Abstract
Germline mutation determines rates of molecular evolution, genetic diversity, and fitness load. In humans, the average point mutation rate is 1.2 × 10−8 per base pair per generation, with every additional year of father’s age contributing two mutations across the genome and males contributing three to four times as many mutations as females. To assess whether such patterns are shared with our closest living relatives, we sequenced the genomes of a nine-member pedigree of Western chimpanzees, Pan troglodytes verus. Our results indicate a mutation rate of 1.2 × 10−8 per base pair per generation, but a male contribution seven to eight times that of females and a paternal age effect of three mutations per year of father’s age. Thus, mutation rates and patterns differ between closely related species.
Accurate determination of the rate of de novo mutation in the germ line of a species is central to the dating of evolutionary events. However, because mutations are rare events, efforts to measure the rate in humans have typically been indirect, calculated from the incidence of genetic disease or sequence divergence (1–4). However, high-throughput sequencing technologies have enabled direct estimates of the mutation rate from comparison of the genome sequence of family members (5–8). Unexpectedly, these studies have indicated a mutation rate of, on average, ~1.2 × 10−8 per base pair per generation, or ~0.5 × 10−9 per base pair per year, approximately half that inferred from phylogenetic approaches (1, 9). Moreover, they have demonstrated a substantial male bias to mutation, such that three to four times as many autosomal mutations occur in the male compared to the female germ line (6, 7). Male bias is largely caused by an increase in the rate of paternal but not maternal mutation with the age of the parent; approximately two additional mutations per year of father’s age at conception (7). This difference is consistent with ongoing cell division in the male germ line but not in females (10).
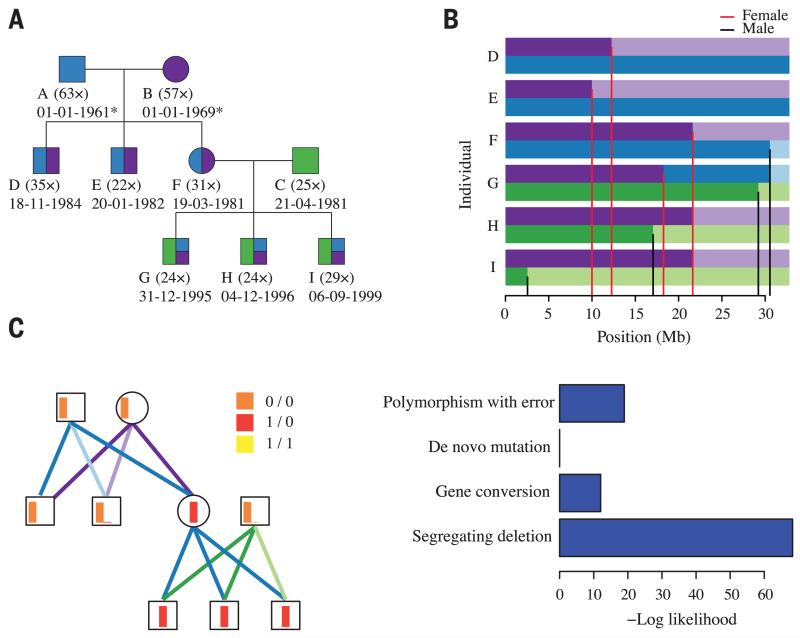
An alternative approach for estimating the extent of male bias is to compare rates of sequence divergence on the autosomes (which spend equal time in the male and female germ lines) and the X chromosome (which spends two-thirds of the time in females) (2, 11). Such indirect approaches broadly agree with direct estimates in humans, but suggest that male bias may be stronger in chimpanzees (12). To test this hypothesis, we sequenced the genomes of nine members of a three-generation pedigree of Western chimpanzees, Pan troglodytes verus (Fig. 1A and fig. S1). One trio was sequenced at high depth (average 51×), while other family members were sequenced to an average of 27× (table S1). We inferred the structure of recombination and transmission across the pedigree (Fig. 1B), which enabled us to detect de novo point mutations in regions of high sequence complexity and to remove artifacts caused by mismapping, sequence that is absent from the reference genome, and reference misassembly (13).
Fig. 1. De novo mutations in a three-generation chimpanzee pedigree.
(A) Structure of the chimpanzee pedigree, indicating sequencing depth and date of birth (estimated for wild founders). Blue, purple, and green colors indicate expected inheritance proportions from founders. (B) Visual representation of transmission across chromosome 21 following color assignment in (A); grandparental origin distinguished by intensity. Red and black lines indicate position of female- and male-specific cross-over events, respectively. (C) Site classification at a candidate de novo mutation on chromosome 2A, showing (left) the relative genotype likelihoods for each individual (as red, orange, and yellow barplots; with height representing relative likelihood and color representing genotype), the resolved transmission structure [as purple, blue, and green lines; following color and intensity assignment in (B)], and (right) the relative likelihoods for different models for the observed genetic variation.
We used a probabilistic approach that, at a given site, compared the likelihood of different models for genetic variation inconsistent with the inferred transmission: genotyping error at a segregating variant, de novo mutation, single-gene conversion event, segregating deletion and erroneous call (Fig. 1C). The design was expected to enable haplotype phasing through transmission for 99.2% of sites that were heterozygous in the founders and 87.5% of de novo mutation events inherited by chimpanzee F (Fig. 1A). Read-based phasing was used to phase de novo events in other offspring, and we performed independent validation to assess the accuracy of de novo variant calls. The false-negative rate was estimated from allele-dropping simulations (13).
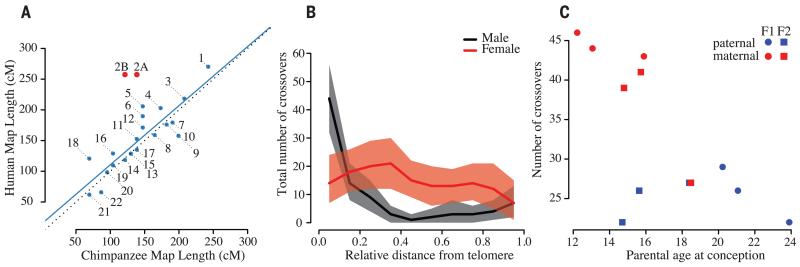
Across the genomes of the nine pedigree members, we called 4.1 million variants [single-nucleotide polymorphisms (SNPs) and short insertions and deletions (indels)] using a mapping-based approach and 3.0 million variants using an assembly-based approach (14). Genotype data confirmed expected pedigree relationships (fig. S2). The intersection of call sets (1.6 million sites with a transition-transversion ratio of 2.2) established the underlying structure of recombination and transmission across the pedigree with a robust version of the Lander-Green algorithm (fig. S3). Briefly, this is a two-stage strategy of identifying dominant inheritance vectors over 1-Mb intervals, followed by fine-mapping of cross-over breakpoints, which guards against problems caused by false-positive variants and genotyping errors (13). Across the pedigree, we identified 375 cross-over events, with a distribution similar to that of human homologs, with the exception of human chromosome 2, which is a fusion of the chimpanzee chromosomes 2A and 2B (15) (Fig. 2A, fig. S4, and tables S2 and S3).
Fig. 2. Parameters of cross-over.
(A) Scatterplot showing the relationship between total genetic map length inferred for chromosomes and their homologs in humans. Black dotted line indicates the identity relationship; blue line indicates best linear fit (excluding chromosomes 2A and 2B). (B) Relationship of recombination rate to relative telomere proximity demonstrates a telomeric effect in males, which is absent in females. (C) Relationship between parental age and the number of cross-over events in mothers and fathers for the two family groups.
Overall, we estimate the sex-averaged auto-somal genetic map length to be 3150 cM [95% equal-tailed probability interval (ETPI) 2850 to 3490], compared to 3505 cM in humans (16, 17). On the X chromosome, we detected nine cross-over events in the non-pseudoautosomal (non-PAR) region, indicating a female-specific genetic map length of 160 cM (95% ETPI 83 to 300), compared to 180 cM in humans. On the pseudoautosomal region (PAR), we detected four male cross-overs, giving a male-specific estimate of 34 cM (95% ETPI 28 to 180; tables S4 and S5), in agreement with estimates in humans (13). Males have 58% of the autosomal cross-over events of females and, unlike females, show an increase in cross-over frequency toward the telomere (Fig. 2B), similar to humans (fig. S5). We also observed a decrease in cross-over frequency with maternal (2.65 cM per year, linear model P = 0.025), but not paternal age (Fig. 2C). However, this observation could be explained by between-female variation (linear model P = 0.13, allowing for a maternal effect). The median interval size to which cross-over events can be localized is 7.0 kb, with 95% of all intervals localized to within 80 kb (excluding complex cross-over events), with cross-over events enriched in regions inferred to have high rates of recombination from patterns of linkage disequilibrium (18) (fig. S6).
Conditional on the inferred transmission, we used a probabilistic approach to identify candidate de novo mutations among all variants called by the mapping approach, incorporating uncertainty in the inferred genotype through the use of genotype likelihoods (13). Across the pedigree, we identified 204 autosomal de novo mutations (2 of which are multinucleotide variants) that pass thresholds for evidence (fig. S7), purity, and consistency (fig. S8 and table S6).
Several lines of evidence indicate a low false-positive rate. First, none of these sites were called as variants in the genomes of 10 unrelated chimpanzees from the same subspecies (18). Second, the transition-transversion ratio of the candidate de novo events (2.16) is comparable to that for segregating variants. Third, the transmission of candidate de novo events in chimpanzee F to her offspring is consistent with expectations. Finally, we used a genotyping platform to validate all de novo events identified in chimpanzees F and I. Of the 61 sites with valid assays (18 failed design), 1 is a false positive, indicating a false discovery rate of ~2% (table S7). To estimate false-negative rates, we used allele-dropping simulations, with empirical distributions of coverage and allele balance. Within the F1 generation, the false-negative rate is estimated to be 3.4%. However, the F2 generation has a higher rate (23%), arising from lower coverage (25.6×) in founder chimpanzee C. False-negative rates were used to correct subsequent regression analyses (table S8). On the X chromosome, we identified three de novo point mutations.
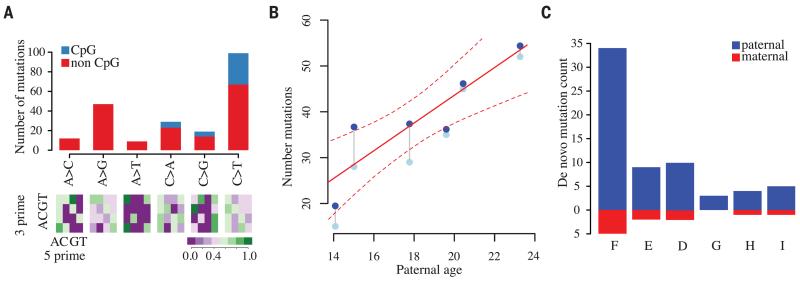
As expected (1), a high fraction of de novo mutations are C>T transitions at CpG sites [24% of all point mutations, compared to 17% in humans (7); likelihood ratio test (LRT), P = 0.03], and even after accounting for such mutations, we see a trend toward AT bases (73 G/C>A/T, 55 A/T>G/C; ratio 1.3:1; LRT, P = 0.11; Fig. 3A). We also found that point mutations tend to cluster within individuals at nearby locations, similar to observations in humans (8). For example, 17% of all point mutations are within 1 Mb of at least one other mutation event in the pedigree, and in 41% of such cases, these all occur in the same individual (compared to an expectation of 13%; permutation P = 0.001). Notably, we validated all variants in clusters of 1 Mb or less in E and F, indicating that these are not false positives. The excess of within-sample clustering extends up to ~200 kb (fig. S9A) and does not correspond to a single mutation type (e.g., CpG mutation). Moreover, the effect remains after increasingly stringent filters for specificity are applied (fig. S9B) (13). The finding of clustered point mutation events, which may potentially arise from correlated exposure to mutagens or variation in the efficacy of DNA repair, implies non-independence in the way novel variation enters a species and has consequences for interpretation of patterns of genetic variation. We do not observe enrichment around genes, repeat elements, or gaps in the assembly (fig. S10).
Fig. 3. Characteristics of de novo mutations.
(A) (Top) Numbers of different types of de novo single-base substitutions (multiple nucleotide events not shown). (Bottom) Relative abundance of each mutation type as a function of the adjacent nucleotides. (B) Relationship between paternal age and total number of de novo mutations; gray points: observed counts; blue points: correction for false-negative rate. The posterior mean and marginal 95% equal-tailed probability intervals for the Bayesian Poisson regression are shown as solid and dotted lines, respectively. (C) The number of de novo mutation events confirmed as paternal or maternal through either transmission to the F2 generation or from direct phasing within read-pairs. Offspring ordered left to right by increasing paternal age.
To assess whether the rate of de novo mutation is affected by parental age, we used Poisson regression, allowing for family effects and separate linear relationships between age and mutation rate for males and females. Despite the small sample size, we find no evidence for either familial or maternal age effects (linear model P > 0.05), but we do find evidence for a paternal age effect (linear model P = 0.006) and consistency in effect on repeat and nonrepeat DNA backgrounds (fig. S11). Although we cannot formally exclude the possibility of familial effects (6), our results are consistent with observations (7) that paternal age explains nearly all variation in mutation rate in humans. Bayesian linear regression, allowing for a paternal effect only and accommodating variation in false-negative rate, indicates a posterior mean paternal age of effect of 3.02 mutations per year (Fig. 3B; 95% ETPI 1.35 to 4.68). In contrast, the paternal age effect in humans is estimated to be 1.95 mutations per year [reanalysis of data from (6); 95% ETPI 1.65 to 2.26].
We ascertained the parent of origin of de novo mutations for chimpanzee F through transmission to the F2 generation, finding that 30 of the 35 autosomal mutations occurred in the paternal lineage. We also found that 25% of de novo events could be phased directly from read-pairs spanning the mutation and a nearby heterozygous site that could be phased through transmission. Across the pedigree, we assigned 31 paternal and 6 maternal autosomal mutations (Fig. 3C). Overall, we estimate the aggregate male-to-female mutation ratio, α = 5.5 (95% ETPI = 3.0 to 10). The point estimate is 40% higher than that reported for humans (7), though it is close to estimates from chimpanzee-specific divergence rates on the X and autosomes (12). In contrast to indirect approaches (3), we find no evidence that different types of mutation have different values of α. For example, α at CpG sites is 5.3, compared to 5.6 at non-CpG sites. Combining data across all mutation types and using available parent-of-origin information in the Bayesian regression model indicates that, on average, mothers contribute 6.7 de novo mutations (95% ETPI = 3.5 to 10.3) and each additional year of paternal age generates 3.0 mutations (95% EPTI = 1.2 to 4.4; fig. S12), with the onset of mutation occurring at 8.1 years of age (95% ETPI = 0 to 12 years), consistent with the onset of spermarche in chimpanzees of 7.5 years (19).
Within the pedigree studied, the average number of autosomal de novo mutations occurring in each generation is 35, lower than current estimates for humans of 74.4 (7, 9). However, the parental ages in the Western chimpanzee pedigree (averages of 18.9 for males and 15.0 for females) are lower than estimates of parental ages in the wild (24.3 for males and 26.3 for females), which are lower than estimates for humans (31.5 for males and 25.6 for females) (20). Using the fitted model for mutation rates, we predict that the average number of autosomal de novo mutations per offspring in the wild should be ~69. We estimate the length of the autosomal genome accessible in our study to be 2360 Mb across the autosomes (table S9), indicating a mutation rate of 1.2 × 10−8 per base pair per generation and α to be 7.8 (table S10).
Under a model in which the mutation rate increases linearly with parental age, the rate of neutral substitution is the ratio of the average number of mutations inherited per generation to the average parental age. We predict the neutral substitution rate to be ~0.46 × 10−9 per base pair (bp) per year in chimpanzees, compared to estimates in humans of ~0.51 × 10−9 bp−1 year−1 (9). These results are consistent with near-identical levels of lineage-specific sequence divergence (12) but surprising given the differences in paternal age effect. In the intersection of the auto-somal genome accessible in this study and regions where human and chimpanzee genomes can be aligned with high confidence, the rate is slightly lower (0.45 × 10−9 bp−1 year−1) and the level of divergence is 1.2% (13), implying an average time to the most common ancestor of 13 million years, assuming uniformity of the mutation rate over this time (95% ETPI 11 to 17 million years; table S11).
Increased male bias can explain low levels of diversity on the chimpanzee X chromosome (21, 22). Taking into account differences in generation time and effective population size, we predict that X-chromosome diversity should be 56% that of autosomes (assuming equal and constant effective population sizes for males and females; table S10), comparable to empirical estimates (21, 22). Similarly, our results predict that the X-chromosome rate of divergence is lower in chimpanzees than humans (74% of the autosomal rate in chimpanzees, 85% of the autosomal rate in humans). Previous explanations for unusual patterns of X-chromosome diversity and divergence include a complex speciation event (23), extensive natural selection on the X chromosome (22), or, as supported by this study, a greater male mutational bias in chimpanzees (12). This is likely related to differences in mating system between the species, with chimpanzees showing higher levels of sperm competition through multiple mating and a higher relative testes mass than humans (0.27% of average adult male weight versus 0.079%) and higher levels of sperm production (24, 25). If differences in male mutational bias are to explain observed patterns of divergence, then gorillas would have a male mutational bias lower than that of humans arising from decreased sperm competition (12). Our results suggest that variation in mating patterns between species can affect the sex bias of mutation and motivate the wider study of mutation rates and relationship to parental age across species.
Supplementary Material
ACKNOWLEDGMENTS
Funded by Wellcome Trust grants 086786/Z/08/Z to O.V. and 090532/Z/09/Z to the Wellcome Trust Centre for Human Genetics and by MRC hub grant G0900747 91070. We thank M. Przeworski and D. Reich for discussion and comments on the manuscript and A. Kong for providing data on request from reference (6). Samples were provided through the Transnational Access Activity in the European Primate Network (EUPRIM-NET) under the Convention on International Trade of Endangered Species (CITES) authorization and a Material Transfer Agreement between the University of Oxford and the Foundation Biomedical Primate Research Centre. Read-level data are accessible under SRA Study accession no. PRJEB5937 from www.ebi.ac.uk/ena/data/view/PRJEB5937. All other project data are available from ftp://birch.well.ox.ac.uk.
Footnotes
www.sciencemag.org/content/344/6189/1272/suppl/DC1
Materials and Methods
Supplementary Text
Figs. S1 to S12
Tables S1 to S11
References (26–40)
REFERENCES AND NOTES
- 1.Nachman MW, Crowell SL. Genetics. 2000;156:297–304. doi: 10.1093/genetics/156.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Makova KD, Li WH. Nature. 2002;416:624–626. doi: 10.1038/416624a. [DOI] [PubMed] [Google Scholar]
- 3.Taylor J, Tyekucheva S, Zody M, Chiaromonte F, Makova KD. Mol. Biol. Evol. 2006;23:565–573. doi: 10.1093/molbev/msj060. [DOI] [PubMed] [Google Scholar]
- 4.Kondrashov FA, Kondrashov AS. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2010;365:1169–1176. doi: 10.1098/rstb.2009.0286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roach JC, et al. Science. 2010;328:636–639. doi: 10.1126/science.1186802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Conrad DF, et al. Nat. Genet. 2011;43:712–714. doi: 10.1038/ng.862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kong A, et al. Nature. 2012;488:471–475. doi: 10.1038/nature11396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Michaelson JJ, et al. Cell. 2012;151:1431–1442. doi: 10.1016/j.cell.2012.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scally A, Durbin R. Nat. Rev. Genet. 2012;13:745–753. doi: 10.1038/nrg3295. [DOI] [PubMed] [Google Scholar]
- 10.Crow JF. Nat. Rev. Genet. 2000;1:40–47. doi: 10.1038/35049558. [DOI] [PubMed] [Google Scholar]
- 11.Miyata T, Hayashida H, Kuma K, Mitsuyasu K, Yasunaga T. Cold Spring Harb. Symp. Quant. Biol. 1987;52:863–867. doi: 10.1101/sqb.1987.052.01.094. [DOI] [PubMed] [Google Scholar]
- 12.Presgraves DC, Yi SV. Trends Ecol. Evol. 2009;24:533–540. doi: 10.1016/j.tree.2009.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Supplementary materials are available on Science Online.
- 14.Iqbal Z, Caccamo M, Turner I, Flicek P, McVean G. Nat. Genet. 2012;44:226–232. doi: 10.1038/ng.1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.IJdo JW, Baldini A, Ward DC, Reeders ST, Wells RA. Proc. Natl. Acad. Sci. U.S.A. 1991;88:9051–9055. doi: 10.1073/pnas.88.20.9051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kong A, et al. Nature. 2010;467:1099–1103. doi: 10.1038/nature09525. [DOI] [PubMed] [Google Scholar]
- 17.Kong A, et al. Nat. Genet. 2002;31:241–247. doi: 10.1038/ng917. [DOI] [PubMed] [Google Scholar]
- 18.Auton A, et al. Science. 2012;336:193–198. doi: 10.1126/science.1216872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marson J, Meuris S, Cooper RW, Jouannet P. Biol. Reprod. 1991;44:448–455. doi: 10.1095/biolreprod44.3.448. [DOI] [PubMed] [Google Scholar]
- 20.Langergraber KE, et al. Proc. Natl. Acad. Sci. U.S.A. 2012;109:15716–15721. doi: 10.1073/pnas.1211740109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Perry GH, Marioni JC, Melsted P, Gilad Y. Mol. Ecol. 2010;19:5332–5344. doi: 10.1111/j.1365-294X.2010.04888.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hvilsom C, et al. Proc. Natl. Acad. Sci. U.S.A. 2012;109:2054–2059. doi: 10.1073/pnas.1106877109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Patterson N, Richter DJ, Gnerre S, Lander ES, Reich D. Nature. 2006;441:1103–1108. doi: 10.1038/nature04789. [DOI] [PubMed] [Google Scholar]
- 24.Short RV. Adv. Stud. Behav. 1979;9:131–158. [Google Scholar]
- 25.Møller AP. J. Hum. Evol. 1988;17:479–488. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.