Abstract
Molecularly Imprinted Polymers (MIPs) are generic alternatives to antibodies in sensors, diagnostics and separations. To displace biomolecules without radical changes in infrastructure in device manufacture, MIPs should share their characteristics (solubility, size, specificity and affinity, localized binding domain) whilst maintaining the advantages of MIPs (low-cost, short development time and high stability) hence the interest in MIP nanoparticles. Herein we report a reusable solid-phase template approach (fully compatible with automation) for the synthesis of MIP nanoparticles and their precise manufacture using a prototype automated UV photochemical reactor. Batches of nanoparticles (30-400 nm) with narrow size distributions imprinted with: melamine (d = 60 nm, Kd = 6.3 × 10−8 m), vancomycin (d = 250 nm, Kd = 3.4 × 10−9 m), a peptide (d = 350 nm, Kd = 4.8 × 10−8 m) and proteins have been produced. Our instrument uses a column packed with glass beads, bearing the template. Process parameters are under computer control, requiring minimal manual intervention. For the first time we demonstrate the reliable re-use of molecular templates in the synthesis of MIPs (≥ 30 batches of nanoMIPs without loss of performance). NanoMIPs are produced template-free and the solid-phase acts both as template and affinity separation medium.
Keywords: Solid-Phase Synthesis, Immobilized Template, Automatic Reactor, Template Re-use, Antibody Replacements
1. Introduction
New tools in nanoscience are required to overcome bottlenecks in the production and characterization of nanomaterials.[1] Indeed the introduction of new methods in chemical biotechnology, such as Merrifield’s solid-phase peptide synthesis (which was recognized by the 1984 Nobel Prize in Chemistry[2]) and similar advances in the solid-supported synthesis of nucleic acid and other molecules[3] were revolutionized by the realization that such processes were compatible with automation. The result was the introduction of a series of new instruments, allowing the automatic synthesis of the same peptides and nucleic acids more reproducibly, more cheaply and more rapidly than ever before. The wide-scale adoption of automated synthesis has resulted in rapid advancements in modern combinatorial chemistry, biotechnology and genetic engineering.[4] Further developments in the automation of DNA sequencing has led to the current ‘genomics era’ and the appearance of sequencing machines capable of decoding gene sequences at the rate of millions of base-pairs per day.[5] Until now there have been no corresponding advances in technologies for the production of molecularly imprinted polymers (MIPs).
MIPs have long held promise as a technology capable of competing with antibodies as the chemically selective species in diagnostic assays,[6] sensors[7] and affinity separations/purifications.[8] MIPs offer the positive advantages of low cost and high stability due to their chemical, as opposed to biochemical, origin.[9] Moreover their synthetic nature readily allows the incorporation of integrated signaling functionalities, modification of surface chemistry and attachment of various labels (colored, fluorescent, electro-active, etc.) during synthesis without affecting molecular recognition. Progress in computational methods has also made the design of MIPs and the selection of functional monomers a routine and reliable process.[10] Despite these benefits the commercial exploitation of MIPs is largely confined to a handful of products for analytical sample preparation by selective solid-phase extraction and a few niche sensor products. Broader uptake of the technology, particularly in the diagnostics sector where antibodies still predominate, is hindered by general skepticism that MIPs can compete in this market, much to the frustration of researchers in the field. Until a reliable and reproducible source of MIPs, capable of directly replacing antibodies (without major changes in infrastructure on the part of device manufacturers) can be provided, exploitation of MIPs in diagnostics will remain a largely academic exercise. In order to act as substitutes for antibodies, MIPs must share key characteristics with their biomolecular counterparts, such as: aqueous solubility, size, affinity and selectivity for the target analyte and localization of the binding domain. The only way to satisfy the first two criteria is to prepare MIPs in the form of nanoparticles (nanoMIPs) in a reproducible, controlled and scalable manner. Only when the barrier of supply is overcome will the advantages of the approach be appreciated and a new generation of MIP-based diagnostics tests and assays reach the market. Moreover recent reports of biological activity[11] and in vivo detoxification[12] using MIP nanoparticles hint at an even greater potential in the pharmaceutical arena.
Ever since the seminal works of Wulff and Mosbach on the synthesis of MIPs for small molecular targets,[13] imprinted polymers have been prepared mostly in the form of bulk materials, for use as adsorbents, in chromatography or as selective coatings in sensors. Despite advances in combinatorial methods and computational tools used in MIP design, as well as improvements in understanding the chemical and physical factors which influence polymer recognition properties, the basic experimental techniques used for bulk polymer preparation remained largely unchanged.[14] Additionally, MIPs prepared in traditional formats (such as monoliths, membranes, films or beads) are perceived to have a number of drawbacks that have hindered their adoption in a variety of practical applications. These include: binding site heterogeneity,[15] leaching of residual template from the polymer [16] and difficulties involved in their integration with sensors and assay protocols. Clearly, MIPs in these formats are not suitable candidates to challenge the dominance of natural receptors in diagnostic applications. A more promising approach in this regard is the synthesis of MIPs as soluble micro- and nano-gels[17] or nanoparticles.[18] Notable work in this context includes the gel particles of Wulff and co-workers, obtained by post-dilution of the polymerization mixture at the point of gelation.[19] However these methods, as well as alternative protocols for the preparation of MIP nanoparticles (e.g. by precipitation,[20] emulsion[21] and miniemulsion[22] polymerization methods and the use of microfluidic reactors[23]) are based on the same monomer recipes as bulk MIPs, employing soluble templates. An exception is the work of Zimmerman and colleagues, who prepared dendrimers, with a template molecule such a porphyrin[24] at the core. The periphery of the dendrimers was decorated with butenyl ether groups which were intramolecularly cross-linked (by thermodynamically-controlled olefin metathesis) to form discrete monomolecular species with one imprint per molecule after removal of the template by hydrolysis. All these methods of preparation are labor-intensive and still suffer from many of the perceived drawbacks of bulk MIPs. In addition they are one-off batch processes not suited for industrial production.
Many of the perceived disadvantages of MIPs can be traced to the template: soluble templates are in motion, both translation and rotation, during the critical stages of binding site formation. The accessibility of binding sites in porous polymers is also a matter of chance – this can range from partial sites on the “walls” of micro- or macro-pores to complete encapsulation, with the ideal lying somewhere in between.[25] These factors are major contributors to the problems of binding site heterogeneity and accessibility, and therefore the “polyclonal” nature of traditional MIPs, as well as for the prolonged retention of template, giving rise to template leaching. Immobilized templates offer a significant advantage in that the template has reduced numbers of degrees of freedom and the formation of the polymer at the interface with the template support means that the imprinted sites will always be accessible. In addition, immobilization allows the orientation of the template to be controlled,[26] such that the binding sites created by imprinting recognize the same sub-structural “epitope” of the template and create a more “monoclonal” MIP. These considerations were behind the development of “hierarchical” imprinted polymer beads[27] where the template is immobilized on porous silica spheres. Filling of the pores with an imprintable monomer mixture, followed by polymerization and dissolution of the silica, resulted in the formation of polymer beads, the structure of which replicated the pore structure of the original silica spheres, with imprint sites located at the surfaces of the newly-formed pores. A more recent related example used silica nanoparticles, with template immobilized on their surfaces, to stabilize a Pickering emulsion polymerization, resulting in micron-sized beads with surface-confined imprints.[28] In each case however the template phase was a sacrificial material, with the silica requiring dissolution in reagents such as HF to reveal the imprints, which destroys the template in the process. It would be far better to work with immobilized templates that can be re-used and that do not require hazardous chemical treatments to separate polymer from template. In fact this becomes feasible when making imprinted nanoparticles, as it is possible to adapt the approach used for affinity separation of nanoparticles developed in our group[29] and subsequently used by others[30] to create a new type of immobilized template. Non-porous glass beads, surface modified with the template molecule, were therefore chosen as the basis of an immobilized template phase for incorporation into a new reactor design. Here we therefore present a significant leap in MIP synthesis with the first successful report of the solid-phase synthesis of molecularly imprinted polymer nanoparticles using a reusable molecular template and with the development of instrumental methods for automating their production which help to shorten the development time and expedite the synthesis of MIP nanoparticles.
2. Results and Discussion
Having proposed the development of an automated synthesizer using an immobilized solid-phase template approach, it became necessary to specify the operational parameters of the new reactor, bearing in mind the aim of controlled, reproducible and automated synthesis of nanoMIPs. The principle behind our proposed method is summarized in Figure 1 with a schematic of the new reactor design presented in Figure 2. In settling on this configuration, the following considerations were influential in the design of the reactor:
MIP synthesis should be performed at moderately low temperature (−30 °C to +10 °C) in an appropriate organic solvent to favor complex formation between monomers and template[31]
The requirement for using low temperatures is best met by initiating the polymerization reaction through photochemical means, since it can be performed at or below room temperature[32]
The synthesis of nanoparticles should be performed under controlled conditions to prevent polymer precipitation[33]
The template should be immobilized to prevent contamination of the synthesized particles[34]
Ideally the template should be capable of being recycled to reduce manufacturing costs
A mechanism for the separation of high-affinity nanoparticles from low-affinity materials and unreacted monomers should be integrated into the reactor design[29, 30]
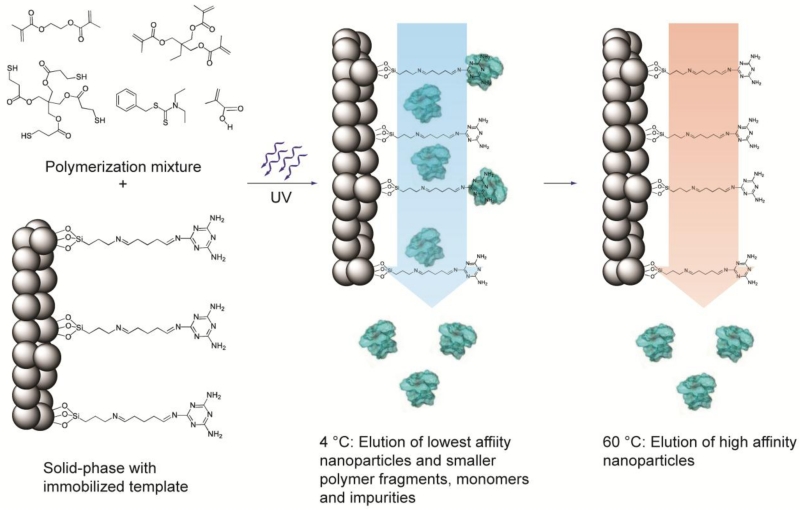
Figure 1.
Schematic representation of the solid-phase synthesis of MIP nanoparticles. The monomer mixture is injected onto the column reactor with immobilized template and polymerization is initiated by UV-irradiation. The low-affinity particles, as well as unreacted monomers, are eluted at low temperature. The temperature is then increased and high-affinity particles are eluted from the column for collection.
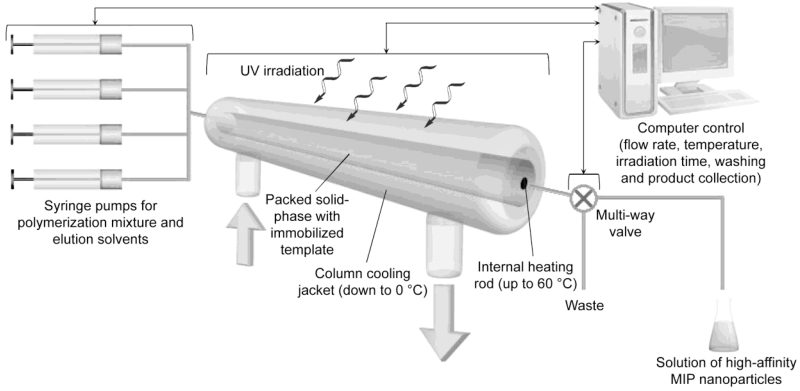
Figure 2.
Schematic diagram showing the mode of operation of the automated solid-phase MIP nanoparticle synthesizer. An engineering schematic is shown in the Supporting Information, Figure S1. Typical operational parameters using melamine as the immobilized target are: operation time: 2 h per cycle; yield of high affinity fraction: 6.6 ± 0.65 mg per cycle; column capacity: 23.5 g derivatized glass beads (solid phase).
Taking the above points into consideration, the ideal method for producing MIP nanoparticles must involve their synthesis using photochemically-initiated polymerization at low temperature in the presence of an immobilized template. The immobilized template will then double as an affinity medium for the subsequent purification and fractionation of the MIP particles.
The following generic protocol has been developed for the automated synthesis and purification of MIP nanoparticles: The first step involves loading the monomer/initiator mixture, dissolved in a suitable solvent, onto a temperature controlled column reactor containing the template immobilized onto a solid support. Once the temperature reaches a pre-determined set point, polymerization is initiated by UV-irradiation of the reactor for the desired reaction time. After polymerization is arrested, the column is washed with fresh solvent at low temperature. At this stage unreacted monomers and other low molecular weight materials are eluted along with low-affinity polymer nanoparticles. This leaves the desired high-affinity particles still bound to the immobilized template phase (Figure 1). These are then collected by increasing the column temperature, in some cases with the addition of auxiliary reagents, such as formic acid. Raising the temperature will increase the rate of exchange of the particles with the template phase, which will assist in eluting the particles, as well as potentially reducing the strength of the association. The benefits of the proposed approach include: i) uniform binding properties, resulting from affinity-based separation on column;[29] ii) eliminating contamination of the product with template; iii) possibility of template re-use; iv) ease of automation and standardization; v) the final product is obtained in a pure form obviating the need for lengthy post-synthesis purification steps; vi) imprints sites are only formed on one “face” of the particle, naturally resulting in a Janus–particle structure. Considering photochemical initiation methods, living polymerization based on iniferter-type initiators[35] is a useful approach in the solid-phase synthesis of nanoMIPs due to its controlled nature. Initiators of this type decompose reversibly to a pair of free radicals; one of which is the active propagating species, while the second is much less reactive and serves to terminate the growing polymer chains in solution, reforming a new initiating species in the process.[36]
This allows better control over the course of the polymerization reaction. Of crucial importance is the capability to initiate polymerization by this approach under mild conditions by near UV irradiation and terminate polymerization by ceasing irradiation.[29] Melamine was initially chosen as the model template with which to demonstrate the solid-phase synthesis of MIP nanoparticles. This molecule is known to form strong electrostatic bonds with methacrylic acid, a popular functional monomer commonly used in molecular imprinting.[29, 37]
The experimental set-up for the automated synthesis of MIP nanoparticles has been developed with the aim of controlling the column temperature, delivery of the monomer mixture and washing solvents, UV-irradiation time and yield of the synthesized material. This comprises a computer-controlled apparatus consisting of a custom-made fluid-jacketed glass reactor with internal heating element, containing the immobilized template, connected to pumps which deliver the reaction mixture, wash and elution solvents. The column is housed in a light-tight box fitted with a UV source that can be activated under software control for a predetermined time to initiate polymerization. The fluid-handling system also employs a multi-way valve post-column to direct the high-affinity nanoparticles to a collection vessel or wash solutions to waste (Figure 2).
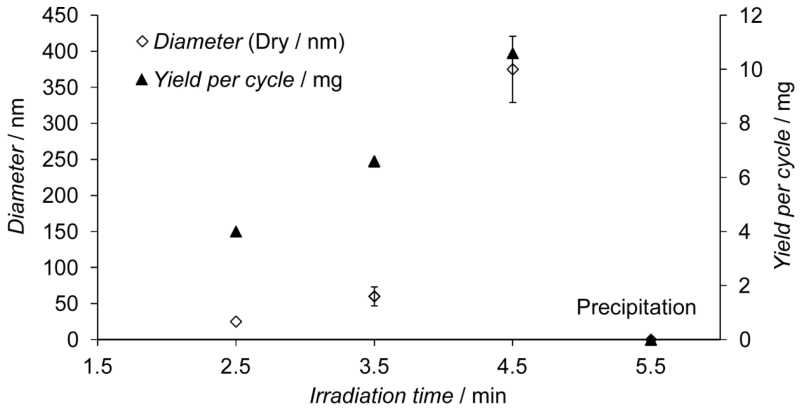
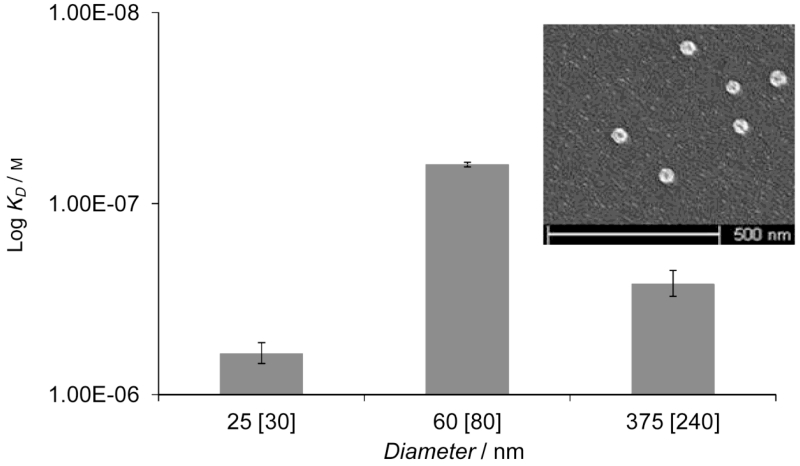
Optimization of the reaction conditions was performed by varying the concentration of reagents[29] and the irradiation time, followed by measuring the yield and properties of the synthesized particles. There was a direct correlation between irradiation time and the yield and diameter of the particles formed (Figure 3). Optimum conditions for the production of high-performance anti-melamine nanoMIPs were determined as follows: UV irradiation for 3.5 min applied to a polymerization solution containing 70/30 monomer mixture/solvent (w/v). The binding properties of the MIPs were assessed by Surface Plasmon Resonance (SPR) experiments using SPR sensor chips modified with the template.[38, 39] The highest affinity was achieved with particles 80 nm in diameter (as measured in acetonitrile) (Figure 4). The poorer performance of particles ≤ 30 nm in diameter may be because they are insufficiently rigid due to low levels of cross-linking or they lack the bulk needed for maintaining the structure of specific binding sites. Particles ≥ 240 nm in diameter may be prevented from forming effective interactions with immobilized template due to steric effects, resulting in a reduction of their measured affinity.[40]
Figure 3.
Influence of the irradiation time on the yield and size of synthesized nanoparticles. Error bars represent ± 1 s.d. (n ≥ 7).
Figure 4.
Influence of the size of the nanoparticles on their affinity (apparent dissociation constant) as determined by SPR. Dry size, measured by SEM and size in acetonitrile (in square brackets) measured by DLS. Inset: SEM of the 60 nm diameter MIP nanoparticles (the full field SEM can be seen in the Supporting Information, Figure S2). Error bars represent ± 1 s.d. (n = 3).
When the synthesizer was run in automatic mode under the optimum conditions, as defined above, we were able to produce three batches of particles per day with practically identical yield (20 ± 2 mg day−1) over five days and very similar binding properties (Kd = 6.3 × 10−8 ± 1.7 × 10−9 m) from a single batch of immobilized template. Evidence that the size (DLS) and SPR response (towards immobilized template and a close structural analogue) of the first and fifth batches of anti-melamine nanoMIPs is comparable is given in the Supporting Information, Figures S3 and S4 respectively. From the calculation of the density of immobilized template versus the quantity of particles produced in one step it is possible to deduce that approximately 560 melamine molecules were required to generate one high affinity particle. The yield of nanoMIPs is limited by the available surface area of the template phase (since template is the limiting reagent in the system) but can be further increased by: optimizing the morphology of the solid support; the use of parallel synthesis; increasing the number of reaction cycles or by using a larger reactor.
That the nanoparticles were selective for melamine was determined by SPR using a sensor chip bearing a structural analogue of the template, desisopropylatrazine. The SPR response of the melamine-MIP nanoparticles to this compound was insignificant (less than six response units for all concentrations of particles, which is similar to the response for solvent alone) and so does not allow for the calculation of a dissociation constant. These results are comparable to those obtained with monoclonal antibodies produced for haptens similar to melamine.[41]
Dispersions of nanoparticles in acetonitrile and in PBS were stable for at least one month at 4 °C. The fractions containing high-affinity particles were analyzed for the presence of residual template by LC-MS, however the template concentration was below the limit of detection for this method. Theoretically the number of binding sites per nanoparticle is proportional to the concentration of template used in MIP preparation. Since the template species in our approach is present only at the surface of the solid phase, it follows that the selective binding sites would only be formed in the MIP at the point of contact with the surface. The silanization of glass surfaces measured by other authors gave probe densities of between 1 × 1012 and 2.6 × 1013 functional units per cm2 (0.01 – 0.26 sites nm−2) for (3-mercaptopropyl)trimethoxysilane[42] and glutaradehyde-modified (3-aminopropyl)trimethoxysilane[43] (as also used in this work). Assuming we have achieved a similar degree of coverage in the melamine-modified glass beads, then in theory, it is likely that the MIP nanoparticles with diameter 30-80 nm would contain only 1-5 binding sites per particle, localized in one region of the nanoparticle surface. This number is in agreement with previously obtained data for MIP nanoparticles synthesized in solution.[29]
It is normal practice in MIP research to prepare a non-imprinted polymer or “blank”, prepared in the same way as the MIP but in the absence of template as an experimental control. The problem arises with the current work that without a template, there was no retention of material on the column following the low temperature elution step; hence it is not possible to prepare a comparable blank polymer. Instead we chose to compare material imprinted with different templates which also served to prove that the reported approach is generic in nature (suitable for small to medium molecular weight organic molecules and peptides etc.). MIP nanoparticles were therefore prepared for other targets, including vancomycin (MW = 1449.3 g mol−1) and a model peptide (TATTSVLG-NH2, MW = 747.9 g mol−1) using the reported automatic solid-phase photo-reactor. The polymerization conditions were identical to those used for the manufacture of MIP nanoparticles for melamine, with the exception of the monomer mixture used. The composition of the monomer mixture used to prepare MIPs for vancomycin and for the peptide was based on a published protocol.[44] Although the latter was optimized for imprinting in an aqueous-based solvent, we have used it in acetonitrile with good results. The binding properties of the synthesized nanoparticles were analyzed by SPR experiments (Biacore) using chips with immobilized templates. The apparent dissociation constants calculated for vancomycin and peptide MIPs were Kd = 3.4 × 10−9 m and Kd = 4.8 × 10−8 m respectively. Particle size was 250 nm for vancomycin MIPs and 350 nm for peptide MIPs (as measured in acetonitrile, see Supporting Information Figure S5 for size distribution results). These experiments have proven that by using automatic solid-phase synthesis it is possible to produce high quality MIP nanoparticles which resemble, in practical terms, monoclonal antibodies. There are a number of reasons why MIP nanoparticles, produced as described above, would have advantages over antibodies for use in assays and sensors (Table 1).
Table 1.
A comparison of the properties of antibodies with those of nanoMIPs.
| Antibodies | MIPs | |
|---|---|---|
| Lifetime | 6-12 months | Years |
| Storage | Freezer | From 6 °C to ambient temperature |
| Regeneration | Problematic, typically <10 times | Easy (e.g. using acid, organic solvent, surfactant) |
| Sterilization | Problematic, typically using γ-irradiation | UV, autoclaving |
| Temperature stability | Denature at ~70 °C | Resistant up to 140 °C |
| Price | 1-1000 $/mg | 0.25-5 $/mg |
3. Conclusions
We have described what we believe to be a major advance in MIP synthesis and in nanoscience; namely the first report of the automated synthesis of imprinted polymer nanoparticles (nanoMIPs) with size, specificity and solubility characteristics comparable to antibodies, suitable for industrial manufacturing. Automation allows the reactor to operate 24 h, eliminating human error and operator fatigue and ensures precisely controlled batch-to-batch reproducibility. Moreover this is the first report of the multiple re-use of molecular templates in the formation of imprinted polymers (more than 30 times without loss of performance) through the use of an immobilized template approach. This could be of great advantage when dealing with templates which are in short supply or are expensive to obtain. The only other reports of template recycling in the creation of imprinted polymers involve the “double imprinting” of immunoglobulin-imprinted nanoparticles, which can be used as “stamps” to imprint sensor chips.[45] The synthesized nanoMIPs are suitable for use as direct replacements for antibodies in assay and sensor development and we are actively pursuing demonstrations of their potential in these applications as well as improvements in reactor instrument design, building on lessons learnt from our first prototype machine.
In order to realize the full scientific and commercial opportunities presented by the technology many more studies remain to be performed covering subjects such as: alternative photoreactor formats, different solid phases for template immobilization, the development of MIPs with integrated functionalities (e.g. combining recognition and signaling functions), etc. It is clear however that even at this early stage in the development of automated methods for the solid-phase synthesis of MIPs that this advance has the potential to usher in a new era in the diagnostics and biotechnology sector in much the same way that solid-phase synthesis enabled combinatorial chemistry and the automation of DNA sequencing has sparked the genomics revolution. With this breakthrough we believe the era of “plastic antibodies” has begun.
4. Experimental
Materials
Formic acid, melamine, desisopropylatrazine, ethylene glycol dimethacrylate (EGDMA), methacrylic acid (MAA), trimethylolpropane trimethacrylate (TRIM), 3-aminopropyltrimethyloxysilane (APTMS), glutaraldehyde (GA), phosphate buffered saline (PBS), N-methyl-2-pyrrolidone (NMP), pentaerythritol-tetrakis-(3-mercaptopropionate), N-isopropylacrylamide (NIPAm), acrylic acid (AA), N,N′-methylene-bis-acrylamide (BIS), N-tert-butylacrylamide (TBAm), vancomycin, methanol, ethanol, toluene and acetone were purchased from Sigma-Aldrich, UK. Acetonitrile (ACN) and sodium hydroxide were obtained from Fisher Scientific (UK). N,N-diethyldithiocarbamic acid benzyl ester was obtained from TCI Europe (Belgium). Double-distilled ultrapure water (Millipore) was used for analysis. All chemicals and solvents were analytical or HPLC grade and were used without further purification. Phosphate buffered saline (PBS) was prepared as directed from PBS buffer tablets (Sigma-Aldrich, Gillingham, UK) and comprised phosphate buffer (0.01 m), potassium chloride (0.0027 m) and sodium chloride (0.137 m), pH 7.4, at 25 °C. Where PBS, pH 7.2 was used, the pH was adjusted by the addition of HCl. The peptide TATTSVLG-NH2 was synthesized and kindly provided by The National Physical Laboratory (UK).
Preparation of template-derivatized glass beads
Glass beads (75 μm diameter from Sigma-Aldrich) were activated by boiling in NaOH (4 m) for 10 min, then washed with double-distilled water followed by acetone and dried at 80 °C. The beads were then incubated in a 2% v/v solution of APTMS in toluene overnight, washed with acetone and subsequently incubated with a 5% v/v solution of GA in PBS buffer pH 7.2 for 2 h, after which they were rinsed with double-distilled water. The surface immobilization of the template was performed by incubating the beads with a solution of the template in PBS, pH 7.2, overnight at 4 °C (concentration: 5 mg mL−1 in the case of melamine or vancomycin; 0.05 mg mL−1 in the case of peptide TATTSVLG-NH2). In the case of melamine, NMP (10% v/v) was also added as co-solvent. Finally, the glass beads were washed with water and dried under vacuum, then stored at 4 °C until used. The procedure has been adapted from that published earlier [46]. The amount of melamine immobilized was calculated by measuring the amount present in the supernatant after immobilization using HPLC-MS.
Quantification of melamine using HPLC-MS
HPLC separation was conducted using a Waters 2975 HPLC system equipped with Luna C18 (2) column (50 × 3 mm, 3 μm, Phenomenex). Mobile phase: methanol with formic acid (0.1%). The flow rate was 0.2 μL min−1; injection volume 10 μL; the column temperature +25 °C. Fragment of melamine m/z 85 was detected by mass-spectrometric detector: Micromass Quattro Micro (Waters, UK) equipped with an ESI interface in positive ion mode. The MS parameters were following: desolvation gas: 850 L h−1, cone gas: 50 L h−1, capillary: 3.5 kV, cone: 35 V, collision energy: 19 eV, source temperature: +120 °C, desolvation temperature: +350 °C, multiplier: 650 V.
Automated synthesis and setup
All system components were purchased from HEL Ltd. (UK). Standard components which were used “off the shelf” include the control software, PC interface, syringe pumps, cooler/heater, thermocouple and multi-way valve. The glass column reactor (6.6 mm internal diameter × 150 mm length) its respective end-adaptors and the UV light source (4 × 8W power) were specially designed and adapted for this project. For melamine MIP nanoparticles, the polymerization mixture was prepared by mixing MAA (0.96 g) as functional monomer, TRIM (1.08 g) and EGDMA (1.08 g) as cross-linkers, N,N-diethyldithiocarbamic acid benzyl ester (0.261 g) as initiator (iniferter) and pentaerythritol-tetrakis-(3-mercaptopropionate) (0.06 g) as chain transfer agent in ACN (3.51 g) (total monomer concentration: 70% w/v). The composition has been adapted from that used for the preparation of MIP nanoparticles formed under homogeneous conditions [29]. The mixture was placed in a 20 mL glass vials and purged with N2 for 20 min. The mixture (4 mL) was automatically injected into the column packed with the immobilized template phase and polymerized under UV irradiation for 3.5 min at 4 °C. After polymerization, the reactor was maintained at 4 °C and flushed with ACN (120 mL, flow rate: 2 mL min−1). Then the temperature was raised to 25 °C, after which ACN (45 mL) with 10 mm formic acid (used as additive) were eluted. Eventually, the same mobile phase (40 mL) was eluted at 60 °C to collect the high-affinity nanoparticles. The yield has been expressed in mg of nanoparticles produced.
For vancomycin and peptide MIP nanoparticles, the polymerization mixture was prepared by mixing NIPAm (0.936 g) as backbone monomer, BIS (0.048 g) as cross-linker, TBAm (0.8 g) as hydrophobic functional monomer, AA (56.8 μL) as negatively-charged functional monomer and N,N-diethyldithiocarbamic acid benzyl ester (0.152 g) as iniferter in ACN (7.87 g) (total monomer concentration: 18% w/v). The composition was adapted from that used by Hoshino et al. [44]. The mixture was placed in a 20 mL glass vials and purged with N2 for 20 min. The mixture (4 mL) was automatically injected into the column packed with the immobilized template phase and then polymerized under UV irradiation for 3.5 min at 20 °C. After polymerization, the reactor was maintained at 20 °C and flushed with ACN (60 mL, flow rate: 2 mL min−1). Then the temperature was raised to 25 °C, after which ACN (45 mL) with 10 mm formic acid (used as additive) were eluted. Eventually, the same mobile phase (40 mL) was eluted at 60 °C to collect the high-affinity nanoparticles.
Size analysis of MIP nanoparticles
To measure the size of the synthesized nanoparticles, the solution of the eluted product was concentrated and sonicated for 20 minutes and then analyzed at 25 °C using a Zetasizer Nano (Nano-S) from Malvern Instruments Ltd (Malvern, UK). For scanning electron microscope analyses, samples were placed on silicon slides and dried in a desiccator overnight then plasma-coated with gold on a Polaron Equipment E5100 SEM plasma coater. The microscope used was a FEI XL30 SFEG.
Surface Plasmon Resonance (SPR) affinity analysis of MIP nanoparticles
Affinity analysis was performed using a Biacore 3000 SPR system (Biacore, Sweden). Au-coated chips (SIA Kit Au, Biacore) were cleaned by immersion in Piranha solution (H2SO4/H2O2, 3:1 v/v) for 5 min, thoroughly rinsed with double-distilled water and left in ethanol overnight. The immobilization of the templates was performed by incubating the chips in a solution of cysteamine in ethanol (0.2 mg mL−1) at 4 °C for 24 h, after which they were washed with ethanol and incubated in a solution of GA (7% v/v) in PBS pH 7.2 for 45 min. After this step, the chips were washed with PBS and immersed in a solution of the respective template (1.2 mg mL−1) in PBS, pH 7.2 for 24 h at 4 °C [38]. In the case of melamine and desisopropylatrazine, methanol (20% v/v) was added as co-solvent. Once the immobilization was completed, the chips were assembled on their holders and stored under argon at 4 °C until used. A sample of the solution of nanoparticles in ACN (10 mL) was concentrated down to a final volume of 1 mL through centrifugal membrane filter units (Amicon Centriplus®, 30 kDa MWCO, Millipore, UK) using PBS pH 7.4. This solution was sonicated for 30 min and used as a stock to prepare 5 further dilutions from 1:2 to 1:32 (Stock concentrations: melamine MIP nanoparticles: 330 nm; peptide MIP nanoparticles: 1094 nm; vancomycin MIP nanoparticles: 135 nm). All the affinity experiments were performed using a flow rate of 35 μL min−1 and a temperature of 25 °C for melamine and vancomycin MIP nanoparticles, and 18 °C for peptide MIP nanoparticles. From each dilution, aliquots (100 μL) were injected and the sensor response was analyzed for 2 min. Kinetic data were fitted using BIAEvaluation Software v4.1 (Biacore, Sweden) which assumes a Langmuir isotherm model.
Supplementary Material
Acknowledgements
The authors thank The Wellcome Trust (UK) for the granting of a Translational Award, The Royal Society for the Brian Mercer Feasibility Award and Cranfield University for additional financial support.
Footnotes
Supporting Information is available online from Wiley InterScience or from the author.
References
- [1].Weiss PS. ACS Nano. 2012;6:1877. doi: 10.1021/nn301088h. [DOI] [PubMed] [Google Scholar]
- [2].a) Merrifield B. [date accessed: 9-Aug-2012];Bruce Merrifield - Nobel Lecture: Solid Phase Synthesis. http://www.nobelprize.org/nobel_prizes/chemistry/laureates/1984/merrifield-lecture.html, Nobelprize.org.; b) Hoeprich PD., Jr. Nat. Biotech. 1996;14:1311. [Google Scholar]; c) Horvath SJ, Firca JR, Hunkapiller T, Hunkapiller MW, Hood L. In: Recombinant DNA Part E. Wu R, Grossman L, editors. Vol. 154. Academic Press; 1987. p. 314. [Google Scholar]
- [3].Kent SH, Hood LE, Beilar H, Meister S, Geiser T. In: Ragnarsson E, editor. High yield chemical synthesis of biologically active peptides on an automated peptide synthesizer of novel design; Peptides 1984: Proceedings of the 18th European Peptide Symposium; Almqvist and Wiksell, Stockholm. 1984.p. 185. [Google Scholar]
- [4].Hunkapiller MW, Hood LE. Science. 1980;207:523. doi: 10.1126/science.7352258. [DOI] [PubMed] [Google Scholar]
- [5].Kambara H, Nishikawa T, Katayama Y, Yamaguchi T. Nat. Biotech. 1988;6:816. [Google Scholar]
- [6].a) Díaz-Díaz G, Antuña-Jiménez D, Blanco-López MC, Lobo-Castañón MJ, Miranda-Ordieres AJ, Tuñón-Blanco P. TrAC, Trends Anal. Chem. 2012;33:68. [Google Scholar]; b) Fang GZ, Lu JP, Pan MF, Li W, Ren L, Wang S. Food Anal. Meth. 2011;4:590. [Google Scholar]; c) Xu ZX, Gao HJ, Zhang LM, Chen XQ, Qiao XG. J. Food Sci. 2011;76:R69–R75. doi: 10.1111/j.1750-3841.2010.02020.x. [DOI] [PubMed] [Google Scholar]
- [7].a) Malitesta C, Mazzotta E, Picca RA, Poma A, Chianella I, Piletsky SA. Anal. Bioanal. Chem. 2012;402:1827. doi: 10.1007/s00216-011-5405-5. [DOI] [PubMed] [Google Scholar]; b) Whitcombe MJ, Chianella I, Larcombe L, Piletsky SA, Noble J, Porter R, Horgan A. Chem. Soc. Rev. 2011;40:1547. doi: 10.1039/c0cs00049c. [DOI] [PubMed] [Google Scholar]; c) Jenik M, Seifner A, Krassnig S, Seidler K, Lieberzeit PA, Dickert FL, Jungbauer C. Biosens. Bioelectron. 2009;25:9. doi: 10.1016/j.bios.2009.01.019. [DOI] [PubMed] [Google Scholar]
- [8].a) Haginaka J. J. Sep. Sci. 2009;32:1548. doi: 10.1002/jssc.200900085. [DOI] [PubMed] [Google Scholar]; b) Liu ZS, Zheng C, Yan C, Gao RY. Electrophoresis. 2007;28:127. doi: 10.1002/elps.200600544. [DOI] [PubMed] [Google Scholar]; c) Ulbricht M. J. Chromatogr. B. 2004;804:113. doi: 10.1016/j.jchromb.2004.02.007. [DOI] [PubMed] [Google Scholar]
- [9].Mayes AG, Whitcombe MJ. Adv. Drug Deliv. Rev. 2005;57:1742. doi: 10.1016/j.addr.2005.07.011. [DOI] [PubMed] [Google Scholar]
- [10].a) Karim K, Breton F, Rouillon R, Piletska EV, Guerreiro A, Chianella I, Piletsky SA. Adv. Drug Deliv. Rev. 2005;57:1795. doi: 10.1016/j.addr.2005.07.013. [DOI] [PubMed] [Google Scholar]; b) Subrahmanyam S, Piletsky SA. In: Combinatorial Methods for Chemical and Biological Sensors. Potyrailo RA, Mirsky VM, editors. Springer; New York: 2009. p. 135. [Google Scholar]
- [11].Cutivet A, Schembri C, Kovensky J, Haupt K. J. Am. Chem. Soc. 2009;131:14699. doi: 10.1021/ja901600e. [DOI] [PubMed] [Google Scholar]
- [12].Hoshino Y, Koide H, Urakami T, Kanazawa H, Kodama T, Oku N, Shea KJ. J. Am. Chem. Soc. 2010;132:6644. doi: 10.1021/ja102148f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].a) Wulff G, Sarhan A. Angew. Chem.,Intl. Ed. Engl. 1972;11:341. [Google Scholar]; b) Arshady R, Mosbach K. Macromol. Chem. Phys. 1981;182:687. [Google Scholar]
- [14].a) Yan H, Row KH. Int. J. Mol. Sci. 2006;7:155. [Google Scholar]; b) Piletska EV, Guerreiro AR, Whitcombe MJ, Piletsky SA. Macromolecules. 2009;42:4921. [Google Scholar]
- [15].Umpleby RJ, Bode M, Shimizu KD. Analyst. 2000;125:1261. [Google Scholar]
- [16].Venn RF, Goody RJ. Chromatographia. 1999;50:407. [Google Scholar]
- [17].Biffis A, Graham NB, Siedlaczek G, Stalberg S, Wulff G. Macromol. Chem. Phys. 2001;202:163. [Google Scholar]
- [18].a) Flavin K, Resmini M. Anal. Bioanal. Chem. 2009;393:437. doi: 10.1007/s00216-008-2496-8. [DOI] [PubMed] [Google Scholar]; b) Poma A, Turner APF, Piletsky SA. Trends Biotechnol. 2010;28:629. doi: 10.1016/j.tibtech.2010.08.006. [DOI] [PubMed] [Google Scholar]
- [19].Wulff G, Chong BO, Kolb U. Angew. Chem. Intl. Ed. 2006;45:2955. doi: 10.1002/anie.200503926. [DOI] [PubMed] [Google Scholar]
- [20].a) Ye L, Weiss R, Mosbach K. Macromolecules. 2000;33:8239. [Google Scholar]; b) Ye L, Mosbach K. Reac. Func. Polym. 2001;48:149. [Google Scholar]; c) Zhang ZC, Cheng ZQ, Zhang CF, Wang HY, Li JF. J. Appl. Polym. Sci. 2012;123:962. [Google Scholar]
- [21].a) Pérez N, Whitcombe MJ, Vulfson EN. J. Appl. Polym. Sci. 2000;77:1851. [Google Scholar]; b) Pérez-Moral N, Mayes AG. Anal. Chim. Acta. 2004;504:15. [Google Scholar]
- [22].a) Vaihinger D, Landfester K, Krauter I, Brunner H, Tovar GEM. Macromol. Chem. Phys. 2002;203:1965. [Google Scholar]; b) Tan CJ, Wangrangsimakul S, Bai R, Tong YW. Chem. Mater. 2008;20:118. [Google Scholar]
- [23].a) Choi KM. Res. Lett. Mater. Sci. 2008 Art. No. 458158. [Google Scholar]; b) Liu XY, Lei JD. Polym. Eng. Sci. 2012;52:2099. [Google Scholar]; c) Zourob M, Mohr S, Mayes AG, Macaskill A, Pérez-Moral N, Fielden PR, Goddard NJ. Lab. Chip. 2006;6:296. doi: 10.1039/b513195b. [DOI] [PubMed] [Google Scholar]
- [24].Zimmerman SC, Wendland MS, Rakow NA, Zharov I, Suslick KS. Nature. 2002;418:399. doi: 10.1038/nature00877. [DOI] [PubMed] [Google Scholar]
- [25].Sellergren B, Hall AJ. In: in Molecularly Imprinted Polymers: Man-Made Mimics of Antibodies and their Applications in Analytical Chemistry. Sellergren B, editor. Vol. 23. Elsevier; Amsterdam: 2001. p. 21. [Google Scholar]
- [26].Liu LJ, Zheng JJ, Fang GJ, Xie WH. Anal. Chim. Acta. 2012;726:85. doi: 10.1016/j.aca.2012.03.026. [DOI] [PubMed] [Google Scholar]
- [27].a) Titirici MM, Hall AJ, Sellergren B. Chem. Mater. 2002;14:21. [Google Scholar]; b) Titirici MM, Sellergren B. Anal. Bioanal. Chem. 2004;378:1913. doi: 10.1007/s00216-003-2445-5. [DOI] [PubMed] [Google Scholar]; c) Volkmann A, Brüggemann O. Reac. Func. Polym. 2006;66:1725. [Google Scholar]
- [28].Shen XT, Ye L. Macromolecules. 2011;44:5631. doi: 10.1021/ma200837n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Guerreiro AR, Chianella I, Piletska E, Whitcombe MJ, Piletsky SA. Biosens. Bioelectron. 2009;24:2740. doi: 10.1016/j.bios.2009.01.013. [DOI] [PubMed] [Google Scholar]
- [30].Hoshino Y, Haberaecker WWI, Kodama T, Zeng ZY, Okahata Y, Shea KJ. J. Am. Chem. Soc. 2010;132:13648. doi: 10.1021/ja1058982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].a) Piletsky SA, Piletska EV, Karim K, Freebairn KW, Legge CH, Turner APF. Macromolecules. 2002;35:7499. [Google Scholar]; b) Allender CJ, Heard CM, Brain KR. Chirality. 1997;9:238. [Google Scholar]
- [32].a) Mijangos I, Navarro-Villoslada F, Guerreiro A, Piletska E, Chianella I, Karim K, Turner A, Piletsky S. Biosens. Bioelectron. 2006;22:381. doi: 10.1016/j.bios.2006.05.012. [DOI] [PubMed] [Google Scholar]; b) O’Shannessy DJ, Ekberg B, Mosbach K. Anal. Biochem. 1989;177:144. doi: 10.1016/0003-2697(89)90029-8. [DOI] [PubMed] [Google Scholar]
- [33].a) Bompart M, Haupt K. Aus. J. Chem. 2009;62:751. [Google Scholar]; b) Gonzato C, Courty M, Pasetto P, Haupt K. Adv. Funct. Mater. 2011;21:3947. [Google Scholar]
- [34].Lorenzo RA, Carro AM, Alvarez-Lorenzo C, Concheiro A. Int. J. Mol. Sci. 2011;12:4327. doi: 10.3390/ijms12074327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].a) Pérez-Moral N, Mayes AG. Macromol. Rapid Commun. 2007;28:2170. [Google Scholar]; b) Li JY, Zu BY, Zhang Y, Guo XZ, Zhang HQ. J. Polym. Sci. A,Polym. Chem. 2010;48:3217. [Google Scholar]
- [36].a) Otsu T, Matsunaga T, Kuriyama A, Yoshioka M. Eur. Polym. J. 1989;25:643. [Google Scholar]; b) Rückert B, Hall AJ, Sellergren B. J. Mater. Chem. 2002;12:2275. [Google Scholar]
- [37].Li M, Zhang LY, Meng ZH, Wang ZY, Wu H. J. Chromatogr B. 2010;878:2333. doi: 10.1016/j.jchromb.2010.07.003. [DOI] [PubMed] [Google Scholar]
- [38].Jiang DC, Tang J, Liu BH, Yang PY, Shen XR, Kong JL. Biosens. Bioelectron. 2003;18:1183. doi: 10.1016/s0956-5663(02)00253-1. [DOI] [PubMed] [Google Scholar]
- [39].Ivanova-Mitseva PK, Guerreiro A, Piletska EV, Whitcombe MJ, Zhou Z, Mitsev PA, Davis F, Piletsky SA. Angew. Chem. Intl. Ed. 2012;51:5196. doi: 10.1002/anie.201107644. [DOI] [PubMed] [Google Scholar]
- [40].Piletska EV, Piletsky SA. Langmuir. 2010;26:3783. doi: 10.1021/la904834y. [DOI] [PubMed] [Google Scholar]
- [41].a) Grant SD, Porter AJ, Harris WJ. J. Agric. Food Chem. 1998;47:340. doi: 10.1021/jf9808574. [DOI] [PubMed] [Google Scholar]; b) Kramer K. Environ. Sci. Technol. 2002;36:4892. doi: 10.1021/es010209s. [DOI] [PubMed] [Google Scholar]
- [42].Halliwell CM, Cass AEG. Anal. Chem. 2001;73:2476. doi: 10.1021/ac0010633. [DOI] [PubMed] [Google Scholar]
- [43].Sheng H, Ye BC. Appl. Biochem. Biotechnol. 2009;152:54. doi: 10.1007/s12010-008-8245-9. [DOI] [PubMed] [Google Scholar]
- [44].Hoshino Y, Kodama T, Okahata Y, Shea KJ. J. Am. Chem. Soc. 2008;130:15242. doi: 10.1021/ja8062875. [DOI] [PubMed] [Google Scholar]
- [45].a) Schirhagl R, Latif U, Dickert FL. J. Mater. Chem. 2011;21:14594. [Google Scholar]; b) Schirhagl R, Lieberzeit PA, Blaas D, Dickert FL. Adv. Mater. 2010;22:2078. doi: 10.1002/adma.200903517. [DOI] [PubMed] [Google Scholar]; c) Schirhagl R, Latif U, Podlipna D, Blumenstock H, Dickert FL. Anal. Chem. 2012;84:3908. doi: 10.1021/ac201687b. [DOI] [PubMed] [Google Scholar]
- [46].Piletsky SA, Guerriero ARL, Whitcombe MJ. Patent WO2011067563 2011
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.