Abstract
Mitochondrial diseases cause a range of clinical manifestations even in patients carrying the same mtDNA mutations. New work reveals that a common disease-associated mtDNA mutation is selectively segregated from wild-type mtDNA during the reprogramming of induced pluripotent stem cells and that high levels of this mutation in differentiated neurons upregulate Parkin-mediated mitophagy.
Multiple copies of mitochondrial DNA (mtDNA) reside in mitochondria and this DNA encodes the tRNA and rRNA machinery required to translate 13 components of all of the complexes of the respiratory chain, except complex II. Mutations in the mtDNA that affect oxidative phosphorylation function are common; it is estimated that 1 in 5,000 children and adults have mitochondrial diseases caused by mtDNA mutations [1]. MtDNA mutations reach varying levels of heteroplasmy — the ratio of wild-type to mutated mtDNA molecules — in different tissues and present a wide range of clinical symptoms, even between patients harboring the same type of mutation. The most common mtDNA mutation, m.3243A>G, disrupts the gene encoding tRNA Leucine(UUR) and causes two distinct mitochondrial diseases: maternally inherited diabetes and deafness (MIDD) and mitochondrial encephalomyopathy, lactic acidosis, and stroke-like episodes (MELAS) [1]. To date, the most puzzling question is how mtDNA mutations affect different cell types to cause different phenotypes and how mutational load is determined in different tissues. Recent work by Hämäläinen et al. [2], published in Proceedings of the National Academy of Sciences, used an induced pluripotent stem cell (iPSC) model to provide mechanistic insight into how mtDNA mutations affect neurons differently from other cell types and how mtDNA mutations segregate in iPSCs to affect their differentiated progeny.
This study from Suomalainen’s group shows that the m.3243A>G mutation causes a defect in respiratory chain complex I in differentiated neurons, but has no detrimental effect on oxidative phosphorylation activity in iPSCs [2]. Disrupting the proof-reading domain of the nuclear gene polymerase γ, which controls mtDNA replication, causes an accelerated accumulation of mtDNA mutations in a premature aging Mutator mouse model. These mtDNA mutations impact mitochondrial function with age, causing Mutator mice to suffer from weight loss, cardiomyopathies, age-related muscle wasting, fur graying, and other phenotypes that mimic human aging [3,4]. Prior work from the Suomalainen group showed that high mtDNA mutational loads in neural stem cells (NSCs) from Mutator mice do not result in a respiratory defect, but lead to oxidative phosphorylation dysfunction in adult neurons later in life [5]. MtDNA mutations in iPSCs or NSCs do not have the same adverse effects on oxidative phosphorylation activity as in other cell types, likely due to the heavy reliance of these stem cells on glycolysis for energy metabolism [6]. However, mtDNA mutations negatively affect the survival and proliferative abilities of stem cells, possibly due to alternative signaling pathways, such as the generation of reactive oxygen species [5]. It remains mysterious how a tRNA Leucine(UUR) mutation selectively impairs complex I in post-mitotic neurons when it is needed for the translation of all mitochondrial genes.
Neurons are complex specialized cell types categorized by location and by the type of neurotransmitters they release. Often, this view itself is simplistic; for example, different subtypes of dopaminergic neurons express different calcium-binding proteins and have distinct baseline neuronal firing oscillations. This is important because different disruptions in mtDNA integrity cause divergent neuroanatomical susceptibilities in the central nervous system [7]. Knocking out the function of complex III or complex IV in the same subset of neurons expressing calcium/calmodulin-dependent protein kinase IIα(CaMKIIα) causes distinct patterns of neurodegeneration, resulting in dissimilar phenotypical consequences [8]. While future work will explore how different neuronal subtypes are dependent on mitochondrial function, it is noteworthy that Hämä lä inen et al. [2] report that mtDNA mutations cause distinct types of mitochondrial dysfunction and compensation mechanisms that are unique to neurons.
Pharmacological and genetic knockout models that dissipate the mitochondrial membrane potential (Δψm) have supported the idea that Parkin, an E3 ubiquitin ligase, is recruited to dysfunctional mitochondria to target the whole organelle for autophagic engulfment and removal — a process termed mitophagy [9]. Hämä lä inen et al. [2] demonstrate that Parkin recruitment and LC3 lipidation (a protein modification that indicates the induction of autophagy) specifically target the faulty complex I components for removal in m.3243A>G differentiated neurons. In agreement with this finding, specific respiratory complex proteins are subject to selective turnover in Drosophila brain mitochondria and this turnover is impeded in Parkin- and autophagy-deficient fly models [10]. Owing to the high respiratory demands of neurons for survival and physiological function, the complete removal of dysfunctional mitochondria may be energetically costly. In fact, cells attempt to compensate for inherited oxidative phosphorylation defects by an increase in mitochondrial proliferation [11]. It would be paradoxical to generate new mitochondria for destruction, so the selective removal of damaged oxidative phosphorylation complexes from otherwise functional mitochondria may yield a refined quality control mechanism that may also occur in other cell types harboring mtDNA mutations. In support of this idea, differentiated neurons generated from heteroplasmic iPSCs harboring mtDNA mutations in the COXI and ND5/ND6 genes disrupting complex IV and complex I, respectively, were able to maintain Δψm, even in the context of a severe loss of oxidative phosphorylation [12]. Thus, the depolarization of mitochondria cannot be the only way to alert the cell that mitochondria are dysfunctional. Consistent with this idea, accumulation of misfolded proteins inside mitochondria can trigger Parkin-mediated mitophagy without membrane depolarization [13]. How damaged subsets of mitochondrial proteins can be segregated for selective disposal remains to be elucidated.
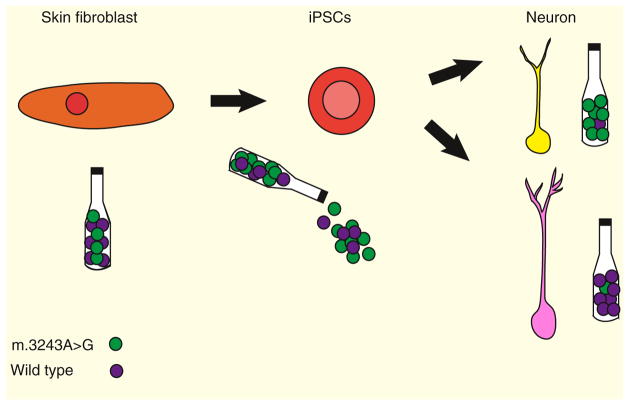
Finally, this study shows that reprogramming of somatic cells to iPSCs segregates heteroplasmic mtDNA mutations differently than in somatic cells, as the m.3243A>G mutation preferentially shifts towards homoplasmy, populating iPSCs with either mainly mutant or mainly wild-type mtDNA molecules (Figure 1) [2]. It appears that cells undergo a genetic bottleneck during dedifferentiation, similar to mtDNA in the female germ line during oogenesis [14]. Contrary to the belief that mtDNA levels are reduced to generate a physical genetic bottleneck to preferentially select one mtDNA variant over another, Hämä lä inen et al. [2] show that mtDNA levels are increased in iPSCs harboring mtDNA mutations during this reprogramming [14]. This new study confirms that mtDNA mutational selection occurs during the acquisition of stem cell fate, although we do not understand how this occurs. This genetic selection and increase in mtDNA levels appears to differ in iPSCs with wild-type mtDNA, as the levels of mtDNA drop as mitochondrial content is reduced, owing to the lack of demand during anaerobic metabolism [15]. These data also could explain the severe mitochondrial defects found in healthy aged human colon crypts that arise from mtDNA mutations in the crypt stem cells, which pass these mutations to their differentiated progeny [16]. Heart, skeletal muscle, and the central nervous system are the most affected tissues in mitochondrial disease patients. It was thought that, due to their post-mitotic state and low cell turnover, these tissues retained high levels of mutant mtDNA. Given the findings reported in this new study, it is possible that the regenerative stem cell pools in these tissues could also contribute to mtDNA mutant load. Yet, it is important to verify that this genetic bottleneck does not only occur during experimental reprogramming and that this heteroplasmic shift occurs in natural stem cell pools.
Figure 1. mtDNA bottleneck for tRNA mutations during iPSC differentiation.
Patient skin fibroblasts harbor a mutant mtDNA content of approximately 30%. During induced pluripotency, mtDNA copy number rises, shifting heteroplasmy to reach levels closer to homoplasmy in the direction of either mostly wild-type or mostly mutant molecules. The shift towards homoplasmy is permanent after differentiation.
The work of Hämä lä inen et al. [2] provides us with a deeper understanding of mtDNA biology in different cell types and gives insight into how mtDNA mutations are transmitted and lead to pathophysiological consequences. This powerful model, using these m.3243A>G iPSCs, can be used to investigate whether other post-mitotic cell types, such as skeletal muscle, also show distinct phenotypes from the parental fibroblast line.
References
- 1.Schaefer AM, Taylor RW, Turnbull DM, Chinnery PF. The epidemiology of mitochondrial disorders - past, present and future. Biochim Biophys Acta Bioenerg. 2004;1659:115–120. doi: 10.1016/j.bbabio.2004.09.005. [DOI] [PubMed] [Google Scholar]
- 2.Hamalainen RH, Manninen T, Koivumaki H, Kislin M, Otonkoski T, Suomalainen A. Tissue- and cell-type-specific manifestations of heteroplasmic mtDNA 3243A>G mutation in human induced pluripotent stem cell-derived disease model. Proc Natl Acad Sci USA. 2013;110:E3622–E3630. doi: 10.1073/pnas.1311660110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Trifunovic A, Wredenberg A, Falkenberg M, Spelbrink JN, Rovio AT, Bruder CE, Bohlooly YM, Gidlof S, Oldfors A, Wibom R, et al. Premature ageing in mice expressing defective mitochondrial DNA polymerase. Nature. 2004;429:417–423. doi: 10.1038/nature02517. [DOI] [PubMed] [Google Scholar]
- 4.Kujoth GC, Hiona A, Pugh TD, Someya S, Panzer K, Wohlgemuth SE, Hofer T, Seo AY, Sullivan R, Jobling WA, et al. Mitochondrial DNA mutations, oxidative stress, and apoptosis in mammalian aging. Science. 2005;309:481–484. doi: 10.1126/science.1112125. [DOI] [PubMed] [Google Scholar]
- 5.Ahlqvist KJ, Hamalainen RH, Yatsuga S, Uutela M, Terzioglu M, Gotz A, Forsstrom S, Salven P, Angers-Loustau A, Kopra OH, et al. Somatic progenitor cell vulnerability to mitochondrial DNA mutagenesis underlies progeroid phenotypes in Polg mutator mice. Cell Metab. 2012;15:100–109. doi: 10.1016/j.cmet.2011.11.012. [DOI] [PubMed] [Google Scholar]
- 6.Folmes CDL, Nelson TJ, Martinez-Fernandez A, Arrell DK, Lindor JZ, Dzeja PP, Ikeda Y, Perez-Terzic C, Terzic A. Somatic oxidative bioenergetics transitions into pluripotency-dependent glycolysis to facilitate nuclear reprogramming. Cell Metab. 2011;14:264–271. doi: 10.1016/j.cmet.2011.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pickrell AM, Fukui H, Wang X, Pinto M, Moraes CT. The striatum is highly susceptible to mitochondrial oxidative phosphorylation dysfunctions. J Neurosci. 2011;31:9895–9904. doi: 10.1523/JNEUROSCI.6223-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Diaz F, Garcia S, Padgett KR, Moraes CT. A defect in the mitochondrial complex III, but not complex IV, triggers early ROS-dependent damage in defined brain regions. Hum Mol Genet. 2012;21:5066–5077. doi: 10.1093/hmg/dds350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Narendra D, Tanaka A, Suen DF, Youle RJ. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J Cell Biol. 2008;183:795–803. doi: 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vincow ES, Merrihew G, Thomas RE, Shulman NJ, Beyer RP, MacCoss MJ, Pallanck LJ. The PINK1-Parkin pathway promotes both mitophagy and selective respiratory chain turnover in vivo. Proc Natl Acad Sci USA. 2013;110:6400–6405. doi: 10.1073/pnas.1221132110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moraes CT, Ricci E, Bonilla E, Dimauro S, Schon EA. The Mitochondrial Tert-Rna Leu(Uur) mutation in mitochondrial encephalomyopathy, lactic-acidosis, and stroke-like episodes (Melas) - genetic, biochemical, and morphological correlations in skeletal-muscle. Am J Hum Genet. 1992;50:934–949. [PMC free article] [PubMed] [Google Scholar]
- 12.Abramov AY, Smulders-Srinivasan TK, Kirby DM, Acin-Perez R, Enriquez JA, Lightowlers RN, Duchen MR, Turnbull DM. Mechanism of neurodegeneration of neurons with mitochondrial DNA mutations. Brain. 2010;133:797–807. doi: 10.1093/brain/awq015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jin SM, Youle RJ. The accumulation of misfolded proteins in the mitochondrial matrix is sensed by PINK1 to induce PARK2/Parkin-mediated mitophagy of polarized mitochondria. Autophagy. 2013 doi: 10.4161/auto.26122. epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cao LQ, Shitara H, Sugimoto M, Hayashi JI, Abe K, Yonekawa H. New evidence confirms that the mitochondrial bottleneck is generated without reduction of mitochondrial DNA content in early primordial germ cells of mice. PLoS Genet. 2009;5 doi: 10.1371/journal.pgen.1000756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu X, Duan S, Yi F, Ocampo A, Liu GH, Izpisua Belmonte JC. Mitochondrial regulation in pluripotent stem cells. Cell Metab. 2013;18:325–332. doi: 10.1016/j.cmet.2013.06.005. [DOI] [PubMed] [Google Scholar]
- 16.Greaves LC, Preston SL, Tadrous PJ, Taylor RW, Barron MJ, Oukrif D, Leedham SJ, Deheragoda M, Sasieni P, Novelli MR, et al. Mitochondrial DNA mutations are established in human colonic stem cells, and mutated clones expand by crypt fission. Proc Natl Acad Sci USA. 2006;103:714–719. doi: 10.1073/pnas.0505903103. [DOI] [PMC free article] [PubMed] [Google Scholar]