Abstract
Ubiquitination, the covalent binding of the small protein modifier ubiquitin to a target protein, is an important and frequently studied posttranslational protein modification. Multiple reports provide useful insights into the plant ubiquitinome, but mostly at the protein level without comprehensive site identification. Here, we implemented ubiquitin combined fractional diagonal chromatography (COFRADIC) for proteome-wide ubiquitination site mapping on Arabidopsis thaliana cell cultures. We identified 3009 sites on 1607 proteins, thereby greatly increasing the number of known ubiquitination sites in this model plant. Finally, The Ubiquitination Site tool (http://bioinformatics.psb.ugent.be/webtools/ubiquitin_viewer/) gives access to the obtained ubiquitination sites, not only to consult the ubiquitination status of a given protein, but also to conduct intricate experiments aiming to study the roles of specific ubiquitination events. Together with the antibodies recognizing the ubiquitin remnant motif, ubiquitin COFRADIC represents a powerful tool to resolve the ubiquitination maps of numerous cellular processes in plants.
THE IMPORTANCE OF UBIQUITINATION IN PLANTS
In the postgenomic era, it is increasingly apparent that the one gene-one function model is not sufficiently broad to fully understand the molecular mechanisms at play within a cell. Numerous levels of complexity, such as protein-protein interactions and posttranslational modifications (PTMs), are essential in determining the life span, localization, and activity of a protein. By affecting activity, structure, complex formation, and subcellular localization of targeted proteins, PTMs dynamically regulate various cellular processes in plants (Guo et al., 2013; Barneche et al., 2014; Seo and Mas, 2014; Banfield, 2015; Furniss and Spoel, 2015; Polyn et al., 2015). An important PTM, not only in plants, but in all eukaryotes, is the conjugation of the small (∼8.5 kD), highly conserved, and abundant protein ubiquitin to substrates. Ubiquitination most often occurs via the formation of an isopeptidyl bond between the flexible C terminus of ubiquitin and the ε-amino group of lysine residues of a substrate (Heride et al., 2014). Besides regulation of protein catabolism through targeted degradation by the ubiquitin proteasome system (UPS), ubiquitination can also alter protein activity, localization, and interactions (Hua and Vierstra, 2011). As ubiquitin can form linear or branched chains by means of linkage of ubiquitin moieties to its own N terminus or internal lysine residues, respectively, a large diversity in ubiquitination types exists, each thought to affect protein fate in a specific manner (Komander and Rape, 2012).
In view of the importance of this PTM in plants, more than 1500 Arabidopsis thaliana genes are predicted to encode components of E3 ligases, proteins responsible for the transfer of ubiquitin to specific targets (Hua and Vierstra, 2011). More specifically, close to 900 F-box-type E3 ligases are annotated in the Arabidopsis genome, which is 10-fold more than in the human genome (Hua et al., 2011). The role of protein ubiquitination by E3 ligases in plants is illustrated by numerous studies, mostly at the single protein level, revealing that this PTM acts in the plant’s response to drought stress, temperature tolerance, and coordination of responses to phytohormones, such as auxin, brassinosteroids, and jasmonates (Cui et al., 2012; Cuéllar Pérez and Goossens, 2013; Guseman et al., 2015). E3 ligases have been shown to be essential regulators of plant immunity, and many microbes even seem to have evolved a way to sabotage the host UPS (Marino et al., 2012). Whereas a growing body of research supports the importance of ubiquitination in plants, a full grasp of the significance and the variety of roles played by this PTM can only be reached via comprehensive mapping of the ubiquitinome.
THE STATE OF THE ART OF UBIQUITIN PROFILING
Trapping Ubiquitinated Proteins in Plants
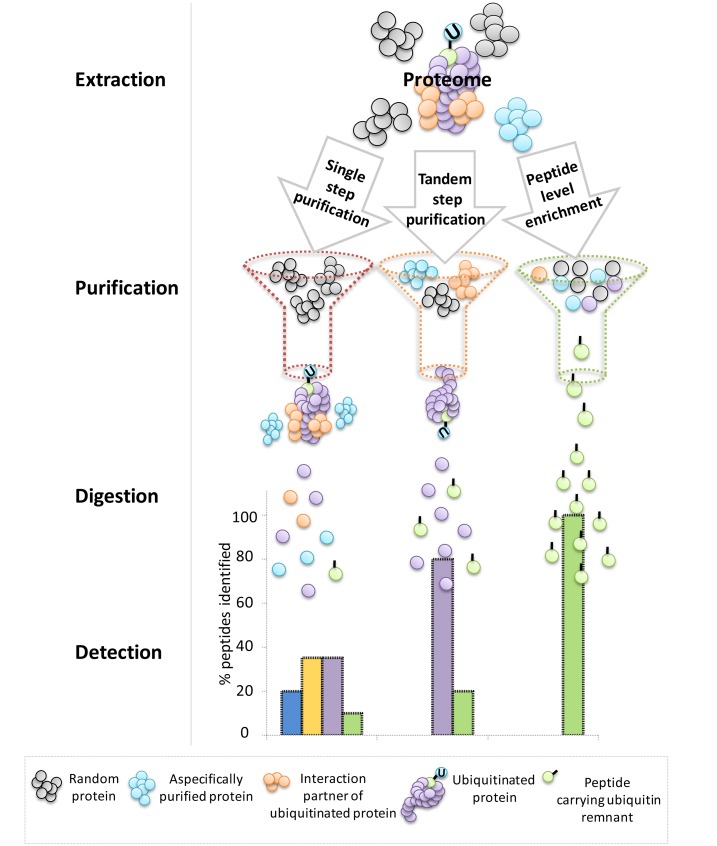
Various strategies have been developed to gain a proteome-wide insight into ubiquitination processes in plants. Pioneering studies relied on single-step purification approaches based on affinity matrices, such as ubiquitin-associated domains, ubiquitin interaction motifs, and monoclonal antiubiquitin antibodies, to enrich for ubiquitin conjugates at the protein level (Maor et al., 2007; Manzano et al., 2008, Igawa et al., 2009). In the most successful case, almost 300 potentially ubiquitinated proteins could be identified in Arabidopsis (Maor et al., 2007). Although these studies represented a major leap for the field at the time, the nondenaturing conditions used were cause for concern. A large number of false positives is potentially generated as it is difficult to distinguish between ubiquitinated proteins and aspecific proteins, such as copurified interaction partners and proteins that aspecifically bound the affinity matrix (Figure 1). To reduce this experimental bias, Saracco et al. (2009) created an Arabidopsis line that overexpresses a His-tagged variant of ubiquitin, which was used in conjunction with a newly developed tandem affinity purification (TAP) protocol. The initial enrichment step of ubiquitin conjugates, based on the ubiquitin binding region from human HHR23A (USU), could now be followed by nickel-chelate affinity chromatography under strong denaturing conditions, thereby reducing the background. As a result, 90 possible ubiquitinated proteins were identified. Due to the extra level of stringency associated with this improved experimental setup, the reduction in the number of proteins identified as potentially ubiquitinated is largely compensated for by increased reliability thanks to a decrease in the number of false positives. More recently, the same transgenic line was used in a more sensitive two-step affinity approach, by means of tandem ubiquitin binding entities (TUBEs) as affinity matrices during the first purification step (Kim et al., 2013). The timely transfer of the TUBE technology, previously established in the mammalian research field, to plant protocols allowed the identification of 950 proteins (Kim et al., 2013). Addition of the proteasomal inhibitor MG132 revealed that the ubiquitination state of more than half of the identified ubiquitinated proteins increased upon treatment, pointing toward the probable proteasomal degradation of these ubiquitination targets. In another TAP approach, Arabidopsis leaves were exposed to the more specific proteasomal inhibitor syringolin (Svozil et al., 2014), thereby bypassing the broad range of action attributed to MG132 that does not solely target the 26S proteasome (Gu et al., 2010).
Figure 1.
Theoretical Comparison of the Peptides Identified via Different Techniques for Ubiquitin Profiling in Plants.
Comparison between single-step purification, TAP approaches, and techniques based on enrichment at the peptide level. The peptide types that will eventually be injected into the mass spectrometer for detection are illustrated.
Are We Being Short-“Sited”?
The most reliable manner to distinguish proteins that are truly ubiquitinated from false positives is by identification of the exact ubiquitination site on the target protein, which also provides opportunities to investigate its functionality. For instance, site-directed mutagenesis of the respective lysine residues can reveal the role of a given ubiquitination event. Table 1 summarizes the results of studies in plants in which ubiquitination sites were examined for specific proteins.
Table 1. Previously Reported Ubiquitination Sites in Arabidopsis.
| Protein | Ubiquitination Sites | Site Identification Technique | Reference |
|---|---|---|---|
| H2B | Lys-143 | K-ε-GG footprint via LC-MS/MS | Zhang et al. (2007) |
| H2B | Lys-143 | K-ε-GG footprint via LC-MS/MS | Sridhar et al. (2007) |
| WRNIP1 | Multiple sites | K-ε-GG footprint via LC-MS/MS | Bish and Myers (2007) |
| IRT1 | Lys-146, Lys-171 | Site-directed mutagenesis K→R | Kerkeb et al. (2008) |
| H2A | Lys-121 | Site-directed mutagenesis K→R | Bratzel et al. (2010) |
| CRY2 | Lys-541, Lys-554 | Site-directed mutagenesis K→R | Zuo et al. (2012) |
| PIN2 | Six sites in hydrophobic loop | Site-directed mutagenesis K→R | Leitner et al. (2012) |
| COI1 | Lys-297 | K-ε-GG footprint via LC-MS/MS and site-directed mutagenesis K→A | Yan et al. (2013) |
| PCNA | Lys-164 | Site-directed mutagenesis K→R | Strzalka et al. (2013) |
| ABI5 | Lys-344 | Site-directed mutagenesis K→A | Liu and Stone (2013) |
| SINAL7 | Lys-23, Lys-124 | Site-directed mutagenesis K→A | Peralta et al. (2013) |
| GL3 | Lys-535, Lys-536 | Site-directed mutagenesis K→R | Patra et al. (2013) |
| EGL3 | Lys-493, Lys-495 | Site-directed mutagenesis K→R | Patra et al. (2013) |
| BOR1 | Lys-590 | Site-directed mutagenesis K→A | Kasai et al. (2014) |
| PHOT1 | Lys-526 | K-ε-GG footprint via LC-MS/MS | Deng et al. (2014) |
| BRI1 | Lys-866 | K-ε-GG footprint via LC-MS/MS and site-directed mutagenesis K→R | Martins et al. (2015) |
| OLE1-4 | Two sites in C terminus | K-ε-GG footprint via LC-MS/MS | Deruyffelaere et al. (2015) |
| JAZ12 | Lys-169 | K-ε-GG footprint via LC-MS/MS | Pauwels et al. (2015) |
| No. of proteins with sites | No. of reported sites | ||
| 56 | 85 | K-ε-GG footprint via LC-MS/MS | Maor et al. (2007) |
| 15 | 13 | K-ε-GG footprint via LC-MS/MS | Saracco et al. (2009) |
| 109 | 120 | K-ε-GG footprint via LC-MS/MS | Kim et al. (2013) |
| 1607 | 3009 | Lys-Gly label via LC-MS/MS | This work |
Three studies that profiled ubiquitinated proteins on a proteome-wide level (Manzano et al., 2008; Igawa et al., 2009; Svozil et al., 2014) were not included because they did not report any actual sites.
With affinity-based methods, in which the enrichment occurs at the protein level, it is not straightforward to reach a comprehensive proteome-wide view of the exact ubiquitination sites. Nevertheless, it is possible to identify a relatively low number of such sites by searching spectra for the typical K-ε-GG motif that remains upon tryptic cleavage of ubiquitinated proteins. Maor et al. (2007) identified 85 of these diglycine footprints on 56 proteins, representing the earliest report on precise protein ubiquitination sites in plants. Subsequently, by means of similar approaches, more exact sites have been identified in Arabidopsis (summarized in Table 1). The modest number of identified ubiquitination sites so far is due to the low stoichiometry of the site-modified peptides in relation to all other unmodified peptides resulting from digestion of the ubiquitinated proteins. Indeed, one of the main limiting factors in all mass spectrometry-based experiments aiming to profile PTMs is the time needed by the instrument to detect modified peptides in complex mixtures. Even in the case of a very successful purification that retains only ubiquitinated proteins, the majority of the resulting peptides supplied to the instrument will be unmodified. These peptides will monopolize the instrument’s acquisition time, leaving little chance for the identification of the more interesting, but less abundant, ubiquitination site-carrying peptides. To achieve the goal of comprehensive site mapping, it is imperative to operate at the peptide level by specific enrichment for ubiquitinated peptides and not simply ubiquitin conjugates (Figure 1).
In the biomedical field, this gold standard has been facilitated by the development of antibodies against the ubiquitin remnant motif K-ε-GG, enabling the specific enrichment of ubiquitination site-carrying peptides. Since then, the number of reported ubiquitination sites has skyrocketed, and now it is no longer rare to identify >10,000 sites in human cell lines (Kim et al., 2011; Wagner et al., 2011; Udeshi et al., 2013). This result is in stark contrast to the total number of ∼200 sites described in Arabidopsis, leaving a gap that needs to be filled in the plant science field (Maor et al., 2007; Manzano et al., 2008; Saracco et al., 2009; Kim et al., 2013) (Table1).
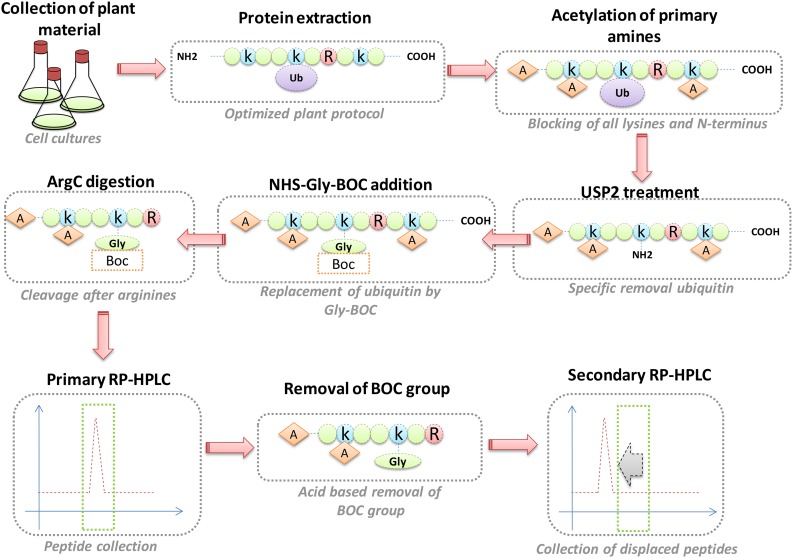
As a complementary alternative to K-ε-GG antibodies, the ubiquitin combined fractional diagonal chromatography (COFRADIC) method allows the identification of exact ubiquitination sites, also through enrichment at the peptide level. This technique has been applied to human cell lysates, yielding over 7000 unique sites (Stes et al., 2014). An overview of this methodology is provided in Figure 2. In short, the protocol starts by blocking all primary amines (lysines and N termini) via chemical acetylation, followed by incubation with a deubiquitinase (DUB), an enzyme that specifically cleaves ubiquitin from target proteins, thereby freeing primary amines on previously modified residues. To these free primary amines, a chemical handle is attached, which is subsequently used to isolate these peptides via two consecutive reverse-phase HPLC (RP-HPLC) runs (Figure 2).
Figure 2.
The Ubiquitin COFRADIC Pipeline.
During ubiquitin COFRADIC, the primary amines of extracted proteins are first chemically acetylated (orange diamonds labeled A for acetylation). USP2cc is then used to specifically cleave off the (now acetylated) ubiquitin, revealing a free primary amine on the previously ubiquitinated lysine. A glycine linked to a hydrophobic tert-butyloxycarbonyl (BOC) group (orange square) is linked to this primary amine, followed by a trypsin digest, which will now cleave C-terminally of arginine (red circle labeled R) but not of lysine (blue circle labeled k). After a first RP-HPLC run, the peptides are collected and pooled into 20 fractions that are treated with TFA to cleave off the BOC group, after which secondary RP-HPLC runs are conducted. All peaks with a hydrophilic shift are collected and identified by mass spectrometry. Ubiquitination sites are marked with a glycine (labeled “Gly”). This scheme only presents ubiquitination sites on lysines, but the same applies for N-terminally linked ubiquitin.
MOVING FORWARD: UBIQUITINATION SITE MAPPING IN PLANTS
K-ε-GG Antibodies
Two groups have successfully utilized K-ε-GG antibodies in plants. First, in rice (Oryza sativa) leaf tissue, 861 ubiquitination sites were identified on 464 proteins (Xie et al., 2015). Although the commonly accepted 10,000-site benchmark in human cells was not reached, the compatibility of the K-ε-GG antibodies with plant material was proven for the first time. This pioneer study was closely followed by the identification of 1500 ubiquitination sites in African rice (Oryza glaberrima) (Li et al., 2015). This differential analysis led to the discovery of the role of the Thermo-tolerance 1 protein in heat stress tolerance.
Ubiquitin COFRADIC
The successful transfer of TUBE technology and K-ε-GG antibodies to the plant research field motivated us to test the ubiquitin COFRADIC technology in Arabidopsis. We adapted several points of the protocol of Stes et al. (2014) both to improve the technology and to make it compatible for plants. A more stringent protein extraction protocol, based on a methanol/chloroform precipitation step, allowed us to greatly increase the overall protein yield and also to more rapidly impede endogenous DUB activity.
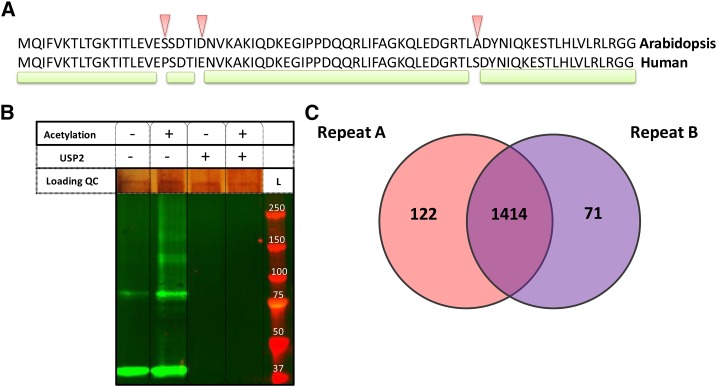
One of our main concerns was the use of the commercially available catalytic core domain of a human DUB, USP2cc, that is used to cleave the isopeptidyl bond between the ubiquitin C terminus and the ɛ-amino group of the ubiquitinated residue. USP2 activity is well documented (Renatus et al., 2006; Shahnawaz et al., 2007; Soboleva and Baker, 2004). Given that UPS2cc specifically recognizes the last five amino acids of ubiquitin (Renatus et al., 2006) and that they are conserved between human and plant ubiquitin (Figure 3A), we anticipated that USP2cc would be functional on plant proteins. Since the first step of the COFRADIC protocol entails a protein N-acetylation step, we also needed to evaluate the USP2cc activity on acetylated plant proteins. To this end, we used protein extracts of seedlings that overexpress UBIQUITIN10 (UBQ10) that is N-terminally tagged with hexahistidine. Protein immunoblot analysis revealed that protein-conjugated ubiquitin, visualized by the antihexahistidine antibody, in both the nonacetylated and acetylated protein extracts, disappeared after USP2cc treatment (Figure 3B). This result demonstrated that USP2cc was active on Arabidopsis protein extracts, independently of protein acetylation, and therefore could be used together with the COFRADIC approach in Arabidopsis.
Figure 3.
The Ubiquitin COFRADIC in Plants.
(A) Alignment of the Arabidopsis and human ubiquitin amino acid sequences. Arrowheads indicate mismatches between the two sequences and the green line marks the portions of shared amino acid sequence.
(B) Immunoblot analysis to test the efficiency of USP2cc to cleave ubiquitin from either acetylated or nonacetylated Arabidopsis proteins. Extracts derived from 35S:HIS6UBQ10 plants were not acetylated (−) or acetylated (+) and subsequently incubated with (+) or without (−) UPS2cc. Ubiquitinated proteins were detected with the anti-HIS antibodies. Loading quality control (QC) is given by silver staining. L, protein size ladder.
(C) Venn diagram displaying protein overlaps for which at least one ubiquitination site was detected in two replicates of the ubiquitin COFRADIC in Arabidopsis cell cultures.
A COFRADIC experiment with two biological replicates (two independent experiments) identified 2277 and 1762 ubiquitination sites on 1536 and 1485 proteins in the first and second experiments, respectively. In total, 3009 unique ubiquitination sites were obtained and mapped to 1607 proteins (Supplemental Data Set 1). Overlaps of 87% (Figure 3C) and 35% were found at the protein and site levels, respectively, between the two experiments. It is noteworthy that this large number of sites was acquired without proteasomal blockers prior to protein extraction, therefore likely providing an unbiased snapshot of the ubiquitinome at endogenous levels in Arabidopsis cell cultures.
Several ubiquitination sites were found on various transcription factors belonging to different families, such as the TEOSINTE BRANCHED1/CYCLOIDEA/PROLIFERATING CELL NUCLEAR ANTIGEN FACTOR (TCP), basic leucine zipper domain, MYB, WRKY, and basic helix-loop-helix families. For example, we identified ubiquitination sites on TCP8 and TCP22, class I members of the TCP family regulating cell proliferation and growth (Martín-Trillo and Cubas, 2010), and on the AUXIN RESPONSE FACTOR19 (ARF19), ARF2, ETHYLENE RESPONSIVE TRANSCRIPTION FACTOR113 (ERF113), ERF115, ABSCISIC ACID RESPONSIVE ELEMENTS BINDING PROTEIN3, and ABSCISIC ACID RESPONSIVE ELEMENTS BINDING FACTOR2, transcription factors involved in auxin, ethylene, brassinosteroid, and abscisic acid signal transduction cascades, respectively (Supplemental Table 1). Regulation of cell proliferation and hormonal signaling pathways has been linked extensively to UPS components, with F-box-containing E3 ubiquitin ligases playing central roles (Chapman and Estelle, 2009; Liu and Stone, 2011). Interestingly, ubiquitination sites on enzymes involved in hormone biosynthesis were found as well, such as 12-OXOPHYTODIENOATE REDUCTASE1 and 3-KETO-ACYL-CoA THIOLASE1, two of the three enzymes needed to convert 12-oxo phytodienoic acid into jasmonic acid in the peroxisome (Wasternack and Hause, 2013).
Ubiquitination sites were also detected on many UPS components, including E1, E2, and many E3 enzymes as well as 26S proteome subunits and deubiquitinating enzymes (Supplemental Table 1). Finally, a ubiquitination site on ubiquitin itself was identified. Of the seven internal lysines, ubiquitination occurred only on Lys-48, a branched ubiquitination type known to give rise to proteasomal degradation. The lack of the other linkage types might be due to their relatively reduced stoichiometry (Kim et al., 2013) and to the ArgC-type digestion used, generating relatively larger peptides that, on average, are more difficult to detect than tryptic peptides.
GRXS17 as a Proof-of-Concept Protein
A ubiquitination site was detected on a redox enzyme, GLUTAREDOXIN S17 (GRXS17), involved in auxin responses (Cheng et al., 2011). GRXS17 is the Arabidopsis ortholog of the human monothiol glutathioredoxin PKCθ-interacting protein (PICOT)/GLRX3, a putative nucleocytoplasmic protein regulating hormone and redox signaling in Arabidopsis (Cheng et al., 2011). Although not reported for any plant homologs yet, a ubiquitination site in human GLRX3 had been identified via antibody-based capture of K-ε-GG-containing peptides (Kim et al., 2011). To confirm ubiquitination of the plant GRXS17 protein detected with the COFRADIC technique, we applied a technique commonly used in the plant ubiquitin research field. An affinity purification on 35S:HIS6UBQ10 plants revealed, in agreement with the COFRADIC results, the ubiquitination of GRXS17 in three independent biological replicates (Supplemental Data Set 2). In light of the Lys-48 ubiquitin linkages found in the COFRADIC experiment, a possible ubiquitin-mediated proteasomal degradation of GRXS17 was tested by recombinant production of a V5-HIS6-tagged protein and by incubation with total protein extract derived from wild-type seedlings. The epitope-tagged GRXS17 was degraded over time in seedling extracts, unless the proteasome inhibitor MG132 was added (Supplemental Figure 1A). Similarly, the level of epitope-tagged GRXS17 from protein extracts of 35S:GRXS17:3xHA seedlings decreased over time, an event that could also be prevented by treatment with MG132 (Supplemental Figure 1B). Taken together, our data suggest that GRXS17 is ubiquitinated and that its ubiquitination leads to proteasomal degradation.
SHARING IS CARING
To make our Arabidopsis ubiquitination map available to the plant community, we constructed The Ubiquitination Site (http://bioinformatics.psb.ugent.be/webtools/ubiquitin_viewer/), an online database that consolidates the identified ubiquitination sites and serves as a searchable knowledge base. The website with all its functions (Supplemental Figure 2) provides the user with a query box in which one or multiple Arabidopsis Genome Initiative locus identifiers can be entered. Ubiquitination sites on corresponding proteins are returned with information concerning the respective splice variant, the modified sequence, the sequence window, as well as the position of the site within the protein. Information on the MaxQuant score and delta score are provided as well. The site is linked to PLAZA (http://plaza.psb.ugent.be/), allowing the user to gain access to more ample information on the protein(s) in question.
OUTLOOK
We Are Ready for “Site”-Seeing in Plants
With the K-ε-GG antibody-based method and now also the ubiquitin COFRADIC established in plants, two complementary techniques are available for the plant science community to investigate protein ubiquitination at the site level. At the biochemical level, several differences should be considered between these methodologies. For instance, because USP2cc recognizes the last five amino acids of ubiquitin and no other ubiquitin-like proteins harbor this sequence at their C termini, this DUB is ubiquitin specific in plants, in contrast to the K-ε-GG antibodies that also recognize the epitope generated upon tryptic cleavage of other small protein modifiers, such as RELATED TO UBIQUITIN (Vierstra, 2012). Currently, it is difficult to assess to which extent this ambiguity is problematic because the degree of rubbylation in plants is unknown, but its functionality has been suggested to supersede that of neddylation, its mammalian PTM counterpart (Hakenjos et al., 2011; Mergner and Schwechheimer, 2014; Mergner et al., 2015).
Different techniques lead to different insights into ubiquitin linkage types. Often, linear or branched ubiquitin chains are formed by conjugating additional ubiquitin moieties to the N terminus or a lysine residue of the initial ubiquitin. These chains are reported to lead to different fates for the target protein. For example, Lys-48-linked chains commonly trigger degradation by the 26S proteasome and chains linked via Lys-63 play a role in DNA repair (Pickart and Fushman, 2004). Hence, the identification of specific linkage types on the target protein can serve as an important hint for the function of a particular ubiquitination event. By means of a TAP approach, several ubiquitination sites could be identified on ubiquitin itself, thereby revealing the types of ubiquitin branches present in the sample, together with their relative abundance (Kim et al., 2013). However, in our data set, only Lys-48 linker chains were retrieved, possibly due to a biochemistry-associated bias. Digestion of acetylated ubiquitin by trypsin (i.e., ArgC digestion type) will generate peptides probably too long or too short to have ideal properties for their identification via mass spectrometry. Although not analyzed in plant ubiquitination studies, the K-ε-GG antibodies should, in principle, not be confronted with this problem and could therefore provide a deeper insight into this aspect of ubiquitin biology.
Ubiquitination beyond Lysine Residues
Another difference between the antibody-based method and the ubiquitin COFRADIC approach is the manner in which the sites are targeted for enrichment. As the antibodies are raised against the K-ε-GG footprint, exclusively lysine ubiquitination is identified. In contrast, USP2cc recognizes ubiquitin on the target protein, independently of the affected residue, thereby rendering the identification of ubiquitination on other residues theoretically possible. Taking this into account, we searched the spectra for proof of alternative ubiquitination types. One possibility was the ubiquitination on protein N termini that, so far, has only been described in non-plant species (Bloom et al., 2003; Ciechanover and Ben-Saadon, 2004; Scaglione et al., 2013; Stes et al., 2014; Vittal et al., 2015). We identified 16 proteins that carry ubiquitin on the utmost N-terminal amino acid, including BRASSINAZOLE RESISTANT1 (BZR1), a key transcription factor involved in control of brassinosteroid-responsive genes (He et al., 2002; Wang et al., 2002). BZR1 is reported to be regulated via various PTMs, including ubiquitination (Gampala et al., 2007; Wang et al., 2013). Moreover, N-terminal ubiquitination was detected on two ubiquitin-conjugating enzyme variants (UEVs), i.e., proteins related to ubiquitin-conjugating (UBC) (E2) enzymes, but lacking a catalytic cysteine. UEVs act in a heterodimeric complex together with the UBC35 or UBC36 to trigger Lys-63-mediated ubiquitination (Tatham et al., 2013). Remarkably, the human Ubc13-UEV1 complex was shown to mediate Lys-63 ubiquitination of its target protein SUMO, only when SUMO is N-terminally ubiquitinated by the E2 ligase Ube2w (Tatham et al., 2013). Based on our data, it is tempting to speculate that UEV proteins are themselves regulated by N-terminal ubiquitination. This hypothesis is further supported by the ability of human Ube2w to mediate ubiquitin conjugation to its own N terminus (Tatham et al., 2013). Together, these results illustrate the power of the ubiquitin COFRADIC as a less biased technique for identification of different ubiquitination types. Nevertheless, for the time being, the protocol allows only detection of ubiquitin sites on lysine and N-terminal residues, because of the currently applied N-acylation steps.
To Be or Not to Be Ubiquitinated
Despite the improved reliability through the use of TAP methods, the estimation of false-positive identifications (i.e., the false discovery rate [FDR] intrinsic to the experimental procedure) is currently missing from most plant ubiquitination profiling literature. To reach the standards of the mammalian PTM field, we recommend reporting the FDR in plant ubiquitinome studies so that the reader can objectively assess the reliability of the methodology and the produced data. In the COFRADIC approach, false positives could arise during the chemical N-acetylation step applied to block all free amine groups: A low acetylation efficiency could potentially leave free lysines and protein N termini that later would be falsely detected as ubiquitination sites. We therefore tested the acetylation efficiency in two separate manners. Initially, the resistance to endoproteinase LysC digestion of N-acetylated protein extracts was checked by SDS-PAGE. Indeed, the chemical modification steps efficiently rendered proteins resistant to LysC digestion (Supplemental Figure 3), indicative of highly efficient N-acetylation. Subsequently, we set up a control experiment in Arabidopsis cell cultures, during which the complete COFRADIC protocol was followed, but without the USP2cc step. As a result, only five ubiquitin sites were found (Supplemental Data Set 1), demonstrating the very high N-acetylation efficiency and an FDR < 0.01% confirming our initial results. As such, the previously reported FDR of 6.7% in Jurkat cells is outperformed (Stes et al., 2014), possibly due to the newly introduced chloroform/methanol precipitation step early in the sample preparation process. Upon protein precipitation, all the endogenous DUBs are immediately and definitively put out of action.
Ubiquitination Hot Spots?
Protein ubiquitination often appears not to be restricted to a single lysine residue, but rather to occur in a specific protein region that might be considered as a “ubiquitination hot spot.” Hence, multiple lysine-to-arginine mutations need to be considered when the ubiquitination status of a particular protein is studied (Bish and Myers, 2007; Leitner et al., 2012). The “ubiquitination hot spot” hypothesis is supported by our observation that on the protein sequences with more than one detected UBC site, 33% of these sites could be found within a sequence window of 10 amino acids from each other. Additionally, this observation is also reflected in the high overlap between the two COFRADIC experiments at the protein level (87%), in contrast to the low overlap at the site level (35%). To fully understand the relative importance of exact sites compared with that of hot spots, a comprehensive mapping of the ubiquitinome in Arabidopsis will be necessary. With this issue in mind, it should be underlined that exact site determination remains, until now, the most reliable way of identifying a protein as being ubiquitinated.
SUMMARY
In contrast to the previous proteome-wide studies that offered insight into the Arabidopsis ubiquitinome on the protein level, two novel technologies are now available that enable ubiquitination mapping at the site level: K-ε-GG antibodies and ubiquitin COFRADIC. Both approaches greatly increase the ubiquitination profiling resolution in plants, moving toward comprehensive ubiquitin site mapping in a quantitative manner. With a large body of plant research currently focusing on E3 ligases (Ni et al., 2014; Song et al., 2014; Kinoshita et al., 2015), both technologies provide suitable solutions for the identification of their targets by the differential analysis of wild-type and mutant plants. An enhanced knowledge of their substrates will certainly lead to significant advances in the understanding of the biochemical mechanisms and cellular processes governed by protein ubiquitination.
METHODS
Ubiquitin COFRADIC
Arabidopsis thaliana (accession Columbia-0) cell suspension cultures (PSB-D) were maintained as described previously (Van Leene et al., 2007) and grown simultaneously in two biological replicates. After 3 weeks of growth under continuous light, 3 g of fresh weight material was harvested, flash-frozen in liquid nitrogen, and manually ground into a fine powder with pestle and mortar. The material was resuspended in 10 mL of homogenization buffer, containing 290 mM sucrose, 50 mM sodium phosphate buffer (pH 8), and 25 mM EDTA in milliQ water, vigorously agitated for resuspension, and five times sonicated on ice with a 1-cm probe for 10 s to disrupt cell walls. Samples were centrifuged at 1500g for 15 min at 4°C to remove debris. Supernatants were collected and a methanol/chloroform precipitation was performed by addition of 3:1:4 parts of methanol, chloroform, and water. Samples were centrifuged for 10 min at 5000g (room temperature). The upper (aqueous) phase was removed and 4 volumes of methanol were added onto the interface and bottom phase remaining in each tube to give rise to a precipitation.
Pellets were washed three times with acetone and resuspended in 4 M guanidinium hydrochloride in 50 mM sodium phosphate buffer (pH 8). Ubiquitin COFRADIC was done as described previously (Stes et al., 2014). To reduce the liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis time, peptide fractions eluting 15 min apart were pooled, dried, and redissolved in 15 μL of 2% (v/v) acetonitrile with 0.1% (v/v) trifluoroacetic acid (TFA). In total, 60 samples were analyzed via LC-MS/MS on an Ultimate 3000 RSLC nano-LC (Thermo Fisher Scientific) in-line connected to a Q Exactive mass spectrometer (Thermo Fisher Scientific). Settings and machine configurations were as described (Stes et al., 2014). MS/MS spectra were searched with MaxQuant 1.4.1.2. Andromeda search engine against The Arabidopsis Information Resource (TAIR10_pep_20101214 containing 27,416 protein-coding genes) database with MaxQuant software (version 1.4.0.3); the precursor mass tolerance was set to 20 ppm for the first search (used for nonlinear mass recalibration) and to 4.5 ppm for the main search. ArgC was selected as enzyme setting because cleavage after lysine residues was obstructed by acylation of their side chains. Cleavages between arginine and proline residues and up to one missed cleavage were allowed. Methionine oxidation and carbamidomethylation of cysteines were searched as fixed modifications, whereas N-terminal protein acetylation, lysine acetylation, N-terminal pyroglutamate, Lys-ε-Gly, and Gly on protein N termini were set as variable modifications. The FDR for peptide, protein, and site identification was set to 1% and the minimum peptide length to 7. The minimum score threshold for both modified and unmodified peptides was set to 30. All mass spectrometry proteomics data have been deposited in the ProteomeXchange Consortium via the PRIDE partner repository with the PRIDE accession PXD002297. All annotated spectra are also downloadable in PDF format from The Ubiquitination Site website.
Generation of HIS6-Tagged UBQ10 Arabidopsis Plants
The UBQ10 (AT4G05320) genomic open reading frame (ORF) was cloned in two consecutive PCR steps. Due to the number of multiple UBQ-encoding genes in Arabidopsis, the 5′-TCTGATTTACAGATGCAGATCTTTG-3′ forward and 5′-GAAACATTGAACTTCTTAAGCATAAC-3′ reverse primers used for the first amplification step included parts of the 5′- and 3′-untranslated regions, respectively, to convey specificity to the UBQ10 ORF. A second PCR with the gene-specific 5′-ATGCAGATCTTTGTTAAGACTCTC-3′ forward and 5′-TTAAGCATAACAGAGACGAGATTTA-3′ reverse primers was used to eliminate the flanking 5′- and 3′-untranslated region sequences. The UBQ10 ORF was recombined after the HBH TAP-tag42 and inserted into the pKNTAP destination vector, generating an N-terminally tagged UBQ, under control of the constitutive cauliflower mosaic virus 35S promoter by means of the Gateway recombination (Invitrogen). Arabidopsis plants were stably transformed with the Agrobacterium tumefaciens-mediated floral dip method. Plants were selected by kanamycin resistance.
Affinity Enrichment of HIS6-Tagged UBQ10
For the affinity enrichment study, 35S:HIS6UBQ10 lines were grown, in three biological repeats, on half-strength Murashige and Skoog agar plates with 1% (w/v) sucrose for 10 d. The material was flash-frozen in liquid nitrogen and manually ground into a fine powder with pestle and mortar. Protein extraction was done as described above. Pellets were resuspended in 50 mM triethyl ammonium bicarbonate (TEAB) buffer. Ni-NTA beads were resuspended in 50 μL of the same buffer and spun down at 2700g for 2 min at 4°C. This washing step was repeated three times. Samples were incubated in the presence of the beads under constant mixing for 2 h at 4°C, spun down at 2700g for 2 min at 4°C, and the supernatant was removed. Beads were washed twice with 500 μL of ice-cold TEAB buffer prior to a 4-h on-bead digestion (37°C) with 1/100 (w/w) trypsin. Samples were centrifuged at 2700g for 2 min at 4°C. The supernatant was transferred prior to a second 4-h digestion with 1 µg trypsin. Samples were dried under vacuum and redissolved in 15 μL of 2% (v/v) acetonitrile with 0.1% (v/v) TFA loading solvent for LC-MS/MS. Settings and machine configurations were as described (Nelissen et al., 2015). MS/MS spectra were searched with the MaxQuant Andromeda search engine against The Arabidopsis Information Resource (TAIR10_pep_20101214 containing 27,416 protein-coding genes) database with MaxQuant software (version 1.4.0.3), with a precursor mass tolerance set to 20 ppm for the first search (used for nonlinear mass recalibration) and set to 4.5 ppm for the main search. Trypsin/P was selected as enzyme setting and up to one missed cleavage was allowed. Variable modifications were set for methionine oxidation, pyro-glutamate formation of N-terminal glutamine, acetylation of the protein N terminus, and the diglycine motif on lysines. The FDR for peptide, protein, and site identification was set to 1% and the minimum peptide length was set to 7. The minimum score threshold for both modified and unmodified peptides was set to 30.
USP2cc Activity Assay
Plant proteins from cell cultures were extracted as for the affinity enrichment protocol described above. For samples destined for acetylation, NHS-acetate was added to samples in two consecutive steps to reach a final concentration of 40 mM followed by a 2-h incubation at 30°C. Subsequently, 80 mM of glycine was added to quench the residual NHS-acetate. For samples destined for USP2cc treatment, the enzyme was added in a 1/100 (w/w) ratio and samples were incubated overnight at 37°C. For protein immunoblotting, an anti-HIS antibody (RGS-HIS; Qiagen) was used as primary antibody, followed by an anti-mouse fluorophore secondary antibody (800 nm) (LI-COR) and visualized on an Odyssey infrared imaging system (LI-COR). Due to interference of protein acetylation with the antibody activity, a classical loading control could not be used. To overcome this problem, equal loading was assessed with a Silver Stain Kit (Pierce) according to the manufacturer’s instructions.
Cell-Free GRXS17 Degradation Assay
Total protein extracts were prepared by resuspending ground tissue of 10-d-old wild-type Arabidopsis seedlings grown under continuous light at 21°C in cold extraction buffer (25 mM Tris-HCl, pH 7.5, 10 mM NaCl, 10 mM MgCl2, 4 mM phenylmethanesulfonyl fluoride, 5 mM DTT, and 10 mM adenosine triphosphate) at a ratio of 1 g tissue/mL extraction buffer and centrifuged twice at 12,000g for 15 min. To test the GRXS17 (AT4G04950) stability, 50 µM MG132 (Boston Biochem) or 1% (v/v) DMSO as a control was added to the total protein extract. Each reaction was incubated at room temperature and samples were harvested at the indicated time points. To stop the reaction, SDS sample buffer was added, followed by boiling for 10 min before SDS-PAGE. When necessary, 500 ng of Escherichia coli recombinant protein GRXS17:V5-HIS was added to the total protein extract just before the incubation at room temperature.
Construction of the Ubiquitination Site Website
The web interface was built with Cake PHP (version 2.6.1) and mysql as back-end. The PLAZA platform (Proost et al., 2015) was used as reference for the various possible names and identifiers associated with genes and loci. By means of the PLAZA platform, we could assert that searching for noncanonical identifiers could still occur, of which the results are reported by a JavaScript Object Notation-based application programming interface.
Supplemental Data
Supplemental Figure 1. Degradation of GRXS17 by the 26S proteasome.
Supplemental Figure 2. The Ubiquitination Site website page.
Supplemental Figure 3. Acetylation efficiency test.
Supplemental Table 1. Identified ubiquitination sites on transcription factors and UPS members.
Supplemental Data Set 1. List of detected modified peptides through the use of ubiquitin COFRADIC.
Supplemental Data Set 2. List of proteins detected through affinity purification of HIS6-tagged UBQ10 in 35S:HIS6UBQ10-expressing Arabidopsis lines.
Supplementary Material
Acknowledgments
We thank Martine De Cock for help in preparing the manuscript. A.W. is the recipient of a VIB International PhD program fellowship. E.S. and L.P. are Postdoctoral Fellows of the Research Foundation-Flanders. S.I. is a Postdoctoral Fellow of the Belgian Science Policy Office (BELSPO).
AUTHOR CONTRIBUTIONS
A.W., E.S., A.N.D., L.P., L.D.V., A.G., I.D.S., S.G., and K.G. designed research. A.W., E.S., N.C., S.I., A.N.D., E.T., J.H., and F.C. performed research. A.W. and E.S. analyzed data. A.W., E.S., I.D.S., S.G., and K.G. wrote the article.
References
- Banfield M.J. (2015). Perturbation of host ubiquitin systems by plant pathogen/pest effector proteins. Cell. Microbiol. 17: 18–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barneche F., Malapeira J., Mas P. (2014). The impact of chromatin dynamics on plant light responses and circadian clock function. J. Exp. Bot. 65: 2895–2913. [DOI] [PubMed] [Google Scholar]
- Bish R.A., Myers M.P. (2007). Werner helicase-interacting protein 1 binds polyubiquitin via its zinc finger domain. J. Biol. Chem. 282: 23184–23193. [DOI] [PubMed] [Google Scholar]
- Bloom J., Amador V., Bartolini F., DeMartino G., Pagano M. (2003). Proteasome-mediated degradation of p21 via N-terminal ubiquitinylation. Cell 115: 71–82. [DOI] [PubMed] [Google Scholar]
- Bratzel F., López-Torrejón G., Koch M., Del Pozo J.C., Calonje M. (2010). Keeping cell identity in Arabidopsis requires PRC1 RING-finger homologs that catalyze H2A monoubiquitination. Curr. Biol. 20: 1853–1859. [DOI] [PubMed] [Google Scholar]
- Chapman E.J., Estelle M. (2009). Mechanism of auxin-regulated gene expression in plants. Annu. Rev. Genet. 43: 265–285. [DOI] [PubMed] [Google Scholar]
- Cheng N.-H., Liu J.-Z., Liu X., Wu Q., Thompson S.M., Lin J., Chang J., Whitham S.A., Park S., Cohen J.D., Hirschi K.D. (2011). Arabidopsis monothiol glutaredoxin, AtGRXS17, is critical for temperature-dependent postembryonic growth and development via modulating auxin response. J. Biol. Chem. 286: 20398–20406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciechanover A., Ben-Saadon R. (2004). N-terminal ubiquitination: more protein substrates join in. Trends Cell Biol. 14: 103–106. [DOI] [PubMed] [Google Scholar]
- Cuéllar Pérez A., Goossens A. (2013). Jasmonate signalling: a copycat of auxin signalling? Plant Cell Environ. 36: 2071–2084. [DOI] [PubMed] [Google Scholar]
- Cui F., Liu L., Zhao Q., Zhang Z., Li Q., Lin B., Wu Y., Tang S., Xie Q. (2012). Arabidopsis ubiquitin conjugase UBC32 is an ERAD component that functions in brassinosteroid-mediated salt stress tolerance. Plant Cell 24: 233–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Z., Oses-Prieto J.A., Kutschera U., Tseng T.-S., Hao L., Burlingame A.L., Wang Z.-Y., Briggs W.R. (2014). Blue light-induced proteomic changes in etiolated Arabidopsis seedlings. J. Proteome Res. 13: 2524–2533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deruyffelaere C., Bouchez I., Morin H., Guillot A., Miquel M., Froissard M., Chardot T., D’Andrea S. (2015). Ubiquitin-mediated proteasomal degradation of oleosins is involved in oil body mobilization during post-germinative seedling growth in Arabidopsis. Plant Cell Physiol. 56: 1374–1387. [DOI] [PubMed] [Google Scholar]
- Furniss J.J., Spoel S.H. (2015). Cullin-RING ubiquitin ligases in salicylic acid-mediated plant immune signaling. Front. Plant Sci. 6: 154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gampala S.S., et al. (2007). An essential role for 14-3-3 proteins in brassinosteroid signal transduction in Arabidopsis. Dev. Cell 13: 177–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu C., Kolodziejek I., Misas-Villamil J., Shindo T., Colby T., Verdoes M., Richau K.H., Schmidt J., Overkleeft H.S., van der Hoorn R.A.L. (2010). Proteasome activity profiling: a simple, robust and versatile method revealing subunit-selective inhibitors and cytoplasmic, defense-induced proteasome activities. Plant J. 62: 160–170. [DOI] [PubMed] [Google Scholar]
- Guo H., Li L., Aluru M., Aluru S., Yin Y. (2013). Mechanisms and networks for brassinosteroid regulated gene expression. Curr. Opin. Plant Biol. 16: 545–553. [DOI] [PubMed] [Google Scholar]
- Guseman J.M., Hellmuth A., Lanctot A., Feldman T.P., Moss B.L., Klavins E., Calderón Villalobos L.I.A., Nemhauser J.L. (2015). Auxin-induced degradation dynamics set the pace for lateral root development. Development 142: 905–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakenjos J.P., Richter R., Dohmann E.M.N., Katsiarimpa A., Isono E., Schwechheimer C. (2011). MLN4924 is an efficient inhibitor of NEDD8 conjugation in plants. Plant Physiol. 156: 527–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J.-X., Gendron J.M., Yang Y., Li J., Wang Z.-Y. (2002). The GSK3-like kinase BIN2 phosphorylates and destabilizes BZR1, a positive regulator of the brassinosteroid signaling pathway in Arabidopsis. Proc. Natl. Acad. Sci. USA 99: 10185–10190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heride C., Urbé S., Clague M.J. (2014). Ubiquitin code assembly and disassembly. Curr. Biol. 24: R215–R220. [DOI] [PubMed] [Google Scholar]
- Hua Z., Vierstra R.D. (2011). The cullin-RING ubiquitin-protein ligases. Annu. Rev. Plant Biol. 62: 299–334. [DOI] [PubMed] [Google Scholar]
- Hua Z., Zou C., Shiu S.-H., Vierstra R.D. (2011). Phylogenetic comparison of F-Box (FBX) gene superfamily within the plant kingdom reveals divergent evolutionary histories indicative of genomic drift. PLoS One 6: e16219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igawa T., Fujiwara M., Takahashi H., Sawasaki T., Endo Y., Seki M., Shinozaki K., Fukao Y., Yanagawa Y. (2009). Isolation and identification of ubiquitin-related proteins from Arabidopsis seedlings. J. Exp. Bot. 60: 3067–3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasai K., Takano J., Fujiwara T. (2014). Analysis of endocytosis and ubiquitination of the BOR1 transporter. Methods Mol. Biol. 1209: 203–217. [DOI] [PubMed] [Google Scholar]
- Kerkeb L., Mukherjee I., Chatterjee I., Lahner B., Salt D.E., Connolly E.L. (2008). Iron-induced turnover of the Arabidopsis IRON-REGULATED TRANSPORTER1 metal transporter requires lysine residues. Plant Physiol. 146: 1964–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D.-Y., Scalf M., Smith L.M., Vierstra R.D. (2013). Advanced proteomic analyses yield a deep catalog of ubiquitylation targets in Arabidopsis. Plant Cell 25: 1523–1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim W., Bennett E.J., Huttlin E.L., Guo A., Li J., Possemato A., Sowa M.E., Rad R., Rush J., Comb M.J., Harper J.W., Gygi S.P. (2011). Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol. Cell 44: 325–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita A., et al. (2015). A plant U-box protein, PUB4, regulates asymmetric cell division and cell proliferation in the root meristem. Development 142: 444–453. [DOI] [PubMed] [Google Scholar]
- Komander D., Rape M. (2012). The ubiquitin code. Annu. Rev. Biochem. 81: 203–229. [DOI] [PubMed] [Google Scholar]
- Leitner J., Petrášek J., Tomanov K., Retzer K., Pařezová M., Korbei B., Bachmair A., Zažímalová E., Luschnig C. (2012). Lysine63-linked ubiquitylation of PIN2 auxin carrier protein governs hormonally controlled adaptation of Arabidopsis root growth. Proc. Natl. Acad. Sci. USA 109: 8322–8327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X.-M., et al. (2015). Natural alleles of a proteasome α2 subunit gene contribute to thermotolerance and adaptation of African rice. Nat. Genet. 47: 827–833. [DOI] [PubMed] [Google Scholar]
- Liu H., Stone S.L. (2011). E3 ubiquitin ligases and abscisic acid signaling. Plant Signal. Behav. 6: 344–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Stone S.L. (2013). Cytoplasmic degradation of the Arabidopsis transcription factor ABSCISIC ACID INSENSITIVE 5 is mediated by the RING-type E3 ligase KEEP ON GOING. J. Biol. Chem. 288: 20267–20279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzano C., Abraham Z., López-Torrejón G., Del Pozo J.C. (2008). Identification of ubiquitinated proteins in Arabidopsis. Plant Mol. Biol. 68: 145–158. [DOI] [PubMed] [Google Scholar]
- Maor R., Jones A., Nühse T.S., Studholme D.J., Peck S.C., Shirasu K. (2007). Multidimensional protein identification technology (MudPIT) analysis of ubiquitinated proteins in plants. Mol. Cell. Proteomics 6: 601–610. [DOI] [PubMed] [Google Scholar]
- Marino D., Peeters N., Rivas S. (2012). Ubiquitination during plant immune signaling. Plant Physiol. 160: 15–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín-Trillo M., Cubas P. (2010). TCP genes: a family snapshot ten years later. Trends Plant Sci. 15: 31–39. [DOI] [PubMed] [Google Scholar]
- Martins S., Dohmann E.M.N., Cayrel A., Johnson A., Fischer W., Pojer F., Satiat-Jeunemaître B., Jaillais Y., Chory J., Geldner N., Vert G. (2015). Internalization and vacuolar targeting of the brassinosteroid hormone receptor BRI1 are regulated by ubiquitination. Nat. Commun. 6: 6151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mergner J., Heinzlmeir S., Kuster B., Schwechheimer C. (2015). DENEDDYLASE1 deconjugates NEDD8 from non-cullin protein substrates in Arabidopsis thaliana. Plant Cell 27: 741–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mergner J., Schwechheimer C. (2014). The NEDD8 modification pathway in plants. Front. Plant Sci. 5: 103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelissen H., et al. (2015). Dynamical changes in ANGUSTIFOLIA3 complex composition reveal a growth regulatory mechanism in the maize leaf. Plant Cell 27: 1605–1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni W., Xu S.-L., Tepperman J.M., Stanley D.J., Maltby D.A., Gross J.D., Burlingame A.L., Wang Z.-Y., Quail P.H. (2014). A mutually assured destruction mechanism attenuates light signaling in Arabidopsis. Science 344: 1160–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patra B., Pattanaik S., Yuan L. (2013). Ubiquitin protein ligase 3 mediates the proteasomal degradation of GLABROUS 3 and ENHANCER OF GLABROUS 3, regulators of trichome development and flavonoid biosynthesis in Arabidopsis. Plant J. 74: 435–447. [DOI] [PubMed] [Google Scholar]
- Pauwels L., et al. (2015). The RING E3 ligase KEEP ON GOING modulates JASMONATE ZIM-DOMAIN12 stability. Plant Physiol. 169: 1405–1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peralta D.A., Araya A., Nardi C.F., Busi M.V., Gomez-Casati D.F. (2013). Characterization of the Arabidopsis thaliana E3 ubiquitin-ligase AtSINAL7 and identification of the ubiquitination sites. PLoS One 8: e73104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickart C.M., Fushman D. (2004). Polyubiquitin chains: polymeric protein signals. Curr. Opin. Chem. Biol. 8: 610–616. [DOI] [PubMed] [Google Scholar]
- Polyn S., Willems A., De Veylder L. (2015). Cell cycle entry, maintenance, and exit during plant development. Curr. Opin. Plant Biol. 23: 1–7. [DOI] [PubMed] [Google Scholar]
- Proost S., Van Bel M., Vaneechoutte D., Van de Peer Y., Inzé D., Mueller-Roeber B., Vandepoele K. (2015). PLAZA 3.0: an access point for plant comparative genomics. Nucleic Acids Res. 43: D974–D981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renatus M., Parrado S.G., D’Arcy A., Eidhoff U., Gerhartz B., Hassiepen U., Pierrat B., Riedl R., Vinzenz D., Worpenberg S., Kroemer M. (2006). Structural basis of ubiquitin recognition by the deubiquitinating protease USP2. Structure 14: 1293–1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saracco S.A., Hansson M., Scalf M., Walker J.M., Smith L.M., Vierstra R.D. (2009). Tandem affinity purification and mass spectrometric analysis of ubiquitylated proteins in Arabidopsis. Plant J. 59: 344–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scaglione K.M., Basrur V., Ashraf N.S., Konen J.R., Elenitoba-Johnson K.S.J., Todi S.V., Paulson H.L. (2013). The ubiquitin-conjugating enzyme (E2) Ube2w ubiquitinates the N terminus of substrates. J. Biol. Chem. 288: 18784–18788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo P.J., Mas P. (2014). Multiple layers of posttranslational regulation refine circadian clock activity in Arabidopsis. Plant Cell 26: 79–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahnawaz M., Thapa A., Park I.-S. (2007). Stable activity of a deubiquitylating enzyme (Usp2-cc) in the presence of high concentrations of urea and its application to purify aggregation-prone peptides. Biochem. Biophys. Res. Commun. 359: 801–805. [DOI] [PubMed] [Google Scholar]
- Soboleva T.A., Baker R.T. (2004). Deubiquitinating enzymes: their functions and substrate specificity. Curr. Protein Pept. Sci. 5: 191–200. [DOI] [PubMed] [Google Scholar]
- Song Y.H., Estrada D.A., Johnson R.S., Kim S.K., Lee S.Y., MacCoss M.J., Imaizumi T. (2014). Distinct roles of FKF1, GIGANTEA, and ZEITLUPE proteins in the regulation of CONSTANS stability in Arabidopsis photoperiodic flowering. Proc. Natl. Acad. Sci. USA 111: 17672–17677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sridhar V.V., Kapoor A., Zhang K., Zhu J., Zhou T., Hasegawa P.M., Bressan R.A., Zhu J.-K. (2007). Control of DNA methylation and heterochromatic silencing by histone H2B deubiquitination. Nature 447: 735–738. [DOI] [PubMed] [Google Scholar]
- Stes E., Laga M., Walton A., Samyn N., Timmerman E., De Smet I., Goormachtig S., Gevaert K. (2014). A COFRADIC protocol to study protein ubiquitination. J. Proteome Res. 13: 3107–3113. [DOI] [PubMed] [Google Scholar]
- Strzalka W., Bartnicki F., Pels K., Jakubowska A., Tsurimoto T., Tanaka K. (2013). RAD5a ubiquitin ligase is involved in ubiquitination of Arabidopsis thaliana proliferating cell nuclear antigen. J. Exp. Bot. 64: 859–869. [DOI] [PubMed] [Google Scholar]
- Svozil J., Hirsch-Hoffmann M., Dudler R., Gruissem W., Baerenfaller K. (2014). Protein abundance changes and ubiquitylation targets identified after inhibition of the proteasome with syringolin A. Mol. Cell. Proteomics 13: 1523–1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatham M.H., Plechanovová A., Jaffray E.G., Salmen H., Hay R.T. (2013). Ube2W conjugates ubiquitin to α-amino groups of protein N-termini. Biochem. J. 453: 137–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udeshi N.D., Svinkina T., Mertins P., Kuhn E., Mani D.R., Qiao J.W., Carr S.A. (2013). Refined preparation and use of anti-diglycine remnant (K-ε-GG) antibody enables routine quantification of 10,000s of ubiquitination sites in single proteomics experiments. Mol. Cell. Proteomics 12: 825–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Leene J., et al. (2007). A tandem affinity purification-based technology platform to study the cell cycle interactome in Arabidopsis thaliana. Mol. Cell. Proteomics 6: 1226–1238. [DOI] [PubMed] [Google Scholar]
- Vierstra R.D. (2012). The expanding universe of ubiquitin and ubiquitin-like modifiers. Plant Physiol. 160: 2–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vittal V., Shi L., Wenzel D.M., Scaglione K.M., Duncan E.D., Basrur V., Elenitoba-Johnson K.S.J., Baker D., Paulson H.L., Brzovic P.S., Klevit R.E. (2015). Intrinsic disorder drives N-terminal ubiquitination by Ube2w. Nat. Chem. Biol. 11: 83–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner S.A., Beli P., Weinert B.T., Nielsen M.L., Cox J., Mann M., Choudhary C. (2011). A proteome-wide, quantitative survey of in vivo ubiquitylation sites reveals widespread regulatory roles. Mol. Cell. Proteomics 10: M111.013284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., et al. (2013). Identification of BZR1-interacting proteins as potential components of the brassinosteroid signaling pathway in Arabidopsis through tandem affinity purification. Mol. Cell. Proteomics 12: 3653–3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z.-Y., Nakano T., Gendron J., He J., Chen M., Vafeados D., Yang Y., Fujioka S., Yoshida S., Asami T., Chory J. (2002). Nuclear-localized BZR1 mediates brassinosteroid-induced growth and feedback suppression of brassinosteroid biosynthesis. Dev. Cell 2: 505–513. [DOI] [PubMed] [Google Scholar]
- Wasternack C., Hause B. (2013). Jasmonates: biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Ann. Bot. (Lond.) 111: 1021–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X., Kang H., Liu W., Wang G.-L. (2015). Comprehensive profiling of the rice ubiquitome reveals the significance of lysine ubiquitination in young leaves. J. Proteome Res. 14: 2017–2025. [DOI] [PubMed] [Google Scholar]
- Yan J., Li H., Li S., Yao R., Deng H., Xie Q., Xie D. (2013). The Arabidopsis F-box protein CORONATINE INSENSITIVE1 is stabilized by SCFCOI1 and degraded via the 26S proteasome pathway. Plant Cell 25: 486–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K., Sridhar V.V., Zhu J., Kapoor A., Zhu J.-K. (2007). Distinctive core histone post-translational modification patterns in Arabidopsis thaliana. PLoS One 2: e1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo Z.-C., Meng Y.-Y., Yu X.-H., Zhang Z.-L., Feng D.-S., Sun S.-F., Liu B., Lin C.-T. (2012). A study of the blue-light-dependent phosphorylation, degradation, and photobody formation of Arabidopsis CRY2. Mol. Plant 5: 726–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.