Abstract
Background
To investigate the effect of feeding a linseed-enriched diet to growing-finishing pigs on gene expression in skeletal muscle, pigs were fed with a linseed-enriched diet for 0, 30, 60 and 90 d. Transcriptional profiles of longissimus dorsi muscle were measured using Affymetrix Genechip.
Results
Results showed that 264 genes were identified as differentially expressed genes (DEGs). The strongest transcriptional response was clearly observed at 30 d. DEGs were assigned to several main functional terms, including transcription, apoptosis, intracellular receptor-mediated signaling, muscle organ development, fatty acid metabolic process, cell motion, regulation of glucose metabolic process, spermatogenesis and regulation of myeloid cell differentiation. We also found that transcriptional changs of several transcription cofactors might contribute to n-3 PUFAs regulated gene expression. In addition, the increased expression of IGF-1, insulin signaling pathway and the metabolism of amino acids might involve in the muscle growth induced by feeding a linseed-enriched diet. The results also provide the new evidence that the expression changes of PTPN1, HK2 and PGC-1α might contribute to the regulation of insulin sensitivity by n-3 PUFAs.
Conclusions
Our finding provided correlative evidence that feeding the linseed enriched diet affact expression of genes involved in insulin signaling pathway and the metabolism of amino acids.
Keywords: Linseed, mRNA expression, n-3 PUFAs, Skeletal muscle
Background
Dietary n-3 polyunsaturated fatty acids (n-3 PUFAs) play an essential role in animal development and maintaining animal health. Ingestion of n-3 PUFAs will lead to their distribution to virtually every cell in the body with effects on neurological development, reproduction, insulin action, innate and acquired immunity [1]. Therefore, majority of the studies aimed at increasing the n-3 PUFAs levels in pork to improve its nutritional quality, by using the diet enriched in either fish oils [2], or plant sources of 18:3 such as linseed [3] and canola oil [4]. Interestingly, Sousa et al. [5] reported that feeding the pigs with a diet including 10 % linseed oil (α-linolenic acid, C18:3n-3) increased the lean tissue in pigs. Similarly, feeding pigs with a linseed-enriched diet was reported to result in a modest increase in skeletal muscle mass in pigs [6]. Moreover, it has been demonstrated that feeding a linseed-enriched diet increased the intramuscular fat content in porcine longissimus muscle [7].
The broad physiological impact of n-3 PUFAs are dependent on their biologically properties of altering membrane structure and function, enzymatic activities, eicosanoid signaling, as well as cellular interactions [8]. More recently, n-3 PUFAs have been demonstrated to influence the activation or expression of various transcription factors, including peroxisome proliferator activated receptors (PPARs), hepatic nuclear-4α (HNF-4α), nuclear factor κB (NF-κB), retinoid X receptor α (RXRα), sterol regulatory element binding protein-1c (SREBP-1c), and liver X receptors (LXR) [9]. Therefore feeding animals with an n-3 PUFAs enriched diet could make dramatic transcriptional changes in animal tissues.
Microarray technology has accelerated our ability to understand the transcriptomes in various species and tissues. Using microarray, several studies have addressed to elucidate the effect of dietary n-3 PUFAs on mRNA expression in central nervous system (CNS) [10], liver [11] and intestine [12]. In the current study, we characterized gene expression signatures in the longissimus dorsi muscle of pigs fed with a linseed-enriched diet for 0, 30, 60 or 90 d using Affymetrix Genechip.
Methods
Diets and animals
Two diets for growing pigs and two diets for finishing pigs were formulated to meet the NRC recommended requirements (1998): the control diets and the linseed-enriched diets containing 10 % linseed. Diets composition was presented in Table 1. The fatty acid composition of the diets was analyzed by gas chromatography and listed in Table 2. The linseed-enriched diet contained up to 30-fold more linoleic acid (C18:3 n3) than the control diet. In addition, 2-fold linoleic acid was observed in the linseed-enriched diet compared with the control diet. The control diet contained 6-fold more palmitic acid (C16:0) than the linseed-enriched diet.
Table 1.
Composition and calculated analysis (as-fed basis) of diets
| Growing phasea | Finishing phaseb | |||
|---|---|---|---|---|
| Item | Control diet | Linseed-enriched diet | Control diet | Linseed-enriched diet |
| Ingredients, % | ||||
| Corn | 48.70 | 60.50 | 52.90 | 65 |
| Wheat middling | 18.00 | |||
| Wheat bran | 20.00 | 1.00 | ||
| Soybean meal | 27.00 | 26.50 | 21.00 | 21.00 |
| Fat powderc | 3.30 | 3.10 | ||
| linseed | 10.00 | 10.00 | ||
| Premixd | 3.00 | 3.00 | 3.00 | 3.00 |
| Calculated analysis | ||||
| CP, % | 18.07 | 18.01 | 16.05 | 16.08 |
| Ether extract, % | 5.59 | 5.87 | 5.56 | 6.02 |
| DE, Mcal/kg | 3.42 | 3.41 | 3.40 | 3.40 |
| Lysine, % | 1.07 | 1.05 | 0.93 | 0.95 |
| Calcium, % | 0.69 | 0.66 | 0.65 | 0.70 |
| Phosphorus, % | 0.50 | 0.51 | 0.49 | 0.55 |
aBody weight range from 30 to 60 kg
bBody weight range from 60 to 115 kg
cFat powder: The fat powder is commercially available (BERGAFAT HTL-306 from Berg & Schmidt Co.) with a main composition of a palm oil fraction and phospholipid and the ether extract content is more than 99.50 %. Fatty acid profile of the fat powder: palmitic acid (16:0), 70–80 %; stearic acid (18:0), 5–10 %; oleic acid (18:1), 8–15 %
dProvided per kg of premix: vitamin A, 11,250 IU; vitamin D3, 2,500 IU; vitamin E, 200 mg; menadione, 2.5 mg; thiamine, 2.5 mg; riboflavin, 6.0 mg; niacin, 25 mg; d-panthothenic acid, 8 mg; vitamin B6, 3.0 mg;vitamin B12, 0.08 mg; d-biotin, 0.1 mg; folic acid, 12.5 mg. copper, 20 mg; iron, 50 mg; manganese, 30 mg; zinc, 80 mg; iodine, 0.8 mg
Table 2.
Fatty acid composition (g/100 g of total fatty acids) of diets
| Growing phasea | Finishing phaseb | |||
|---|---|---|---|---|
| Item | Control diet | Linseed diet | Control diet | Linseed diet |
| C14:0 | 1.09 | 0.11 | 1.10 | 0.11 |
| C16:0 | 69.50 | 11.40 | 69.60 | 10.54 |
| C16:1 | 0.10 | 0.38 | 0.11 | 0.39 |
| C18:0 | 4.00 | 4.76 | 4.09 | 4.25 |
| C18:1 n-9 | 11.73 | 24.81 | 10.74 | 24.18 |
| C18:2 n-6 | 12.49 | 24.17 | 11.96 | 25.90 |
| C18:3 n-3 | 0.97 | 30.34 | 1.08 | 32.59 |
| C20:0 | 0.19 | 0.21 | 0.18 | 0.21 |
| C20:1 | 0.06 | 0.06 | 0.04 | 0.23 |
| C22:0 | 0.04 | 0.12 | 0.05 | 0.11 |
| C22:6 n-3 | 0.03 | 0.39 | 0.09 | 0.20 |
| SFAc | 74.82 | 16.6 | 75.02 | 15.22 |
| PUFAsd | 13.49 | 54.9 | 13.13 | 58.69 |
| n-3 PUFAse | 1.08 | 30.78 | 1.24 | 32.85 |
| n-3/n-6 ratiof | 0.09 | 1.27 | 0.1 | 1.27 |
aBody weight range from 30 to 60 kg
bBody weight range from 60 to 115 kg
cSaturated fatty acids percentage is sum of C14:0, C16:0, C18:0, C20:0 and C22:0
dPolyunsaturated fatty acids percentage is sum of C18:2 n-6, C22:6 n-3 and 18:3 n-3 (α-linolenic acid)
en-3 polyunsaturated fatty acids (n-3 PUFAs) percentage is the sum of C18:3 n-3 (α-linolenic acid) and C22:6 n-3
fn-3/n-6 ratio was calculated as the ratio between the total n-3 PUFAs and the total n-6 PUFAs
Twenty-four 80 d age barrows (Landrace × Yorkshire, Huazhong Agricultural University) weighing 35 ± 3.7 kg were randomly assigned to 1 of 4 groups. Pigs in those 4 groups were fed with linseed diet for 0, 30, 60, 90 d before slaughter, respectively. The 0 d and 90 d groups were fed with the control or linseed growing diets for 30 d and then fed with the corresponding finishing diets for the following 60 d. Following feeding the control growing diet for 30 d and control finishing diet for 30 d, pigs in the 30 d group were fed with the linseed finishing diet for 30 d before slaughter. Following feeding the control growing diet for 30 d, pigs in the 60 d group were fed with the linseed finishing diet for 60 d before slaughter. Throughout the experimental period, pigs were housed individually and fed ad libitum during the trial period. At the age of 170 d, all the pigs were humanely slaughtered after 24 h fasting. The longissimus dorsi muscle samples were rapidly removed. Following removing the intermuscular adipose tissue, the muscle samples were frozen in liquid nitrogen to be stored at −80 °C. The study was carried out according to Huazhong Agriculture University Animal Care and Use Committee guidelines.
RNA extraction
RNA samples from three biological replicates for every treatment were extracted using TRIzol (Invitrogen Corp., Carlsbad, CA, USA) as described by the manufacturer, followed by DNase digestion using DNAfree kit (Applied Biosystems, Foster City, CA, USA) according to the manufacturer’s instructions and quantitated by spectrophotometric (Eppendorf, Hamburg, Germany) absorbance at 260 nm. RNA quality was evaluated using A260/A280 ratio (>1.8) and the RNA 6000 Nano Chips in the Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA, USA).
Microarray hybridization and analysis
Hybridisation with Affymetrix GeneChip® Porcine Genome Arrays was performed at CapitalBio Corporation (Beijing, China). The model-based expression index algorithm implemented in dCHIP software was used for data pre-processing [13]. Probe sets were filtered using two criteria: (a) P call rate in arrays ≥20 % and (b) high-intensity (≥55) rate in arrays ≥75 %. The extraction and analysis of differential gene expression algorithm [14] was used for differential gene expression analysis. Q < 0.05 was selected to designate differentially expressed genes [15].
Affymetrix Porcine Annotation V5 [16] was used to annotate the differentially expressed genes. Genes represented by multiple probe sets were determined based on previous studies [17]. Briefly, if a gene is represented by a _at probe set and multiple _s_at probe sets or _x_at probe sets simultaneously, the _at probe set is selected. We filtered the genes that exhibited multiple expression patterns at the probe set level. The average expression value was then calculated to represent the expression of probe sets. After removing the represented probe sets, cluster analysis was performed using the self-organizing tree algorithm (SOTA) implemented in Multiple Array Viewer software V.4.81 [18]. We selected the candidate genes involved in muscle mass regulation from the differentially expressed genes as follows. We constructed a synthesized gene set containing genes related to muscle mass regulation based on gene ontology (GO) annotation and literature mining. Pubgene [19] and GOGene [20] are literature mining tools that can retrieve genes involved in skeletal muscle growth in our data. Database for Annotation, Visualization and Integrated Discovery software [21] was used to annotate and analyze GO and pathway.
Quantitative -PCR
Synthesis of cDNA from skeletal muscle was performed as previously described [22]. Differential expression was checked by Q-PCR for some genes, e.g. peroxisome proliferators-activated receptor-γcoactivator 1α (PGC-1α), protein tyrosine phosphatase non-receptor type 1 (PTPN1), baculoviral IAP repeat-containing protein 2 (BIRC2), insulin-like growth factor 1 (IGF-1), Myocyte-specific enhancer factor 2C (MEF2C) and signal transducer and activator of transcription 5 (STAT5A). The primers were designed using Primer5 software. Relative mRNA levels of genes were quantified using Q-PCR with an iQ™127 5 Real Time PCR Detection System (BioRad, Hercules, CA, USA). The β-actin mRNA levels were similarly measured and served as the reference gene. Of each cDNA, 0.1 μg were added to Q-PCR reagent mixture, SYBR® qPCR Mix (Toyobo, Fukui, Japan), with the sense and antisense primers (500 nmol/L each). The PCR parameters were as follows: denaturation at 95 °C for 3 min followed by 40 cycles of denaturation at 94 °C for 20 s, annealing for 20 s, and extension at 72 °C for 20 s and reading plate for 10s. All sample mRNA levels were normalized to the values of β-actin and the results expressed as fold changes of threshold cycle (Ct) value relative to controls using the 2-ΔΔCt method [23]. All samples were measured in triplicate.
Results
Response of global transcriptome in longissimus dorsi muscle to feeding a linseed-enriched diet to growing pigs
The analysis revealed that 368 probe sets were differentially expressed through the course of trial period (Q < 0.05). After removing the redundant probe sets, 264 probe sets were remained. Among these 264 probe sets, 244 (92.4 %) were assigned to known genes. For each DEGs, the maximal FC between the different time point was larger than 1.5. Additionally, the expression of 87 DEGs (29.5 %) were changed ≥ 2.0-fold throughout the course of trial period. The top 10 up-regulated and down-regulated genes were listed in Tables 3 and 4.
Table 3.
Top 10 up-regulated genes
| Gene | Product | Log intensitya | Maximal FCb | |||
|---|---|---|---|---|---|---|
| 0 dc | 30 d | 60 d | 90 d | |||
| RSAD2 | Viperin | 6.151 | 6.52 | 8.01 | 6.39 | 6.28 |
| HLA-A3 | HLA class I histocompatibility antigen, A-3 alpha chain precursor | 7.47 | 9.46 | 8.16 | 6.73 | 4.58 |
| PGC-1α | Peroxisome proliferators-activated receptor-γcoactivator 1α | 6.95 | 7.69 | 7.18 | 8.99 | 4.24 |
| NNAT | Neuronatin | 6.51 | 7.41 | 8.03 | 8.16 | 3.25 |
| PTGES3 | Telomerase-binding protein p23 | 8.94 | 8.36 | 10.01 | 8.68 | 3.14 |
| KCNQ5 | Potassium voltage-gated channel subfamily KQT member 5 | 4.80 | 6.20 | 4.81 | 4.58 | 3.06 |
| SNX21 | Sorting nexin 21 | 6.54 | 6.03 | 7.30 | 7.57 | 3.01 |
| HK2 | Hexokinase, type II | 7.34 | 8.74 | 8.29 | 8.10 | 2.70 |
| PAF1 | Polymerase associated factor | 7.52 | 8.77 | 7.49 | 7.65 | 2.44 |
| TRIM35 | Tripartite motif-containing 35 isoform 2 | 5.42 | 6.61 | 5.64 | 5.41 | 2.41 |
aLog intensity is calculated from Log2 (normalized intensity)
bMaximal FC (Fold change) is calculated from the ratio of maximum intensity/minimum intensity among the time points
cPigs in 0 d, 30 d, 60 d and 90 d groups (80 d age) were first fed with the control diet for 90, 60, 30 and 0 d and then fed the linseed diet for 0, 30, 60 and 90 d, respectively. At 110 d of age, pigs switched from grower to finisher diet
Table 4.
Top 10 down-regulated genes
| Gene | Product | Log intensitya | Maximal FCb | |||
|---|---|---|---|---|---|---|
| 0 dc | 30 d | 60 d | 90 d | |||
| FAM134B | Family with sequence similarity 134, member | 9.97 | 8.75 | 9.54 | 10.78 | −3.91 |
| SLC19A2 | Thiamine transporter 1 | 8.34 | 7.71 | 8.29 | 9.50 | −3.71 |
| CREM | CAMP responsive element modulator | 9.18 | 8.57 | 9.09 | 10.38 | −3.69 |
| FAM13A1 | Protein FAM13A1 | 9.08 | 7.79 | 8.92 | 9.39 | −2.98 |
| SEMA6A | Semaphorin 6A precursor | 7.11 | 5.96 | 7.55 | 7.09 | −2.97 |
| TNFRSF12A | Tumor necrosis factor receptor superfamily member Fn14 precursor | 11.01 | 9.87 | 10.78 | 11.40 | −2.93 |
| SDC2 | Syndecan-2 precursor | 8.33 | 7.42 | 8.60 | 8.84 | −2.66 |
| TRAC | T-cell receptor alpha chain C region | 6.27 | 4.88 | 6.20 | 5.98 | −2.62 |
| ABRA | Striated muscle activator of Rho-dependent signaling | 11.78 | 10.96 | 11.71 | 12.33 | −2.61 |
| NQO1 | NAD(P)H dehydrogenase | 10.25 | 8.86 | 9.77 | 9.62 | −2.55 |
aLog intensity is calculated from Log2 (normalized intensity)
bMaximal FC (Fold change) is calculated from the ratio of maximum intensity/minimum intensity among the time points
cPigs in 0 d, 30 d, 60 d and 90 d groups (80 d age) were first fed with the control diet for 90, 60, 30 and 0 d and then fed the linseed diet for 0, 30, 60 and 90 d, respectively. At 110 d of age, pigs switched from grower to finisher diet
In order to validate the results of microarray, six genes (PTPN1, PGC-1α, BIRC2, MEF2C, IGF-1 and STAT5) were chosen for Q-PCR confirmation. Comparison of the Q-PCR results with the microarray data demonstrated that these genes had statistically significant expression patterns which were similar to those seen in the microarray data (Fig. 1), indicating that our microarray data are highly reliable and accurate.
Fig. 1.
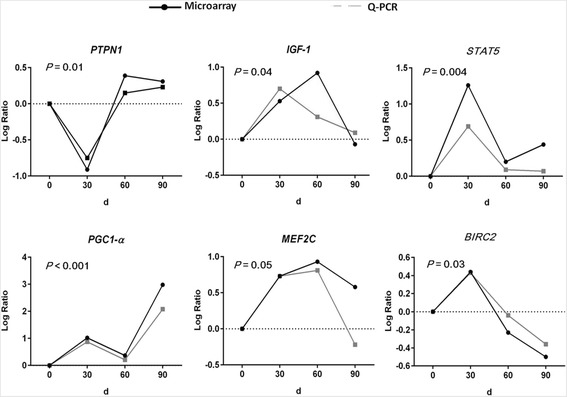
Validation of differentially expressed genes by q-PCR. Pigs in 0 d, 30 d, 60 d and 90 d groups (80 d age) were first fed with the control diet for 90, 60, 30 and 0 d and then fed the linseed diet for 0, 30, 60 and 90 d, respectively. At 110 d of age, pigs switched from grower to finisher diet. Transcriptional profiles of longissimus dorsi muscle were measured using Affymetrix Genechip. Data from q-PCR and microarray were transformed to LogRatio value. The LogRatio value was the ratio of logarithm (base 2) of the mean value of the time point to the mean value of 0 d level. P-values were calculated from q-PCR results
Clustering of DEGs
To extract clearer and meaningful expression patterns, we performed SOTA algorithm for DEGs clustering analysis. The results showed that the DEGs were assigned to 13 clusters (Fig. 2). Interesting, most DEGs (229) were assigned to two main clusters. In cluster 1, there was 155 (58 %) DEGs, of which the expression level was significantly down-regulated at 30 d but raised their expression at 60 and 90 d comparable to those at 30 d. The cluster 2, which contained 74 (28 %) DEGs, had a inverse expression pattern to that of the cluster 1, that genes were significantly induced at 30 d but recovered at 60 and 90 d. The numbers of gene in other clusters except cluster 1 and cluster 2 were all below of 10. DEGs in the cluster 8, cluster 10 and cluster 12 were up-regulated throughout the trail period. These results showed that a strong transcriptional response was clearly observed at 30 d (Fig. 2).
Fig. 2.
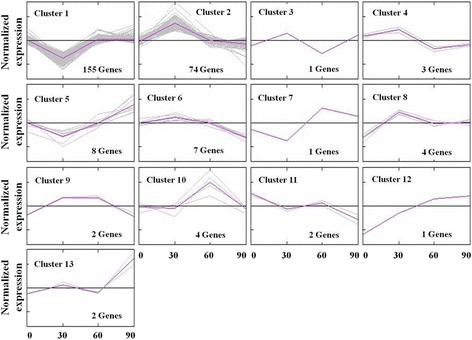
Clustering of differentially expressed genes. Pigs in 0 d, 30 d, 60 d and 90 d groups (80 d age) were first fed with the control diet for 90, 60, 30 and 0 d and then fed the linseed diet for 0, 30, 60 and 90 d, respectively. At 110 d of age, pigs switched from grower to finisher diet. Transcriptional profiles of longissimus dorsi muscle were measured using Affymetrix Genechip. SOTA algorithm was performed for DEGs clustering analysis. Each gene was normalized around zero and then clustered using self-organizing tree algorithm. The purple line was the average expression pattern
Go and pathway analysis of DEGs
To understand the biological functions of the DEGs, GO and pathway analysis were performed in the current study. The results showed that a significant number (p < 0.05) of DEGs were annotated as being involved in transcription (32 genes), apoptosis (21 genes), intracellular receptor-mediated signaling (7 genes), muscle organ development (10 genes), fatty acid metabolic process (9 genes), cell motion (15 genes), regulation of glucose metabolic process (4 genes), spermatogenesis (9 genes) and regulation of myeloid cell differentiation (5 genes) (Table 5).
Table 5.
GO (gene ontology) enrichment of differentially expressed genes
| Category | Count | P Valuea |
|---|---|---|
| Transcription, DNA-dependent | 32 | 0.001 |
| Apoptosis | 21 | 0.002 |
| Intracellular receptor-mediated signaling | 7 | 0.003 |
| Muscle organ development | 10 | 0.011 |
| Fatty acid metabolic process | 8 | 0.017 |
| Cell motion | 15 | 0.021 |
| Regulation of glucose metabolic process | 4 | 0.025 |
| Spermatogenesis | 9 | 0.035 |
| Regulation of myeloid cell differentiation | 5 | 0.035 |
a P value was calculated by hypergeometric distribution test. Enrichment was considered significant at P < 0.05
Pathway analysis revealed that a total of 9 pathways were above the significance threshold (Q < 0.01), including regulation of actin cytoskeleton, amino acids metabolism, biosynthesis of steroids, IκB kinase NF − κΒ cascade related genes, cell adhesion molecules (CAMs), IGF-1/insulin signaling pathway, ubiquitin mediated proteolysis, Wnt signaling pathway and neuroactive ligand-receptor interaction (Table 6). We highlighted the interested GO and pathways with the associated genes as following.
Table 6.
Pathway enrichment of differentially expressed genes
| Pathway | Count | Q-Valuea |
|---|---|---|
| Regulation of actin cytoskeleton | 7 | 0.000 |
| Amino acids metabolism | 3 | 0.001 |
| Biosynthesis of steroids | 3 | 0.002 |
| IκB kinase NF-κB cascade related genes | 6 | 0.003 |
| Cell adhesion molecules (CAMs) | 4 | 0.008 |
| Insulin signaling pathway | 5 | 0.008 |
| Ubiquitin mediated proteolysis | 4 | 0.008 |
| Wnt signaling pathway | 5 | 0.010 |
| Neuroactive ligand-receptor interaction | 5 | 0.010 |
aQ value was calculated by hypergeometric distribution test. Enrichment was considered significant at Q < 0.01
Muscle organ development
The expression of other genes involved in regulation of muscle organ development were also altered, including MEF2C, COUP transcription factor 2 (NR2F2) and Zinc finger protein GLI1 (Gli1 leptin receptor) et al. Our results showed a significant increase in mRNA abundance of MEF2c and NR2F2, but a significant decrease in Gli1 mRNA abundance after feeding a linseed-enriched diet for 30 d.
Fatty acid metabolic process
Eight genes involved in fatty acid metabolic process were changed their mRNA expression level in response to feeding a linseed-enriched diet. The expression of fatty acid synthase (FASN), elovel fatty acid elongase 6 (ELOVL6), Estradiol 17-beta-dehydrogenase 1 (HSD17B1) and mevalonate kinase (MVK) was decreased after feeding pigs with a linseed-enriched diet for 30 d. However, the expression of glycerol-3-phosphate acyltransferase 1, mitochondrial (GPAM) and prostaglandin-endoperoxide synthase 2 (PTGS2) was increased at 30 d. In addition, feeding pigs with a linseed-enriched diet induced the expression of PGC-1α at 30 and 90 d. Moreover, the expression intensity of prostaglandin E synthase 3 (PTGES3) was decreased from 8.9 to 8.4 at 30 d, but then raised to 10.0 at 60 d.
Regulation of glucose metabolic process
Three genes (HK2, PGC-1α and ALDOC) involved in regulating glucose metabolic process were differently expressed in the current study. The expression of HK2 was induced 2.4-fold at 30 d and remained up-regulated at the later time points. For ALDOC, feeding a linseed-enriched diet inhibited its expression at 30 d.
Amino acids metabolism
Notrworthly, expression of amino acids metabolism related genes were also influenced by feeding pigs with a linseed-enriched diet. For example, expression of solute carrier family 38 member 1 (SLC38A1), which is an amino acid transporter, and glutamate-cysteine ligase catalytic subunit (GCLC), were up-regulated at 30 d, while arginase type 2, a gene involved in arginine catabolism, was down-regulated at 30 d.
IκB kinase NF − κΒ cascade
Concerning the top one enriched pathway, the IκB kinase NF-κB cascade, 6 genes were identified as candidate genes. Among them, 4 stimulators of the IκB kinase NF-κB cascade solute carrier family 20, member 1 (SLC20A1), TRAF3 interacting protein 2 (TRAF3IP2), tumor necrosis factor receptor superfamily member Fn14 precursor (TNFRSF12A), and toll-like receptor 9 precursor (TLR9), were down-regulated at 30 d (fold change ≥1.8). BIRC2, an inhibitor of IκB kinase, was up-regulated at 30 d with a 1.7-fold expression change. Interestingly, Tumor protein p53 inducible nuclear protein 1 (TP53INP1), another stimulator of the IκB kinase NF-κB cascade, was down-regulated at 90 d with a 2-fold expression change.
IGF-1/insulin signaling
Four genes involved in IGF-1/insulin signaling were differentially expressed. Among them, STAT5, a transcription factor involved in growth hormone (GH) signaling, was up-regulated in response to feeding pigs with a linseed-enriched diet at 30 d. In addition, significantly lower level of PTPN1, a gene involved in regulation of GH signaling and tyrosine kinase receptor signaling was detected after feeding pigs with a linseed-enriched diet at 30 d. However, both PGC-1α and sorbin and SH3 domain-containing protein 1 (SORBS1), the positive regulators of insulin signaling, were up-regulated at 30 d.
Wnt signaling pathway
The results from the current study suggested that feeding pigs with a linseed enriched-diet influenced the mRNA expression of several genes coding the elements in Wnt pathway, including FBXW11, APC, LEF1, LDB1, MED1 and KAT5. The expression of FBXW11 and KAT5 in porcine skeletal muscle was increased after feeding a linseed-enriched diet for 30 d. However, the expression of APC, LEF1 and LDB1 was decreased at 30 d.
Discussion
It is well known that n-3 PUFAs play a key role in CNS development and liver metabolism by regulating of genes expression [10, 11]. To investigate the effect of dietary n-3 PUFAs on gene expression in skeletal muscle, we focused on the whole transcriptional response to feeding pigs with a linseed-enriched diet in porcine skeletal muscle in the present study.
Several studies have shown that n-3 PUFAs can act as ligands to influence gene expression by regulation of activation or/and expression of transcription factors [9]. In the current study, we showed that 32 (13.1 %) DEGs were related to gene expression regulation (Table 4). These 32 DEGs contained not only transcriptional factors, such as LEF1, but also transcriptional coactivator and corepressor, i.e., PGC-1α, LDB1, SIRT1, SP100, NR2F2, ABRA, HTATIP and ARNT, suggesting that the transcription cofactors might also contribute to n-3 PUFAs regulated gene expression.
In the past decade, dietary n-3 PUFAs has been demonstrated to influence energy metabolism in skeletal muscle, in part by acting on PPARs [24]. In the current study, the GO analysis of the DEGs showed that feeding pigs with a linseed-enriched diet significantly influence the glucose metabolic process. Both HK2 and PGC-1α were in the top 10 up-regulated genes list, suggesting a remarkable change of glucose metabolism might be induced in skeletal muscle by feeding pigs with a linseed-enriched diet. The expression of HK2 was induced 2.4-fold at 30 d and remained up-regulated at the later time points. A hexokinase is an enzyme that phosphorylates hexoses to form hexose phosphate. In most organisms, glucose is the most important substrate of hexokinases, while glucose-6-phosphate is the most important product [25]. Hexokinase has three different isoforms. In muscle cells, HK2 is the predominant isoform [26]. For the significantly decreased activation and expression of HK2 in type II diabetes, it was investigated as a promising candidate gene for type II diabetes.
PGC-1α is another up-regulated gene involved in regulating of glucose metabolic process in the current study. It was a later-induced gene, whose expression was increased 4.3-fold at d 90. PGC-1α is a transcriptional coactivator with an important role in regulation of energy homeostasis [27]. In skeletal muscle, PGC-1α promotes glucose uptake and expression of genes involved in the electron transport chain and oxidative phosphorylation [28]. Therefore our finding provide new evidence that diet n-3 PUFA may improve insulin sensitivity in skeletal muscle.
Our result showed that feeding pigs with a linseed enriched diet influenced the expression of 8 genes involved in fatty acids metabolic process, including PTGES3, GPAM, PTGS2, FASN, HSD17B1, MVK, ELOVL6 and PGC-1α. The main function of fatty acid synthase is to catalyze the synthesis of palmitate from acetyl-CoA and malonyl-CoA. Elongation of long chain fatty acids 6 (ELOVL6) is an elongase that catalyzes the conversion of palmitate to stearate [29]. In the current study, the decreased expression of FASN, the gene coding fatty acid synthase, and ELOVL6 implicated that feeding pigs a linseed-enriched diet inhibit the synthesis of palmitate and stearate in skeletal muscle. In addition, the increased expression of GPAM at 30 d suggested an increased biosynthesis of phospholipid. Moreover, the expression changes of PTGES3, HSD17B1 and MVK indicated that feeding pigs with a linseed-enriched diet might influence the biosynthesis of steroids. Although the intermuscular adipose tissue was removed from muscle samples in the current study, the skeletal muscle also contains the intramuscular fat. That might be why FASN, a highly expressed gene in adipose tissue, was detected in the current study.
It is also of interest to note that mRNA abundance of several genes involved in amino acids metabolism was altered. SLC38A1 is a transporter of neutral amino acids, including glycine, alanine, serine, cysteine, glutamine, asparagine, histidine, methionine, threonine, proline, tyrosine and valine [30]. Therefore the increased expression of SLC38A1 at 30 d in the current study might result in the increased uptake of amino acid in muscle cells. Additionally, feeding pigs with a linseed-enriched diet decreased the expression of ARG2, a gene involved in arginine catabolism. This result provide new evidence suggesting that feeding pigs with a n-3 PUFA enriched diet decrease the whole-body amino acids oxidation [31]. Moreover, the increased expression of GCLC, a rate limiting enzyme in glutathione synthesis, suggested that feeding pigs with a linseed-enriched diet might increase the glutathione level in porcine skeletal muscle. These results suggested that feeding pigs with a linseed-enriched diet might increase the available amino acids in skeletal muscle, especially for arginine and glutathione.
Arginine has been demonstrated to promote skeletal muscle growth by generation of nitric oxide and/or activation of mTOR Signaling [32]. Zheng et al. reported that there is a positive correlation between the expression of GCLC and the postnatal growth rate of skeletal muscle [33]. Therefore the expression changes of GCLC and ARG2 might contribute to the growth of skeletal muscle induced by feeding pigs with a linseed-enriched diet. Actually, we have previously reported that muscle mass in pigs fed a linseed-enriched diet is increased [6].
The expression of several genes involved in muscle organ development was changed in the current study. MEF2c and NR2F2 are well characterized to be essential for myoblast differentiation [34, 35], whereas, Gli1 blocks myoblast differentiation by inhibiting MyoD activation in skeletal muscle [36]. In the current study, the increased expression of MEF2c and NR2F2 and a decreased expression of Gli1 at d 30 suggested that feeding pigs with a linseed-enriched diet might increase the differentiation of myogenic cells in porcine skeletal muscle. However, further studies are needed to confirm these results.
IGF-1 is essential for normal growth in numerous mammalian. Although more than 80 % IGF-1 appearing in the circulation is produced by liver, locally produced IGF-1 in muscle is more important than circulating IGF-1 in controlling muscle growth [37]. In the current study, the pigs fed the linseed-enriched diet for 30 and 60 d had a higher expression of IGF-1 in porcine skeletal muscle than those only fed the control diet, suggesting that feeding a linseed-enriched diet may improve the growth of skeletal muscle.
We also showed that feeding pigs with linseed-enriched diet increased mRNA expression of STAT5A, an essential mediator of GH-induced IGF-1 expression in skeletal muscle [38]. In addition, PTPN1 attenuates growth hormone-mediated JAK2-STAT5 signaling [39], and thus, the suppressed PTPN1 expression might also contribute to the increased IGF-1 expression in this study. In the present study, the decreased expression of PTPN1 corresponded to another microarray study, where a significant inhibition of PTPN1 expression is detected in liver of mice fed an n-3 PUFAs-enriched diet compared with those fed an n-6 PUFAs-enriched diet [11].
PTPN1 can also act as a key negative regulator of insulin signaling by dephosphorylation of IR and IRS-1 [40]. In addition, the expression of two positive regulators of insulin signaling, PGC-1α and SORBS1, was increased at 30 d and/or 90 d. It has been demonstrated that insulin could stimulate skeletal muscle protein synthesis by activating of mTOR signaling pathway in young animals [41]. Therefore those results suggested that feeding pigs with a linseed-enriched diet might increase the insulin-induced protein synthesis in skeletal muscle. This hypothesis is in coincidence with a previous study, in which increased insulin response is associated with higher muscle activation of mTOR pathway and higher whole-body disposal of amino acids in growing bovine fed with a n-3 PUFA-enriched diet [42].
Wnt signaling pathway is implicated in the control of cell proliferation and differentiation. When Wnt signaling is turned on, it will promote the proliferation and inhibit the differentiation, otherwise the proliferation will be inhibited [43]. In the canonical Wnt pathway, Wnt binding leads to the stabilization of the transcription factor β-catenin, which enters the nucleus to active LEF1 and regulates the expression of Wnt pathway target genes. LDB1, MED1 and KAT5 are transcriptional coactivators of Pitx2, which is a target gene of LEF1 and plays an important role in regulation of cell proliferation. APC is a blocker of Wnt signaling which can stabilize the β-catenin complex, while FBXW11 can targets phosphorylation-dependent degradation of β-catenin [44]. In the current study, the increased expression of FBXW11 and decreased expression of LDB1 and MED1 indicated that feeding pigs with a linseed-enriched diet might increase the degradation of β-catenin and inhibit the Pitx2-induced cell proliferation. However, the results of the decreased expression of APC and the increased expression of KAT5 did not support the inference that activation of Wnt signaling pathway was decreased in the current study. The controlled studies are needed to explore whether n-3 PUFA inhibit the activation of Wnt signaling pathway.
In the current study, we showed that linseed-enriched diet up-regulated the expression of genes (SLC20A1, TRAF3IP2, TP53INP1, TNFRSF12A, TLR9 and BIRC2) which involved in IκB inhibition in porcine skeletal muscle, and might lead to decrease NF-κB activation (Table 5). Recent studies indicated that activation of NF-κB is inhibited by n-3 PUFAs in lymphomonocyte [45], and the decreased IKK activation is likely to contribute to the suppressed NF-κB activation [46]. Our results provided an alternative explanation why n-3 PUFAs could suppress NF-κB activation.
NF-κB is a transcription factor that play important role in regulation of inflammatory processes. The expression of many pro-inflammatory cytokines such as IL-1, IL-6 and TNF-α, are induced by NF-κB activation in various tissues [47]. Moreover, NF-κB mediates the anti-inflammation properties of n-3 PUFA partially [48]. Although our microarray results implicated that feeding pigs with a linseed-enriched diet decreased the activation of NF-κB, the expression of IL-1, IL-6 and TNF-α were unfortunately not detected in our microarray experiment and thus filtered for further analysis. This might be cause by low expression level of these genes in skeletal muscles and low sensitivity of microarray. But it is previously reported that expression of IL-1, IL-6 and TNF-α in skeletal muscle were inhibited by feeding a linseed-enriched diet using transcriptase polymerase chain reaction technique to assess the samples in this study [22].
The previous study showed that, by inhibiting the activation of NF-κB, EPA (20C: 5 n3) decreases the expression of muscle RING-finger protein-1 (MuRF1), which plays an important role in controlling protein degradation in skeletal muscle [49]. Unfortunately, we did not find any oligonucleotide set representing MuRF1 gene on the chip used in the current study. Interestingly, PTPN1 has been shown to act as a target gene of NF-κB. Its expression level can be induced by NF-κB activation [50]. Therefore the decreased expression of PTPN1 might cause by the inhibited activation of NF-κB.
Conclusion
Genome-wide investigation of transcriptional response to feeding pigs with a linseed-enriched diet in porcine skeletal muscle provides systematic information for understanding the effect of dietary n-3 PUFA on the gene expression in skeletal muscle. Our finding suggested that the increased expression of IGF-1, insulin signaling pathway and the metabolism of amino acids might involve in the muscle growth induced by feeding a linseed-enriched diet. Our results also provide the new evidence that the expression changes of PTPN1, HK2 and PGC-1α might contribute to the regulation of insulin sensitivity by n-3 PUFAs. However, the effect of dietary n-3 PUFA on the activation of signaling pathways needs to be confirmed by protein and enzyme activity studies.
Acknowledgments
Contract grant sponsor: National Program on Key Basic research Project (No.2012CB127305), Hubei Provincial Creative Team Project of Agricultural Science and Technology (No.2007-620), Hubei Provincial Natural Science Foundation (No.2013CFA010) and Fundamental Research Funds for the Central Universities (2013PY047 and 2013QC004).
Abbreviations
- BIRC2
baculoviral IAP repeat-containing protein 2
- CAMs
cell adhesion molecules
- CNS
central nervous system
- DEGs
differentially expressed genes
- ELOVL6
elovel fatty acid elongase 6
- FASN
fatty acid synthase
- GH
growth hormone
- GO
gene ontology
- GPAM
lycerol-3-phosphate acyltransferase 1, mitochondrial
- HNF-4α
hepatic nuclear-4α
- HSD17B1
Estradiol 17-beta-dehydrogenase 1
- IGF-1
insulin-like growth factor 1
- LXR
liver X receptors
- MEF2C
Myocyte-specific enhancer factor 2C
- MVK
mevalonate kinase
- n-3 PUFAs
n-3 polyunsaturated fatty acids
- NF-κB
nuclear factor κB
- NR2F2
COUP transcription factor 2
- PGC-1α
peroxisome proliferators-activated receptor-γcoactivator 1α
- PPARs
peroxisome proliferator activated receptors
- PTGES3
prostaglandin E synthase 3
- PTGS2
and prostaglandin-endoperoxide synthase 2
- PTPN1
protein tyrosine phosphatase non-receptor type 1
- RXRα
retinoid X receptor α
- SLC20A1
solute carrier family 20, member 1
- SREBP-1c
sterol regulatory element binding protein-1c
- STAT5A
signal transducer and activator of transcription 5
- SORBS1
orbin and SH3 domain-containing protein 1
- SOTA
self-organizing tree algorithm
- TNFRSF12A
tumor necrosis factor receptor superfamily member Fn14 precursor
- TLR9
toll-like receptor 9 precursor
- TP53INP1
Tumor protein p53 inducible nuclear protein 1
- TRAF3IP2
TRAF3 interacting protein 2
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
Jian Peng, Siwen Jiang and Hong-Kui Wei designed research; Hong-Kui Wei, Feiruo Huang, Yuanfei Zhou and Shuzhong Jiang conducted research; Hong-Kui Wei analyzed data; Hong-Kui Wei and Jian Peng wrote the paper. Siwen Jiang and Jian Peng had primary responsibility for final content. All authors read and approved the final manuscript.
Contributor Information
Hongkui Wei, Email: weihongkui@mail.hzua.edu.cn.
Yuanfei Zhou, Email: zhouyuanfei@webmail.hzau.edu.cn.
Shuzhong Jiang, Email: jiangshuzhong@webmail.hzau.edu.cn.
Feiruo Huang, Email: huangfeiruo@mail.hzau.edu.cn.
Jian Peng, Email: pengjian@mail.hzau.edu.cn.
Siwen Jiang, Email: jiangsiwen@mail.hzau.edu.cn.
References
- 1.Jump DB. The Biochemistry of n-3 Polyunsaturated Fatty Acids. J Biol Chem. 2002;277:8755–8. doi: 10.1074/jbc.R100062200. [DOI] [PubMed] [Google Scholar]
- 2.Bryhni EA, Kjos NP, Ofstad R, Hunt M. Polyunsaturated fat and fish oil in diets for growing-finishing pigs: effects on fatty acid composition and meat, fat, and sausage quality. Meat Sci. 2002;62:1–8. doi: 10.1016/S0309-1740(01)00211-X. [DOI] [PubMed] [Google Scholar]
- 3.Enser M, Richardson RI, Wood JD, Gill BP, Sheard PR. Feeding linseed to increase the n-3 PUFA of pork: fatty acid composition of muscle, adipose tissue, liver and sausages. Meat Sci. 2000;55:201–12. doi: 10.1016/S0309-1740(99)00144-8. [DOI] [PubMed] [Google Scholar]
- 4.Rhee KS, Davidson TL, Knabe DA, Cross HR, Ziprin YA, Rhee KC. Effect of dietary high-oleic sunflower oil on pork carcass traits and fatty acid profiles of raw tissues. Meat Sci. 1988;24:249–60. doi: 10.1016/0309-1740(88)90038-1. [DOI] [PubMed] [Google Scholar]
- 5.Sousa RV, Fialho ET, Lima JAF, Alvarez-Leite JI, Cortez WC, Ferreira MSS. Effect of different oils in diets for finishing pigs: performance, carcass traits and fatty acid profile of the meat. Anim Prod Sci. 2010;50:863–8. doi: 10.1071/AN09138. [DOI] [Google Scholar]
- 6.Huang FR, Zhan ZP, Luo J, Liu ZX, Peng J. Duration of dietary linseed feeding affects the intramuscular fat, muscle mass and fatty acid composition in pig muscle. Livest Sci. 2008;118:132–9. doi: 10.1016/j.livsci.2008.01.002. [DOI] [Google Scholar]
- 7.Luo HF, Wei HK, Huang FR, Zhou Z, Jiang SW, Peng J. The effect of linseed on intramuscular fat content and adipogenesis related genes in skeletal muscle of pigs. Lipids. 2009;44:999–1010. doi: 10.1007/s11745-009-3346-y. [DOI] [PubMed] [Google Scholar]
- 8.Reseland JE, Haugen F, Hollung K, Solvoll K, Halvorsen B, Brude IR, et al. Reduction of leptin gene expression by dietary polyunsaturated fatty acids. J Lipid Res. 2001;42:743–50. [PubMed] [Google Scholar]
- 9.Jump DB. Dietary polyunsaturated fatty acids and regulation of gene transcription. Curr Opin Lipidol. 2002;13:155–64. doi: 10.1097/00041433-200204000-00007. [DOI] [PubMed] [Google Scholar]
- 10.Puskas LG, Kitajka K. Nutrigenomic approaches to study the effects of n-3 PUFA diet in the central nervous system. Nutr and Health. 2006;18:227–32. doi: 10.1177/026010600601800305. [DOI] [PubMed] [Google Scholar]
- 11.Berger A, Mutch DM, German JB, Roberts MA. Dietary effects of arachidonate-rich fungal oil and fish oil on murine hepatic and hippocampal gene expression. Lipids Health Dis. 2002;1:2. doi: 10.1186/1476-511X-1-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van Schothorst E, Flachs P, Franssen-van Hal N, Kuda O, Bunschoten A, Molthoff J, et al. Induction of lipid oxidation by polyunsaturated fatty acids of marine origin in small intestine of mice fed a high-fat diet. BMC Genomics. 2009;10:110. doi: 10.1186/1471-2164-10-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li C, Wong WH. Model-based analysis of oligonucleotide arrays: Expression index computation and outlier detection. Proc Natl Acad Sci U S A. 2001;98:31–6. doi: 10.1073/pnas.98.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leek JT, Monsen E, Dabney AR, Storey JD. EDGE: extraction and analysis of differential gene expression. Bioinformatics. 2006;22:507–8. doi: 10.1093/bioinformatics/btk005. [DOI] [PubMed] [Google Scholar]
- 15.Storey JD. A direct approach to false discovery rates. J Roy Stat Soc B. 2002;64:479–98. doi: 10.1111/1467-9868.00346. [DOI] [Google Scholar]
- 16.Tsai S, Cassady JP, Freking BA, Nonneman DJ, Rohrer GA, Piedrahita JA. Annotation of the Affymetrix porcine genome microarray. Anim Genet. 2006;37:423–4. doi: 10.1111/j.1365-2052.2006.01460.x. [DOI] [PubMed] [Google Scholar]
- 17.Gormley M, Dampier W, Ertel A, Karacali B, Tozeren A. Prediction potential of candidate biomarker sets identified and validated on gene expression data from multiple datasets. BMC Bioinformatics. 2007;8:415. doi: 10.1186/1471-2105-8-415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, et al. TM4: a free, open-source system for microarray data management and analysis. Biotechniques. 2003;34:374–8. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- 19.The pubgene database. www.pubgene.org. Accessed 20 Jun 2012.
- 20.The gogene database. http://projects.biotec.tu-dresden.de/gogene/gogene/. Accessed 21 Jun 2012.
- 21.Dennis G, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, et al. DAVID: Database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4(5):P3. doi: 10.1186/gb-2003-4-5-p3. [DOI] [PubMed] [Google Scholar]
- 22.Zhan ZP, Huang FR, Luo J, Dai JJ, Yan XH, Peng J. Duration of feeding linseed diet influences expression of inflammation-related genes and growth performance of growing-finishing barrows. J Anim Sci. 2009;87:603–11. doi: 10.2527/jas.2007-0177. [DOI] [PubMed] [Google Scholar]
- 23.Livak KJ, Schmittgen TD. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2− ΔΔCT Method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 24.Le NH, Shin S, Tu TH, Kim C-S, Kang J-H, Tsuyoshi G, et al. Diet Enriched with Korean Pine Nut Oil Improves Mitochondrial Oxidative Metabolism in Skeletal Muscle and Brown Adipose Tissue in Diet-Induced Obesity. J Agr Food Chem. 2012;60:11935–41. doi: 10.1021/jf303548k. [DOI] [PubMed] [Google Scholar]
- 25.Wilson J. Hexokinases. In: Nilius B, Gudermann Th, Jahn R, Lill R, Petersen OH, de Tombe PP, editors. Reviews of Physiology, Biochemistry and Pharmacology. Berlin: Springer; 1995. p. 65–198. [DOI] [PubMed]
- 26.Heikkinen S, Suppola S, Malkki M, Deeb SS, Jänne J, Laakso M. Mouse hexokinase II gene: structure, cDNA, promoter analysis, and expression pattern. Mamm Genome. 2000;11:91–6. doi: 10.1007/s003350010019. [DOI] [PubMed] [Google Scholar]
- 27.Lin J, Handschin C, Spiegelman BM. Metabolic control through the PGC-1 family of transcription coactivators. Cell Metab. 2005;1:361–70. doi: 10.1016/j.cmet.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 28.Finck BN, Kelly DP. PGC-1 coactivators: inducible regulators of energy metabolism in health and disease. J Clin Invest. 2006;116:615–22. doi: 10.1172/JCI27794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matsuzaka T, Shimano H, Yahagi N, Kato T, Atsumi A, Yamamoto T, et al. Crucial role of a long-chain fatty acid elongase, Elovl6, in obesity-induced insulin resistance. Nat Med. 2007;13:1193–202. doi: 10.1038/nm1662. [DOI] [PubMed] [Google Scholar]
- 30.Albers A, Broer A, Wagner CA, Setiawan I, Lang PA, Kranz EU, et al. Na + transport by the neural glutamine transporter ATA1. Pflug Arch Eur J Phy. 2001;443:92–101. doi: 10.1007/s004240100663. [DOI] [PubMed] [Google Scholar]
- 31.Bergeron K, Julien P, Davis TA, Myre A, Thivierge MC. Long-chain n-3 fatty acids enhance neonatal insulin-regulated protein metabolism in piglets by differentially altering muscle lipid composition. J Lipid Res. 2007;48:2396–410. doi: 10.1194/jlr.M700166-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu G. Functional Amino Acids in Growth, Reproduction, and Health. Adv Nutr. 2010;1:31–7. doi: 10.3945/an.110.1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zheng S, Yumei F, Chen A. De novo synthesis of glutathione is a prerequisite for curcumin to inhibit hepatic stellate cell (HSC) activation. Free Radical Bio Med. 2007;43:444–53. doi: 10.1016/j.freeradbiomed.2007.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Potthoff MJ, Olson EN. MEF2: a central regulator of diverse developmental programs. Development. 2007;134:4131–40. doi: 10.1242/dev.008367. [DOI] [PubMed] [Google Scholar]
- 35.Lee CT, Li L, Takamoto N, Martin JF, DeMayo FJ, Tsai M-J, et al. The nuclear orphan receptor COUP-TFII is required for limb and skeletal muscle development. Mol Cell Biol. 2004;24:10835–43. doi: 10.1128/MCB.24.24.10835-10843.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gerber AN, Wilson CW, Li YJ, Chuang PT. The hedgehog regulated oncogenes Gli1 and Gli2 block myoblast differentiation by inhibiting MyoD-mediated transcriptional activation. Oncogene. 2006;26:1122–36. doi: 10.1038/sj.onc.1209891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Velloso CP. Regulation of muscle mass by growth hormone and IGF-I. Brit J Pharmacol. 2008;154:557–68. doi: 10.1038/bjp.2008.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang Y, Jiang HL. Identification of a distal STAT5-binding DNA region that may mediate growth hormone regulation of insulin-like growth factor-I gene expression. J Biol Chem. 2005;280:10955–63. doi: 10.1074/jbc.M412808200. [DOI] [PubMed] [Google Scholar]
- 39.Gu F, Dube N, Kim JW, Cheng A, Ibarra-Sanchez Mde J, Tremblay ML, et al. Protein tyrosine phosphatase 1B attenuates growth hormone-mediated JAK2-STAT signaling. Mol Cell Biol. 2003;23:3753–62. doi: 10.1128/MCB.23.11.3753-3762.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Goldstein BJ, Bittner-Kowalczyk A, White MF, Harbeck M. Tyrosine dephosphorylation and deactivation of insulin receptor substrate-1 by protein-tyrosine phosphatase 1B - Possible facilitation by the formation of a ternary complex with the GRB2 adaptor protein. J Biol Chem. 2000;275:4283–9. doi: 10.1074/jbc.275.6.4283. [DOI] [PubMed] [Google Scholar]
- 41.Davis TA, Suryawan A, Orellana RA, Fiorotto ML. Postnatal ontogeny of skeletal muscle protein synthesis in pigs. J Anim Sci. 2008;86(suppl 14):E13–8. doi: 10.2527/jas.2007-0419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gingras AA, White PJ, Chouinard PY, Julien P, Davis TA, Dombrowski L, et al. Long-chain omega-3 fatty acids regulate bovine whole-body protein metabolism by promoting muscle insulin signalling to the Akt-mTOR-S6K1 pathway and insulin sensitivity. J Physiol-London. 2007;579:269–84. doi: 10.1113/jphysiol.2006.121079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Watson CJ, Khaled WT. Mammary development in the embryo and adult: a journey of morphogenesis and commitment. Development. 2008;135:995–1003. doi: 10.1242/dev.005439. [DOI] [PubMed] [Google Scholar]
- 44.Logan CY, Nusse R. The Wnt signaling pathway in development and disease. Annu Rev Cell Dev Biol. 2004;20:781–810. doi: 10.1146/annurev.cellbio.20.010403.113126. [DOI] [PubMed] [Google Scholar]
- 45.Zhao Y, Chen LH. Eicosapentaenoic acid prevents lipopolysaccharide-stimulated DNA binding of activator protein-1 and c-Jun N-terminal kinase activity. J Nutr Biochem. 2005;16:78–84. doi: 10.1016/j.jnutbio.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 46.Takase O, Hishikawa K, Kamiura N, Nakakuki M, Kawano H, Mizuguchi K, et al. Eicosapentaenoic acid regulates IκBα and prevents tubulointerstitial injury in kidney. Eur J Pharmacol. 2011;669:128–35. doi: 10.1016/j.ejphar.2011.07.043. [DOI] [PubMed] [Google Scholar]
- 47.Tak PP, Firestein GS. NF-κB: a key role in inflammatory diseases. J Clin Invest. 2001;107:7–11. doi: 10.1172/JCI11830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chapkin RS, Davidson LA, Ly L, Weeks BR, Lupton JR, McMurray DN. Immunomodulatory effects of (n-3) fatty acids: putative link to inflammation and colon cancer. J Nutr. 2007;137(suppl 1):200–4. doi: 10.1093/jn/137.1.200S. [DOI] [PubMed] [Google Scholar]
- 49.Bodine SC, Latres E, Baumhueter S, Lai VK-M, Nunez L, Clarke BA, et al. Identification of ubiquitin ligases required for skeletal muscle atrophy. Sci Signal. 2001;294:1704. doi: 10.1126/science.1065874. [DOI] [PubMed] [Google Scholar]
- 50.Zabolotny JM, Kim YB, Welsh LA, Kershaw EE, Neel BG, Kahn BB. Protein-tyrosine phosphatase 1B expression is induced by inflammation in vivo. J Biol Chem. 2008;283:14230–41. doi: 10.1074/jbc.M800061200. [DOI] [PMC free article] [PubMed] [Google Scholar]


