Powdery mildew attack induces arrest of mitochondria at sites of attempted invasion, peripheral accumulation of the myrosinase PEN2, and redox imbalances in the matrix of the immobilized mitochondria.
Abstract
The atypical myrosinase PENETRATION2 (PEN2) is required for broad-spectrum invasion resistance to filamentous plant pathogens. Previous localization studies suggested PEN2-GFP association with peroxisomes. Here, we show that PEN2 is a tail-anchored protein with dual-membrane targeting to peroxisomes and mitochondria and that PEN2 has the capacity to form homo-oligomer complexes. We demonstrate pathogen-induced recruitment and immobilization of mitochondrial subpopulations at sites of attempted fungal invasion and show that mitochondrial arrest is accompanied by peripheral accumulation of GFP-tagged PEN2. PEN2 substrate production by the cytochrome P450 monooxygenase CYP81F2 is localized to the surface of the endoplasmic reticulum, which focally reorganizes close to the immobilized mitochondria. Exclusive targeting of PEN2 to the outer membrane of mitochondria complements the pen2 mutant phenotype, corroborating the functional importance of the mitochondrial PEN2 protein subpool for controlled local production of PEN2 hydrolysis products at subcellular plant-microbe interaction domains. Moreover, live-cell imaging shows that mitochondria arrested at these domains exhibit a pathogen-induced redox imbalance, which may lead to the production of intracellular signals.
INTRODUCTION
Plants have evolved a complex multilayered defense machinery, which is effective in reducing the invasion and colonization by nonadapted pathogens. Preformed physical and chemical barriers, such as rigid cell walls and phytoanticipins, constitute the first obstacle for potential intruders (Thordal-Christensen, 2003). If a pathogen is able to overcome this constitutive layer of defense, it is exposed to recognition by plasma membrane-anchored pattern recognition receptors, which detect conserved microbe-associated molecular patterns (MAMPs) and trigger active defense responses (Dodds and Rathjen, 2010; Macho and Zipfel, 2014). In the case of a compatible interaction, these immune responses are part of the basal resistance, which limits the severity of the plant disease.
Powdery mildew fungi are obligate biotrophic pathogens that depend on living host cells and cause disease in a wide range of mono- and dicotyledonous plants, including economically important crops. They are characterized by ectoparasitic host plant colonization that is restricted to the epidermal cell layer (Lipka et al., 2008; Micali et al., 2008). To obtain nutrients required for growth and reproduction, powdery mildew conidiospores germinate and form appressoria, which penetrate the epidermal cell wall. Upon successful invasion, haustoria are established which function both as feeding organs and sites of effector secretion (Micali et al., 2008). Concerted and cell-autonomous plant defense responses aim to terminate fungal entry attempts. Typically, active defense is accompanied by a profound reorganization of the cellular infrastructure, including focal rearrangements of cytoskeletal transport routes, local accumulation of cytoplasm, translocation of the nucleus and other organelles toward the penetration site, and a directed deposition of callose and cell wall components leading to papilla formation and cell wall reinforcement (Lipka et al., 2008; Hückelhoven and Panstruga, 2011).
Previously, PENETRATION1 (PEN1), PEN2, and PEN3 were identified as molecular components of powdery mildew entry control that are involved in secretion, activation, and transport of defense-related molecules to the site of attempted penetration (Collins et al., 2003; Lipka et al., 2005; Stein et al., 2006; Kwon et al., 2008; Bednarek et al., 2009). All PEN proteins show focal accumulation patterns at powdery mildew interaction sites, and mutations in the corresponding Arabidopsis thaliana genes result in enhanced entry success of the nonadapted powdery mildew Blumeria graminis f. sp hordei (Bgh), a pathogen of the monocot crop barley (Hordeum vulgare). PEN1 is a plasma membrane-localized SNARE (soluble N-ethylmaleimide-sensitive factor attachment protein receptor) protein (Collins et al., 2003), which is required for localized formation of ternary SNARE complexes (Kwon et al., 2008), exosome secretion, and timely appearance of papilla at fungal entry sites (Meyer et al., 2009; Nielsen and Thordal-Christensen, 2013).
A second, PEN1-independent pathogen entry control mechanism is maintained by the concerted action of PEN2 and PEN3. PEN2 encodes a family 1 β-glycoside hydrolase that is associated with the periphery of mobile spherical organelles that were originally identified as peroxisomes (Lipka et al., 2005). PEN2 is involved in the pathogen-induced enzymatic activation of indole glucosinolates (IGs) (Lipka et al., 2005; Bednarek et al., 2009; Clay et al., 2009). Glucosinolates are secondary metabolites produced by members of the order Brassicales that constitute preformed repellents. To minimize the risk of self-intoxication, glucosinolates and their corresponding glycoside hydrolases are typically separated in different cellular compartments. Thus, glucosinolates are often stored in the vacuole as inactive glycoside precursors of biologically active mustard oils (Halkier and Gershenzon, 2006). Only in cases of tissue disruption, e.g., upon insect feeding or necrotrophic pathogen attack, are the substrate and enzyme brought into spatial proximity and release the toxic reaction products (Halkier and Gershenzon, 2006). In contrast to this classical mustard oil bomb, the pathogen-induced PEN2-dependent hydrolysis of IGs has been suggested to represent an active cell-autonomous defense mechanism (Lipka et al., 2005; Stein et al., 2006; Bednarek et al., 2009; Clay et al., 2009), which requires a complex and fine-tuned subcellular sequence of nontoxic substrate generation, hydrolytic activation, and targeting of the toxic product(s) at the intruder.
Genetic analysis revealed that the ATP binding cassette (ABC) transporter protein PEN3 acts in the same pathway as PEN2 because, in contrast to pen1 pen3 double mutants, pen2 pen3 did not show enhanced Bgh penetration rates in comparison to the single mutants (Lipka et al., 2005; Stein et al., 2006). Notably, pen3 mutants exhibit an enhanced, salicylic acid-dependent resistance against the adapted powdery mildew Golovinomyces cichoracearum, which might be the consequence of an intracellular overaccumulation of potentially toxic IG hydrolysis products (Stein et al., 2006). Analyses with functional GFP fusions revealed a MAMP-induced recruitment of PEN3 into plasma membrane microdomains at plant-microbe interaction sites (Underwood and Somerville, 2013). Together, these data support a scenario in which PEN3 mediates the ATP-dependent transport of glucosinolates that were enzymatically activated by PEN2 and are potentially toxic for invading pathogens (Stein et al., 2006). PEN2 was shown to hydrolyze indol-3-ylmethylglucosinolate (I3G) and 4-methoxyindol-3-ylmethylglucosinolate (4MI3G) (Bednarek et al., 2009) and to be required for production of 4-O-β-d-glucosyl-indol-3-yl formamide (Lu et al., 2015). Deglucosylated precursor molecules of the latter have recently been proposed to represent the critical PEN3 transport substrate in powdery mildew entry control (Lu et al., 2015). As I3G and 4MI3G contain S-glycosidic bonds, PEN2 represents a thioglucosidase (myrosinase) but was designated as an atypical myrosinase due to an unusual amino acid composition within the catalytic center. Importantly, only unidentified hydrolysis products of 4-substituted I3G derivatives, which are synthesized by the cytochrome P450 monooxygenase CYP81F2, seem to be relevant for PEN2-dependent penetration resistance (Bednarek et al., 2009; Clay et al., 2009). Consistent with this hypothesis, cyp81f2 mutants show a pen2-like reduction in penetration resistance, which is not further decreased in pen2 cyp81F2 double mutants (Bednarek et al., 2009). Notably, the core biosynthetic pathway of glucosinolates requires oxidation reactions that are catalyzed by CYP79 and CYP83 monooxygenases at the endoplasmic reticulum (ER)-cytosol interface (Yan and Chen, 2007; Grubb and Abel, 2006). So far, our knowledge about the subcellular localization of CYP81F2 (i.e., the site of PEN2 substrate production) and our understanding of the cellular mechanisms that orchestrate localized PEN2-dependent hydrolysis of 4-substituted I3G derivatives and subsequent energy-dependent discharge of toxic products by PEN3 clearly lags behind.
In this study, we show that the atypical myrosinase PEN2 is both localized to peroxisomes and mitochondria. We demonstrate that PEN2 forms homodimers and pathogen-induced multimeric aggregates that are exclusively associated with the periphery of a subset of mitochondria, which become immobilized at sites of attempted pathogen entry. Confocal live-cell imaging analyses of glutathione redox potential suggest that the immobilized mitochondrial subpopulation shows an oxidative shift. Moreover, we show that exclusive targeting of functional PEN2 to the outer membrane of mitochondria is sufficient to restore its function in penetration resistance. Furthermore, we show pathogen-induced and cell-autonomous expression of CYP81F2, its localization to the ER, and a tight association of the ER and mitochondria at sites of fungal invasion attempts, thus bringing together all the elements of a highly localized toxin synthesis, activation and export machinery.
RESULTS
PEN2 Shows Pathogen-Induced Accumulation in the Periphery of Immobilized Subcellular Compartments
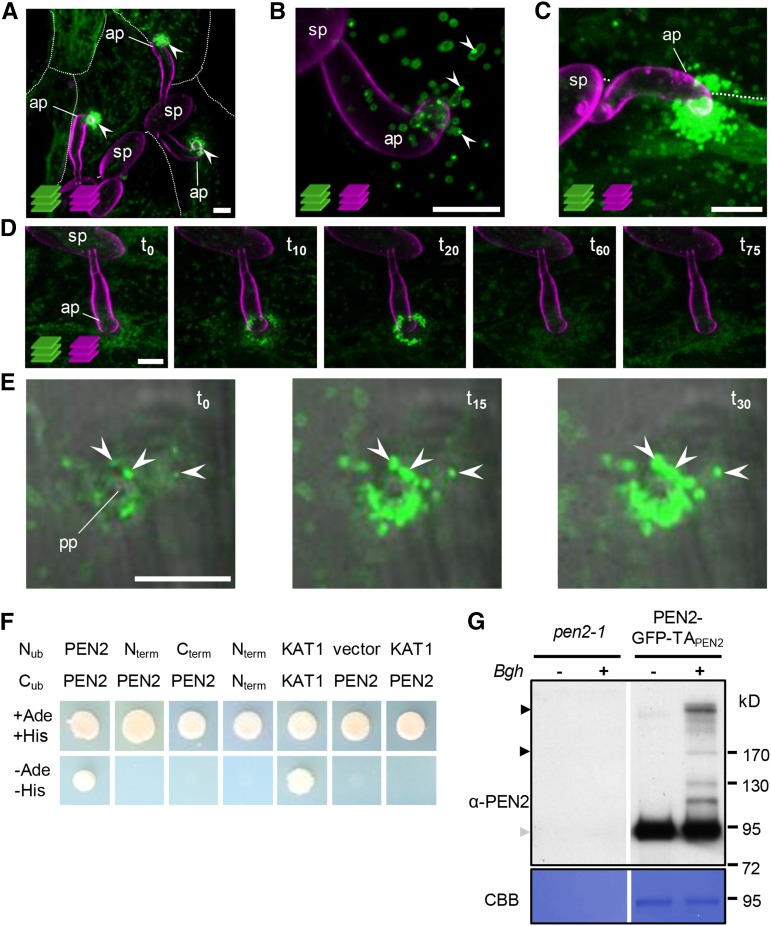
We previously showed that GFP-tagged PEN2 is associated with mobile subcellular compartments that focally accumulate at sites of attempted fungal invasion (Lipka et al., 2005). It is important to note that it was necessary to insert GFP between the globular enzymatic part of the protein and its C-terminal extension (PEN2-GFP-TAPEN2; Supplemental Figure 1A) to produce a functional protein with the capacity to complement the pen2 mutant phenotype (Lipka et al., 2005) and that we used endogenous 5′ regulatory sequences to control expression in the stable transgenic lines in our earlier work (Lipka et al., 2005) and throughout this study. Here, we analyzed the subcellular behavior of functional PEN2-GFP-TAPEN2 in more detail. To this end, we inoculated pen2 mutant plants expressing PEN2-GFP-TAPEN2 with Bgh conidiospores and analyzed the subcellular localization of PEN2-GFP-TAPEN2 by confocal laser scanning microscopy (CLSM). These analyses confirmed our earlier observations, but also revealed distinct levels of fluorescence intensity at individual plant-microbe interaction sites 20 h postinoculation (hpi) (Figure 1A). Closer examination of interaction sites at this time point showed that some sites were characterized by only a few fluorescent structures with comparatively low overall intensities of peripheral GFP fluorescence and occasional peripheral foci with elevated fluorescence intensity (Figure 1B). Other sites had high numbers of individual subcellular compartments that showed highly localized accumulation with intense overall GFP fluorescence (Figure 1C). It is important to note that powdery mildew inoculations do not result in fully synchronized conidiospore germination and cell wall penetration attempts. Thus, we hypothesized that the observed differences reflect distinct stages of attempted invasion and therefore analyzed incipient penetration sites by time-lapse CLSM. Indeed, these experiments confirmed an increase in GFP fluorescence around the tips of fungal appressoria (Figure 1D, t0 to t20). Moreover, higher magnification revealed that a subpopulation of GFP-tagged compartments become immobilized at Bgh invasion sites, whereas other compartments retained their mobility (Figure 1E; Supplemental Movie 1). The arrested compartments in particular developed peripheral patches of intense GFP fluorescence. Within 20 to 30 min, the hyperfluorescence foci expand, intensify, and ultimately decorate the entire compartment. We frequently observed disappearance of intense fluorescence at later time points (Figure 1D, t60 and t75), which may reflect a successful defense response and early termination of fungal invasion.
Figure 1.
PEN2 Exhibits Pathogen-Induced Accumulation at Sites of Attempted Invasion.
(A) to (D) Maximum z-projections of CLSM images of Bgh-challenged epidermal leaf cells expressing PEN2-GFP-TAPEN2 under control of the endogenous promoter in the pen2-1 background. It is important to note that powdery mildew inoculations do not result in fully synchronized conidiospore germination and cell wall penetration attempts. Fungal structures were stained with FM4-64.
(A) Distinct levels of PEN2-GFP-TAPEN2 fluorescence intensity at three individual plant-microbe interaction sites at 20 hpi (arrowheads). White lines indicate cell borders (z-stack 26 µm).
(B) Close-up view of an individual interaction site at 20 hpi shows foci of elevated PEN2-GFP-TAPEN2 fluorescence intensity at the periphery of a membrane compartment (arrowheads; z-stack 12 µm).
(C) Another interaction site at 20 hpi shows subcellular compartments with an intense overall PEN2-GFP-TAPEN2 fluorescence accumulating below the fungal appressorium (z-stack 27 µm).
(D) Time-lapse CLSM of an individual interaction site starting at 20 hpi demonstrates both accumulation of intense PEN2-GFP-TAPEN2 fluorescence at fungal invasion sites and subsequent disappearance within 75 min over time (z-stack 21 µm).
(E) Higher magnification of single CLSM images show mobile GFP-tagged compartments and an immobilized subpopulation developing peripheral patches of higher PEN2-GFP-TAPEN2 fluorescence from 19 to 19.5 hpi with Bgh (arrowheads). ap, appressorium; pp, penetration peg; sp, spore; t, time (min) with t0 = 19 hpi. Bars = 10 µm.
(F) Split-ubiquitin yeast two-hybrid assays indicate homomerization capacity only of full-length PEN2 protein. Neither the globular part (Nterm) nor the C-terminal part (Cterm) of PEN2 interact with the full-length protein and enable yeast growth on selective medium (-Ade, -His). KAT1 dimerization was used as a positive control.
(G) Immunoblot analyses of immunoprecipitated eluate from Bgh challenged and unchallenged pen2-1 mutant and PEN2-GFP-TAPEN2 plants using GFP-Trap coupled to agarose beads confirm specific pull-down of GFP fusion proteins and pathogen-induced PEN2 homomerization 18 hpi with Bgh. Equal volumes of each eluate were subjected to SDS-PAGE. Black arrowheads indicate protein dimers/oligomers, whereas the gray arrowhead marks monomers. Experiments were repeated three times with similar results. CBB, Coomassie Brilliant Blue.
Pathogen Attack Induces Formation of PEN2 Dimers and Oligomers of Higher Order
PEN2 orthologs tend to form homo- or hetero-oligomers and large heteromeric protein aggregates (Kittur et al., 2007; Yamada et al., 2011). Oligomerization and aggregate formation have been shown to control hydrolytic activity (Kwak et al., 2009; Yamada et al., 2011). To analyze the homomerization capacity of PEN2, we first conducted split-ubiquitin yeast two-hybrid assays (Figure 1F). These experiments demonstrated that only full-length PEN2 can homodimerize, whereas the N-terminal globular part (M1-D498) or the C-terminal extension of PEN2 (Q496-N560) alone fail to interact with full-length PEN2. To validate homomer formation of full-length PEN2 in planta, we performed blue native polyacrylamide gel electrophoresis with extracts of wild-type, pen2 mutant, and PEN2-GFP-TAPEN2-producing plants that were challenged with Bgh or not. Subsequent immunoblot analyses using a PEN2-specific antibody confirmed in planta formation of PEN2 homodimers in unchallenged and challenged wild-type and PEN2-GFP-TAPEN2-producing plants (Supplemental Figure 1B). However, these experiments neither provided evidence for larger PEN2 homo-oligomers nor did they support pathogen-induced homo-oligomerization. It is important to note that the detection sensitivity may be limited when analyzing cell-autonomous pathogen-induced responses of individual Arabidopsis epidermal pavement cells in crude plant leaf extracts. To increase detection sensitivity, we immunoprecipitated PEN2-GFP-TAPEN2 protein complexes from extracts of control and Bgh-challenged plants using GFP-Trap coupled to agarose beads (Supplemental Figure 1C). Immunoblot analyses with α-PEN2 confirmed the specific pull-down of GFP fusion proteins and a pathogen-induced formation of SDS-resistant PEN2 dimers and oligomers of higher order (Figure 1G; Supplemental Figure 1D).
The C Terminus of PEN2 Has Tail Anchor Function
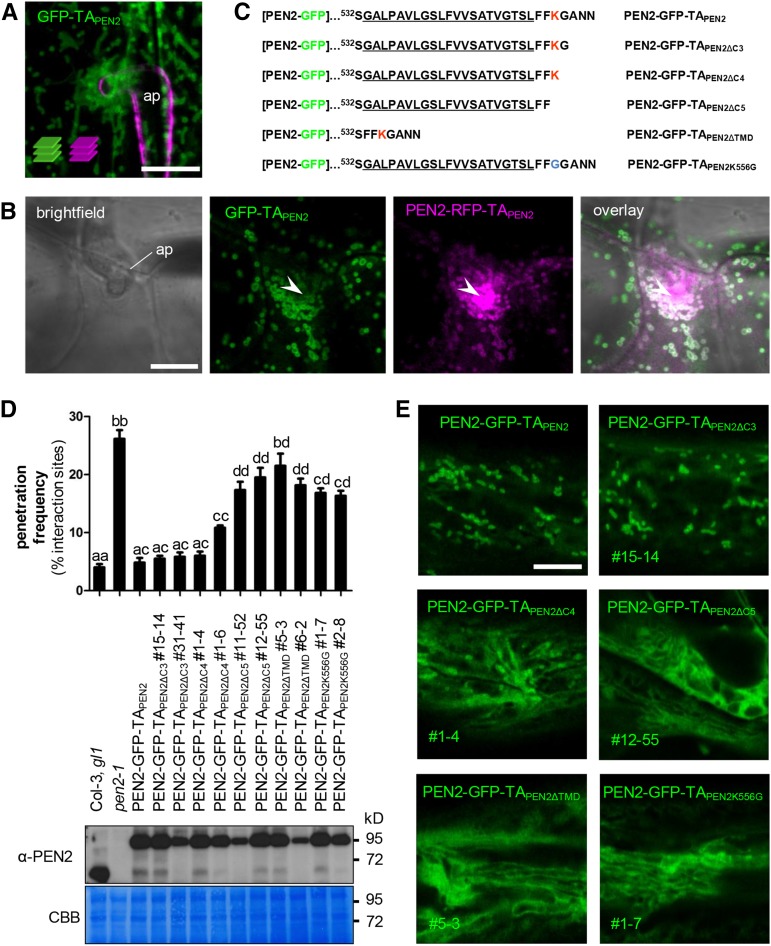
The unique C-terminal extension of PEN2 was previously shown to be required for functionality in plant innate immunity (Lipka et al., 2005), suggesting that it may control enzymatic activity and/or subcellular localization. Indeed, the C terminus of PEN2 harbors a stretch of hydrophobic amino acids with a predicted α-helical structure (Lipka et al., 2005), which resembles classical C-terminal tail anchors (TAs). TAs are characterized by a single transmembrane domain (TMD), which functions as a posttranslational targeting sequence and mediates insertion in intracellular membranes, thereby positioning the large N-terminal protein portion toward the cytosol (Abell and Mullen, 2011). To analyze whether or not the C terminus of PEN2 has TA function, we generated transgenic pen2-1 mutant plants that express GFP fused to PEN2’s C-terminal extension (Q496-N560; GFP-TAPEN2; Supplemental Figure 1A) under control of the native PEN2 promoter sequence. CLSM revealed that GFP-TAPEN2 copied the subcellular localization pattern of full-length PEN2-GFP-TAPEN2 and tagged the periphery of subcellular membrane compartments that were either mobile or focally accumulating at plant-microbe interaction sites (Figures 2A and 2B). This supports the idea that the C terminus has TA function and is sufficient to determine the subcellular localization of the PEN2 protein. However, we were unable to detect hyperfluorescence typical for full-length PEN2-GFP-TAPEN2 decorating the immobilized organelle subpopulation at plant-microbe interaction domains. This suggests that the N-terminal globular part is required for pathogen-induced protein aggregate formation and also rules out the possibility that aggregate formation and hyperfluorescence are the result of unspecific fluorescence protein interactions. Experiments in which we stably coexpressed GFP-TAPEN2 and full-length PEN2-RFP-TAPEN2 support this conclusion, as we were able to detect a clear overall colocalization pattern of GFP and RFP signals, but only hyperfluorescent signals of RFP at sites of attempted fungal invasion (Figure 2B). To learn more about TA functionality and its contribution to subcellular protein localization behavior, we generated a series of PEN2 TA deletion constructs (Figure 2C) that we used for pen2-1 mutant complementation analyses and localization by CLSM. Constructs in which the last three C-terminal amino acids were deleted (PEN2-GFP-TAPEN2∆C3) retained full complementation capacity (Figure 2D) and showed a subcellular localization pattern that was indistinguishable from PEN2-GFP-TAPEN2 (Figure 2E). Similarly, constructs missing the last four amino acids (PEN2-GFP-TAPEN2∆C4) showed a dosage-dependent capacity to complement the compromised pathogen entry control phenotype of the pen2 mutant background (Figure 2D). However, CLSM utilizing the corresponding transgenic Arabidopsis lines suggested a comparatively lower level of fluorescence intensity associated with intracellular membrane compartments and higher fluorescence intensity in the cytosol (Figure 2E). A discrete peripheral membrane association was impossible to detect with constructs that lack the last five C-terminal amino acids (PEN2-GFP-TAPEN2∆C5) or the entire predicted C-terminal TMD (PEN2-GFP-TAPEN2∆TMD) or when the positively charged amino acid lysine (fifth from last) is exchanged with a neutral glycine residue (PEN2-GFP-TAPEN2K556G) (Figure 2E). Instead, CLSM shows diffuse distribution in the cytoplasm and, possibly, reticulate ER localization. These mistargeted proteins are not able to fully restore the compromised pathogen entry defense of pen2 mutant plants (Figure 2D), underpinning the functional importance of the membrane compartment-associated PEN2 protein subpool.
Figure 2.
The C-Terminal Extension of PEN2 Is Important for Subcellular Localization and Functionality.
(A) Maximum z-projection of CLSM images from a transgenic leaf epidermal cell expressing GFP-TAPEN2 reveal localization of the truncated protein to the periphery of subcellular membrane compartments 24 hpi with Bgh. Fungal structures were stained with FM4-64 (z-stack 22 µm). ap, appressorium. Bar = 10 µm.
(B) Single CLSM image of transgenic leaf epidermal cells coexpressing GFP-TAPEN2 and full-length PEN2-RFP-TAPEN2 shows colocalization of both proteins in the same membrane compartment, but GFP-TAPEN2 does not contribute to PEN2-RFP-TAPEN2 hyperfluorescence underneath the fungal appressorium 22 hpi with Bgh. Arrowheads point to PEN2-RFP-TAPEN2 hyperfluorescence at the site of attempted fungal invasion. ap, appressorium. Bar = 10 µm.
(C) Protein sequence of the C-terminal extension of different PEN2 TA deletion constructs. The predicted transmembrane domain is underlined. Positively charged amino acid lysine is marked in red. Number indicates protein position in the native PEN2 protein.
(D) and (E) pen2-1 mutant complementation analyses and CLSM of PEN2-GFP TA deletion constructs indicate full complementation capacity only occurs for proteins that associate with membrane compartments.
(D) Upper part: Frequency of invasive growth at Bgh interaction sites 72 hpi on Col-3, gl1, pen2-1, and plants expressing full-length PEN2-GFP-TAPEN2 or different deletion proteins. Different letters indicate significantly different classes (99% confidence intervals) determined by one-way ANOVA with Tukey’s post test. Depicted results represent two biological replicates with 100 interaction sites analyzed on three different leaves each. Error bars indicate se of the mean. Lower part: Corresponding immunoblot analyses of all plant lines using 30 µg protein and the PEN2-specific antibody show different levels of protein expression. CBB, Coomassie Brilliant Blue staining. Experiments were repeated three times with similar results.
(E) Single CLSM images of transgenic plants show subcellular localization of PEN2-GFP-TAPEN2 and different PEN2-GFP TA deletion constructs. The deletion proteins PEN2-GFP-TAPEN2∆C5, PEN2-GFP-TAPEN2∆TMD, and PEN2-GFP-TAPEN2K556G show diffuse distribution in the cytoplasm and, possibly, reticulate ER localization. Bar = 10 µm.
PEN2 Is Dually Targeted to Peroxisomes and Mitochondria, but Only Mitochondria Are Immobilized at Plant-Microbe Interaction Sites
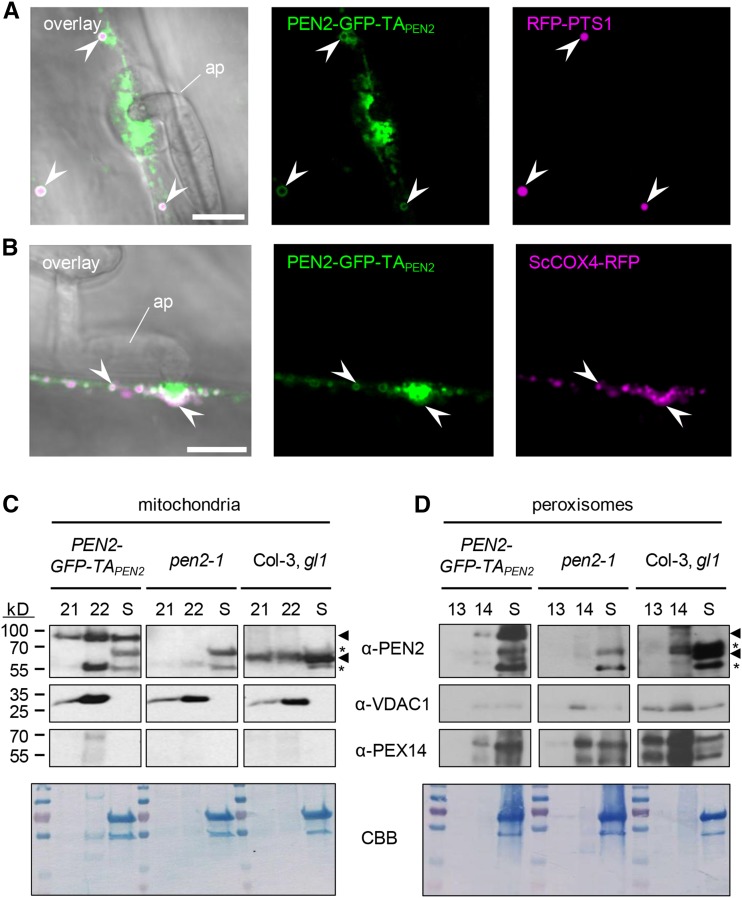
Previous experiments, in which uninfected Arabidopsis plants expressing PEN2-GFP-TAPEN2 were used for colocalization studies with transiently expressed fluorescent organelle markers, suggested that a cellular subpool of PEN2 is associated with the periphery of peroxisomes (Lipka et al., 2005). Here, we generated stable transgenic pen2-1 mutant lines coexpressing PEN2-GFP-TAPEN2 together with the peroxisomal matrix marker RFP-PEROXISOME TARGETING SIGNAL1 (RFP-PTS1) (Nelson et al., 2007). CLSM corroborated our earlier finding that PEN2-GFP-TAPEN2 is associated with the peroxisome membrane (Figure 3A). However, we did not observe stable immobilization of RFP-tagged peroxisomes at plant microbe-interaction sites or PEN2 aggregate formation in the periphery of RFP-tagged peroxisomes. Although CLSM time-lapse experiments showed transport to interaction sites, accumulation of peroxisomes was only transient (Supplemental Figures 2A and 2B). Notably, PEN2-GFP-TAPEN2 aggregate formation was restricted to other, smaller membrane compartments, which were immobilized in the immediate proximity of the tips of fungal appressoria. Similar analyses with stable transgenic pen2-1 mutant lines coexpressing PEN2-GFP-TAPEN2 and the mitochondrial matrix marker ScCOX4-RFP (Saccharomyces cerevisiae CYTOCHROME C OXIDASE IV fused to RFP) (Nelson et al., 2007) suggested that these organelles are mitochondria (Figure 3B). Indeed, our time-course analyses showed that PEN2-GFP-TAPEN2 also resides in the periphery of mobile mitochondria, that a subpopulation of mitochondria is focally positioned and locked at pathogen entry sites, and that PEN2 aggregate formation occurs at these arrested mitochondria (Supplemental Figures 2C and 2D). To assess mitochondrial mobility and PEN2-GFP-TAPEN2 fluorescence intensities in a quantitative manner, we systematically analyzed eight time-lapse CLSM data sets of pen2-1 mutant plants coexpressing PEN2-GFP-TAPEN2 and ScCOX4-RFP at 24 hpi with Bgh (Supplemental Figure 3 and Supplemental Movie 2). First, we generated pseudo-color-coded probability maps for the presence of individual mitochondria over the observation time using ScCOX4-RFP as a mitochondrial marker (Supplemental Figure 3A), with the rationale of low mobility to correlate with high probability of presence and vice versa. These analyses clearly indicated low mobility at the site of attempted fungal penetration (red, high probability of presence/low mobility; blue, low probability of presence/high mobility). High values around the attempted penetration site are in agreement with visually apparent immobilization of mitochondria in the corresponding video sequences (Supplemental Movie 2). In a separate approach, we measured the average probability for the presence of individual mitochondria as a function of distance from the site of attempted invasion (Supplemental Figure 3B), which confirmed high values close to the penetration site tailing off at a distance of 10 to 15 µm. We next used the same time series to quantify the average fluorescence intensities of PEN2-GFP-TAPEN2 and the mitochondrial marker ScCOX4-RFP per single mitochondrion as a function of distance to the plant-microbe interaction site (Supplemental Figure 3C). GFP fluorescence intensity per mitochondrion was consistently increased close to the penetration site in the individual time series and the corresponding average analysis (Supplemental Movie 2 and Supplemental Figure 3D). By contrast, the RFP signal was relatively constant per mitochondrion and showed no distance-related trend. Together, these quantitative data support the idea of PEN2 aggregate formation at arrested mitochondria close to sites of attempted powdery mildew invasion. Inspection of wild-type Arabidopsis-Bgh interaction sites with transmission electron microscopy supports focal accumulation of mitochondria together with ER, Golgi stacks, and secretory vesicles beneath pathogen-induced cell wall appositions (Supplemental Figure 2E). Moreover, CLSM with Bgh-challenged wild-type and pen2-1 mutant plants expressing ScCOX4-RFP alone substantiate the above findings and exclude that transgenic expression of PEN2-GFP-TAPEN2 or lack of PEN2 wild-type protein affect the subcellular behavior of mitochondria (Supplemental Figures 2F and 2G). To verify the dual organelle localization of PEN2 to the membrane of peroxisomes and the outer mitochondrial membrane (OMM), we employed a biochemical approach. To this end, we isolated peroxisomes and mitochondria from leaves of wild-type, pen2-1 mutant, and PEN2-GFP-TAPEN2-producing plants using previously published methods (Kruft et al., 2001; Reumann et al., 2007; Sweetlove et al., 2007). Comparative immunoblot experiments with specific antibodies for PEN2, the peroxisomal marker protein PEROXIN14 (PEX14) (Hayashi et al., 2000) and the OMM protein VOLTAGE-DEPENDENT ANION CHANNEL1 (VDAC1) (Tateda et al., 2011), showed an enrichment of mitochondria and peroxisomes, respectively, and support in vivo targeting of PEN2 to the OMM and the membrane of peroxisomes (Figures 3C and 3D).
Figure 3.
PEN2 Is Localized to the Periphery of Peroxisomes and Mitochondria.
(A) CLSM image shows association of PEN2-GFP-TAPEN2 with the membrane of RFP-PTS1-marked peroxisomes in double transgenic leaf epidermal cells 20 hpi with Bgh. Arrowheads point to the localization of PEN2-GFP-TAPEN2 in the periphery of peroxisomes. Bar = 10 µm.
(B) CLSM image of double transgenic leaf epidermal cells expressing PEN2-GFP-TAPEN2 and mitochondrial marker ScCOX4-RFP confirms association of PEN2 with the membrane of mitochondria 22 hpi with Bgh. Arrowheads point to colocalization of mitochondria with PEN2-GFP-TAPEN2 aggregates and to association of PEN2-GFP-TAPEN2 with the periphery of this organelle. ap, appressorium. Bar = 10 µm.
(C) and (D) Immunoblot analyses of mitochondria-enriched (C) and peroxisome-enriched (D) fractions from Col-3, gl1 wild-type, pen2-1 knockout, and transgenic pen2-1 mutant plants expressing PEN2-GFP-TAPEN2. Following chloroplast sedimentation, the supernatant (S) was enriched for mitochondria or peroxisomes using Percoll (fractions 21 and 22 of 24 1.5-mL fractions) or sucrose (fractions 13 and 14 of 14 1-mL fractions) density gradients, respectively. Using a PEN2-specific antibody allowed the detection of endogenous PEN2 (∼65 kD, lower arrowhead) in wild-type plants and the detection of PEN2-GFP-TAPEN2 (∼95 kD, upper arrowhead) in transgenic pen2-1 mutant plants producing PEN2-GFP-TAPEN2 in both mitochondria (C) and peroxisome (D) fractions. Asterisks indicate unspecific signals detected by α-PEN2. Antibodies against the mitochondrial marker protein VDAC1 and the peroxisome-specific PEX14 were used to observe enrichment of the respective organelles. Note that due to small sample size and low protein concentration (∼30 µg), the same mitochondria (C) or peroxisome (D) fraction containing blot was used for probing with the indicated antibodies (involving stripping and reprobing). The Coomassie Brilliant Blue (CBB) staining shows loading on the respective membrane.
Exclusive Targeting of Functional PEN2 to Outer Mitochondrial Membranes Mediates PEN2-Dependent Pathogen Entry Resistance
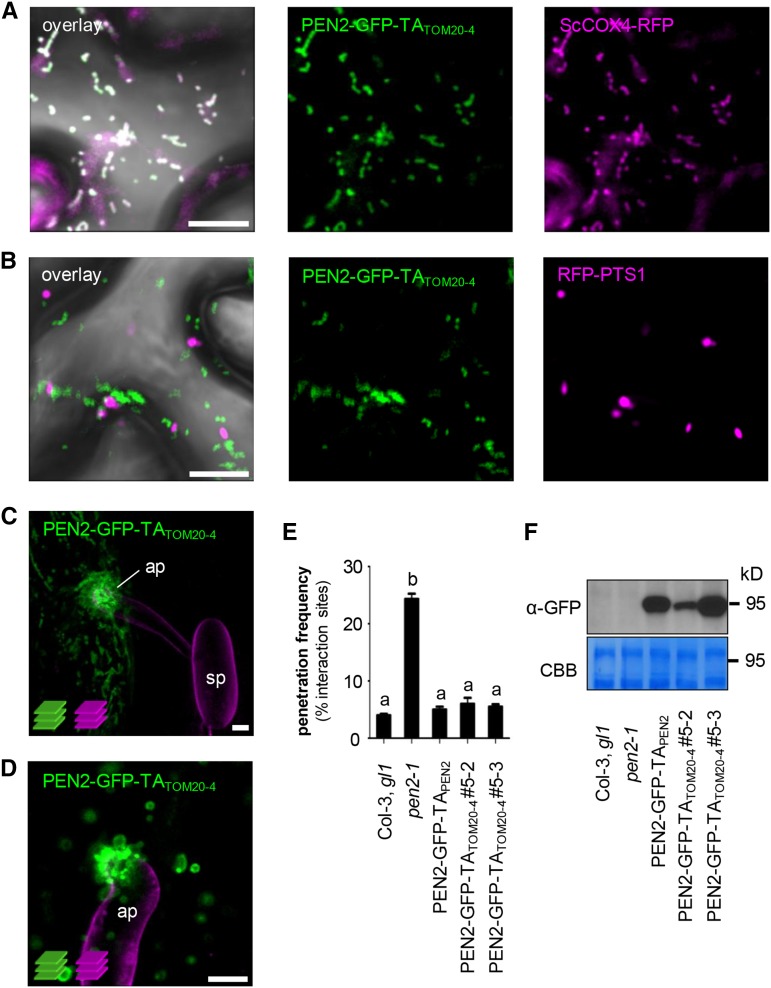
Next, we addressed the question of whether or not the mitochondrial PEN2 subpool alone is sufficient to mediate PEN2-dependent entry control. For this purpose, we looked for a plant protein TA that provides exclusive targeting to the OMM, which could be used to replace the promiscuous TA of PEN2 in transgenic complementation experiments. One TA that we considered likely to meet this criterion was the C terminus of Arabidopsis TRANSLOCASE OF OUTER MITOCHONDRIAL MEMBRANE 20-4 (TOM20-4), a subunit of the mitochondrial outer membrane import complex (Abell and Mullen, 2011). Indeed, in transgenic plants coexpressing RFP-tagged TOM20-4 (RFP-TOM20-4; Supplemental Figure 4A) and GFP/CFP-markers for either mitochondria (ScCOX4-GFP) or peroxisomes (CFP-PTS1), we detected specific labeling of mitochondria and not of peroxisomes (Supplemental Figures 4B and 4C). Moreover, when we analyzed transgenic plants coexpressing RFP-TOM20-4 and PEN2-GFP-TAPEN2 by CLSM we observed colocalization and noticed organelle accumulation and immobilization at pathogen interaction sites and peripheral PEN2 aggregate formation as before (Supplemental Figure 4D). As a next step, we made PEN2-GFP fusion constructs in which the genuine TA of PEN2 was replaced by the C-terminal TA of TOM20-4 (PEN2-GFP-TATOM20-4; Supplemental Figure 4A). CLSM analyses of Nicotiana benthamiana leaf epidermal cells transiently coexpressing either ScCOX4-RFP or RFP-PTS1 together with PEN2-GFP-TATOM20-4 confirmed specific targeting of the latter to the periphery of mitochondria (Figures 4A and 4B). Finally, we generated stably transformed pen2-1 mutant plants producing PEN2-GFP-TATOM20-4 under control of native 5′ regulatory sequences. Subsequent Bgh inoculation experiments corroborated mitochondrial recruitment to pathogen interaction sites and revealed aggregate formation of PEN2-GFP-TATOM20-4 at the periphery of immobilized mitochondria that was identical to the pattern observed for PEN2-GFP-TAPEN2 (Figures 4C and 4D). In addition, expression of PEN2-GFP-TATOM20-4 complemented the pen2 mutant phenotype in a dosage-dependent manner and had the capacity to fully restore pathogen entry control (Figures 4E and 4F). Together, our data suggest that the TA of TOM20-4 can functionally replace the genuine TA of PEN2 and that the mitochondrial PEN2 protein subpool alone is sufficient to provide PEN2-dependent penetration resistance against Bgh. We also aimed to generate analogous transgenic pen2 mutant plants producing GFP-tagged PEN2 protein that is exclusively localized tovperoxisomal membranes. To this end, we used the TAs of ASCORBATE PEROXIDASE3 (TAAPX3) or MONODEHYDROASCORBATE REDUCTASE4 (TAMDAR4), which were recently described to provide exclusive targeting to peroxisomal membranes (Lisenbee et al., 2005; Narendra et al., 2006). However, all transgenic lines generated with these two different TAs showed promiscuous localization to both peroxisomes and mitochondria. This precluded reciprocal experiments to analyze the contribution of the peroxisomal PEN2 protein subpool to pathogen entry control directly.
Figure 4.
The C-Terminal TA of TOM20-4 Can Functionally Replace PEN2’s TA for Proper Subcellular Localization and Functionality.
(A) and (B) CLSM images of N. benthamiana leaf epidermal cells, transiently coexpressing PEN2-GFP-TATOM20-4 and either RFP-tagged mitochondrial marker (A) or RFP-labeled peroxisomes (B), show that PEN2-GFP-TATOM20-4 is exclusively associated with mitochondria and not peroxisomes 3 d after Agrobacterium tumefaciens infiltration.
(C) and (D) Two examples of z-projections of CLSM images from transgenic leaf epidermal cells expressing PEN2-GFP-TATOM20-4 exhibit the same subcellular behavior and aggregate formation as PEN2-GFP-TAPEN2 20 hpi with Bgh. Fungal structures were stained with FM4-64 (z-stack 22 and 9 µm). ap, appressorium; sp, spore. Bars = 10 µm.
(E) Complementation analyses of pen2-1 mutant by PEN2-GFP-TATOM20-4 indicate full complementation capacity of the chimeric construct. Results were scored 72 hpi with Bgh. Different letters indicate significantly different classes (99% confidence intervals) determined by one-way ANOVA with Tukey’s post test. Depicted results represent two biological replicates with 100 interaction sites analyzed on three different leaves, each. Error bars indicate se of the mean.
(F) Corresponding immunoblot analyses of all plant lines using 30 µg protein extract and a GFP-specific antibody show different levels of protein expression. Experiments have been repeated three times with similar results. CBB, Coomassie Brilliant Blue staining.
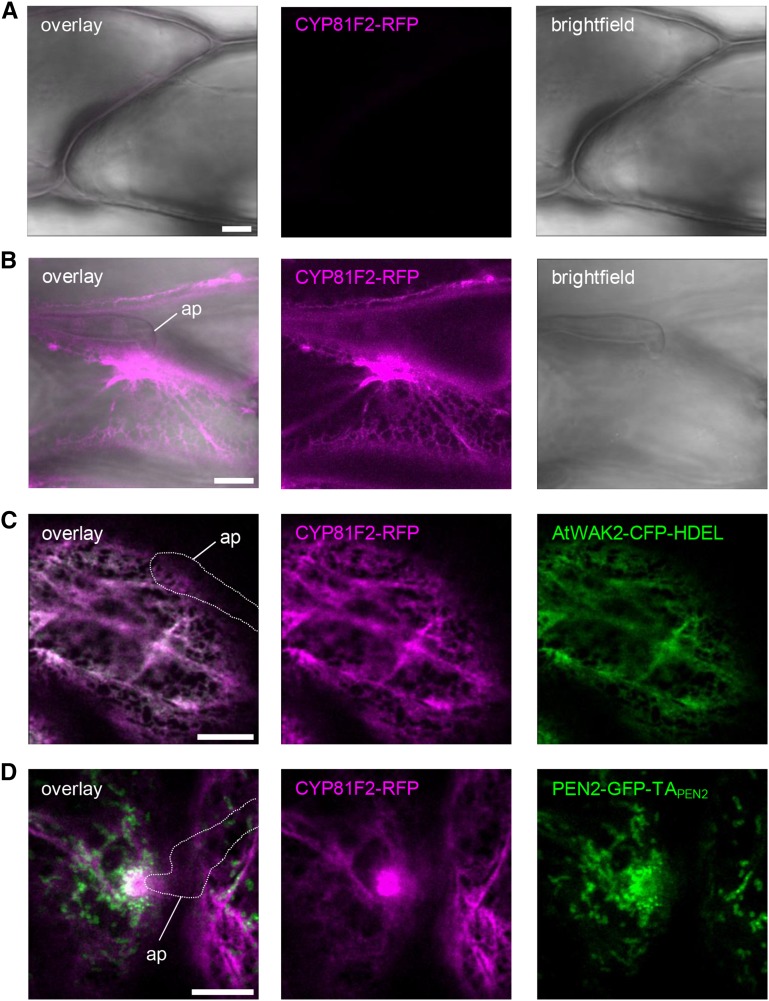
PEN2 Substrate Production Is Localized to the Surface of the ER, Which Focally Reorganizes toward Sites of Pathogen Attack
Recent work by Bednarek et al. (2009) and Clay et al. (2009) showed that the cytochrome P450 monooxygenase CYP81F2 is required for the pathogen-induced production of 4-substituted I3G substrates of PEN2 and subsequent resistance to powdery mildew entry. In general, metabolite production requires a highly coordinated compartmentalization of pathway enzymes in order to channel precursors to their target enzymes, optimize flux efficiency, and sequester pathway intermediates and products (Sweetlove and Fernie, 2013). This applies particularly to the production of toxic metabolites, which are potentially harmful for the host organism, as in the case of the PEN2 pathway products (Stein et al., 2006). To learn more about the subcellular site of PEN2 substrate production, we generated transgenic Arabidopsis plants expressing RFP-tagged CYP81F2 under the control of endogenous promoter sequences in the cyp81f2-2 mutant background. As CYP81F2 was predicted to contain an N-terminal membrane anchor (Pfalz et al., 2009), we decided to use a C-terminal RFP fusion construct (CYP81F2-RFP). The transgenic plants obtained show pathogen-induced expression of functional CYP81F2, as indicated by immunoblot analysis of leaf protein extracts and restoration of resistance to fungal entry (Supplemental Figures 5A to 5C). Cell-autonomous expression of CYP81F2-RFP in response to pathogen attack was confirmed by CLSM, as we observed strong RFP fluorescence only in epidermal cells that were subject to attempted fungal invasion (Figures 5A and 5B), suggesting that the enhanced CYP81F2-RFP detected in our immunoblot analysis results from a dramatic pathogen-induced production in these individual epidermal cells. In these cells, RFP fluorescence showed a reticulate distribution and focal accumulation pattern at plant-microbe interaction sites (Figure 5B), suggesting a potential association of CYP81F2 with the ER and its recruitment to the contact zone between the plant and the invader. To test this hypothesis, we generated stable transgenic cyp81f2 mutant plants coexpressing functional CYP81F2-RFP together with the ER-marker AtWAK2-CFP-HDEL (Arabidopsis WALL-ASSOCIATED KINASE2 at the N terminus and ER retention signal His-Asp-Glu-Leu at its C terminus; Nelson et al., 2007). CLSM experiments showed a clear colocalization (Figure 5C), supporting the idea that CYP81F2, like most other eukaryotic cytochrome P450 monooxygenases (Grubb and Abel, 2006), is N-terminally anchored in the ER membrane positioning its catalytic site facing the cytoplasm. Further CLSM analyses in which we analyzed pathogen-challenged transgenic Arabidopsis plants coexpressing CYP81F2-RFP and PEN2-GFP-TAPEN2 confirmed a network-like RFP distribution pattern coalescing into a lamellar sheath of bright RFP fluorescence at sites of attempted fungal ingress (Figure 5D). These areas of strong RFP fluorescence are in close proximity to the immobilized mitochondria that are characterized by PEN2-GFP-TAPEN2 aggregate formation (Figure 5D).
Figure 5.
The ER-Localized Fusion Protein CYP81F2-RFP Focally Accumulates at Plant-Microbe Interaction Sites.
(A) and (B) CLSM images from transgenic leaf epidermal cells expressing CYP81F2-RFP under control of the endogenous promoter show no detectable RFP fluorescence in unchallenged epidermal cells (A) but strong fluorescence particularly under the infection site in pathogen treated ones 20 hpi with Bgh (B).
(C) CLSM image of transgenic cells coexpressing CYP81F2-RFP and CFP-labeled ER lumen demonstrates colocalization of both proteins to the ER 18 hpi with Bgh.
(D) CLSM analysis of transgenic leaf epidermal cells coexpressing CYP81F2-RFP and PEN2-GFP-TAPEN2 indicates close proximity of ER accumulation and PEN2 aggregation at plant-microbe interaction sites 20 hpi with Bgh.
White line indicates fungal appressorium. ap, appressorium. Bars = 10 µm.
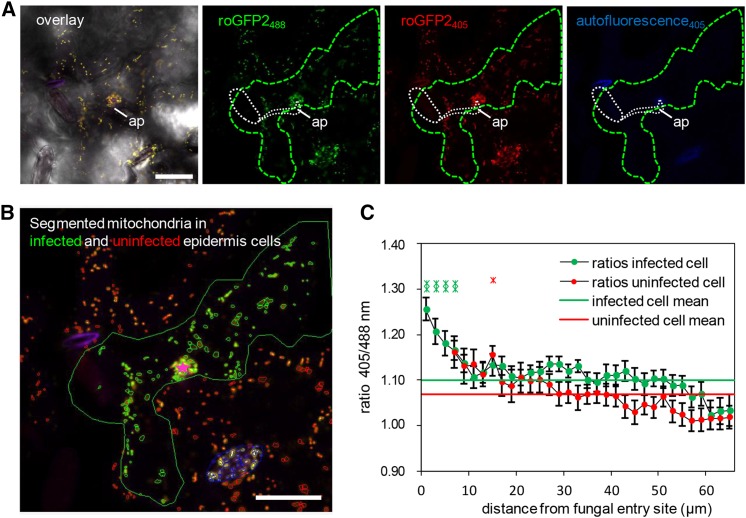
Mitochondria in Close Proximity to Sites of Pathogen Attack Exhibit Redox Imbalances
It is important to note that the dynamic cellular responses induced by microbial attack such as cytoskeletal rearrangements, motor protein-dependent organelle and vesicle transport, as well as SNARE and ABC transporter-mediated secretion are highly energy-dependent processes. Thus, it is possible that the recruitment of plant mitochondria to interaction sites with invading microbes may also supply ATP for energy-dependent transport and secretion at the plasma membrane. Moreover, as the respiratory machinery is very sensitive to changing redox conditions, mitochondria are also known to function as sensors for cellular functional imbalance (Schwarzländer and Finkemeier, 2013; Ng et al., 2014). Thus, changes in the mitochondrial redox status can induce signaling events leading to direct effects on mitochondrial function and dynamics and to adaptations in gene expression via retrograde signaling between mitochondria and the nucleus. Recently, mitochondrial matrix-targeted redox-sensitive GFP sensors have become available for in vivo measurement of glutathione redox potential in Arabidopsis by ratiometric determination of GFP fluorescence emission captured at 500 to 540 nm after excitation at 405 versus 488 nm (Schwarzländer et al., 2008; Albrecht et al., 2014). Here, we used these sensor lines to test whether or not barley powdery mildew attack induces oxidative perturbation in epidermal mitochondria. CLSM time-lapse scans of Bgh-challenged plants expressing mt-roGFP2 confirmed the dynamic movement of mitochondria in Arabidopsis epidermal cells and the clustered immobilization of individual mitochondria at sites of attempted fungal penetration (Supplemental Movie 3; Figure 6A). Next, we determined the thiol redox status of individual mitochondria in attacked and directly adjacent epidermal cells dependent on their distance from the fungal entry site (Figures 6B and 6C) using quantitative redox imaging software recently developed by Fricker (2015). These analyses demonstrated that mitochondria within 5 µm of the infection site show significantly increased sensor oxidation than the rest of the mitochondrial population. Interestingly, there also appears to be a trend toward slightly higher levels of oxidation in mitochondria of uninfected cells that are close to the interaction site. Together, these data corroborate the idea that pathogen attack induces the formation of distinct mitochondrial subpopulations within single epidermal cells. Moreover, the thiol redox status of individual mitochondria changes as a function of distance from the actual site of fungal invasion. Consequently, mitochondria that are subject to immobilization at sites of attempted fungal ingress are characterized by a significantly higher sensor oxidation. This reflects an imbalance in the matrix thiol-redox homeostasis as a likely consequence of localized release of oxidizing molecules such as hydrogen peroxide.
Figure 6.
Ratiometric Analysis of Mitochondrial roGFP2 Fluorescence Reveals Enhanced Matrix Oxidation in Bgh-Infected Cells.
(A) and (B) Representative single frames of a typical CLSM time lapse (Supplemental Movie 3) of transgenic mt-roGFP2 in epidermal cells infected by Bgh (green outline) and uninfected neighboring epidermal cells that show an accumulation of roGFP2-expressing mitochondria below the fungal appressorium (ap; white dashed line). In (A), an overlay image of bright-field mt-roGFP2 fluorescence with excitation at 405 nm (red) and 488 nm (green) and autofluorescence (blue), as well as the respective single-channel images, are shown. Germinated fungal structures including the appressorium (ap) are indicated by white dashed line. Ratiometric analysis was performed on segmented mitochondria in infected (green) and uninfected (red) cells, and their distance was measured relative to the attempted penetration site (pink star) (B). Mitochondria localized in stomata (blue outlines) were excluded from the analysis presented in (C). Bars = 25 µm.
(C) Plot of the mt-roGFP2 fluorescence intensity ratio indicates higher 405/488-nm ratios and, therefore, higher oxidative state of mitochondria in Bgh-infected cells, especially close to the attempted penetration site. Data points represent average (±se) 405/488-nm ratios of each mitochondria present in concentric 2-µm annuli centered on the attempted penetration site and measured over a time frame of 01:26 min (34 frames) for 24 individual penetration events (i.e., a total number of 816 analyzed frames). Additionally, whole-cell 405/488-nm ratios were averaged and indicated for infected (green line) and uninfected (red line) cells.
DISCUSSION
With this study, we provide evidence for the formation and functional importance of a mitochondrial subpopulation in individual leaf epidermal cells that are under attack by nonadapted powdery mildew fungi. This pathogen-induced mitochondrial subpopulation is characterized by recruitment to and immobilization at sites of attempted fungal invasion. Mitochondrial translocation is mediated by myosin motor protein-dependent transport along actin filaments (Yang et al., 2014), which are known to focally reorganize toward sites of pathogen attack as a consequence of mechanical stimulation (Hardham et al., 2008) and/or MAMP-induced signaling (Li et al., 2015). Thus, focal reorganization of the actin cytoskeleton may indeed serve the local recruitment of mitochondria and other organelles. Double fluorescence labeling experiments conducted in our study showed that immobilization of mitochondria is accompanied by a focal rearrangement of the ER coalescing into a lamellar sheath in close proximity to this mitochondrial subpopulation. Whether or not mitochondrial arrest is a consequence of the structural ER reorganization remains to be determined. In Arabidopsis, clustering and immobilization of mitochondria has been observed in response to UV light exposure (Gao et al., 2008) or upon application of methyl jasmonate (Zhang and Xing, 2008), the oxylipin 9-hydroxy-10,12,15-octadecatrienoic acid (Vellosillo et al., 2013), or reactive oxygen species (ROS) (Scott and Logan, 2008) and was suggested to be controlled by the CLUSTERED MITOCHONDRIA-type FRIENDLY gene product (El Zawily et al., 2014). Thus, it will be interesting to test friendly mutant plants in regard to pathogen-induced mitochondrial arrest, PEN2 recruitment patterns, and subsequent disruption of pathogen entry resistance. We postulate that the pathogen-induced concerted structural assembly of ER and mitochondria observed in this study may even result in a functionally specialized subcellular cytosolic microcompartment. Notably, the observed structural rearrangements coincide with a simultaneous and cell-autonomous transcriptional induction and protein recruitment of both CYP81F2 and PEN2 to the cytoplasmic surface of the ER and the OMM, respectively. These findings support the idea of metabolon formation as a conceptual framework for the spatio-temporal organization of metabolism in plant cells (Sweetlove and Fernie, 2013). Indeed, one critical function of the postulated microcompartment may be the generation of a spatio-temporally controlled IG metabolon that channels metabolites for biosynthesis and activation of 4-substituted I3Gs. It is also not difficult to imagine that concerted organelle activities create a specific cytosolic reaction milieu with regard to pH, ATP, glutathione, and ion concentrations; H2O2 and other ROS; and thiol redox potential that affects the activity of essential enzymes like CYP81F2, PEN2, and PEN3, thus fine-tuning the control of metabolite production, activation, and discharge. It is important to note in this context that homo- or heteromeric aggregate formation, as shown here for PEN2, is known to control enzymatic activity of related glycosyl hydrolases (Kwak et al., 2009; Yamada et al., 2011). It is likely that similar regulatory mechanisms apply for PEN2 and provide additional control for metabolite flux and release of bioactive hydrolysis products. Future efforts should address this question and aim to identify aggregate complex partners and regulatory proteins that mediate both the formation and, as observed in this study, disassembly of PEN2-containing aggregate complexes. Another challenging task for the future will be to directly demonstrate whether the observed homomerization capacity of PEN2 contributes to the accumulation of PEN2-GFP fluorescence in the periphery of mitochondria. Finally, in wild-type Arabidopsis plants, energy-dependent discharge of the PEN2 hydrolysis products by the ABC transporter PEN3, which is subject to MAMP-induced recruitment into an adjacent PM microdomain (Stein et al., 2006; Underwood and Somerville, 2013), is likely to guarantee direct sequestration of the toxic pathway end products from the interior of the plant cell. Thus, and in contrast to the classical mustard oil bomb, which involves cellular compartmentalization of myrosinases and glucosinolate glucosides in distinct cells and is triggered upon tissue disruption by insect feeding or necrotrophic fungi (Halkier and Gershenzon, 2006), glucosinolate-based defense against biotrophic fungi is based on cell-autonomous subcellular compartmentalization and concerted metabolon activity. This involves dynamic cell polarization and structural reorganization accompanied by organelle repositioning, transcriptional reprogramming, and dynamic changes in subcellular protein localization.
In this study, we have shown that PEN2 is dually localized to the membrane of peroxisomes and the OMM and provided evidence that this promiscuous localization pattern is mediated by the TA of PEN2. Promiscuous TA-mediated targeting to peroxisomes and mitochondria has been observed for only a few Arabidopsis proteins, all of which play a role in fission of both peroxisomes and mitochondria (e.g., FISSION1A [FIS1A] and FIS1B [Zhang and Hu, 2009]; PEROXISOMAL AND MITOCHONDRIAL DIVISION FACTOR1 [PMD1; Aung and Hu, 2011]). Specific targeting to the OMM alone, as observed for the TA of TOM20-4 in this study, has been suggested to be either dependent on a dibasic targeting motif within the TA or on the presence of a TA in combination with upstream protein features and lipid composition of the OMM (Marty et al., 2014). In our study, exclusive targeting of PEN2 to the OMM by replacing the genuine TA with the TA of TOM20-4 was sufficient to restore entry control against nonadapted Bgh conidiospores. Together with our observation that PEN2 homo-/heteromeric aggregate formation is restricted to immobilized mitochondria, this may suggest that the peroxisomal subpool of PEN2 is dispensable for penetration resistance in general. This begs the question why PEN2 associates with peroxisome membranes at all. In this context, it is important to note that PEN2 mutations affect a remarkably broad spectrum of interactions with various microbes, including other fungal pathogens such as Plectosphaerella cucumerina (Sanchez-Vallet et al., 2010), Magnaporthe oryzae (Maeda et al., 2009), Leptosphaeria maculans (Elliott et al., 2008), and Colletotrichum species (Hiruma et al., 2010), oomycetes (e.g., Phytophthora brassicae; Schlaeppi et al., 2010), and a growth-promoting endophytic fungus (Piriformospora indica; Jacobs et al., 2011). An important question for the future will be whether or not the peroxisomal PEN2 protein subpool is critical for resistance against these pathogens and what the subcellular behavior of peroxisomes, mitochondria, and the PEN2 protein is like in these interactions.
It is noteworthy that pathogen-induced cell polarization, accompanied by recruitment of organelles and protein translocation to sites of interaction with microbial pathogens, is a general response of monocot and dicot plants (Schmelzer, 2002). In particular, mitochondria have also been shown to accumulate at powdery mildew penetration sites in the monocot crop barley (Kunoh and Ishizaki, 1973), reinforcing the idea that recruitment of this organelle represents an evolutionarily ancient and conserved phenomenon. By contrast, the pathogen-inducible production of IGs is restricted to the dicot lineage of the Brassicales (Bednarek et al., 2011; Hofberger et al., 2013) and thus evolved after the monocot-dicot split (Chaw et al., 2004; Kagale et al., 2014). In conclusion, we propose that recruitment and arrest of mitochondria serves another function in plant innate immunity that, from an evolutionary perspective, predates IG metabolon formation and was co-opted for PEN2-dependent activation of toxic products at plant-microbe interaction sites. Multiple lines of evidence recently suggested fundamental roles of mitochondria in plant innate immunity (reviewed in Colombatti et al., 2014). In particular, exemplary studies have emphasized the importance of salicylic acid-triggered mitochondrial ROS production (Gleason et al., 2011), controlled mitochondrial protein import (Huang et al., 2013), and mitochondrial retrograde signaling control (Vellosillo et al., 2013) in defense against microbial pathogens. These finding are supported by recent Arabidopsis transcriptome meta-analyses suggesting that upregulation of defense gene expression might be under mitochondrial retrograde control (Schwarzländer et al., 2012) and that transcripts of alternative respiration, representing marker genes for retrograde signaling, are specifically induced by Bgh attack (Schwarzländer and Finkemeier, 2013). Consequently, it is not surprising that microbial pathogens evolved effector molecules that target mitochondria in order to manipulate their functionality (Block et al., 2010). It is conceivable to postulate that this may be particularly important for biotrophic pathogens as mitochondria are not only master regulators of danger signaling and programmed cell death in animals (Galluzzi et al., 2012) but have also been shown to play an active role in plant hypersensitive response-related cell death (Amirsadeghi et al., 2007; Colombatti et al., 2014). Using Arabidopsis lines expressing the mitochondrial redox sensor mt-roGFP2, we were able to show here that mitochondria immobilized at plant-microbe interaction sites exhibit a redox shift toward oxidation as a likely consequence of localized generation of ROS. Regardless of whether mitochondria are the prime source of ROS, or a target for oxidative damage, or whether perturbation of mitochondrial function and dynamics is the cause or consequence of the observed elevated oxidation status, it is important to note that mitochondrial ROS can trigger retrograde signaling and hypersensitive cell death in plants (Schwarzländer and Finkemeier, 2013; Ng et al., 2014). Thus, it is tempting to speculate that the original evolutionary function of the mitochondrial subpopulation observed in this study was to contribute to pathogen-induced retrograde signaling to the nucleus and transcriptional reprogramming. Ultimately, excess perturbation of the mitochondrial oxidation status, as for example upon successful pathogen entry, may then trigger cell death execution, which typically accompanies postinvasion resistance to Bgh (Lipka et al., 2005). Future research combining pathogen challenge experiments with in vivo redox imaging and analysis of mutants with defects in either cell death execution or mitochondrial function and dynamics is needed to test this hypothesis.
METHODS
Plant Lines and Growth Conditions
The following Arabidopsis thaliana wild-type, mutant, and transgenic lines were used in this study: Col-0, Col-3 gl1, cyp81F2-2 (Bednarek et al., 2009), pen2-1, pen2-2, pen2-1 expressing PEN2-GFP-TAPEN2 (Lipka et al., 2005), AtWAK2-CFP-HDEL (Nelson et al., 2007), ScCOX4-GFP (Nelson et al., 2007), and CFP-PTS1 (Nelson et al., 2007).
Arabidopsis, Nicotiana benthamiana, and barley (Hordeum vulgare) seeds were sown on soil (Fruhstorfer Erde Typ T 25 fein from Hawita) and grown in growth chambers at 18 to 22°C with an 8-h photoperiod (Arabidopsis) and 16-h photoperiod (N. benthamiana and barley), respectively.
Construction of Transgenic Lines
All constructs were cloned into the binary vector pAMPAT-MCS or the derivative pAM-MCS-NotI possessing kanamycin resistance instead of BASTA (Lipka et al., 2005). To create the PEN2-RFP-TAPEN2 fusion construct, GFP was replaced in the original PEN2-GFP-TAPEN2 plasmid described by Lipka et al. (2005) by monomeric RFP using the primers listed in Supplemental Table 1. For the deletion construct lacking the globular part of PEN2, the GFP-TAPEN2 fragment was constructed by PCR amplification and integrated via restriction sites into pAMPAT-MCS. The C-terminal deletion constructs were obtained from PEN2-GFP-TAPEN2 plasmid by PCR using primers listed in Supplemental Table 1 by replacement of the original sequence with the mutated fragments via restriction and ligation. The chimeric construct PEN2-GFP-TATOM20-4 was generated by exchange of the wild-type sequence with a PCR fragment encoding the last 34 amino acids of TOM20-4. RFP-TOM20-4 was constructed by amplifying the open reading frame of TOM20-4 and integration into the plasmid pAM-MCS-NotI containing a 35S promoter and RFP. CYP81F2-RFP was constructed by amplifying CYP81F2 open reading frame from cDNA and RFP from plasmid pSAT6-DEST-RFP-N1 (GenBank: EF212309.1) and integration of a 1998-bp native promoter fragment starting immediately upstream of the ATG start codon from genomic DNA. All constructs were confirmed by sequencing and transformed by electroporation into Agrobacterium tumefaciens strain GV3101 (pMP90RK).
Transformation of Arabidopsis
The floral dip method described by Clough and Bent (1998) was used to create stable transgenic lines. Depending on the binary vector, either the Agrobacterium strain GV3101 or GV3101 pMP90RK was used. Bacteria were grown at 28°C for 48 h in dYT medium (16 g/L tryptone, 10 g/L yeast extract, and 10 g/L NaCl) containing 50 mg/L of kanamycin, 50 mg/L of rifampicin, and, for selection of transformed GV3101 pMP90RK cells, 25 mg/L of carbenicillin. Bacterial suspensions were centrifuged for 15 min at 4000g and resuspended in dipping solution (5% sucrose and 0.05% Silwet L-77). Transgenic plant selection was either performed on half-strength MS medium (2.2 g/L Murashige and Skoog salt, 0.5 g/L MES, pH 5.7, and 14 g/L phyto agar) containing 50 mg/L of kanamycin or by spraying BASTA (Bayer CropScience) on soil-grown seedlings.
Transient Transformation of N. benthamiana
Three-week-old N. benthamiana plants were used for transient expression of fluorescent protein-tagged constructs. Two-day-old Agrobacterium GV3101 suspensions with and without the helper plasmid pMP90RK were centrifuged and resuspended in infiltration medium (10 mM MgCl2, 5 mM MES, pH 5.3, and 150 µM acetosyringone). A bacterial solution at OD600 = 1.0 was infiltrated into leaf panels with a syringe. The peroxisomal and mitochondrial marker proteins were described by Nelson et al. (2007) and were obtained from TAIR. Coinfiltration of the gene-silencing inhibitor p19 (Popescu et al., 2007) was done to enhance construct expression.
Inoculation of Arabidopsis and Pathogenicity Assay
Barley powdery mildew (Blumeria graminis f. sp hordei isolate K1) was grown on H. vulgare cv Ingrid (line I-10) for 10 to 14 d and inoculated onto 4- to 5-week-old Arabidopsis plants using a settling tower (Lipka et al., 2005). Quantification of fungal invasive growth was performed as described by Lipka et al. (2005).
Microscopy
CLSM was conducted with a Leica DM6000B/TCS SP5 microscope (Leica Microsystems) with the following excitation wavelengths: aniline blue, 405 nm; CFP, 458 nm; GFP, 488 nm; RFP and FM4-64 (Invitrogen), 561 nm. For visualization of fungal structures, infected leaves were incubated for 30 s in FM4-64 (Invitrogen) staining solution (2 mM FM4-64 in water). Images, overlays, and z-stacks were acquired and processed using the Leica analysis software LAS AF2.2.1.
Ratiometric and positional analyses of individual mitochondria were performed using custom software as described by Fricker (2015). In brief, mitochondria were initially separated from each other into nonoverlapping domains using watershed segmentation of an inverted template image calculated from the maximum of both fluorescent channels in the case of mt-roGFP2 or just the ScCOX4-RFP channel in dual-labeling experiments with PEN2-GFP-TAPEN2. Each mitochondrion was refined using a local intensity-based threshold set at the 50% value between the local maximum in each watershed domain and the background to accommodate differing overall mitochondrial intensities. In parallel, the (nonconfocal) bright-field transmission image was processed to highlight cell boundaries and calculate the distance from the attempted penetration site. Mitochondrial distribution patterns and physiological responses were typically averaged over 2 µm concentric annuli from the penetration site. To estimate the degree of mitochondrial immobilization in the vicinity of the penetration site, the probability of finding a mitochondrion at each pixel was calculated for each pixel in each frame of a 5-min time sequence taken at 5-s intervals and presented as a pseudo-color-coded image ranging from blue (low probability) to red (high probability). A high value was taken to indicate that either a single mitochondrion was immobilized at that point, or there was a sufficiently high density of organelle traffic such that different mitochondria were detected at high frequency.
For transmission electron microscopy, leaf segments were fixed with 3% glutaraldehyde (Sigma-Aldrich) in sodium cacodylate buffer (SCB), pH 7.2, for 3 h at room temperature, washed with SCB, postfixed with 1% osmium tetroxide (Carl Roth) in SCB, dehydrated in a graded ethanol series, and embedded in epoxy resin (Spurr, 1969). After polymerization, the material was sectioned with an Ultramicrotome S (Leica). Ultrathin sections (80 nm) were transferred to formvar-coated grids and poststained with uranyl acetate and lead citrate. The sections were observed with an EM 900 transmission electron microscope (Zeiss SMT) at an acceleration voltage of 80 kV. Electron micrographs were taken with a slow scan camera (Variospeed SSCCD camera SM-1k-120; TRS).
Split-Ubiquitin Y2H Assays
Split-ubiquitin yeast two-hybrid experiments were performed as described by Obrdlik et al. (2004). For interaction screening, full-length PEN2 cDNA, the N-terminal globular part (nucleotides 1 to 1494), and C-terminal extension of PEN2 (nucleotides 1486 to 1683) were cloned into vectors carrying either Nub or Cub in the N-terminal position (Supplemental Table 1). Vectors were subsequently transformed into yeast strains THY.AP5 and THY.AP4. After interaction assays, yeast cells were selected on media lacking the amino acid adenine and histidine.
Immunoprecipitation
Protein extractions for immunoprecipitations were performed as described by Moffett et al. (2002) with minor modification. For precipitation, GFP-Trap coupled to agarose beads (Chromotek) was used according to the manufacturer’s instructions. Equal volumes of immunoprecipitated protein were analyzed by immunoblot analysis.
Immunoblot Analysis
Protein concentration was either measured with standard Bradford or Lowry protein assays. Samples were separated on 10% SDS-PAGE gels or on NativePAGE Novex 3-12% Bis-Tris gels (Invitrogen), blotted to a polyvinylidene fluoride membrane (Millipore), and probed with the corresponding antibody diluted in Tris-buffered saline with Tween 20 (TBST) (20 mM Tris-HCl, pH 7.5, 500 mM NaCl, and 0.1% Tween 20) with 0.2% skim milk powder. PEN2 protein was detected using the polyclonal antibody α-PEN2 (Lipka et al., 2005) diluted 1:10,000 in TBST. GFP or RFP tag was detected with α-GFP and α-RFP monoclonal antibody diluted 1:5000 (ChromoTek), respectively. PEX14 protein was detected using the α-PEX14 polyclonal antibody (Agrisera) diluted 1:10,000. VDAC1 protein was detected using the α-VDAC1 polyclonal antibody (Agrisera) diluted 1:5000.
Peroxisome and Mitochondria Enrichment from Arabidopsis Leaf Extracts
For organelle enrichment, Arabidopsis plants were grown for 5 to 6 weeks under short-day conditions and the green parts were used for the extractions. Peroxisome-enriched extracts were obtained following the protocol described by Reumann et al. (2007). Mitochondria-enriched extracts were obtained following the protocols described by Kruft et al. (2001) and Sweetlove et al. (2007).
Statistical Analysis
Statistics were evaluated by one-way ANOVA with Tukey’s post test.
Accession Numbers
Sequence data for the genes described in this article can be found in the Arabidopsis Genome Initiative database under the following accession numbers: PEN2, AT2G44490; KAT1, AT5G46240; TOM20-4, AT5G40930; CYP81F2, AT5G57220.
Supplemental Data
Supplemental Figure 1. PEN2 shows homomerization capacity.
Supplemental Figure 2. PEN2-GFP-TAPEN2 aggregate formation is restricted to an immobilized subpopulation of mitochondria.
Supplemental Figure 3. PEN2-GFP-TAPEN2 forms mitochondrial aggregates underneath attempted Bgh penetration sites that exhibit a high fluorescence intensity and reduced mobility.
Supplemental Figure 4. The C-terminal anchored protein TOM20-4 is specifically localized in the outer membrane of mitochondria.
Supplemental Figure 5. The CYP81F2-RFP fusion protein complements the cyp81F2 penetration phenotype.
Supplemental Table 1. Oligonucleotide primers used in this study
Supplemental Movie 1. Formation of PEN2-GFP-TAPEN2 aggregates at attempted fungal penetration site 20 hpi with Bgh.
Supplemental Movie 2. Mitochondrial mobility and PEN2-GFP-TAPEN2 distribution underneath an attempted Bgh penetration site.
Supplemental Movie 3. Mitochondria expressing roGFP2 accumulate and form immobile aggregates at attempted fungal penetration site at 1 dpi with Bgh.
Supplementary Material
Acknowledgments
We thank Holger Eubel (Leibniz University of Hannover) for help with the isolation of mitochondria and Gabi Schauermann for excellent technical support. This work was supported by The Gatsby Charitable Foundation and by a Deutsche Forschungsgemeinschaft (DFG) grant to V.L. (LI1317/2-1). M.S. was supported by the DFG through the Emmy-Noether program (SCHW 1719/1-1).
AUTHOR CONTRIBUTIONS
R.F., M.K., M.S., and V.L. conceived and designed the experiments. R.F., M.K., C.K., and G.H. performed the experiments. R.F., M.K., G.H., M.D.F., A.J.M., M.S., and V.L. analyzed the data. M.K. and V.L. wrote the article with contributions from all authors.
Glossary
- MAMP
microbe-associated molecular pattern
- Bgh
Blumeria graminis f. sp hordei
- IG
indole glucosinolate
- ER
endoplasmic reticulum
- CLSM
confocal laser scanning microscopy
- hpi
hours postinoculation
- TA
tail anchor
- OMM
outer mitochondrial membrane
- ROS
reactive oxygen species
- SCB
sodium cacodylate buffer
References
- Abell B.M., Mullen R.T. (2011). Tail-anchored membrane proteins: exploring the complex diversity of tail-anchored-protein targeting in plant cells. Plant Cell Rep. 30: 137–151. [DOI] [PubMed] [Google Scholar]
- Albrecht S.C., Sobotta M.C., Bausewein D., Aller I., Hell R., Dick T.P., Meyer A.J. (2014). Redesign of genetically encoded biosensors for monitoring mitochondrial redox status in a broad range of model eukaryotes. J. Biomol. Screen. 19: 379–386. [DOI] [PubMed] [Google Scholar]
- Amirsadeghi S., Robson C.A., Vanlerberghe G.C. (2007). The role of the mitochondrion in plant responses to biotic stress. Physiol. Plant. 129: 253–266. [Google Scholar]
- Aung K., Hu J. (2011). The Arabidopsis tail-anchored protein PEROXISOMAL AND MITOCHONDRIAL DIVISION FACTOR1 is involved in the morphogenesis and proliferation of peroxisomes and mitochondria. Plant Cell 23: 4446–4461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bednarek P., Pislewska-Bednarek M., Svatoš A., Schneider B., Doubsky J., Mansurova M., Humphry M., Consonni C., Panstruga R., Sanchez-Vallet A., Molina A., Schulze-Lefert P. (2009). A glucosinolate metabolism pathway in living plant cells mediates broad-spectrum antifungal defense. Science 323: 101–106. [DOI] [PubMed] [Google Scholar]
- Bednarek P., Piślewska-Bednarek M., Ver Loren van Themaat E., Maddula R.K., Svatoš A., Schulze-Lefert P. (2011). Conservation and clade-specific diversification of pathogen-inducible tryptophan and indole glucosinolate metabolism in Arabidopsis thaliana relatives. New Phytol. 192: 713–726. [DOI] [PubMed] [Google Scholar]
- Block A., Guo M., Li G., Elowsky C., Clemente T.E., Alfano J.R. (2010). The Pseudomonas syringae type III effector HopG1 targets mitochondria, alters plant development and suppresses plant innate immunity. Cell. Microbiol. 12: 318–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaw S.M., Chang C.C., Chen H.L., Li W.H. (2004). Dating the monocot-dicot divergence and the origin of core eudicots using whole chloroplast genomes. J. Mol. Evol. 58: 424–441. [DOI] [PubMed] [Google Scholar]
- Clay N.K., Adio A.M., Denoux C., Jander G., Ausubel F.M. (2009). Glucosinolate metabolites required for an Arabidopsis innate immune response. Science 323: 95–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Collins N.C., Thordal-Christensen H., Lipka V., Bau S., Kombrink E., Qiu J.-L., Hückelhoven R., Stein M., Freialdenhoven A., Somerville S.C., Schulze-Lefert P. (2003). SNARE-protein-mediated disease resistance at the plant cell wall. Nature 425: 973–977. [DOI] [PubMed] [Google Scholar]
- Colombatti F., Gonzalez D.H., Welchen E. (2014). Plant mitochondria under pathogen attack: a sigh of relief or a last breath? Mitochondrion 19: 238–244. [DOI] [PubMed] [Google Scholar]
- Dodds P.N., Rathjen J.P. (2010). Plant immunity: towards an integrated view of plant-pathogen interactions. Nat. Rev. Genet. 11: 539–548. [DOI] [PubMed] [Google Scholar]
- Elliott C.E., Harjono, Howlett B.J. (2008). Mutation of a gene in the fungus Leptosphaeria maculans allows increased frequency of penetration of stomatal apertures of Arabidopsis thaliana. Mol. Plant 1: 471–481. [DOI] [PubMed] [Google Scholar]
- El Zawily A.M., et al. (2014). FRIENDLY regulates mitochondrial distribution, fusion, and quality control in Arabidopsis. Plant Physiol. 166: 808–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fricker M.D. (2015). Quantitative redox imaging software. Antioxid. Redox Signal., in press. [DOI] [PubMed] [Google Scholar]
- Gao C., Xing D., Li L., Zhang L. (2008). Implication of reactive oxygen species and mitochondrial dysfunction in the early stages of plant programmed cell death induced by ultraviolet-C overexposure. Planta 227: 755–767. [DOI] [PubMed] [Google Scholar]
- Galluzzi L., Kepp O., Kroemer G. (2012). Mitochondria: master regulators of danger signalling. Nat. Rev. Mol. Cell Biol. 13: 780–788. [DOI] [PubMed] [Google Scholar]
- Gleason C., Huang S., Thatcher L.F., Foley R.C., Anderson C.R., Carroll A.J., Millar A.H., Singh K.B. (2011). Mitochondrial complex II has a key role in mitochondrial-derived reactive oxygen species influence on plant stress gene regulation and defense. Proc. Natl. Acad. Sci. USA 108: 10768–10773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grubb C.D., Abel S. (2006). Glucosinolate metabolism and its control. Trends Plant Sci. 11: 89–100. [DOI] [PubMed] [Google Scholar]
- Halkier B.A., Gershenzon J. (2006). Biology and biochemistry of glucosinolates. Annu. Rev. Plant Biol. 57: 303–333. [DOI] [PubMed] [Google Scholar]
- Hardham A.R., Takemoto D., White R.G. (2008). Rapid and dynamic subcellular reorganization following mechanical stimulation of Arabidopsis epidermal cells mimics responses to fungal and oomycete attack. BMC Plant Biol. 8: 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi M., Nito K., Toriyama-Kato K., Kondo M., Yamaya T., Nishimura M. (2000). AtPex14p maintains peroxisomal functions by determining protein targeting to three kinds of plant peroxisomes. EMBO J. 19: 5701–5710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiruma K., Onozawa-Komori M., Takahashi F., Asakura M., Bednarek P., Okuno T., Schulze-Lefert P., Takano Y. (2010). Entry mode-dependent function of an indole glucosinolate pathway in Arabidopsis for nonhost resistance against anthracnose pathogens. Plant Cell 22: 2429–2443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y., Chen X., Liu Y., Roth C., Copeland C., McFarlane H.E., Huang S., Lipka V., Wiermer M., Li X. (2013). Mitochondrial AtPAM16 is required for plant survival and the negative regulation of plant immunity. Nat. Commun. 4: 2558. [DOI] [PubMed] [Google Scholar]
- Hückelhoven R., Panstruga R. (2011). Cell biology of the plant-powdery mildew interaction. Curr. Opin. Plant Biol. 14: 738–746. [DOI] [PubMed] [Google Scholar]
- Hofberger J.A., Lyons E., Edger P.P., Chris Pires J., Eric Schranz M. (2013). Whole genome and tandem duplicate retention facilitated glucosinolate pathway diversification in the mustard family. Genome Biol. Evol. 5: 2155–2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs S., Zechmann B., Molitor A., Trujillo M., Petutschnig E., Lipka V., Kogel K.H., Schäfer P. (2011). Broad-spectrum suppression of innate immunity is required for colonization of Arabidopsis roots by the fungus Piriformospora indica. Plant Physiol. 156: 726–740. Erratum. Plant Physiol. 157: 531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagale S., Robinson S.J., Nixon J., Xiao R., Huebert T., Condie J., Kessler D., Clarke W.E., Edger P.P., Links M.G., Sharpe A.G., Parkin I.A. (2014). Polyploid evolution of the Brassicaceae during the Cenozoic era. Plant Cell 26: 2777–2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kittur F.S., Lalgondar M., Yu H.Y., Bevan D.R., Esen A. (2007). Maize beta-glucosidase-aggregating factor is a polyspecific jacalin-related chimeric lectin, and its lectin domain is responsible for beta-glucosidase aggregation. J. Biol. Chem. 282: 7299–7311. [DOI] [PubMed] [Google Scholar]
- Kruft V., Eubel H., Jänsch L., Werhahn W., Braun H.P. (2001). Proteomic approach to identify novel mitochondrial proteins in Arabidopsis. Plant Physiol. 127: 1694–1710. [PMC free article] [PubMed] [Google Scholar]
- Kunoh H., Ishizaki H. (1973). Incorporation of host mitochondria into the haustorial encapsulation of barley powdery mildew. Ann. Phytopath. Soc. Japan 39: 42–48. [Google Scholar]
- Kwak S.N., Kim S.Y., Choi S.R., Kim I.S. (2009). Assembly and function of AsGlu2 fibrillar multimer of oat beta-glucosidase. Biochim. Biophys. Acta 1794: 526–531. [DOI] [PubMed] [Google Scholar]
- Kwon C., et al. (2008). Co-option of a default secretory pathway for plant immune responses. Nature 451: 835–840. [DOI] [PubMed] [Google Scholar]
- Li J., Henty-Ridilla J.L., Staiger B.H., Day B., Staiger C.J. (2015). Capping protein integrates multiple MAMP signalling pathways to modulate actin dynamics during plant innate immunity. Nat. Commun. 6: 7206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipka U., Fuchs R., Lipka V. (2008). Arabidopsis non-host resistance to powdery mildews. Curr. Opin. Plant Biol. 11: 404–411. [DOI] [PubMed] [Google Scholar]
- Lipka V., et al. (2005). Pre- and postinvasion defenses both contribute to nonhost resistance in Arabidopsis. Science 310: 1180–1183. [DOI] [PubMed] [Google Scholar]
- Lisenbee C.S., Lingard M.J., Trelease R.N. (2005). Arabidopsis peroxisomes possess functionally redundant membrane and matrix isoforms of monodehydroascorbate reductase. Plant J. 43: 900–914. [DOI] [PubMed] [Google Scholar]
- Lu X., Dittgen J., Piślewska-Bednarek M., Molina A., Schneider B., Svatoš A., Doubský J., Schneeberger K., Weigel D., Bednarek P., Schulze-Lefert P. (2015). Mutant allele-specific uncoupling of PEN3 functions reveals engagement of the ABC transporter in distinct tryptophan metabolic pathways. Plant Physiol. 168: 814–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macho A.P., Zipfel C. (2014). Plant PRRs and the activation of innate immune signaling. Mol. Cell 54: 263–272. [DOI] [PubMed] [Google Scholar]
- Maeda K., Houjyou Y., Komatsu T., Hori H., Kodaira T., Ishikawa A. (2009). AGB1 and PMR5 contribute to PEN2-mediated preinvasion resistance to Magnaporthe oryzae in Arabidopsis thaliana. Mol. Plant Microbe Interact. 22: 1331–1340. [DOI] [PubMed] [Google Scholar]
- Marty N.J., Teresinski H.J., Hwang Y.T., Clendening E.A., Gidda S.K., Sliwinska E., Zhang D., Miernyk J.A., Brito G.C., Andrews D.W., Dyer J.M., Mullen R.T. (2014). New insights into the targeting of a subset of tail-anchored proteins to the outer mitochondrial membrane. Front. Plant Sci. 5: 426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer D., Pajonk S., Micali C., O’Connell R., Schulze-Lefert P. (2009). Extracellular transport and integration of plant secretory proteins into pathogen-induced cell wall compartments. Plant J. 57: 986–999. [DOI] [PubMed] [Google Scholar]
- Micali C., Göllner K., Humphry M., Consonni C., Panstruga R. (2008). The powdery mildew disease of Arabidopsis: A paradigm for the interaction between plants and biotrophic fungi. Arabidopsis Book 6: e0115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffett P., Farnham G., Peart J., Baulcombe D.C. (2002). Interaction between domains of a plant NBS-LRR protein in disease resistance-related cell death. EMBO J. 21: 4511–4519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendra S., Venkataramani S., Shen G., Wang J., Pasapula V., Lin Y., Kornyeyev D., Holaday A.S., Zhang H. (2006). The Arabidopsis ascorbate peroxidase 3 is a peroxisomal membrane-bound antioxidant enzyme and is dispensable for Arabidopsis growth and development. J. Exp. Bot. 57: 3033–3042. [DOI] [PubMed] [Google Scholar]
- Nelson B.K., Cai X., Nebenführ A. (2007). A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J. 51: 1126–1136. [DOI] [PubMed] [Google Scholar]
- Ng S., De Clercq I., Van Aken O., Law S.R., Ivanova A., Willems P., Giraud E., Van Breusegem F., Whelan J. (2014). Anterograde and retrograde regulation of nuclear genes encoding mitochondrial proteins during growth, development, and stress. Mol. Plant 7: 1075–1093. [DOI] [PubMed] [Google Scholar]
- Nielsen M.E., Thordal-Christensen H. (2013). Transcytosis shuts the door for an unwanted guest. Trends Plant Sci. 18: 611–616. [DOI] [PubMed] [Google Scholar]
- Obrdlik P., et al. (2004). K+ channel interactions detected by a genetic system optimized for systematic studies of membrane protein interactions. Proc. Natl. Acad. Sci. USA 101: 12242–12247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfalz M., Vogel H., Kroymann J. (2009). The gene controlling the indole glucosinolate modifier1 quantitative trait locus alters indole glucosinolate structures and aphid resistance in Arabidopsis. Plant Cell 21: 985–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popescu S.C., Popescu G.V., Bachan S., Zhang Z., Seay M., Gerstein M., Snyder M., Dinesh-Kumar S.P. (2007). Differential binding of calmodulin-related proteins to their targets revealed through high-density Arabidopsis protein microarrays. Proc. Natl. Acad. Sci. USA 104: 4730–4735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reumann S., Babujee L., Ma C., Wienkoop S., Siemsen T., Antonicelli G.E., Rasche N., Lüder F., Weckwerth W., Jahn O. (2007). Proteome analysis of Arabidopsis leaf peroxisomes reveals novel targeting peptides, metabolic pathways, and defense mechanisms. Plant Cell 19: 3170–3193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Vallet A., Ramos B., Bednarek P., López G., Piślewska-Bednarek M., Schulze-Lefert P., Molina A. (2010). Tryptophan-derived secondary metabolites in Arabidopsis thaliana confer non-host resistance to necrotrophic Plectosphaerella cucumerina fungi. Plant J. 63: 115–127. [DOI] [PubMed] [Google Scholar]
- Schlaeppi K., Abou-Mansour E., Buchala A., Mauch F. (2010). Disease resistance of Arabidopsis to Phytophthora brassicae is established by the sequential action of indole glucosinolates and camalexin. Plant J. 62: 840–851. [DOI] [PubMed] [Google Scholar]
- Schmelzer E. (2002). Cell polarization, a crucial process in fungal defence. Trends Plant Sci. 7: 411–415. [DOI] [PubMed] [Google Scholar]
- Schwarzländer M., Finkemeier I. (2013). Mitochondrial energy and redox signaling in plants. Antioxid. Redox Signal. 18: 2122–2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzländer M., Fricker M.D., Müller C., Marty L., Brach T., Novak J., Sweetlove L.J., Hell R., Meyer A.J. (2008). Confocal imaging of glutathione redox potential in living plant cells. J. Microsc. 231: 299–316. [DOI] [PubMed] [Google Scholar]
- Schwarzländer M., König A.C., Sweetlove L.J., Finkemeier I. (2012). The impact of impaired mitochondrial function on retrograde signalling: a meta-analysis of transcriptomic responses. J. Exp. Bot. 63: 1735–1750. [DOI] [PubMed] [Google Scholar]
- Scott I., Logan D.C. (2008). Mitochondrial morphology transition is an early indicator of subsequent cell death in Arabidopsis. New Phytol. 177: 90–101. [DOI] [PubMed] [Google Scholar]
- Spurr A.R. (1969). A low-viscosity epoxy resin embedding medium for electron microscopy. J. Ultrastruct. Res. 26: 31–43. [DOI] [PubMed] [Google Scholar]
- Stein M., Dittgen J., Sánchez-Rodríguez C., Hou B.-H., Molina A., Schulze-Lefert P., Lipka V., Somerville S. (2006). Arabidopsis PEN3/PDR8, an ATP binding cassette transporter, contributes to nonhost resistance to inappropriate pathogens that enter by direct penetration. Plant Cell 18: 731–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweetlove L.J., Fernie A.R. (2013). The spatial organization of metabolism within the plant cell. Annu. Rev. Plant Biol. 64: 723–746. [DOI] [PubMed] [Google Scholar]
- Sweetlove L.J., Taylor N.L., Leaver C.J. (2007). Isolation of intact, functional mitochondria from the model plant Arabidopsis thaliana. Methods Mol. Biol. 372: 125–136. [DOI] [PubMed] [Google Scholar]
- Tateda C., Watanabe K., Kusano T., Takahashi Y. (2011). Molecular and genetic characterization of the gene family encoding the voltage-dependent anion channel in Arabidopsis. J. Exp. Bot. 62: 4773–4785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thordal-Christensen H. (2003). Fresh insights into processes of nonhost resistance. Curr. Opin. Plant Biol. 6: 351–357. [DOI] [PubMed] [Google Scholar]
- Underwood W., Somerville S.C. (2013). Perception of conserved pathogen elicitors at the plasma membrane leads to relocalization of the Arabidopsis PEN3 transporter. Proc. Natl. Acad. Sci. USA 110: 12492–12497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vellosillo T., Aguilera V., Marcos R., Bartsch M., Vicente J., Cascón T., Hamberg M., Castresana C. (2013). Defense activated by 9-lipoxygenase-derived oxylipins requires specific mitochondrial proteins. Plant Physiol. 161: 617–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan X., Chen S. (2007). Regulation of plant glucosinolate metabolism. Planta 226: 1343–1352. [DOI] [PubMed] [Google Scholar]
- Yang L., Qin L., Liu G., Peremyslov V.V., Dolja V.V., Wei Y. (2014). Myosins XI modulate host cellular responses and penetration resistance to fungal pathogens. Proc. Natl. Acad. Sci. USA 111: 13996–14001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada K., Hara-Nishimura I., Nishimura M. (2011). Unique defense strategy by the endoplasmic reticulum body in plants. Plant Cell Physiol. 52: 2039–2049. [DOI] [PubMed] [Google Scholar]
- Zhang L., Xing D. (2008). Methyl jasmonate induces production of reactive oxygen species and alterations in mitochondrial dynamics that precede photosynthetic dysfunction and subsequent cell death. Plant Cell Physiol. 49: 1092–1111. [DOI] [PubMed] [Google Scholar]
- Zhang X., Hu J. (2009). Two small protein families, DYNAMIN-RELATED PROTEIN3 and FISSION1, are required for peroxisome fission in Arabidopsis. Plant J. 57: 146–159. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.