of Arabidopsis were obtained from TAIR and screened for the number of occurrences and positions of the respective motif, using a statistical significance cutoff value of >4.5. To test for overrepresentation of the predicted motifs in the 49 core HRGs promoters versus all Arabidopsis promoters, the hypergeometric distribution was calculated.
Plasmid Construction
All DNA constructs, vectors, and their origins as well as all primers used for cloning are listed in Supplemental Data Set 5. N-terminal HA-tagged effector constructs for the protoplast transfection, the effector control p35S:HA-GFP, and the LUC normalization vector p70SRUC have been described before (Stahl et al., 2004; Wehner et al., 2011; Klecker et al., 2014). For the construction of p35S:(MA)RAP2.2-hormone binding domain (HBD), the RAP2.2 coding sequence was amplified with a forward primer that replaced the second codon, the N-terminal Cys (C2), with Ala to remove susceptibility to N-end rule degradation. The reverse primer was designed to allow for a C-terminal translational fusion. The final product was recombined with pDONR211 via BP Clonase (Invitrogen). The resulting entry clone was recombined with p35S:rfA-HBD, a kind gift from Monika Tomar, using LR Clonase (Invitrogen). LUC reporter constructs for the protoplast transfection system were derivatives of pBT10GAL4-upstream activation sequence (UAS) (Wehner et al., 2011). Upstream sequences of LBD41 and PCO1 were amplified from Col-0 genomic DNA using specific primers (Supplemental Data Set 5). Promoter sequences with highlighted binding sites for forward primers can be found in Supplemental Figure 8. Both inserts and pBT10GAL4UAS were digested with NcoI and BamHI before ligation, thereby removing the GAL4UAS sequence. Single nucleotide substitutions were introduced using primer pairs with site-specific mismatches and de novo synthesis of reporter vectors with Phusion DNA Polymerase (Thermo Scientific). Before transformation in Escherichia coli, the methylated template vector was digested with DpnI. For the construction of pmin:LUC, specific primers were used to amplify a minimal promoter region of 52 bp from the CaMV 35S promoter of p35S:HF-GFP-RPL18 (Mustroph et al., 2009). Here, restriction enzymes NcoI and BamHI were used for digestion. LBD41 promoter deletions were introduced into pmin:LUC via two BamHI sites. Artificial promoters with triplicated sequences were constructed as follows: single-stranded, partially overlapping oligo DNAs were synthesized (Invitrogen) and dissolved to 100 µM in water, and 20 μL of plus- and minus-strand oligos were mixed, heated to 94°C for 3 min, and cooled to room temperature over 30 min for annealing. Ends were filled with Taq Polymerase at 72°C for 1 h. Double-stranded promoter concatemers were ligated into pmin:LUC after BamHI digestion. For protoplast experiments, plasmids were purified with a NucleoBond PC 500 Midi Kit (Macherey-Nagel) and stored at –20°C.
LUC and GFP reporters for stable plant genome integration were constructed from destination vectors pBGWL7 (Karimi et al., 2005) and pGATA:HF-GFP-RPL18 (Mustroph et al., 2009), respectively. LBD41-deleted promoters, 35S promoter, and the 3x33bpmin cassette were amplified with specific Gateway recombination-compatible primers. Products were recombined into pDONR211 via BP Clonase, and confirmed entry vectors were used for LR reaction for recombination with destination vectors (Invitrogen). ERF-VII overexpression constructs for Agrobacterium tumefaciens-mediated plant transformation were constructed using of entry clones containing coding sequences (Wehner et al., 2011) recombined into the destination vector p35S:HF-GATA (Mustroph et al., 2010).
All yeast cloning vectors (pMW#2, pMW#3, pMW#5, and pDEST-AD) and control vectors (PromB0507.1-HIS, PromB0507.1-LAC, and AD-CES-1) were a kind gift from A.J.M. Walhout (Walhout et al., 2000; Deplancke et al., 2006; Reece-Hoyes et al., 2009). For generating yeast AD-TF fusions, entry clones containing coding sequences from ERF-VIIs (Wehner et al., 2011) were recombined with pDEST-AD (Walhout et al., 2000). Double-stranded BamHI-cut promoter concatemers were ligated into the GW donor pMW#5 and recombined with the HIS reporter pMW#2 and LacZ reporter pMW#3 by LR reaction (Invitrogen). All clones were purified, digested, and confirmed by sequencing (LGC Genomics).
Protoplast Isolation, Transient Transformation, and Treatment
Detailed protoplast isolation, transformation procedure, and LUC activity measurement were described previously (Klecker et al., 2014). A total of 12,000 protoplasts were used for each transformation. For protein gel blot analysis and RNA isolation from transformed protoplasts, the amount of plasmid DNA was increased to 20 µg per reaction, and volumes of protoplast suspensions and transformation reagents were doubled. For protein gel blot analyses, the incubation of transformed protoplasts was performed under LD conditions overnight for 22 h before nitrogen freezing. For drug treatments, after 18 h LD incubation cell suspensions were mixed with 100 µM cycloheximide or 0.16% (v/v) of the solvent DMSO. After 30 min, 1 µM Dex or 0.83% (v/v) of the solvent ethanol was added to the suspension and incubated for an additional 4 h under LD conditions. Cells were frozen in liquid nitrogen and stored at −80°C until use.
Stable Plant Transformation
To generate stable A. thaliana reporter lines, binary vectors were introduced into the Agrobacterium strain GV3101 (Koncz et al., 1984) using a freeze/thaw shock transformation method (Chen et al., 1994). The germ cell line of A. thaliana Col-0 was transformed by floral dip into 5% (w/v) sucrose/0.02% (v/v) Silwet L-77/Agrobacteria suspension (Clough and Bent, 1998). Seeds were harvested and positive transformants were identified by growth characteristics on MS plates (see above) supplied with either kanamycin or phosphinothricin. Resistant individuals of segregating progeny were selected and the homozygous T3 generation was used for experiments.
Measurement of ADH Enzymatic Activity
Pulverized plant material (7-d-old seedlings) was homogenized on ice in 300 μL extraction buffer (50 mM HEPES-KOH, pH 6.8, 5 mM Mg acetate, 15% [w/v] glycerin, 1 mM EDTA, 1 mM EGTA, 0.1 mM Pefabloc [Sigma-Aldrich], and 5 mM β-mercaptoethanol [β-ME]) and centrifuged for 15 min at 4°C and 12,000g, and the supernatant was recovered. In each assay, 20 μL of supernatant was incubated with 960 μL 50 mM TES buffer, pH 7.5, and 10 μL 17 mM NADH for 5 min. Change in absorbance (340 nm) was monitored for 5 min (Specord 200 PLUS; Analytik Jena) before and after addition of 10 μL 1 M acetaldehyde. Protein quantification was done using Bradford assay (Roti Quant; Carl Roth). Enzyme activity was calculated as the change of absorption divided by whole protein content.
Protein Gel Blot Analysis and in Vivo Bioluminescence Imaging
For immunodetection of HA-tagged proteins from protoplasts, frozen cells were lysed in 30 μL 3× urea buffer (4 M urea, 5% [v/v] β-ME, 5% [w/v] SDS, 16.5% [v/v] glycerol, and Bromophenol blue) and heated to 95°C for 10 min before SDS-PAGE and blotting to a polyvinylidene fluoride membrane (Roth) (Sambrook et al., 1989). For immunodetection, murine monoclonal primary HA-antibody (Sigma-Aldrich; H9658, 1:3000) and polyclonal secondary anti-mouse horseradish peroxidase antibody (Carl Roth; 4759.1; 1:10,000) were used, along with horseradish peroxidase substrate solution Roti-Lumin (Roth). Chemiluminescence of bands was detected with a CCD camera of a low-light imaging system (Intas) using a shutter time of 5 min. For LUC activity imaging, intact plants or cut leaves were evenly sprayed with 2 mM luciferin (PJK) + 0.1% (v/v) Triton X-100. After 2 min dark incubation, the plants were photographed in a low-light imaging system (Intas) with a camera shutter time of 10 min.
RNA Isolation and Transcript Quantitation Analyses
RNA isolation from plant tissue, cDNA synthesis, reverse transcription standard, and quantitative PCR (RT-qPCR) were described previously in detail (Klecker et al., 2014) using primers in Supplemental Data Set 5. Briefly, RT-qPCR was performed on a MyiQ single-color real-time PCR detection system (Bio-Rad) using SensiFAST SYBR and Fluroescein Mix (Bioline). Data were analyzed via the iQ5 Optical System Software (Bio-Rad). Three biological replicates were measured, each with three technical repetitions.
Y1H Assay
All Y1H vectors are listed in Supplemental Data Set 5. To generate bait strains, linearized pMW#2/pMW#3 constructs were cotransformed into Y1H-aS2 for stable genome integration (Green and Sambrook, 2012). Positive transformants were selected on synthetic complete (SC) dropout medium –Ura –His (1.7 g/L yeast nitrogen base without amino acids and ammonium sulfate, 5 g/L ammonium sulfate, 20 g/L glucose, 0.8 g/L dropout mix, and 1% [w/v] agar, pH 5.6, with NaOH). Equally diluted colonies (3.5 µL) were spotted onto SC-Ura-His plates supplied with 10 to 80 mM 3-AT and incubated for 5 d at 30°C, with 40 mM 3-AT suppressing background activity of all HIS baits tested. To test TF-DNA interaction, a 2-μL volume of 40 ng AD-TF prey vector was transformed in bait strains (Green and Sambrook, 2012). To control for autoactivty of bait strains, water was added instead of DNA. To control for autoactivation capacity of the AD domain, AD expressing empty pDEST-AD vector was transformed. Transformation reactions were diluted to the same optical density (600 nm), subsequently pipetted on SC-Ura-His-Trp plates with or without 40 mM 3-AT, and incubated for 5 d before documentation. Five-day-old yeast colony spots grown on SC-Ura-His-Trp-3-AT with highest initial yeast concentration were used for β-galactosidase colony-lift filter assays: Colonies were transferred to a nitrocellulose membrane, frozen in liquid nitrogen for 10 s, and membranes thawed and placed, colony side up, on a nitrocellulose filter presoaked with Z-buffer/X-gal solution (X-gal solution [1.67 mL]: 2 mg/mL 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside in N,N-dimethylformamide; Z-buffer [100 mL]: 16 g/L Na2HPO47H2O, 5.5 g/L NaH2PO4H2O, 0.75 g/L KCl, 0.246 g/L MgSO47H2O, pH 7; β-ME [0.27 mL]). Plates were incubated for up to 4 h at 30°C before photo-optical documentation of β-galactosidase activity.
ChIP and Directed RT-qPCR
ChIP was performed according to Deal and Henikoff (2010) with minor modifications. Seven-day-old seedlings (developmental stage) grown on 0.5× MS salts, 0.8% (w/v) agar, and 1% (w/v) Suc, pH 5.7, were pretreated with 100 µM Calpain inhibitor IV (American Peptide) and 1% (w/v) DMSO for 4 h and cross-linked in 1% (v/v) formaldehyde nuclei purification buffer (NPB) (20 mM MOPS, pH 7.0, 40 mM NaCl, 90 mM KCl, 2 mM EDTA, and 0.5 mM EGTA) for 10 min under vacuum. The reaction was quenched by the addition of glycine to 125 mM by infiltration for 5 min; seedlings were washed three times, blotted dry, and then pulverized under liquid N2. To isolate nuclei, 0.5 g tissue was thawed to 4°C in 10 mL NPB that additionally contained 0.5 mM spermidine, 0.2 mM spermine, and 1× Plant Protease Inhibitor Cocktail (Sigma-Aldrich P9599). The nuclei were pelleted by centrifugation in 4°C at 1200g for 10 min, resuspended in 500 μL NPB, and lysed with the addition of 120 μL nuclei lysis buffer (50 mM Tris, pH 8.0, 10 mM EDTA, 1% [w/v] SDS, and 1× Plant Protease Inhibitor Cocktail) by vortexing for 2 min at room temperature. The chromatin was sheared into 200- to 600-bp fragments by sonication (Diagenode) with 40 cycles of 30 s ON and 30 s OFF at 4°C. The sample was cleared by centrifugation at 16,000g at 4°C for 2 min. The supernatant was diluted to 1.2 mL with dilution buffer (16.7 mM Tris, pH 8.0, 167 mM NaCl, 1.1% [v/v] Triton X-100, and 1.2 mM EDTA). Prior to the immunopurification, the sample was precleared by incubation with uncoupled magnetic agarose beads (Chromotek) for 30 min followed by collection of the supernatant. Three hundred microliters of the input chromatin was incubated with 30 μL magnetic anti-FLAG beads (Sigma-Aldrich) overnight, after which time the beads were magnetically collected, washed for 5 min each with low NaCl wash buffer (20 mM Tris, pH 8.0, 150 mM NaCl, 0.1% [w/v] SDS, 1% [v/v] Triton X-100, and 2 mM EDTA), high NaCl wash buffer (20 mM Tris, pH 8.0, 500 mM NaCl, 0.1% [w/v] SDS, 1% [v/v] Triton X-100, and 2 mM EDTA), LiCl wash buffer (10 mM Tris, pH 8.0, 250 mM LiCl, 1% [w/v] sodium deoxycholate, 1% [v/v] Nonidet P-40, and 1 mM EDTA), and standard TE buffer (10 mM Tris, pH 8.0, and 1 mM EDTA). Following this series of washes, the chromatin was eluted from the anti-FLAG beads by heating to 65°C in elution buffer (100 mM NaHCO3 and 1% [w/v] SDS) for 15 min. The eluted chromatin was then reverse cross-linked by the addition of 20 μL 5 M NaCl with heating at 65°C overnight. Following reverse cross-linking, 1 µg of RNase A (Sigma-Aldrich) was added and the sample incubated at 37°C for 15 min. Proteinase K (0.8 units; New England Biolabs) was then added and the sample incubated at 55°C for 15 min. The final ChIP-DNA sample was purified using Qiagen MinElute columns according to the manufacturer’s instructions and eluted with 12 μL of elution buffer. Immunopurified chromatin was assayed by qPCR using primers listed in Supplemental Data Set 5.
Accession Numbers
Arabidopsis accession numbers of the genes studied in this article can be found in Supplemental Data Set 5. The 49 core HRGs are presented in Supplemental Data Set 3.
Supplemental Data
Supplemental Figure 1. T-DNA insertion mutations studied were null alleles (transcript undetectable).
Supplemental Figure 2. Submergence phenotypes of rap2.2 and rap2.12 single and double mutants.
Supplemental Figure 3. Comparative phylogenetic footprinting can detect expected cis-elements.
Supplemental Figure 4. Normalized promoter activities for Figures 5 and 6.
Supplemental Figure 5. ERF-VIIs do not activate unrelated Y1H reporters.
Supplemental Figure 6. Comparison of the HRPE and AREs.
Supplemental Figure 7. Promoter of ADH1 from Arabidopsis and maize with marked motifs, as described in the legend.
Supplemental Figure 8. Promoters of LBD41 and PCO1 from Arabidopsis with marked motifs and underlined primers, as described in the legend.
Supplemental Table 1. Overview of group VII ERF T-DNA insertion lines used in the literature
Supplemental Table 2. Majority of C9motif-containing core genes are early N-end rule targets.
Supplemental Data Set 1. Overview over homologous genes of the 49 core hypoxia response genes in 25 species from the PLAZA 2.5 database
Supplemental Data Set 2. Overview over position-specific scoring matrices identified in this article.
Supplemental Data Set 3. Occurrence of the nine motifs in the 49 core hypoxia response genes, together with published microarray data.
Supplemental Data Set 4. Occurrence and overlap of the HRPE and the GT-rich region of At ARE (Hoeren et al., 1998) in the 49 core hypoxia response genes, and in Zm LBD41 and Zm Adh1.
Supplemental Data Set 5. List of constructs, primers, and AGI codes used in this article.
Supplementary Material
Acknowledgments
We thank Wolfgang Dröge-Laser, Nora Wehner, and Christoph Weiste for constructs and help with the protoplast transactivation system. This work was supported by the U.S. National Science Foundation (Grant IOS-1121626) and the U.S. Department of Agriculture, National Institute of Food and Agriculture-Agriculture and Food Research Initiative (Grant 2011-04015) to J.B.-S. and by the DFG (German Research Foundation, MU 2755/4-1) and the Stifterverband für die Deutsche Wissenschaft (H140 5409 9999 15625) to A.M. and P.G.
AUTHOR CONTRIBUTIONS
P.G., J.B.-S., and A.M. planned and designed the experiments. P.G., J.T.M., M.F., T.L., and A.M. performed experiments. P.G., J.B-S., and A.M. wrote the manuscript.
Glossary
- HRG
hypoxia-responsive gene
- ChIP
chromatin immunopurification
- ARE
anaerobic-responsive element
- HRPE
hypoxia-responsive promoter element
- Chx
cycloheximide
- Dex
dexamethasone
- PSSM
position-specific scoring matrix
- Y1H
yeast one-hybrid
- 3-AT
3-amino-1,2,4-triazole
- amiRNA
artificial microRNA
- MS
Murashige and Skoog
- LD
long-day
- SD
short-day
- β-ME
β-mercaptoethanol
- NPB
nuclei purification buffer
References
- Abbas M., Berckhan S., Rooney D.J., Gibbs D.J., Vicente Conde J., Sousa Correia C., Bassel G.W., Marín-de la Rosa N., León J., Alabadí D., Blázquez M.A., Holdsworth M.J. (2015). Oxygen sensing coordinates photomorphogenesis to facilitate seedling survival. Curr. Biol. 25: 1483–1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey T.L., Elkan C. (1994). Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 2: 28–36. [PubMed] [Google Scholar]
- Bailey-Serres J., Voesenek L.A.C.J. (2008). Flooding stress: acclimations and genetic diversity. Annu. Rev. Plant Biol. 59: 313–339. [DOI] [PubMed] [Google Scholar]
- Bernard B., Thorsson V., Rovira H., Shmulevich I. (2012). Increasing coverage of transcription factor position weight matrices through domain-level homology. PLoS One 7: e42779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brudno M., Malde S., Poliakov A., Do C.B., Couronne O., Dubchak I., Batzoglou S. (2003). Glocal alignment: finding rearrangements during alignment. Bioinformatics 19 (suppl. 1): i54–i62. [DOI] [PubMed] [Google Scholar]
- Brudno M., Steinkamp R., Morgenstern B. (2004). The CHAOS/DIALIGN WWW server for multiple alignment of genomic sequences. Nucleic Acids Res. 32: W41–W44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buhler J., Tompa M. (2002). Finding motifs using random projections. J. Comput. Biol. 9: 225–242. [DOI] [PubMed] [Google Scholar]
- Bui L.T., Giuntoli B., Kosmacz M., Parlanti S., Licausi F. (2015). Constitutively expressed ERF-VII transcription factors redundantly activate the core anaerobic response in Arabidopsis thaliana. Plant Sci. 236: 37–43. [DOI] [PubMed] [Google Scholar]
- Bustos R., Castrillo G., Linhares F., Puga M.I., Rubio V., Pérez-Pérez J., Solano R., Leyva A., Paz-Ares J. (2010). A central regulatory system largely controls transcriptional activation and repression responses to phosphate starvation in Arabidopsis. PLoS Genet. 6: e1001102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Büttner M., Singh K.B. (1997). Arabidopsis thaliana ethylene-responsive element binding protein (AtEBP), an ethylene-inducible, GCC box DNA-binding protein interacts with an ocs element binding protein. Proc. Natl. Acad. Sci. USA 94: 5961–5966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Nelson R.S., Sherwood J.L. (1994). Enhanced recovery of transformants of Agrobacterium tumefaciens after freeze-thaw transformation and drug selection. Biotechniques 16: 664–668, 670. [PubMed] [Google Scholar]
- Cheng M.-C., Liao P.-M., Kuo W.-W., Lin T.-P. (2013). The Arabidopsis ETHYLENE RESPONSE FACTOR1 regulates abiotic stress-responsive gene expression by binding to different cis-acting elements in response to different stress signals. Plant Physiol. 162: 1566–1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi H., Hong J., Ha J., Kang J., Kim S.Y. (2000). ABFs, a family of ABA-responsive element binding factors. J. Biol. Chem. 275: 1723–1730. [DOI] [PubMed] [Google Scholar]
- Christianson J.A., Llewellyn D.J., Dennis E.S., Wilson I.W. (2010). Comparisons of early transcriptome responses to low-oxygen environments in three dicotyledonous plant species. Plant Signal. Behav. 5: 1006–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cliften P., Sudarsanam P., Desikan A., Fulton L., Fulton B., Majors J., Waterston R., Cohen B.A., Johnston M. (2003). Finding functional features in Saccharomyces genomes by phylogenetic footprinting. Science 301: 71–76. [DOI] [PubMed] [Google Scholar]
- Cliften P.F., Hillier L.W., Fulton L., Graves T., Miner T., Gish W.R., Waterston R.H., Johnston M. (2001). Surveying Saccharomyces genomes to identify functional elements by comparative DNA sequence analysis. Genome Res. 11: 1175–1186. [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Cooper S.J., Trinklein N.D., Anton E.D., Nguyen L., Myers R.M. (2006). Comprehensive analysis of transcriptional promoter structure and function in 1% of the human genome. Genome Res. 16: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooks G.E., Hon G., Chandonia J.-M., Brenner S.E. (2004). WebLogo: a sequence logo generator. Genome Res. 14: 1188–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das M.K., Dai H.-K. (2007). A survey of DNA motif finding algorithms. BMC Bioinformatics 8 (suppl. 7): S21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davuluri R.V., Sun H., Palaniswamy S.K., Matthews N., Molina C., Kurtz M., Grotewold E. (2003). AGRIS: Arabidopsis gene regulatory information server, an information resource of Arabidopsis cis-regulatory elements and transcription factors. BMC Bioinformatics 4: 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bruxelles G.L., Peacock W.J., Dennis E.S., Dolferus R. (1996). Abscisic acid induces the alcohol dehydrogenase gene in Arabidopsis. Plant Physiol. 111: 381–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deal R.B., Henikoff S. (2010). A simple method for gene expression and chromatin profiling of individual cell types within a tissue. Dev. Cell 18: 1030–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deplancke B., et al. (2006). A gene-centered C. elegans protein-DNA interaction network. Cell 125: 1193–1205. [DOI] [PubMed] [Google Scholar]
- Dolferus R., Jacobs M., Peacock W.J., Dennis E.S. (1994). Differential interactions of promoter elements in stress responses of the Arabidopsis Adh gene. Plant Physiol. 105: 1075–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drew M.C. (1997). Oxygen deficiency and root metabolism: Injury and acclimation under hypoxia and anoxia. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48: 223–250. [DOI] [PubMed] [Google Scholar]
- Ferl R.J. (1990). ARF-B(2): A protein complex that specifically binds to part of the anaerobic response element of maize Adh 1. Plant Physiol. 93: 1094–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferl R.J., Nick H.S. (1987). In vivo detection of regulatory factor binding sites in the 5′ flanking region of maize Adh1. J. Biol. Chem. 262: 7947–7950. [PubMed] [Google Scholar]
- Franco-Zorrilla J.M., López-Vidriero I., Carrasco J.L., Godoy M., Vera P., Solano R. (2014). DNA-binding specificities of plant transcription factors and their potential to define target genes. Proc. Natl. Acad. Sci. USA 111: 2367–2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazer K.A., Pachter L., Poliakov A., Rubin E.M., Dubchak I. (2004). VISTA: computational tools for comparative genomics. Nucleic Acids Res. 32: W273–W279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelfand M.S., Koonin E.V., Mironov A.A. (2000). Prediction of transcription regulatory sites in Archaea by a comparative genomic approach. Nucleic Acids Res. 28: 695–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs D.J., Lee S.C., Isa N.M., Gramuglia S., Fukao T., Bassel G.W., Correia C.S., Corbineau F., Theodoulou F.L., Bailey-Serres J., Holdsworth M.J. (2011). Homeostatic response to hypoxia is regulated by the N-end rule pathway in plants. Nature 479: 415–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs D.J., et al. (2014). Nitric oxide sensing in plants is mediated by proteolytic control of group VII ERF transcription factors. Mol. Cell 53: 369–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giuliano G., Pichersky E., Malik V.S., Timko M.P., Scolnik P.A., Cashmore A.R. (1988). An evolutionarily conserved protein binding sequence upstream of a plant light-regulated gene. Proc. Natl. Acad. Sci. USA 85: 7089–7093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giuntoli B., Lee S.C., Licausi F., Kosmacz M., Oosumi T., van Dongen J.T., Bailey-Serres J., Perata P. (2014). A trihelix DNA binding protein counterbalances hypoxia-responsive transcriptional activation in Arabidopsis. PLoS Biol. 12: e1001950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodstein D.M., Shu S., Howson R., Neupane R., Hayes R.D., Fazo J., Mitros T., Dirks W., Hellsten U., Putnam N., Rokhsar D.S. (2012). Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 40: D1178–D1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green, M.R., and Sambrook, J. (2012). Molecular Cloning: A Laboratory Manual, 4th ed. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press). [Google Scholar]
- Guiltinan M.J., Marcotte W.R. Jr., Quatrano R.S. (1990). A plant leucine zipper protein that recognizes an abscisic acid response element. Science 250: 267–271. [DOI] [PubMed] [Google Scholar]
- Harbison C.T., et al. (2004). Transcriptional regulatory code of a eukaryotic genome. Nature 431: 99–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess N., Klode M., Anders M., Sauter M. (2011). The hypoxia responsive transcription factor genes ERF71/HRE2 and ERF73/HRE1 of Arabidopsis are differentially regulated by ethylene. Physiol. Plant. 143: 41–49. [DOI] [PubMed] [Google Scholar]
- Higo K., Ugawa Y., Iwamoto M., Korenaga T. (1999). Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res. 27: 297–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinz M., Wilson I.W., Yang J., Buerstenbinder K., Llewellyn D., Dennis E.S., Sauter M., Dolferus R. (2010). Arabidopsis RAP2.2: an ethylene response transcription factor that is important for hypoxia survival. Plant Physiol. 153: 757–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeren F.U., Dolferus R., Wu Y., Peacock W.J., Dennis E.S. (1998). Evidence for a role for AtMYB2 in the induction of the Arabidopsis alcohol dehydrogenase gene (ADH1) by low oxygen. Genetics 149: 479–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu F.-C., Chou M.-Y., Peng H.-P., Chou S.-J., Shih M.-C. (2011). Insights into hypoxic systemic responses based on analyses of transcriptional regulation in Arabidopsis. PLoS One 6: e28888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaglo-Ottosen K.R., Gilmour S.J., Zarka D.G., Schabenberger O., Thomashow M.F. (1998). Arabidopsis CBF1 overexpression induces COR genes and enhances freezing tolerance. Science 280: 104–106. [DOI] [PubMed] [Google Scholar]
- Jarillo J.A., Leyva A., Salinas J., Martinez-Zapater J.M. (1993). Low temperature induces the accumulation of alcohol dehydrogenase mRNA in Arabidopsis thaliana, a chilling-tolerant plant. Plant Physiol. 101: 833–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juntawong P., Girke T., Bazin J., Bailey-Serres J. (2014). Translational dynamics revealed by genome-wide profiling of ribosome footprints in Arabidopsis. Proc. Natl. Acad. Sci. USA 111: E203–E212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M., De Meyer B., Hilson P. (2005). Modular cloning in plant cells. Trends Plant Sci. 10: 103–105. [DOI] [PubMed] [Google Scholar]
- Klecker M., Gasch P., Peisker H., Dörmann P., Schlicke H., Grimm B., Mustroph A. (2014). A shoot-specific hypoxic response of Arabidopsis sheds light on the role of the phosphate-responsive transcription factor PHOSPHATE STARVATION RESPONSE1. Plant Physiol. 165: 774–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klok E.J., Wilson I.W., Wilson D., Chapman S.C., Ewing R.M., Somerville S.C., Peacock W.J., Dolferus R., Dennis E.S. (2002). Expression profile analysis of the low-oxygen response in Arabidopsis root cultures. Plant Cell 14: 2481–2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koncz C., Kreuzaler F., Kalman Z., Schell J. (1984). A simple method to transfer, integrate and study expression of foreign genes, such as chicken ovalbumin and alpha-actin in plant tumors. EMBO J. 3: 1029–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosmacz M., Parlanti S., Schwarzländer M., Kragler F., Licausi F., Van Dongen J.T. (2015). The stability and nuclear localization of the transcription factor RAP2.12 are dynamically regulated by oxygen concentration. Plant Cell Environ. 38: 1094–1103. [DOI] [PubMed] [Google Scholar]
- Lee S.C., Mustroph A., Sasidharan R., Vashisht D., Pedersen O., Oosumi T., Voesenek L.A.C.J., Bailey-Serres J. (2011). Molecular characterization of the submergence response of the Arabidopsis thaliana ecotype Columbia. New Phytol. 190: 457–471. [DOI] [PubMed] [Google Scholar]
- Li H.-Y., Chye M.-L. (2004). Arabidopsis Acyl-CoA-binding protein ACBP2 interacts with an ethylene-responsive element-binding protein, AtEBP, via its ankyrin repeats. Plant Mol. Biol. 54: 233–243. [DOI] [PubMed] [Google Scholar]
- Li H.-Y., Xiao S., Chye M.-L. (2008). Ethylene- and pathogen-inducible Arabidopsis acyl-CoA-binding protein 4 interacts with an ethylene-responsive element binding protein. J. Exp. Bot. 59: 3997–4006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licausi F., Kosmacz M., Weits D.A., Giuntoli B., Giorgi F.M., Voesenek L.A.C.J., Perata P., van Dongen J.T. (2011a). Oxygen sensing in plants is mediated by an N-end rule pathway for protein destabilization. Nature 479: 419–422. [DOI] [PubMed] [Google Scholar]
- Licausi F., van Dongen J.T., Giuntoli B., Novi G., Santaniello A., Geigenberger P., Perata P. (2010). HRE1 and HRE2, two hypoxia-inducible ethylene response factors, affect anaerobic responses in Arabidopsis thaliana. Plant J. 62: 302–315. [DOI] [PubMed] [Google Scholar]
- Licausi F., Weits D.A., Pant B.D., Scheible W.-R., Geigenberger P., van Dongen J.T. (2011b). Hypoxia responsive gene expression is mediated by various subsets of transcription factors and miRNAs that are determined by the actual oxygen availability. New Phytol. 190: 442–456. [DOI] [PubMed] [Google Scholar]
- Liu F., Vantoai T., Moy L.P., Bock G., Linford L.D., Quackenbush J. (2005). Global transcription profiling reveals comprehensive insights into hypoxic response in Arabidopsis. Plant Physiol. 137: 1115–1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu G., Paul A.L., McCarty D.R., Ferl R.J. (1996). Transcription factor veracity: is GBF3 responsible for ABA-regulated expression of Arabidopsis Adh? Plant Cell 8: 847–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lüscher B., Eisenman R.N. (1990). New light on Myc and Myb. Part II. Myb. Genes Dev. 4: 2235–2241. [DOI] [PubMed] [Google Scholar]
- MacIsaac K.D., Wang T., Gordon D.B., Gifford D.K., Stormo G.D., Fraenkel E. (2006). An improved map of conserved regulatory sites for Saccharomyces cerevisiae. BMC Bioinformatics 7: 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahony S., Benos P.V. (2007). STAMP: a web tool for exploring DNA-binding motif similarities. Nucleic Acids Res. 35: W253–W258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marín-de la Rosa N., Sotillo B., Miskolczi P., Gibbs D.J., Vicente J., Carbonero P., Oñate-Sánchez L., Holdsworth M.J., Bhalerao R., Alabadí D., Blázquez M.A. (2014). Large-scale identification of gibberellin-related transcription factors defines group VII ETHYLENE RESPONSE FACTORS as functional DELLA partners. Plant Physiol. 166: 1022–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama K., Sakuma Y., Kasuga M., Ito Y., Seki M., Goda H., Shimada Y., Yoshida S., Shinozaki K., Yamaguchi-Shinozaki K. (2004). Identification of cold-inducible downstream genes of the Arabidopsis DREB1A/CBF3 transcriptional factor using two microarray systems. Plant J. 38: 982–993. [DOI] [PubMed] [Google Scholar]
- McGuire A.M., Hughes J.D., Church G.M. (2000). Conservation of DNA regulatory motifs and discovery of new motifs in microbial genomes. Genome Res. 10: 744–757. [DOI] [PubMed] [Google Scholar]
- Medina-Rivera A., et al. (2015). RSAT 2015: Regulatory Sequence Analysis Tools. Nucleic Acids Res. 43: W50–W56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menke F.L., Champion A., Kijne J.W., Memelink J. (1999). A novel jasmonate- and elicitor-responsive element in the periwinkle secondary metabolite biosynthetic gene Str interacts with a jasmonate- and elicitor-inducible AP2-domain transcription factor, ORCA2. EMBO J. 18: 4455–4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohanty B., Krishnan S.P.T., Swarup S., Bajic V.B. (2005). Detection and preliminary analysis of motifs in promoters of anaerobically induced genes of different plant species. Ann. Bot. (Lond.) 96: 669–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moses A.M., Chiang D.Y., Kellis M., Lander E.S., Eisen M.B. (2003). Position specific variation in the rate of evolution in transcription factor binding sites. BMC Evol. Biol. 3: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustroph A., Lee S.C., Oosumi T., Zanetti M.E., Yang H., Ma K., Yaghoubi-Masihi A., Fukao T., Bailey-Serres J. (2010). Cross-kingdom comparison of transcriptomic adjustments to low-oxygen stress highlights conserved and plant-specific responses. Plant Physiol. 152: 1484–1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustroph A., Zanetti M.E., Jang C.J.H., Holtan H.E., Repetti P.P., Galbraith D.W., Girke T., Bailey-Serres J. (2009). Profiling translatomes of discrete cell populations resolves altered cellular priorities during hypoxia in Arabidopsis. Proc. Natl. Acad. Sci. USA 106: 18843–18848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narsai R., Rocha M., Geigenberger P., Whelan J., van Dongen J.T. (2011). Comparative analysis between plant species of transcriptional and metabolic responses to hypoxia. New Phytol. 190: 472–487. [DOI] [PubMed] [Google Scholar]
- Ohme-Takagi M., Shinshi H. (1995). Ethylene-inducible DNA binding proteins that interact with an ethylene-responsive element. Plant Cell 7: 173–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olive M.R., Peacock W.J., Dennis E.S. (1991). The anaerobic responsive element contains two GC-rich sequences essential for binding a nuclear protein and hypoxic activation of the maize Adh1 promoter. Nucleic Acids Res. 19: 7053–7060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olive M.R., Walker J.C., Singh K., Dennis E.S., Peacock W.J. (1990). Functional properties of the anaerobic responsive element of the maize Adh1 gene. Plant Mol. Biol. 15: 593–604. [DOI] [PubMed] [Google Scholar]
- Papdi C., Abrahám E., Joseph M.P., Popescu C., Koncz C., Szabados L. (2008). Functional identification of Arabidopsis stress regulatory genes using the controlled cDNA overexpression system. Plant Physiol. 147: 528–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papdi C., Pérez-Salamó I., Joseph M.P., Giuntoli B., Bögre L., Koncz C., Szabados L. (2015). The low oxygen, oxidative and osmotic stress responses synergistically act through the ethylene response factor VII genes RAP2.12, RAP2.2 and RAP2.3. Plant J. 82: 772–784. [DOI] [PubMed] [Google Scholar]
- Para A., et al. (2014). Hit-and-run transcriptional control by bZIP1 mediates rapid nutrient signaling in Arabidopsis. Proc. Natl. Acad. Sci. USA 111: 10371–10376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash A., Blanchette M., Sinha S., Tompa M. (2004). Motif discovery in heterogeneous sequence data. Pac. Symp. Biocomput. 2004: 348–359. [DOI] [PubMed] [Google Scholar]
- Proost S., Van Bel M., Sterck L., Billiau K., Van Parys T., Van de Peer Y., Vandepoele K. (2009). PLAZA: a comparative genomics resource to study gene and genome evolution in plants. Plant Cell 21: 3718–3731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reece-Hoyes J.S., Deplancke B., Barrasa M.I., Hatzold J., Smit R.B., Arda H.E., Pope P.A., Gaudet J., Conradt B., Walhout A.J.M. (2009). The C. elegans Snail homolog CES-1 can activate gene expression in vivo and share targets with bHLH transcription factors. Nucleic Acids Res. 37: 3689–3698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reece-Hoyes J.S., Diallo A., Lajoie B., Kent A., Shrestha S., Kadreppa S., Pesyna C., Dekker J., Myers C.L., Walhout A.J.M. (2011). Enhanced yeast one-hybrid assays for high-throughput gene-centered regulatory network mapping. Nat. Methods 8: 1059–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riber W., Müller J.T., Visser E.J.W., Sasidharan R., Voesenek L.A.C.J., Mustroph A. (2015). The greening after extended darkness1 is an N-end rule pathway mutant with high tolerance to submergence and starvation. Plant Physiol. 167: 1616–1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rombauts S., Déhais P., Van Montagu M., Rouzé P. (1999). PlantCARE, a plant cis-acting regulatory element database. Nucleic Acids Res. 27: 295–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio V., Linhares F., Solano R., Martín A.C., Iglesias J., Leyva A., Paz-Ares J. (2001). A conserved MYB transcription factor involved in phosphate starvation signaling both in vascular plants and in unicellular algae. Genes Dev. 15: 2122–2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rushton P.J., Torres J.T., Parniske M., Wernert P., Hahlbrock K., Somssich I.E. (1996). Interaction of elicitor-induced DNA-binding proteins with elicitor response elements in the promoters of parsley PR1 genes. EMBO J. 15: 5690–5700. [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press). [Google Scholar]
- Sessions A., et al. (2002). A high-throughput Arabidopsis reverse genetics system. Plant Cell 14: 2985–2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha S., Blanchette M., Tompa M. (2004). PhyME: a probabilistic algorithm for finding motifs in sets of orthologous sequences. BMC Bioinformatics 5: 170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl D.J., Kloos D.U., Hehl R. (2004). A sugar beet chlorophyll a/b binding protein promoter void of G-box like elements confers strong and leaf specific reporter gene expression in transgenic sugar beet. BMC Biotechnol. 4: 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stålberg K., Ellerstöm M., Ezcurra I., Ablov S., Rask L. (1996). Disruption of an overlapping E-box/ABRE motif abolished high transcription of the napA storage-protein promoter in transgenic Brassica napus seeds. Planta 199: 515–519. [DOI] [PubMed] [Google Scholar]
- Steffens N.O., Galuschka C., Schindler M., Bülow L., Hehl R. (2004). AthaMap: an online resource for in silico transcription factor binding sites in the Arabidopsis thaliana genome. Nucleic Acids Res. 32: D368–D372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaresan V., Springer P., Volpe T., Haward S., Jones J.D., Dean C., Ma H., Martienssen R. (1995). Patterns of gene action in plant development revealed by enhancer trap and gene trap transposable elements. Genes Dev. 9: 1797–1810. [DOI] [PubMed] [Google Scholar]
- Tagle D.A., Koop B.F., Goodman M., Slightom J.L., Hess D.L., Jones R.T. (1988). Embryonic epsilon and gamma globin genes of a prosimian primate (Galago crassicaudatus). Nucleotide and amino acid sequences, developmental regulation and phylogenetic footprints. J. Mol. Biol. 203: 439–455. [DOI] [PubMed] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28: 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran L.-S.P., Nakashima K., Sakuma Y., Simpson S.D., Fujita Y., Maruyama K., Fujita M., Seki M., Shinozaki K., Yamaguchi-Shinozaki K. (2004). Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell 16: 2481–2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urao T., Yamaguchi-Shinozaki K., Urao S., Shinozaki K. (1993). An Arabidopsis myb homolog is induced by dehydration stress and its gene product binds to the conserved MYB recognition sequence. Plant Cell 5: 1529–1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dongen J.T., Fröhlich A., Ramírez-Aguilar S.J., Schauer N., Fernie A.R., Erban A., Kopka J., Clark J., Langer A., Geigenberger P. (2009). Transcript and metabolite profiling of the adaptive response to mild decreases in oxygen concentration in the roots of arabidopsis plants. Ann. Bot. (Lond.) 103: 269–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varala K., Li Y., Marshall-Colón A., Para A., Coruzzi G.M. (2015). “Hit-and-Run” leaves its mark: catalyst transcription factors and chromatin modification. BioEssays 37: 851–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vashisht D., Hesselink A., Pierik R., Ammerlaan J.M.H., Bailey-Serres J., Visser E.J.W., Pedersen O., van Zanten M., Vreugdenhil D., Jamar D.C.L., Voesenek L.A.C.J., Sasidharan R. (2011). Natural variation of submergence tolerance among Arabidopsis thaliana accessions. New Phytol. 190: 299–310. [DOI] [PubMed] [Google Scholar]
- Voesenek L.A.C.J., Bailey-Serres J. (2015). Flood adaptive traits and processes: an overview. New Phytol. 206: 57–73. [DOI] [PubMed] [Google Scholar]
- Voesenek L.A.C.J., Bailey-Serres J. (2013). Flooding tolerance: O2 sensing and survival strategies. Curr. Opin. Plant Biol. 16: 647–653. [DOI] [PubMed] [Google Scholar]
- Walhout A.J., Sordella R., Lu X., Hartley J.L., Temple G.F., Brasch M.A., Thierry-Mieg N., Vidal M. (2000). Protein interaction mapping in C. elegans using proteins involved in vulval development. Science 287: 116–122. [DOI] [PubMed] [Google Scholar]
- Walker J.C., Howard E.A., Dennis E.S., Peacock W.J. (1987). DNA sequences required for anaerobic expression of the maize alcohol dehydrogenase 1 gene. Proc. Natl. Acad. Sci. USA 84: 6624–6628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T., Stormo G.D. (2003). Combining phylogenetic data with co-regulated genes to identify regulatory motifs. Bioinformatics 19: 2369–2380. [DOI] [PubMed] [Google Scholar]
- Wang T., Stormo G.D. (2005). Identifying the conserved network of cis-regulatory sites of a eukaryotic genome. Proc. Natl. Acad. Sci. USA 102: 17400–17405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Haberer G., Mayer K.F.X. (2009). Discovery of cis-elements between sorghum and rice using co-expression and evolutionary conservation. BMC Genomics 10: 284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasserman W.W., Palumbo M., Thompson W., Fickett J.W., Lawrence C.E. (2000). Human-mouse genome comparisons to locate regulatory sites. Nat. Genet. 26: 225–228. [DOI] [PubMed] [Google Scholar]
- Wehner N., Hartmann L., Ehlert A., Böttner S., Oñate-Sánchez L., Dröge-Laser W. (2011). High-throughput protoplast transactivation (PTA) system for the analysis of Arabidopsis transcription factor function. Plant J. 68: 560–569. [DOI] [PubMed] [Google Scholar]
- Weits D.A., Giuntoli B., Kosmacz M., Parlanti S., Hubberten H.-M., Riegler H., Hoefgen R., Perata P., van Dongen J.T., Licausi F. (2014). Plant cysteine oxidases control the oxygen-dependent branch of the N-end-rule pathway. Nat. Commun. 5: 3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsch R., Maass D., Voegel T., Dellapenna D., Beyer P. (2007). Transcription factor RAP2.2 and its interacting partner SINAT2: stable elements in the carotenogenesis of Arabidopsis leaves. Plant Physiol. 145: 1073–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue G.-P. (2003). The DNA-binding activity of an AP2 transcriptional activator HvCBF2 involved in regulation of low-temperature responsive genes in barley is modulated by temperature. Plant J. 33: 373–383. [DOI] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K., Shinozaki K. (1994). A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low-temperature, or high-salt stress. Plant Cell 6: 251–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C.-Y., Hsu F.-C., Li J.-P., Wang N.-N., Shih M.-C. (2011). The AP2/ERF transcription factor AtERF73/HRE1 modulates ethylene responses during hypoxia in Arabidopsis. Plant Physiol. 156: 202–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S., Wang S., Liu X., Yu Y., Yue L., Wang X., Hao D. (2009). Four divergent Arabidopsis ethylene-responsive element-binding factor domains bind to a target DNA motif with a universal CG step core recognition and different flanking bases preference. FEBS J. 276: 7177–7186. [DOI] [PubMed] [Google Scholar]
- Zou C., Sun K., Mackaluso J.D., Seddon A.E., Jin R., Thomashow M.F., Shiu S.-H. (2011). Cis-regulatory code of stress-responsive transcription in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 108: 14992–14997. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.