Plant-specific RNA binding proteins RZ-1B and RZ-1C interact with a number of highly conserved serine/arginine-rich proteins to regulate RNA splicing and gene expression in Arabidopsis.
Abstract
Nuclear-localized RNA binding proteins are involved in various aspects of RNA metabolism, which in turn modulates gene expression. However, the functions of nuclear-localized RNA binding proteins in plants are poorly understood. Here, we report the functions of two proteins containing RNA recognition motifs, RZ-1B and RZ-1C, in Arabidopsis thaliana. RZ-1B and RZ-1C were localized to nuclear speckles and interacted with a spectrum of serine/arginine-rich (SR) proteins through their C termini. RZ-1C preferentially bound to purine-rich RNA sequences in vitro through its N-terminal RNA recognition motif. Disrupting the RNA binding activity of RZ-1C with SR proteins through overexpression of the C terminus of RZ-1C conferred defective phenotypes similar to those observed in rz-1b rz-1c double mutants, including delayed seed germination, reduced stature, and serrated leaves. Loss of function of RZ-1B and RZ-1C was accompanied by defective splicing of many genes and global perturbation of gene expression. In addition, we found that RZ-1C directly targeted FLOWERING LOCUS C (FLC), promoting efficient splicing of FLC introns and likely also repressing FLC transcription. Our findings highlight the critical role of RZ-1B/1C in regulating RNA splicing, gene expression, and many key aspects of plant development via interaction with proteins including SR proteins.
INTRODUCTION
Several RNA binding proteins (RBPs) play pivotal roles in regulating gene expression, both cotranscriptionally and posttranscriptionally. The defining feature of RBPs is the presence of putative RNA binding domains, including RNA recognition motifs (RRMs), K homology domains, zinc fingers, DEAD/DEAH boxes, Pumilio/FBF domains, and pentatricopeptide repeat domains (Lorković, 2009). Also present in plant RBPs are modular auxiliary domains rich in glycine, arginine, and serine, which exist in a variety of arrangements. The Arabidopsis thaliana genome encodes more than 600 putative RBPs, but the detailed functions of the vast majority of these RBPs remain unclear.
Glycine-rich RNA binding proteins (GRPs), a small group of plant RBPs, contain an N-terminal RRM domain and a C-terminal glycine-rich stretch. GRPs in Arabidopsis include GRP1-8 and RZ-1A-C. GRP7 and GRP8 are important regulators of circadian oscillations (Schöning et al., 2007; Schmal et al., 2013), flowering time (Streitner et al., 2008), responses to plant pathogens (Nicaise et al., 2013), and cold stress (Kim et al., 2008; Kwak et al., 2011). RZ-1A-C has a zinc finger motif between the RRM domain and C terminus (Lorković and Barta, 2002). RZ-1, which was first reported in the tobacco (Nicotiana sylvestris), belongs to a subgroup of GRPs and binds to a large ribonucleoprotein particle localized in the nucleus (Hanano et al., 1996). Homologs of RZ-1 in Arabidopsis were shown to have RNA chaperone activity in Escherichia coli (Kim et al., 2005). Overexpression of RZ-1A, but not homologous genes RZ-1B and RZ-1C, confers freezing tolerance in transgenic Arabidopsis (Kim et al., 2007; Kim et al., 2010a; Kim et al., 2010b). However, there is no direct genetic evidence to support the apparently important roles of these GRPs in plant growth and development and in responses to environmental stimuli.
Serine/arginine-rich (SR) proteins and heterogeneous ribonucleoproteins (hnRNPs) are among the most important groups of RBPs. In animals, SR proteins are mostly known for their function in regulating splicing. Mammalian SR proteins bind to cis-elements in exons (exon splicing enhancers) through the RRM domain and promote splicing by recruiting spliceosome components or inhibiting negative regulators, including hnRNPs. In addition to splicing, several additional putative functions of SR proteins have been reported. For example, SR proteins and the exon junction complex cooperate to promote mRNA packaging and compaction, protecting long mRNA stretches from nuclease digestion (Singh et al., 2012). Moreover, human SR protein SC35 facilitates productive elongation by releasing the CTDSer2 Kinase P-TEFb from the 7SK inhibitory complex and binding to nascent RNA at the promoter-proximal region (Ji et al., 2013).
Because of extensive gene duplication, plant genomes harbor many more SR proteins than are harbored by mammalian genomes. Among the 19 SR proteins in Arabidopsis, only a few have been studied functionally. Gain-of-function analyses of SR protein genes RS2Z33 (Kalyna et al., 2003) and SR30 (Lopato et al., 1999), as well as loss-of-function analyses of SR45 (Ali et al., 2007), SCL33, SCL33a (Thomas et al., 2012), RS40, and RS41 (Chen et al., 2013), have revealed that the proteins encoded by these genes regulate alternative splicing of their own pre-mRNA and that of other SR genes. SR45 has also been linked to RNA-directed DNA methylation, although a detailed mechanism has not been reported (Ausin et al., 2012). Currently, the degree to which plant SR proteins regulate gene expression at the whole-genome level is unclear. In addition, it is unclear whether plant SR proteins have functions beyond splicing.
In this work, we provide functional evidence that Arabidopsis RZ-1B and RZ-1C represent a unique group of GRPs. rz-1b rz-1c double mutants displayed a wide variety of defects, including delayed germination, retarded development of root and shoot meristems, late flowering, reduced stature, and serrated leaves. RZ-1B and RZ-1C interact with SR proteins through their C-terminal domains, which are also essential for nuclear speckle localization. High-throughput RNA-seq analysis of rz-1b rz-1c double mutants revealed perturbation of the splicing of many genes. Defective splicing was also observed in plants overexpressing the C-terminal domain of RZ-1C, confirming that RZ-1B/1C regulates splicing via interaction with SR proteins. RZ-1B and RZ-1C were found to be required for maintaining the optimal expression levels of more than 3000 genes, including many developmental regulators. Finally, using FLOWERING LOCUS C (FLC) as an example, we studied the mode of action of RZ-1C. RZ-1C targeted FLC directly, promoting splicing of FLC and repressing FLC transcription. Taken together, these findings reveal the essential roles of RZ-1B and RZ-1C as regulators of plant development, splicing, and general gene expression via interaction with proteins including SR proteins.
RESULTS
Arabidopsis RZ-1B and RZ-1C Represent a Group of Unique GRP Proteins in Plants
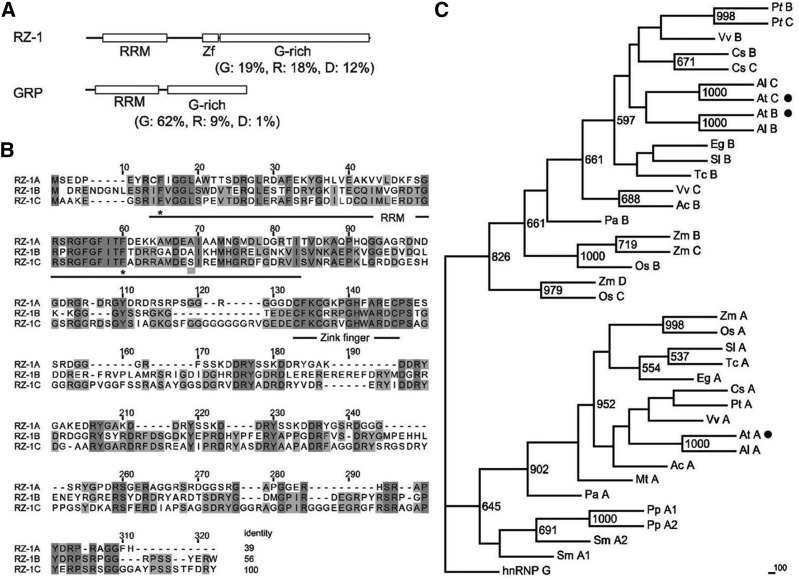
Typical GRPs contain an N-terminal RRM domain and a C-terminal glycine-rich stretch with low sequence complexity. GRPs play pivotal roles in flowering time control and responses to cold stress and pathogens. Arabidopsis has 11 GRP-encoding genes, three of which, RZ-1A, RZ-1B, and RZ-1C, form a distinct and independent clade in the phylogenetic tree generated using At GRP sequences (Supplemental Figure 1) (Lorković and Barta, 2002). At RZ-1A, At RZ-1B, and At RZ-1C possess C termini rich in glycine, arginine, and aspartic acid, as well as a zinc finger motif between the RRM domain and C terminus (Figures 1A and 1B) (Lorković and Barta, 2002). The unique structural features of Arabidopsis RZ-1A, RZ-1B, and RZ-1C imply that these RZ-1 proteins have functions distinct from those of other GRPs. Phylogenetic analysis showed that At RZ-1B, At RZ-1C, and their homologs in seed plants were grouped together, while At RZ-1A and its homologs were in a separate clade (Figure 1C; Supplemental Table 1), suggesting that At RZ-1B and At RZ-1C may have distinct and specialized, but redundant, functions in seed plants.
Figure 1.
Domain Architecture, Sequence Alignment, and Phylogenetic Tree of RZ-1s and Related Proteins.
(A) Domain arrangement and amino acid composition of RZ-1 and GRP7. The percentage of glycine (G), arginine (R), and aspartic acid (D) residues of the C terminus after the zinc finger are shown in parentheses. Zf, zinc finger.
(B) Protein sequence alignment of At RZ-1s. The positions of RRM and zinc fingers are marked with lines and letters below the sequence. The asterisks indicate key residues that are required for RNA binding in the RRM domain. The identity (%) of each protein sequence relative to At RZ-1C is shown at the end of each sequence.
(C) Phylogenetic tree of At RZ-1-related proteins from different plant species. The tree consists of two large clades. The number at each node indicates the bootstrap value (1000 replicates). Three At RZ-1 proteins are marked with black circles. At A, At B, and At C refer to At RZ-1A, At RZ-1B,and At RZ-1C, respectively. The abbreviations of RZ-1 proteins and species are explained in Supplemental Table 1.
RZ-1B and RZ-1C Are Required for Normal Plant Development
To assess the physiological functions of RZ-1B and RZ-1C in Arabidopsis, we analyzed the effects of T-DNA insertions in both genes. T-DNA insertions disrupted the production of full-length transcripts of RZ-1B and RZ-1C (Supplemental Figure 2). Neither the rz-1b single mutant (SALK_001328) nor the rz-1c single mutant (GABI_030b12) showed obvious morphological alterations in comparison with the wild-type plant (Col-0). Therefore, we generated rz-1b rz-1c double mutants through genetic crossing.
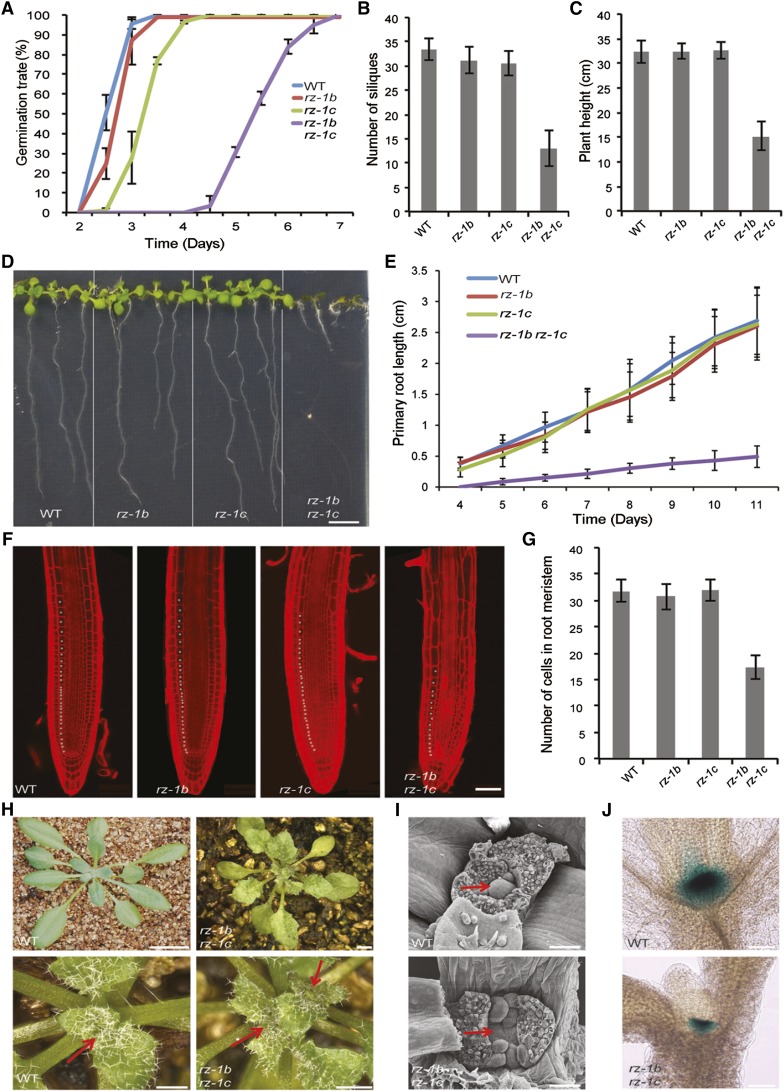
The rz-1b rz-1c double mutant displayed pleiotropic phenotypes, including delayed seed germination (Figure 2A) and severe retardation of root growth (Figures 2D and 2E) with a reduced number of dividing cells in the root meristem (Figures 2F and 2G). The aerial parts of the rz-1b rz-1c double mutants showed dwarfism, fewer flowers per inflorescence (Figures 2B and 2C; Supplemental Figure 3A), reduced leaf size, and serrated leaves (Supplemental Figure 3B). Interestingly, we also observed that 52% (n = 61) of the rz-1b rz-1c double mutants formed multiple rosettes (Figure 2H) with small, flattened shoot apical meristems (SAMs) (Figures 2I and 2J), although leaf primordia were initiated from multiple locations (Figure 2I; Supplemental Figure 3C).
Figure 2.
Pleotropic Defects of the rz-1b rz-1c Double Mutant.
(A) Statistics of the germination (i.e., radical emergence) rate of the wild type, rz-1b, rz-1c, and rz-1b rz-1c. Germination rates were examined at different time points after imbibition and are shown in the graph. Error bars indicate the sd obtained from three plates, each having 80 to 120 seeds per genotype.
(B) Statistics of the number of siliques produced from the main stem in different genotypes. Data were collected after the plants stopped growing. Error bars represent sd (n = 20).
(C) Statistics of final plant height from different genotypes. Data were collected after the plants stopped growing. Error bars represent sd (n = 20).
(D) Image of seedlings grown vertically for 10 d after germination. Bar = 0.5 cm.
(E) Statistics of the primary root length from day 4 to day 11 after germination. Error bars represent sd (n = 20).
(F) Confocal image of root meristems from different genotypes. The image shows the representative root meristems from seedlings at 4 d after germination. The roots were stained with propidium iodide. Asterisks indicate the dividing cells in the endodermis. Bar = 50 μm.
(G) Statistics of endodermal cells from different genotypes within the meristematic regions. Error bars represent sd (n = 15).
(H) rz-1b rz-1c generates multiple SAMs during vegetative growth. The images show wild-type and rz-1b rz-1c seedlings 3 weeks after germination. The lower panels show magnified images of the upper panels. Arrows indicate the positions of SAMs. Bars = 5 mm (upper panel) and 1 mm (lower panel).
(I) Scanning electron microscopy images of the SAMs of the wild type and rz-1b rz-1c. Arrows indicate the SAM in the wild type and the corresponding region in rz-1b rz-1c. Bar = 50 μm.
(J) Histochemical staining of CLAVATA3:GUS reporter in wild-type and rz-1b rz-1c backgrounds. Bar = 80 μm.
To confirm that the defective phenotypes were indeed caused by mutations in RZ-1B and RZ-1C, we transformed a construct containing the 6-kb genomic region encoding RZ-1C fused with GFP at its N terminus into the rz-1b rz-1c double mutant. The transformed mutants showed restored seed germination, plant stature, and root length in many independent lines (Supplemental Figure 4), suggesting that RZ-1B and RZ-1C are redundantly required for these developmental processes.
RZ-1C Is Widely Expressed during Plant Development
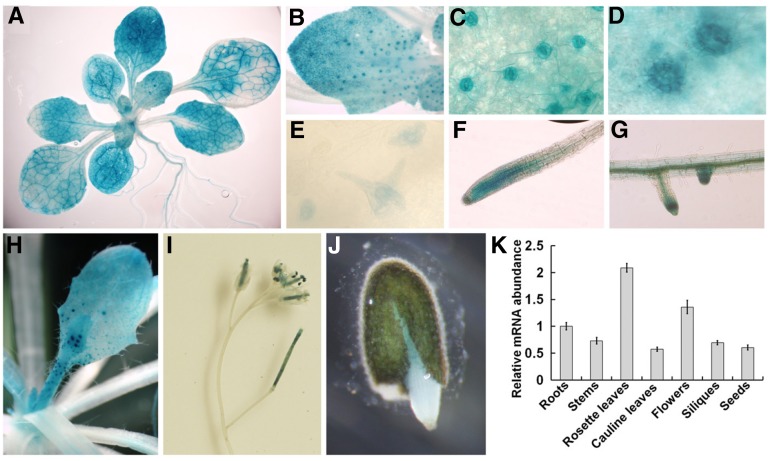
To investigate the expression pattern of RZ-1C, we generated a construct harboring the GUS reporter gene driven by the 2-kb promoter region of RZ-1C and transformed it into wild-type Arabidopsis. Four independent transgenic lines showed similar GUS staining patterns. GUS activity was ubiquitously detected, especially in highly proliferative tissues, including the embryo and endosperm of germinating seeds, as well as in newly initiated leaves (Figures 3A to 3J). In mature leaves, GUS staining was enriched in leaf veins, stomata, developing trichomes, and trichome socket cells. In roots, strong GUS staining was observed in the root tips, lateral root caps, and vascular tissues. At the reproductive stage, staining was observed in developing stems, cauline leaves, inflorescences, and young siliques. The ubiquitous expression pattern of RZ-1C was further confirmed using qRT-PCR analysis (Figure 3K).
Figure 3.
Expression Pattern of RZ-1C.
(A) to (J) GUS staining pattern of wild-type plants that harbor the RZ-1C:GUS reporter.
(A) Two-week-old seedlings.
(B) Young leaf.
(C) Stomata.
(D) Trichome socket cells.
(E) Developing trichomes.
(F) Root tip.
(G) Lateral root cap.
(H) Cauline leaf and developing stem.
(I) Young siliques and inflorescence.
(J) Germinating seeds (48 h after imbibition).
(K) qRT-PCR results indicating the expression levels of RZ-1C in different organs relative to its expression in roots. Error bars represent se acquired from three biological repeats.
RZ-1C Is Localized to Nuclear Speckles through Its C Terminus and Tightly Bound to Chromatin
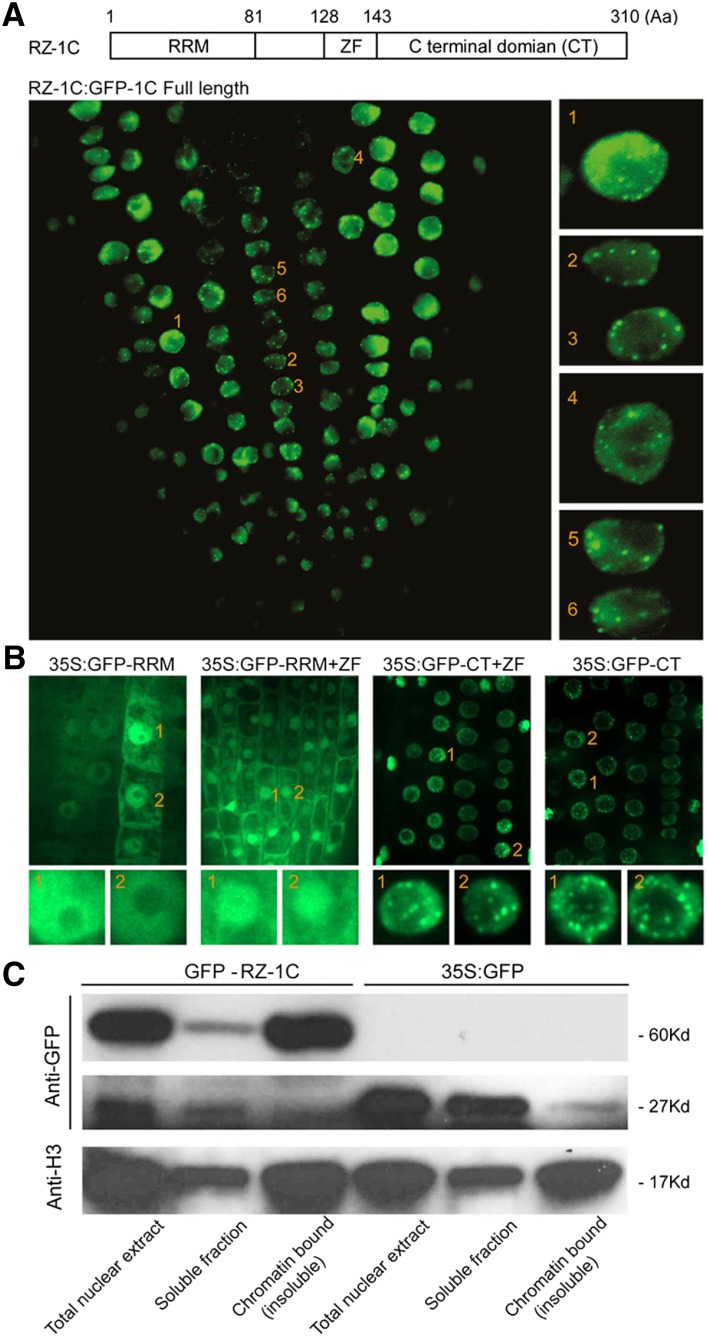
To investigate the subcellular localization of RZ-1B and RZ-1C, we conducted transient expression assays in onion epidermal cells by introducing GFP-tagged RZ-1 protein. RZ-1B and RZ-1C were localized in the nucleus, primarily in nuclear speckles (Supplemental Figure 5). Speckle localization was also observed when GFP-RZ-1C expression was driven in its native genomic context in Arabidopsis (Figure 4A). To determine the domain responsible for subcellular localization of RZ-1B and RZ-1C, a series of deletions were generated in RZ-1C, after which subcellular localization was assessed (Figure 4B; Supplemental Figure 5). The C-terminal region was found to be essential for nuclear localization of RZ-1C in onion (Supplemental Figure 5) and Arabidopsis cells (Figure 4B).
Figure 4.
RZ-1C Localizes to Nuclear Speckles and Binds to Chromatin.
(A) The nucleus localization of GFP-RZ-1C, driven by its native genomic context. The domain arrangement within RZ-1C and the number of amino acids of corresponding domains are depicted in the schematic at the top. The right panel shows magnified images of the selected individual cells (number 1 to 6), each with nuclear speckles. Aa, amino acids; ZF, zinc finger.
(B) The localization of different domains of RZ-1C in Arabidopsis root tips. Different domains of RZ-1C shown in (A) were fused with GFP and expressed under the control of the CaMV 35S promoter. The lower panel shows magnified images of the selected individual nuclei (number 1 and 2), with the four images on the right showing nuclear speckles.
(C) RZ-1C exists mostly as a chromatin-bound protein. The distribution of GFP-RZ-1C (driven in its native genomic context), GFP alone (driven by a 35S promoter), and Histone H3 in different fractions of the nucleus was examined through protein gel blotting using anti-GFP antibody.
We also examined the distribution of RZ-1C in several nuclear fractions. Transcription machinery (e.g., histone H3 and RNA polymerase II) known to be tightly associated with chromatin appears in the insoluble fraction during nucleus preparation (Kimura et al., 1999). We found that GFP-RZ-1C, driven in its native genomic context in Arabidopsis (Supplemental Figure 4), was mostly detected in the insoluble fraction, with a much smaller, but significant, amount of protein detected in the soluble fraction (Figure 4C). Similarly, histone H3 was also detected mostly in the insoluble fraction, consistent with its chromatin-bound nature. In contrast, GFP alone, driven by the CaMV 35S promoter, was detected mostly in the soluble nucleoplasmic fractions (Figure 4C). Therefore, our results indicate that RZ-1C is a chromatin-bound protein.
RZ-1B/1C Proteins Interact with SR Proteins through the C Termini of RZ-1B/1C
To study the functions of RZ-1B and RZ-1C, we investigated the proteins interacting with RZ-1C by screening a yeast two-hybrid cDNA library constructed from Arabidopsis seedlings. Five of eight positive clones were identified as SR proteins: SCL30 (two clones), SCL33, RSZ21, and RSZ22. To determine whether other SR proteins also interacted with RZ-1B or RZ-1C, we used yeast two-hybrid assays to assess binding of RZ-1B and RZ-1C to RZ-1A, U1-70K (a key subunit of U1 snRNP complex and a reported SR-interacting protein; Golovkin and Reddy, 1999), and six selected SR proteins, RS31, RS2Z33, SR34, SCL28, SCL30a, and SR45, which represent different SR protein subgroups (Barta et al., 2010). RZ-1B and RZ-1C interacted with the eight identified/selected SR proteins in yeast cells, with the exception of U1-70k, RS2Z33, and SR34 (Table 1; Supplemental Figure 6). We also detected interactions between RZ-1C and RZ-1B, RZ-1B and RZ-1B, as well as RZ-1C and RZ-1C (Table 1; Supplemental Figure 6). Interestingly, RZ-1A did not interact with any of these identified/selected SR proteins, consistent with the functional divergence of RZ-1A from RZ-1B and RZ-1C.
Table 1. Interaction of RZ-1 Proteins with SR Proteins in Yeast.
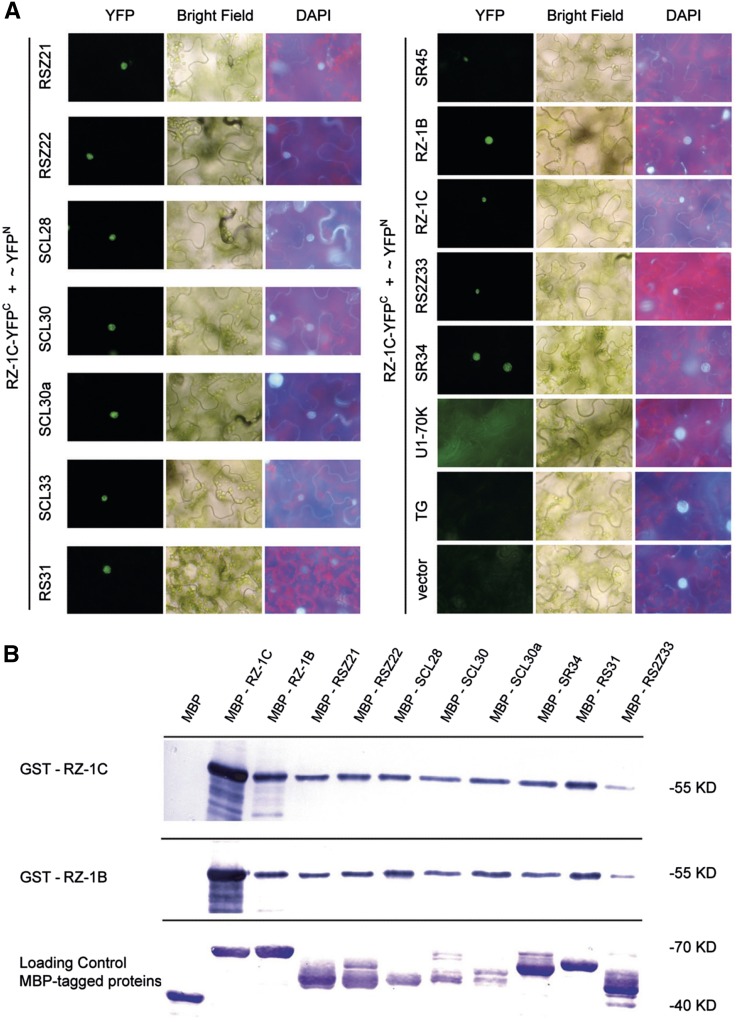
Next, the interactions between RZ-1B/RZ-1C and SR proteins were confirmed in wild tobacco (Nicotiana benthamiana) leaf epidermal cells, with U1-70K, an empty vector, and TRANSLUCENT GREEN (TG), an unrelated nuclear protein (Zhu et al., 2014), used as negative controls. The recombined YFP fluorescence signal was detected in the nucleus for all interaction pairs (Figure 5A). Interestingly, the interaction of RZ-1B and RZ-1C with SR34 and RSZ33 was also observed in wild tobacco leaf epidermal cells. In in vitro pull-down assays, GST-fused RZ-1B and RZ-1C strongly interacted with MBP-tagged SR proteins, with the exception of RSZ33 (Figure 5B). Formation of RZ-1B/RZ-1C heterodimer and their corresponding homodimers was also confirmed (Figure 5B). These results show that RZ-1B and RZ-1C are able to interact with a wide range of SR proteins, with preference for RSZ and SC/SCL subfamily members (Barta et al., 2010).
Figure 5.
Interaction of RZ-1B and RZ-1C with SR Proteins in N. benthamiana Leaf Cells and in Vitro System.
(A) BiFC assay in wild tobacco epidermal cells. RZ-1C fused with YFPC. SR proteins, U1-70 K, and nuclear localized control protein TG were fused with YFPN. Different panels show YFP fluorescence, bright field, and DAPI fluorescence, respectively.
(B) Pull-down assay of RZ-1B and RZ-1C with several SR proteins. Amylose beads anchored to MBP-tagged SR proteins were used to pull down the GST-tagged RZ-1B and RZ-1C from solution. The images show the results of protein gel blotting using anti-GST antibody (upper and middle panels) and anti-MBP antibody (lower panel), respectively.
Next, we identified the RZ-1B/RZ-1C domain responsible for SR protein interaction using RZ-1C as an example. We generated a series of truncated versions of RZ-1C and tested their interactions with SR proteins using yeast two-hybrid assays. The glycine/arginine/aspartic acid-rich stretch at the C terminus of RZ-1C was required and sufficient for SR protein interaction (Table 2; Supplemental Figure 7). Therefore, the C terminus of RZ-1C is responsible for its nuclear localization and SR protein interaction.
Table 2. Interaction of Truncated RZ-1C Proteins with SR Proteins in Yeast.
| Proteins Tested | RRM | RRM+ZF | CT+ZF | CT |
|---|---|---|---|---|
| RSZ21 | − | − | +++ | +++ |
| RSZ22 | − | − | +++ | +++ |
| SCL28 | − | − | +++ | +++ |
| SCL30 | − | − | +++ | +++ |
| SCL30a | − | − | +++ | +++ |
| SCL33 | − | − | +++ | +++ |
| RS31 | − | − | +++ | +++ |
| SR45 | − | +++ | +++ | +++ |
| RZ-1B | − | − | +++ | ++ |
| RZ-1C | − | − | +++ | + |
| Positive control | ++++ | ++++ | ++++ | ++++ |
| Negative control | − | − | − | − |
ZF, zinc finger; CT, C-terminal domain.
RZ-1C Binds a Purine-Rich Element through Its N-Terminal RRM Domain
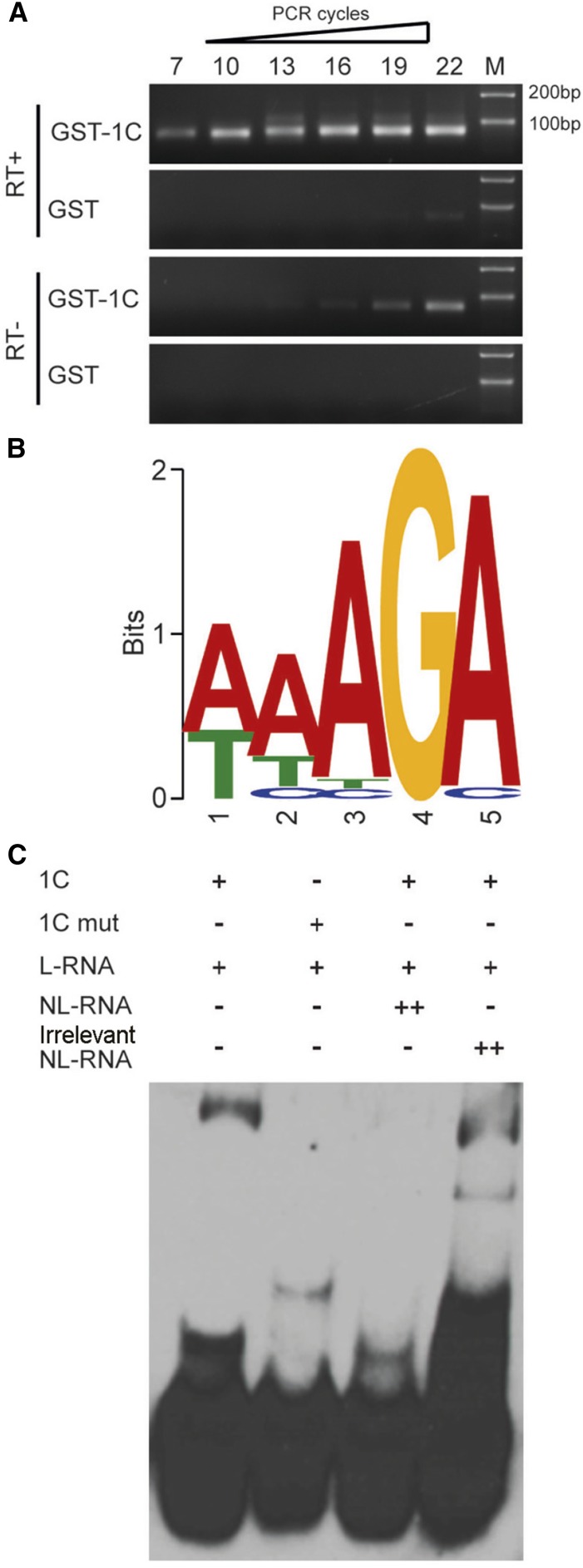
Because RZ-1B and RZ-1C both have an RRM domain (a putative RNA binding domain), we determined whether RZ-1C binds to RNA and determined its sequence preference for binding by performing systematic evolution of ligands by exponential enrichment (SELEX) experiments. RZ-1C was expressed as a GST fusion protein and selected against random RNA sequences. The resulting sequences were reverse transcribed into cDNA, after which they were transcribed in vitro into RNA and used for the next round of selection. The RNA enriched after seven rounds of selection was converted into cDNA and sequenced. The resulting sequence was highly enriched in purine and contained a short AAAGA motif (Figures 6A and 6B; Supplemental Figure 8). We verified the interaction of RZ-1C with this purine-rich RNA in gel shift assays using a sequence of four repeats of 5′-AAAGAAG-3′ as a probe. Migration of the probe through the gel was retarded by GST-RZ-1C; however, the retarded migration of the probe was abolished by the addition of more unlabeled probe, but not by the addition of another unrelated unlabeled RNA, i.e., four repeats of 5′-UUUCUUC-3′, indicating the binding specificity of RZ-1C to the probe (Figure 6C). To test whether the RRM domain was required for binding, we generated point mutations (F10A and F50A) in the RRM domain according to a published structural analysis (Schmitzová et al., 2012). These mutations abolished the ability of GST-RZ-1C to bind the probe (Figure 6C), indicating that the RRM domain is required for the binding of RZ-1C to the purine-rich RNA motif AAAGA.
Figure 6.
RZ-1C Protein Binds to the AAAGA Motif.
(A) Gel electrophoresis of the enriched fragment after seven rounds of selection in the SELEX experiments. M, size markers.
(B) Sequence logo of the enriched motif. The y axis (letter size) indicates the information content (bits) of each position.
(C) In vitro binding of RZ-1C protein to the AAAGA motif in the EMSA assay. 1C, RZ-1C protein; 1C mut, RZ-1C protein with mutations F10AF50A; L-RNA, biotin-labeled RNA; NL-RNA, nonlabeled RNA; Irrelevant NL-RNA, four repeats of nonlabeled 5′-UUUCUUC-3′.
Overexpression of the RZ-1C C Terminus Phenocopied the rz-1b rz-1c Double Mutant
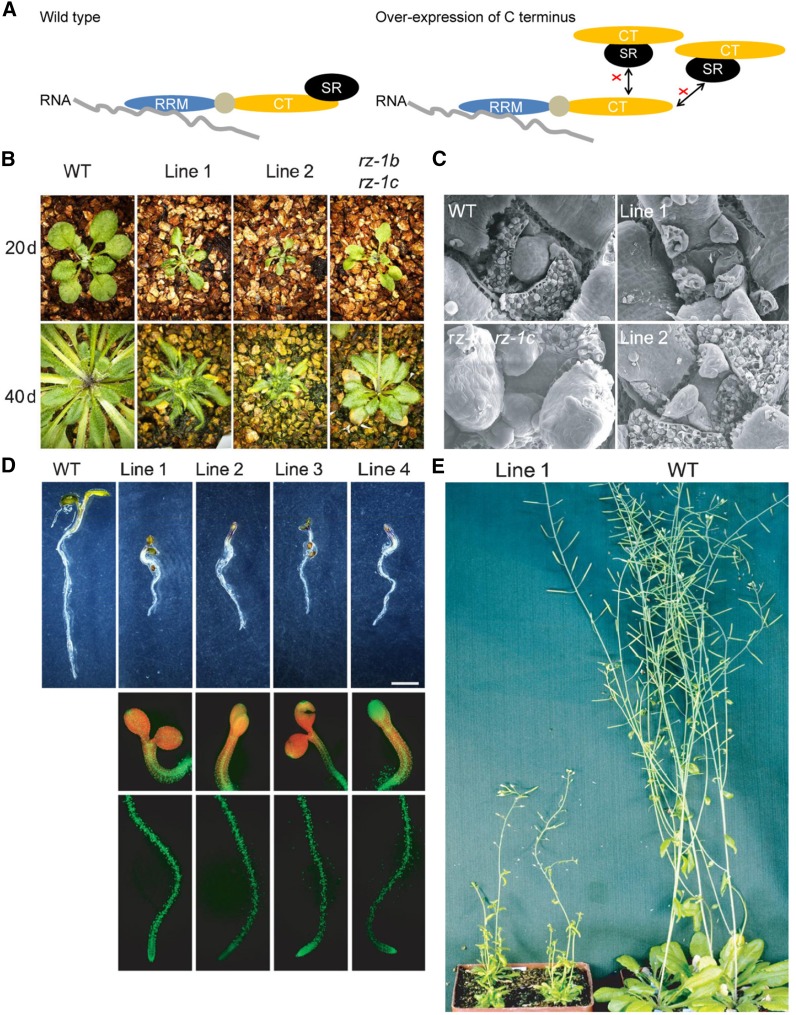
The results described above showed that the N terminus and C terminus of RZ-1C were responsible for its RNA binding and SR protein binding activities, respectively. We hypothesized that overexpression of the C terminus (35S:CT) of RZ-1C in wild-type plants could disrupt binding between endogenous RZ-1B/RZ-1C and SR proteins, thus disconnecting RNA binding activity (conferred by the N-terminal RRM) from SR protein and/or other protein binding (conferred by the C-terminal domain) (Figure 7A). As expected, defective phenotypes were observed in 35S:CT plants, including smaller cotyledons and true leaves (Figure 7B), defective SAMs (Figure 7C), shorter roots (Figure 7D), and dwarfism (Figure 7E), which were reminiscent of the rz-1b rz-1c double mutant phenotype. In contrast, no morphological phenotype was observed in wild-type plants overexpressing the full length RZ-1C transcript, with the exception of accelerated flowering time (see below). These results indicate that the connection between the RRM and C terminus of RZ-1C is critical for its function. Moreover, these findings suggest that C terminus-mediated interactions with proteins including, but not limited to, SR proteins are critical for the functioning of RZ-1B and RZ-1C in plant development.
Figure 7.
Overexpression of the RZ-1C C Terminus Mimics the Phenotype of the rz-1b rz-1c Double Mutant.
(A) Schematic illustration of the experimental design. Overexpression of C terminus of RZ-1C (35S:CT) would disconnect the RNA binding activity from the SR protein binding activity of endogenous RZ-1C.
(B) Plant stature of the wild type, 35S:CT lines 1 and 2, and rz-1b rz-1c after growth in soil for 20 and 40 d after germination.
(C) Scanning electron microscope images of the SAMs of the wild type, 35S:CT lines 1 and 2, and rz-1b rz-1c.
(D) Comparison of seedling root length of wild-type and 35S:CT lines. YFP signals from the transgenic lines are shown below. Bar = 2 mm.
(E) Comparison of plant stature between the wild type and 35S:CT line 1 at 55 d after germination.
RZ-1B and RZ-1C Have Global Impacts on Gene Expression
To elucidate the pleiotropic effects of mutations in RZ-1B and RZ-1C, we performed RNA-seq analysis on wild-type and rz-1b rz-1c double mutant seedlings at 2 weeks of age. We found 3176 differentially expressed genes (difference > 1.5-fold, P < 0.01) in the wild-type and double mutant seedlings (Supplemental Data Set 1), revealing the global roles of RZ-1B and RZ-1C as regulators of gene expression.
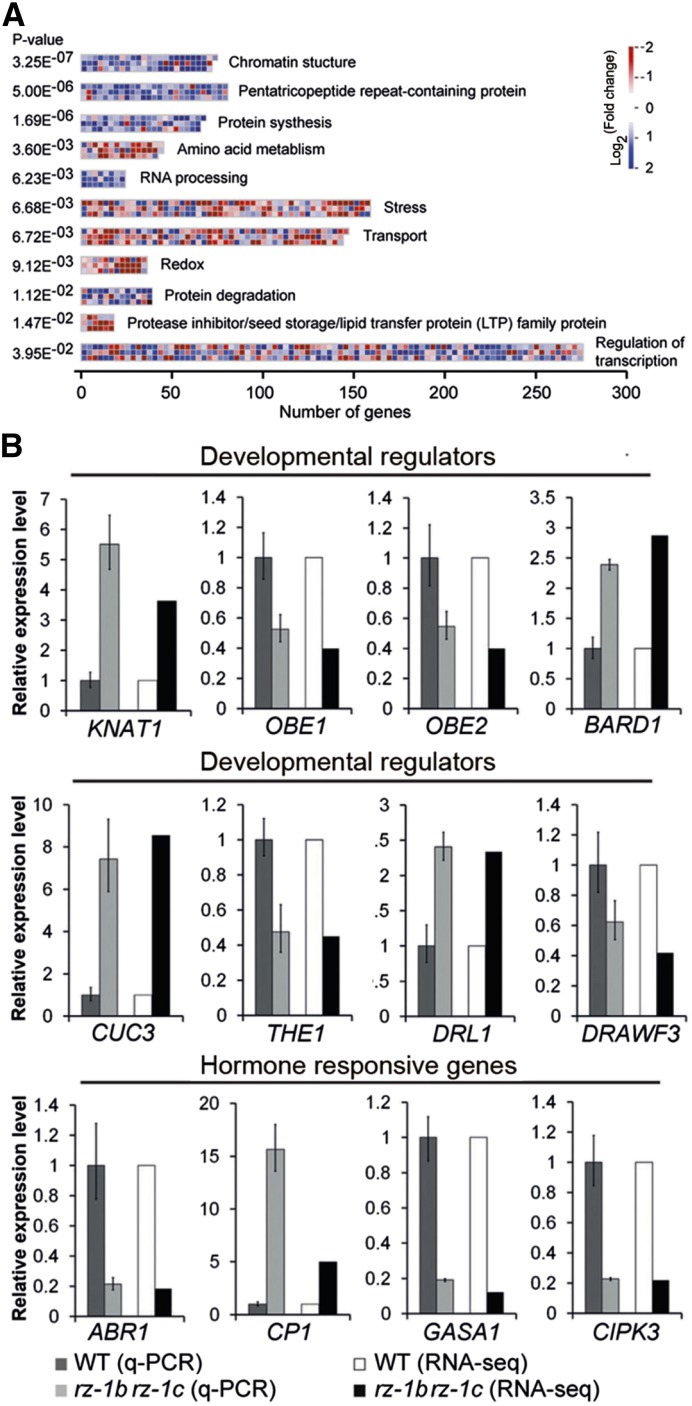
Assessing the set of 3176 differentially expressed genes using MapMan pathway analysis (Thimm et al., 2004) showed that the set was enriched in genes involved in essential processes, including chromatin structure, RNA processing, protein synthesis and degradation, lipid metabolism, stress, and regulation of transcription (Figure 8A; Supplemental Data Set 2). We also found correlations between the phenotypes of rz-1b rz-1c double mutants and expression changes in key regulators (Figure 8B). For example, upregulation of the homeobox gene KNAT1 might explain the leaf serration phenotype (Chuck et al., 1996); downregulation of OBERON1 (OBE1)/OBE2 and upregulation of BARD1, CUP SHAPED COTYLEDON3, and DEFORMED ROOTS AND LEAVES1 might underlie the defect in SAM development (Chuck et al., 1996; Nelissen et al., 2003; Vroemen et al., 2003; Hibara et al., 2006; Han et al., 2008; Saiga et al., 2008); and reduced expression of THESEUS1 and DWARF3 could contribute to the defect in root growth (Szekeres et al., 1996; Hématy et al., 2007). In addition, many hormone-related genes were differentially expressed (Figure 8B; Supplemental Data Set 3). Altered expression of hormone-related genes could be a reflection of changes in hormone homeostasis and might contribute to the pleotropic phenotypes observed in the double mutant plants. Therefore, the multiple growth defects of the rz-1b rz-1c double mutant plants were likely a consequence of changes in the expression levels of many developmental regulators and hormone-related genes.
Figure 8.
The Effect of the Loss of Function of RZ-1B and RZ-1C on the Transcriptome.
(A) Pathways and functional groups that were enriched among the 3176 genes (analyzed with MapMan software). The figure shows the enriched groups with corrected P value < 0.05 (Benjamini Hochberg correction). The folds of alteration (log2 (rz-1b rz-1c/WT)) are indicated by different levels of red (≤0.585) and blue (>0.585) coloring.
(B) Confirmation of selected genes with altered expression in rz-1b rz-1c by qRT-PCR. Different genes are grouped according to their published functions. The fold changes from the RNA-seq are shown in parallel with the qRT-PCR data. Values mean ± se from three biological replicates.
RZ-1B and RZ-1C Regulate Pre-mRNA Splicing of Many Genes
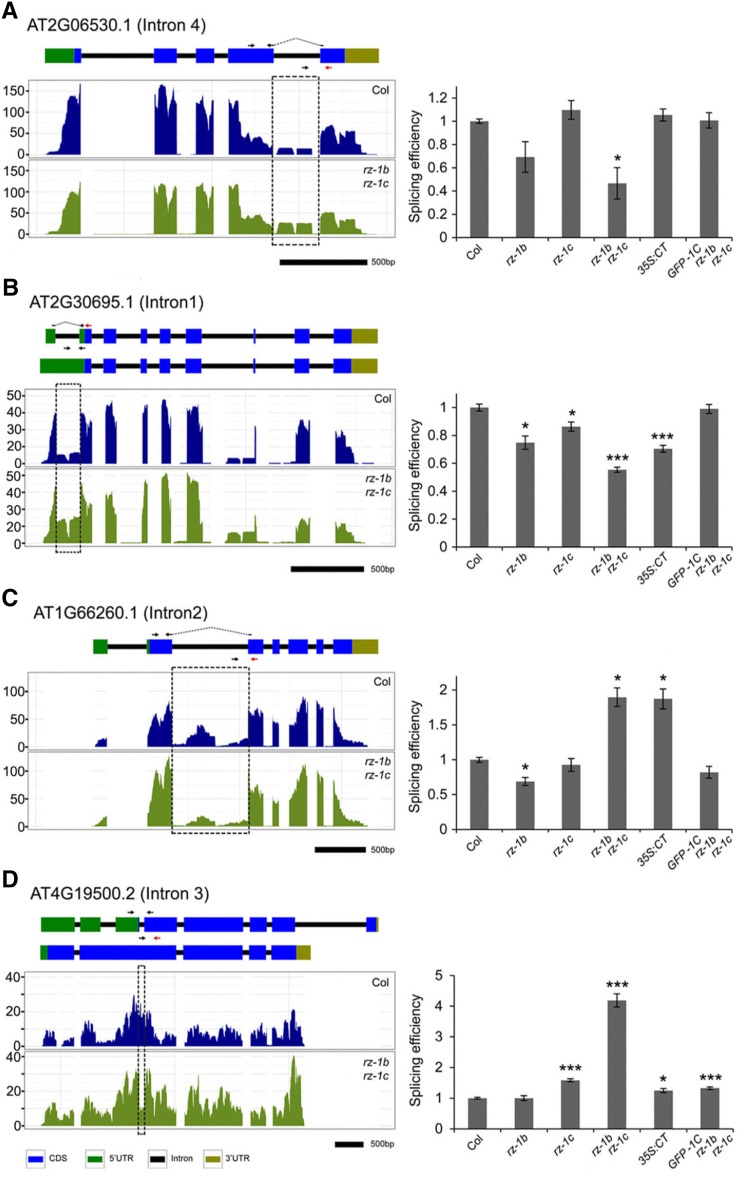
The interactions of RZ-1B and RZ-1C with SR proteins involved in regulating splicing suggest that RZ-1B and RZ-1C might regulate splicing; therefore, we searched our RNA-seq data for splicing defects. We examined introns that were differentially expressed relative to their adjacent exons, as previously reported (Deng et al., 2010). We found that, in comparison with those in wild-type plants, 52 introns (from 50 genes) were more efficiently spliced, while 134 introns (from 109 genes) were less efficiently spliced (P < 0.01, intron reads >5 reads per kilobase of exon model per million mapped reads [RPKM], intron coverage >95%) in the rz-1b rz-1c double mutant plants (Supplemental Data Set 4). Next, eight introns were selected for further validation of the splicing defects via measurement of splicing efficiency (the ratio of spliced RNA over unspliced RNA) with qRT-PCR. The splicing defects in seven introns were validated by qRT-PCR (Figure 9; Supplemental Figure 9). Moreover, five of the seven introns exhibited splicing defects similar to those that were observed in 35S:CT plants (Line 1, Figure 7), suggesting that C terminus-mediated protein interactions are important for the proper splicing of those introns. These results suggest that RZ-1B and RZ-1C regulate pre-mRNA splicing of many genes, consistent with the interaction of RZ-1B and RZ-1C with SR proteins.
Figure 9.
RZ-1B and RZ-1C Regulate Pre-mRNA Splicing of Many Genes.
RNA-seq and qPCR validation of four selected genes which were differentially spliced in the rz-1b rz-1c double mutant. The left panel shows wiggle plots of RNA-seq data with the schematic of genes at the top. The arrows indicate the locations of primers used to measure the splicing efficiency of introns. The spliced and unspliced primers are shown at the top and bottom of each diagram, respectively. The spliced primers that cross exon-exon junctions are shown with dashed lines over the introns. The primers in red were used for reverse transcription. The dashed boxes indicate introns which are differentially spliced. Values on the y axis indicate reads per 10 million (RP10M). The right panel shows the splicing efficiency of the selected introns in different genetic backgrounds. The splicing efficiencies were calculated as the level of spliced RNA normalized to the level of unspliced RNA, with qPCR primers shown on the left panel. Values are mean ± se from at least three biological replicates. Asterisks indicate statistical significance: *P < 0.05, **P < 0.01, and ***P < 0.001, two-sided unpaired t test. Three other examples of altered splicing observed in the rz-1b rz-1c mutant are presented in Supplemental Figure 9. GFP-1C represents GFP-RZ-1C (driven in its native genomic context).
RZ-1B and RZ-1C Regulate FLC Intron Splicing and Transcription
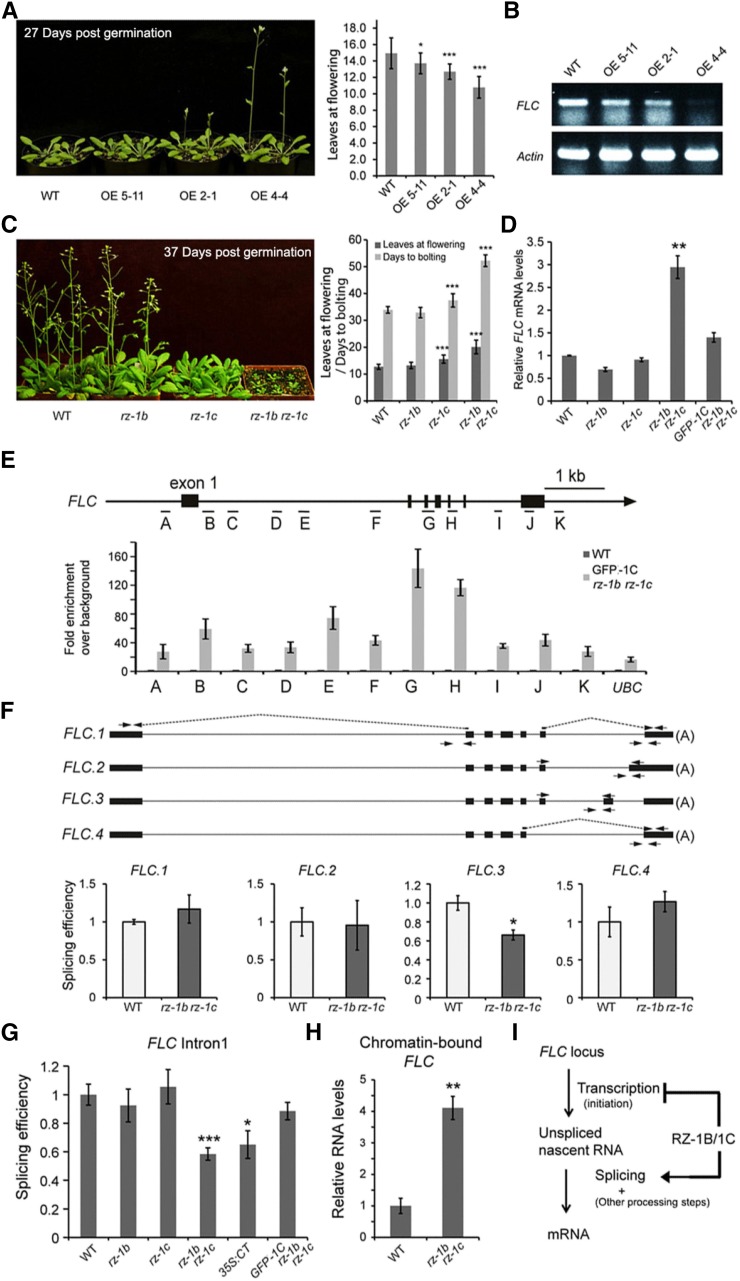
Because overexpression of full-length RZ-1C resulted in early flowering (Figure 10A), we determined whether FLC expression was altered using qRT-PCR. The mRNA level of FLC was reduced in the RZ-1C-overexpressor plants, implying that RZ-1C suppresses the expression of FLC (Figure 10B). Downregulation of FLC expression by RZ-1C was further supported by the late flowering phenotype and FLC upregulation exhibited by the rz-1b rz-1c double mutants (Figures 10C and 10D). Using chromatin immunoprecipitation (ChIP), we found that RZ-1C bound to FLC chromatin with the highest enrichment over the distal exon patches (Figure 10E), suggesting that FLC is a direct target of RZ-1C. Because we found that RZ-1B and RZ-1C are involved in regulation of splicing, we determined whether FLC splicing was altered in the rz-1b rz-1c double mutant plants. First, the splicing efficiency of featured introns was assessed for four different isoforms of FLC. The splicing efficiency of intron 6 of isoform 3 was significantly reduced in the double mutant plants, while that of the other tested introns remained unchanged (Figure 10F). Because FLC.1, which encodes a functional FLC protein, is the most abundantly expressed isoform among the four different isoforms, while the others are expressed in much lower abundance (Supplemental Figure 10A), changes in splicing efficiency of FLC.3 intron 6 would only have a minor influence on the level of FLC.1 mRNA. In order to investigate the reason why FLC mRNA levels increased, we further analyzed the splicing efficiency of FLC intron 1, a constitutively spliced intron. The splicing efficiency of FLC intron 1 was reduced in rz-1b rz-1c double mutant and 35S:CT plants (Figure 10G). Therefore, RZ-1B and RZ-1C are required for efficient intron splicing of FLC.
Figure 10.
RZ-1B/1C Regulate FLC Intron Splicing and Transcription.
(A) Early flowering phenotype was observed in RZ-1C overexpressing plants. Right panel shows flowering time of three independent overexpression (OE) lines under long-day conditions. Error bars represent sd (n = 14 to 16).
(B) Reduced FLC levels in different overexpression lines as examined by one-step RT-PCR. Actin was used as an internal control.
(C) Late flowering phenotype was observed in rz-1b rz-1c double mutants. Right panel shows flowering time in different genetic backgrounds. Error bars represent sd (n = 18 to 25).
(D) FLC mRNA level are increased in rz-1b rz-1c double mutants, as examined by qPCR. Values are mean ± se from two or three biological repeats.
(E) RZ-1C associates with FLC chromatin. ChIP analysis was conducted in complementation line 1 (Supplemental Figure 4) using anti-GFP antibody. A schematic of FLC and primer locations is shown. Error bars represent se from four biological repeats.
(F) Examination of splicing efficiency of featured introns associated with different isoforms of FLC in rz-1b rz-1c mutants. A schematic of FLC isoforms is shown at the top; primer locations are indicated by arrows (same as in Figure 9). Values are mean ± se from three biological repeats.
(G) RZ-1B/1C promote the efficient splicing of FLC intron 1. Splicing efficiency of FLC intron 1 was examined in different background. Primer locations are indicated in (F). Values are mean ± se from three to five biological repeats.
(H) Increased level of nascent FLC in rz-1b rz-1c mutants. qPCR was performed on chromatin-bound RNA using primers binding to FLC exon 7. Values are mean ± se from three or four biological repeats.
(I) A proposed model of how RZ-1B/1C regulate FLC. RZ-1B/1C promote the splicing of FLC introns while inhibiting its transcription.
(A) to (H) Asterisks indicate statistical significance: *P < 0.05, **P < 0.01, and ***P < 0.001, two-sided unpaired t test. GFP-1C represented GFP-RZ-1C (driven in its native genomic context).
The relationship between altered FLC splicing and expression was investigated. It is reasonable to predict that decreased splicing efficiency of FLC intron 1 would result in an increased level of unspliced RNA but result in a reduced level of spliced/mature RNA. However, we found that the abundance of spliced FLC mRNA was increased in the rz-1b rz-1c double mutant plants (Figure 10D), suggesting that, although splicing efficiency was reduced, either FLC transcription was enhanced or the mRNA turnover rate was reduced. Next, nascent FLC abundance in the chromatin-bound RNA fraction was measured to test whether the increased FLC mRNA abundance was due to increased transcription in compensation for the decrease in splicing efficiency. Nucleoplasmic RNA was removed by washing the nuclei with urea; however, RNA that was mostly RNA polymerase II-bound and undergoing transcription was retained (Wuarin and Schibler, 1994; Khodor et al., 2011; Bhatt et al., 2012; Bentley, 2014; Mayer et al., 2015; Wu et al., 2016). As expected, the abundance of unspliced FLC RNA was greatly enriched in the chromatin-bound RNA fraction (Supplemental Figure 10B). The abundance of nascent FLC at exon 7 (the last exon) was increased in the rz-1b rz-1c double mutant plants (Figure 10H), suggesting an increased level of transcription. Taken together, these results demonstrate that RZ-1B and RZ-1C promote efficient splicing of FLC introns and repress FLC transcription (Figure 10I).
DISCUSSION
In this study, we provided evidence that two Arabidopsis proteins, RZ-1B and RZ-1C, represent a unique group of RNA binding proteins that play important roles in plant development, pre-mRNA splicing, and general gene expression. Loss of function of RZ-1B and RZ-1C conferred severe defects in various aspects of development, including delayed germination, retarded root and shoot meristems, late flowering, reduced stature, and serrated leaves. RZ-1B and RZ-1C have a similar nuclear localization pattern and similar binding activity to a group of SR proteins. RZ-1C binds to purine-rich RNA sequences through its N-terminal RRM, while interacting with SR proteins through its C terminus. Dominant-negative analysis revealed that the C terminus-mediated interactions of RZ-1C are essential for RZ-1C function. Simultaneous inactivation of RZ-1B and RZ-1C produced global perturbation of the expression levels of thousands of genes, including those of many key developmental regulators and hormone-related genes. RZ-1B and RZ-1C were required for faithful pre-mRNA splicing of many genes, consistent with their interactions with SR proteins. Furthermore, RZ-1B and RZ-1C promote efficient splicing of FLC intron 1 while repressing FLC transcription.
The hnRNPs are a group of loosely defined proteins that bind hnRNA but do not belong to well-defined RNP groups (e.g., snRNPs). At RZ-1 proteins are homologs of tobacco (N. sylvestris) hnRNP protein RZ-1, which was identified as a putative homolog of hnRNP G based on sequence similarity and bioinformatics analysis (Wang and Brendel, 2004). Indeed, At RZ-1 proteins have amino acid compositions similar to that of human hnRNP G at the C terminus, while RZ-1 proteins and human hnRNP G both feature a single RRM domain at the N terminus. However, At RZ-1s feature a CCHC-type zinc finger that is not present in hnRNP G; moreover, the amino acid sequences of the C termini of the two protein types are poorly aligned. Interestingly, according to our phylogenetic analysis, the overall domain structure and amino acid sequences of At RZ-1B and At RZ-1C are highly conserved across many plant species. Therefore, we propose that At RZ-1B and At RZ-1C represent a plant-specific type of RNA binding protein with a unique domain arrangement and C terminus amino acid sequence. It is worth noting that hnRNP G has been reported to interact with SR protein SRP75 (Wang et al., 2011). Therefore, it is possible that the C termini of At RZ-1B and At RZ-1C might adopt a 3D structure similar to that of hnRNP G, despite differences in their amino acid sequences.
Complex interaction networks between SR proteins have been reported (Lopato et al., 2002). Several SR members interact with components of the U1 and U2 subunits of the spliceosome, including U170K (Golovkin and Reddy, 1999). In this study, we did not observe interaction between RZ-1B/1C and U170K, suggesting that RZ-1B and RZ-1C are probably not directly associated with the core spliceosome. Using a yeast two-hybrid system, a plant transient expression system (bimolecular fluorescence complementation [BiFC]), and in vitro pull-down assays, we found that RZ-1B and RZ-1C have the potential to interact with almost all of the SR proteins tested in this study. Overexpression of the C terminus of RZ-1C, a domain responsible for interaction with SR proteins, resulted in severe developmental defects similar to those observed in the rz-1b rz-1c double mutants. Therefore, proper interactions between RZ-1B/1C and SR proteins are likely critical for their functions. The interactions between RZ-1B/1C and SR proteins in vivo are probably highly dynamic, possibly limited by the distribution of SR proteins in different tissues and at different developmental stages, and regulated by posttranslational modification such as phosphorylation. Currently, we cannot distinguish whether RZ-1B/1C and their interacting SR proteins form one complex or multiple complexes. RZ-1B/1C may form a large complex with several different SR proteins, as well as their RNA targets, which involve higher order interactions, as reported recently for mammalian SR proteins (Singh et al., 2012).
Through RNA-seq analysis, we observed altered pre-mRNA splicing of more than 100 genes and global changes in gene expression, including those of many key developmental regulators, in rz-1b rz-1c double mutants. Therefore, the pleiotropic phenotypes observed in rz-1b rz-1c double mutants might be a collective effect of disrupted expression of developmental regulators at the molecular level. Interestingly, among the 3176 differentially expressed genes, only 31 were differentially spliced (Supplemental Data Set 1), suggesting that RZ-1B/1C may regulate a large number of genes in a manner independent of splicing. However, it is worth noting that, as indicated in this study, mRNA/RNA-seq is most suited for determining differences in alternative splicing, rather than in constitutive splicing, because of the generally very low (mostly null) signal of most constitutively spliced introns in comparison with those of their adjacent exons (Khodor et al., 2011; this study). Therefore, RZ-1B and RZ-1C might regulate the splicing of constitutive introns to affect gene expression. Related to this hypothesis, another question not yet answered is the number of genes directly targeted by RZ-1B/1C. In this study, we found that RZ-1C preferentially binds to the purine-rich RNA motif AAAGA, similar to SR proteins in mammals (Long and Caceres, 2009). The degenerative and widespread nature of such a motif makes it difficult to make a genome-wide prediction of the direct targets of RZ-1B/1C. However, for genes that were less efficiently spliced in rz-1b rz-1c double mutants, the AAAGA motif was more enriched in 5′-untranslated regions (UTRs) and introns and less enriched in 3′-UTRs and exons (Supplemental Table 2), suggesting that RZ-1B and RZ-1C are closely associated with the promotion of splicing of such genes. It is worth noting that the consensus binding sequence may not be sufficient (and in some cases not essential) for in vivo binding, as reported in vivo binding of SR proteins to their targets in mammals is context-specific and highly dynamic (Pandit et al., 2013; Fu and Ares, 2014). We speculate that the direct targets of RZ-1C are likely to be widespread, given its broad effect on gene expression and its tight association with chromatin. Future studies will test these hypotheses by investigating nascent RNA and RZ-1B/1C localization through deep sequencing.
RZ-1B and RZ-1C are redundantly required for suppression of the expression of FLC, one of the central regulators of flowering time in Arabidopsis. Intron splicing of FLC antisense RNA COOLAIR was found to be important for regulation of FLC involving alterations in chromatin (Marquardt et al., 2014), in which case intron splicing of the sense FLC transcript was not affected. In this study, we found that RZ-1B and RZ-1C promoted efficient splicing of FLC sense large intron 1, adding another layer to FLC regulation. Apart from FLC intron 1, other constitutive introns are also likely to be spliced less efficiently in the rz-1b rz-1c double mutant, since the ratio of unspliced FLC RNA (across introns 2 and 3) to spliced FLC RNA (introns 4, 5, and 6 spliced) is higher in the double mutant (Supplemental Figure 10B). This is consistent with the observation that RZ-1C is distributed over the entire FLC locus, with the highest enrichment in the small exon patches. Therefore, we speculate that RZ-1B and RZ-1C may function redundantly as cotranscriptional regulators of FLC pre-mRNA splicing. In addition to promoting FLC splicing, we found that RZ-1B and RZ-1C repress FLC transcription; therefore, upregulation of FLC mRNA in rz-1b rz-1c double mutants is a combined consequence of increased transcription and decreased splicing (Figure 10). Currently it is not clear whether the observed effects on transcription and splicing are internally coupled (e.g., through altered transcriptional initiation and elongation) or happen as two independent events. In addition to splicing, mammalian SR proteins also regulate gene transcription by altering the transition of RNA polymerase II from the 5′ pause to productive elongation (Ji et al., 2013). Future studies will test whether there is a general link between transcription and splicing as a function of RZ-1B and RZ-1C and their interacting SR proteins.
In this study, functional analysis was used to show that two SR-interacting and hnRNP-like proteins, RZ-1B and RZ-1C, are important regulators of plant development, pre-mRNA splicing, and gene expression. Our work sheds light on the functions of RZ-1 proteins in plants.
METHODS
Phylogenetic Analysis
Homologs of RZ-1 proteins were identified in Phytozome (www.phytozome.net). Protein sequences were adjusted to ESTs using tBLASRn. Protein sequence alignment was performed in ClustalX 1.86. The alignment (Supplemental File 1) was manually edited using Jalview 2.4 software. The aligned sequences were analyzed using PHYLIP 3.68 with default parameters. The rooted tree was constructed using the neighbor-joining algorithm; human hnRNP G was taken as the outgroup. The bootstrap values were calculated with 1000 replicates. The phylogenetic tree was visualized using TreeView 1.6.6.
Plant Material and Growth Conditions
Arabidopsis thaliana ecotype Columbia served as the wild type. All transgenic lines used in this study were of the Columbia ecotype. Plants were grown as previously described (Qin et al., 2005). Seeds were surface sterilized, sown on 0.5× Murashige and Skoog (MS) plates (1% sucrose and 0.8% phyto agar), stratified for 3 d at 4°C, and transferred to a Percival LT-36VL growth chamber (16-h white light [Philips F17T8/TL841] and 8-h-dark cycles at 22°C with light intensity of ∼170 μmol/m2/s). The plated seedlings were then used for phenotypic analysis or transferred to soil at 2 weeks postgermination for yield seeds. For the flowering time assay and FLC-related experiments, seeds were sown and germinated in MS plates (3% glucose and 0.8% phyto agar). The rz-1b mutant (SALK_001328) was obtained from the ABRC, while the rz-1c mutant (GABI-030b12) was obtained from the GABI-KAT (www.gabi-kat.de/). The insertion sites of each mutant were confirmed by PCR and sequencing.
Genotyping of the T-DNA Insertion Mutants
Primers Salk001328-F and Salk001328-R (Supplemental Data Set 5) were used to amplify the RZ-1B wild-type allele. The rz-1b mutant allele was detected using SALK T-DNA-specific primer LBb1.3 and Salk001328-R. Primers GABI-030B12-F and GABI-030B12-R were used to amplify the RZ-1C wild-type allele. GABI-TDNA-LB and GABI-030B12-F were used to detect the T-DNA insertion in the rz-1c mutant allele.
Germination Assay
For the seed germination assay, wild-type, rz-1b, rz-1c, and rz-1b rz-1c double mutant siliques were ripened naturally and dried naturally in a chamber (22°C, 50% humidity) for 3 months. In triplicate experiments, 80 to 120 seeds of each genotype were surface-sterilized and sown individually on 0.5× MS plates (minus sucrose), after which the plates were transferred immediately to a growth chamber (16 h light and 8 h dark, 22°C) without stratification. The germination rate (radical emergence) was determined at different time point under a dissecting microscope.
Microscopy
For imaging of the shoot apical meristem, seedlings were harvested from 0.5× MS plates 7 d postgermination, fixed and dehydrated as previously described (Guo et al., 2009), and CO2 critical-point dried for 3 h (Leica EM CPD030). The samples were then attached to the sample plate with the SAM region facing upward. The SAM region was exposed by removing excess leaves under a dissecting microscope. The samples were coated with platinum powder and photographed under a scanning electron microscope (JSM 6610 LV; JEOL).
With the exception of the BiFC assay, all fluorescence imaging was performed using a Leica SPE confocal microscope. The excitation laser was set at 405 nm for 4′,6-diamidino-2-phenylindole (DAPI) staining, 488 nm for GFP/YFP, and 532 nm for propidium iodide staining of the root cells. Root tips from vertically growing plants were collected 5 d postgermination for propidium iodide staining by following a published protocol (http://sites.duke.edu/benfey/protocols/confocal-imaging-of-roots/).
Complementation Assay
The RZ-1C promoter, open reading frame plus 3′-UTR, and eGFP with overlap extension were amplified individually with primers listed in the primer list (Supplemental Table 2). Two complementary primers, VecGenome-F and VecGenome-R, were annealed to form a double-strand DNA, which was transferred into TOPO vectors using a TOPO PCR cloning kit (Invitrogen). Primers LivGenomie-F and LivGenomie-R were used to linearize the vector by PCR, after which the promoter, eGFP, and open reading frame plus 3′-UTR were amplified. Four fragments were added into a system capable of generating a vector using circular polymerase extension cloning, as previously described (Quan and Tian, 2011). The destination vector was generated from the pEntry clone and Gateway vector pH7FWG0 by an LR reaction. The construct was transformed into Agrobacterium tumefaciens GV3101 and transformed into the rz-1b rz-1c double mutant plants by floral dip (Liu et al., 2013). The transformants were identified through antibiotic selection and PCR confirmation, after which the T2 generation was generated and several independent lines were used for phenotype analysis.
Isolation of Total Nuclear, Nucleoplasmic, and Chromatin-Bound Proteins
Chromatin-bound protein was isolated based on a previous report (Uchiyama and Wang, 2004). Seedlings grown for two weeks were harvested and cross-linked (1% formaldehyde), followed by nucleus isolation using Honda buffer, as described previously (Wierzbicki et al., 2008). Three volumes of 1% SDS (10 mM Tris, pH 7.5, 2 mM EDTA, and 1× proteinase inhibitor) were added to the nucleus pellet, which was vortexed for 1 min, followed by centrifugation at 14,000g for 10 min. The supernatant containing soluble nucleoplasmic proteins was collected. The remaining pellet was resuspended in 3 volumes of 1% SDS and sonicated, followed by centrifugation at 14,000g for 10 min. The supernatant containing chromatin-bound proteins was collected. Total nuclear protein was isolated by adding 3 volumes of 1% SDS directly to the nucleus pellet, followed by boiling at 95°C for 10 min. Equal volumes of each fraction were mixed with loading buffer, boiled, gel-separated, and subjected to protein gel blot analysis. Anti-H3 (ab1791, lot GR135321-1) (1:5000 dilution) and anti-GFP (ab290, lot GR240324-1) (1:2500 dilution) antibodies from Abcam were used in this assay.
Yeast Two-Hybrid Screening and Verification
The yeast two-hybrid screening procedure was described previously (Wu et al., 2011). Briefly, the full-length cDNA of RZ-1C was cloned into the pGBKT7 vector (Clontech) and transformed into yeast strain AH109. Transformed cells were further transformed with plasmids from a cDNA library constructed from 14-d-old Arabidopsis seedlings. The resulting transformants (∼5 × 106) were screened on SD-Trp-Leu-His-Ade medium supplemented with 2.5 mM 3-amino-1,2,4-triazole (a concentration that suppresses the self-activation activity of GAL4BD-RZ-1C). Surviving clones were replated on the same medium, and LacZ activity was assayed. Positive clones were sequenced. For the confirmation assay, coding sequences (CDSs) of RZ-1C, RZ-1B, and RZ-1A, as well as different fragments of RZ-1C, were amplified from cDNA and inserted into the pGBK-T7 (bait) through EcoRI and SalI restriction recognition sites. The CDS of different SR protein coding genes was amplified from cDNA and inserted into the pACT-2 vector (prey) using restriction enzymes (see Supplemental Table 2 for restriction sites). The prey and bait were then cotransformed into yeast strain AH109. Transformants were picked from SD-Trp-Leu medium, gradient-diluted, and plated on SD-Trp-Leu-His-Ade medium supplemented with 2.5 mM 3-amino-1,2,4-triazole. Results were obtained after 5 d of growth at 30°C.
BiFC Assays
In order to produce a construct for BiFC, the coding regions of RZ-1B/1C and different SR protein-coding genes were amplified and inserted into p35S-SPYNE and p35S-SPYCE (Bracha-Drori et al., 2004), respectively, using various restriction enzymes to cut the restriction sites that were introduced into the primers (Supplemental Data Set 5). The constructs for fusion protein production were verified by sequencing. BiFC assays were performed in 6-week-old wild tobacco (Nicotiana benthamiana) leaves as described previously (Bracha-Drori et al., 2004; Ou et al., 2011). Briefly, combinations of Agrobacterium hosting different vectors were coinfiltrated into wild tobacco leaves. Images were collected 2 d after infiltration. The BiFC signal was detected using the channel for YFP florescence. DAPI staining was also performed to illustrate the nuclear localization of the BiFC signal.
In Vitro Pull-Down
In order to prepare GST-fused RZ-1B/1C and MBP-fused SR proteins, CDSs of RZ-1B/1C were cloned into pGEX4T-1 (GE Healthcare) and CDSs of different SR protein-coding genes were cloned into pMAL-c2x (NEB) using restriction enzymes, after which the resulting constructs were transformed into Escherichia coli BL21 (DE3) cells for protein expression. Cells were grown in Terrific Broth medium at 37°C until the OD600 reached 1.0, followed by induction of expression at 30°C for 4 h in the presence of 1 mM isopropyl β-d-1-thiogalactopyranoside. Soluble MBP-SR proteins and GST-RZ-1C/1B were purified using amylose beads (NEB) and Glutathione Sepharose 4 Fast Flow (GE Healthcare Life Science) separately according to the manufacturer’s instructions. Roughly 200 ng of each MBP-SR protein sample was then rebound to 5 μL (bed volume) of amylose beads individually, after which the beads were incubated with 1 µg purified GST-RZ-1B, GST-RZ-1C, or GST alone in 200 μL of binding buffer (10 mM Tris, pH 7.5, 200 mM NaCl, 0.1% Triton X-100, and 1× C-complete proteinase inhibitor) for 30 min at 20°C. The beads were then washed four times with 1 mL binding buffer and proteins were released by boiling the beads in SDS loading buffer. The released proteins were then detected by protein gel blotting using either monoclonal anti-GST (34860; Qiagen; 1:5000 dilution) or anti-MBP (E8032; NEB, 1:10,000 dilution) antibodies.
Systematic Evolution of Ligands by Exponential Enrichment
SELEX was performed as described previously (Cavaloc et al., 1999) with modifications. To generate the RNA library, 10 µM JK251 primer, which harbors 20 random DNA bases, was annealed with 10 µM JK252 to generate a double-stranded T3 promoter. The hybrid mixture was used as a template and subjected to large-scale in vitro transcription by T3 RNA polymerase (RiboMax kit; Promega). Following the reaction, the DNA template was digested using the Turbo DNase-free kit from Ambion. The resulting RNA (around 50 nucleotides) was precipitated and purified from 12% TBE-PAGE gel with 8 M urea, after which 100 µg of purified RNA was dissolved in 100 μL binding buffer (20 mM HEPES, pH 7.9, 150 mM KCl, 0.1% Triton X-100, 1 mM DTT, and 500 ng/μL tRNA). Next, 1 µg of the purified GST fusion protein was bound to 5 μL of glutathione Sepharose beads, which were washed and stored in 3 bed volumes of binding buffer. The RNA was heat-denatured at 80°C, after which the temperature was slowly reduced to 22°C to allow maximum formation of secondary structures. The binding reaction was performed by mixing the RNA with the beads in the presence of 1 units/μL RNAase Out (Life Science Technology) at 22°C for 30 min with shaking at 1000 rpm. The beads were then washed four times with binding buffer (250 mM KCl) and resuspended in 1× PK buffer (100 mM Tris-Cl, pH 7.5, 50 mM NaCl, and 10 mM EDTA), after which associated RNA was released through proteinase K (200 ng/µL) treatment for 30 min at 37°C. The RNA was precipitated, dissolved in 10 μL RNase-free water, and subjected to reverse transcription with primer JK253 using M-MLV RTase (Takara). Several control samples, including RNA bound to GST alone and a sample without reverse transcriptase, were included at this step. The resulting product was diluted 10 times and subjected to PCR amplification using primers JK252 and JK253. The PCR product was checked after different numbers of cycles. The minimum cycle number that gave a visible band was used for further amplification. The PCR product was gel-purified, after which 500 ng of the product was used as a template for the next round of in vitro transcription. After 7 and 10 rounds of selection, the PCR product was cloned into the pGEM-T vector (Promega) and at least 30 clones were sequenced. The sequences were aligned and analyzed to identify potential consensus sequences using MEME (http://meme.nbcr.net/meme/).
Electrophoretic Mobility Shift Assay
Mutated RZ-1C (F10AF50A) was generated from pMAL-C2X-RZ-1C using a fast mutagenesis system (Transgen). MBP-RZ-1C and MBP-RZ-1C mutant proteins were prepared as described in a previous section. Biotin-labeled and -unlabeled RNAs (5′-AAAGAAG-3′)4 were obtained from Invitrogen. For the binding reaction, 3 μg of different recombinant proteins was incubated with 2 pmol of biotinylated probe in 20 μL of 1× buffer containing 10 mM HEPES (pH 7.3), 20 mM KCl, 1 mM MgCl2, and 1 mM DTT at room temperature for 10 min. For the competition assay, the binding reaction was performed in the presence of 20 pmol (10×) of the unlabeled probe. The RNA-protein mixture was resolved on 10% 1× TAE acrylamide gel under 100 V for 50 min, followed by electrophoretic transfer to positively charged nylon membranes (GE Healthcare). The biotinylated RNA on the membrane was UV cross-linked and detected using chemiluminescent nucleic acid detection following the manufacturer’s instructions (Thermo).
Overexpression of RZ-1C and Its Domains
The RZ-1C C terminus-encoding (144 to 310 amino acids) and full-length RZ-1C gene were amplified and cloned into pK7YWG2 (https://gateway.psb.ugent.be/), respectively (Supplemental Data Set 5). Constructs were transformed into Agrobacterium GV3101 and further transformed into wild-type plants via floral dip. Multiple lines in the T2 generation were randomly selected and subjected to phenotype analysis. For subcellular localization (Figure 4B), fragments of RZ-1C, including the RRM, RRM plus zinc finger, and C terminus plus zinc finger, were cloned to generate transgenic lines. For the flowering time assay, the full-length RZ-1C gene was cloned into pJim19-HA-FLAG (Zhu et al., 2014) to generate transgenic plants in which HA-FLAG-RZ-1C was driven by the CaMV 35S promoter.
ChIP
ChIP was conducted according to a previously described protocol (Wierzbicki et al., 2008; Wang et al., 2014). Chromatin was prepared from cross-linked material using Honda buffer, sonicated, diluted, and subjected to immunoprecipitation using anti-GFP (Abcam; ab290, lot GR240324-1) antibodies and Dynabeads Protein A. The immunoprecipitation was performed at 4°C for 3 h, after which the beads were washed four times with ChIP dilution buffer and twice with 1× TE buffer (10 min each time). Immunoprecipitated DNA was eluted after reverse cross-linking by boiling at 95°C for 10 min, followed by treatment with proteinase K for 1 h at 55°C. For each primer pair, qRT-PCR data were normalized to 1% of the input, whereas the level of the wild type was set to one. As a control for the ChIP, the same chromatin was also used for an H3K4me3 ChIP experiment. The expected profiles of H3K4me3 were observed at FLC, confirming the proper functioning of the ChIP procedure (Supplemental Figure 11).
Quantitative RT-PCR
For qRT-PCR, RNA was extracted from 2-week-old seedlings using the RNeasy Plant Mini Kit (Qiagen). After DNase treatment, 2.5 μg RNA was reverse transcribed with Superscript III (Invitrogen) and oligo d(T)18 or gene-specific primers (for splicing efficiency and FLC expression). cDNA was diluted and used for quantitative PCR using a Roche Lightcycler 480 and SYBR Green Master Mix. Data were presented as average fold changes over the wild type from three biological repeats.
Measurement of Splicing Efficiency
Splicing efficiency measurement was conducted as previously reported (Marquardt et al., 2014). In brief, the splicing efficiency was calculated by determining the level of spliced RNA normalized to the level of unspliced RNA for each intron tested. The unspliced primers were designed strictly to span the intron-exon junction to avoid amplification of the intron lariat. The spliced primers, where possible, were designed such that one primer crossed the exon-exon junction to ensure specific amplification of spliced RNA. The primers located next to the next exon and 2.5 μg RNA were used in the reverse transcription. qPCR assays were performed on diluted cDNA with spliced or unspliced primers. Data are presented as average fold change over the splicing efficiency in wild-type plants from three biological repeats.
Determination of the Nascent RNA Level of FLC
Chromatin-bound/nascent RNA was isolated as previously described with minor modifications (Wuarin and Schibler, 1994; Bhatt et al., 2012; Wu et al., 2016). In brief, nuclei from 2-g seedlings were prepared using the protocol used for the ChIP assay with Honda buffer in the presence of 50 ng/μL tRNA, 20 units/mL RNase inhibitor (SUPERase In; Life Technologies) and 1× Complete Protease Inhibitor (Roche). The nuclei pellet was resuspended in an equal volume of resuspension buffer (50% glycerol, 0.5 mM EDTA, 1 mM DTT, 25 mM Tris-HCl, pH 7.5, and 100 mM NaCl). The nuclei were washed twice with 2 volumes of urea wash buffer (25 mM Tris-HCl, pH 7.5, 300 mM NaCl, 1 M urea, 0.5 mM EDTA, 1 mM DTT, and 1% Tween 20). The chromatin-bound RNA was extracted from the remaining pellet by phenol/chloroform extraction followed by precipitation with isopropanol. The RNA was treated twice with Turbo DNase (Ambion) and used as a template (500 ng) for reverse transcription with the reverse primer for FLC exon 7 and the EF1A reverse primer (1 pmol each). The qPCR data were normalized and analyzed as described above.
High-Throughput RNA Sequencing and Bioinformatics Analysis
Total RNA was extracted from 2-week-old seedlings using TRIzol (Invitrogen) according to the manufacturer’s instructions. RNA quality was monitored using an Agilent 2100 Bioanalyzer (Agilent Technologies). RNA was purified using the PolyATtract mRNA Isolation System (Promega) following the manufacturer’s instructions. Strand-specific RNA-seq libraries generated from the wild-type and double mutant plants were constructed according to the dUTP method (replicate 1) or the Illumina Directional mRNA-Seq library prep protocol v1.5 (replicate 2) and sequenced on an Illumina GAII. Replicate 1 was paired-end sequenced, while replicate 2 was single-ended sequenced. The RNA-seq reads were aligned to the TAIR10 Arabidopsis Genome using TopHat v2.0 (Trapnell et al., 2009). A summary of read alignments is shown in Supplemental Table 3. Only uniquely mapped reads were retained for subsequent analysis. The expression levels for gene models from the TAIR10 Arabidopsis Genome were measured and normalized as RPKM (Mortazavi et al., 2008). Next, P values for each gene were calculated by DEGseq based on the MA-plot-based method with random sampling model (Wang et al., 2010). Genes with more than 1.5-fold change and P < 0.01 were regarded as differentially expressed genes. For pathway analysis, the list of differential expressed genes and their corresponding log2 fold changes were analyzed using MapMan (Thimm et al., 2004). Differentially spliced introns were detected as previously reported (Deng et al., 2010). In brief, Fisher's exact test was performed to identify introns with differential splicing between wild-type plants and double mutants using read counts from each intron and its two flanking exons. In total, introns with more than 95% read coverage, intron RPKM > 5, and a two-sided P value < 0.01 were regarded as differentially spliced introns.
Accession Numbers
RNA-seq data have been deposited in the Gene Expression Omnibus database (GSE75959). Sequence data from this article can be found in the Arabidopsis Genome Initiative and GenBank/EMBL databases under the following accession numbers: RZ-1B (At1G60650), RZ-1C (At5G04280), RZ-1A (At3G26420), RSZ21 (At1G23860), RSZ22 (At4G31580), SCL28 (At5G18810), SCL30 (At3G55460), SCL30a (At3G13570), SCL33 (At1G55310), RS31 (At3G61860), SR45 (At1G16610), RS2Z33 (At2G37340), SR34 (At1G02840), TG (At1G36060), and U1-70K (At31G50670). Accession numbers of gene loci used for the phylogenetic analysis are listed in Supplemental Table 1.
Supplemental Data
Supplemental Figure 1. Phylogenetic structure of GRPs and RZ-1s in Arabidopsis.
Supplemental Figure 2. Molecular characterization of rz-1b and rz-1c mutants.
Supplemental Figure 3. Comparison of inflorescence, leaf morphology, and SAMs in the wild type and rz-1b rz-1c double mutant.
Supplemental Figure 4. Phenotype of rz-1b rz-1c mutant can be rescued by RZ-1C genomic fragment.
Supplemental Figure 5. RZ-1B and RZ-1C localize to the nuclear speckles in onion epidermal cells.
Supplemental Figure 6. Interaction of RZ-1s and SR proteins in yeast.
Supplemental Figure 7. C terminus of RZ-1C protein is responsible for interaction with SR proteins in yeast cells.
Supplemental Figure 8. Alignment of sequences cloned from SELEX PCR products.
Supplemental Figure 9. qPCR validation of the splicing defects revealed by RNA-seq
Supplemental Figure 10. Relative abundance of FLC isoforms in total RNA and enrichment of unspliced FLC RNA in the chromatin-bound RNA fraction.
Supplemental Figure 11. Expected pattern of H3K4me3 was observed in the GFP-RZ-1C ChIP experiment.
Supplemental Table 1. Accession numbers of gene loci used for the phylogenetic analysis.
Supplemental Table 2. Distribution of AAAGA motif in genes which are differentially spliced in the rz-1b rz-1c double mutant.
Supplemental Table 3. Summary of high-throughput RNA-seq alignment.
Supplemental Data Set 1. Differentially expressed gene (difference >1.5 fold, P < 0.01) between the wild-type and the rz-1b rz-1c double mutant over two biological replicates.
Supplemental Data Set 2. Detailed information about the pathways affected in the rz-1b rz-1c double mutants, as analyzed using MapMan software.
Supplemental Data Set 3. RZ-1B/1C regulate hormone-related genes.
Supplemental Data Set 4. Differentially spliced gene between the wild type and the rz-1b rz-1c double mutant based on RNA-seq.
Supplemental Data Set 5. Primers used in this work.
Supplemental File 1. Alignment used for phylogenetic analysis.
Supplementary Material
Acknowledgments
We thank Xing Wang Deng (Peking University, China) and Yunde Zhao (University of California, San Diego) for their valuable comments. This work was supported by the National Natural Science Foundation of China (Grant 31230006 to L.-J.Q. and NSFC-JSPS 31211140041) and partially by the 111 Project.
AUTHOR CONTRIBUTIONS
L.-J.Q., H.G., T.A., C.D., and X.C. designed research. Z.W., D.Z., X.L., J.M., X.D., Q.Y., K.S., and D.Z. performed research. Z.W., D.Z., X.L., J.M., L.G., C.D., T.A., T.T., and L.-J.Q. analyzed data. Z.W., D.Z., X.L., and L.-J.Q. wrote the article.
Glossary
- RBP
RNA binding protein
- RRM
RNA recognition motif
- hnRNP
heterogeneous ribonucleoprotein
- SAM
shoot apical meristem
- SELEX
systematic evolution of ligands by exponential enrichment
- ChIP
chromatin immunoprecipitation
- BiFC
bimolecular fluorescence complementation
- UTR
untranslated region
- MS
Murashige and Skoog
- DAPI
4′,6-diamidino-2-phenylindole
- CDS
coding sequence
- RPKM
reads per kilobase of exon model per million mapped reads
References
- Ali G.S., Palusa S.G., Golovkin M., Prasad J., Manley J.L., Reddy A.S.N. (2007). Regulation of plant developmental processes by a novel splicing factor. PLoS One 2: e471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausin I., Greenberg M.V.C., Li C.F., Jacobsen S.E. (2012). The splicing factor SR45 affects the RNA-directed DNA methylation pathway in Arabidopsis. Epigenetics 7: 29–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barta A., Kalyna M., Reddy A.S.N. (2010). Implementing a rational and consistent nomenclature for serine/arginine-rich protein splicing factors (SR proteins) in plants. Plant Cell 22: 2926–2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley D.L. (2014). Coupling mRNA processing with transcription in time and space. Nat. Rev. Genet. 15: 163–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatt D.M., Pandya-Jones A., Tong A.J., Barozzi I., Lissner M.M., Natoli G., Black D.L., Smale S.T. (2012). Transcript dynamics of proinflammatory genes revealed by sequence analysis of subcellular RNA fractions. Cell 150: 279–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracha-Drori K., Shichrur K., Katz A., Oliva M., Angelovici R., Yalovsky S., Ohad N. (2004). Detection of protein-protein interactions in plants using bimolecular fluorescence complementation. Plant J. 40: 419–427. [DOI] [PubMed] [Google Scholar]
- Cavaloc Y., Bourgeois C.F., Kister L., Stévenin J. (1999). The splicing factors 9G8 and SRp20 transactivate splicing through different and specific enhancers. RNA 5: 468–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T., Cui P., Chen H., Ali S., Zhang S., Xiong L. (2013). A KH-domain RNA-binding protein interacts with FIERY2/CTD phosphatase-like 1 and splicing factors and is important for pre-mRNA splicing in Arabidopsis. PLoS Genet. 9: e1003875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuck G., Lincoln C., Hake S. (1996). KNAT1 induces lobed leaves with ectopic meristems when overexpressed in Arabidopsis. Plant Cell 8: 1277–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X., Gu L., Liu C., Lu T., Lu F., Lu Z., Cui P., Pei Y., Wang B., Hu S., Cao X. (2010). Arginine methylation mediated by the Arabidopsis homolog of PRMT5 is essential for proper pre-mRNA splicing. Proc. Natl. Acad. Sci. USA 107: 19114–19119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu X.D., Ares M. Jr. (2014). Context-dependent control of alternative splicing by RNA-binding proteins. Nat. Rev. Genet. 15: 689–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golovkin M., Reddy A.S. (1999). An SC35-like protein and a novel serine/arginine-rich protein interact with Arabidopsis U1-70K protein. J. Biol. Chem. 274: 36428–36438. [DOI] [PubMed] [Google Scholar]
- Guo Y., Qin G., Gu H., Qu L.-J. (2009). Dof5.6/HCA2, a Dof transcription factor gene, regulates interfascicular cambium formation and vascular tissue development in Arabidopsis. Plant Cell 21: 3518–3534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han P., Li Q., Zhu Y.-X. (2008). Mutation of Arabidopsis BARD1 causes meristem defects by failing to confine WUSCHEL expression to the organizing center. Plant Cell 20: 1482–1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanano S., Sugita M., Sugiura M. (1996). Isolation of a novel RNA-binding protein and its association with a large ribonucleoprotein particle present in the nucleoplasm of tobacco cells. Plant Mol. Biol. 31: 57–68. [DOI] [PubMed] [Google Scholar]
- Hématy K., Sado P.-E., Van Tuinen A., Rochange S., Desnos T., Balzergue S., Pelletier S., Renou J.-P., Höfte H. (2007). A receptor-like kinase mediates the response of Arabidopsis cells to the inhibition of cellulose synthesis. Curr. Biol. 17: 922–931. [DOI] [PubMed] [Google Scholar]
- Hibara K., Karim M.R., Takada S., Taoka K., Furutani M., Aida M., Tasaka M. (2006). Arabidopsis CUP-SHAPED COTYLEDON3 regulates postembryonic shoot meristem and organ boundary formation. Plant Cell 18: 2946–2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji X., Zhou Y., Pandit S., Huang J., Li H., Lin C.Y., Xiao R., Burge C.B., Fu X.-D. (2013). SR proteins collaborate with 7SK and promoter-associated nascent RNA to release paused polymerase. Cell 153: 855–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalyna M., Lopato S., Barta A. (2003). Ectopic expression of atRSZ33 reveals its function in splicing and causes pleiotropic changes in development. Mol. Biol. Cell 14: 3565–3577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodor Y.L., Rodriguez J., Abruzzi K.C., Tang C.H., Marr M.T. II, Rosbash M. (2011). Nascent-seq indicates widespread cotranscriptional pre-mRNA splicing in Drosophila. Genes Dev. 25: 2502–2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.S., Jung H.J., Lee H.J., Kim K.A., Goh C.-H., Woo Y., Oh S.H., Han Y.S., Kang H. (2008). Glycine-rich RNA-binding protein 7 affects abiotic stress responses by regulating stomata opening and closing in Arabidopsis thaliana. Plant J. 55: 455–466. [DOI] [PubMed] [Google Scholar]
- Kim J.Y., Kim W.Y., Kwak K.J., Oh S.H., Han Y.S., Kang H. (2010a). Glycine-rich RNA-binding proteins are functionally conserved in Arabidopsis thaliana and Oryza sativa during cold adaptation process. J. Exp. Bot. 61: 2317–2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.Y., Park S.J., Jang B., Jung C.-H., Ahn S.J., Goh C.-H., Cho K., Han O., Kang H. (2007). Functional characterization of a glycine-rich RNA-binding protein 2 in Arabidopsis thaliana under abiotic stress conditions. Plant J. 50: 439–451. [DOI] [PubMed] [Google Scholar]
- Kim W.Y., Kim J.Y., Jung H.J., Oh S.H., Han Y.S., Kang H. (2010b). Comparative analysis of Arabidopsis zinc finger-containing glycine-rich RNA-binding proteins during cold adaptation. Plant Physiol. Biochem. 48: 866–872. [DOI] [PubMed] [Google Scholar]
- Kim Y.-O., Kim J.S., Kang H. (2005). Cold-inducible zinc finger-containing glycine-rich RNA-binding protein contributes to the enhancement of freezing tolerance in Arabidopsis thaliana. Plant J. 42: 890–900. [DOI] [PubMed] [Google Scholar]
- Kimura H., Tao Y., Roeder R.G., Cook P.R. (1999). Quantitation of RNA polymerase II and its transcription factors in an HeLa cell: little soluble holoenzyme but significant amounts of polymerases attached to the nuclear substructure. Mol. Cell. Biol. 19: 5383–5392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak K.J., Park S.J., Han J.H., Kim M.K., Oh S.H., Han Y.S., Kang H. (2011). Structural determinants crucial to the RNA chaperone activity of glycine-rich RNA-binding proteins 4 and 7 in Arabidopsis thaliana during the cold adaptation process. J. Exp. Bot. 62: 4003–4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Zhong S., Guo X., Hao L., Wei X., Huang Q., Hou Y., Shi J., Wang C., Gu H., Qu L.J. (2013). Membrane-bound RLCKs LIP1 and LIP2 are essential male factors controlling male-female attraction in Arabidopsis. Curr. Biol. 23: 993–998. [DOI] [PubMed] [Google Scholar]
- Long J.C., Caceres J.F. (2009). The SR protein family of splicing factors: master regulators of gene expression. Biochem. J. 417: 15–27. [DOI] [PubMed] [Google Scholar]
- Lopato S., Forstner C., Kalyna M., Hilscher J., Langhammer U., Indrapichate K., Lorković Z.J., Barta A. (2002). Network of interactions of a novel plant-specific Arg/Ser-rich protein, atRSZ33, with atSC35-like splicing factors. J. Biol. Chem. 277: 39989–39998. [DOI] [PubMed] [Google Scholar]
- Lopato S., Kalyna M., Dorner S., Kobayashi R., Krainer A.R., Barta A. (1999). atSRp30, one of two SF2/ASF-like proteins from Arabidopsis thaliana, regulates splicing of specific plant genes. Genes Dev. 13: 987–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorković Z.J. (2009). Role of plant RNA-binding proteins in development, stress response and genome organization. Trends Plant Sci. 14: 229–236. [DOI] [PubMed] [Google Scholar]
- Lorković Z.J., Barta A. (2002). Genome analysis: RNA recognition motif (RRM) and K homology (KH) domain RNA-binding proteins from the flowering plant Arabidopsis thaliana. Nucleic Acids Res. 30: 623–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquardt S., Raitskin O., Wu Z., Liu F., Sun Q., Dean C. (2014). Functional consequences of splicing of the antisense transcript COOLAIR on FLC transcription. Mol. Cell 54: 156–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer A., di Iulio J., Maleri S., Eser U., Vierstra J., Reynolds A., Sandstrom R., Stamatoyannopoulos J.A., Churchman L.S. (2015). Native elongating transcript sequencing reveals human transcriptional activity at nucleotide resolution. Cell 161: 541–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortazavi A., Williams B.A., McCue K., Schaeffer L., Wold B. (2008). Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 5: 621–628. [DOI] [PubMed] [Google Scholar]
- Nelissen H., Clarke J.H., De Block M., De Block S., Vanderhaeghen R., Zielinski R.E., Dyer T., Lust S., Inzé D., Van Lijsebettens M. (2003). DRL1, a homolog of the yeast TOT4/KTI12 protein, has a function in meristem activity and organ growth in plants. Plant Cell 15: 639–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicaise V., Joe A., Jeong B.R., Korneli C., Boutrot F., Westedt I., Staiger D., Alfano J.R., Zipfel C. (2013). Pseudomonas HopU1 modulates plant immune receptor levels by blocking the interaction of their mRNAs with GRP7. EMBO J. 32: 701–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou B., Yin K.-Q., Liu S.-N., Yang Y., Gu T., Wing Hui J.M., Zhang L., Miao J., Kondou Y., Matsui M., Gu H.Y., Qu L.J. (2011). A high-throughput screening system for Arabidopsis transcription factors and its application to Med25-dependent transcriptional regulation. Mol. Plant 4: 546–555. [DOI] [PubMed] [Google Scholar]
- Pandit S., Zhou Y., Shiue L., Coutinho-Mansfield G., Li H., Qiu J., Huang J., Yeo G.W., Ares M. Jr., Fu X.-D. (2013). Genome-wide analysis reveals SR protein cooperation and competition in regulated splicing. Mol. Cell 50: 223–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin G., Gu H., Zhao Y., Ma Z., Shi G., Yang Y., Pichersky E., Chen H., Liu M., Chen Z., Qu L.J. (2005). An indole-3-acetic acid carboxyl methyltransferase regulates Arabidopsis leaf development. Plant Cell 17: 2693–2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quan J., Tian J. (2011). Circular polymerase extension cloning for high-throughput cloning of complex and combinatorial DNA libraries. Nat. Protoc. 6: 242–251. [DOI] [PubMed] [Google Scholar]
- Saiga S., Furumizu C., Yokoyama R., Kurata T., Sato S., Kato T., Tabata S., Suzuki M., Komeda Y. (2008). The Arabidopsis OBERON1 and OBERON2 genes encode plant homeodomain finger proteins and are required for apical meristem maintenance. Development 135: 1751–1759. [DOI] [PubMed] [Google Scholar]
- Schmal C., Reimann P., Staiger D. (2013). A circadian clock-regulated toggle switch explains AtGRP7 and AtGRP8 oscillations in Arabidopsis thaliana. PLOS Comput. Biol. 9: e1002986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitzová J., Rasche N., Dybkov O., Kramer K., Fabrizio P., Urlaub H., Lührmann R., Pena V. (2012). Crystal structure of Cwc2 reveals a novel architecture of a multipartite RNA-binding protein. EMBO J. 31: 2222–2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schöning J.C., Streitner C., Page D.R., Hennig S., Uchida K., Wolf E., Furuya M., Staiger D. (2007). Auto-regulation of the circadian slave oscillator component AtGRP7 and regulation of its targets is impaired by a single RNA recognition motif point mutation. Plant J. 52: 1119–1130. [DOI] [PubMed] [Google Scholar]
- Singh G., Kucukural A., Cenik C., Leszyk J.D., Shaffer S.A., Weng Z., Moore M.J. (2012). The cellular EJC interactome reveals higher-order mRNP structure and an EJC-SR protein nexus. Cell 151: 750–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streitner C., Danisman S., Wehrle F., Schöning J.C., Alfano J.R., Staiger D. (2008). The small glycine-rich RNA binding protein AtGRP7 promotes floral transition in Arabidopsis thaliana. Plant J. 56: 239–250. [DOI] [PubMed] [Google Scholar]
- Szekeres M., Németh K., Koncz-Kálmán Z., Mathur J., Kauschmann A., Altmann T., Rédei G.P., Nagy F., Schell J., Koncz C. (1996). Brassinosteroids rescue the deficiency of CYP90, a cytochrome P450, controlling cell elongation and de-etiolation in Arabidopsis. Cell 85: 171–182. [DOI] [PubMed] [Google Scholar]
- Thimm O., Bläsing O., Gibon Y., Nagel A., Meyer S., Krüger P., Selbig J., Müller L.A., Rhee S.Y., Stitt M. (2004). MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 37: 914–939. [DOI] [PubMed] [Google Scholar]
- Thomas J., Palusa S.G., Prasad K.V.S.K., Ali G.S., Surabhi G.-K., Ben-Hur A., Abdel-Ghany S.E., Reddy A.S.N. (2012). Identification of an intronic splicing regulatory element involved in auto-regulation of alternative splicing of SCL33 pre-mRNA. Plant J. 72: 935–946. [DOI] [PubMed] [Google Scholar]
- Trapnell C., Pachter L., Salzberg S.L. (2009). TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25: 1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchiyama M., Wang T.S. (2004). The B-subunit of DNA polymerase alpha-primase associates with the origin recognition complex for initiation of DNA replication. Mol. Cell. Biol. 24: 7419–7434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vroemen C.W., Mordhorst A.P., Albrecht C., Kwaaitaal M.A.C.J., de Vries S.C. (2003). The CUP-SHAPED COTYLEDON3 gene is required for boundary and shoot meristem formation in Arabidopsis. Plant Cell 15: 1563–1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B.-B., Brendel V. (2004). The ASRG database: identification and survey of Arabidopsis thaliana genes involved in pre-mRNA splicing. Genome Biol. 5: R102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Feng Z., Wang X., Wang X., Zhang X. (2010). DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 26: 136–138. [DOI] [PubMed] [Google Scholar]
- Wang Y., Wang J., Gao L., Stamm S., Andreadis A. (2011). An SRp75/hnRNPG complex interacting with hnRNPE2 regulates the 5′ splice site of tau exon 10, whose misregulation causes frontotemporal dementia. Gene 485: 130–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z.W., Wu Z., Raitskin O., Sun Q., Dean C. (2014). Antisense-mediated FLC transcriptional repression requires the P-TEFb transcription elongation factor. Proc. Natl. Acad. Sci. USA 111: 7468–7473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wierzbicki A.T., Haag J.R., Pikaard C.S. (2008). Noncoding transcription by RNA polymerase Pol IVb/Pol V mediates transcriptional silencing of overlapping and adjacent genes. Cell 135: 635–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu R., Li S., He S., Wassmann F., Yu C., Qin G., Schreiber L., Qu L.-J., Gu H. (2011). CFL1, a WW domain protein, regulates cuticle development by modulating the function of HDG1, a class IV homeodomain transcription factor, in rice and Arabidopsis. Plant Cell 23: 3392–3411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z., Ietswaart R., Liu F., Yang H., Howard M., Dean C. (2016). Quantitative regulation of FLC via coordinated transcriptional initiation and elongation. Proc. Natl. Acad. Sci. USA 113: 218–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuarin J., Schibler U. (1994). Physical isolation of nascent RNA chains transcribed by RNA polymerase II: evidence for cotranscriptional splicing. Mol. Cell. Biol. 14: 7219–7225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu D., Wu Z., Cao G., Li J., Wei J., Tsuge T., Gu H., Aoyama T., Qu L.-J. (2014). TRANSLUCENT GREEN, an ERF family transcription factor, controls water balance in Arabidopsis by activating the expression of aquaporin genes. Mol. Plant 7: 601–615. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.